Summary
The research article describing the discovery of ribosomal frameshifting in the bacterial CopA gene also reported the occurrence of frameshifting in the expression of the human ortholog ATP7B based on assays using dual luciferase reporters. An examination of the publicly available ribosome profiling data and the phylogenetic analysis of the proposed frameshifting site cast doubt on the validity of this claim and prompted us to reexamine the evidence. We observed similar apparent frameshifting efficiencies as the original authors using the same type of vector that synthesizes both luciferases as a single polyprotein. However, we noticed anomalously low absolute luciferase activities from the N-terminal reporter that suggests interference of reporter activity or levels by the ATP7B test cassette. When we tested the same proposed ATP7B frameshifting cassette in a more recently developed reporter system in which the reporters are released without being included in a polyprotein, no frameshifting was detected above background levels.
Keywords: ribosomal frameshifting, ATP7B, artifacts, dual reporters, ribosome profiling, translation, mRNA decoding
Highlights
-
•
Reported ribosomal frameshifting in the expression of human ATP7B is unlikely to occur
-
•
Care is needed to avoid artifacts with dual reporters fused with test sequences
-
•
StopGo-containing vectors help avoid fusion artifacts
-
•
No human gene of non-viral origin is known to use −1 frameshifting in their expression
Loughran et al. provide evidence arguing against previously reported efficient ribosomal frameshifting in the expression of human ATP7B. Loughran et al. show that this previous report was based on an infrequent artifact of fused reporters that can be mitigated against by introducing StopGo.
Introduction
Meydan et al. reported the discovery and functional characterization of efficient ribosomal frameshifting in the Escherichia coli (E. coli) copA gene (Meydan et al., 2017). This finding is well supported by external ribosome profiling data that prompted their investigation (Li et al., 2014; Atkins et al., 2017) and was subsequently confirmed by an independent study (Drees et al., 2017). Along with this important discovery, the authors reported that ribosomal frameshifting occurs in the APT7B gene, which is a human ortholog of copA. Their supporting evidence relied on the expression of an alleged ATP7B frameshifting site within a dual luciferase reporter system (Grentzmann et al., 1998).
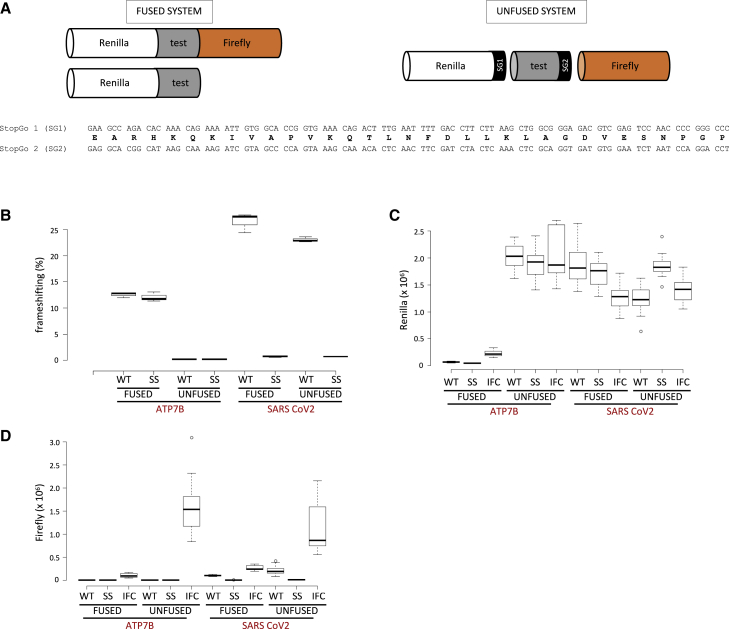
Frameshifting efficiency is often measured with fused dual luciferase reporter systems. The upstream reporter (often Renilla luciferase [R-luc]) monitors zero-frame translation, and the downstream reporter (Firefly luciferase [F-luc]) monitors an alternative reading frame. Ribosomes that frameshift synthesize an R-luc-F-luc fusion protein, whereas those that do not yield R-luc alone (Grentzmann et al., 1998). Frameshifting efficiency is calculated as the ratio of F-luc/R-luc activities, normalized to the ratio expressed from an in-frame control (IFC) reporter expressing an identical protein sequence as the product generated by frameshifting. However, although R-luc activity expressed from the IFC vector is derived solely from the R-luc-F-luc fusion protein, the R-luc activity from the test vector is derived from both the R-luc-F-luc fusion (generally minor product) as well as the R-luc termination product (usually major). Estimations of frameshifting efficiency can be problematic if the R-luc activities from the fusion and termination product are dissimilar, which can arise when the test sequence product influences reporter activity or stability. To address this, we recently generated an “unfused” dual reporter vector that insulates the test sequence from the reporter sequence by introducing flanking StopGo (SG) elements derived from foot and mouth disease virus. SG elements allow the co-translational hydrolysis of a specific peptidyl-tRNA linkage without disrupting continued ribosome elongation. Therefore, luciferase reporters are co-translationally separated from the test sequence product and have the same amino acid sequence irrespective of the test sequence (Loughran et al., 2017). Hereafter, we term this dual luciferase reporter system “unfused reporter” to differentiate it from the earlier “fused reporter” iteration employed by Meydan et al. (2017) (Figure 1A).
