Abstract
Recent breakthroughs in high-throughput technologies, transcriptomics and advances in our understanding of gene regulatory networks have enhanced our perspective on the complex interplay between parasite and host. Non-coding RNA molecules have been implicated in critical roles covering a broad range of biological processes in the Apicomplexa. Processes that are affected range from parasite development to host-parasite interactions and include interactions with epigenetic machinery and other regulatory factors. Here we review recent progress involving non-coding RNAs and their functions in the Apicomplexa with a focus on three parasites: Plasmodium, Toxoplasma, and Cryptosporidium. We discuss the limitations and challenges of current methods applied to apicomplexan non-coding RNA study and discuss future directions in this exciting field.
Keywords: ncRNA, Apicomplexa, lncRNA, sncRNA
The emerging importance of ncRNAs
With little to no protein-coding capacity, non-coding RNA (ncRNA) (see Glossary) is an essential transcriptome component detected across all domains of life [1]. Although initially considered transcriptional noise (e.g., read-through or non-specific transcription), ncRNAs have been shown to play critical roles in gene expression regulation at the levels of transcription, RNA processing, and translation [2]. Ribosomal RNAs (rRNAs) and transfer RNAs (tRNAs) were first identified in the 1950s, followed by the discovery of small nuclear RNAs (snRNAs) and small nucleolar RNAs (snoRNAs) [2]. The first ncRNAs to be characterized were generally small (< 300 nt except for rRNAs), contain stable secondary structure(s) and often operate as components of conserved RNA-protein complexes (see Table 1). The ncRNA world blossomed in the early 2000s with advances in sequencing technologies. Since then, various long non-coding RNAs (lncRNAs), microRNA (miRNAs) and more recently tRNA and snoRNA derived small ncRNAs (18–40 nt) have been discovered [3, 4]. ncRNAs are classified based on transcript length, secondary structure, genomic and cellular localization [5] (Table 1). The abundance and variety of these molecules have reshaped our understanding of ncRNAs as fundamental transcriptional and post-transcriptional regulators.
Table 1.
Categories, properties and occurrence of ncRNA types in the Apicomplexa
| Category | Abbrev. | Size (nt) |
Main Functions | P.f.a | T.g.b | C.p.c | Refs | ||
|---|---|---|---|---|---|---|---|---|---|
| ncRNA | long ncRNA (>200 nt) | structual/ function based | circRNA | 100s ~1000s | miRNA sponge | ✓d | ◯e | ◯ | [98] |
| SRP RNA | ~300 | Associate with the ribosome and target nascent proteins to the endoplasmic reticulum for secretion or membrane insertion | ✓ | ✓ | ✓ | [5] | |||
| RNase MRP RNA | 100s ~1000s | Initiate mitochondrial DNA replication and process precursor rRNA in nucleus | ✓ | ✓ | ✓ | [5] | |||
| position based | TERRA | 100s ~1000s | Maintain telomeres structure and functions | ✓ | ◯ | ◯ | [58] | ||
| NAT | 100s ~1000s | Heterogeneous functions in wide range of biological process | ✓ | ✓ | ◯ | [22] | |||
| Intronic lncRNA | ✓ | ✓ | ◯ | [5] | |||||
| lincRNA | ✓ | ✓ | ◯ | [96] | |||||
| Sense lncRNA | ✓ | ✓ | ◯ | [5, 23] | |||||
| Bidirectional lncRNA | ✓ | ◯ | ◯ | [5] | |||||
| short ncRNA (<200 nt) | structual/ function based | tRNA | 76 – 90 | Confer an amino acid to ribosome as directed by genetic codons in mRNA | ✓ | ✓ | ✓ | [5] | |
| snoRNA | 60–300 | Component of small nucleolar ribonucleoprotein (snoRNP) and guide snoRNP to chemically modify pre-rRNA to form mature rRNA | ✓ | ✓ | ✓ | [4, 100] | |||
| snRNA | ~150 | Component of small nuclear ribonucleoprotein (snRNPs) and involved in RNA splicing | ✓ | ✓ | ✓ | [5] | |||
| miRNA | ~22 | Operate in the RNA interference (RNAi) pathway, bind to target mRNA and mediate mRNA degradation or translation inhibition. | ✘f | ✓ | ◯ | [76, 77] | |||
| siRNA | 20–25 | Similar to miRNA, operating in the RNA interference (RNAi) pathway, bind to target mRNA and mediate mRNA degradation or translation inhibition. | ✘ | ✓ | ◯ | [71] | |||
| piRNA | 24–32 | Associate with piwi proteins involved in epigenetic and post-transcriptional silencing of transposons | ✘ | ◯ | ◯ | [70] | |||
| position based | centromere associated small RNA | <200 | Incorporate into centromeric chromatin and associated with kinetochore | ✓ | ◯ | ◯ | [102] | ||
| tsRNA | 14–50 | Interact with Ago and Piwi proteins, potentially regulate gene expression | ✓ | ◯ | ◯ | [3, 74] |
P.f., Plasmodium falciparum;
T.g.,Toxoplasma gondii;
C.p., Cryptosporidium parvum;
✓, detected;
◯, status unknown;
✘, Not detected
We emphasize lncRNAs in this review because this class of ncRNA is very heterogeneous and participates in an incredibly diverse set of processes. Most lncRNAs share many similarities with mRNAs, such as RNA polymerase II-mediated transcription, a 5′ 7-methylguanosine cap and a 3′ poly(A) tail [6]. Comparative analyses of lncRNAs reveal that they are not well conserved across species [7] and usually have greater tissue- or development-specific expression patterns than mRNAs [7, 8]. By interacting with protein, DNA, or RNA molecules, lncRNAs participate in multiple layers of gene regulation including transcriptional, post-transcriptional, chromatin modification and nuclear architecture conformation alterations (Box 1). The misregulation of lncRNAs in multi-cellular eukaryotes has been shown to lead to tumor genesis [9, 10], cardiovascular disease [11], and neurodegenerative dysfunction [12] and thus can be used as biomarkers for diagnosis.
Box 1. lncRNA regulatory functions and mechanisms of action in eukaryotes.
Transcriptional regulation
lncRNAs can target chromatin modifiers such as chromatin-modifying enzymes and nucleosome-remodeling factors in cis or trans to the target promoter resulting in activation or repression of gene expression. This process is usually sequence-dependent [34] (Figure IA). In some cases, the lncRNA sequence itself may not be essential, rather it is transcriptional activation of the particular lncRNA via recruitment of RNA polymerase II (Pol II) and alteration of the local transcriptional environment that affects transcription of neighboring genes [88] (Figure IB). This mechanism is also seen in lncRNAs transcribed from enhancer regions thus assisting neighboring gene transcription but independent of the lncRNA transcript itself [89]. LncRNA transcription may also suppress neighboring gene expression by transcriptional interference [90], such as competing for transcription-related molecules [91] and limiting available space leading to transcriptional machinery collision [92, 93] (Figure IC).
Post-transcriptional regulation
In eukaryotes, pre-mRNAs can undergo several processes including intron splicing, nuclear export, localization, translation and decay. lncRNA can regulate post-transcriptional processes via direct or indirect interaction with the factors involved in the processes, such as RNA binding proteins (RBPs) and miRNAs. For example, polyadenylated lncRNAs with Alu elements have been shown to form imperfect base-pairing with mRNAs harboring Alu elements in the 3′ UTR and trigger mRNA decay [94]. mRNA can also be stabilized by lncRNAs, especially NATs by forming an RNA duplex thus controlling the interaction with RNA decay factors [95] (Figure ID). Translation can be activated [96] or suppressed [96] by interactions between lncRNAs and translation factors (Figure IE). In the RNA interference (RNAi) pathway, lncRNAs can compete with miRNA for target mRNA or act as an miRNA sponge like circRNAs, resulting in an increased target mRNA level [97, 98] (Figure IF). lncRNAs are also considered as mRNA splicing regulators through interactions with splicing modulators, or as protectors of particular introns [99, 100]. Notably, a class of snoRNA-ended lncRNAs (sno-lncRNAs) were detected in humans, which were derived from an intron and processed on both ends by the snoRNA machinery. Sno-lncRNA can interact with Fox family splicing regulators and alter splicing patterns in cells [100] (Figure IG).
Spatial organization
lncRNAs are involved in nuclear organization (Figure IH) by helping to correctly localize co-activation or co-repression of gene loci dependent on their spatial proximity [101].
Figure I (in Box 1). Multiple dimensions of gene regulation by lncRNAs.
