Abstract
Unlike the ς70-controlled P2 promoter for the osmotically regulated proU operon of Escherichia coli and Salmonella enterica serovar Typhimurium, the ςs-controlled P1 promoter situated further upstream appears not to contribute to expression of the proU structural genes under ordinary growth conditions. For S. enterica proU P1, there is evidence that promoter crypticity is the result of a transcription attenuation phenomenon which is relieved by the deletion of a 22-base C-rich segment in the transcript. In this study, we have sought to identify growth conditions and trans-acting mutations which activate in vivo expression from proU P1. The cryptic S. enterica proU P1 promoter was activated, individually and additively, in a rho mutant (which is defective in the transcription termination factor Rho) as well as by growth at 10°C. The E. coli proU P1 promoter was also cryptic in constructs that carried 1.2 kb of downstream proU sequence, and in these cases activation of in vivo expression was achieved either by a rho mutation during growth at 10°C or by an hns null mutation (affecting the nucleoid protein H-NS) at 30°C. The rho mutation had no effect at either 10 or 30°C on in vivo expression from two other ςs-controlled promoters tested, those for osmY and csiD. In cells lacking the RNA-binding regulator protein Hfq, induction of E. coli proU P1 at 10°C and by hns mutation at 30°C was still observed, although the hfq mutation was associated with a reduction in the absolute levels of P1 expression. Our results suggest that expression from proU P1 is modulated both by nucleoid structure and by Rho-mediated transcription attenuation and that this promoter may be physiologically important for proU operon expression during low-temperature growth.
The ProU transporter in Escherichia coli and Salmonella enterica serovar Typhimurium is a binding-protein-dependent transport system that mediates the cytoplasmic accumulation of compatible solutes such as glycine betaine, l-proline, and related compounds during growth of cells in media of elevated osmolarity (9, 10). The subunit polypeptides of the transporter are encoded by three genes, proV, proW, and proX, which together constitute (in that order) the proU operon (16).
Transcription of proU in both E. coli and S. enterica is activated several-hundredfold in cultures grown in high-osmolarity media, but the mechanism of osmotic induction of the operon is not fully understood (reviewed in references 10, 19, and 29). Two cis regulatory elements that have been identified (see Fig. 1) include a ς70-driven promoter whose transcription start site is situated 60 bases upstream of proV (16, 24, 28, 54) and a negative regulatory element (NRE) approximately 500 bp long, which is situated downstream of the promoter (overlapping the proV coding region) and whose deletion results in partial derepression of proU at low osmolarity (11, 24, 37, 38). Mutations in hns, the gene encoding an abundant nucleoid protein, H-NS, also result in partial derepression of proU expression (for a review of H-NS, see reference 58); two regions of curved DNA exist in the proU cis regulatory region, one falling within the proU NRE and the other located about 200 bp upstream of the promoter (16, 38, 50, 51), to both of which H-NS exhibits preferential binding (30, 38, 50).
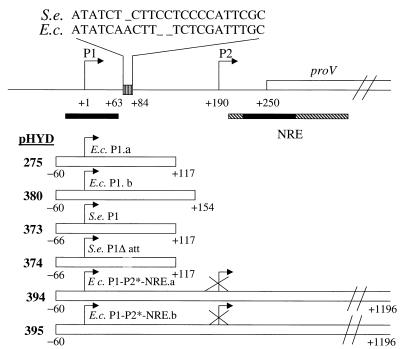
FIG. 1.
Description of plasmids used for studies of in vivo expression from the proU P1 promoter. The line on top depicts the proximal region of the proU operon, and the positions of the cis regulatory elements (namely, P1, P2, and the NRE) are marked relative to that of its first structural gene, proV. Nucleotide number designations are given, with the start site of P1 transcription taken as +1. Solid bars, in the vicinity of P1 and within the NRE, mark the positions of bent-DNA motifs in this region. The box with vertical stripes depicts the location of a 22-base C-rich segment on the P1-initiated transcript of S. enterica (S.e.). The sequence of this segment from S. enterica and from E. coli (E.c.) is shown above the line. Beneath are represented (by the individual open bars) the extents of DNA from the proU regulatory region carried upstream of the lacZ reporter gene in the single-copy-number plasmids (whose pHYD number designations, and abbreviated descriptions, are given alongside) that were used for the in vivo expression studies. Deletion of the 22-base region in pHYD374 is denoted by the interrupted line segment. Note that the P2 promoter in each of the plasmids pHYD394 and pHYD395 has been mutationally inactivated.
Also situated upstream is a second promoter, whose role (if any) in proU regulation is still enigmatic (17). For convenience, the upstream and downstream promoters are designated P1 and P2, respectively (Fig. 1). The two promoters are 190 bp apart and are oriented to transcribe in the same direction, that is, toward the proU structural genes. Data from in vivo and in vitro studies have shown that P1 transcription is moderately osmoresponsive and absolutely dependent on the stationary-phase sigma factor RpoS or ςs (11, 16, 31, 42, 43). However, the physiological function of P1 is unclear for the reasons that (i) cis constructs that carry P2 with the NRE but have P1 deleted continue to exhibit normal osmotic regulation of reporter gene expression (28), (ii) cis- or trans-acting mutations that affect P2 activity abolish all expression from constructs that carry both P1 and P2 (34, 60, 61), and (iii) proU osmotic regulation is unaffected in rpoS mutants that lack RpoS (31).
One clue to the paradox of the P1 promoter has been the finding that, at least in S. enterica, the promoter is rendered cryptic in vivo because of transcription attenuation occurring in the leader region between P1 and P2. Attenuation was not observed in a defined in vitro transcription system, leading to the suggestions that the phenomenon is factor dependent and therefore that, under particular culture conditions (hitherto unidentified), attenuation is relieved and P1 may be able to transcribe the proU structural genes (42).
In this study, we have sought to identify growth conditions as well as trans-acting mutations that relieve in vivo crypticity of the proU P1 promoter. Reporter gene expression from constructs carrying P1 was shown to be increased in an RpoS-specific manner by mutations in rho (the gene for transcription termination factor Rho [reviewed in references 21, 44, and 56]) and hns and by the growth of cultures at 10°C. Our results suggest that the P1 promoter is involved in expression of the proU operon during cold stress and provide additional support for the hypothesis that transcription initiated from the promoter is regulated by transcription attenuation.
MATERIALS AND METHODS
Bacterial strains and plasmids.
E. coli K-12 strains that were used are listed in Table 1. The high-copy-number plasmid vector used was pBluescript II KS (Stratagene, La Jolla, Calif.). Derivatives of the IncW-based single-copy-number plasmid vectors pMU575 and pMU2385 (both of which encode trimethoprim resistance and carry the lacZ reporter gene downstream of a multiple cloning site [MCS] region) (2, 18) were used to measure in vivo expression of lacZ from various proU P1-carrying fragments that had been cloned into the MCS region of the vectors. The extent of proU DNA (relative to the start site of P1 transcription, taken as +1) carried on each of the plasmids is shown in Fig. 1.
