Abstract
Rhodococcus ruber (formerly Gordonia terrae) IFP 2001 is one of a few bacterial strains able to degrade ethyl tert-butyl ether (ETBE), which is a major pollutant from gasoline. This strain was found to undergo a spontaneous 14.3-kbp chromosomal deletion, which results in the loss of the ability to degrade ETBE. Sequence analysis of the region corresponding to the deletion revealed the presence of a gene cluster, ethABCD, encoding a ferredoxin reductase, a cytochrome P-450, a ferredoxin, and a 10-kDa protein of unknown function, respectively. The EthB and EthD proteins could be easily detected by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and were induced by ETBE in the wild-type strain. Upstream of ethABCD lies ethR, which codes for a putative positive transcriptional regulator of the AraC/XylS family. Transformation of the ETBE-negative mutant by a plasmid carrying the ethRABCD genes restored the ability to degrade ETBE. Complementation was abolished if the plasmid carried ethRABC only. The eth genes are located in a DNA fragment flanked by two identical direct repeats of 5.6 kbp. The ETBE-negative mutants carry a single copy of this 5.6-kbp repeat, suggesting that the 14.3-kbp chromosomal deletion resulted from a recombination between the two identical sequences. The 5.6-kbp repeat is a class II transposon carrying a TnpA transposase, a truncated form of the recombinase TnpR, and a terminal inverted repeat of 38 bp. The truncated TnpR is encoded by an IS3-interrupted tnpR gene.
Methyl tert-butyl ether (MTBE) and ethyl tert-butyl ether (ETBE) are used as additives in unleaded gasoline. These ethers enhance the octane number of gasoline and are thought to improve combustion efficiency, thereby reducing emissions of unburned hydrocarbons to the atmosphere. Typically, up to 15% (vol/vol) MTBE can be used in oxygenated gasoline, making MTBE one of the main organic chemicals produced in the United States (33). ETBE is used in some European countries, and its interest resides in its potential to increase the market for ethanol, as ETBE is manufactured from ethanol and isobutene. ETBE has additional advantages over MTBE, in terms of lower vapor pressure and higher octane number (M. Iborra, J. F. Izquierdo, J. Tejero, and F. Cunill, Chemtech, p. 120–122, February 1988).
The widespread use of ethers in gasoline has resulted in their introduction from leaky tanks and spills into groundwater, exposing the general public to low levels of ethers from drinking water (4). Compared with other compounds in gasoline, ethers are relatively nontoxic. However, their unpleasant taste and odor at very low concentrations render water unfit for drinking, making these xenobiotic compounds important pollutants.
To develop bioremediation of these compounds, studies on the biodegradability of MTBE and ETBE have been undertaken (31). Bacteria capable of using MTBE as the sole carbon and energy source have been isolated (15, 27). To date, the enzymatic mechanism used by these bacteria to degrade MTBE has not been elucidated. Several microorganisms which cannot use MTBE as the sole carbon and energy source can degrade MTBE during or after growth on an inducer substrate. Using pentane as the source of carbon and energy, Pseudomonas aeruginosa was shown to degrade MTBE (13). The filamentous fungus Graphium sp. and Pseudomonas putida degrade MTBE after growth on n-butane and camphor, respectively (16, 37). Propane-oxidizing bacteria, including Mycobacterium vaccae JOB5, were shown to degrade MTBE or ETBE after growth on propane (37). MTBE and ETBE were oxidized to tert-butyl alcohol (TBA), which was further oxidized to products not effective for growth of the propane oxidizers. Oxidation of both MTBE and TBA involves a soluble cytochrome P-450, which most likely corresponds to the propane monooxygenase. Comamonas testoteroni E1 and the gram-positive bacterial strain E2 have been recently isolated as ether fuel degraders (21). Metyrapone inhibition and spectrophotometric analysis strongly suggested that degradation of ETBE by both strains involves a cytochrome P-450.
The actinomycetes Rhodococcus ruber (formerly Gordonia terrae) IFP 2001 and Rhodococcus zopfii (formerly Rhodococcus equi) IFP 2002 are the first isolated bacteria capable of using ETBE as the sole source of carbon and energy (10). Both strains convert stoichiometrically ETBE into TBA, which accumulates in the culture medium. R. ruber is unable to use MTBE or tert-amyl methyl ether (TAME) as the sole carbon and energy source but can degrade MTBE and TAME to TBA and tert-amyl alcohol, respectively, after growth on ETBE (17). One mole of oxygen is consumed per mole of ETBE degraded, which suggests that scission of the ether bond proceeds through hydroxylation by a monooxygenase, yielding a hemiacetal intermediate which spontaneously dismutates into TBA and acetaldehyde. The most likely monooxygenase candidate is an inducible cytochrome P-450, which was detected as a peak at 447 nm in the carbon monoxide difference spectrum of reduced crude extracts of R. ruber grown on ETBE (17).
