Abstract
Chimeric transcription factors drive lineage-specific oncogenesis but are notoriously difficult to target. Alveolar rhabdomyosarcoma (aRMS) is an aggressive childhood soft tissue sarcoma transformed by the pathognomonic PAX3–FOXO1 fusion protein, which governs a core regulatory circuitry transcription factor (CRC TF) network. Here we show that the histone lysine demethylase KDM4B is a therapeutic vulnerability for PAX3–FOXO1+ RMS. Genetic and pharmacologic inhibition of KDM4B significantly delays tumor growth by disrupting the expression of CRC TFs caused by epigenetic alterations of PAX3–FOXO1-governed super enhancers. Combining KDM4B inhibition with cytotoxic chemotherapy leads to significant tumor regression in preclinical PAX3–FOXO1+ RMS models. In summary, we have identified a targetable mechanism required for maintenance of the PAX3-FOXO1-related CRC TF network, which may translate to a therapeutic approach for fusion-positive RMS.
One Sentence Summary:
KDM4 is a therapeutic target in PAX3-FOXO1 positive rhabdomyosarcoma.
INTRODUCTION
Dysregulation of gene transcription is a hallmark of cancer and leads to dependencies of cancer cells on aberrant transcriptomic programs (1). In particular, a small set of interconnected lineage-specific transcription factors (TFs) form a core regulatory circuitry (CRC) that establishes and maintains cell identity (2-4), and grants selective dependencies of distinct cancer types (5-10). Cancer genomic sequencing has identified hundreds of fusion genes that may have regulatory functions in gene transcription by creating neomorphic chimeric TFs (https://ccsm.uth.edu/FusionGDB/), many of which are known oncogenic drivers (11, 12). While the functions of chimeric TFs in core regulatory circuitry remain largely unknown, there is strong evidence that PAX3-FOXO1 is a chimeric TF that plays an essential role in constructing a CRC TF network in cancer (13). Although CRC TFs are essential to cancer cell survival (5, 6, 9, 10, 13-15), selective pharmacologic targeting of CRC TFs is very challenging.
Rhabdomyosarcoma (RMS) is the most common soft tissue sarcoma in children (16, 17), and is classified into two main histologic subtypes, alveolar RMS (aRMS) and embryonal RMS (eRMS). The cell of origin of aRMS may derive from fetal myoblasts, whose transformation is primarily driven by the pathognomonic fusion oncoprotein PAX3–FOXO1 or its variant PAX7–FOXO1 (18-20). PAX3–FOXO1 fusion-positive aRMS is more aggressive than fusion-negative RMS and has a higher rate of metastasis and poorer prognosis (16, 21, 22). Although current treatment modalities have steadily improved the survival of patients with low-risk RMS, the outcome for those with high-risk disease, fusion-positive RMS, in particular, remains dismal (13). Among the few patients who are cured, aggressive chemotherapy and radiotherapy often lead to long-term adverse effects, including secondary malignancies (17). Thus, there is an urgent need to better understand the biology of fusion-positive RMS in order to develop more effective therapies.
As an oncogenic transcription factor, PAX3–FOXO1 is essential for the development, proliferation and survival of aRMS. It regulates the expression of multiple oncogenes, including IGF2, FGFR4, MYCN, MET, and ALK (7, 19, 23, 24), by reprograming the cis-regulatory landscape via induction of de novo super enhancers (7). Recent studies have further defined the role of PAX3-FOXO1 as a CRC TF that, together with MYOD1, MYOG, MYCN, and SOX8, acts in the CRC TF network that directs the myogenic program, with each of them being essential for aRMS cell survival (7, 10, 25). While therapies targeting molecules downstream of PAX3–FOXO1 have been tested in preclinical models and clinical trials (26-32), additional mediators of aRMS growth, e.g., HES3 and YAP (33-35) may also provide new targets for aRMS, but effective and safe strategies to target the CRC TF network governed by PAX3-FOXO1 have not been identified.
Compared with fusion-negative RMS, the fusion-positive RMS has fewer genetic mutations (36, 37) and a worse clinical outcome (21), suggesting that epigenetic modifiers are implicated in disease progression. The chromatin remodeler CHD4 interacts with PAX3–FOXO1 and regulates a subset of PAX3–FOXO1 target genes (38). PAX3–FOXO1 also recruits BRD4 to super-enhancers, thereby creating dependency on BRD4 and causing sensitivity to JQ-1, a bromodomain inhibitor (7). Histone deacetylases (HDAC) help regulate PAX3–FOXO1 activity and HDAC inhibitors show antitumor activity in PAX3–FOXO1 aRMS mouse models (20, 25, 39, 40). Recent studies indicate that appropriate acetylation established by HDAC are required for core regulatory transcription (10, 25). These data suggest that epigenetic machinery is involved in the regulation of activity of CRC TFs that are direct targets of PAX3–FOXO1.
Histone lysine demethylase (KDM) activity is dysregulated in many cancers driven by chimeric TFs. For example, KDM1A interacts with EWS–FLI1 to mediate transcriptional repression of tumor suppressors in Ewing sarcoma (41) and sustains the oncogenic potential of MLL–AF9 leukemia stem cells (42). The SS18–SSX oncoprotein is recruited by KDM2B to drive synovial sarcoma (43) and KDM4 subfamily members are required for MLL–AF9-mediated leukemogenesis (44). MLL–GAS7 and MOZ–TIF2 mediate epigenetic reprogramming and transformation of acute myeloid leukemia via KDM4C (45). Together, these data suggest that oncogenic fusion TFs require KDMs for tumorigenesis. However, whether KDMs are involved in the maintenance of a CRC TF network that is governed by chimeric TFs is not clear. Here, we show that the dual H3K9me3/me2 and H3K36me3/me2 demethylase (46), KDM4B, is a targetable regulator of CRC TFs in PAX3–FOXO1 driven aRMS, thereby revealing the molecular mechanism of a druggable histone demethylase in maintaining oncogenic CRC TF in aRMS.
RESULTS
KDM4B expression is dysregulated in aRMS and regulated by PAX3–FOXO1
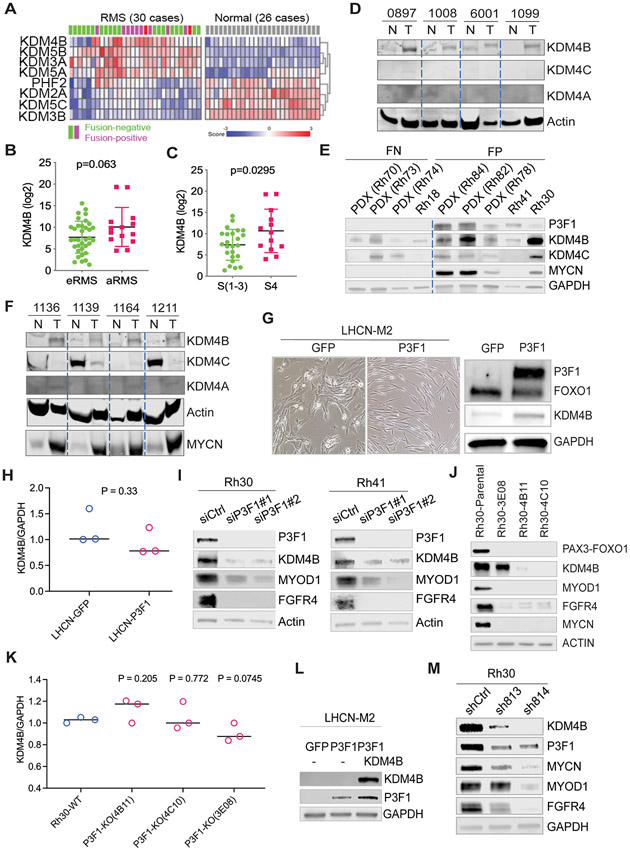
The poor survival rate of patients with PAX3–FOXO1 aRMS underscores the need to further unravel the mechanisms involved in tumorigenesis and disease progression driven by this chimeric TF. Reprogramming of normal cells into neoplastic counterparts by oncogenic drivers is often accompanied by dysregulation of epigenetic modifiers (47). Re-analysis of gene transcriptomes from primary patient samples and normal muscle tissues (Schafer Welle RMS cohort data set: https://hgserver1.amc.nl/cgi-bin/r2/main.cgi) showed that four KDMs (KDM4B, KDM3A, KDM5A, and KDM5B) were expressed significantly higher in RMS compared to normal muscle tissues (p<0.05) (Fig. 1A). Analysis of the St. Jude RMS cohort showed that KDM4B expression tended to be higher in aRMS (Fig. 1B), although the p value was not significant probably due to small sample size. However, KDM3A, KDM5A, and KDM5B were higher in eRMS (fig. S1A). KDM4B, but not the other three KDMs, was also highly expressed in stage IV high-risk RMS (Fig. 1C, fig. S1, B and C). Western blot analysis of primary human RMS tissues and matched normal muscle tissues confirmed that KDM4B was highly expressed in tumors (Fig. 1D), whereas the other two KDM4 members, KDM4A and KDM4C, were barely detectable (Fig. 1D). KDM4D, which only bears the catalytic domain and is expressed at very low amount of mRNA in RMS, was not assessed. Western blot analyses of patient-derived xenograft (PDX) tissues and RMS cell lines showed that KDM4B tended to be expressed at high in fusion–positive RMS samples (Fig. 1E, fig.S1D). These data suggested that KDM4B is dysregulated in PAX3–FOXO1 aRMS. To further validate this dysregulation, we analyzed expression of the KDM4 family members in tumor tissues and the matched normal skeletal muscle tissues using a genetic murine RMS model driven by PAX3–FOXO1 in a p53–null genetic background (20). The results showed that KDM4B was up-regulated in PAX3–FOXO1-driven tumor tissues in comparison with the matched normal tissues (Fig. 1F), consistent with the induction of MYCN, a well-known target of PAX3–FOXO1 in aRMS (23, 48). Interestingly, KDM4C was highly expressed in normal murine muscle tissues, but greatly repressed in transformed murine tumor tissues (Fig. 1F). Consistently, retroviral transduction of PAX3–FOXO1 into LHCN-M2, a human normal muscle cell line, induced KDM4B expression in addition to cellular morphological changes (Fig. 1G). We also transduced PAX3–FOXO1 into RD, a fusion-negative eRMS cell line, which resulted in elevated KDM4B expression (fig. S1E). We then performed RNA-seq after overexpression of PAX3–FOXO1 in LHCN-M2 cells but did not see changes in KDM4B transcript (fig. S1F), although transcription of other PAX3–FOXO1 target genes (MYOD1 and FGFR4) was induced. Quantitative real-time reverse transcription polymerase chain reaction (qRT-PCR) validated that PAX3-FOXO1 overexpression had no significant effect on KDM4B mRNA (Fig. 1H). These data indicate that PAX3–FOXO1 likely regulates KDM4B expression at the posttranscriptional level. To further corroborate that PAX3–FOXO1 regulates KDM4B expression, we used siRNAs that specifically target the fusion sequence of PAX3–FOXO1 in two fusion-positive RMS cell lines, Rh30 and Rh41. The resultant depletion of PAX3–FOXO1 protein from Rh30 and Rh41 cells reduced KDM4B expression (Fig. 1I), which was verified by CRISPR-Cas9–mediated knockout of PAX3-FOXO1 (Fig. 1J). Again, qRT-PCR showed that knockout of PAX3-FOXO1 had no significant negative effect on KDM4B mRNA expression (Fig. 1K). Taken together, these data strongly suggest that cancer cells transformed by the PAX3–FOXO1 display dysregulated KDM4B expression at the posttranscriptional level.
Fig. 1. KDM4B expression is dysregulated in RMS and regulated by PAX3–FOXO1.
A. Heatmap of differentially expressed KDMs (p<0.01) in RMS and normal muscle tissues (Schäfer Welle cohort).
B. The expression of KDM4B (RNA-seq) in eRMS and aRMS (St Jude cohort). The p-value was calculated using the Student t-test.
C. The expression of KDM4B (RNA-seq) in stage IV and other stages of RMS (St Jude cohort). The p-value was calculated using the Student t-test.
D. Western blot showing the expression of KDM4A, KDM4B, and KDM4C in human primary RMS and matched normal muscle tissues.