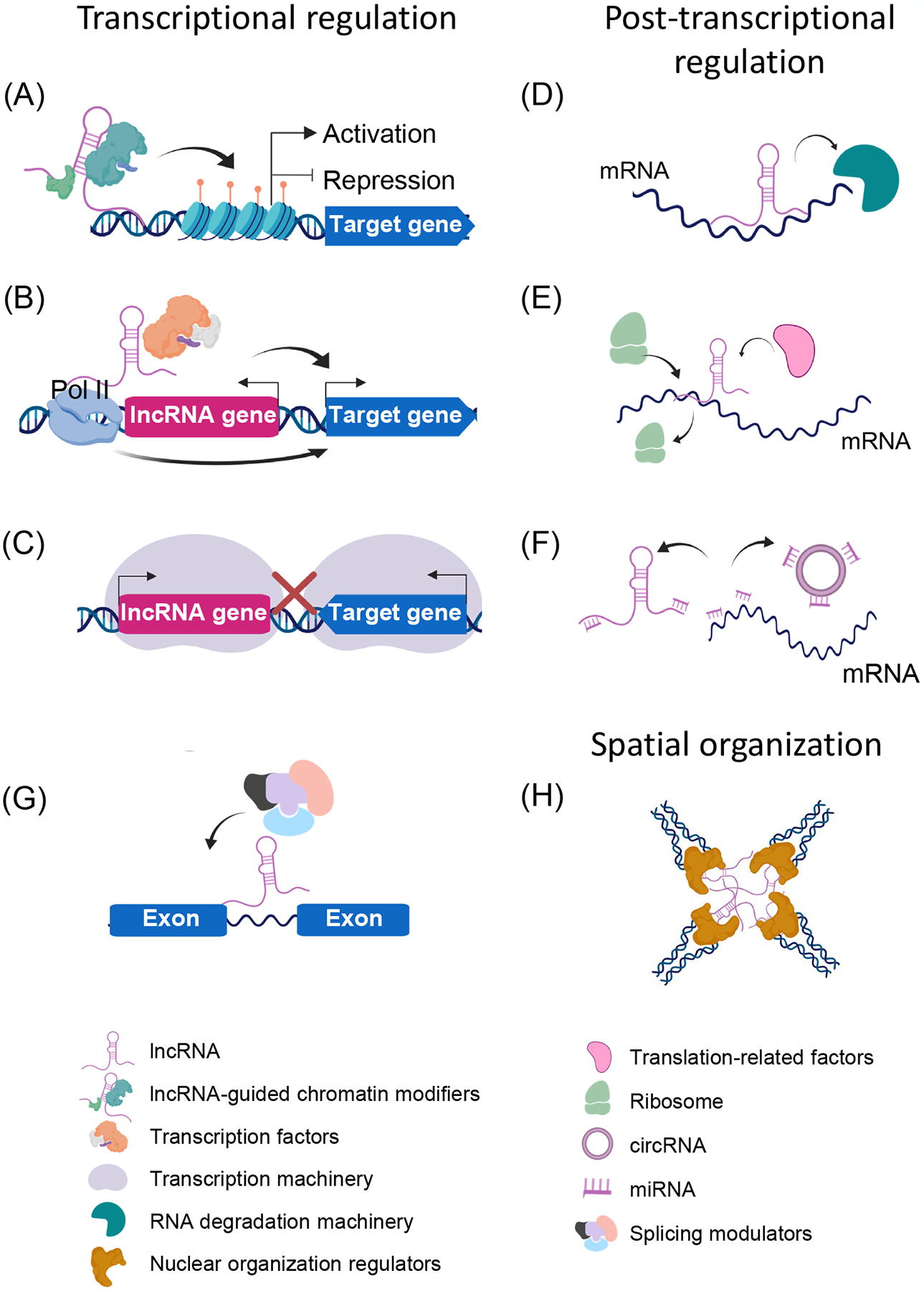
lncRNAs can regulate gene expression via different mechanisms. The major processes influenced by lncRNAs are: (A-C) Transcriptional level modes of interaction include: (A) interaction with or recruitment of epigenetic modulators such as chromatin modifiers to activate or repress the target gene expression in cis or trans; (B) changing the local transcriptional environment such as recruitment of transcriptional factors and Pol II to activate or repress neighboring gene expression which can be lncRNA sequence-independent; (C) transcriptional interference via competition for transcription-related molecules and spatial limitation. (D-G) Post-transcriptional level modes of lncRNA interaction include: (D) triggering or preventing mRNA decay process through interaction with RNA decay machinery; (E) activation or repression or translational processes through interaction with translational factors and likely influencing the ribosome RNA drop-off rate; (F) circRNAs acting as an miRNA sponge and lncRNAs competing with miRNA for target mRNAs; (G) regulation of intron splicing through interaction with splicing modulators. (H) Higher-order structure effects include: lncRNAs mediate intra- or inter-chromosomal interaction through interactions with nuclear organization factors.
lncRNAs in the Apicomplexa
The phylum Apicomplexa is a large and diverse clade of protist parasites responsible for devastating diseases in animals and humans. Overall, lncRNAs in the Apicomplexa are difficult to differentiate from neighboring mRNAs and read-through transcriptional noise because of the compact nature of their genomes (8–130 Mb) [13–15]. Deep RNA sequencing has provided a glimpse into the abundance of lncRNAs and other ncRNAs in Plasmodium and Toxoplasma [14, 16–19]. There is evidence of ncRNA participation in a remarkably broad spectrum of apicomplexan biological processes including parasite development and gene expression regulation [14, 20–22]. Experimental functional validation of lncRNAs in intracellular parasites is challenging. Some regulatory roles of apicomplexan lncRNAs are likely shared with model organism species (Box 1). However, others may function in parasitic-specific ways including interactions with the host. Additional studies targeting the functional roles of lncRNAs and examination of how they contribute to gene expression regulation will provide much needed insight into parasite developmental regulation.
Here, we review recent ncRNA discoveries in Plasmodium, Toxoplasma and Cryptosporidium. We review the types and functional roles that have been discovered to date including transcriptional regulation, epigenetic associations, and host-parasite interactions. Given the dearth of ncRNA information outside of the genus Plasmodium, we emphasize challenges related to the discovery of novel ncRNAs by RNA sequencing (RNA-Seq). The limitations and challenges of current methods applied to apicomplexan ncRNA discovery, functional characterization and future directions in this exciting field are discussed.
lncRNAs as regulators of gene expression in the malaria parasite Plasmodium falciparum
ncRNAs are best characterized in P. falciparum where they have been shown to play a prominent role in gene regulation [20]. The most famous and complex example is related to the P. falciparum var gene which is an ~60 member multigene family that encodes the important virulence factor, erythrocyte membrane protein 1. A var gene is composed of a variable exon 1, a conserved exon 2 and a conserved intron between them. var genes have been shown to have mutually exclusive expression (MEE) patterns associated with immune evasion in humans [20]. Studies of MEE in this system have led to a variety of discoveries, including a prominent role for two lncRNAs. These lncRNAs were discovered in elegant experiments designed to ascertain how one var gene is activated while the others are silenced. The experiments implicate the conserved intron. We now know, the lncRNAs are transcribed from a bidirectional promoter located within the conserved var gene intron giving to one antisense lncRNA complementary to the first exon and a second sense lncRNA that extends into the second exon [23] (Figure 1A). The lncRNAs are reported to be transcribed by Pol II [24]; capped, but not polyadenylated; remain in the nucleus; localize to discrete perinuclear foci and incorporated into chromatin [22, 23].
Figure 1. Illustration of lncRNA functions in apicomplexan parasite.