In order to identify the genes involved in the degradation of ETBE, we characterized spontaneous mutants of R. ruber unable to use ETBE as the sole source of carbon and energy. Loss of the ability to degrade ETBE was shown to result from a chromosomal deletion secondary to a recombination between direct repeats. The deletion led to the removal of a putative operon encoding a cytochrome P-450 system whose expression was induced by ETBE. Complementation of the mutant using ethRABCD genes was successful, demonstrating the involvement of the Eth cytochrome P-450 system in the degradation of ETBE.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
Strain IFP 2001 (10), formerly identified as G. terrae, has been subjected to 16S RNA analysis. Since 100% identity was observed with the type strain of R. ruber (GenBank X80625) (unpublished data), G. terrae IFP 2001 has been renamed R. ruber IFP 2001. R. ruber was grown at 30°C in Luria-Bertani (LB) medium (2) or in minimal medium MM1 which contained 50 mM KH2PO4, 50 mM K2HPO4, 0.16 mM MgSO4, 1.9 mM Na2HPO4, 28 mM NH4Cl, 0.27 mM CaCl2, 4.4 μM FeCl3, 200 μg of biotin per liter, 50 μg of riboflavin per liter, 50 μg of nicotinic acid per liter, 50 μg of calcium pantothenate per liter, 50 μg of p-aminobenzoic acid per liter, 20 μg of folic acid per liter, 15 μg of thiamine hydrochloride per liter, and 1.5 μg of cyanocobalamin per liter, with 10 mM ETBE (Aldrich Chemical Co.) or 0.5% (vol/vol) ethanol as the sole carbon source. For cultivation on solid medium, ETBE was supplied in the gas phase in sealed glass petri dishes containing MM1 medium with 1.5% (wt/vol) agar. Escherichia coli TG1 (14) was grown at 37°C in LB medium. E. coli cells were transformed by electroporation using the conventional procedure (2), with selection on LB agar plates containing either ticarcillin (100 μg/ml) or kanamycin (20 μg/ml).
Transformation of R. ruber.
R. ruber was grown in LB medium to a turbidity (optical density at 600 nm [OD600]) of 1 to 2. Cells were washed twice in cold water and once in cold 10% (vol/vol) glycerol. Cells were resuspended in cold 10% glycerol (1/1,000 volume of the culture) and kept at −80°C. Before use, cells were washed and resuspended in cold 10% glycerol at an OD600 between 40 and 80. Electrocompetent cells (100 μl) were mixed with 1 μg of plasmid in a 0.1-cm gap cuvette. The electroporation conditions were 2.5 kV, 25 μF, and 800 Ω. Electroporated cells were recovered in 1 ml of SOC medium (2) and incubated for 4 h at 30°C with shaking. Using the E. coli/R. equi shuttle vector pRE-7 (39), transformants of R. ruber were selected on LB agar plates containing kanamycin (100 μg/ml) and were obtained with an efficiency of 2 to 10 per μg. The presence of plasmids pRE-7, pGT222, and pGT223 in R. ruber transformants was verified by recovering these plasmids from E. coli TG1 transformed by an alkaline-sodium dodecyl sulfate (SDS) lysate of R. ruber.
Isolation of spontaneous mutants unable to degrade ETBE.
R. ruber was plated on MM1 agar with ETBE vapor as the carbon source, and independent clones were transferred to liquid LB medium. After growth to saturation, clones were diluted into fresh LB medium and the procedure was repeated for 60 generations. Cultures were then plated on LB plates, and individual colonies were patched on LB plates and ETBE-containing MM1 plates, including wild-type controls. After 8 to 10 days, clones showing markedly reduced growth on ETBE plates were selected. TBA production was assayed in cell-free culture supernatants using a Peri-2000 gas chromatograph (Perichrom) fitted with a 3-m-long free fatty acid phase column (Perichrom).
Pulsed-field gel electrophoresis.
R. ruber was grown in 40 ml of LB medium to an OD600 of ∼1. Plugs were prepared as described previously (30) and were digested with 3 U of XbaI per ml. After digestion, plugs were loaded in a 1% (wt/vol) agarose gel. Pulsed-field gel electrophoresis was performed in a contour-clamped homogeneous electric field apparatus (Bio-Rad, Munich, Germany) in which the electrode distribution was such that the reorientation angle of DNA molecules was 120°C. Large restriction fragments were separated at 14°C with a pulse ramp of 1.6 to 21.3 s for 23 h.
Chromosomal DNA extraction.
An R. ruber culture (400 ml) at an OD600 of ∼1.3 was harvested for 15 min at 5,000 × g. Cells were resuspended in 15 ml of 0.1 M Tris-HCl (pH 8)–0.1 M EDTA–0.15 M NaCl supplemented with 150 μl of Triton X-100 and 100 mg of lysozyme and incubated overnight at 37°C with agitation. The lysate was further incubated for 1 h at 60°C in the presence of 1.3 mg of RNase A per ml, followed by treatment with 0.6 mg of proteinase K per ml and 2% (wt/vol) SDS at 40°C for 2 h. Chromosomal DNA was extracted with phenol and chloroform. Ethanol-precipitated DNA (about 800 μg) was recovered by spooling on the tip of a Pasteur pipette.
Construction of genomic libraries and colony screening.
Chromosomal DNA was digested with BamHI and fragments of the appropriate size were cloned into pUC18. Colonies of transformed E. coli were transferred to nylon filters and lysed by incubation on an absorbent filter paper soaked in 2× SSC (0.3 M NaCl, 0.03 M sodium citrate)–5% (wt/vol) SDS for 10 min. DNA fixation and denaturation were carried out by exposing the dried filters to 650-W microwaves for 2 min. Filters were washed in 5× SSC–0.1% SDS at 65°C for 30 min. Lysate left on the surface of the filters was scratched up with a gloved finger. Membranes were rinsed in 2× SSC and hybridized at 65°C in Rapid-Hyb buffer (Amersham Pharmacia Biotech). Probes were obtained from purified DNA fragments labeled with [32P]dCTP using a random prime labeling system (Rediprime II; Amersham Pharmacia Biotech). Unspecific hybridizations were removed by washing the membranes twice in 1× SSC–0.1% SDS at 65°C for 30 min.
DNA sequencing.
The pGT200 and pGT220 plasmids, containing 7.4- and 16.3-kbp inserts, respectively, were fragmented by nebulization, and gel-purified fragments in the range of 1 to 2 kbp were cloned in the pcDNA2.1 vector using the nonpalindromic cloning method as described before (11). The inserts of randomly chosen clones were sequenced from both ends using a Perkin-Elmer ABI 3700 automated sequencer. The sequences were assembled using the Phred, Phrap, and Consed software tools (8, 9). A product overlapping the BamHI site between the two contiguous sequences was amplified by PCR. The complete sequence was obtained as a single contiguous sequence of 23,696 bases.