E. Western blot showing the expression of PAX3-FOXO1, KDM4B, KDM4C, and MYCN in RMS PDXs (Rh72, Rh73, Rh74, Rh78, Rh82, Rh84) and RMS cell lines (Rh18, Rh30, and Rh41).
F. Western blot showing the expression of KDM4A, KDM4B, and KDM4C in primary RMS and adjacent normal muscle tissues in a murine RMS genetic mouse model driven by PAX3–FOXO1.
G. Microscopic images of the human myoblast cell line LHCN-M2 after retroviral transduction with GFP or PAX3–FOXO1 vectors (left). Cell lysates were then subjected to Western blotting with the indicated antibodies (right).
H. Real-time quantitative PCR to determine the expression of KDM4B in LHCN-M2 cells transduced with GFP and PAX3-FOXO1 retroviral vectors, respectively. The p-value was calculated using the Student t-test.
I. siRNA knockdown of PAX3-FOXO1 in Rh30 and Rh41 cells followed by immunoblotting of cell lysates with the indicated antibodies.
J. CRISPR-cas9 mediated PAX3-FOXO1 knockout in Rh30 cells yielded three knockout clones (3E08, 4B11, 4C10), which were analyzed by western blot with indicated antibodies.
K. Real-time quantitative PCR to determine the expression of KDM4B in Rh30 PAX3-FOXO1 knockout cells. P value was calculated by the Student t test.
L. LHCN-M2 cells were transduced with retroviral vectors to overexpress KDM4B and/or PAX3–FOXO1 followed by immunoblotting of the cell lysates with the indicated antibodies.
M. Rh30 cells were transduced with lentiviral based shRNAs to knockdown KDM4B, followed by immunoblotting of the cell lysates with the indicated antibodies.
Physical association of PAX3–FOXO1 and KDM4B
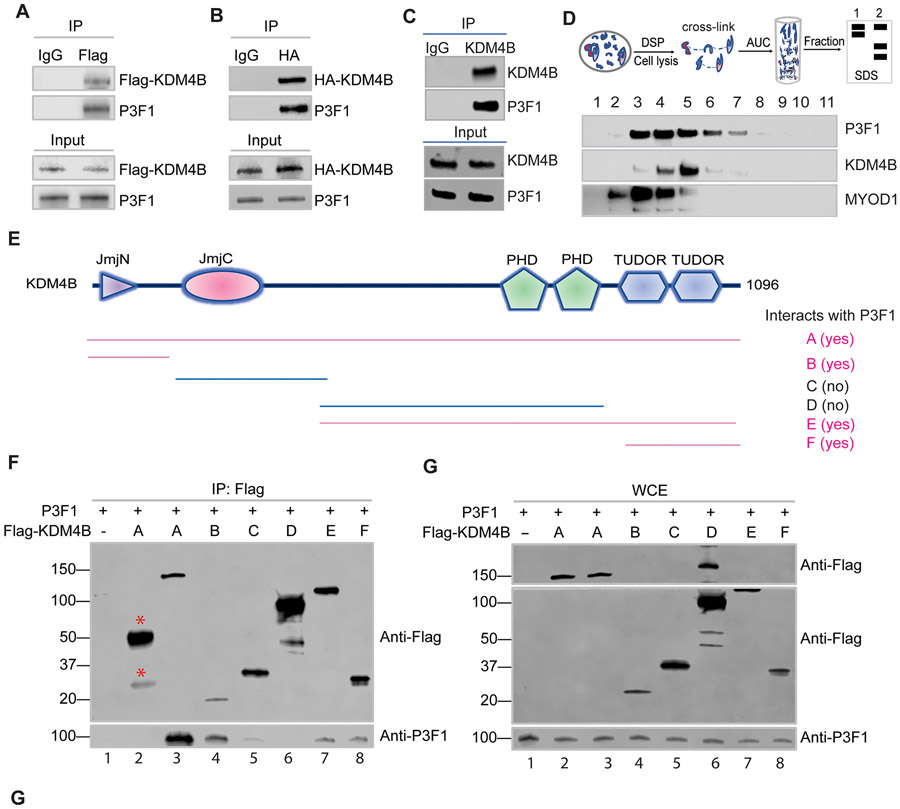
Intriguingly, introduction of KDM4B in LHCN-M2 cells reciprocally enhanced PAX3–FOXO1 expression (Fig. 1L). Conversely, knockdown of KDM4B using previously validated lentiviral shRNAs (sh-813 and sh-814) (49) greatly reduced PAX3–FOXO1 expression (Fig. 1M). qRT-PCR with fusion specific primers showed that PAX3–FOXO1 messenger RNA was about 2-fold reduced after KDM4B knockdown (fig. S1G), indicating that KDM4B at least partly regulates PAX3–FOXO1 gene transcription. The mutual regulation of expression indicates that PAX3–FOXO1 and KDM4B are highly interdependent. We and others previously demonstrated that KDM4B interacts with the lineage-specific transcription factors ERα and MYCN (49-52). We then tested whether PAX3–FOXO1 and KDM4B physically interact, given that protein-protein interaction is one of the key mechanisms mediating stabilization of interacting protein networks. We used Flag-tagged KDM4B, which was transiently transfected into HEK293T cells, together with PAX3–FOXO1, and showed by immunoprecipitation that KDM4B and PAX3–FOXO1 do indeed form a complex (Fig. 2A). We also used HA-tagged KDM4B and obtained similar results (Fig. 2B), indicating that the interaction of KDM4B and PAX3–FOXO1 was not due to the tags. We further confirmed that endogenous KDM4B and PAX3–FOXO1 formed a complex in Rh30 cells (Fig. 2C). However, we did not detect the co-immunoprecipitation of endogenous PAX3–FOXO1 with KDM4A or KDM4C in Rh30 cells (fig. S2A). The physical association between KDM4B and PAX3–FOXO1 was also evidenced by analytical ultracentrifugation (AUC) analysis (Fig. 2D). We used dithiobis succinimidyl propionate (DSP) to crosslink adjacent proteins within 12 Å, and then used AUC to fractionate the cellular components. Western blot analysis from different fractions showed that KDM4B and PAX3–FOXO1 co-localized in a subset of fractions (Fig. 2D). Interestingly, we also detected the CRC TF MYOD1 in the same fractions, supporting a previous study that PAX3–FOXO1 and MYOD1 co-localize at a subset of enhancers (25) and that KDM4B and MYOD1 physically interact (53). To determine which domain(s) of KDM4B interact(s) with PAX3–FOXO1, we tested a series of KDM4B-truncation constructs that bear different functional domains (Fig. 2E). Immunoprecipitation results indicated that the Jumonji N (JmjN) and the tandem Tudor domains of KDM4B might be key sites for interaction with PAX3–FOXO1 (Fig. 2, E-G). In comparison with the full-length KDM4B, the tandem Tudor domains or JmjN domain showed a weaker interaction with PAX3–FOXO1, suggesting that both are needed for maximal binding to PAX3–FOXO1. To test if these domains exert a dominant-negative effect on that could affect endogenous KDM4B and PAX3-FOXO1, we transfected these fragments into Rh30 cells and performed western blot analysis to assess the expression of PAX3-FOXO1 and KDM4B. Indeed, overexpression of the Tudor domain or the tandem PHD-Tudor domains greatly reduced the expression of endogenous KDM4B and PAX3-FOXO1, and of PAX3-FOXO1 targets (fig. S2B). These data suggest that disruption of PAX3-FOXO1 and KDM4B interaction by these fragments may lead to a destabilization of the PAX3-FOXO1/KDM4B complex. Taken together, these data reveal that KDM4B and PAX3–FOXO1 form a physical complex in aRMS cells.
Fig. 2. PAX3–FOXO1 and KDM4B physically interact.
A-B. HEK293T cells were transiently co-transfected with retroviral Flag-tagged KDM4B and PAX3–FOXO1 vectors. Cell lysates were immunoprecipitated with Flag antibody, followed by immunoblotting of the precipitated complex with the indicated antibodies.
B. HEK293T cells were transiently co-transfected with retroviral based Hemagglutinin (HA)-tagged KDM4B and PAX3–FOXO1 vectors. Cell lysates were immunoprecipitated with Flag antibody, followed by immunoblotting of the precipitated material with the indicated antibodies.
C. Immunoprecipitation of Rh30 cell lysates was performed using anti-KDM4B antibody and the immunoprecipitated complex was subjected to Western blotting with the indicated antibodies.
D. Cellular proteins were crosslinked with DSP, followed by analytical ultracentrifugation (AUC) and each fraction was immunoblotted with the indicated antibodies.
E. Diagram of Flag-tagged fragments of KDM4B in pCMV vectors. Pink color indicates physical interaction with PAX3–FOXO1 from Panel (F).
F. HEK293T cells were transiently transfected with Flag-tagged KDM4B fragments together with PAX3–FOXO1. Cell lysates were immunoprecipitated using anti-Flag antibody to pull down the KDM4B fragments and the associated proteins (lanes 1, and 3-8). Lane 1 served as a negative control that was not transfected with Flag-tagged KDM4B. Lane 2, cell lysate was immunoprecipitated with a non-relevant mouse IgG antibody (lane 2). The immunoprecipitated complex was immunoblotted with the indicated antibodies. Red stars indicate the IgG heavy and light chains in lane 2, serving as the negative control.
G. Whole-cell lysates (WCEs) used for immunoprecipitation in Panel (F) were subjected to Western blotting with the indicated antibodies. The upper panel in G shows full length of KDM4B protein. The upper and lower panels are from the same western blot representing different exposure times, which deconvolutes the overwhelmingly high signal of the KDM4B fragment, spilling over into the adjacent lanes.
KDM4B drives cell proliferation and tumorigenesis of PAX3–FOXO1 aRMS
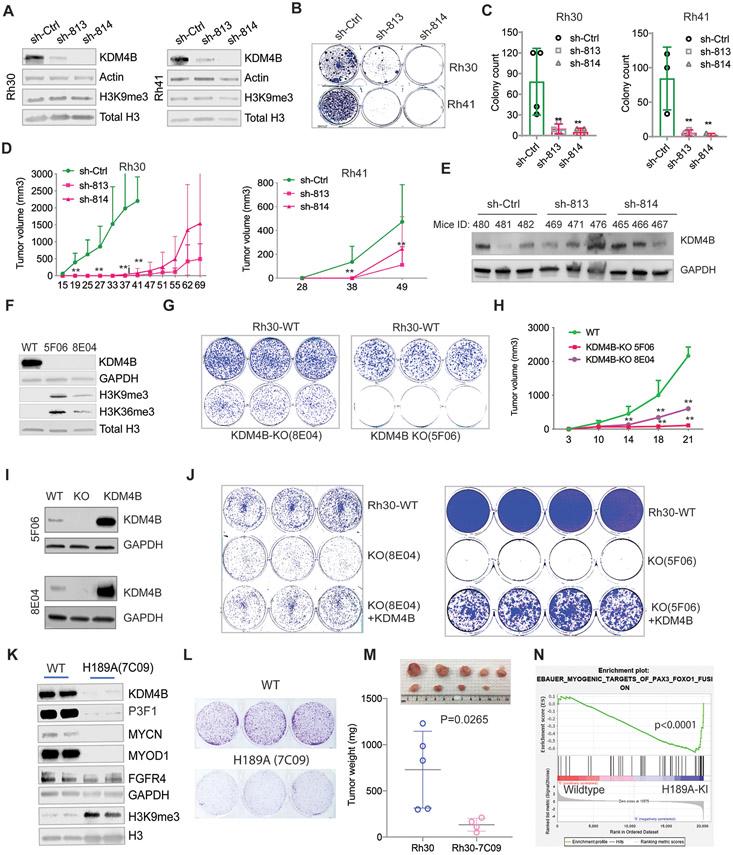
To investigate the functions of KDM4B in PAX3–FOXO1 aRMS, we used lentiviral shRNAs (sh-813 and sh-814) to knockdown KDM4B in Rh30 and Rh41 cells (Fig. 3A). Acute knockdown of KDM4B had no effect on global H3K9me3, what was seen in breast cancer and neuroblastoma cells (49, 50). However, genetic depletion of KDM4B in Rh30 and Rh41 significantly reduced their capacity to form colonies (Fig. 3, B and C). In addition, the loss of KDM4B significantly delayed tumor growth of Rh30 and Rh41 xenografts in mice (Fig. 3D). We analyzed KDM4B expression in three Rh30 xenografts per group that grew and found that KDM4B was re-expressed in these knockdown tumor samples comparable to that of controls (Fig. 3E). This suggests that loss-of-function of KDM4B by shRNA knockdown conferred a selective pressure to silence shRNAs or it was due to a selection of rare clones that were not transduced by KDM4B shRNA. To further validate the importance of KDM4B in PAX3–FOXO1 aRMS, we generated two Rh30 KDM4B-knockout clones using CRISPR-Cas9 technology (Fig. 3F). Again, genetic deletion of KDM4B greatly affected the colony growth in vitro (Fig. 3G) and tumor growth in vivo (Fig. 3H). Flow cytometry analysis of both clones showed that KDM4B deletion induced a remarkable cell death in clone 5F06 but only a modest G1 cell cycle arrest in clone 8E04 (fig. S3). This reflected the more profound growth inhibitory effect of KDM4B knockout in 5F06 than 8E04 (Fig. 3G-H), suggesting that cellular heterogeneity may affect the response to KDM4B deletion. Importantly, re-introduction of a retroviral KDM4B expression construct largely rescued the cell proliferation deficit (Fig. 3, I and J), demonstrating that the observed phenotype is KDM4B-specific. To test whether the demethylase activity of KDM4B was essential for its tumor promoting activity, we replaced the histidine at 189 with alanine (H189A), which disrupts the Fe2+ binding activity of KDM4B and consequently leading to loss of demethylase function (54), by using a CRISPR knock-in approach (fig. S4A). Interestingly, KDM4B(H189A) caused a dramatic reduction of endogenous KDM4B expression, accompanied by downregulation of PAX3-FOXO1 and its downstream targets (Fig. 3K). These data suggest that KDM4B(H189A) may cause KDM4B destabilization. Colony formation assays of the KDM4B(H189A) mutant cells showed reduced colony size, suggesting impaired cell proliferation (Fig. 3L). Intramuscular implantation of KDM4B(H189A) mutant Rh30 cells into NSG mice revealed that tumor growth was significantly delayed (Fig. 3M). To assess the downstream effects of KDM4B(H189A) expression, we performed GSEA pathway analysis after RNA-seq. KDM4B(H189A) expression resulted in a significant upregulation of the muscle differentiation program that is suppressed by PAX3-FOXO1 (Fig. 3N). This suggests that KDM4B plays an important role in maintaining the block in differentiation observed in aRMS.