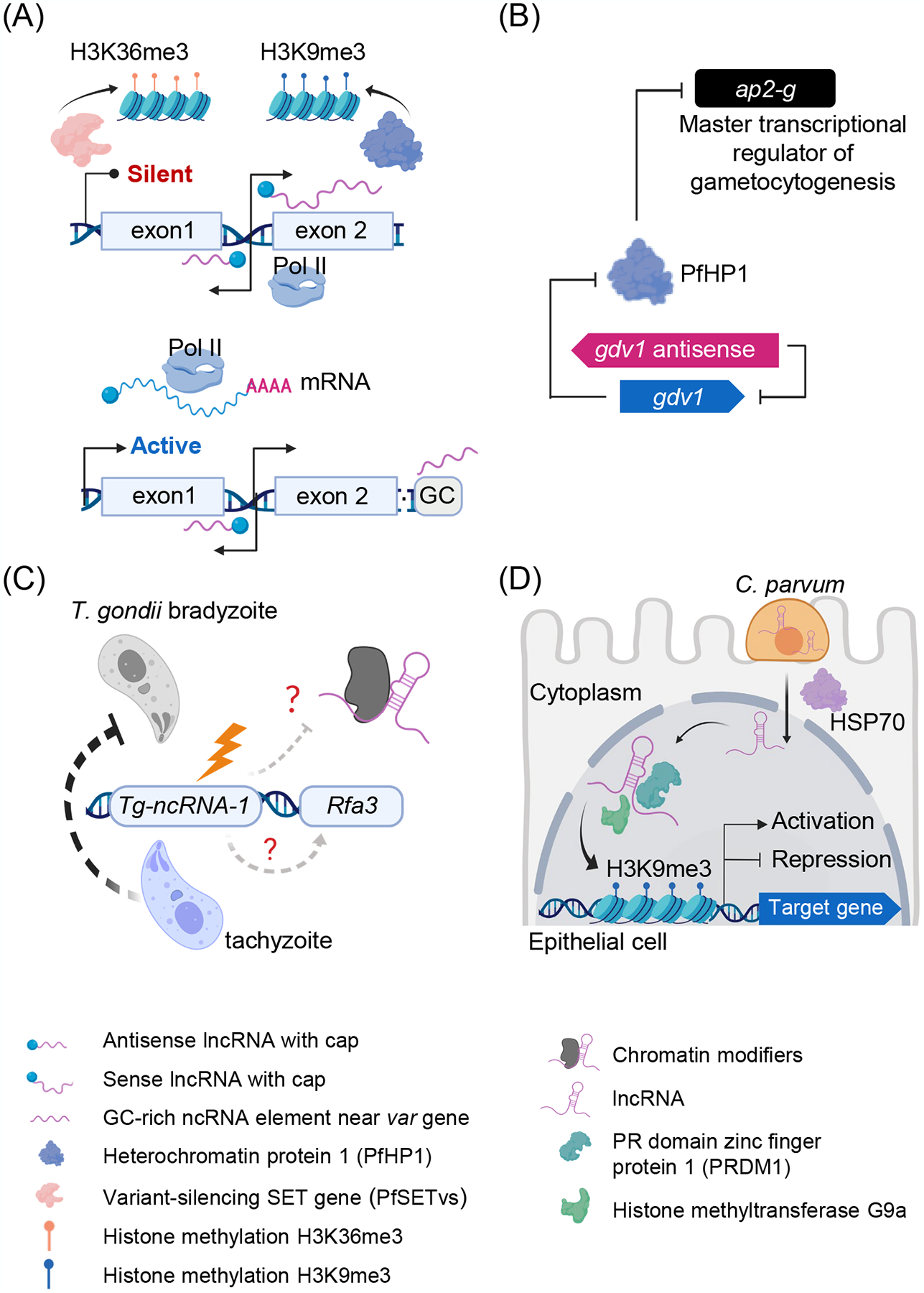
(A) ncRNAs as one of the critical regulators of the virulence gene expression in P. falciparum. The important intron structure of var gene, lncRNAs transcribed it, and histone modifications have been associated with var gene silencing; The antisense transcribed from the intron and ncRNA from a proximal GC-rich element of internal var genes have been associated with var gene activation.
(B) lncRNA associated with P. falciparum sexual commitment. P. falciparum gametocyte development 1 (GDV1) induces sexual commitment by antagonizing HP1-dependent gene silencing. gdv1 expression is repressed by its antisense.
(C) lncRNA Tg-ncRNA-1 as a regulator during T. gondii development. Tachyzoites with a mutation in Tg-ncRNA-1 fail to develop into bradyzoites.
(D) Some lncRNAs from C. parvum are found in the host cell nucleus and have been shown to hijack the hosts epigenetic machinery including protein PRDM1 and G9a to manipulate host gene expression. Transport of the parasite lncRNA from host cell cytoplasm into the host nucleus is believed to be assisted by HSP70.
The regulation of var gene expression is a multi-dimensional and complex process. Many studies have indicated a role for the var gene intron in silencing the remaining, non-activated, var genes [25, 26], though the mechanism remains to be elucidated. This area is under active investigation with conflicting results emerging from studies of non-homologous var genes using different experimental approaches. Given the importance of the var gene intron, Bryant et al., used the CRISPR/Cas9 system to knock out the var2csa intron resulting in an upregulation of transcription of the var2csa gene in ring-stage parasites while not affecting the subsequent transcriptional silencing of the var gene in trophozoites [27]. These results imply the intron is not essential. The authors point out, rightly, that the conflicting results may be due to functional differences between internally located versus subtelomeric var genes, or the existence of as of yet unidentified collaborative regulators. The differences may also result from experimental differences between artificial and endogenous experimental systems.
Recent studies have associated var gene activation with the intron derived antisense lncRNA (Figure 1A). Expression of antisense lncRNAs from plasmid transfections was seen to activate a silent var gene in a sequence- and dose-dependent manner [22]. In another study, exogenous artificial antisense lncRNAs transcribed from an episome could activate the homologous var gene and co-express it with the previously dominant var gene in the same parasite nucleus as observed by RNA fluorescence in situ hybridization (FISH) probes [28], thus indicating an override of MEE.
ncRNAs of 136 bp derived from GC-rich elements that are interspersed among the internal chromosomal var gene clusters also contribute to var gene regulation [29] (Figure 1A). They are localized to the perinuclear expression sites of both internally located and subtelomeric var genes in trans as shown by FISH [29]. Overexpression of distinct GC-rich elements resulted in the activation of a specific subset of var genes, escaping MEE control [29]. Transcriptional repression of all GC-rich members by CRISPR interference (CRISPRi) led to downregulation of the entire var gene family in ring-stage parasites [30]. Thus, these GC-rich ncRNAs are hypothesized to play a role in var gene activation. [30]. These GC-rich ncRNAs are hypothesized to play a role in var gene activation.
The full picture of how ncRNAs regulate var gene expression is still not clear. One hypothesis states that ncRNAs are important for the site-specific targeting of epigenetic regulation to the var genes [20]. Histone modifications, as in other organisms, are essential for orchestrating gene expression in Plasmodium [31]. A key epigenetic factor, the P. falciparum variant-silencing SET gene (PfSETvs), which controls histone H3 lysine 36 trimethylation (H3K36me3) on var genes, is reported to play a key role in var gene silencing (Figure 1A) [32]. PfSETvs was shown to be recruited by Pol II to the var gene region [33]. Since Pol II transcribes the lncRNAs, it raises the possibility that expression of the var gene intron represses var gene expression via the lncRNA transcription process itself [33].
lncRNA-mediated nucleosome positioning has been reported in many organisms [34]. A nucleosome occupancy study showed that general var gene expression trends are consistent with the chromatin status of the var gene intron [20]. Another study identified clonally variant chromatin accessibility via ATAC-seq associated with two GC-rich elements flanking an active var gene. A nucleosome occupancy study showed that general var gene expression trends are consistent with the chromatin status of the var gene intron [20]. Another study identified clonally variant accessibility via ATAC-seq linked to two GC-rich elements flanking an active var gene [35]. Although lncRNAs have been implicated in nucleosome positioning in other organisms, there is not, as of yet, any direct interaction or mechanism detected in the Apicomplexa.
lncRNAs are also implicated in gametocyte differentiation in Plasmodium. AP2-G is the master transcriptional regulator of gametocytogenesis that triggers sexual commitment [36]. The heterochromatin protein 1 (HP1) prevents sexual conversion by silencing ap2-g. The P. falciparum gametocyte development 1 protein (GDV1) in turn, targets heterochromatin and triggers HP1 eviction thus permitting sexual conversion. The gdv1 gene has an antisense lncRNA transcript that negatively regulates GDV1 expression, probably via gdv1 mRNA transcription, stability, or translation [37] (Figure 1B). The antisense (as) lncRNA has 5 exons, and the 4th exon overlaps gdv1 in entirety. A knock-out of the aslncRNA by removing the 5′ end of the gene led to an increase in gametocytes [37]. Interestingly, HP1 is also important in var gene regulation. Conditional depletion of HP1, which has been shown to associate with the repressive histone mark H3K9me3 on silenced var genes, revealed that var gene repression and cluster colocalization were lost when HP1 is removed [38, 39] (Figure 1A).
lncRNAs are emerging as regulators of developmental transitions in Toxoplasma gondii
Toxoplasma gondii has two asexual developmental forms: proliferating tachyzoites and latent bradyzoite-cyst forms. Bradyzoites can remain dormant in the host for years. Upon immune suppression, bradyzoites can transition back into proliferating tachyzoites causing disease. Currently, there is no available drug to eliminate bradyzoites, leading to a lifetime risk of recrudescence [40]. Elucidation of factors that drive the transition from tachyzoite to bradyzoite is critical for improving medical treatment. Disruptions of the genetic locus upstream of the gene TGME49_238110 (Replication factor A protein 3, Rfa3) disrupt the transition to bradyzoites. This region harbors Tg-ncRNA-1, an alternatively spliced gene that gives rise to two lncRNA transcripts, one is 2601 bp long and the other is 940 bp long [21, 41] (Figure 1B). The function of this non-coding gene and its transcripts is unclear. The relationship between the lncRNA and the neighboring replication factor gene remains to be elucidated. It was hypothesized that Tg-ncRNA-1 might help recruit a histone modification complex [21] to regulate developmental gene expression during bradyzoite formation [42]. Recently, a master regulator of this differentiation process, a Myb-like transcription factor (BFD1) was discovered. The relationship, if any, between Tg-ncRNA-1 and BFD1 remains unclear.