Plasmid construction.
The 4,923-bp NheI/SacI fragment from pGT220 carrying ethRABCD was subcloned between XbaI and SacI sites of pRE-7 (39), yielding the pGT222 plasmid. A deletion derivative of ethD was constructed by first amplifying a 307-bp fragment through PCR using oligonucleotides SacI (5′-TTGGAGCTCGCTCGTGGTGAA-3′) and StyI-2 (5′-CGACCGGCCAAGGTGTGCGCGACGATGGGAAACATGCTGCACC-3′). This fragment, containing the putative transcription terminator of the eth genes, was cloned into pCR2.1-TOPO (TOPO TA cloning kit; Invitrogen). The nucleotide sequence of the 307-bp insert was verified. In a second step, the 2,956-bp StyI/SphI fragment of pGT222, carrying ethABC, and the 7,190-bp SphI/SacI fragment of pGT222, corresponding to the pRE-7 vector and the ethR gene, were purified. StyI is located 8 nucleotides upstream of the open reading frame (ORF) of ethD. Finally, the 297-bp SacI/StyI fragment, carrying the eth terminator, the 2,956-bp StyI/SphI fragment, carrying ethABC, and the 7,190-bp SphI/SacI fragment of pGT222 were ligated together to give the pGT223 plasmid.
Crude extract preparation and analysis.
R. ruber cells in exponential growth phase were harvested for 15 min at 5,000 × g. Pellets were resuspended in 50 mM Tris (pH 7.5), and cells were disrupted three times through a prechilled French pressure cell at 200 MPa (SLM-Aminco). Cell debris were removed by centrifugation at 27,000 × g for 15 min. Total proteins of the supernatant were assayed with the Coomassie blue reagent (Bio-Rad) and analyzed by denaturing 10-to-15% (wt/vol) polyacrylamide gradient gel electrophoresis.
Peptide sequencing of two ETBE-induced proteins.
Crude extracts of ETBE-induced R. ruber were centrifuged for 1 h at 100,000 × g, and the supernatant was subjected to SDS-polyacrylamide gel electrophoresis. A major ETBE-induced band of 43 kDa was cut from a Tris-glycine gel containing 7.5% polyacrylamide (23), and a minor ETBE-induced band of 10 kDa was cut from a Tris-Tricine gel containing 20% polyacrylamide (35). The bands were subsequently digested with trypsin, and peptides were separated by DEAE-C18 reverse-phase chromatography using an acetonitrile gradient in the presence of 0.1% (vol/vol) trifluoroacetic acid. Selected peptides were sequenced by the Edman method, using a model 473A sequencer (Applied Biosystems).
ETBE-degrading activity of resting cells of R. ruber.
R. ruber was grown in MM1 medium containing either 18 mM ETBE or 18 mM ETBE plus 0.5% (wt/vol) glucose. Transformants carrying pRE-7, pGT222, or pGT223 were grown in the presence of 100 μg of kanamycin per ml. Cells in late exponential phase were centrifuged and were washed once in 50 mM Tris-HCl, pH 7.5. The pellet was recovered in 50 mM Tris-HCl (pH 7.5) to reach an OD600 of approximately 100. The cell suspension (0.4 ml) and 90 mM ETBE (0.1 ml) were incubated at 30°C. Samples (100 μl) were mixed with 5 μl of 10% (vol/vol) phosphoric acid to stop the reaction at different times. Cells were pelleted and the TBA production was measured in the supernatant by gas chromatography analysis.
Nucleotide sequence accession number.
The nucleotide sequences presented here have been assigned accession no. AF333761 by GenBank.
RESULTS
Identification of R. ruber proteins induced in the presence of ETBE.
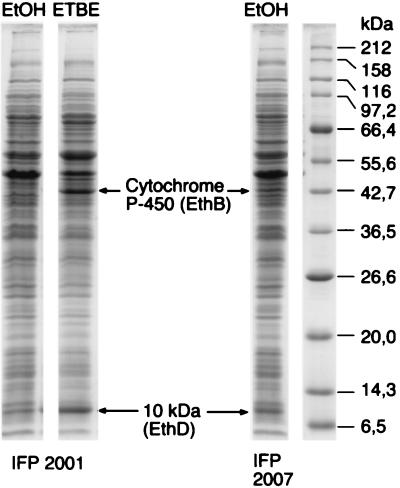
Figure 1 shows an SDS-polyacrylamide gel analysis of crude extracts prepared from R. ruber cells grown on ethanol and on ETBE. Two polypeptides of 43 and 10 kDa are clearly induced in the wild-type strain upon growth on ETBE. They are also present, although less abundant, in cells of a previously isolated mutant, IFP 2007, which constitutively produces ETBE-degrading activity (17). Peptide microsequencing yielded the partial sequences HALGDWQTFSSAQGI, FDSVAQWFTR, and SVSNTEMIALWTELG for the 43-kDa protein and GQPTDTEAFDTYYS for the 10-kDa protein. The first sequence, HALGDWQTFSSAQGI, was 66% identical to a putative cytochrome P-450 from Mycobacterium tuberculosis H37Rv (Genpept Z177137_5), suggesting that the 43-kDa polypeptide may be the inducible cytochrome P-450 observed in ETBE-grown R. ruber cells by Hernandez-Perez et al. (17). The GQPTDTEAFDTYYDS sequence was 47% identical to the orf4 product from Rhodococcus erythropolis (Genpept U17130_4). The R. erythropolis orf4 gene is part of a cytochrome P-450 gene cluster, suggesting that the inducible 10-kDa polypeptide is related to a cytochrome P-450 system. Neither of the two other sequences showed significant similarity with any characterized proteins in the databases.