Fig. 3. KDM4B is required for cell proliferation and tumorigenesis of PAX3–FOXO1 aRMS.
A. Western blotting indicating the efficiency of KDM4B shRNA knockdown in Rh30 and Rh41 cells.
B. Colony formation assay of Rh30 and Rh41 cells after KDM4B knockdown.
C. Quantification of the number of colonies from Panel (B). ** p<0.05, calculated using biological triplicates.
D. Growth curve of Rh30 and Rh41 tumor xenografts implanted in CB17 SCID mice. Wilcoxon rank sum test was used to compare the outcome of the two groups at each time point. **p<0.05
E. Western blot showing KDM4B expression in tumor tissues shown in Panel (D).
F. Western blot showing KDM4B expression in two Rh30 KDM4B-knockout clones (8E04 and 5F06) after CRISPR-Cas9 knockout.
G. The KDM4B-knockout clones (8E04 and 5F06) and parental Rh30 cells were seeded at a low cell density (1,000 cells per well) in triplicates. The cells were cultured for 10 days and stained by crystal violet.
H. Growth curve of Rh30 parental and KDM4B-knockout (8E04 and 5F06) xenograft tumors in NSG mice. Wilcoxon rank sum test was used to compare the outcome of the two groups at each time point. **p<0.05
I. Western blot showing KDM4B expression after introduction of KDM4B retroviral vectors into the KDM4B-knockout clones (8E04 and 5F06).
J. The Rh30 parental cells, KDM4B knockout clones, and KDM4B re-introduction knockout clones were seeded at a low cell density (2000 cells per well) in triplicates or quadruplicates. The cells were cultured for 15 days and stained with crystal violet.
K. Cell lysates of Rh30 cells carrying a CRISPR-Cas9-generated knock-in of KDM4B(H189A) were immunoblotted with the indicated antibodies.
L. Colony forming assay of Rh30 wildtype and the KDM4B(H189A) mutant cells.
M. Tumor volume of wildtype Rh30 and the KDM4B(H189A) knock-in mutant cells orthotopically implanted in the right quadriceps muscle of NSG mice. Tumors were dissected and weighted. P value was calculated using the Student t test.
N. GSEA analysis of the differentially expressed genes of Rh30 and the KDM4B(H189A) mutant cells, as determined by RNA-seq.
As the KDM4B(H189A) mutant knock-in cells expressed low KDM4B and PAX3-FOXO1 protein, we used a gain-of-function approach to test whether exogenous overexpression of KDM4B(H189A) had any impact on PAX3-FOXO1 function. In comparison with overexpression of wildtype KDM4B, the KDM4B(H189A) also increased the expression of PAX3-FOXO1, albeit to a lesser extent (fig. S4B). However, the expression of PAX3-FOXO1 targets was not substantially affected (fig. S4B). Then we transduced KDM4B(H189A) into KDM4B KO Rh30 cells. While the wildtype KDM4B largely rescued the cell viability of KDM4B KO cells (fig. S4C), the overexpression of KDM4B(H189A) mutant also had a partial rescue (fig. S4C), indicating that both the demethylase-dependent and demethylase-independent functions of KDM4B are involved in its tumor promoting activities.
Given that KDM4B and KDM4C are hypoxia inducible (55), we tested whether KDM4C is also critical to tumorigenesis of PAX3–FOXO1 aRMS. We generated two KDM4C-knockout Rh30 cell lines (fig. S4D). KDM4C knockout did not significantly affect the growth of Rh30 xenograft tumors (fig. S4E), although tumor growth seemed to be delayed to some degree. Interestingly, loss of KDM4C induced KDM4B expression (fig. S4D), suggesting that KDM4B might compensate for the loss of KDM4C function. Taken together, our data indicate that KDM4B is a therapeutic vulnerability to PAX3–FOXO1 aRMS.
KDM4B drives tumor growth of fusion negative RMS
Some eRMS tumors expressed KDM4B comparable to that in aRMS, suggesting that KDM4B might also play a role in these tumors. To test this possibility, we used JR1, a fusion-negative RMS cell line, and generated two KDM4B knockout cell lines (fig. S5A). Interestingly, while loss of KDM4B minimally affected colony formation capacity of JR1 cells (fig. S5B), xenograft studies showed that KDM4B knockout significantly delayed JR1 tumor growth (fig. S5C). We noticed that KDM4B deletion led to a remarkable reduction in the expression of MYOD1 but not FGFR4 (fig. S5A). These data indicate that KDM4B may have distinct functions in fusion positive and fusion negative RMS. In both scenarios, KDM4B can drive in vivo tumor growth.
The small molecule QC6352 is a potent KDM4B inhibitor
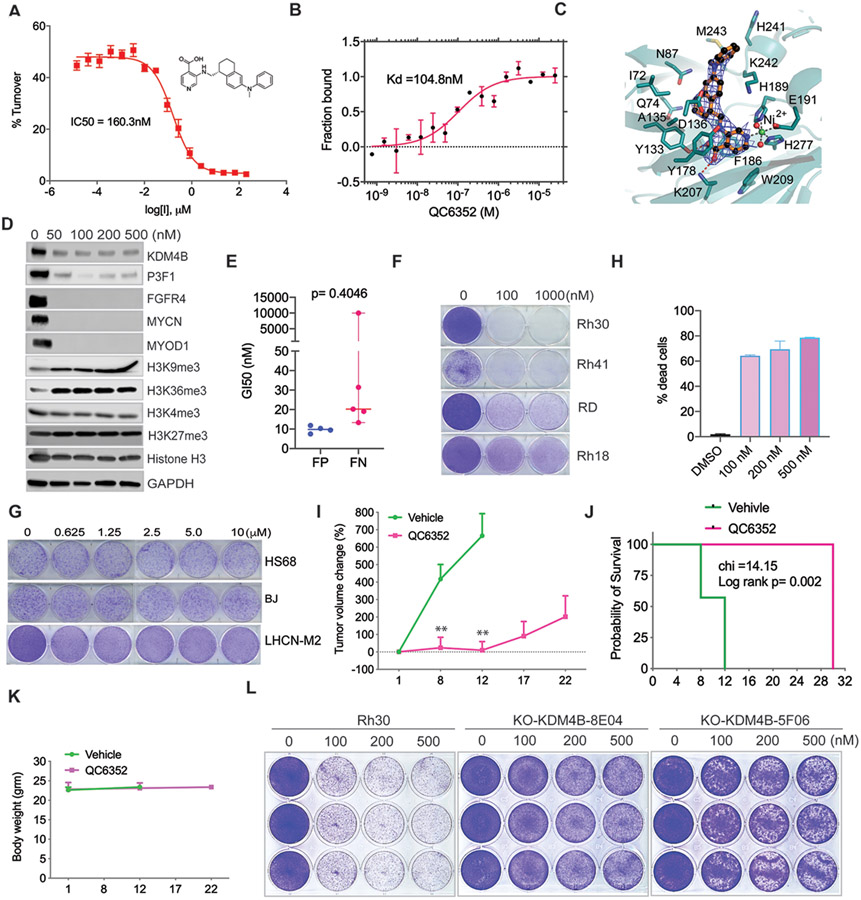
Currently, there are no Jumonji KDM inhibitors available for clinical application. We recently reported that ciclopirox is a KDM4 inhibitor that shows anticancer activity in neuroblastoma by targeting KDM4B (49). Unfortunately, ciclopirox exhibits poor in vivo pharmacokinetics (56) and may not be suitable in a clinical setting. Celgene recently developed a potent, selective, and cell permeable KDM4 inhibitor, QC6352, which is about 15-fold more selective to KDM4 members than KDM5 family members and more than 70-fold selective to other members of the KDM subfamily, with pharmacokinetics parameters suitable for oral administration (57). To validate that QC6352 can also inhibit KDM4B, we established an in vitro mass spectrometry-based assay, Matrix-Assisted Laser Desorption/Ionization-Fourier Transform Ion Cyclotron Resonance mass spectrometry (MALDI-FTICR MS), by using purified KDM4B catalytic domain. MALDI-FTICR MS validated QC6352 as a potent KDM4B inhibitor (IC50 = 160 nM) by measuring H3K9me2 converted from H3K9me3 (Fig. 4A). Next, we used Microscale Thermophoresis (MST) to measure the binding of QC6352 to KDM4B; the dissociation constant (Kd) was 104.8 nM (Fig. 4B). We then determined the crystal structure of the complex (Fig. 4C). We crystallized the JmjN and JmjC domains of KDM4B (1-366aa) in the absence of the H3K9me3 peptide and determined the structure complexed with QC6352 at 2.7 Å resolution (Table S1). Curiously, two molecules in the asymmetric unit were connected by a disulfide bond (Cys165A-Cys165B), but this was clearly a crystal artifact because the purified protein behaves as a monomer in solution. The structure revealed that QC6352 binds to KDM4B in a very similar fashion to how it binds KDM4A (Fig. 4C), which is unsurprising because the binding sites are almost identical in the two related proteins. Briefly, the pyridine ring stacks onto Phe186 and engages the active site nickel ion (replaces the iron ion in the crystal structure) via the ring nitrogen and Tyr133 and Lys207 via the carboxyl substituent. The central tetrahydronaphthalene moiety occupies the pocket bound by Ile72, Gln74, Asn87, Tyr133, Asp136, and Lys242. In the KDM4A complex, a structural zinc ion organizes two loop regions that loosely engage the distal phenyl group of QC6352 (57). In the KDM4B complex, the zinc is not present in the structure, which causes the two loop regions (231-236 and 306-313) to be disordered and to lose the interaction with the phenyl group.
Fig. 4. Pharmacologic inhibition of KDM4B by QC6352 exhibits anticancer activity in PAX3–FOXO1 aRMS.
A. Dose-response curve of QC6352 showing inhibition of KDM4B with an IC50 of 160.3 nM, measured by MALDI-FTICR. KDM4B inhibition plot in the presence of QC6352. Value of the dissociation constant, Ki, was calculated upon fitting the plot with the Morrison equation. Data are shown as mean ± standard deviation (n = 3). Y-axis indicates the turnover rate of H3K9me3 to H3K9me2, catalyzed by KDM4B. X-axis indicates the concentration of QC6352.
