Abstract
Developing broad-spectrum, host-directed antiviral therapeutics can be adapted to combat emerging viruses. In this issue of Cell Chemical Biology, Maarifi and colleagues implement a Nano luciferase reporter-based protein complementation assay to screen for small molecules and identify Gilteritinib, which enhances interferon induction and antagonizes virus replication.
Developing broad-spectrum, host-directed antiviral therapeutics can be adapted to combat emerging viruses. In this issue of Cell Chemical Biology, Maarifi and colleagues implement a Nano luciferase reporter-based protein complementation assay to screen for small molecules and identify Gilteritinib, which enhances interferon induction and antagonizes virus replication.
Main text
Drug development, from discovery to clinical application, is a lengthy process that creates a barrier to treat emerging viruses. The limited information known about novel viruses upon their emergence further precludes timely identification and introduction of virus-targeted therapeutics. A potential strategy to eclipse the delay in the development of virus-specific antivirals is the identification of host-directed therapeutics. Targeting the host’s immune responses could be an effective strategy to treat infection with various viruses as well as reduce the likelihood of escape mutant evolving. The type I interferon (IFN-I) response is a logical target. IFN-I induction and signaling pathways are innate signaling cascades critical for mounting an effective antiviral response. In this issue of Cell Chemical Biology, Maarifi et al. (2022) identify Gilteritinib as a broad-spectrum antiviral that facilitates IFN-I induction.
In response to virus infection, pathogen-associated molecules stimulate the activation of the IFN-I pathway. Viral nucleic acids commonly act as agonists for host pathogen-recognition receptors (PRRs), including the cytoplasmic retinoic acid-inducible gene I (RIG-I)-like receptors (RLRs) and endosomal Toll-like receptors (TLRs) (McNab et al., 2015). After the PRRs bind to their viral agonist, a signaling cascade is triggered. Adaptor molecules transduce the signal facilitating the assembly of complexes that promote TANK-binding kinase 1 (TBK1) and IκB kinase ε (IKKε)-mediated phosphorylation and activation of the IFN regulator factor (IRF)3 and/or IRF7 transcription factors (Figure 1 ). Phosphorylated IRF3 and/or IRF7 translocate to the nucleus and bind to the positive regulatory domain I and III regions of IFN-stimulated regulator elements in the promoter of IFN-I genes to induce their expression (Honda et al., 2006). For optimal IFN-β expression, other transcription factors, including nuclear factor-κB and AP-1 (a heterodimer comprised of heterodimer of activating transcription factor 2 and c-Jun) are recruited to PRDs II and IV, respectively. In addition, depending on the cell type and the viral agonist, other IRFs (e.g., IRF-1, IRF-5) can be activated to induce optimal IFN-I levels. The assembly of these transcription factors at the IFN-I gene promoters enhances their expression through the recruitment of histone acetyl transferases including CREB binding protein (CBP). Once synthesized, IFN-I binds to the IFN-I receptor (IFNAR) to activate an antiviral transcriptional repertoire comprised of IFN-stimulated genes (ISGs). ISGs are the effector proteins that restrict virus growth at various stages of their replication cycles. IFN-I signaling also induces the expression of negative feedback regulators such as suppressors of cytokine signaling (SOCS), which can antagonize both IFN-I induction and signaling (Yoshimura et al., 2005) to minimize IFN-I-associated damage to the host. An important consideration when developing host-directed therapeutics targeting the IFN-I pathways is how to specifically activate the antiviral factors without coincidentally enhancing the expression of pro-inflammatory factors.