TABLE 1.
List of E. coli K-12 strainsa
| Strain | Mutation(s) |
|---|---|
| GJ11 | proU224::lac |
| GJ862 | Nil |
| GJ863 | rho |
| GJ866 | hns |
| GJ867 | rho hns |
| GJ870 | rpoS |
| GJ871 | rho rpoS |
| GJ872 | hns rpoS |
| GJ873 | rho hns rpoS |
| GJ884 | csiD::lac |
| GJ885 | csiD::lac rho |
| GJ887 | csiD::lac rho rpoS |
| GJ888 | osmY::lac |
| GJ889 | osmY::lac rho |
| GJ891 | osmY::lac rho rpoS |
| GJ2733 | csiD::lac rpoS |
| GJ2734 | osmY::lac rpoS |
| GJ2739 | csiD::lac hns |
| GJ2741 | osmY::lac hns |
| GJ2743 | proU224::lac rpoS |
| GJ2745 | hfq-2 |
| GJ2746 | hfq-1 |
| GJ2748 | hfq-1 hns |
| GJ2750 | csiD::lac hns rpoS |
| GJ2751 | osmY::lac hns rpoS |
| GJ2752 | hfq-1 hns rpoS |
All listed strains are F− and, with the exception of GJ11 (15), were constructed in this study. All strains are derivatives of MC4100 (15), and mutations for each in addition to those present in this strain are listed. Genotype designations are as in the work of Berlyn (6). The complete genotype of MC4100 is Δ(argF-lac)U169 rpsL 150 relA1 araD139 flbB5301 deoC1 ptsF25. Allele numbers of the listed mutations and the strains (sources) from which they were transferred by P1 transduction are as follows: rho, rho-4(Am) from CGSC5072 (E. coli Genetic Stock Center); hns, hns-206::Ap from PD32 (12); rpoS, rpoS359::Tn10 from RH90 (5); csiD::lac, csiD::λplacMu15 from DW12 (5); osmY::lac (previously called csi-5::lac), osmY::λplacMu55 from RO151-a (5); hfq-1, hfq-1::Ω from TX2822 (M. E. Winkler strain derived from TX2808 [53]); and hfq-2, hfq-2::Ω from TX2765 (M. E. Winkler strain derived from TX2758 [53]).
Two derivatives of the vector pMU2385 and four derivatives of pMU575 were employed in this study. The proU insert in each of the pMU2385-derived plasmids pHYD394 and pHYD395 is a modification of that in pMU6441 (18), which carries the entire wild-type E. coli proU cis regulatory region from −60 to +1196 (relative to P1), including the P1 and P2 promoters and the downstream NRE. The insert of pMU6441 (subcloned into an M13 phage vector) was modified at the P2 promoter by site-specific mutagenesis using either of two different mismatched primers corresponding to the bottom-strand sequence, 5′-ACTTTTTTCTACCCGGACATACTGAGAATC-3′ or 5′-TAGTCACTTTTTTCGGCCCTAACATACTGA-3′ (mismatches italicized), so that the −10 region of P2 was changed from TAGGGTA to CCGGGTA or TAGGGCC, respectively. The modified proU inserts were then cloned into the MCS region of vector pMU2385 to generate plasmids pHYD394 and pHYD395, respectively. Therefore, plasmids pHYD394 and pHYD395 each carry the entire proU cis regulatory region (from −60 to +1196) with a site-specific inactivation of promoter P2 and are referred to as E. coli P1-P2*-NRE.a and E. coli P1-P2*-NRE.b, respectively (Fig. 1).
Of the four pMU575-derived plasmids that were used, three (pHYD275, pHYD373, and pHYD374 [Fig. 1]) have been described earlier (11, 42). Plasmids pHYD275 (E. coli P1.a) and pHYD373 (S. enterica P1) carry the isolated P1 promoters from E. coli (−60 to +117) and S. enterica (−66 to +117), respectively. Plasmid pHYD374 (S. enterica P1Δatt) is a derivative of pHYD373 with a 22-bp deletion from +63 to +84 which results in relief of attenuation of the transcripts initiated from P1. Plasmid pHYD380, which is a pMU575 derivative carrying E. coli proU P1 from −60 to +154 (E. coli P1.b), was constructed as follows. A pBluescript II KS derivative carrying the wild-type proU insert (−60 to +1196) of pMU6441 was used as a template in PCR with a pair of primers, 5′-TGTAGAGATCTGATGGCAAATGTGG-3′ and 5′-TGTAGAGATCTTTTCTATTGCATGGC-3′. The primers were designed to read outwards from within the proU insert such that the entire plasmid except the region between +154 and +240 (indicated by the bases marked in boldface for the bottom strand on the first primer and the top strand on the second primer, respectively) was amplified by PCR. Digestion of the PCR product with BglII (recognition sites in the two primer sequences italicized) and its circularization by ligation yielded a plasmid derivative (pHYD401) with a proU insert carrying P1 and the NRE and bearing a unique BglII site at the site of P2 deletion between nucleotides +154 and +240. The presence of the new BglII site was exploited to subclone a proU fragment, extending from −60 to +154, from pHYD401 into the MCS region of pMU575 so as to generate pHYD380.
Media and growth conditions.
For routine experiments, Luria-Bertani (LB) medium (35) and glucose-minimal A medium (35) were used as the nutrient and defined media, respectively, and the incubation temperature for growth was 37°C. Unless otherwise indicated, cultures for β-galactosidase assays were grown with shaking at 10 or 30°C (as specified) in media that were based on either LBON (which is LB medium with NaCl omitted [11]) or a modified version of low-osmolarity K medium (15) in which 0.5% Casamino Acids had been replaced by 1% Bacto Tryptone (Difco) (K-tryptone); in either growth medium, typical culture doubling times at 10°C were around 24 h for rho+ strains and 48 h for rho mutant strains. When required, K-tryptone medium was supplemented with NaCl to 0.3 M. Concentrations of antibiotics used were as earlier described (31).
The procedure for growth of biofilm cultures was essentially as described previously (41). The bacterial strain was inoculated in 2 ml of K-tryptone medium in a sterile 35-mm-diameter polystyrene petri plate and incubated without shaking for 40 h at 37°C. The liquid medium containing the free-living bacterial forms was carefully removed, and the sessile biofilm bacteria growing on the inner surface of the plate were then harvested in a small volume of fresh medium by repeated and vigorous pipetting.
Experimental techniques.