FIG. 1.
SDS–10-to-15% polyacrylamide gradient gel electrophoresis. R. ruber crude extracts of the wild type (IFP 2001) and the constitutive mutant (IFP 2007) were analyzed after growth in the presence of ethanol (EtOH) or ETBE as the sole source of carbon. The migration of molecular size markers is indicated on the right.
Isolation of independent ETBE-negative mutants.
In an attempt to verify the stability of the ETBE-positive phenotype, five independent clones of R. ruber were cultivated in LB broth for 60 generations. Then, cultures were screened for the presence of mutants unable to grow in the presence of ETBE as the sole source of energy and carbon. Of the clones tested, 20 to 100% were unable to degrade ETBE. Five independent mutants, derived from the five original wild-type clones, were further characterized. When grown to saturation in minimal medium containing 0.5% glucose and 18 mM ETBE, none of the mutants converted more than 0.3 mM ETBE into TBA, whereas under the same conditions, 10.6 mM TBA was produced by the wild-type strain. The reversion to the ETBE+ phenotype was not detectable (no positive colony out of at least 3 × 107 viable cells plated), suggesting the occurrence of an irreversible genetic rearrangement. Wild-type and mutant strains were compared after growth in the presence of 0.5% glucose plus 18 mM ETBE. Mutant resting cells grown in the presence of 0.5% glucose plus 18 mM ETBE displayed less than 1% of the ETBE-, MTBE-, and TAME-degrading activities observed with wild-type cells grown under the same conditions. SDS-polyacrylamide gel analysis of crude extracts showed that, in contrast to the wild type, none of the mutants produced the induced 43- and 10-kDa proteins (results not shown).
Evidence for a 15-kbp chromosomal deletion in ETBE-negative mutants.
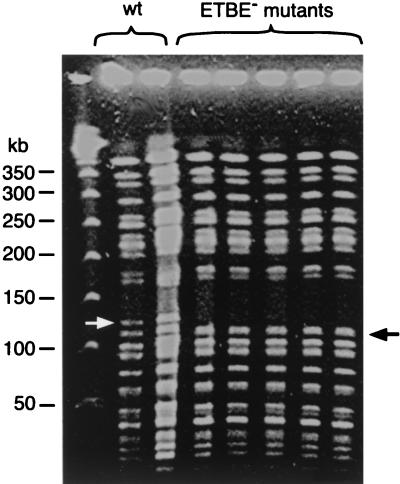
XbaI-genomic digests of wild-type and mutant strains were analyzed by pulsed-field gel electrophoresis (Fig. 2). A 125-kbp fragment was present in the wild-type strain and was absent in the ETBE-negative mutants. In addition, a 110-kbp fragment was observed in the ETBE-negative mutants only. Southern blot hybridization revealed that the wild-type 125-kbp fragment used as a probe hybridized with the mutant 110-kbp fragment (data not shown), showing that the 110-kbp fragment was a deletion-containing form of the 125-kbp fragment. This result indicated that ETBE-negative mutants resulted from an ∼15-kbp chromosomal deletion. Since all independent mutants showed the same genotype, a single mutant, termed IFP 2006, was used for further investigation.
FIG. 2.
Pulsed-field gel electrophoresis of XbaI-digested chromosomal DNA from the wild type (IFP 2001) and ETBE-negative mutants of R. ruber. The arrows designate the 125-kb band of the wild-type (wt) strain and the 110-kb band of the mutants. The migration of the 50-kb concatemers of lambda DNA (Biolabs) is indicated on the left.
Cloning of the wild-type DNA region corresponding to the deletion.
The wild-type XbaI-fragment of 125 kbp was purified from a pulsed-field gel electrophoresis gel and was used as a probe in Southern blot analysis. Hybridization of the 125-kb XbaI probe with BamHI genomic digests showed that a 7.4-kbp band and a 16.3-kbp band, present in the wild-type strain, disappeared in the ETBE-negative mutant. Conversely, a new 9.3-kbp band, which was absent in the wild-type strain, was detected in the ETBE-negative mutant (data not shown). This demonstrated that the 15-kbp deletion identified by pulsed-field gel electrophoresis involved the two BamHI fragments of 7.4 and 16.3 kbp, which were reshuffled into a new BamHI fragment of 9.3 kbp. In order to determine the sequence of the region corresponding to the deletion, the two wild-type BamHI-fragments of 7.4 and 16.3 kbp were cloned. The 7.4-kbp BamHI fragment was selected by colony hybridization using the 125-kbp XbaI fragment as a probe. The cloned 7.4-kbp fragment was then used as a probe in a Southern blot hybridization. In addition to self hybridization, the 7.4-kb BamHI probe also hybridized with the 16.3-kbp BamHI fragment of the wild type and with the 9.3-kbp BamHI fragment of the ETBE-negative mutant. This indicated that the wild-type 7.4-kbp fragment carried a sequence that was also present in these two fragments. Thus, the wild-type 16.3-kbp BamHI fragment and the mutant 9.3-kbp BamHI fragment were cloned by colony hybridization using the 7.4-kb BamHI fragment as a probe.