Host-parasite interaction: lncRNAs from Cryptosporidium parvum manipulate host gene expression
Cryptosporidial infection causes significant changes in host biochemical pathways, including pro-inflammatory reactions, cytoskeleton rearrangement, cell proliferation, and apoptosis (both induced [43] and inhibited [44]). Host lncRNAs respond to C. parvum infection and have been implicated in Hedgehog (Hg) and Wnt signaling pathways [43–45]. The parasite appears to be using lncRNAs to control its environment. Several putative lncRNAs are selectively delivered into intestinal epithelial cells during C. parvum infection [46]. One candidate, Cdg7_FLc_0990, was shown to translocate into the host cell nucleus with the help of host HSP70. Cdg7_FLc_0990 is believed to regulate transcriptional suppression of host genes through recruitment of the H3K9 methylation protein complex G9a/PRDM1 to the promoter region of the target genes [47]. lncRNA may also regulate host gene expression mediated by G9a, but independent of PRDM1 [48]. Another candidate, lncRNA Cdg7_FLc_1000, was reported to suppress several genes related to cell migration and adherence, resulting in attenuation of intestinal epithelial cell migration [49–51] (Figure 1C). How the lncRNAs are transported into the host and the mechanisms used to target specific genes remains unknown. No substantial evidence for base-pairing between the lncRNA and the target gene promoter sequence was observed. It was speculated that the lncRNA and the promoter region might form an RNA/DNA triplex [47].
Other lncRNAs in the Apicomplexa
To date, many lncRNAs have been identified, but few have been validated experimentally. This is due in part to a lack of homology with multicellular eukaryotes and difficult experimental systems involving intracellular parasites. Thousands of lncRNAs have been reported in P. falciparum and P. vivax using sequence-based transcriptomic methods including Serial Analysis of Gene Expression (SAGE) tags [16] and microarrays [52, 53] initially and more recently with RNA-Seq [14, 17, 54]. Some transcripts are processed via splicing and/or the addition of polyadenylated tails [14, 17, 54, 55]. Natural antisense transcripts (NATs) are an important type of lncRNA in Plasmodium, believed to be synthesized by Pol II [24]. NAT introns overlapping sense intron sequence were observed more than would be expected by chance [54]. The expression relationship between antisense and sense transcripts varied under different conditions. According to a SAGE tag analysis in P. falciparum, NATs were inversely correlated to the nearest gene’s sense transcription [16]. A similar antisense-sense pair relationship was seen using RNA-Seq with significantly more negatively correlated sense-antisense pairs than random mRNA pairs, while the transcript-level relationship between long intergenic noncoding RNAs (lincRNAs) and neighboring mRNAs was significantly more positive [17]. However, it was also found that many sense-antisense RNA pairs exhibited positively co-regulated expression profiles during intraerythrocytic development using real-time PCR [56]. A positive correlation between the expression of sense-antisense pairs in both P. vivax and P. falciparum was observed in parasite RNA using isolates taken from patients [53]. A third pattern was found in an RNA-seq study of intraerythrocytic developmental stages, where most NAT expression was independent of sense mRNA transcription, and a significant subset was correlated with neighboring mRNA transcript levels [14]. These observations suggests that both bidirectional and cryptic promoters contribute to lncRNA transcription in Plasmodium [20] and that the results depend on parasite culture conditions, the subset of genes that are analyzed and the resolution of the technology employed. The expression correlation properties of intergenic lncRNAs and NATs with neighboring mRNAs are likely to be different [17].
To infer lncRNA function in silico, one common approach is to look at the functional annotation of the sense mRNA or the nearest neighbor mRNA and assess its transcriptional correlation to the mRNA. NATs in Plasmodium have been associated with a variety of biological processes using this approach [53, 57]. In asexual stages, NATs are over-represented near genes related to translation and proteolysis, perhaps indicating a regulatory role during rapid replication [16]. Ultimately, functional determination requires experimental evidence.
Additional types of lncRNAs are emerging in P. falciparum. Strand-specific, non-polyA-selected RNA sequences reveal hundreds of intriguing P. falciparum circular RNAs (circRNAs), some with experimental validation [17]. Significant human miRNA binding sites were predicted of the circRNAs, giving rise to a possibility of functioning in host-parasite interaction [17]. Another family of lncRNAs encodes telomeric- and subtelomeric-associated lncRNAs whose transcripts are spatially concentrated at the nuclear periphery. It has been hypothesized that these telomere-associated lncRNAs are involved in telomere maintenance [58]. They are grouped into two classes: (i) an ~4 kb transcript class derived from TARE-3 elements; and (ii) a >6 kb transcript class composed of 21-bp repeats from TARE-6 elements [58, 59]. TARE-6 lncRNA 21-bp repeats are predicted to form a stable and repetitive hairpin structure that is able to bind histones and perhaps function as a histone chaperone related to assembly and/or disassembly of subtelomeric heterochromatin [59]. RNA-Seq data demonstrate that subtelomeric lncRNA expression peaks sharply during the asexual parasite invasion stage [17]. This expression pattern is shared with some var lncRNAs [23], leading the authors to suggest a possible unknown coordinated function between them [59].
In T. gondii ME49, NATs were first reported in 2005 in a SAGE analysis of tachyzoite transcripts. A strong inverse relationship between antisense transcript abundance and the corresponding level of sense transcript was observed [60]. This relationship holds true in multiple T. gondii developmental stages [18, 60]. Using strand-specific RNA-Seq technology, hundreds of novel NATs, lincRNAs, and UTRs were computationally predicted. The UTRs of the T. gondii VEG strain are quite long, almost four times longer than other model eukaryotes including AP2-family transcription factors [18]. Since the genome is fairly compact, the long UTRs, especially the 5’ UTR, suggests critical post-transcriptional regulation in T. gondii [18].
Unfortunately, a systematic study of lncRNA in Cryptosporidium spp. is lacking. No genome-wide annotation and analysis of ncRNA exist. Given the new C. parvum IOWA-ATCC reference genome sequence released in CryptoDBi and emerging transcriptome data, this knowledge gap is expected to be filled soon, facilitating comparative studies on the roles of lncRNAs across the Apicomplexa.
Small ncRNAs in apicomplexan parasites
Early studies using homology searches and comparative genomics identified a variety of structurally conserved small ncRNAs (sncRNA) in the Apicomplexa, including snoRNAs, snRNAs and tRNAs [5]. miRNAs are detected in T. gondii, but the apicomplexan RNAi pathway is significantly different from other eukaryotes. Plasmodium and Cryptosporidium are RNAi-deficient based on bioinformatics and functional analysis [61, 62] and the RNAi mechanism in T. gondii is atypical. In recent years, many novel small RNAs and new functions for sncRNAs are emerging as a result of small RNA sequencing and extracellular vesicle (EV) research [63–66]. These findings suggest additional gene regulation strategies are employed by protist pathogens in their interactions with the host.
Although Plasmodium lacks endogenous miRNAs, in the case of hosts with sickle cell anemia, miRNAs from the host sickle cell erythrocytes can be translocated into the parasite and inhibit the parasite translation process by impairing ribosomal loading thus contributing to the host resistance to malaria observed in these individuals [65]. EVs, which include exosomes and microvesicles, have been shown to be important in cell-cell communications. The information exchange and resulting gene modulation can be multiway including host cell-to-cell, host-to-parasite, parasite-to-host and parasite-to-parasite [63–66]. It is noteworthy that the host miRNA-Argonaute 2 complex has been detected in EVs [66] and has been shown to target and regulate gene expression in P. falciparum in one study [67]. These findings raise the intriguing hypothesis that the parasite might utilize host Argonaute 2 for its own gene regulation [68].