The procedures for phage P1 transduction (15) and recombinant DNA manipulations (45) were as described previously. Mutations in hns, hfq, and rpoS were introduced by P1 transduction, with the aid of antibiotic resistance markers (to ampicillin, kanamycin, and tetracycline, respectively) that were 100% linked to them. The chromosomal osmY::lac and csiD::lac fusions were also transduced by selecting for the kanamycin resistance marker situated adjacent to each of them. The rho-4 mutation was introduced by cotransduction with the ilv-3164::Tn10Kan marker from the collection of Singer et al. (46), following which the ilv marker was crossed out in a second transduction to prototrophy. Site-directed mutagenesis was performed with the aid of a kit from United States Biochemical Corp. and was based on the method of Vandeyar et al. (55).
β-Galactosidase assays were performed by the method of Miller (35), and enzyme specific activity values are reported in Miller units. The component of ςs-specific expression for any particular combination of promoter-lacZ fusion, trans-acting chromosomal mutations, and culture growth conditions was calculated as the difference between the β-galactosidase specific activity value for the rpoS+ strain and that for an isogenic rpoS mutant.
RESULTS
Reporter gene expression from S. enterica proU P1: effects of rho and hns mutations.
In an earlier study (42), we had shown that the in vivo expression from an S. enterica proU P1 promoter fragment extending from −66 to +117 (S. enterica P1) of the lacZ reporter gene borne on a very low-copy-number plasmid (pHYD373) is prevented because of transcription attenuation occurring some distance downstream of the site of transcription initiation. Attenuation was relieved, and lacZ expression was consequently observed, in strains carrying a mutant plasmid derivative (pHYD374) that had suffered a 22-bp deletion between nucleotides +63 and +84 relative to the transcription start site (S. enterica P1Δatt). The deleted stretch of nucleotides is C rich on the strand corresponding to the mRNA transcript (Fig. 1), a feature which suggested that it may be the site for loading on mRNA of the transcription termination factor Rho (21, 44). Furthermore, such a C-rich stretch is absent at the corresponding site downstream of the E. coli P1 promoter which (when present on a similar fragment extending from −60 to +117) is active for reporter gene expression in vivo (E. coli P1.a [Fig. 1]).
Based on these considerations, we examined the effect of a rho mutation on expression from proU P1 of S. enterica. As explained above, all lac expression values were obtained in, and are reported for, pairs of isogenic rpoS+ and rpoS derivatives, whose difference has been taken to represent the in vivo activity of the RpoS-dependent proU P1 promoter under the particular test conditions; by these criteria, the plasmid vectors pMU575 and pMU2385 carrying the promoterless lacZ reporter gene displayed insignificant RpoS-dependent lac expression under any of the conditions tested in this study (data not shown). It may also be noted that the in vivo proU P1 activation studies were not amenable to analysis by mRNA primer-extension experiments, in light of our earlier findings (16, 42) that even the cryptic promoter in both S. enterica and E. coli exhibits normal transcription initiation.
With plasmid pHYD373 (S. enterica P1), lac expression was absent in the wild-type strain, as expected; in the rho mutant, however, there was a marked (at least 12-fold-induced) level of RpoS-dependent lac expression which was further elevated moderately in cultures grown with 0.3 M NaCl supplementation (Table 2). With plasmid pHYD374 (S. enterica P1Δatt), we observed a 15-fold increase in expression (over pHYD373) even in the wild-type strain and there was only an additional 2-fold effect of the rho mutation under these conditions. These results suggest that the Rho factor is involved, directly or indirectly, in rendering the S. enterica P1 promoter cryptic and that the nucleotide stretch downstream of the promoter that is identified by the deletion in pHYD374 mediates this effect of Rho.
TABLE 2.
proU P1-lac expression at 30 and 10°Ca
| Temp and GJ rpoS+ strain no.d | Mutation(s) | β-Galactosidase sp act for P1-lac-bearing plasmid (description) on rpoS+ strainb
|
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| pHYD373 (S. enterica P1)
|
pHYD374 (S. enterica P1Δatt)
|
pHYD275 (E. coli P1.a)
|
pHYD380 (E. coli P1.b)
|
pHYD394 (E. coli P1-P2∗- NRE.a)
|
pHYD395 (E. coli P1-P2∗- NRE.b)
|
||||||||
| −NaCl | +NaCl | −NaCl | +NaCl | −NaCl | +NaCl | −NaCl | +NaCl | −NaCl | +NaCl | −NaCl | +NaCl | ||
| 30°C | |||||||||||||
| 862 (870) | Nil | 4 (<1) | 4 (<1) | 60 (2) | 111 (<1) | 63 (4) | 275 (8) | 51 (5) | 277 (8) | 3 (2) | 6 (3) | 2 (1) | 6 (3) |
| 863 (871) | rho | 51 (<1) | 69 (3) | 136 (5) | 159 (4) | 126 (15) | 322 (18) | 73 (13) | 291 (16) | 4 (3) | 11 (4) | 3 (2) | 8 (6) |
| 866 (872) | hns | 5 (3) | 7 (2) | 391 (3) | 301 (4) | 407 (16) | 620 (22) | 349 (35) | NDc | 75 (8) | 112 (14) | 58 (10) | 168 (15) |
| 867 (873) | rho hns | 58 (5) | 82 (2) | 472 (7) | 331 (5) | 393 (27) | 500 (38) | ND | ND | 106 (27) | 207 (41) | 80 (23) | 251 (35) |
| 10°C | |||||||||||||
| 862 (870) | Nil | 54 (2) | 22 (2) | 728 (6) | 288 (4) | 962 (16) | 665 (16) | ND | ND | 5 (4) | 8 (7) | 5 (1) | 10 (1) |
| 863 (871) | rho | 463 (35) | 277 (20) | 917 (67) | 427 (38) | 885 (69) | 667 (47) | ND | ND | 114 (17) | 136 (13) | 136 (22) | 175 (13) |
Isogenic rpoS+ and rpoS cultures were grown in trimethoprim-supplemented K-tryptone medium without NaCl (−NaCl) or with 0.3 M NaCl (+NaCl) at 30 or 10°C for β-galactosidase assays. Enzyme specific activity values are reported in Miller units (35). Values in parentheses are for rpoS derivatives.
Plasmid descriptions are as explained in Fig. 1 and its legend.
ND, not determined.
Strain numbers outside and within parentheses designate isogenic rpoS+ and rpoS derivatives, respectively.
As described above, the nucleoid protein H-NS binds with high affinity to two regions in proU, one of which overlaps the P1 promoter region (30, 38, 50). Furthermore, cells lacking H-NS have elevated ςs levels (5, 59). We therefore examined the effect of hns mutations, alone or in combination with rho, on S. enterica P1 expression. The wild-type P1 promoter on plasmid pHYD373 was unaffected by hns in either the rho+ or rho mutant strains (Table 2). On the other hand, the deletion derivative pHYD374 displayed a sixfold increase in P1 expression in strains lacking H-NS, and there was a marginal additivity with the rho mutation (Table 2). These results are further discussed below.