ethABCD code for a cytochrome P-450 system
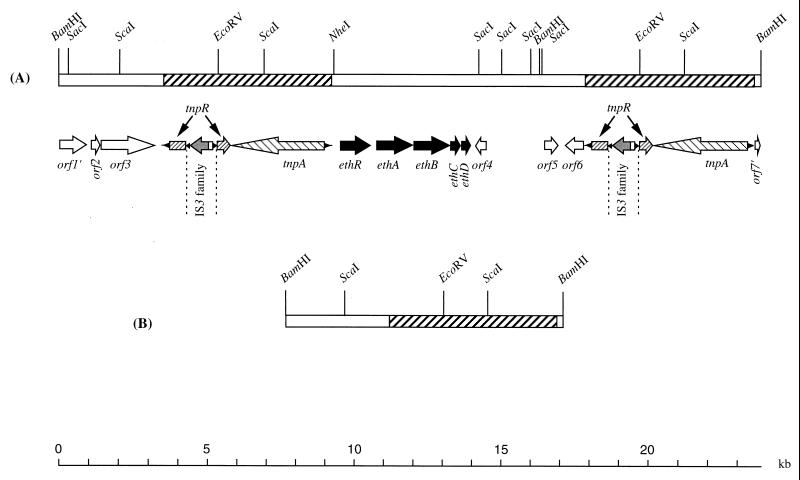
The features of the 23.7-kbp region covered by the two wild-type BamHI fragments are shown in Fig. 3. A cluster of four ORFs with the same orientation and named ethABCD was identified. Based on protein alignments, ethABC could be assigned to individual components of a P-450-containing monooxygenase system.
FIG. 3.
Wild-type genetic organization of the 23.7-kbp region carrying the genes involved in ETBE degradation (A) and restriction map of the 9.3-kbp BamHI fragment of the ETBE-negative mutant (B). The eth genes encode a putative transcriptional activator (ethR), a ferredoxin reductase (ethA), the ETBE-inducible cytochrome P-450 (ethB), a ferredoxin (ethC), and an ETBE-inducible unknown protein (ethD). Based on protein alignments, the other ORFs shown correspond to a resolvase (tnpR), transposase (tnpA), a two-component system response regulator (truncated orf1′), integral membrane protein (orf2), sodium:solute symporter (orf3), membrane protein (truncated orf7′), and three unidentified proteins (orf4, orf5, and orf6).
EthA (412 amino acids) is similar to glutathione reductase-like ferredoxin reductases. It contains amino acids typical of two ADP-binding βαβ folds which encompass the completely conserved consensus motif GXGXXG (38). The N-terminal ADP-binding site (Val-1 to Asp-31) may constitute the FAD-binding site, and the centrally located ADP-binding site (Arg-144 to Asp-172) may constitute the NAD+-binding site. The ferredoxin reductase ThcD of R. erythropolis (Genpept U17130_6) is the closest relative to EthA (47% identity).
EthB (400 amino acids) corresponds to the ETBE-induced protein of 43 kDa, since it contains the three peptides that were sequenced. In addition, EthB is similar to cytochromes P-450, which suggests that EthB is the ETBE-induced cytochrome P-450 (17) likely to catalyze the oxidation of ETBE. EthB carries a cysteine residue at position 349, which is strictly conserved in all cytochromes P-450. This residue is part of the consensus FGXGXHXCXG and possibly provides anchoring of the heme in the active site of the cytochrome P-450. The highest similarity score of EthB (33% identity) was found with a putative cytochrome P-450 from the phenanthrene-degrading actinomycete Nocardioides sp. (20). Among the characterized cytochromes P-450, EthB shows highest similarity (25% identity) to the Pseudomonas sp. cytochrome P-450terp, which hydroxylates the monoterpene α-terpineol as a step in its catabolic assimilation (29). Two actinomycetal cytochromes P-450 also show 25% identity to EthB: NikQ from the antibiotic nikkomycin-producing Streptomyces tendae (24) and the orf3 product from the mitomycin C-producing Streptomyces lavendulae (25).
EthC (106 amino acids) is similar to putidaredoxin-type [2Fe-2S] ferredoxins and probably serves as an electron carrier between the NADH-dependent ferredoxin reductase (EthA) and the cytochrome P-450 (EthB). The four cysteine residues located at positions 40, 46, 49, and 76 of EthC correspond to the perfectly conserved residues that are required for coordinating the prosthetic group. The greatest extent of similarity (48% identity) was found with the ferredoxin ThcC of R. erythropolis (Genpept U17130_5).
EthD (103 amino acids) corresponds to the ETBE-induced protein of 10 kDa, since it carries the sequenced peptide. EthD is similar to products of four ORFs of unknown function: orfY product from Pseudonocardia sp. (Genpept AJ296087_1), an ORF from Bacillus halodurans (Genpept AP001507_200), an ORF from Mesorhizobium loti (Genpept AP002998_321), and orf4 product from R. erythropolis (Genpept U17130_4), which are 40, 34, 31, and 40% identical to EthD, respectively. R. erythropolis orf4 belongs to the thc gene cluster, which encodes a cytochrome P-450 system catalyzing the N-dealkylation of thiocarbamates (28).
ethR encodes a transcriptional regulator of the AraC/XylS family.
The ethR gene lies 183 bp upstream of ethA (Fig. 3). EthR (331 amino acids) is highly similar to positive transcriptional regulators of the AraC/XylS family. The highly conserved C-terminal domain encompassing the bipartite DNA-binding domain (12, 22, 34) is located between amino acids 250 and 325 of EthR. The most closely related member is R. erythropolis ThcR (Genpept U17130_2), which is 31% identical to EthR.
Transposon repeats flanking eth genes.
Two directly identical sequences of 5.6 kbp flank the eth genes (Fig. 3). The first repeat ends 880 bp upstream of ethR, and the second repeat starts 3,908 bp downstream of ethD. Three potential coding regions (orf4, orf5, and orf6) were identified in the 3,908-bp region using the heuristic approach of the GeneMark program (3). Amino acid comparison of products of orf4, orf5, and orf6 using the Blast program (1) did not show any significant similarity with the bacterial Genpept database.