B. Microscale Thermophoresis assay of direct binding of KDM4B and QC6352. Y-axis represents the fraction of KDM4B protein bound by QC6352. X-axis represents QC6352 concentration. Kd = dissociation constant.
C. Crystal structure of the KDM4B-QC6352 complex (resolution 2.7 Å, PDB 7JM5). The 2Fo-Fc electron density map of the compound is shown as a blue mesh contoured at 1.0 σ. The ball-and-stick model of QC6352 is colored with gold-colored bonds, black carbon atoms, red oxygen atoms and blue nitrogen atoms. The KDM4B cartoon representation is colored in teal. Note that the active site iron atom is replaced by a nickel atom in this structure (green).
D. Rh30 cells were treated with QC6352 at the indicated concentrations for 48 hours and cell lysates were subjected to immunoblotting with the indicated antibodies.
E. GI50 of QC6352 in PAX3-FOXO1 fusion-positive and fusion-negative RMS cell lines measured by the PrestoBlue assay. The p-value was calculated using the Student t-test.
F. Colony forming assay of PAX3-FOXO1 fusion-positive (Rh30 and Rh41) and fusion-negative (Rh18 and RD) RMS cells after treatment with DMSO and QC6352 for 8 days.
G. Colony forming assay of normal human fibroblast (BJ and HS68) and myoblasts (LHCN-M2) cells after treatment with DMSO and QC6352 for 8 days.
H. Cell death analysis of Rh30 cells as determined by propidium iodide (PI) staining/flow cytometry following treatment with DMSO or QC6352 for 8 days. Results are expressed as percentages of dead cells (n = 3).
I. Growth curve of Rh30 PDX tumors in NSG mice treated with vehicle (n=6) or QC6352 (n=10) for 3 weeks. Welch’s two-sample t-test (two-sided) was used to compare the outcome of the two groups at each time point. **p<0.05
J. Kaplan-Meier analysis of survival of mice treated with vehicle or QC6352. Log-rank test for comparison of survival of each group.
K. Mouse body weight during treatment course.
L. Colony forming assay for Rh30 and the KDM4B-knockout clones (8E04 and 5F06) after treatment with DMSO or QC6352 for 8 days.
Pharmacologic inhibition of KDM4B has anticancer activity in PAX3–FOXO1 aRMS
Once we validated QC6352 as a KDM4B inhibitor, we next assessed QC6352’s activity against tumor cells. Treatment of PAX3–FOXO1 Rh30 and Rh41 aRMS cells, with QC6352 increased the amount of the KDM4 substrates, H3K9me3 and H3K36me3 (Fig. 4D, fig. S6A). However, we did not see obvious changes in the amount of H3K4me3 and H3K27me3 marks, indicating that QC6352 does not inhibit KDM5 and KDM6 family members at the tested concentrations, respectively, both of which share high sequence homology with the KDM4 family of lysine demethylases. Western blot analysis showed that the inhibitor not only greatly reduced the amounts of KDM4B and PAX3–FOXO1, but also that of key PAX3–FOXO1 targets, such as FGFR4, MYCN, and MYOD1 (Fig. 4D, fig. S6A). The remarkable reduction of KDM4 protein by QC6352 suggests that this small molecule induces the proteasomal protein degradation, a notion we verified by addition of MG132 proteasome inhibitor to the culture medium, which rescued the KDM4B protein expression (fig. S6B).
Since KDM4B critically regulates tumor growth of PAX3–FOXO1 aRMS, we tested whether pharmacological inhibition of KDM4B would have therapeutic efficacy. Overall, PAX3–FOXO1 fusion-positive RMS cell lines were about 2-fold more sensitive to QC6352 than PAX3–FOXO1 fusion-negative RMS cell lines (Fig. 4E). While the proliferation of most RMS cell lines was significantly inhibited by 100 nM of QC6352, the human normal muscle cell line LHCN-M2 as well as fibroblast cell lines (HS68 and BJ) were completely resistant (Fig. 4, F and G). These data suggest that treatment of non-cancerous human cells with QC6352 is safe and that cancer cells are addicted to KDM4B for survival. Fluorescence-activated cell sorting (FACS) analysis revealed that long-term treatment of PAX3–FOXO1 fusion-positive Rh30 cells with QC6352 induced cell death in at least 60% of cells (Fig. 4H). Likewise, QC6352 significantly inhibited tumor growth of Rh30 xenografts (fig. S6C) and significantly extended mouse survival (fig. S6D) without overt toxicity, as measured by maintenance of body weight during the treatment (fig. S6E). Epigenetic profiles may change when cells are cultured, which may impact their response to epigenetic agents (58), and therefore we tested QC6352 in a Rh30 PDX model that was solely propagated in mice. Like the results with the Rh30 cell line-based xenografts, QC6352 significantly delayed Rh30 PDX growth and extended mouse survival (Fig. 4, I and J). Again, mice maintained their body weight, showing that the drug is not toxic (Fig. 4K). To test whether QC6352 exerts its anticancer activity through KDM4B, we treated Rh30 parental and stable KDM4B-knockout clones with QC6352 (Fig. 4L). The data showed that loss of KDM4B conferred resistance to QC6352 treatment, suggesting that KDM4B is the main therapeutic target of QC6352.
Since KDM4 demethylates H3K9me3/me2 and H3K36me3/me2 (46), we investigated whether the methylation of H3K9 and H3K36 is critical to rhabdomyosarcoma by overexpressing wildtype histone H3.3(WT), and mutants that replaced the lysine with methionine, including H3.3(K9M), H3.3(K36M), H3.3(K9M/K36M) in Rh30 cells. Western blot revealed that K9M and K36M mutants led to a global demethylation of H3K9me3 and H3K36me3, respectively (fig. S6F), consistent with a previous report (59). While the colony formation of these cells was greatly halted by the global demethylation of H3K9 or H3K36, they eventually adapted to survive. Next, we treated these cells with QC6352 and found that H3.3(K36M) cells were resistant to KDM4 inhibition while the double H3.3(K9M/K36M) mutant cells were even more resistant (fig. S6G), suggesting that methylation on both K9 and K36 could be involved in mediating the anticancer activity of QC6352.
As knockout of PAX3-FOXO1 led to a great reduction of KDM4B expression, we tested whether loss of PAX3-FOXO1 would affect QC6352 efficacy. Indeed, while wildtype Rh30 showed remarkable sensitivity to QC6352, all three PAX3-FOXO1 knockout clones were very resistant to QC6352 (fig. S6H), which supports our hypothesis that PAX3-FOXO1/KDM4B complex is critical for mediating the anticancer activity of QC6352 in aRMS.
KDM4B inhibition impacts the transcriptome of PAX3–FOXO1 and its related CRC TFs
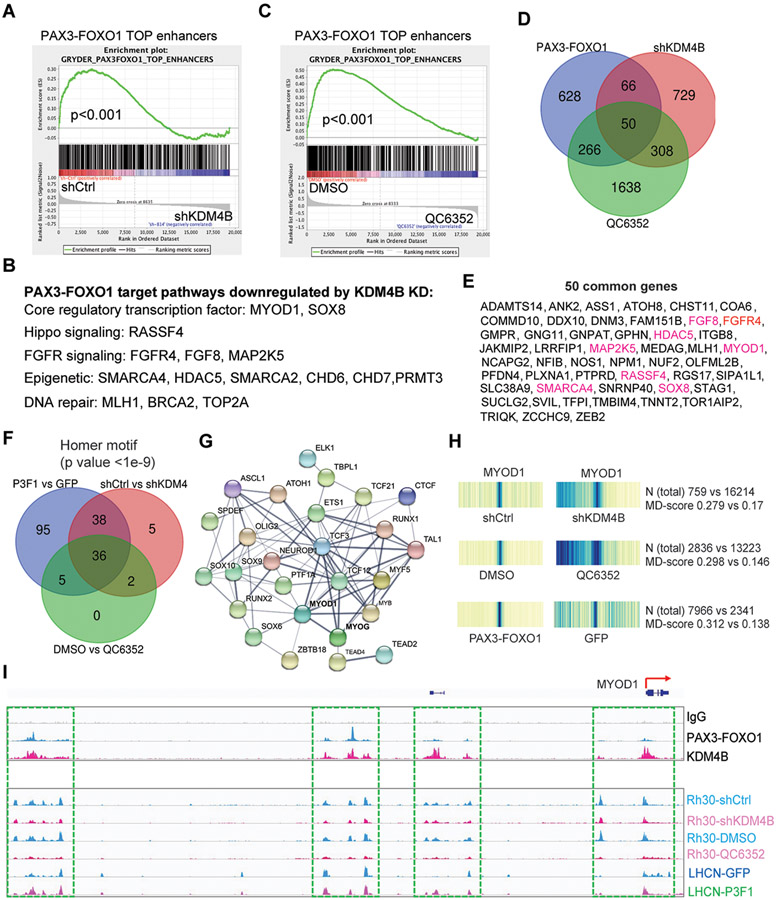
To investigate the working mechanism of KDM4B in PAX3–FOXO1-driven aRMS, we identified KDM4B targets by performing a microarray study comparing Rh30 cells with Rh30 KDM4B shRNA knockdown cells and analyzed the transcriptome. Differential gene expression showed that 896 genes were upregulated, and 1172 genes were downregulated (Table S2). Gene set enrichment analysis (GSEA) showed that the downregulated genes were enriched in chromatin maintenance, cell cycle, and DNA repair genes (fig. S7A). Genes involved in suppressing cell proliferation (e.g., GDF15, CDKN1A, and GADD45A) were upregulated by KDM4B knockdown (Table S2). Genes involved in muscle differentiation, such as MYF5, were also upregulated (Table S2). The downregulated genes were significantly associated with a PAX3–FOXO1 target gene signature (60) (p<0.0001) (Fig. 5A). The common target genes of KDM4B and PAX3–FOXO1 mediate key biological functions in aRMS (Fig. 5B), such as the CRC TFs MYOD1 and SOX8 (7), Hippo-signaling molecule RASSF4, and tyrosine kinase–signaling molecules FGF8 and FGFR4. A recent study showed that the expression of MYF5 and MYOD1 is mutually exclusive and required for sustained tumor growth of eRMS in a zebrafish model (60). One study showed that PAX3–FOXO1 promotes tumorigenesis through RASSF4, which inhibits the Hippo pathway (34). FGFR signaling is critical to RMS tumor growth (27, 61-63). Epigenetic modifiers or remodelers (e.g., SMARCA4, HDAC5, CHD6, CHD7, and PRMT3) and genes involved in DNA repair (MLH1, BRCA2, and TOP2A) were also downregulated by KDM4B knockdown (Fig. 5B, Table S2). SMARCA4 has been reported to regulate PAX3–FOXO1 expression (39). However, knockdown of KDM4C showed no impact on PAX3-FOXO1 pathway (fig. S7C), These data indicate that KDM4B is required to regulate genes involved in cancer genomic integrity, DNA repair, proliferation, and survival of PAX3-FOXO1 aRMS cells.
Fig. 5. KDM4B regulates expression of PAX3–FOXO1 target genes and the chromatin accessibility of myogenic program genes governed by MYOD1.
A. Pathway enrichment analysis by GSEA for the genes downregulated by depletion of KDM4B in Rh30 cells.
B. Functions for genes downregulated by KDM4B knockdown in Rh30 cells.
C. Pathway enrichment analysis by GSEA for the genes downregulated by QC6352 treatment of Rh30 cells.
D. Venn diagram for PAX3–FOXO1 target genes and those downregulated by knockdown of KDM4B and QC6352 treatment of Rh30 cells.
E. The 50 common genes listed in Panel (D). Genes highlighted in red color indicate those described in the pathways descirbed in Figure 5D.
F. Venn diagram showing the common transcription factor motifs based on ATAC-seq peaks among those increased by PAX3–FOXO1 in LHCN-M2 cells and those downregulated by knockdown of KDM4B or QC6352 treatment of Rh30 cells.
G. STRING analysis showing the protein-protein interaction (PPI) network of the common transcription factors in Panel (F). PPI enrichment p-value <1.0-e16.
H. Heatmap showing the motif displacement distribution of MYOD1 (increasingly dark blue indicates increasing frequencies of the motif), MD-score and the number of this motif within 1.5 kb of an ATAC-seq peak before and after KDM4B knockdown or QC6352 treatment in Rh30 cells, or after ectopic overexpression of PAX3–FOXO1 in LHCN-M2 cells.