Figure 1.
Frameshifting efficiency of ATP7B
(A) Illustration comparing the protein products expressed from typical fused and unfused dual luciferase reporter systems. The lower panel indicates the nucleotide and amino acid sequence of StopGo 1 (SG1) and StopGo 2 (SG2).
(B) Frameshifting efficiencies (%) of wild-type (WT) ATP7B and SARS-CoV-2 frameshift cassettes calculated by both fused and unfused dual luciferase reporter systems transfected into HEK293T cells. SS refers to cassettes in which the frameshifting slippery site was destroyed without altering the encoded amino acid sequence.
(C) Absolute R-luc activities (IFC, in-frame control).
(D) Absolute F-luc activities. n = 12 (4 technical relicates from 3 biological samples) for (B)–(D). Box plots: the central line indicates the median, the box limits indicate the interquartile area, whiskers indicate 1.5 × interquartile range, and outliers (if they occur) are indicated with dots.
Results
We cloned the same frameshifting cassette reported in Meydan et al. (2017) as producing >11% frameshifting efficiency into both fused and unfused reporter systems and tested each for frameshifting efficiency. Whereas a control construct containing the −1 frameshifting site from SARS-CoV-2 showed frameshifting activities at the expected level (∼25%), apparent −1 frameshifting rates from the ATP7B −1 frameshifting construct were ∼12% when determined using fused reporters but <0.5% when determined using unfused reporters (Figure 1B). Furthermore, a slippery site mutant (SS) of the ATP7B −1 frameshifting site in which frameshifting should be abolished showed a similarly high frameshifting efficiency indistinguishable from the wild-type (WT) ATP7B. Examination of the absolute luciferase activities from both R-luc and F-luc revealed a dramatic reduction in R-luc activities in cells transfected with fused ATP7B reporters compared with the unfused ATP7B reporters and the SARS-CoV-2 controls (Figure 1C). F-luc activities for IFCs from both ATP7B and SARS-CoV-2 fused vectors are also greatly reduced compared with their unfused counterparts (Figure 1D). For ATP7B, a greater decrease in absolute F-luc relative to absolute R-luc led to the increase in F-luc/R-luc ratios and thus inflated the estimated −1 frameshifting efficiency (Figures 1C and 1D).