The 5.6-kbp repeat consists of a class II transposon containing a terminal inverted repeat of 38 bp, a tnpA gene, encoding a putative transposase, and an insertion sequence (IS)-interrupted tnpR gene. Discounting the entire IS sequence, the intact tnpR gene may encode a putative resolvase of 311 amino acids. TnpA (1,008 amino acids) and TnpR (311 amino acids) show very high amino acid similarity to TnpA and the orf5 product, respectively, of the Streptomyces fradiae Tn4556 transposon (36). The TnpA transposase of S. fradiae is the closest relative of R. ruber TnpA, with 49% identity. The orf5 product of S. fradiae Tn4556 is a potential resolvase of Tn4556 whose similarity with the R. ruber TnpR extends into the upstream region of this ORF, disregarding a TAG stop codon as mentioned by De Mot et al. (6). The deduced polypeptide of 324 residues is 31% identical to TnpR of R. ruber. The closest relative of R. ruber TnpR is R. erythropolis PmrA (62% identity), which is a site-specific recombinase of the integrase family and may be involved in stabilization of the cryptic plasmid pFAJ2600 (6). Amino acid comparisons revealed that other proteins related to R. ruber TnpR are almost exclusively site-specific recombinases of the integrase family. This suggests that, unlike most resolvases of class II transposons, TnpR belongs to the integrase family and not to the resolvase-invertase family of site-specific recombinases.
The region coding for TnpR is interrupted by an insertion of 1,409 bp at codon 180, introducing a stop codon at position 181. This 1,409-bp insertion displays all structural characteristics of mobile elements of the IS3 family. Imperfect 45-bp inverted repeats flank a single ORF with a translational frameshift. The predicted protein is 420 amino acids long and shows extended similarity to several transposases of the IS3 family. The most closely related is the transposase of Mycobacterium avium IS999 (Genpept AF232829_2), which is 40% identical to the IS3-type transposase of R. ruber. The region coding for the R. ruber transposase of 420 amino acids overlaps two ORFs in phase 0 and −1 encoding, respectively, the N-terminal (108 amino acids) and the C-terminal (312 amino acids) regions of the potential transposase. As in other members of IS3 family, the translational frameshift may be a means of producing several proteins using the same coding region (5).
Genetic rearrangement generating ETBE-negative mutants.
To elucidate the molecular mechanism responsible for the 14.3-kbp deletion, we cloned the 9.3-kbp BamHI fragment which is specific for ETBE-negative mutants. The genetic organization of the 9.3-kbp BamHI fragment was determined by sequencing each end of the fragment (68 and 452 nucleotides, respectively) and by restriction analysis (Fig. 3). The 9.3-kbp BamHI fragment corresponds to the wild-type 23.7 kbp region with one copy of the 5.6-kbp transposon and the intergenic region between the two copies of the transposon deleted. This deletion encompasses the eth gene cluster, which is involved in ETBE degradation. Thus, the genetic organizations of the wild type and ETBE-negative mutants suggest that spontaneous loss of the ability to degrade ETBE results from a homologous recombination between the two identical direct repeats of the 5.6-kbp transposon.
Complementation of the ETBE-negative mutant of R. ruber.
The NheI/SacI fragment carrying ethRABCD (Fig. 3A) was subcloned into the pRE-7 vector, yielding the pGT222 plasmid. The ETBE-negative mutant IFP 2006(pGT222) had the same doubling time as the wild-type strain IFP 2001 on a minimal medium containing ETBE as the sole source of carbon and energy (Table 1). In addition, strains IFP 2001 and IFP 2006(pGT222) produced similar amounts of TBA using resting cells incubated in the presence of ETBE. Together, these results demonstrate that the ethRABCD genes are sufficient to complement the ETBE-negative mutant of R. ruber. In contrast, complementation was almost totally abolished in the absence of the ethD gene: plasmid pGT223, which carries the ethRABC genes only, failed to restore the ability to grow on a minimal medium containing ETBE as the sole source of carbon and energy (Table 1). Production of TBA by resting cells of R. ruber IFP 2006(pGT223) grown on minimal medium containing glucose and ETBE was detectable but severely decreased compared to that of wild-type cells and mutant cells complemented by pGT222. Mutant cells harboring the control plasmid pRE-7 showed no detectable activity.
TABLE 1.
Complementation of the ETBE-negative mutant IFP 2006 by the pGT222 and pGT223 plasmids carrying the ethRABCD genes and the ethRABC genes, respectively
| R. ruber strain | Doubling time on minimal medium + ETBE (h) | ETBE-degrading activity (pmol min−1
OD600−1) of resting cells grown on:
|
|
|---|---|---|---|
| ETBE | Glucose + ETBE | ||
| IFP 2001 | 10 | 1,930 | 372 |
| IFP 2006(pRE-7) | ≥150 | NDa | <1 |
| IFP 2006(pGT222) | 10 | 1,341 | 140 |
| IFP 2006(pGT223) | ≥150 | ND | 12 |
ND, not done.
DISCUSSION
R. ruber easily loses the ability to degrade ETBE. This loss coincides with the deletion of a chromosomal DNA segment carrying ethRABCD. The ethABC genes encode a ferredoxin reductase, a cytochrome P-450, and a ferredoxin, respectively, which are typical of bacterial cytochrome P-450 systems. The deletion mutant also lost the ability to cleave MTBE and TAME, indicating that the same system also accounts for the degradation of these compounds. Complementation with pGT222 demonstrated that the ethRABCD genes were sufficient to restore the ability to degrade ETBE. Further evidence that the eth cluster is involved in ether fuel degradation was provided by demonstrating that the synthesis of the proteins EthB and EthD was induced by ETBE.