Deep sequencing of RNA from intraerythrocytic P. falciparum developmental stages has revealed a collection of novel intermediate-size ncRNAs including novel snoRNAs and unclassified small RNAs. Many of these unclassified RNAs are conserved among different Plasmodium species and are differentially expressed between early and late intraerythrocytic stages [69]. Additionally, a potential novel class of sncRNAs derived from tRNA fragments was revealed in P. falciparum [68]. tRNA-derived small RNAs (tsRNAs) have been reported in several protist organisms, including Tetrahymena [70], Giardia lamblia [71], Trypanosoma cruzi [72], and in Leishmania donovani exosomes [73]. In humans, tsRNAs have been associated with cancer, neurodegenerative disorders, viral infection, and other pathological conditions [3]. The mechanism of tsRNA function is still unclear. tRFs harbor similarity with miRNAs but may use an alternative pathway to RNAi [74, 75]. The function of tsRNAs in Plasmodium remains uncharacterized.
In T. gondii, thousands of miRNAs have been detected via deep sequencing and computational prediction. T. gondii miRNAs related to 2 metazoan miRNA families have been reported [19]. Many of the putative miRNA target genes are associated with T. gondii virulence or invasion [76]. It is also speculated that T. gondii may export miRNAs into its hosts via extracellular vesicles to manipulate its host [77, 78]. Computational analyses reveal a binding capacity for some Toxoplasma miRNAs to host mRNAs, but this has not been experimentally confirmed [77]. Intriguingly, T. gondii has a chimeric RNAi mechanism with plant/fungal-like machinery and a metazoan-like Argonaute [79]. Significant effort has been directed at understanding how T. gondii utilizes its miRNAs to achieve RNA silencing. T. gondii argonaute (TgAgo) lacks the canonical DDE/H catalytic triad and displays weak target RNA cleavage activity [80]. In general, protozoan miRNAs do not share high similarity with other eukaryotes [63].
Based on available genome annotation for Cryptosporidium sppsi, RNAi-related genes are absent, suggesting that the canonical RNAi pathway is lost. However, the possibility that Cryptosporidium has alternative RNAi pathways cannot be ruled out. Systematic analyses of lncRNA and sncRNA are needed in this and other understudied parasite species.
Challenges and limitations to the study of ncRNA in apicomplexan parasites
There are two significant challenges that face most ncRNA studies in Apicomplexan and other parasites. The first significant challenge is the identification of the ncRNA itself. Not all non-coding or low-coding potential RNA sequences represent classes of ncRNA. Developmental time course or differential condition gene expression data in addition to comparative genomic analyses are often needed to identify some classes on ncRNA. The second significant challenge is the determination of the ncRNA’s function. Currently, the different apicomplexan parasite communities are at very different stages with respect to these challenges, with most communities still struggling to identify ncRNAs in their parasite’s genome. Thus, we focus more heavily on the limitations and challenges to the identification. However, equally daunting challenges exist for functional characterization of ncRNAs.
Challenges for lncRNA study
The intrinsic features of lncRNA that facilitate plasticity in regulatory roles also challenge lncRNA detection and study. lncRNAs function by both sequence- and structure-based mechanisms. lncRNA structures, like tRNA structure, can be conserved without maintaining primary sequence conservation [7]. Unlike mRNAs harboring coding sequences (CDS) that can be ascertained directly and easily from the transcript, structural and putative functional domains in lncRNAs cannot be inferred solely based on primary sequence information. Thus, lncRNA detection is quite difficult and needs special attention.
In organisms with compact genome sequences, a characteristic of most apicomplexan parasites, disambiguation of lncRNA boundaries from mRNAs and transcriptional noise is a further challenge. Typical pipelines to identify lncRNAs utilize two steps: (i) transcript assembly and (ii) lncRNA discovery. High gene density can lead to artificially fused transcripts during assembly due to overlapping transcripts from the same strand. This phenomenon increases the rate of both false positive and false negative lncRNA predictions. For the second step, two approaches have been developed to separate lncRNAs from mRNAs, alignment-based and alignment-free. Alignment-based approaches search databases of known mRNAs and look for transcripts without a match (e.g. the tool Coding Potential Calculator CPCii) or apply comparative sequence analysis with related organisms to look for transcripts without coding sequence evolution pressure (e.g. the tool PhyloCSFiii). These approaches are subject to false-positive lncRNA prediction as a result of misassembled transcripts (missing introns, hybrid transcripts, or uncalled mRNAs resulting from gaps present in the genome sequence used to map transcripts); incompleteness of the mRNA databases for each species; and a lack of sufficient genome sequences and data from related species. Alignment-free tools, such as CPAT (Coding Potential Assessment Tooliv) and PLEK (a predictor of lncRNAs and messenger RNAs based on an improved k-mer schemev), are fast and less affected by transcript integrity. The accuracy of alignment-free tools relies on the high quality of training data or similarity with the species that the tool was designed for. The default training data are often from model eukaryotes, significantly limiting their use in protist studies. Since we lack extensive knowledge of lncRNA in apicomplexan parasites and because most protists are distantly related to most model species, interpretation of computational predictions requires caution. The best features to distinguish mRNA from lncRNA might not be the same as those identified by the algorithm. Popular features such as GC content, the Fickett TESTCODE statistic, and hexameriv usage bias may not work well in organisms with compact genomes or skewed GC content.
The biggest challenge is lncRNA function prediction. Currently, computational inference of lncRNA associated biological processes often occurs by assessing the functions of neighboring genes and co-expressed mRNAs [81]. However, this method cannot identify the specific role of a lncRNA and is limited in apicomplexan and other parasites due to the high percentage of uncharacterized proteins. Another strategy is to infer functionality from homology with known lncRNAs [7]. Because of high sequence divergence, classic homology search approaches have minimal power to detect conserved biological domains in lncRNAs across species and phyla. Conservation of stem and loop structures can facilitate classification and provide insights into potential functions [7, 82]. However, accurate prediction of folding for long RNA molecules is difficult due to the enormous number of possible spatial structures that can form under different environmental conditions. Additionally, methods for comparing computational results remain sparse. Experimental validation of lncRNA structure is the gold standard, but it too can be difficult, although CRISPR is making it easier, except for some antisense transcripts where alterations to the antisense will also affect the sense transcript. The large evolutionary distance that protists have from most model eukaryotes results in poorly conserved and novel lncRNAs and along with evolution of differing repertoires of interacting partners. The general lack of knowledge concerning most lncRNA functions and mechanisms of action hinders the interpretation of related apicomplexan lncRNAs.
Finally, lncRNAs tend to be much less stable and abundant than mRNAs [83]. Even routine RNA isolation methods in the same lab may result in variable lncRNA transcript yields. While many lncRNAs appear to be poly-adenylated and detectable in poly(A)-enriched libraries, determination of the full repertoire of lncRNAs requires an analysis of ribosomal RNA depleted total RNA libraries. However, no commercial kits specific for apicomplexan or other protist parasite rRNA removal are available for this purpose and the existing ribosomal RNA depletion methods are insufficient and leave high levels of rRNA, effectively reducing non-rRNA reads [14, 17]. Challenges exist at every step of lncRNA study.
Challenges for sncRNA study
New approaches and algorithms have been developed recently for sncRNA detection. Tools like snoReportvi and RNAsnoopvii were developed to predict snoRNAs based on support vector machine (SVM) approaches whose accuracy of prediction depends on how similar the sncRNA structures are to the data used to train the algorithm. Both high false positives and false negatives are possible when protist RNA is studied. Novel small RNA genes that are species- and parasite-specific are likely to be under detected and due to their lack of homology or sequence divergence e.g. snoRNA variants.
Small RNA-Seq is considered to be the most effective and efficient approach to detect small RNA expression. However, separation of parasitic small RNA transcripts <~25 bp from environmental bacterial or host contamination and RNA degradation products remains a challenge due to algorithm limitations in short read alignment.
Finally, new ncRNA types and associated functions emerge often. Recent studies demonstrated that ncRNA, including lncRNAs, circRNAs and primary miRNAs (pri-miRNAs) can also produce small peptides or proteins, some of which are experimentally validated as functional [6, 84]. The use of short open reading frames as an identifier for ncRNA needs further consideration. The blurring boundaries between ncRNA and mRNA makes the study of ncRNA more challenging and exciting.