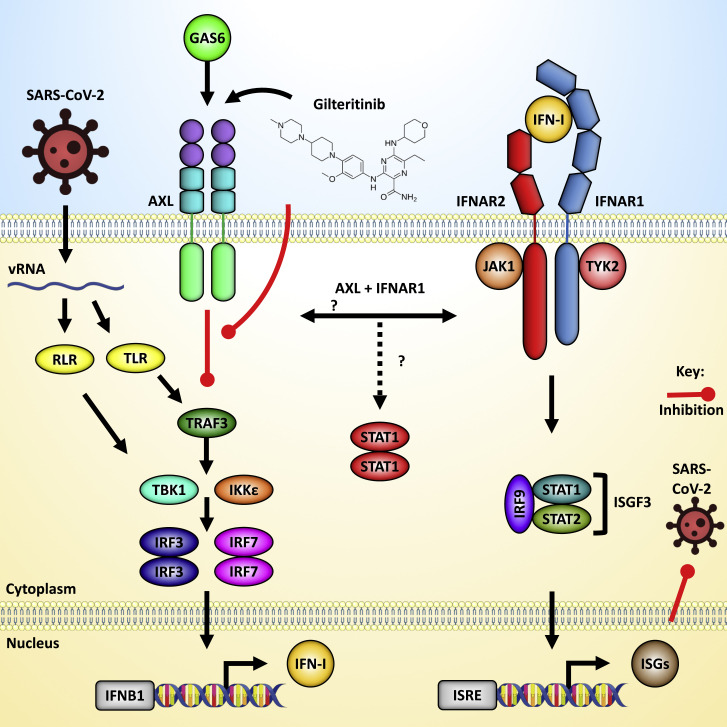
Figure 1.
The innate antiviral type I IFN response and interactions with the AXL receptor
Infection with SARS-CoV-2, and other RNA viruses, results in viral RNA (vRNA) in the cytoplasm or endosomes that triggers the activation of RLRs and TLRs, respectively. Downstream of vRNA-RLR interactions, RLRs interact with MAVS at mitochondrial membranes to facilitate the assembly of a signalosome where TBK1 and IKKε are recruited. TLR signaling activates TNF receptor-associated factor (TRAF)3 for downstream activation of TBK1/IKKε. These kinases then phosphorylate IRFs 3 and 7, which translocate to the nuclear to induce the expression of IFN-I. IFN-I signals through the IFNAR1 to simulate the phosphorylation of STAT1 and STAT2, resulting in the formation of ISGF3, which translocates to the nucleus to bind ISRE-containing promoters and facilitate the expression of ISGs. Gas6 is AXL’s natural ligand and activates AXL signaling that negatively regulates the IFN-I pathway. AXL is able to interact with IFNAR1, and this interaction results in the preferential formation of STAT1 homodimers. STAT1 activation then leads to the induction of SOCS 1 and 3, which inhibit TRAF3 and JAK1, resulting in reduced activation of IFN-I induction and signaling pathways. Gilteritinib is a small molecule that inhibits AXL’s tyrosine kinase activity allowing for enhanced IFN-I induction following SARS-CoV-2 infection.
Maarifi et al. (2022) apply a reporter protein complementation strategy to screen for kinase inhibitors that induce IRF-induced gene expression to identify host-directed antivirals. Specifically, they adapted their luminance-based Alpha Centauri assay, first developed to identify inhibitors of the human immunodeficiency virus-1 (HIV-1) integrase (Fernandez et al., 2021), to monitor IRF nuclear translocation and localization to gene promoters. They fused the 13 amino acid α component of Nano luciferase to IRFs and the complementary fragment, Cen, to CBP, so that the enzymatically active luciferase would assemble when the IRF and CBP co-localize. The luminance resulting from the Nano luciferase-mediated cleavage of furimazine is the readout. The use of CBP as the complementary protein, versus a nuclear localization signal, increases the specificity of the assay beyond IRF nuclear localization and toward the formation of an active transcriptional complex. To validate this method, they over-expressed RIG-I-2CARD, a constitutively active form of RIG-I that contains only the caspase recruitment domains, to stimulate IFN-I induction and monitor the recruitment of the different IRFs to their promoter. Treatment with MRT67307 to block IRF kinases TBK1 and IKKε prevents IRF3 phosphorylation and nuclear translocation, and they observed diminished luminance signal following RIG-I-2CARD expression. The authors also investigated which IRFs are induced following infection with a variety of viruses. They found that different groups of IRFs engage with CBP at their promoters following virus infections. For example, they found that infection with Sendai virus activates IRFs 1, 3, and 7, Influenza A virus induces IRFs 3 and 7, but severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) does not induce translocation of any IRFs. The authors hypothesized that virus-encoded innate immune antagonists, such as the papain-like protease (PLP), prevent the activation of IRFs. In support of this, treatment with PLP inhibitor GRL-0617 during SARS-CoV-2 infection allowed for the activation of IRFs 3, 5, and 7.