I. Snapshot of using the IGV program displaying the PAX3–FOXO1 and KDM4B binding peaks in Rh30 cells by CUT&RUN (top), and ATAC-seq peaks at the MYOD1 genomic locus before and after KDM4B knockdown or QC6352 treatment of Rh30 cells, or ectopic overexpression of PAX3–FOXO1 in LHCN-M2 cells (bottom).
We further investigated whether pharmacologic inhibition of KDM4B phenocopied genetic knockdown of KDM4B. We performed transcriptome analysis after Rh30 cells were treated with 200 nM of QC6352 for 48 h. The inhibitor upregulated and downregulated the expression of 2572 and 2329 genes, respectively (Table S3). GSEA results showed that genes downregulated by QC6352 were also associated with high-confidence PAX3–FOXO1 target genes (7) (Fig. 5C) and various pathways implicated in chromatin maintenance, cell cycle, and DNA repair (fig. S7B), phenocopying the KDM4B knockdown. The CRC TF genes, including MYOD1, MYOG, SOX8, MYCN, and PAX3–FOXO1 itself, were all downregulated (Table S3, fig. S7D). Nearly 33% of the genes downregulated by the KDM4B knockdown were in common with those downregulated by QC6352 (Fig. 5, D and E). The key players in aRMS (i.e., FGFR4, MYOD1, SOX8, and RASSF4) and those in other cancers (e.g., SMARCA4, STAG1, and MLH1) were all in common (Fig. 5E). Similar results were obtained from Rh41 cells treated with QC6352 (fig. S7, E and F), which further confirmed that pharmacologic inhibition of KDM4B negatively influences expression of PAX3–FOXO1 target genes including CRC TFs.
KDM4B regulates the chromatin accessibility of myogenic program
Genome-wide analysis revealed that PAX3–FOXO1-binding sites are mostly distal to transcription start sites (48), suggesting that PAX3-FOXO1 locates at enhancer regions. This idea is reinforced by a recent study showing that PAX3–FOXO1 reprograms the cis-regulatory landscape by inducing de novo super enhancers (7). MYOD1 is a CRC TF in aRMS and is a direct target of PAX3–FOXO1, which is essential to PAX3–FOXO1-mediated cell survival (10). Targeting KDM4B led to inhibition of MYOD1 expression. To more fully determine the impact of KDM4B on PAX3–FOXO1-mediated transcription, we used ATAC-seq (assay for transposase-accessible chromatin using sequencing) (64) to compare the changes of TF binding motifs after transduction of PAX3–FOXO1 into LHCN-M2 cells, knockdown of KDM4B and QC6352 treatment of Rh30 cells. Homer motif analysis for increased DNA accessibility by overexpression of PAX3–FOXO1 and decreased DNA accessibility by genetic/pharmacologic KDM4B inhibition showed that 36 common TF binding motifs were significantly altered (Fig. 5F), including those of the CRC TFs MYOD1 and MYOG (Fig. 5H, fig.S7G, and data file S3). STRING (Search Tool for the Retrieval of Interacting Genes/Proteins) analysis of these 36 TFs revealed that most of them formed an interacting network (Fig. 5G). Thus, these ATAC-seq data demonstrated that PAX3–FOXO1 most significantly enhanced the chromatin accessibility of genes that bear binding motifs of MYOD1, and other related TFs and this activity is significantly repressed when KDM4B function is inhibited. Notably, chromatin accessibility of PAX3–FOXO1 to the MYOD1 locus was greatly reduced, exemplified by the fact that the promoter and the potential enhancer regions of MYOD1, showed increased chromatin accessibility to PAX3–FOXO1 after its introduction into LHCN-M2 cells (Fig. 5I, upper panel). Conversely, genetic depletion of KDM4B via shRNA knockdown or pharmacologic inhibition with QC6352 inhibited the chromatin accessibility at these regions (Fig. 5I, lower panel). We performed CUT&RUN (Cleavage Under Targets and Release Using Nuclease) analysis, a method that precisely maps global DNA binding sites for any protein of interest (65), to determine the genomic binding sites of PAX3–FOXO1 and KDM4B. The results showed that PAX3–FOXO1 and KDM4B co-bind at the MYOD1 locus (Fig. 5I), indicating that KDM4B and PAX3–FOXO1 together directly control MYOD1 expression.
KDM4 inhibition leads to repression of CRC TF super enhancers governed by PAX3-FOXO1
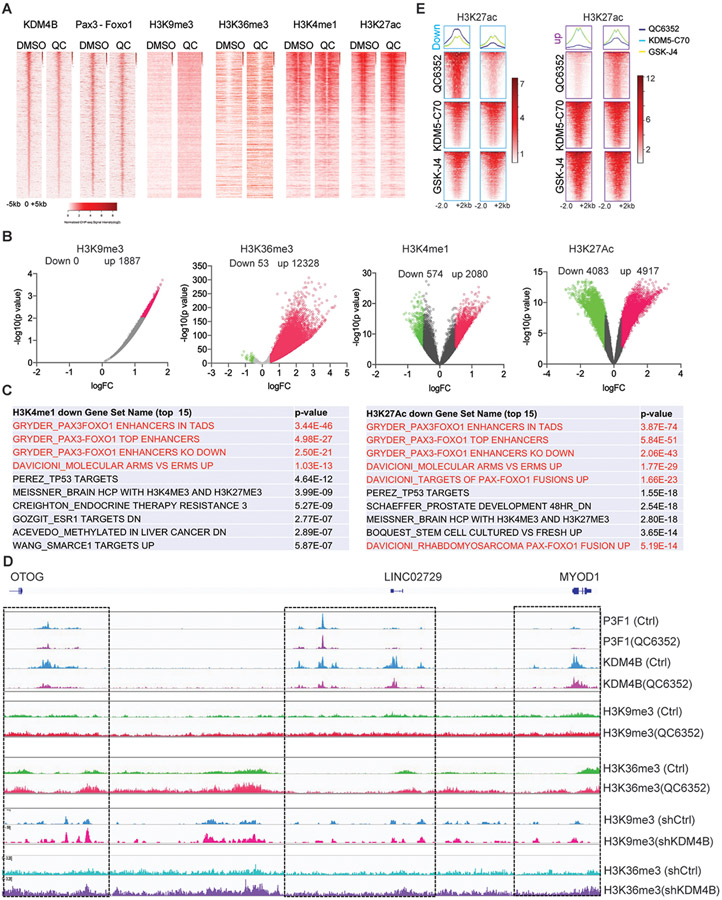
To further understand the mechanism by which QC6352 downregulates PAX3–FOXO1 function, we performed epigenetic analysis to investigate the changes of the KDM4 substrates H3K9me3 and H3K36me3 marks (46), as well as the enhancer marks H3K4me1 and H3K27ac, given that PAX3–FOXO1 exclusively binds at enhancer regions (7). While binding of KDM4B to chromatin tended to be reduced by QC6352 treatment, PAX3–FOXO1 was not greatly impacted at this timepoint, suggesting that KDM4B is the primary target of QC6352. Homer motif analysis of KDM4B and PAX3-FOXO1 binding sites showed that KDM4B uniquely bound at MYOD1/MYOG/E-box motif sites, which were greatly diminished by QC6352 treatment (fig. S8A). These results were consistent with the ATAC-seq data (Fig. 5H). The PAX3-FOXO1 binding sites were also enriched in CRC TF motifs of MYOD1/MYOG, which were modestly decreased by QC6352 (fig. S8B). Correspondingly, the H3K9me3 and H3K36me3 peaks appeared to be increased and this reflected the level of KDM4B binding (Fig.6A). However, there were no obvious pattern changes at PAX3–FOXO1 binding sites for the other histone marks queried (Fig. 6A). Nevertheless, QC6352 treatment mainly induced a global increase in H3K9me3 peaks (down 0 vs up 1887) and H3K36me3 peaks (down 53 vs up 12328) (Fig. 6B), indicating that QC6352 hits the targets in cells. The enhancer marker H3K4me1 also showed increased chromatin peaks (down 574 vs up 2080). However, the number of increased and decreased H3K27ac peaks were comparable (down 4083 vs up 4917) (Fig. 6B). These data suggest that KDM4 inhibition by QC6352 resulted in a global reprogramming of enhancers. To determine whether these enhancer alterations were associated with PAX3–FOXO1, we performed GSEA pathway analysis and found that the annotated genes marked by reduced H3K4me1 or H3K27Ac were significantly enriched in enhancers that are governed by PAX3–FOXO1 or PAX3–FOXO1 target genes (Fig. 6C), such as MYOG, MYCN and FGFR4 that showed a similar pattern in that reduction in H3K4me1 and H3K27ac marks was accompanied by an increase in H3K9me3 and H3K36me3 marks (fig. S9, A-C). QC6352 also induces a broad induction of H3K36me3 marks at the regulatory regions of MYOD1 (Fig. 6D). These data were consistent with those of the ATAC-seq, which showed decreased chromatin accessibility at these sites (Fig. 5I). While it is generally accepted that H3K9me3 is a heterochromatic mark that represses active gene transcription, the impact on H3K36me3 by QC6352 was not clear. We correlated the aberrant accumulation of H3K36me3 with the enhancer mark H3K27ac. After excluding the promoter regions (transcription start site ± 1kb), we found a negative correlation between the two marks (correlation coefficient −0.27, adjusted p<0.05) (fig. S9D) , suggesting that an increase in H3K36me3 at these regulatory regions might interfere with enhancer activity marked by H3K27ac. The induction of H3K9me3 and H3K36me3 by QC6352 at these loci was consistent with the effect of KDM4B depletion, which led to an increase in peaks of H3K9me3 and H3K36me3, as evidenced by the CUT&TAG-seq and ChIP-seq analyses (Fig. 6D, fig. S10A-D). This suggests that QC6352’s effect is directly linked to KDM4B, at least for part of the PAX3-FOXO1 target genes. To further verify that the epigenetic impact induced by QC6352 is KDM4-specific, we performed CUT&TAG-seq to investigate the impact on H3K27ac mark by the KDM5-C70 and GSK-J4, inhibitors of KDM5 (66) and KDM6 (67), respectively. Within the KDM family members, KDM4, KDM5 and KDM6 have the highest sequence homology(68). Distinct from the QC6352 treatment which induced different H3K27ac profiles and mainly downregulated the H3K27ac peaks of PAX3-FOXO1 target genes (Fig. 6C, 6E), pathway enrichment analysis revealed that the PAX3-FOXO1 enhancer regions or PAX3-FOXO1 target genes were not significantly affected by the inhibition of KDM5 and KDM6 (fig. S11). This is consistent with our previous study showing that KDM5-C50 and GSK-J4 had no effect on Rh30 cell proliferation (69). These data suggest that KDM4 inhibition selectively targets the PAX3-FOXO1 pathway through epigenetic reprogramming.
Fig. 6. KDM4B inhibition leads to repression of PAX3–FOXO1 super enhancers.
A. CUT&RUN-seq (KDM4B, PAX3-FOXO1) and ChIP-seq (H3K9me3, H3K36me3, H3K4me1, H3K27ac) density heatmaps of Rh30 cells after 48 hours treatment with DMSO control or 200 nM QC6352, ranked by KDM4B read intensity within ± 5kb of peak summits.
B. Volcano plot showing the genomic loci with significant changes of H3K9me3, H3K36me3, H3K4me1, and H3K27ac marks by QC6352 treatment.
C. GSEA showing the top 10 gene signatures associated with downregulation of H3K4me1 and H3K27ac.
D. Snapshot of the IGV program displaying the PAX3–FOXO1, KDM4B, H3K9me3, H3K36me3, at the MYOD1 genomic locus impacted by QC6352 treatment and KDM4B knockdown.
E. Heatmap indicating the signal intensity of H3K27ac peaks downregulated (left panel) and upregulated (right panel) in Rh30 cells by QC6352 (200nM, 48h), KDM5-C70 (5μM, 48h), GSK-J4 (2.5μM, 48h), respectively.