ETBE, MTBE, and TAME are xenobiotics, which raises the issue of the origin of the enzyme systems that degrade them. Although the primary substrate of the Eth system is unknown at this stage, its original function may be related to ether metabolism, since ethB and ethD are induced by ether fuels. This contrasts with the cytochromes P-450 presumably involved in the degradation of MTBE by the fungus Graphium, M. vaccae JOB5, and P. putida (16, 37). In these cases, the system is induced by n-butane, propane, and camphor, respectively. Furthermore, although cytochrome P-450 systems often show a broad specificity, cleavage of ether fuels is not a universal feature of cytochrome P-450 monooxygenases. As reported by Steffan et al. (37), Rhodococcus rhodochrous, which produces two P-450 monooxygenases, does not oxidize MTBE.
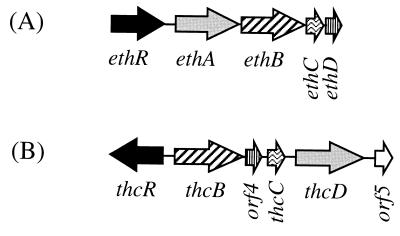
The Eth cytochrome P-450 system of R. ruber displays remarkable similarity, in terms of genetic organization and primary structure of the individual components, to the Thc system of R. erythropolis. The Thc system was proposed to catalyze the N-dealkylation of the thiocarbamate herbicide S-ethyl dipropylthiocarbamate (EPTC) to propionaldehyde and N-dipropyl EPTC (28), which represents a reaction similar to the O-dealkylation of ETBE by the Eth system. Each eth gene has a homologue in the thc cluster (Fig. 4). The electron-supplying system shows the highest degree of conservation (47 and 48% amino acid identities). Most strikingly, both the R. ruber eth and R. erythropolis thc cytochrome P-450 clusters contain a gene (ethD and orf4, respectively) encoding a similar 10-kDa protein. Such proteins have not been detected in the numerous other cytochrome P-450 systems studied so far, and their exact function is unknown. However, EthD clearly participates in the degradation of ETBE, since it was induced by ETBE and since the deleted mutant was poorly complemented by pGT223, which carries ethRABC but not ethD. The similarity between the R. ruber eth and R. erythropolis thc clusters extends to the putative regulatory gene. In both bacteria, the first gene of the cluster codes for a putative positive transcriptional regulator of the AraC/XylS family (12). The amino acid similarity between EthR and ThcR is remarkable, since ThcR is the only member of the AraC/XylS family which shows significant similarity with EthR outside the conserved C-terminal domain of the family.
FIG. 4.
Genetic organization of R. ruber (A) and
R. erythropolis (B) (28) cytochrome P-450
systems. Transcriptional activators (▪), cytochromes P-450 (▨),
ferredoxin reductases (░⃞), ferredoxins
( ) and unknown proteins
(▥) are 31, 24, 47, 48, and 40% identical, respectively.
) and unknown proteins
(▥) are 31, 24, 47, 48, and 40% identical, respectively.
Two identical copies of a class II transposon were found to flank the ethRABCD genes. This may result from the formation of a cointegrate that could not be resolved, since the resolvase gene tnpR was inactivated by the IS3 insertion. No additional copies of the 5.6-kbp transposon were found in the R. ruber genome, as shown by Southern blot hybridization using a transposon-containing fragment as a probe (data not shown).
The eth gene cluster can be lost by spontaneous chromosomal deletion of a specific 14.3-kbp fragment. This deletion most probably occurs by homologous recombination between the two identical direct repeats of the 5.6-kbp transposon. Similar events were previously reported for the loss of the ability to utilize other sources of carbon such as toluene in P. putida (26, 32), citrate in E. coli (18, 19), and isopropylbenzene in P. putida (7). In the process of the 14.3-kbp deletion by homologous recombination, a circular element carrying the eth gene cluster is believed to be generated in R. ruber cells. Since this element would possess a transposase gene, it could potentially be integrated into other replicons and, therefore, be involved in the horizontal transfer of eth genes.
The identification of R. ruber eth genes provides new insights into the biodegradation of gasoline ethers, offering new opportunities for effective bioremediation strategies for these recalcitrant pollutants. Probes may be developed to monitor survival and spreading of strains harboring eth genes and to assess the biodegradation potential of contaminated soils. Preliminary experiments suggest that highly similar cytochrome P-450 systems account for the degradation of ETBE by other actinomycetes (unpublished data). Finally, cloning of eth genes offers the opportunity to generate new strains able to degrade ETBE.
ACKNOWLEDGMENTS
We thank M. Schwartz for its continuing interest and support. We are indebted to C. Rusniok and P. Glaser for the nucleotidic sequencing facilities, to J. F. Prescott for the gift of plasmid pRE-7, and to A. Varnerot for 16S RNA analysis. We thank G. Guglielmi, F. Monot, H. Bedouelle, and J.-P. Vandecasteele for helpful discussion. We are grateful to J. d'Alayer for sequencing peptides from ETBE-induced proteins.