Environmental stimulus for P1 activation in S. enterica: low-temperature growth.
In order to determine whether any environmental stimuli could activate expression from the cryptic P1 promoter on plasmid pHYD373, we tested two candidate culture conditions, namely, growth in biofilms and growth at low temperature (10°C). The rationale for undertaking these tests was (i) implication of a role for RpoS in biofilm physiology (1) and a recent report which had suggested that E. coli proU is induced in biofilms (41) and (ii) evidence that an untranslated RNA, DsrA, acts to increase RpoS levels during exponential growth at low temperature (48).
No activation of lac expression from pHYD373 was observed when cells were grown as biofilms by the protocol described in the earlier report (41); the lac expression values for strain GJ862/pHYD373 were 3 and 4 Miller units after growth as free-living cells and as biofilms, respectively. On the other hand, a 12-fold induction was obtained when the strain carrying pHYD373 was cultivated at 10°C (Table 2). This low-temperature induction appeared to be mediated by a mechanism different from that leading to the 12-fold induction at 30°C in the rho mutant described above, because the same rho mutation also conferred an additional 8-fold activation of P1 expression at 10°C (Table 2).
With the deletion-bearing plasmid pHYD374, growth at 10°C resulted in a remarkably high level of RpoS-dependent lac expression (nearly 200-fold more than that for the cryptic wild-type promoter at 30°C) in the rho+ strain, which was again only marginally elevated by introduction of the rho mutation (Table 2). The hns effect on promoter P1 activity at low temperature could not be tested because hns mutants are inviable at 10°C (12).
Wild-type E. coli P1 behaves like deletion-bearing S. enterica P1 in vivo.
As noted above, the E. coli proU regulatory sequence lacks the C-rich segment whose presence downstream of P1 in S. enterica is correlated with transcription attenuation in the latter. Two different plasmids, pHYD275 and pHYD380, which carry the E. coli proU P1 promoter sequences from −60 to +117 (E. coli P1.a) and from −60 to +154 (E. coli P1.b) upstream of the vector-borne lacZ reporter gene, respectively, behaved virtually identically to the S. enterica deletion plasmid derivative pHYD374 in experiments testing the effects of various conditions on P1 expression in vivo. Thus, at 30°C, each of the two plasmids with E. coli P1 showed modest expression in the wild-type strain that was induced around sevenfold in the hns mutant and around twofold in the rho derivative (Table 2). We had earlier found that expression from pHYD275 (E. coli P1.a) is in fact reduced in another hns mutant (11, 43), but the latter had carried an uncharacterized missense mutation in hns whereas the present results have been obtained with a true null hns allele.
Cultures of rho+ and rho strains with plasmid pHYD275 were also tested after growth at 10°C, and very pronounced lac expression was observed (as for pHYD374) even in the rho+ hns+ background (Table 2).
Expression from E. coli P1 in the presence of other proU cis regulatory elements.
The studies above had been done with the isolated P1 promoters of either S. enterica or E. coli, in the absence of the other proU cis regulatory sequences downstream of around +120 (relative to P1). In order to study how E. coli proU P1 expression in vivo might be affected by the presence of additional downstream elements such as the NRE (which is also known to bind H-NS with high affinity [30, 38]), we constructed two lac expression derivatives, pHYD394 (E. coli P1-P2*-NRE.a) and pHYD395 (E. coli P1-P2*-NRE.b), each of which carried the proU sequences from −60 to +1196 (that is, encompassing P1, P2, and the NRE) with site-specific mutations in the −10 region of P2 that knocked out the activity of this promoter.
At 30°C, both plasmids pHYD394 and pHYD395 conferred nil lac expression in wild-type or rho strains; RpoS-dependent expression was induced around 50-fold in the hns mutant, and there was a further marginal elevation in the hns rho double mutant (Table 2). At 10°C, there was a 30- to100-fold induction of RpoS-dependent expression in the rho mutant but none in the wild-type strain (Table 2). Once again, the hns effect could not be studied at 10°C because of the problem of inviability of the mutant strains (12).
Effects of rho and hns mutations and low-temperature growth on other ςs-dependent promoters.
In order to determine whether the conditions activating proU P1 were specific for this promoter or common to other ςs-controlled promoters, we tested the effects of rho and hns mutations, and of growth at 10°C, on activity in vivo of the promoters for csiD and osmY (both of which are known to be ςs dependent).
RpoS-dependent expression of osmY-lac in LBON medium was unaffected by rho but was increased around 6-fold by hns (comparable to that reported earlier [5]) and around 25-fold by growth at 10°C (Table 3). The magnitude of osmY expression was growth medium dependent, and the activating effects of hns and low temperature were not as pronounced in K-tryptone medium that had been used by us for the proU P1 studies (Table 3). On the other hand, the described effects of rho, hns, and low temperature on proU P1 were largely independent of the growth medium used (data not shown). It may also be noted from the data in Table 3 that there is an unexpectedly high component of ςs-independent lac expression in the osmY-lac rho strain in K-tryptone medium at 10°C, but the basis for this is not known.
TABLE 3.
lac expression from control promoters osmY and csiDa
| lac fusion | β-Galactosidase sp act for rpoS+ and rpoS strains in medium at tempb:
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| K-tryptone
|
LBON
|
|||||||||
| 30°C
|
10°C
|
30°C
|
10°C
|
|||||||
| w.t. | rho | hns | w.t. | rho | w.t. | rho | hns | w.t. | rho | |
| osmY::lac | 376 (14) | 171 (30) | 287 (NDc) | 587 (20) | 334 (216) | 43 (12) | 78 (ND) | 257 (12) | 835 (36) | 489 (ND) |
| csiD::lac | 522 (64) | 195 (12) | 372 (ND) | 251 (33) | 258 (18) | 110 (10) | 80 (4) | 96 (4) | 37 (10) | 152 (39) |
The following GJ strains were used in the experiment, given in the order wild type (w.t.), rho mutant, and hns mutant for each with the isogenic rpoS derivatives indicated in parentheses: osmY::lac, 888 (2734), 889 (891), and 2741 (2751); csiD::lac, 884 (2733), 885 (887), and 2739 (2750).
Cultures of the various rpoS+ and isogenic rpoS strains were grown in trimethoprim-supplemented K-tryptone or LBON medium at 30 or 10°C for β-galactosidase assays. Enzyme specific activity values are reported in Miller units (35). Values in parentheses are for rpoS derivatives.
ND, not determined.
RpoS-dependent expression of csiD-lac was not affected by any of the conditions, that is, by rho or hns mutations or growth at low temperature (Table 3). Once again, we observed a medium dependence in the absolute values of lac expression (including a reduction in expression in the wild-type background for LBON medium at 10°C), but the conclusion concerning the absence of effect was valid for both media tested.