Combination of QC6352 and conventional chemotherapy significantly enhances the efficacy against PAX3–FOXO1 aRMS
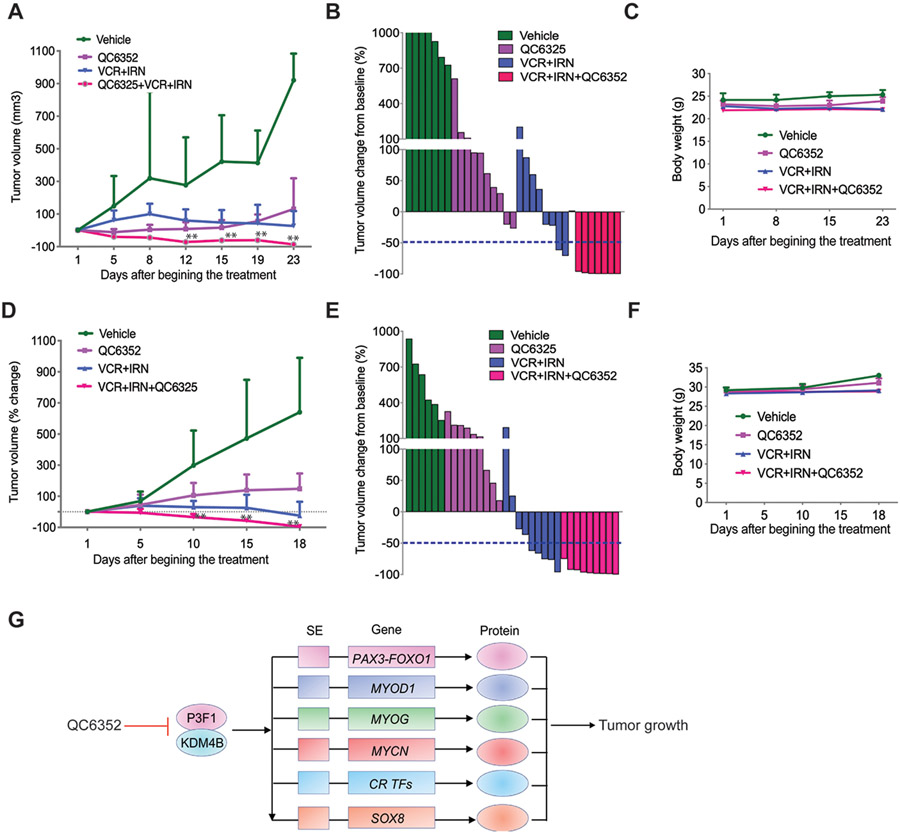
Genetic and pharmacologic inhibition of KDM4B significantly delays tumor growth, inhibits expression of CRC TFs and key PAX3–FOXO1 target genes (e.g., MYOD1, SOX8, MYCN, FGFR4, and RASSF4), and alters their chromatin accessibility and enhancer activity. Collectively, these data indicate that inhibition of KDM4B is a sound approach to targeting PAX3–FOXO1 and CRC TFs in aRMS. We further explored the therapeutic efficacy using QC6352 and extended our study by combining QC6352 with conventional chemotherapy using PAX3–FOXO1 aRMS models. QC6352 inhibits not only PAX3–FOXO1 target genes that are key to aRMS cell survival but also genes involved in DNA repair (e.g., MLH1, BRCA2, and TOP2A). Given that KDM4 family members are involved in DNA repair (70, 71), we hypothesized that QC6352 treatment would not only target PAX3–FOXO1 function but also enhance the tumor response to DNA damaging chemotherapeutics, and thus enhance the therapeutic efficacy of standard-of-care regimens. Therefore, we treated the Rh41 xenografts and Rh30R PDX tumors with a combination of QC6352 (25 mg/kg, BID) and vincristine/irinotecan (VCR/IRN, 0.38 mg/kg for VCR, once weekly; 1.25 mg/kg for IRN, once daily) (72), which is used in the clinical treatment of relapsed RMS. Three weeks of treatment with QC6352 or VCR/IRN alone significantly delayed tumor growth, but the combination of the three drugs caused significant tumor regression (Fig. 7, A, B, D, E, Table S5-6). Animals in the combination treatment arm (QC6352 and VCR/IRN) experienced 87.5% and 100% complete response in Rh41 and Rh30R aRMS models, respectively, while those in the VCR/IRN group had only 25% and 55% complete response, respectively (Fig. 7, B and E). In addition, we did not observe overt toxicity of the combination therapy as indicated by maintenance of the mouse body weight (Fig.7, C and F). These data indicate that QC6352 enhances the effect of conventional chemotherapy, suggesting it may be useful to incorporated in the standard-of-care regimen for high-risk aRMS.
Fig. 7. Combination of QC6352 and conventional chemotherapy significantly enhances efficacy against PAX3–FOXO1 aRMS.
A. Growth curve of Rh41 tumors in NSG mice treated with vehicle (n=7), QC6352 (n=10), VCR/IRN (n=8), or QC6352/VCR/IRN (n=7) for 3 weeks. Wilcoxon rank sum test (two-sided) was used to compare outcome of the two groups at each time point. ** p<0.05 when comparing QC6352/VCR/IRN with QC6352 or VCR/IRN.
B. Waterfall plot of response to treatment. The blue dotted line indicates complete response (CR).
C. Mouse body weight during treatment course.
D. Growth curve of Rh30R PDX tumors in NSG mice treated with vehicle (n=6), QC6352 (n=9), VCR/IRN (n=9), or QC6352/VCR/IRN (n=9) for 3 weeks. Wilcoxon rank sum test (two-sided) was used to compare outcome of the two groups at each time point. ** p<0.05 when comparing QC6352/VCR/IRN with QC6352 or VCR/IRN.
E. Waterfall plot of response to treatment. The blue dotted line indicates complete response (CR).
F. Mouse body weight during treatment course.
G. Model of how KDM4B regulates the PAX3–FOXO1-driven core regulatory transcription factor network.
As QC6352 also showed anticancer activity in eRMS, and KDM4B knockout reduced the tumor growth of JR1 eRMS cells, we investigated whether addition of QC6352 would also enhance the efficacy of chemotherapy to JR1 and RD tumor xenografts. QC6352 showed no significant effect on both JR1 (80% progressive disease, 20% stable disease) and RD (100% progressive disease) xenografts, although JR1 tumors responded slightly better (fig. S12). JR1 xenografts were also resistant to chemotherapy (60% progressive disease, 40% stable disease) while RD xenografts showed significant tumor growth delay (20% progressive disease, 80% stable disease). In contrast, the QC6352/VCR/IRN combination led to a significant tumor growth inhibition (fig. S12A, C), with 40% stable disease in JR1 model, 60% stable disease and 40% partial response in the RD model (fig, S12B, D). Although a synergistic effect of QC6352/standard of care combination therapy was observed in fusion negative tumors, the response of fusion positive tumors to this therapy was much superior (Fig. 7).
DISCUSSION
The oncogenic CRC TFs are attractive therapeutic targets but effective therapies to safely drug CRC TFs are lacking. Epigenetic dysregulation is a common feature of most cancers and numerous therapeutic epigenetic modulators have entered clinical development (58). Chromatin modification complexes play a key role in chimeric TF-driven malignancies (12). Targeting chromatin-based mechanisms may provide new therapeutic opportunities for treating fusion TF-driven cancers (12). PAX3–FOXO1 is an aberrant TF derived from the translocation of t(2;13) (q35;q14) (18, 73) and functions as an oncogenic driver in high-risk aRMS (74-77). Here we show that the histone demethylase KDM4B is an epigenetic vulnerability of PAX3–FOXO1 positive aRMS in which genetic or pharmacologic inhibition of KDM4B negatively affects the functions of PAX3–FOXO1. This leads to downregulation of CRC TFs which elicits potent anticancer activity. Our findings indicate that KDM4B regulates several aspects of PAX3–FOXO1 function. First, KDM4B and PAX3–FOXO1 physically interact to form a complex that binds to chromatin and regulates the expression of CRC TFs as well as PAX3–FOXO1 target genes such as MYOD1. Second, PAX3–FOXO1 stabilizes KDM4B expression, while increased KDM4B upregulated PAX3–FOXO1 protein expression. On the other hand, loss of KDM4B results in decreased PAX3–FOXO1 expression and consequently impacts the epigenetic landscape of PAX3–FOXO1 target genes. Pharmacologic inhibition of KDM4B with a selective KDM4 inhibitor (QC6352) disrupts the PAX3–FOXO1 oncogenic transcriptome, coincides with elevated H3K9me3 and H3K36me3 marks, both of which are known KDM4 substrates. PAX3–FOXO1 is a pioneering factor, shaping the formation of super enhancers at myogenic gene loci including MYOD1. This we confirmed by showing that introduction of PAX3–FOXO1 into a human muscle cell line, LHCN-M2, significantly increased the presence of the fusion protein at MYOD1 binding loci. Conversely, KDM4B inhibition greatly downregulated this activity. Studies from our laboratory and others indicate that KDMs are important conduits for lineage-specific oncogenic TF activity. These include the estrogen receptor in breast cancer (50, 51, 55, 78), the androgen receptor in prostate cancer (79), MYCN in neuroblastoma (49), and other pathognomonic fusion oncoproteins in pediatric cancers (41-45). Therefore, inactivation of these KDMs may disrupt the lineage developmental programs, such as that of MYOD1-mediated transcriptional activity in muscle, which plays an essential role in PAX3–FOXO1-positive or TWIST2-amplified RMS (7, 60, 80).
PAX3–FOXO1, MYOD1, SOX8 and MYCN are CRC TFs in aRMS and the gene loci of these CRC TFs are governed by super enhancers that drive active gene transcription (10, 25). CRC TFs orchestrate expression of lineage identity-specific gene networks that play key roles in normal physiology and cancer (1, 4, 81, 82). RMS shows critical CRC TF dependencies (10, 25). However, targeting CRC TFs is challenging as numerous studies tested the ability of bromodomain inhibitors to block active transcription but unwanted adverse effects dampened the enthusiasm for their application (83-85). Downregulation of RMS CRC TFs by KDM4B inhibition suggests that KDM4B is required to maintain the CRC TF circuitry for survival (Figure 7G), supporting the notion that that KDM4B is a RMS therapeutic target. Interestingly, some of the fusion positive and fusion negative RMS lack KDM4B overexpression, which calls for further study to better understand the clinical behavior of these tumors. This may determine why these patient samples had low KDM4B expression, opening the possibility that other KDM4s or KDM family members are involved in tumorigenesis in these cases.
By exploring the activity of a KDM4 inhibitor, QC6352, we found that it exerts its anticancer activity mainly via KDM4B in PAX3–FOXO1+ aRMS; QC6352 induces a similar phenotype as loss-of-function of KDM4B and knockout of KDM4B confers resistance of cancer cells to QC6352 treatment. We validated that QC6352 directly binds KDM4B and inhibits the activity of KDM4B as measured by multiple biophysical approaches. While several KDM4 inhibitors have been reported (86), QC6352 is perhaps the best-in-class compound that we have tested in terms of KDM selectivity and cellular activity. Importantly, QC6352 showed no overt toxicity in vitro (normal human fibroblast and muscle cells) and in vivo (PAX3-FOXO1 aRMS mouse models). The animal safety results published by Celgene further corroborate our observation (57). We further assessed the therapeutic efficacy of QC6352 in combination with conventional chemotherapy (VCR/IRN), which spurred enhanced tumor response, suggesting that KDM4B inhibitors may be incorporated into the standard-of-care regimens in the future. Thus, our findings indicate that selectively inhibiting KDM4B by small molecules has translational potential in the context of a disease that is driven by a currently undruggable fusion oncoprotein.
Despite the fact that most eRMS cells were about 2-fold less responsive than the fusion-positive RMS cells, we surmise that KDM4B inhibition could have a broad therapeutic effect on both types of RMS. Indeed, we found that QC6352 also inhibited key survival genes of fusion-negative RMS cells such as MYOD1. This was corroborated by KDM4B knockout in the eRMS cell line JR1, which led to a dramatic reduction of MYOD1 expression. Intriguingly, KDM4B deletion showed minimal impact on cell colony formation of JR1 in vitro, but significantly delays tumor growth in vivo. Considering that KDM4B is a hypoxia-inducible, it is likely that KDM4B could be involved in regulation of tumor metabolism and angiogenesis, warranting further studies. A recurrent neomorphic mutation in MYOD1, which mimics MYC function (87), defines a clinically aggressive subset of eRMS and sclerosing/spindle cell RMS (87-90). Understanding the potential regulation of the MYOD1 program by KDM4B in these RMS subtypes may provide additional intervention avenues but awaits further investigation.