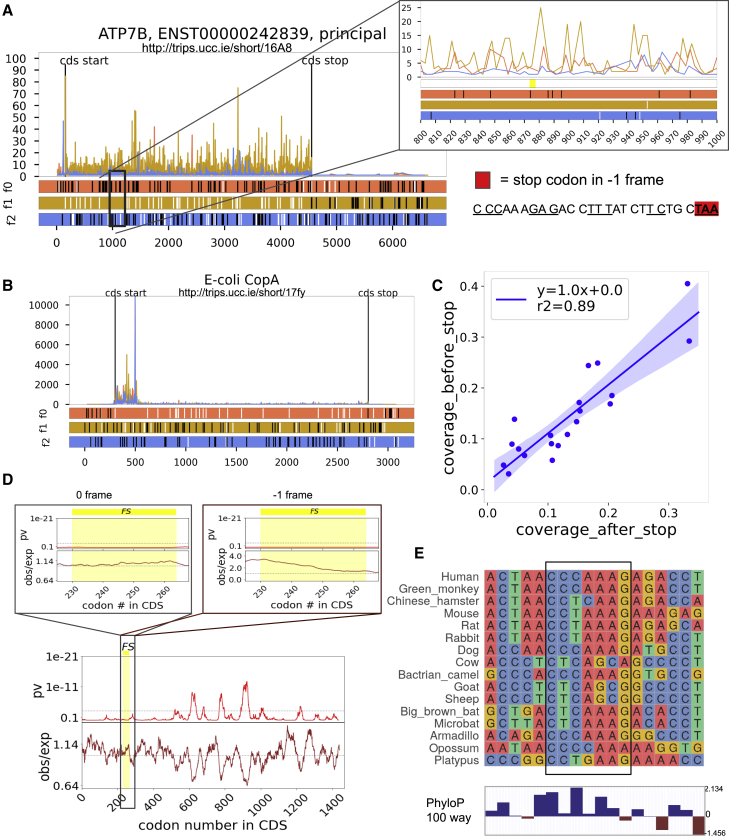
In addition to reexamining the data obtained with dual luciferase reporters, we explored publicly available ribosome profiling data available in Trips-Viz (Kiniry et al., 2019, 2021) for evidence of ATP7B frameshifting. No drop-off in ribosome density is observed downstream of the −1 frame premature stop codon in the aggregated data (Figure 2A), unlike in the case of E. coli copA mRNA (Figure 2B). Examination of the ribosome profiling data from individual datasets does not differ from what is expected in the absence of frameshifting (Figure 2C). The reported frameshifting in ATP7B has been proposed to be stimulated by a predicted RNA pseudoknot. If the sequence of the proposed pseudoknot was evolutionary important, we would expect increased conservation of synonymous positions of ATP7B overlapping with the pseudoknot. However, the analysis of nucleotide substitutions with SynPlot2 (Firth, 2014) does not reveal any reduction in the evolutionary rates of nucleotide substitutions in the corresponding regions (Figure 2D). The sequence of frameshifting pattern is also highly variable across mammals (Figure 2E).
Figure 2.
Analysis of publicly available data
(A) Trips-Viz ribosome profiling data aligned to ATP7B mRNA. The colors are matched to the reading frames in the ORF plot at the bottom; AUGs are shown as white and STOPs as black dashes. The region with frameshifting site and stop codon (yellow bar), and pseudoknot is zoomed.
(B) Same as (A) but the data are aligned to the E. coli copA gene.
(C) Comparison of Ribo-seq read coverage upstream and downstream of the stop codon (TAA) in −1 frame downstream of the proposed frameshift site. Productive frameshifting is expected to result in a drop in ribosome footprint density and the slope of the curve <1, which is not the case (R2 = 0.89, y = 1x + 0).
(D) Synonymous site conservation in the ATP7B coding region for the vertebrates. The 0 frame and −1 frame are zoomed. The brown line is the ratio of the synonymous substitutions within a 25 codons window to the number expected under a null model of neutral evolution at synonymous sites, and the red line showing the corresponding p value. The horizontal gray dashed line indicates a p = 0.05 threshold after an approximate correction for multiple testing. The yellow bar shows frameshifting site, stop codon, and pseudoknot.
(E) Multiple sequence alignment for ATP7B frameshifting site (in frame). At the bottom: 100 vertebrates’ PhyloP score.
Discussion
The −1 ribosomal frameshifting is often utilized in the expression of viral genes or transposable elements but occurs extremely rarely in the expression of host genes where frameshifting origin can often be traced to viruses (Atkins et al., 2016). The only non-retroviral-derived cases of −1 frameshifting reported in human genes are CCR5 (Belew et al., 2014) and ATP7B (Meydan et al., 2017). In previous work, we demonstrated that frameshifting in CCR5 was a misinterpretation of the results obtained with dual luciferase reporters (Khan et al., 2022), and here, we show that this is also the case for ATP7B.
Although there are cases of conserved +1 ribosomal frameshifting in cellular genes unrelated to viruses (Ivanov and Atkins, 2007) and increased frameshifting has been associated with certain pathological conditions (Bartok et al., 2021; Saffert et al., 2016), we deduce that there are currently no known validated non-viral-derived cellular genes in humans that functionally utilize efficient −1 frameshifting for their expression. Knowledge of the extent to which −1 frameshifting is used in human gene expression is of paramount importance for ongoing studies of compounds targeting viral −1 frameshifting (Bhatt et al., 2021; Sun et al., 2021).