In a related context, we also tested the effect of low-temperature growth on activity of the ς70-dependent P2 promoter of proU, by measuring lac expression from the chromosomal proU-lac fusion strain GJ11 and its rpoS derivative GJ2743. For both strains, β-galactosidase specific activity values were around 3 and 11 Miller units after growth at 30 and 10°C, respectively, in the low-osmolarity K-tryptone medium, suggesting that the P2 promoter is also not significantly stimulated by cold stress.
Effect of hfq on proU P1.
The RNA-binding protein Hfq has earlier been shown to be a positive regulator of ςs synthesis at the level of translation (reviewed in references 14, 20, and 23). Mutations in hfq have been reported previously to be epistatic to hns (36) and dsrA (49) with reference to rpoS regulation, suggesting that the regulatory effects of both H-NS and DsrA are mediated indirectly via Hfq. In light of these reports, we tested the effects of hfq insertion mutations in our proU P1 assay systems.
The chromosomal hfq gene is part of a complex operon, and one needs to distinguish between a true hfq mutant effect and that caused by polarity of the hfq insertion on downstream genes in the operon. For this purpose, we employed the strategy of comparing the phenotypic effects of two different insertion mutations, one near the 5′ end of hfq (hfq-1::Ω) and the other just beyond its 3′ end (hfq-2::Ω), as suggested earlier by Tsui et al. (53). When lac expression from the isolated E. coli proU P1 promoter on plasmid pHYD275 was examined, the hfq-2::Ω mutation had little effect at either 30 or 10°C, whereas the hfq-1::Ω mutation was associated with a three- to fivefold reduction in expression at both growth temperatures (Table 4). Importantly, however, the magnitude of cold induction of the P1 promoter was roughly similar in all the three strains. We conclude that Hfq (i) affects the absolute level of proU P1 promoter activity in vivo but (ii) is not required to mediate its induction during low-temperature growth.
TABLE 4.
lac expression from pHYD275 (E. coli P1.a) in hfq mutantsa
| Strain | Mutation | β-Galactosidase sp act at temp:
|
|
|---|---|---|---|
| 30°C | 10°C | ||
| GJ862 | Nil | 123 | 839 |
| GJ2746 | hfq-1 | 24 | 243 |
| GJ2745 | hfq-2 | 85 | 750 |
Cultures of the indicated strains carrying plasmid pHYD275 were grown in trimethoprim-supplemented K-tryptone medium at 30 or 10°C for β-galactosidase assays. Enzyme specific activity values are reported in Miller units (35).
We also tested the epistatic interaction, if any, of the hfq-1::Ω insertion on activation by hns of E. coli proU P1 borne on plasmid pHYD275. In LB medium-grown cultures, the specific activity values of β-galactosidase for the pHYD275 derivatives of GJ862 (wild type), GJ866 (hns), GJ2746 (hfq-1), and GJ2748 (hns hfq-1) were 70, 301, 21, and 153 Miller units, respectively, and in all four instances the measured expression was shown to be predominantly ςs dependent (data not shown). Similar results were also obtained for cultures grown in LBON or glucose-minimal A medium (data not shown). Our results indicate that hns-mediated derepression of an RpoS-controlled promoter occurs even in an hfq-1 mutant and therefore are at apparent variance with the conclusion from an earlier report (36) that the latter is epistatic to the former with regard to regulation of ςs synthesis.
DISCUSSION
In this study, we have identified several conditions in which expression in trans from the cryptic ςs-controlled P1 promoter of proU in E. coli and S. enterica is activated or enhanced. In general, such enhancement could be envisaged as occurring at either the level of ςs synthesis itself (whose regulation is known to be extremely complex [14, 20, 23]) or the more local level of the proU P1 cis regulatory region. In order to distinguish between these alternatives, we have examined whether each of the conditions that activates reporter gene expression from proU P1 also does so from other ςs-controlled promoters such as those for osmY or csiD. Based on the results with the osmY-lac strain (Table 3), it appears that the activating effect of the rho mutation on proU P1 occurs at the local level, whereas that of low-temperature growth occurs at the more upstream level of ςs synthesis. The latter conclusion is consistent with the findings of an earlier report (48). The absence of effect of low-temperature growth (or of hns [see below]) on csiD expression may perhaps be explained on the grounds that this promoter requires another transcriptional activator (Crp) for its expression (32) and therefore that an increase in ςs levels alone may not be sufficient for its activation.
H-NS and proU P1.
As discussed below, our data suggest that the hns mutant effect on proU P1 is exerted at both the local and the upstream levels. It is known that ςs synthesis is derepressed about sixfold in strains lacking H-NS (5, 59), and this could explain the moderate increase in proU P1-lac expression from plasmids pHYD275 (E. coli P1.a) and pHYD374 (S. enterica P1Δatt) as well as the increase in osmY-lac expression. On the other hand, the hns mutant effect on proU P1-lac expression from plasmids pHYD394 and pHYD395 (which carry more than 1 kb of DNA downstream of P1 including the NRE and inactivated P2) is around 50-fold, which is very much more pronounced than can be accounted for by the upstream effect on ςs synthesis alone; it is likely, therefore, that in this case H-NS is also acting locally, perhaps by binding to the high-affinity binding sites at the NRE and around P1 (30, 38, 50), to repress reporter gene expression.
A null mutation in stpA, the gene encoding the H-NS-like protein StpA, which is believed to represent a molecular backup of H-NS (58), had no effect on proU P1 expression either by itself or in combination with other mutations such as rho and hns (data not shown).
Rho and proU P1.
The activating effect of the rho mutation on proU P1 is most prominent in two situations (where the promoter is otherwise cryptic), namely, on S. enterica P1 (plasmid pHYD373) at 30°C and on E. coli P1-P2*-NRE (plasmids pHYD394 and pHYD395) at 10°C. The magnitude of rho-mediated P1 activation on pHYD373 at 10°C is also quite significant. Although the mechanism by which P1 activation occurs in the rho mutants is not known, two lines of evidence suggest that it may be related to the relief of attenuation of transcripts initiated from P1. First, such a mechanism will be consistent with the previously characterized activities and functions of the Rho protein (21, 44, 56). Second, the rho mutant effect is considerably diminished for S. enterica P1Δatt (plasmid pHYD374), which bears the deletion of a 22-base C-rich segment (on the coding strand) and which has previously been shown (42) to be defective in attenuation. It is reasonable, therefore, to postulate that the C-rich target segment on the S. enterica P1-initiated transcript serves as a site for Rho factor loading and consequent termination of transcription. The precise site of occurrence of the latter remains to be determined. This scheme is reminiscent of the mechanism by which Rho factor autoregulates its own synthesis by transcription attenuation (4, 25, 33). Our results would also suggest, by analogy, that Rho-dependent attenuation occurs for native E. coli P1-associated expression (that is, in the presence of the long downstream sequence), at least during growth at 10°C, but once again the cis site of action is not known.