Limitations of the study
It is important to point out several limitations of our study. First, QC6352 is a KDM4 inhibitor that also targets other KDM4 members. While the genetic and epigenetic data support the notion that QC6352 activity is directly linked to KDM4B inhibition, the antitumor effect of QC6352 could be a combined effect of inhibition of multiple or all KDM4 members. Second, the function of each individual KDM4 family member in RMS needs to be further defined, which may provide insight into how PAX3-FOXO1 “hijacks” the normal activities of these demethylases, a question remains to be answered.
MATERIALS AND METHODS
Study design
This study tested the hypothesis that KDM4B is a potential therapeutic target for PAX3-FOXO1-driven rhabdomyosarcoma. Therefore, we tested the therapeutic efficacy of the KDM4 inhibitor QC6352 as a single agent in both cell line–based xenograft and PDX RMS models. We also determined the efficacy of QC5352 in combination with vincristine and irinotecan in RMS xenograft models. Sample sizes were determined based on previous experimental experience and on the growth profiles of the individual models (n = 5 to 10). For all experiments, tumors were measured every 2 to 3 days in two dimensions using electronic calipers, and tumor volume was calculated as π/6 × d3 where d is the mean of two diameters taken at right angles. Both genders of animals were included and randomly assigned to each study group. Investigators were not blinded to the experiments. Animals were euthanized if they lost 20% of their weight or exhibited signs of malaise impaired health. Mice were euthanized when the tumor volume exceeded ~2000 mm3.
Statistical analysis
Given that data are not normally distributed, and sample size is relatively small (less than 10) for the animal experiments, t-test is not appropriate and Wilcoxon rank sum test (which is more robust to data structure) is used to compare the outcome of the two groups at each time point. If the data do not violate the normal assumption, but the variances between the groups at each time point are very different, Welch’s two-sample t-test (which does not rely on equal variance) is used to compare the outcome of the two groups at each time point. P-values are adjusted for multiple comparison using the Benjamini-Hochberg (1) method. The Benjamini-Krieger-Yekutieli (2) two-stage procedure used by Prism is more powerful but is too aggressive for data with dependence (as is our case), and error control is not guaranteed. On the other hand, the Benjamini-Hochberg method tolerates data that are positively dependent. We ran both BH and BKY methods on the data and found out that the adjusted p-values from these two methods differ only slightly, and still arrive at the same conclusions. Therefore, we suggest the Benjamini-Hochberg method here, which is the standard and most widely accepted procedure for multiple comparison correction. Statistical analyses were performed using R version 4.0.2. To determine statistical significance for other experiments, the unpaired, two-tailed Student t test was calculated using the t test calculator available on GraphPad Prism 8.0 software. A p value of less than 0.05 was considered statistically significant. Kaplan-Meier survival analysis was calculated using log-rank (Mantel-Cox) method in GraphPad Prism 8.0 software.
Supplementary Material
ACKNOWLEDGMENTS.
We thank Christopher Morton for PDX models, Steven Henikoff for CUT&RUN reagents and protocols. We thank the St. Jude Hartwell Center for genomic sequencing, the Animal Research Center for mouse work, the Flow Cytometry and Cell Sorting Shared Resource for FACS analysis, the Molecular Interaction Analysis for AUC analysis. We thank Liang Cao and Javed Khan from NHGRI kindly providing anti-PAX3-FOXO1 monoclonal antibody.
Funding:
This work was partly supported by American Cancer Society-Research Scholar (130421-RSG-17-071-01-TBG, J.Y.) and National Cancer Institute (R03CA212802-01A1, J.Y.; 1R01CA229739-01, J.Y.). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
Competing interest: All authors declare no other competing interests.
Data and materials availability: All data associated with this study are present in the paper or the Supplementary Materials. Any data and materials that can be shared will be released via a material transfer agreement by contacting J.Y. Microarray data are available at the National Center for Biotechnology Information Gene Expression Omnibus (NCBI GEO) accession number: GSE158394. RNA-seq, ChIP-seq, CUT&RUN, CUT&TAG sequencing data are available at the National Center for Biotechnology Information Gene Expression Omnibus (NCBI GEO) accession number GSE157095 series. The co-crystal structure of KDM4B-QC6352 is available in RCSB database with accession number 7JM5. Softwares and programs used for data analysis are described and referenced in the corresponding Supplementary Materials and Methods.
References
- 1.Bradner JE, Hnisz D, Young RA, Transcriptional Addiction in Cancer. Cell 168, 629–643 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen X et al. , Integration of external signaling pathways with the core transcriptional network in embryonic stem cells. Cell 133, 1106–1117 (2008). [DOI] [PubMed] [Google Scholar]
- 3.Neph S et al. , Circuitry and dynamics of human transcription factor regulatory networks. Cell 150, 1274–1286 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Saint-Andre V et al. , Models of human core transcriptional regulatory circuitries. Genome Res 26, 385–396 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ott CJ et al. , Enhancer Architecture and Essential Core Regulatory Circuitry of Chronic Lymphocytic Leukemia. Cancer Cell 34, 982–995 e987 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sanda T et al. , Core transcriptional regulatory circuit controlled by the TAL1 complex in human T cell acute lymphoblastic leukemia. Cancer Cell 22, 209–221 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gryder BE et al. , PAX3-FOXO1 Establishes Myogenic Super Enhancers and Confers BET Bromodomain Vulnerability. Cancer Discov 7, 884–899 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ran L et al. , FOXF1 Defines the Core-Regulatory Circuitry in Gastrointestinal Stromal Tumor. Cancer Discov 8, 234–251 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Durbin AD et al. , Selective gene dependencies in MYCN-amplified neuroblastoma include the core transcriptional regulatory circuitry. Nat Genet 50, 1240–1246 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gryder BE et al. , Histone hyperacetylation disrupts core gene regulatory architecture in rhabdomyosarcoma. Nat Genet 51, 1714–1722 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kim P, Zhou X, FusionGDB: fusion gene annotation DataBase. Nucleic Acids Res 47, D994–D1004 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brien GL, Stegmaier K, Armstrong SA, Targeting chromatin complexes in fusion protein-driven malignancies. Nat Rev Cancer 19, 255–269 (2019). [DOI] [PubMed] [Google Scholar]
- 13.Yohe ME et al. , Insights into pediatric rhabdomyosarcoma research: Challenges and goals. Pediatr Blood Cancer 66, e27869 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Decaesteker B et al. , TBX2 is a neuroblastoma core regulatory circuitry component enhancing MYCN/FOXM1 reactivation of DREAM targets. Nat Commun 9, 4866 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang L et al. , ASCL1 is a MYCN- and LMO1-dependent member of the adrenergic neuroblastoma core regulatory circuitry. Nat Commun 10, 5622 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hettmer S et al. , Rhabdomyosarcoma: current challenges and their implications for developing therapies. Cold Spring Harb Perspect Med 4, a025650 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Skapek SX et al. , Rhabdomyosarcoma. Nat Rev Dis Primers 5, 1 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Barr FG, Gene fusions involving PAX and FOX family members in alveolar rhabdomyosarcoma. Oncogene 20, 5736–5746 (2001). [DOI] [PubMed] [Google Scholar]
- 19.Linardic CM, PAX3-FOXO1 fusion gene in rhabdomyosarcoma. Cancer Lett 270, 10–18 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Abraham J et al. , Lineage of origin in rhabdomyosarcoma informs pharmacological response. Genes Dev 28, 1578–1591 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Skapek SX et al. , PAX-FOXO1 fusion status drives unfavorable outcome for children with rhabdomyosarcoma: a children's oncology group report. Pediatr Blood Cancer 60, 1411–1417 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sorensen PH et al. , PAX3-FKHR and PAX7-FKHR gene fusions are prognostic indicators in alveolar rhabdomyosarcoma: a report from the children's oncology group. J Clin Oncol 20, 2672–2679 (2002). [DOI] [PubMed] [Google Scholar]
- 23.Mercado GE et al. , Identification of PAX3-FKHR-regulated genes differentially expressed between alveolar and embryonal rhabdomyosarcoma: focus on MYCN as a biologically relevant target. Genes Chromosomes Cancer 47, 510–520 (2008). [DOI] [PubMed] [Google Scholar]
- 24.Ginsberg JP, Davis RJ, Bennicelli JL, Nauta LE, Barr FG, Up-regulation of MET but not neural cell adhesion molecule expression by the PAX3-FKHR fusion protein in alveolar rhabdomyosarcoma. Cancer Res 58, 3542–3546 (1998). [PubMed] [Google Scholar]
- 25.Gryder BE et al. , Chemical genomics reveals histone deacetylases are required for core regulatory transcription. Nat Commun 10, 3004 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wierdl M et al. , Targeting ALK in pediatric RMS does not induce antitumor activity in vivo. Cancer Chemother Pharmacol 82, 251–263 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li SQ et al. , Targeting wild-type and mutationally activated FGFR4 in rhabdomyosarcoma with the inhibitor ponatinib (AP24534). PLoS One 8, e76551 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Szewczyk B, Skrzypek K, Majka M, Targeting MET Receptor in Rhabdomyosarcoma: Rationale and Progress. Curr Drug Targets 18, 98–107 (2017). [DOI] [PubMed] [Google Scholar]
- 29.Nguyen TH, Barr FG, Therapeutic Approaches Targeting PAX3-FOXO1 and Its Regulatory and Transcriptional Pathways in Rhabdomyosarcoma. Molecules 23, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wan X, Helman LJ, Levels of PTEN protein modulate Akt phosphorylation on serine 473, but not on threonine 308, in IGF-II-overexpressing rhabdomyosarcomas cells. Oncogene 22, 8205–8211 (2003). [DOI] [PubMed] [Google Scholar]
- 31.Pappo AS et al. , A phase 2 trial of R1507, a monoclonal antibody to the insulin-like growth factor-1 receptor (IGF-1R), in patients with recurrent or refractory rhabdomyosarcoma, osteosarcoma, synovial sarcoma, and other soft tissue sarcomas: results of a Sarcoma Alliance for Research Through Collaboration study. Cancer 120, 2448–2456 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Malempati S et al. , The addition of cixutumumab or temozolomide to intensive multiagent chemotherapy is feasible but does not improve outcome for patients with metastatic rhabdomyosarcoma: A report from the Children's Oncology Group. Cancer 125, 290–297 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kendall GC et al. , PAX3-FOXO1 transgenic zebrafish models identify HES3 as a mediator of rhabdomyosarcoma tumorigenesis. Elife 7, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Crose LE et al. , Alveolar rhabdomyosarcoma-associated PAX3-FOXO1 promotes tumorigenesis via Hippo pathway suppression. J Clin Invest 124, 285–296 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Oristian KM et al. , Loss of MST/Hippo Signaling in a Genetically Engineered Mouse Model of Fusion-Positive Rhabdomyosarcoma Accelerates Tumorigenesis. Cancer Res 78, 5513–5520 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen X et al. , Targeting oxidative stress in embryonal rhabdomyosarcoma. Cancer Cell 24, 710–724 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shern JF et al. , Comprehensive genomic analysis of rhabdomyosarcoma reveals a landscape of alterations affecting a common genetic axis in fusion-positive and fusion-negative tumors. Cancer Discov 4, 216–231 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bohm M et al. , Helicase CHD4 is an epigenetic coregulator of PAX3-FOXO1 in alveolar rhabdomyosarcoma. J Clin Invest 126, 4237–4249 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bharathy N et al. , The HDAC3-SMARCA4-miR-27a axis promotes expression of the PAX3:FOXO1 fusion oncogene in rhabdomyosarcoma. Sci Signal 11, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bharathy N et al. , Preclinical rationale for entinostat in embryonal rhabdomyosarcoma. Skelet Muscle 9, 12 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sankar S et al. , Mechanism and relevance of EWS/FLI-mediated transcriptional repression in Ewing sarcoma. Oncogene 35, 6155–6156 (2016). [DOI] [PubMed] [Google Scholar]
- 42.Harris WJ et al. , The histone demethylase KDM1A sustains the oncogenic potential of MLL-AF9 leukemia stem cells. Cancer Cell 21, 473–487 (2012). [DOI] [PubMed] [Google Scholar]