Possible solutions and future directions
To better characterize lncRNA from transcriptome data, strand-specific approaches are essential to disambiguate sense and NAT transcription. When using Illumina approaches, paired-end sequencing is also highly recommended to increase the likelihood of distinguishing neighboring gene boundaries. Current long-read approaches such as Iso-Seq (Pacific Biosystems) and single molecule pore-sequencing approaches (Oxford Nanopore Technologies, ONT) can provide full-length transcripts without assembly, but some correction to the base calls may be needed. lncRNA boundaries and isoforms can easily be identified and some RNA modifications may be discernable from long-read single-molecule platforms (ONT). Adjustment of parameters during transcript assembly can also increase the accuracy of transcriptome assembly. De novo assembly tools such as Trinityviii provide a parameter to decrease the fusion of transcripts in compact, gene-dense genomes and genome-based assemblers like StringTieix control the minimum gap distance between two proximal transcripts.
Since lncRNAs are usually less abundant and stable than mRNAs, deeper sequencing than a typical RNA-Seq experiment is recommended for their discovery. To achieve better depletion of rRNA in non-model species, a customized rRNA depletion method can be deployed. Efficient and highly-specific rRNA removal approaches using biotinylated DNA oligos have been tested successfully in trypanosomatid rRNAs [85] and can be applied to other species.
To obtain better inference of lncRNA function, genomic technologies such as ChIRP-Seq (Chromatin Isolation by RNA Purification) and LIGR-seq (LIGation of interacting RNA followed by high-throughput sequencing) can help identify lncRNA interactions with DNA, RNA, and protein. Additionally, new strategies to computationally infer lncRNA functionalities are emerging. One of these is a kmer-based method to predict biological clustering and functional domains [86]. Also, a synteny assisted ncRNA ortholog search strategy has been successfully applied to detect lncRNA homology between mammalian and insect lncRNAs [87]. Increasing the representation of apicomplexan and evolutionary diverse lncRNAs in established RNA and sequence repositories would particularly facilitate the discovery of lncRNA families and functional domains/mechanisms. Finally, advanced molecular techniques including RNA-FISH and CRISPR/CAS9 when possible will help reveal the subcellular location and help reveal the function of the ncRNA targets.
With respect to sncRNA research (targeting small RNAs other than miRNA), longer read lengths, e.g. 150 bp single-end (SE) and pair-end (PE) would help to increase the confidence for identification of full-length small RNA transcripts and thus separate them from RNA degradation products. Specifically, replicates should be used to improve the power of sncRNA discovery. Sequencing with multiple methods e.g., 75bp SE and 150bp SE would help to detect and reduce technical bias.
Concluding remarks
ncRNAs play vital roles in apicomplexan parasite biology. They participate in both parasite developmental processes and host-parasite interactions. Advances in sequencing technologies and functional characterization have revealed many novel ncRNAs and implicated several in aspects of gene regulation. However, most ncRNA candidates require greater characterization in order to discern their function (see Outstanding Questions). Careful RNA-Seq design and customized data analyses are necessary to identify new ncRNAs. Genetic manipulation explicitly targeting ncRNAs and suspected molecular partners is needed in order to decipher their numerous biological roles.
Outstanding Questions.
How much crosstalk between the host and parasite happens at the ncRNA level?
How are apicomplexan ncRNAs transported into parasites and out to host cells? What is the recognition signal?
What additional apicomplexan proteins interact with ncRNAs?
How do sncRNAs compensate for the lack of miRNAs in translation repression regulation in apicomplexan parasites?
Can a sufficient number of features be identified to permit the computational detection of putative lncRNAs from mRNA in the Apicomplexa or other protist pathogens?
Is there a correlation between the genomic location of lncRNAs and their function? Does their genomic position matter?
Highlights.
Recent advances in experimental and sequencing technologies have revealed new classes and several new functions for non-coding RNA (ncRNA) in the Apicomplexa.
Some ncRNAs have been shown to be associated with the epigenetic machinery and participate in parasite development and manipulation of host gene expression.
ncRNAs remain understudied in the Apicomplexa. Experimental and algorithmic methodologies need to be optimized to better understand ncRNA in these highly divergent, non-model species.
Acknowledgements and funding information
This work was supported in part by funding from NIH/NIAID 1R21AI144779-01A1.
Glossary
- Bidirectional lncRNA
A category of lncRNAs transcribed from a bidirectional promoter.
- Circular RNA (circRNA)
A type of closed ncRNA, in which the 5′ and 3′ termini are covalently linked by back-splicing (head-to-tail splicing).
- Cryptic promoter
An epigenetically silenced and normally inactive promoter which can be activated by genetic or extraneous alterations.
- Intronic lncRNA
A category of lncRNAs that are transcribed from intronic regions of other genes.
- Long intergenic noncoding RNA (lincRNA)
A group of ncRNAs that do not overlap protein-coding genes.
- Long non-coding RNA (lncRNA)
A type of ncRNA that is > 200 nucleotides.
- MicroRNA (miRNA)
A class of sncRNA that is 18–25 nucleotides and plays key roles in post-transcriptional gene regulation.
- Natural antisense transcript (NAT)
A category of lncRNAs that are transcribed from the opposite strand to a sense protein-coding with partial or complete complementarity.
- Non-coding RNA (ncRNA)
An RNA molecule transcribed from DNA but not translated into a protein.
- Read-through transcription
Occurs when RNA polymerases fail to terminate properly and continue transcribing beyond the canonical termination site.
- RNA secondary structure
The structure formed by intramolecular hydrogen bonding between bases within an RNA molecule resulting in folding into stem and loop or psequdoknot structures.
- RNase MRP RNA
The RNA subunit of the RNase for mitochondrial RNA processing (MRP) enzyme complex.
- Sense lncRNA
A group of lncRNAs that are transcribed from the same DNA strand as the sense protein-coding gene with partial or complete complementarity.
- Short non-coding RNA (sncRNA)
Defined as ncRNA that is < 200 nucleotides.
- Small nuclear ribonucleoprotein (snRNP)
An RNA-protein complex that accumulates in the nucleus and participates in RNA splicing in the splicesome.
- Small nuclear RNA (snRNA)
A class of sncRNAs that forms snRNPs associated with intron splicing and other RNA processing.
- Small nucleolar ribonucleoprotein (snoRNP)
An RNA-protein complex that guides sequence-specific 2′-O-ribose methylation and psuedouridylation of other RNAs, mainly ribosomal RNAs.
- Small nucleolar RNA (snoRNA)
A class of sncRNA that forms snoRNPs. Two main classes are C/D box and H/ACA box, associated with methylation and pseudouridylation, respectively.
- Telomeric repeat-containing RNA (TERRA)
A category of ncRNA that is transcribed from telomeres.
- Transcriptional noise
Aberrant, or unexplained transcription of unspecified origin.