Because the strongest luminance signals detected during SARS-CoV-2 infection resulted from IRF7-α-CBP-Cen interaction, this experimental set-up was used to screen a panel of 21 kinase inhibitors for immunomodulatory activity. Using this assay, the authors identified the AXL and fms-like tyrosine kinase 3 (FLT3) inhibitor Gilteritinib (Dhillon, 2019), which enhances IRF7-CRB interaction in response to infection with SARS-CoV-2, stimulates expression of IFNα4, IFNβ, and CXCL10, and inhibits SARS-CoV-2 replication. Importantly, they found that Gilteritinib specifically stimulated the antiviral arm without altering expression of pro-inflammatory IFN-γ, IL-6, or TNF-α. In the absence of virus infection, Gilteritinib had no immunostimulatory effect, implying that the molecule does not directly activate IRF7. Treatment with an IFNAR antibody that blocks IFN-I signaling prevented immunostimulation by Gilteritinib and partially reversed its antiviral effects during SARS-CoV-2 infection. Using gene knockdown experiments, they showed that Gilteritinib’s activity was mainly mediated through blocking AXL but not its other target FLT3.
The partial retention of antiviral activity in the absence of IFN-I signaling and following AXL knockdown suggests that Gilteritinib may also have an IFN-I signaling- and AXL-independent mechanism for the inhibition of SARS-CoV-2. AXL is a member of the TAM (Tyro3, Axl, and Mer) receptor tyrosine kinase (RTK) family. This group of RTKs typically functions to facilitate the phagocytosis of apoptotic cells and/or membranes with detectable phosphatidylserine (PS) (Lemke, 2013). In order to recognize apoptotic cells, growth arrest-specific protein 6 (Gas6) binds to PS and the extracellular immunoglobulin domains of TAMs, mainly AXL, to trigger their activation (Figure 1). Several enveloped viruses exploit TAMs to facilitate virus entry when PS is incorporated into their membrane. Further work is needed to determine whether SARS-CoV-2 can similarly hijack AXL and/or the other TAMs to facilitate entry.
In addition to a potential role for facilitating virus entry, blocking AXL’s enzymatic activity could bolster IFN-I signaling. Investigation in dendritic cells has demonstrated that AXL negatively regulates both IFN-I and pro-inflammatory cytokine production (Rothlin et al., 2007; Sharif et al., 2006). AXL signaling facilitates the expression of transcriptional repressors Twist 1 and 2 following IFN-I signaling to antagonize the expression TNFα, IL-8, and COX2 (Sharif et al., 2006). Although Maarifi et al. did not elucidate the precise molecular mechanism by which Gilteritinib promotes the IFN-mediated antiviral response, it could involve crosstalk between AXL and the IFN receptor (Figure 1). Studies have shown that AXL and IFNAR1 interact following IFN-I signaling (Rothlin et al., 2007). The AXL-IFNAR crosstalk results in the formation of STAT1 homodimers, which stimulates gene expression of SOCS1 and SOCS3 that negatively regulate TLRs, RLRs, and IFNAR through blocking critical adaptor molecules such as TRAFs 3 and 6 (activated by TLR agonists) and JAK1 (activated downstream the IFN-I receptor) (Rothlin et al., 2007). Because of AXL’s key role as a negative regulator of pro-inflammatory pathways and in the maintenance of tissue homeostasis through the clearance of apoptotic cells to prevent autoinflammatory diseases (Lemke, 2013), it will be important to evaluate the antiviral therapeutic potential for Gilteritinib in phagocytic cells and characterize the role of AXL in regulating innate antiviral immune responses in multiple cell types.