- 43.Banito A et al. , The SS18-SSX Oncoprotein Hijacks KDM2B-PRC1.1 to Drive Synovial Sarcoma. Cancer Cell 34, 346–348 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Agger K et al. , Jmjd2/Kdm4 demethylases are required for expression of Il3ra and survival of acute myeloid leukemia cells. Genes Dev 30, 1278–1288 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cheung N et al. , Targeting Aberrant Epigenetic Networks Mediated by PRMT1 and KDM4C in Acute Myeloid Leukemia. Cancer Cell 29, 32–48 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Whetstine JR et al. , Reversal of histone lysine trimethylation by the JMJD2 family of histone demethylases. Cell 125, 467–481 (2006). [DOI] [PubMed] [Google Scholar]
- 47.Baylin SB, Jones PA, Epigenetic Determinants of Cancer. Cold Spring Harb Perspect Biol 8, (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cao L et al. , Genome-wide identification of PAX3-FKHR binding sites in rhabdomyosarcoma reveals candidate target genes important for development and cancer. Cancer Res 70, 6497–6508 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yang J et al. , The role of histone demethylase KDM4B in Myc signaling in neuroblastoma. J Natl Cancer Inst 107, djv080 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yang J et al. , The histone demethylase JMJD2B is regulated by estrogen receptor alpha and hypoxia, and is a key mediator of estrogen induced growth. Cancer Res 70, 6456–6466 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kawazu M et al. , Histone demethylase JMJD2B functions as a co-factor of estrogen receptor in breast cancer proliferation and mammary gland development. PLoS One 6, e17830 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shi L et al. , Histone demethylase JMJD2B coordinates H3K4/H3K9 methylation and promotes hormonally responsive breast carcinogenesis. Proc Natl Acad Sci U S A 108, 7541–7546 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Choi JH, Song YJ, Lee H, The histone demethylase KDM4B interacts with MyoD to regulate myogenic differentiation in C2C12 myoblast cells. Biochem Biophys Res Commun 456, 872–878 (2015). [DOI] [PubMed] [Google Scholar]
- 54.Fodor BD et al. , Jmjd2b antagonizes H3K9 trimethylation at pericentric heterochromatin in mammalian cells. Genes Dev 20, 1557–1562 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yang J et al. , Role of hypoxia-inducible factors in epigenetic regulation via histone demethylases. Ann N Y Acad Sci 1177, 185–197 (2009). [DOI] [PubMed] [Google Scholar]
- 56.Minden MD et al. , Oral ciclopirox olamine displays biological activity in a phase I study in patients with advanced hematologic malignancies. Am J Hematol 89, 363–368 (2014). [DOI] [PubMed] [Google Scholar]
- 57.Chen YK et al. , Design of KDM4 Inhibitors with Antiproliferative Effects in Cancer Models. ACS Med Chem Lett 8, 869–874 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mohammad HP, Barbash O, Creasy CL, Targeting epigenetic modifications in cancer therapy: erasing the roadmap to cancer. Nat Med 25, 403–418 (2019). [DOI] [PubMed] [Google Scholar]
- 59.Lewis PW et al. , Inhibition of PRC2 activity by a gain-of-function H3 mutation found in pediatric glioblastoma. Science 340, 857–861 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tenente IM et al. , Myogenic regulatory transcription factors regulate growth in rhabdomyosarcoma. Elife 6, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wu C et al. , Insight into ponatinib resistance mechanisms in rhabdomyosarcoma caused by the mutations in FGFR4 tyrosine kinase using molecular modeling strategies. Int J Biol Macromol 135, 294–302 (2019). [DOI] [PubMed] [Google Scholar]
- 62.McKinnon T et al. , Functional screening of FGFR4-driven tumorigenesis identifies PI3K/mTOR inhibition as a therapeutic strategy in rhabdomyosarcoma. Oncogene 37, 2630–2644 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Crose LE et al. , FGFR4 blockade exerts distinct antitumorigenic effects in human embryonal versus alveolar rhabdomyosarcoma. Clin Cancer Res 18, 3780–3790 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Buenrostro JD, Wu B, Chang HY, Greenleaf WJ, ATAC-seq: A Method for Assaying Chromatin Accessibility Genome-Wide. Curr Protoc Mol Biol 109, 21 29 21–29 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Skene PJ, Henikoff S, An efficient targeted nuclease strategy for high-resolution mapping of DNA binding sites. Elife 6, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Horton JR et al. , Structural Basis for KDM5A Histone Lysine Demethylase Inhibition by Diverse Compounds. Cell Chem Biol 23, 769–781 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kruidenier L et al. , A selective jumonji H3K27 demethylase inhibitor modulates the proinflammatory macrophage response. Nature 488, 404–408 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Shi Y, Whetstine JR, Dynamic regulation of histone lysine methylation by demethylases. Mol Cell 25, 1–14 (2007). [DOI] [PubMed] [Google Scholar]
- 69.Singh S et al. , 17-DMAG dually inhibits Hsp90 and histone lysine demethylases in alveolar rhabdomyosarcoma. iScience 24, 101996 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Young LC, McDonald DW, Hendzel MJ, Kdm4b histone demethylase is a DNA damage response protein and confers a survival advantage following gamma-irradiation. J Biol Chem 288, 21376–21388 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Khoury-Haddad H et al. , PARP1-dependent recruitment of KDM4D histone demethylase to DNA damage sites promotes double-strand break repair. Proc Natl Acad Sci U S A 111, E728–737 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Stewart E et al. , Identification of Therapeutic Targets in Rhabdomyosarcoma through Integrated Genomic, Epigenomic, and Proteomic Analyses. Cancer Cell 34, 411–426 e419 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Galili N et al. , Fusion of a fork head domain gene to PAX3 in the solid tumour alveolar rhabdomyosarcoma. Nat Genet 5, 230–235 (1993). [DOI] [PubMed] [Google Scholar]
- 74.Scheidler S, Fredericks WJ, Rauscher FJ 3rd, Barr FG, Vogt PK, The hybrid PAX3-FKHR fusion protein of alveolar rhabdomyosarcoma transforms fibroblasts in culture. Proc Natl Acad Sci U S A 93, 9805–9809 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Rudzinski ER et al. , Histology, fusion status, and outcome in metastatic rhabdomyosarcoma: A report from the Children's Oncology Group. Pediatr Blood Cancer, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Pandey PR et al. , PAX3-FOXO1 is essential for tumour initiation and maintenance but not recurrence in a human myoblast model of rhabdomyosarcoma. J Pathol 241, 626–637 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Nishijo K et al. , Credentialing a preclinical mouse model of alveolar rhabdomyosarcoma. Cancer Res 69, 2902–2911 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Hahm ER et al. , Withaferin A-induced apoptosis in human breast cancer cells is mediated by reactive oxygen species. PLoS One 6, e23354 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Coffey K et al. , The lysine demethylase, KDM4B, is a key molecule in androgen receptor signalling and turnover. Nucleic Acids Res 41, 4433–4446 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Li S et al. , Twist2 amplification in rhabdomyosarcoma represses myogenesis and promotes oncogenesis by redirecting MyoD DNA binding. Genes Dev 33, 626–640 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Boyer LA et al. , Core transcriptional regulatory circuitry in human embryonic stem cells. Cell 122, 947–956 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Loven J et al. , Selective inhibition of tumor oncogenes by disruption of super-enhancers. Cell 153, 320–334 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Piha-Paul SA et al. , First-in-Human Study of Mivebresib (ABBV-075), an Oral Pan-Inhibitor of Bromodomain and Extra Terminal Proteins, in Patients with Relapsed/Refractory Solid Tumors. Clin Cancer Res 25, 6309–6319 (2019). [DOI] [PubMed] [Google Scholar]
- 84.Amorim S et al. , Bromodomain inhibitor OTX015 in patients with lymphoma or multiple myeloma: a dose-escalation, open-label, pharmacokinetic, phase 1 study. Lancet Haematol 3, e196–204 (2016). [DOI] [PubMed] [Google Scholar]
- 85.Stathis A et al. , Clinical Response of Carcinomas Harboring the BRD4-NUT Oncoprotein to the Targeted Bromodomain Inhibitor OTX015/MK-8628. Cancer Discov 6, 492–500 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Lee DH et al. , Advances in histone demethylase KDM4 as cancer therapeutic targets. FASEB J 34, 3461–3484 (2020). [DOI] [PubMed] [Google Scholar]
- 87.Kohsaka S et al. , A recurrent neomorphic mutation in MYOD1 defines a clinically aggressive subset of embryonal rhabdomyosarcoma associated with PI3K-AKT pathway mutations. Nat Genet 46, 595–600 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.A MYOD1 mutation cooperates with PI3K pathway activation in ERMS. Cancer Discov 4, OF19 (2014). [DOI] [PubMed] [Google Scholar]
- 89.Szuhai K, de Jong D, Leung WY, Fletcher CD, Hogendoorn PC, Transactivating mutation of the MYOD1 gene is a frequent event in adult spindle cell rhabdomyosarcoma. J Pathol 232, 300–307 (2014). [DOI] [PubMed] [Google Scholar]
- 90.Agaram NP et al. , Recurrent MYOD1 mutations in pediatric and adult sclerosing and spindle cell rhabdomyosarcomas: evidence for a common pathogenesis. Genes Chromosomes Cancer 53, 779–787 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Zhu CH et al. , Cellular senescence in human myoblasts is overcome by human telomerase reverse transcriptase and cyclin-dependent kinase 4: consequences in aging muscle and therapeutic strategies for muscular dystrophies. Aging Cell 6, 515–523 (2007). [DOI] [PubMed] [Google Scholar]
- 92.Connelly JP, Pruett-Miller SM, CRIS.py: A Versatile and High-throughput Analysis Program for CRISPR-based Genome Editing. Sci Rep 9, 4194 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Darawshe S, Rivas G, Minton AP, Rapid and accurate microfractionation of the contents of small centrifuge tubes: application in the measurement of molecular weight of proteins via sedimentation equilibrium. Anal Biochem 209, 130–135 (1993). [DOI] [PubMed] [Google Scholar]
- 94.Skene PJ, Henikoff JG, Henikoff S, Targeted in situ genome-wide profiling with high efficiency for low cell numbers. Nat Protoc 13, 1006–1019 (2018). [DOI] [PubMed] [Google Scholar]
- 95.Jin H et al. , ChIPseqSpikeInFree: a ChIP-seq normalization approach to reveal global changes in histone modifications without spike-in. Bioinformatics 36, 1270–1272 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Whyte WA et al. , Master transcription factors and mediator establish super-enhancers at key cell identity genes. Cell 153, 307–319 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Buenrostro JD, Giresi PG, Zaba LC, Chang HY, Greenleaf WJ, Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat Methods 10, 1213–1218 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Buenrostro JD, Wu B, Chang HY, Greenleaf WJ, ATAC-seq: A Method for Assaying Chromatin Accessibility Genome-Wide. Curr Protoc Mol Biol 109, 21 29 21–21 29 29 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Li H, Durbin R, Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics 26, 589–595 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Li H et al. , The Sequence Alignment/Map format and SAMtools. Bioinformatics 25, 2078–2079 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Zhang Y et al. , Model-based analysis of ChIP-Seq (MACS). Genome Biol 9, R137 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Quinlan AR, Hall IM, BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26, 841–842 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Heinz S et al. , Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell 38, 576–589 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Thorvaldsdottir H, Robinson JT, Mesirov JP, Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief Bioinform 14, 178–192 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Kulakovskiy IV et al. , HOCOMOCO: towards a complete collection of transcription factor binding models for human and mouse via large-scale ChIP-Seq analysis. Nucleic Acids Res 46, D252–D259 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Grant CE, Bailey TL, Noble WS, FIMO: scanning for occurrences of a given motif. Bioinformatics 27, 1017–1018 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Tripodi IJ, Allen MA, Dowell RD, Detecting Differential Transcription Factor Activity from ATAC-Seq Data. Molecules 23, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Otwinowski Z, Minor W, Processing of X-ray diffraction data collected in oscillation mode. Methods in enzymology 276, 307–326 (1997). [DOI] [PubMed] [Google Scholar]
- 109.Afonine PV et al. , Towards automated crystallographic structure refinement with phenix.refine. Acta crystallographica. Section D, Biological crystallography 68, 352–367 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Emsley P, Lohkamp B, Scott WG, Cowtan K, Features and development of Coot. Acta crystallographica. Section D, Biological crystallography 66, 486–501 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.