Low-temperature growth and proU P1.
As argued above, at least one component of the activation of proU P1 during low-temperature growth may be accounted for by an upstream effect at the level of ςs synthesis. At the same time, a synergism is apparent between low-temperature growth and loss of Rho activity with respect to the ability of each to activate lac expression from P1 of both S. enterica (on pHYD373) and E. coli (on pHYD394 and pHYD395). It is possible that transcription termination events in the cell generally become more critically Rho dependent at the low temperature; for example, Bae et al. (3) have reported that transcriptional readthrough at Rho-independent terminator sites is increased in cultures grown at low temperature. Alternatively, Rho might influence temperature-responsive changes in DNA topology, as suggested by Tobe et al. (52).
Sledjeski et al. (48, 49) have reported that the induction of ςs synthesis during low-temperature growth is mediated by an untranslated RNA, DsrA, and that Hfq is required for DsrA to stimulate the translation of RpoS. On the other hand, our own data demonstrating that hfq mutants are not defective in cold induction of proU P1 do not readily fit into the model proposed by Sledjeski et al. We were unable to obtain the dsrA-null strain to continue these studies.
Physiological role of proU P1 promoter?
The role of the proU P1 promoter in enterobacterial physiology has so far remained obscure (17). This study has identified at least three factors which in an interactive manner may be involved in stimulating expression from this promoter, namely, growth at low temperature and inactivation of the Rho and H-NS proteins. Thus, the in vivo activity of the isolated wild-type E. coli P1 promoter in cells grown at 10°C (Table 2) marks it as one of the very strong and substantially regulated promoters under these conditions. However, the mechanisms of interaction among the three identified factors, and the physiological relevance of such interactions, remain to be determined.
It is possible that under certain growth conditions the activity of Rho or of H-NS is reduced or antagonized even in a rho+ hns+ strain. For example, (i) other factors involved in transcription elongation and termination such as NusA and NusG are known to modulate Rho function (7, 8, 21, 39, 44); (ii) phage-encoded proteins Psu (from P4) and gp5.5 (from T7) are physiological antagonists of Rho and H-NS, respectively (26, 27); and (iii) the cellular functions of Rho and H-NS are antagonized by overexpression of the chromosomally encoded yaeO (40) and dsrA (47) genes, respectively. There is also in vitro evidence that the activities of H-NS (43, 54) and of Rho (39, 57) are sensitive to the potassium salt concentration, which is known to vary within the cell with changes in osmolarity of the growth medium (10).
Finally, an overlap between adaptation to osmotic stress and to cold stress has been demonstrated earlier in plants and other bacteria. In Listeria monocytogenes, intracellular accumulation of glycine betaine is necessary for growth both at low temperature and in media of elevated osmolarity and occurs via an active uptake mechanism (13). Holmstrom et al. (22) have reported that synthesis of glycine betaine in transgenic tobacco lines is associated with improved tolerance to both salinity and low temperature. It is, therefore, possible that the proU operon in E. coli or S. enterica mediates adaptive accumulation of glycine betaine in response to both osmotic stress and cold stress and that the P2 and P1 promoters are primarily responsible for transcription of the operon under the respective conditions.
ACKNOWLEDGMENTS
We thank Mary Berlyn, Erhard Bremer, Carol Gross, Regine Hengge-Aronis, and James Pittard for making available various strains and plasmids that were used in this study, and we thank Richard Hayward and Akira Ishihama for valuable suggestions. We also acknowledge the assistance of N. Nagesh for oligonucleotide synthesis.
J.G. is an honorary faculty member of the Jawaharlal Nehru Centre for Advanced Scientific Research.
REFERENCES
- 1.Adams J L, McLean R J C. Impact of rpoS deletion on Escherichia coli biofilms. Appl Environ Microbiol. 1999;65:4285–4287. doi: 10.1128/aem.65.9.4285-4287.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Andrews A E, Lawley B, Pittard A J. Mutational analysis of repression and activation of the tyrP gene in Escherichia coli. J Bacteriol. 1991;173:5068–5078. doi: 10.1128/jb.173.16.5068-5078.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bae W, Xia B, Inouye M, Severinov K. Escherichia coli CspA-family RNA chaperones are transcription antiterminators. Proc Natl Acad Sci USA. 1999;97:7784–7789. doi: 10.1073/pnas.97.14.7784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barik S, Bhattacharya P, Das A. Autogenous regulation of transcription termination factor Rho. J Mol Biol. 1985;182:495–508. doi: 10.1016/0022-2836(85)90236-0. [DOI] [PubMed] [Google Scholar]
- 5.Barth M, Marschall C, Muffler A, Fischer D, Hengge-Aronis R. Role for the histone-like protein H-NS in growth phase-dependent and osmotic regulation of ςs and many ςs-dependent genes in Escherichia coli. J Bacteriol. 1995;177:3455–3464. doi: 10.1128/jb.177.12.3455-3464.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Berlyn M K B. Linkage map of Escherichia coli K-12, edition 10: the traditional map. Microbiol Mol Biol Rev. 1998;62:814–984. doi: 10.1128/mmbr.62.3.814-984.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Burns C M, Richardson L V, Richardson J P. Combinatorial effects of NusA and NusG on transcription elongation and Rho-dependent termination in Escherichia coli. J Mol Biol. 1998;278:307–316. doi: 10.1006/jmbi.1998.1691. [DOI] [PubMed] [Google Scholar]
- 8.Burova E, Gottesman M E. NusG overexpression inhibits Rho-dependent termination in Escherichia coli. Mol Microbiol. 1995;17:633–641. doi: 10.1111/j.1365-2958.1995.mmi_17040633.x. [DOI] [PubMed] [Google Scholar]
- 9.Cosquer A, Pichereau V, Pocard J-A, Minet J, Cormier M, Bernard T. Nanomolar levels of dimethylsulfoniopropionate, dimethylsulfonioacetate, and glycine betaine are sufficient to confer osmoprotection to Escherichia coli. Appl Environ Microbiol. 1999;65:3304–3311. doi: 10.1128/aem.65.8.3304-3311.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Csonka L N, Epstein W. Osmoregulation. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C.: ASM Press; 1996. pp. 1210–1223. [Google Scholar]