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Chemicals, peptides, and recombinant proteins | ||
| Lipofectamine 2000 | Invitrogen | 11668-027 |
| Optimem | Invitrogen | 51985-026 |
| L-glutamine | Sigma | G7513 |
| Pen/Strep | Sigma | P4333 |
| FBS | Sigma | F7524 |
| DMEM | Sigma | D6429 |
| Restriction enzyme XhoI | NEB | R0146L |
| Restriction enzyme BglII | NEB | R0144L |
| Restriction enzyme BamHI-HF | NEB | R3136L |
| Phusion® High-Fidelity DNA Polymerase | NEB | M0530L |
| T4 DNA ligase | NEB | M0202S |
| Passive Lysis Buffer | Promega | E1941 |
| Half-area 96-well white luminometer plate | Fisher | DPS-150-010W |
| Experimental models: Cell lines | ||
| HEK293T | ATCC | CRL-3216 |
| Oligonucleotides | ||
|
ATP7B WT XhoI sense ATAACTCGAG CCCAAAGGACCTTTATCTTCTG |
IDT | NA |
|
ATP7B SS XhoI sense ATAACTCGAG TCCTAAGGACCTTTATCTTCTG |
IDT | NA |
|
ATP7B IFC XhoI sense ATAACTCGAG TCCTAAAGGACCTTTATCTTCTG |
IDT | NA |
|
ATP7B BglII antisense TTATAGATCT CCCGGGGGATCCGTGCATTCC |
IDT | NA |
| SARS CoV2 sense XhoI ATAACTCGA GACCAACTTGTGCTAATGACCC |
IDT | NA |
| SARS CoV2 antisense BamHI TTATGGA TCCATTGTAGATGTCAAAAGCC |
IDT | NA |
| Software and algorithms | ||
| Trips-viz browser | Kiniry et al., 2021 | https://trips.ucc.ie/ |
| Synplot2 | Firth, 2014 | https://github.com/AndrewFirth12/synplot2 |
| Mafft | Katoh et al., 2019 | https://mafft.cbrc.jp/alignment/software/ |
| NCBI Orthologs annotation pipeline | Thibaud-Nissen et al., 2016 | https://www.ncbi.nlm.nih.gov/gene/540/ortholog/?scope=7776&term=ATP7B |
Resource availability
Lead contact
Please direct any requests for further information or reagents to the lead contact, Pavel V. Baranov (p.baranov@ucc.ie).
Materials availability
This study did not generate new unique reagents.
Experimental model and subject details
Cell culture and transfections
HEK293T cells (ATCC) were maintained in DMEM supplemented with 10% FBS, 1 mM L-glutamine and antibiotics. Cells were transfected with Lipofectamine 2000 reagent (Invitrogen), using the 1-day protocol in which suspended cells are added directly to the DNA complexes in half-area 96-well plates. The following were added to each well: 25 ng of each plasmid plus 0.2 μl Lipofectamine 2000 in 25 μl Opti-Mem (Gibco). The transfecting DNA complexes in each well were incubated with 4 × 104 cells suspended in 50 μl DMEM + 10% FBS at 37°C in 5% CO2 for 20 hr.
Method details
Plasmids
ATP7B dual luciferase expression constructs were generated by PCR on HEK293T genomic DNA using primer sequences outlined in key resources table which incorporated 5’ XhoI and 3’ BglII restriction sites. PCR amplicons were digested with XhoI / BglII and cloned into XhoI / BglII digested pDLuc (Fixsen and Howard, 2010) to generate fused luciferases, and PspXI / BglII digested pSGDlucV3.0 (Addgene 119760) to generate unfused luciferases.
Unfused SARS CoV2 dual luciferase constructs were generated previously (Bhatt et al.,2021). Fused SARS CoV2 dual luciferase constructs were generated by PCR on the unfused SARS CoV2 constructs using primer sequences outlined in key resources table which incorporated 5′ XhoI and 3’ BamHI restriction sites. PCR amplicons were digested with XhoI / BamHI and cloned into XhoI / BglII digested pDLuc. All clones were verified by Sanger sequencing (Figure S1).
Dual luciferase assay
Relative light units were measured on a Veritas Microplate Luminometer with two injectors (Turner Biosystems). Transfected cells were lysed in 15 μl of 1 × passive lysis buffer (PLB: Promega) and light emission was measured following injection of 50 μl of either Renilla or firefly luciferase substrate (Dyer et al., 2000).