References
- 1.Kalvari I et al. (2018) Rfam 13.0: shifting to a genome-centric resource for non-coding RNA families. Nucleic Acids Res 46 (D1), D335–D342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cech TR and Steitz JA (2014) The noncoding RNA revolution-trashing old rules to forge new ones. Cell 157 (1), 77–94. [DOI] [PubMed] [Google Scholar]
- 3.Anderson P and Ivanov P (2014) tRNA fragments in human health and disease. FEBS Lett 588 (23), 4297–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Falaleeva M and Stamm S (2013) Processing of snoRNAs as a new source of regulatory non-coding RNAs: snoRNA fragments form a new class of functional RNAs. Bioessays 35 (1), 46–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Matrajt M (2010) Non-coding RNA in apicomplexan parasites. Mol Biochem Parasitol 174 (1), 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Quinn JJ and Chang HY (2016) Unique features of long non-coding RNA biogenesis and function. Nat Rev Genet 17 (1), 47–62. [DOI] [PubMed] [Google Scholar]
- 7.Diederichs S (2014) The four dimensions of noncoding RNA conservation. Trends Genet 30 (4), 121–3. [DOI] [PubMed] [Google Scholar]
- 8.Ulitsky I et al. (2011) Conserved function of lincRNAs in vertebrate embryonic development despite rapid sequence evolution. Cell 147 (7), 1537–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tsoi LC et al. (2015) Analysis of long non-coding RNAs highlights tissue-specific expression patterns and epigenetic profiles in normal and psoriatic skin. Genome Biol 16, 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Necsulea A et al. (2014) The evolution of lncRNA repertoires and expression patterns in tetrapods. Nature 505 (7485), 635–40. [DOI] [PubMed] [Google Scholar]
- 11.Tang R et al. (2019) LncRNA GAS5 regulates vascular smooth muscle cell cycle arrest and apoptosis via p53 pathway. Biochim Biophys Acta Mol Basis Dis 1865 (9), 2516–2525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang L et al. (2018) LncRNA SNHG1 regulates cerebrovascular pathologies as a competing endogenous RNA through HIF-1alpha/VEGF signaling in ischemic stroke. J Cell Biochem 119 (7), 5460–5472. [DOI] [PubMed] [Google Scholar]
- 13.Kissinger JC and DeBarry J (2011) Genome cartography: charting the apicomplexan genome. Trends Parasitol 27 (8), 345–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Siegel TN et al. (2014) Strand-specific RNA-Seq reveals widespread and developmentally regulated transcription of natural antisense transcripts in Plasmodium falciparum. BMC Genomics 15, 150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Blazejewski T et al. (2015) Systems-based analysis of the Sarcocystis neurona genome identifies pathways that contribute to a heteroxenous life cycle. mBio 6 (1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gunasekera AM et al. (2004) Widespread distribution of antisense transcripts in the Plasmodium falciparum genome. Mol Biochem Parasitol 136 (1), 35–42. [DOI] [PubMed] [Google Scholar]
- 17.Broadbent KM et al. (2015) Strand-specific RNA sequencing in Plasmodium falciparum malaria identifies developmentally regulated long non-coding RNA and circular RNA. BMC Genomics 16, 454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ramaprasad A et al. (2015) Comprehensive evaluation of Toxoplasma gondii VEG and Neospora caninum LIV genomes with tachyzoite stage transcriptome and proteome defines novel transcript features. PLoS One 10 (4), e0124473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang J et al. (2012) A comparative study of small RNAs in Toxoplasma gondii of distinct genotypes. Parasit Vectors 5, 186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vembar SS et al. (2014) Noncoding RNAs as emerging regulators of Plasmodium falciparum virulence gene expression. Curr Opin Microbiol 20, 153–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Patil V et al. (2012) Disruption of the expression of a non-coding RNA significantly impairs cellular differentiation in Toxoplasma gondii. Int J Mol Sci 14 (1), 611–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Amit-Avraham I et al. (2015) Antisense long noncoding RNAs regulate var gene activation in the malaria parasite Plasmodium falciparum. Proc Natl Acad Sci U S A 112 (9), E982–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Epp C et al. (2009) Chromatin associated sense and antisense noncoding RNAs are transcribed from the var gene family of virulence genes of the malaria parasite Plasmodium falciparum. RNA 15 (1), 116–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Militello KT et al. (2005) RNA polymerase II synthesizes antisense RNA in Plasmodium falciparum. RNA 11 (4), 365–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gannoun-Zaki L et al. (2005) A silenced Plasmodium falciparum var promoter can be activated in vivo through spontaneous deletion of a silencing element in the intron. Eukaryot Cell 4 (2), 490–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Frank M et al. (2006) Strict pairing of var promoters and introns is required for var gene silencing in the malaria parasite Plasmodium falciparum. J Biol Chem 281 (15), 9942–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bryant JM et al. (2017) CRISPR/Cas9 Genome Editing Reveals That the Intron Is Not Essential for var2csa Gene Activation or Silencing in Plasmodium falciparum. MBio 8 (4). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jing Q et al. (2018) Plasmodium falciparum var Gene Is Activated by Its Antisense Long Noncoding RNA. Front Microbiol 9, 3117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Guizetti J et al. (2016) Trans-acting GC-rich non-coding RNA at var expression site modulates gene counting in malaria parasite. Nucleic Acids Res 44 (20), 9710–9718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Barcons-Simon A et al. (2020) CRISPR Interference of a Clonally Variant GC-Rich Noncoding RNA Family Leads to General Repression of var Genes in Plasmodium falciparum. mBio 11 (1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Voss TS et al. (2014) Epigenetic memory takes center stage in the survival strategy of malaria parasites. Curr Opin Microbiol 20, 88–95. [DOI] [PubMed] [Google Scholar]
- 32.Jiang L et al. (2013) PfSETvs methylation of histone H3K36 represses virulence genes in Plasmodium falciparum. Nature 499 (7457), 223–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ukaegbu UE et al. (2014) Recruitment of PfSET2 by RNA polymerase II to variant antigen encoding loci contributes to antigenic variation in P. falciparum. PLoS Pathog 10 (1), e1003854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bohmdorfer G and Wierzbicki AT (2015) Control of Chromatin Structure by Long Noncoding RNA. Trends Cell Biol 25 (10), 623–632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ruiz JL et al. (2018) Characterization of the accessible genome in the human malaria parasite Plasmodium falciparum. Nucleic Acids Res 46 (18), 9414–9431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kafsack BF et al. (2014) A transcriptional switch underlies commitment to sexual development in malaria parasites. Nature 507 (7491), 248–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Filarsky M et al. (2018) GDV1 induces sexual commitment of malaria parasites by antagonizing HP1-dependent gene silencing. Science 359 (6381), 1259–1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bunnik EM et al. (2018) Changes in genome organization of parasite-specific gene families during the Plasmodium transmission stages. Nat Commun 9 (1), 1910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Brancucci NMB et al. (2014) Heterochromatin protein 1 secures survival and transmission of malaria parasites. Cell Host Microbe 16 (2), 165–176. [DOI] [PubMed] [Google Scholar]
- 40.Weiss LM and Kim K (2000) The development and biology of bradyzoites of Toxoplasma gondii. Front Biosci 5, D391–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lescault PJ et al. (2010) Genomic data reveal Toxoplasma gondii differentiation mutants are also impaired with respect to switching into a novel extracellular tachyzoite state. PLoS One 5 (12), e14463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Behnke MS et al. (2008) The transcription of bradyzoite genes in Toxoplasma gondii is controlled by autonomous promoter elements. Mol Microbiol 68 (6), 1502–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Deng M et al. (2004) Cryptosporidium parvum regulation of human epithelial cell gene expression. Int J Parasitol 34 (1), 73–82. [DOI] [PubMed] [Google Scholar]
- 44.Mirhashemi ME et al. (2018) Transcriptome analysis of pig intestinal cell monolayers infected with Cryptosporidium parvum asexual stages. Parasit Vectors 11 (1), 176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liu TL et al. (2018) Expression Profiles of mRNA and lncRNA in HCT-8 Cells Infected With Cryptosporidium parvum IId Subtype. Front Microbiol 9, 1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang Y et al. (2017) Delivery of Parasite RNA Transcripts Into Infected Epithelial Cells During Cryptosporidium Infection and Its Potential Impact on Host Gene Transcription. J Infect Dis 215 (4), 636–643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang Y et al. (2017) Delivery of parasite Cdg7_Flc_0990 RNA transcript into intestinal epithelial cells during Cryptosporidium parvum infection suppresses host cell gene transcription through epigenetic mechanisms. Cell Microbiol 19 (11). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhao GH et al. (2018) Nuclear delivery of parasite Cdg2_FLc_0220 RNA transcript to epithelial cells during Cryptosporidium parvum infection modulates host gene transcription. Vet Parasitol 251, 27–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ming Z et al. (2017) Involvement of Cryptosporidium parvum Cdg7_FLc_1000 RNA in the Attenuation of Intestinal Epithelial Cell Migration via Trans-Suppression of Host Cell SMPD3. J Infect Dis 217 (1), 122–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ming Z et al. (2018) Trans-suppression of defense DEFB1 gene in intestinal epithelial cells following Cryptosporidium parvum infection is associated with host delivery of parasite Cdg7_FLc_1000 RNA. Parasitol Res 117 (3), 831–840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ming Z et al. (2018) Trans-suppression of host CDH3 and LOXL4 genes during Cryptosporidium parvum infection involves nuclear delivery of parasite Cdg7_FLc_1000 RNA. Int J Parasitol 48 (6), 423–431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Boopathi PA et al. (2013) Revealing natural antisense transcripts from Plasmodium vivax isolates: evidence of genome regulation in complicated malaria. Infect Genet Evol 20, 428–43. [DOI] [PubMed] [Google Scholar]