- 11.Dattananda C S, Rajkumari K, Gowrishankar J. Multiple mechanisms contribute to osmotic inducibility of proU operon expression in Escherichia coli: demonstration of two osmoresponsive promoters and of a negative regulatory element within the first structural gene. J Bacteriol. 1991;173:7481–7490. doi: 10.1128/jb.173.23.7481-7490.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dersch P, Kneip S, Bremer E. The nucleoid-associated DNA-binding protein H-NS is required for the efficient adaptation of Escherichia coli to a cold environment. Mol Gen Genet. 1994;245:255–259. doi: 10.1007/BF00283274. [DOI] [PubMed] [Google Scholar]
- 13.Gerhardt P N M, Smith L T, Smith G M. Osmotic and chill activation of glycine betaine porter II in Listeria monocytogenes membrane vesicles. J Bacteriol. 2000;182:2544–2550. doi: 10.1128/jb.182.9.2544-2550.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Goodrich-Blair H, Uria-Nickelsen M, Kolter R. Regulation of gene expression in stationary phase. In: Lin E C C, Lynch A S, editors. Regulation of gene expression in E. coli. Georgetown, Tex: Landes Bioscience Publishers; 1996. pp. 571–583. [Google Scholar]
- 15.Gowrishankar J. Identification of osmoresponsive genes in Escherichia coli: evidence for participation of potassium and proline transport systems in osmoregulation. J Bacteriol. 1985;164:434–445. doi: 10.1128/jb.164.1.434-445.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gowrishankar J. Nucleotide sequence of the osmoregulatory proU operon of Escherichia coli. J Bacteriol. 1989;171:1923–1931. doi: 10.1128/jb.171.4.1923-1931.1989. . (Erratum, 172:1165, 1990.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gowrishankar J. Ploughing a lonely furrow: the curious case of the P1 promoter in the osmotically regulated proU operon of Escherichia coli. J Indian Inst Sci. 1999;79:41–47. [Google Scholar]
- 18.Gowrishankar J, Pittard A J. Superimposition of TyrR protein-mediated regulation on osmoresponsive transcription of Escherichia coli proU in vivo. J Bacteriol. 1998;180:6743–6748. doi: 10.1128/jb.180.24.6743-6748.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gowrishankar J, Manna D. How is osmotic regulation of transcription of the Escherichia coli proU operon achieved? A review and a model. Genetica. 1996;97:363–378. doi: 10.1007/BF00055322. [DOI] [PubMed] [Google Scholar]
- 20.Hengge-Aronis R. Regulation of gene expression during entry into stationary phase. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C.: ASM Press; 1996. pp. 1497–1512. [Google Scholar]
- 21.Henkin T M. Control of transcription termination in prokaryotes. Annu Rev Genet. 1996;30:35–57. doi: 10.1146/annurev.genet.30.1.35. [DOI] [PubMed] [Google Scholar]
- 22.Holmstrom K O, Somersalo S, Mandal A, Palva T E, Welin B. Improved tolerance to salinity and low temperature in transgenic tobacco producing glycine betaine. J Exp Bot. 2000;51:177–185. doi: 10.1093/jexbot/51.343.177. [DOI] [PubMed] [Google Scholar]
- 23.Ishihama A. Functional modulation of Escherichia coli RNA polymerase. Annu Rev Microbiol. 2000;54:499–518. doi: 10.1146/annurev.micro.54.1.499. [DOI] [PubMed] [Google Scholar]
- 24.Jordi B J A M, Higgins C F. The downstream regulatory element of the proU operon of Salmonella typhimurium inhibits open complex formation by RNA polymerase at a distance. J Biol Chem. 2000;275:12123–12128. doi: 10.1074/jbc.275.16.12123. [DOI] [PubMed] [Google Scholar]
- 25.Kung H-F, Bekesi E, Gutterman S K, Gray J E, Traub L, Calhoun D H. Autoregulation of the rho gene of Escherichia coli K-12. Mol Gen Genet. 1984;193:210–213. doi: 10.1007/BF00330669. [DOI] [PubMed] [Google Scholar]
- 26.Linderoth N A, Calendar R L. The Psu protein of bacteriophage P4 is an antitermination factor for Rho-dependent transcription termination. J Bacteriol. 1991;173:6722–6731. doi: 10.1128/jb.173.21.6722-6731.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu Q, Richardson C C. Gene 5.5 protein of bacteriophage T7 inhibits the nucleoid protein H-NS of Escherichia coli. Proc Natl Acad Sci USA. 1993;90:1761–1765. doi: 10.1073/pnas.90.5.1761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lucht J M, Bremer E. Characterization of mutations affecting the osmoregulated proU promoter of Escherichia coli and identification of 5′ sequences required for high-level expression. J Bacteriol. 1991;173:801–809. doi: 10.1128/jb.173.2.801-809.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lucht J M, Bremer E. Adaptation of Escherichia coli to high osmolarity environments: osmoregulation of the high-affinity glycine betaine transport system ProU. FEMS Microbiol Rev. 1994;14:3–20. doi: 10.1111/j.1574-6976.1994.tb00067.x. [DOI] [PubMed] [Google Scholar]
- 30.Lucht J M, Dersch P, Kempf B, Bremer E. Interactions of the nucleoid-associated DNA-binding protein H-NS with the regulatory region of the osmotically controlled proU operon of Escherichia coli. J Biol Chem. 1994;269:6578–6586. [PubMed] [Google Scholar]
- 31.Manna D, Gowrishankar J. Evidence for involvement of proteins HU and RpoS in transcription of the osmoresponsive proU operon in Escherichia coli. J Bacteriol. 1994;176:5378–5384. doi: 10.1128/jb.176.17.5378-5384.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Marschall C, Labrousse V, Kreimer M, Weichart D, Kolb A, Hengge-Aronis R. Molecular analysis of the regulation of csiD, a carbon starvation-inducible gene in Escherichia coli that is exclusively dependent on ςs and requires activation by cAMP-CRP. J Mol Biol. 1998;276:339–353. doi: 10.1006/jmbi.1997.1533. [DOI] [PubMed] [Google Scholar]
- 33.Matsumoto Y, Shigesada K, Hirano M, Imai M. Autogenous regulation of the gene for transcription factor Rho in Escherichia coli: localization and function of its attenuators. J Bacteriol. 1986;166:945–958. doi: 10.1128/jb.166.3.945-958.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mellies J, Brems R, Villarejo M. The Escherichia coli proU promoter element and its contribution to osmotically signaled transcription activation. J Bacteriol. 1994;176:3638–3645. doi: 10.1128/jb.176.12.3638-3645.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Miller J H. A short course in bacterial genetics: a laboratory manual and handbook for Escherichia coli and related bacteria. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1992. [Google Scholar]
- 36.Muffler A, Fischer D, Hengge-Aronis R. The RNA-binding protein HF-I, known as a host factor for phage Qβ RNA replication, is essential for rpoS translation in Escherichia coli. Genes Dev. 1996;10:1143–1151. doi: 10.1101/gad.10.9.1143. [DOI] [PubMed] [Google Scholar]