Frameshifting efficiencies were determined by calculating relative luciferase activities (firefly/Renilla) from test constructs and dividing by relative luciferase activities from replicate wells of matched in-frame-control constructs. Three replicate biological samples were assayed each with four technical repeats.
Quantification and statistical analysis
Analysis of publicly available data
282 orthologous sequences of ATP7B gene in vertebrates were retrieved using NCBI Orthologs annotation pipeline. CDSs were translated and aligned using mafft (Katoh et al., 2019), then back-translated using custom python scripts to create codon alignments; columns with gaps in reference (human) sequence were removed. Synonymous site conservation was assessed using Synplot2 (Firth, 2014).
Processed ribo-seq data were downloaded from Trips-viz (Kiniry et al., 2019, 2021). Ribo-seq read coverage is calculated as the number of mapped reads divided by length of the region. Multiple sequence alignment is performed using mafft (Katoh et al., 2019). Linear regression was calculated for Ribo-seq coverage before stop codon vs after stop using python, package scipy v1.5.3.
Acknowledgments
We wish to acknowledge support from the SFI-HRB-Wellcome Trust Biomedical Research Partnership Investigator Award in Science (210692/Z/18/) to P.V.B., the Irish Research Council Advanced Laureate (IRCLA/2019/74) to J.F.A., and the Science Foundation Ireland (SFI Centre for Research Training in Genomics Data Science) to A.D.F. (18/CRT/6214).
Author contributions
G.L., J.F.A., and P.V.B. conceived and conceptualized the study. G.L. carried out the experimental analysis. P.V.B. and A.D.F. performed the analysis of available ribosome profiling data and the phylogenetic analysis. Y.A.K. provided valuable information regarding dual luciferase assays used in the original study. J.F.A. and P.V.B. provided funding and supervision. G.L. and P.V.B. drafted the manuscript, and all the authors participated in the final version editing.
Declaration of interests
G.L. and P.V.B. are cofounders and shareholders of EIRNA Bio Ltd.
Published: September 16, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.molcel.2017.01.002.
Contributor Information
Gary Loughran, Email: g.loughran@ucc.ie.
Pavel V. Baranov, Email: p.baranov@ucc.ie.
Supplemental information
Data and code availability
-
•
All data reported in this paper will be shared by the lead contact upon request.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- Atkins J.F., Loughran G., Baranov P.V. A [Cu]rious ribosomal profiling pattern leads to the discovery of ribosomal frameshifting in the synthesis of a copper chaperone. Mol. Cell. 2017;65:203–204. doi: 10.1016/j.molcel.2017.01.002. [DOI] [PubMed] [Google Scholar]
- Atkins J.F., Loughran G., Bhatt P.R., Firth A.E., Baranov P.V. Ribosomal frameshifting and transcriptional slippage: from genetic steganography and cryptography to adventitious use. Nucleic Acids Res. 2016;44:7007–7078. doi: 10.1093/nar/gkw530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartok O., Pataskar A., Nagel R., Laos M., Goldfarb E., Hayoun D., Levy R., Körner P.R., Kreuger I.Z.M., Champagne J., et al. Anti-tumour immunity induces aberrant peptide presentation in melanoma. Nature. 2021;590:332–337. doi: 10.1038/s41586-020-03054-1. [DOI] [PubMed] [Google Scholar]
- Belew A.T., Meskauskas A., Musalgaonkar S., Advani V.M., Sulima S.O., Kasprzak W.K., Shapiro B.A., Dinman J.D. Ribosomal frameshifting in the CCR5 mRNA is regulated by miRNAs and the NMD pathway. Nature. 2014;512:265–269. doi: 10.1038/nature13429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhatt P.R., Scaiola A., Loughran G., Leibundgut M., Kratzel A., Meurs R., Dreos R., O'Connor K.M., McMillan A., Bode J.W., et al. Structural basis of ribosomal frameshifting during translation of the SARS-CoV-2 RNA genome. Science. 2021;372:1306–1313. doi: 10.1126/science.abf3546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drees S.L., Klinkert B., Helling S., Beyer D.F., Marcus K., Narberhaus F., Lübben M. One gene, two proteins: coordinated production of a copper chaperone by differential transcript formation and translational frameshifting in Escherichia coli. Mol. Microbiol. 2017;106:635–645. doi: 10.1111/mmi.13841. [DOI] [PubMed] [Google Scholar]