- 53.Subudhi AK et al. (2014) Natural antisense transcripts in Plasmodium falciparum isolates from patients with complicated malaria. Exp Parasitol 141, 39–54. [DOI] [PubMed] [Google Scholar]
- 54.Sorber K et al. (2011) RNA-Seq analysis of splicing in Plasmodium falciparum uncovers new splice junctions, alternative splicing and splicing of antisense transcripts. Nucleic Acids Res 39 (9), 3820–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kim A et al. (2017) Characterization of P. vivax blood stage transcriptomes from field isolates reveals similarities among infections and complex gene isoforms. Sci Rep 7 (1), 7761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Raabe CA et al. (2010) A global view of the nonprotein-coding transcriptome in Plasmodium falciparum. Nucleic Acids Res 38 (2), 608–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Subudhi AK et al. (2014) An in vivo transcriptome data set of natural antisense transcripts from Plasmodium falciparum clinical isolates. Genom Data 2, 393–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Broadbent KM et al. (2011) A global transcriptional analysis of Plasmodium falciparum malaria reveals a novel family of telomere-associated lncRNAs. Genome Biol 12 (6), R56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sierra-Miranda M et al. (2012) Two long non-coding RNAs generated from subtelomeric regions accumulate in a novel perinuclear compartment in Plasmodium falciparum. Mol Biochem Parasitol 185 (1), 36–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Radke JR et al. (2005) The transcriptome of Toxoplasma gondii. BMC Biol 3, 26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mueller AK et al. (2014) RNAi in Plasmodium. Curr Pharm Des 20 (2), 278–83. [DOI] [PubMed] [Google Scholar]
- 62.Abrahamsen MS et al. (2004) Complete genome sequence of the apicomplexan, Cryptosporidium parvum. Science 304 (5669), 441–5. [DOI] [PubMed] [Google Scholar]
- 63.Bayer-Santos E et al. (2017) Non-coding RNAs in Host-Pathogen Interactions: Subversion of Mammalian Cell Functions by Protozoan Parasites. Front Microbiol 8, 474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Regev-Rudzki N et al. (2013) Cell-cell communication between malaria-infected red blood cells via exosome-like vesicles. Cell 153 (5), 1120–33. [DOI] [PubMed] [Google Scholar]
- 65.LaMonte G et al. (2012) Translocation of sickle cell erythrocyte microRNAs into Plasmodium falciparum inhibits parasite translation and contributes to malaria resistance. Cell Host Microbe 12 (2), 187–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Mantel PY et al. (2016) Infected erythrocyte-derived extracellular vesicles alter vascular function via regulatory Ago2-miRNA complexes in malaria. Nat Commun 7, 12727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wang Z et al. (2017) Red blood cells release microparticles containing human argonaute 2 and miRNAs to target genes of Plasmodium falciparum. Emerg Microbes Infect 6 (8), e75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wang Z et al. (2019) Genome-wide identification and characterization of transfer RNA-derived small RNAs in Plasmodium falciparum. Parasit Vectors 12 (1), 36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wei C et al. (2014) Deep profiling of the novel intermediate-size noncoding RNAs in intraerythrocytic Plasmodium falciparum. PLoS One 9 (4), e92946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Couvillion MT et al. (2010) A growth-essential Tetrahymena Piwi protein carries tRNA fragment cargo. Genes Dev 24 (24), 2742–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Liao JY et al. (2014) Both endo-siRNAs and tRNA-derived small RNAs are involved in the differentiation of primitive eukaryote Giardia lamblia. Proc Natl Acad Sci U S A 111 (39), 14159–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Reifur L et al. (2012) Distinct subcellular localization of tRNA-derived fragments in the infective metacyclic forms of Trypanosoma cruzi. Mem Inst Oswaldo Cruz 107 (6), 816–9. [DOI] [PubMed] [Google Scholar]
- 73.Lambertz U et al. (2015) Small RNAs derived from tRNAs and rRNAs are highly enriched in exosomes from both old and new world Leishmania providing evidence for conserved exosomal RNA Packaging. BMC Genomics 16, 151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kim HK et al. (2017) A transfer-RNA-derived small RNA regulates ribosome biogenesis. Nature 552 (7683), 57–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kuscu C et al. (2018) tRNA fragments (tRFs) guide Ago to regulate gene expression post-transcriptionally in a Dicer-independent manner. RNA 24 (8), 1093–1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Xu MJ et al. (2013) Comparative characterization of microRNA profiles of different genotypes of Toxoplasma gondii. Parasitology 140 (9), 1111–8. [DOI] [PubMed] [Google Scholar]
- 77.Sacar MD et al. (2014) Computational prediction of microRNAs from Toxoplasma gondii potentially regulating the hosts’ gene expression. Genomics Proteomics Bioinformatics 12 (5), 228–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Silva VO et al. (2018) Extracellular vesicles isolated from Toxoplasma gondii induce host immune response. Parasite Immunol 40 (9), e12571. [DOI] [PubMed] [Google Scholar]
- 79.Braun L et al. (2010) A complex small RNA repertoire is generated by a plant/fungal-like machinery and effected by a metazoan-like Argonaute in the single-cell human parasite Toxoplasma gondii. PLoS Pathog 6 (5), e1000920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Musiyenko A et al. (2012) PRMT1 methylates the single Argonaute of Toxoplasma gondii and is important for the recruitment of Tudor nuclease for target RNA cleavage by antisense guide RNA. Cell Microbiol 14 (6), 882–901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Liao Q et al. (2014) Genome-wide identification and functional annotation of Plasmodium falciparum long noncoding RNAs from RNA-seq data. Parasitol Res 113 (4), 1269–81. [DOI] [PubMed] [Google Scholar]
- 82.Smith MA et al. (2013) Widespread purifying selection on RNA structure in mammals. Nucleic Acids Res 41 (17), 8220–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Clark MB et al. (2012) Genome-wide analysis of long noncoding RNA stability. Genome Res 22 (5), 885–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Wu P et al. (2020) Emerging role of tumor-related functional peptides encoded by lncRNA and circRNA. Mol Cancer 19 (1), 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kraus AJ et al. (2019) Efficient and specific oligo-based depletion of rRNA. Sci Rep 9 (1), 12281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Kirk JM et al. (2018) Functional classification of long non-coding RNAs by k-mer content. Nat Genet 50 (10), 1474–1482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Quinn JJ et al. (2016) Rapid evolutionary turnover underlies conserved lncRNA-genome interactions. Genes Dev 30 (2), 191–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Engreitz JM et al. (2016) Local regulation of gene expression by lncRNA promoters, transcription and splicing. Nature 539 (7629), 452–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Allen MA et al. (2014) Global analysis of p53-regulated transcription identifies its direct targets and unexpected regulatory mechanisms. Elife 3, e02200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Shearwin KE et al. (2005) Transcriptional interference--a crash course. Trends Genet 21 (6), 339–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Kino T et al. (2010) Noncoding RNA gas5 is a growth arrest- and starvation-associated repressor of the glucocorticoid receptor. Sci Signal 3 (107), ra8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Kindgren P et al. (2018) Transcriptional read-through of the long non-coding RNA SVALKA governs plant cold acclimation. Nat Commun 9 (1), 4561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Hobson DJ et al. (2012) RNA polymerase II collision interrupts convergent transcription. Mol Cell 48 (3), 365–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Gong C and Maquat LE (2011) lncRNAs transactivate STAU1-mediated mRNA decay by duplexing with 3’ UTRs via Alu elements. Nature 470 (7333), 284–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Carrieri C et al. (2012) Long non-coding antisense RNA controls Uchl1 translation through an embedded SINEB2 repeat. Nature 491 (7424), 454–7. [DOI] [PubMed] [Google Scholar]
- 96.Yoon JH et al. (2012) LincRNA-p21 suppresses target mRNA translation. Mol Cell 47 (4), 648–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Cesana M et al. (2011) A long noncoding RNA controls muscle differentiation by functioning as a competing endogenous RNA. Cell 147 (2), 358–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Hansen TB et al. (2013) Natural RNA circles function as efficient microRNA sponges. Nature 495 (7441), 384–8. [DOI] [PubMed] [Google Scholar]
- 99.Tripathi V et al. (2010) The nuclear-retained noncoding RNA MALAT1 regulates alternative splicing by modulating SR splicing factor phosphorylation. Mol Cell 39 (6), 925–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Yin QF et al. (2012) Long noncoding RNAs with snoRNA ends. Mol Cell 48 (2), 219–30. [DOI] [PubMed] [Google Scholar]
- 101.Kopp F and Mendell JT (2018) Functional Classification and Experimental Dissection of Long Noncoding RNAs. Cell 172 (3), 393–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Li F et al. (2008) Nuclear non-coding RNAs are transcribed from the centromeres of Plasmodium falciparum and are associated with centromeric chromatin. J Biol Chem 283 (9), 5692–8. [DOI] [PubMed] [Google Scholar]
Resources
- i. https://cryptodb.org/
- ii. http://cpc.gao-lab.org/
- iii. https://github.com/mlin/PhyloCSF/wiki .
- iv. http://lilab.research.bcm.edu/cpat/
- v. https://sourceforge.net/projects/plek/files/
- vi. https://github.com/joaovicers/snoreport2 .
- vii. http://www.bioinf.uni-leipzig.de/~htafer/RNAsnoop/RNAsnoop.html .
- viii. https://github.com/trinityrnaseq/trinityrnaseq/wiki .
- ix. https://ccb.jhu.edu/software/stringtie/