- 37.Overdier D G, Csonka L N. A transcriptional silencer downstream of the promoter in the osmotically controlled proU operon of Salmonella typhimurium. Proc Natl Acad Sci USA. 1992;89:3140–3144. doi: 10.1073/pnas.89.7.3140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Owen-Hughes T A, Pavitt G D, Santos D S, Sidebotham J M, Hulton C S J, Hinton J C D, Higgins C F. The chromatin-associated protein H-NS interacts with curved DNA to influence DNA topology and gene expression. Cell. 1992;71:255–265. doi: 10.1016/0092-8674(92)90354-f. [DOI] [PubMed] [Google Scholar]
- 39.Pasman Z, von Hippel P H. Regulation of rho-dependent transcription termination by NusG is specific to the Escherichia coli elongation complex. Biochemistry. 2000;39:5573–5585. doi: 10.1021/bi992658z. [DOI] [PubMed] [Google Scholar]
- 40.Pichoff S, Alibaud L, Guédant A, Castanié M-P, Bouché J-P. An Escherichia coli gene (yaeO) suppresses temperature-sensitive mutations in essential genes by modulating Rho-dependent transcription termination. Mol Microbiol. 1998;29:859–869. doi: 10.1046/j.1365-2958.1998.00981.x. [DOI] [PubMed] [Google Scholar]
- 41.Prigent-Combaret C, Vidal O, Dorel C, Lejeune P. Abiotic surface sensing and biofilm-dependent regulation of gene expression in Escherichia coli. J Bacteriol. 1999;181:5993–6002. doi: 10.1128/jb.181.19.5993-6002.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rajkumari K, Ishihama A, Gowrishankar J. Evidence for transcription attenuation rendering cryptic a ςs-dependent promoter of the osmotically regulated proU operon of Salmonella typhimurium. J Bacteriol. 1997;179:7169–7173. doi: 10.1128/jb.179.22.7169-7173.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rajkumari K, Kusano S, Ishihama A, Mizuno T, Gowrishankar J. Effects of H-NS and potassium glutamate on ςS- and ς70-directed transcription in vitro from osmotically regulated P1 and P2 promoters of proU in Escherichia coli. J Bacteriol. 1996;178:4176–4181. doi: 10.1128/jb.178.14.4176-4181.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Richardson J P, Greenblatt J. Control of RNA chain elongation and termination. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C.: ASM Press; 1996. pp. 822–848. [Google Scholar]
- 45.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 46.Singer M, Baker T A, Schnitzler G, Deischel S M, Goel M, Dove W, Jaacks K J, Grossman A D, Erickson J W, Gross C A. A collection of strains containing genetically linked alternating antibiotic resistance elements for genetic mapping of Escherichia coli. Microbiol Rev. 1989;53:1–24. doi: 10.1128/mr.53.1.1-24.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sledjeski D, Gottesman S. A small RNA acts as an antisilencer of the H-NS-silenced rcsA gene of Escherichia coli. Proc Natl Acad Sci USA. 1995;92:2003–2007. doi: 10.1073/pnas.92.6.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sledjeski D D, Gupta A, Gottesman S. The small RNA, DsrA, is essential for the low temperature expression of rpoS during exponential growth in Escherichia coli. EMBO J. 1996;15:3993–4000. [PMC free article] [PubMed] [Google Scholar]
- 49.Sledjeski D D, Whitman C, Zhang A. Hfq is necessary for regulation by the untranslated RNA DsrA. J Bacteriol. 2001;183:1997–2005. doi: 10.1128/JB.183.6.1997-2005.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tanaka K, Muramatsu S, Yamada H, Mizuno T. Systematic characterization of curved DNA segments randomly cloned from Escherichia coli and their functional significance. Mol Gen Genet. 1991;226:367–376. doi: 10.1007/BF00260648. [DOI] [PubMed] [Google Scholar]
- 51.Tanaka K, Ueguchi C, Mizuno T. Importance of stereospecific positioning of the upstream cis-acting DNA element containing a curved DNA structure for the functioning of the Escherichia coli protein proV promoters. Biosci Biotechnol Biochem. 1994;58:1097–1101. doi: 10.1271/bbb.58.1097. [DOI] [PubMed] [Google Scholar]
- 52.Tobe T, Yoshikawa M, Sasakawa C. Deregulation of temperature-dependent transcription of the invasion regulatory gene, virB, in Shigella by rho mutation. Mol Microbiol. 1994;12:267–276. doi: 10.1111/j.1365-2958.1994.tb01015.x. [DOI] [PubMed] [Google Scholar]
- 53.Tsui T H-C, Leung H-C E, Winkler M E. Characterization of broadly pleiotropic phenotypes caused by an hfq insertion mutation in Escherichia coli K-12. Mol Microbiol. 1994;13:35–49. doi: 10.1111/j.1365-2958.1994.tb00400.x. [DOI] [PubMed] [Google Scholar]
- 54.Ueguchi C, Mizuno T. The Escherichia coli nucleoid protein H-NS functions directly as a transcriptional repressor. EMBO J. 1993;12:1039–1046. doi: 10.1002/j.1460-2075.1993.tb05745.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Vandeyar M A, Weiner M P, Hutton C J, Batt C A. A simple and rapid method for the selection of oligodeoxynucleotide-directed mutants. Gene. 1988;65:129–133. doi: 10.1016/0378-1119(88)90425-8. [DOI] [PubMed] [Google Scholar]
- 56.von Hippel P H, Rees W A, Rippe K, Wilson K S. Specificity mechanisms in the control of transcription. Biophys Chem. 1996;59:231–246. doi: 10.1016/0301-4622(96)00006-3. [DOI] [PubMed] [Google Scholar]
- 57.Walstrom K M, Dozono J M, von Hippel P H. Effects of reaction conditions on RNA secondary structure and on the helicase activity of Escherichia coli transcription termination factor Rho. J Mol Biol. 1998;279:713–726. doi: 10.1006/jmbi.1998.1814. [DOI] [PubMed] [Google Scholar]
- 58.Williams R M, Rimsky S. Molecular aspects of the E. coli nucleoid protein, H-NS: a central controller of gene regulatory networks. FEMS Microbiol Lett. 1997;156:175–185. doi: 10.1111/j.1574-6968.1997.tb12724.x. [DOI] [PubMed] [Google Scholar]
- 59.Yamashino T, Ueguchi C, Mizuno T. Quantitative control of the stationary phase-specific sigma factor, ςs, in Escherichia coli: involvement of the nucleoid protein H-NS. EMBO J. 1995;14:594–602. doi: 10.1002/j.1460-2075.1995.tb07035.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yim H H, Brems R L, Villarejo M. Molecular characterization of the promoter of osmY, an rpoS-dependent gene. J Bacteriol. 1994;176:100–107. doi: 10.1128/jb.176.1.100-107.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhang X, Fletcher S A, Csonka L N. Site-directed mutational analysis of the osmotically regulated proU promoter of Salmonella typhimurium. J Bacteriol. 1996;178:3377–3379. doi: 10.1128/jb.178.11.3377-3379.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]