- Dyer B.W., Ferrer F.A., Klinedinst D.K., Rodriguez R. A noncommercial dual luciferase enzyme assay system for reporter gene analysis. Anal. Biochem. 2000;282:158–161. doi: 10.1006/abio.2000.4605. [DOI] [PubMed] [Google Scholar]
- Firth A.E. Mapping overlapping functional elements embedded within the protein-coding regions of RNA viruses. Nucleic Acids Res. 2014;42:12425–12439. doi: 10.1093/nar/gku981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fixsen S.M., Howard M.T. Processive selenocysteine incorporation during synthesis of eukaryotic selenoproteins. J. Mol. Biol. 2010;399:385–396. doi: 10.1016/j.jmb.2010.04.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grentzmann G., Ingram J.A., Kelly P.J., Gesteland R.F., Atkins J.F. A dual-luciferase reporter system for studying recoding signals. RNA. 1998;4:479–486. [PMC free article] [PubMed] [Google Scholar]
- Ivanov I.P., Atkins J.F. Ribosomal frameshifting in decoding antizyme mRNAs from yeast and protists to humans: close to 300 cases reveal remarkable diversity despite underlying conservation. Nucleic Acids Res. 2007;35:1842–1858. doi: 10.1093/nar/gkm035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K., Rozewicki J., Yamada K.D. MAFFT online service: multiple sequence alignment, interactive sequence choice and visualization. Brief. Bioinform. 2019;20:1160–1166. doi: 10.1093/bib/bbx108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan Y.A., Loughran G., Steckelberg A.L., Brown K., Kiniry S.J., Stewart H., Baranov P.V., Kieft J.S., Firth A.E., Atkins J.F. Evaluating ribosomal frameshifting in CCR5 mRNA decoding. Nature. 2022;604:E16–E23. doi: 10.1038/s41586-022-04627-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiniry S.J., Judge C.E., Michel A.M., Baranov P.V. Trips-viz: an environment for the analysis of public and user-generated ribosome profiling data. Nucleic Acids Res. 2021;49:W662–W670. doi: 10.1093/nar/gkab323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiniry S.J., O'Connor P.B.F., Michel A.M., Baranov P.V. Trips-viz: a transcriptome browser for exploring Ribo-Seq data. Nucleic Acids Res. 2019;47:D847–D852. doi: 10.1093/nar/gky842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G.W., Burkhardt D., Gross C., Weissman J.S. Quantifying absolute protein synthesis rates reveals principles underlying allocation of cellular resources. Cell. 2014;157:624–635. doi: 10.1016/j.cell.2014.02.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loughran G., Howard M.T., Firth A.E., Atkins J.F. Avoidance of reporter assay distortions from fused dual reporters. RNA. 2017;23:1285–1289. doi: 10.1261/rna.061051.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meydan S., Klepacki D., Karthikeyan S., Margus T., Thomas P., Jones J.E., Khan Y., Briggs J., Dinman J.D., Vázquez-Laslop N., et al. Programmed ribosomal frameshifting generates a copper transporter and a copper chaperone from the same gene. Mol. Cell. 2017;65:207–219. doi: 10.1016/j.molcel.2016.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saffert P., Adamla F., Schieweck R., Atkins J.F., Ignatova Z. An expanded CAG repeat in huntingtin causes +1 frameshifting. J. Biol. Chem. 2016;291:18505–18513. doi: 10.1074/jbc.M116.744326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y., Abriola L., Niederer R.O., Pedersen S.F., Alfajaro M.M., Silva Monteiro V., Wilen C.B., Ho Y.C., Gilbert W.V., Surovtseva Y.V., et al. Restriction of SARS-CoV-2 replication by targeting programmed -1 ribosomal frameshifting. Proc. Natl. Acad. Sci. USA. 2021;118 doi: 10.1073/pnas.2023051118. e2023051118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thibaud-Nissen F., DiCuccio M., Hlavina W., Kimchi A., Kitts P.A., Murphy T.D., Pruitt K.D., Souvorov A. P8008 the NCBI eukaryotic genome annotation pipeline. J. Anim. Sci. 2016;94:184. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
All data reported in this paper will be shared by the lead contact upon request.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.