The Alpha Centauri approach taken by Maarifi and colleagues could potentially be applied beyond therapeutics screening. An important finding of their study was the distinct IRF activation profiles following infection with different virus species. A potential permutation of their approach could be fusing the components of a split enzyme proximity labeling system (Qin et al., 2021) to identify transcriptional regulators recruited to IFN-I gene promoters in response to unique viruses. This type of technology could be applied to emerging viruses to aid in understanding which innate immune pathways are viable targets for host-directed therapeutics. Although this approach does present some limitations, especially considering that it needs over-expression or transduction of the Alpha Centauri components, the system could be adapted to elucidate how IRFs and other transcription factors differentially regulate cytokine production in a cell type specific manner. Overall, Alpha Centauri could provide an accelerated approach for the identification of host-targeted antivirals and at the same time providing a tool to study mechanisms of action.
Acknowledgments
The Rajsbaum lab is supported by NIH/NIAID grants R01 AI166668, R01 AI134907, R01 AI155466, and P01 AI150585. S.v.T was supported by grants T32-AI060549 and 1F31- AI15242201A1, and A.H. was supported by T32-AI007526.
Declaration of interests
The authors declare no competing interests.
References
- Dhillon S. Gilteritinib: first Global Approval. Drugs. 2019;79:331–339. doi: 10.1007/s40265-019-1062-3. [DOI] [PubMed] [Google Scholar]
- Fernandez J., Hassen-Khodja C., Georget V., Rose T., Jacob Y., Janin Y.L., Nisole S., Vidalain P.O., Arhel N.J. Measuring the subcellular compartmentalization of viral infections by protein complementation assay. Proceedings of the National Academy of Sciences of the United States of America. 2021;118 doi: 10.1073/pnas.2010524118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honda K., Takaoka A., Taniguchi T. Type I Inteferon gene induction by the interferon regulatory factor Family of transcription factors. Immunity. 2006;25:349–360. doi: 10.1016/j.immuni.2006.08.009. [DOI] [PubMed] [Google Scholar]
- Lemke G. Biology of the TAM receptors. Cold Spring Harbor Perspect. Biol. 2013;5:a009076. doi: 10.1101/cshperspect.a009076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maarfi G., Martin M.F., Zebboudj A., Boulay A., Nouaux P., Fernandez J., Lagisquet J., Garcin D., Gaudin R., Arhel N.J., et al. Identifying enhancers of innate immune signaling as broad-spectrum antivirals active against emerging viruses. Cell Chemical Biology. 2022 doi: 10.1016/j.chembiol.2022.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNab F., Mayer-Barber K., Sher A., Wack A., O'Garra A. Type I interferons in infectious disease. Nat. Rev. Immunol. 2015;15:87–103. doi: 10.1038/nri3787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin W., Cho K.F., Cavanagh P.E., Ting A.Y. Deciphering molecular interactions by proximity labeling. Nat. Methods. 2021;18:133–143. doi: 10.1038/s41592-020-01010-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothlin C.V., Ghosh S., Zuniga E.I., Oldstone M.B., Lemke G. TAM receptors are pleiotropic inhibitors of the innate immune response. Cell. 2007;131:1124–1136. doi: 10.1016/j.cell.2007.10.034. [DOI] [PubMed] [Google Scholar]
- Sharif M.N., Sosic D., Rothlin C.V., Kelly E., Lemke G., Olson E.N., Ivashkiv L.B. Twist mediates suppression of inflammation by type I IFNs and Axl. J. Exp. Med. 2006;203:1891–1901. doi: 10.1084/jem.20051725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimura A., Nishinakamura H., Matsumura Y., Hanada T. Negative regulation of cytokine signaling and immune responses by SOCS proteins. Arthritis Res. Ther. 2005;7:100. doi: 10.1186/ar1741. [DOI] [PMC free article] [PubMed] [Google Scholar]