C. Le Dantec is the recipient of a fellowship from Novotech (Lyonnaise des Eaux).
REFERENCES
- 1.Altschul S F, Madden T L, Schaffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Gapped blast and psi-blast: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K. Current protocols in molecular biology. New York, N.Y: Greene Publishing and Wiley Interscience; 1990. [Google Scholar]
- 3.Besemer J, Borodovsky M. Heuristic approach to deriving models for gene finding. Nucleic Acids Res. 1999;27:3911–3920. doi: 10.1093/nar/27.19.3911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brown S L. Atmospheric and potable water exposures to methyl tert-butyl ether (MTBE) Regul Toxicol Pharmacol. 1997;25:256–276. doi: 10.1006/rtph.1997.1104. [DOI] [PubMed] [Google Scholar]
- 5.Chandler M, Fayet O. Translational frameshifting in the control of transposition in bacteria. Mol Microbiol. 1993;7:497–503. doi: 10.1111/j.1365-2958.1993.tb01140.x. [DOI] [PubMed] [Google Scholar]
- 6.De Mot R, Nagy I, De Schrijver A, Pattanapipitpaisal P, Schoofs G, Vanderleyden J. Structural analysis of the 6 kb cryptic plasmid pFAJ2600 from Rhodococcus erythropolis NI86/21 and construction of Escherichia coli-Rhodococcusshuttle vectors. Microbiology. 1997;143:3137–3147. doi: 10.1099/00221287-143-10-3137. [DOI] [PubMed] [Google Scholar]
- 7.Eaton R W, Timmis K N. Spontaneous deletion of a 20-kilobase DNA segment carrying genes specifying isopropylbenzene metabolism in Pseudomonas putidaRE204. J Bacteriol. 1986;168:428–430. doi: 10.1128/jb.168.1.428-430.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ewing B, Green P. Base-calling of automated sequencer traces using phred. II. Error probabilities. Genome Res. 1998;8:186–194. [PubMed] [Google Scholar]
- 9.Ewing B, Hillier L, Wendl M C, Green P. Base-calling of automated sequencer traces using phred. I. Accuracy assessment. Genome Res. 1998;8:175–185. doi: 10.1101/gr.8.3.175. [DOI] [PubMed] [Google Scholar]
- 10.Fayolle F, Hernandez G, Le Roux F, Vandecasteele J-P. Isolation of two aerobic bacterial strains that degrade efficiently ethyl t-butyl ether (ETBE) Biotechnol Lett. 1998;20:283–286. [Google Scholar]
- 11.Frangeul L, Nelson K E, Buchrieser C, Danchin A, Glaser P, Kunst F. Cloning and assembly strategies in microbial genome projects. Microbiology. 1999;145:2625–2634. doi: 10.1099/00221287-145-10-2625. [DOI] [PubMed] [Google Scholar]
- 12.Gallegos M-T, R. S, Bairoch A, Hofmann K, Ramos J L. AraC/XylS family of transcriptional regulators. Microbiol Mol Biol Rev. 1997;61:393–410. doi: 10.1128/mmbr.61.4.393-410.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Garnier P M, Auria R, Augur C, Revah S. Cometabolic biodegradation of methyl t-butyl ether by Pseudomonas aeruginosagrown on pentane. Appl Microbiol Biotechnol. 1999;51:498–503. doi: 10.1007/s002530051423. [DOI] [PubMed] [Google Scholar]
- 14.Gibson T J. Studies on the Epstein-Barr virus genome. Ph.D. dissertation. Cambridge, United Kingdom: University of Cambridge; 1984. [Google Scholar]
- 15.Hanson J R, Ackerman C E, Scow K M. Biodegradation of methyl tert-butyl ether by a bacterial pure culture. Appl Environ Microbiol. 1999;65:4788–4792. doi: 10.1128/aem.65.11.4788-4792.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hardison L K, Curry S S, Ciuffetti L M, Hyman M R. Metabolism of diethyl ether and cometabolism of methyl tert-butyl ether by a filamentous fungus, a Graphiumsp. Appl Environ Microbiol. 1997;63:3059–3067. doi: 10.1128/aem.63.8.3059-3067.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hernandez-Perez G, Fayolle F, Vandecasteele J-P. Biodegradation of ethyl t-butyl ether (ETBE), methyl t-butyl ether (MTBE) and t-amyl methyl ether (TAME) by Gordonia terrae. Appl Microbiol Biotechnol. 2001;55:117–121. doi: 10.1007/s002530000482. [DOI] [PubMed] [Google Scholar]
- 18.Ishiguro N, Sato G. Nucleotide sequence of insertion sequence IS3411, which flanks the citrate utilization determinant of transposon Tn3411. J Bacteriol. 1988;170:1902–1906. doi: 10.1128/jb.170.4.1902-1906.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ishiguro N, Sato G. Spontaneous deletion of citrate-utilizing ability promoted by insertion sequences. J Bacteriol. 1984;160:642–650. doi: 10.1128/jb.160.2.642-650.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Iwabuchi T, Harayama S. Biochemical and genetic characterization of 2-carboxybenzaldehyde dehydrogenase, an enzyme involved in phenanthrene degradation by Nocardioidessp. strain KP7. J Bacteriol. 1997;179:6488–6494. doi: 10.1128/jb.179.20.6488-6494.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kharoune M, Kharoune L, Lebeault L M, Pauss A. Isolation and characterization of two aerobic bacterial strains that completely degrade ethyl tert-butyl ether (ETBE) Appl Microbiol Biotechnol. 2001;55:348–353. doi: 10.1007/s002530000528. [DOI] [PubMed] [Google Scholar]
- 22.Kwon H J, Bennik M H J, Demple B, Ellenberger T. Crystal structure of the Escherichia coliRob transcription factor in complex with DNA. Nat Struct Biol. 2000;7:424–430. doi: 10.1038/75213. [DOI] [PubMed] [Google Scholar]
- 23.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 24.Lauer B, Russwurm R, Bormann C. Molecular characterization of two genes from Streptomyces tendaeTue901 required for the formation of the 4-formyl-4-imidazolin-2-one containing nucleoside moiety of the peptidyl nucleoside antibiotic nikkomycin. Eur J Biochem. 2000;267:1698–1706. doi: 10.1046/j.1432-1327.2000.01162.x. [DOI] [PubMed] [Google Scholar]
- 25.Mao Y, Varoglu M, Sherman D H. Molecular characterization and analysis of the biosynthetic gene cluster for the antitumor antibiotic mitomycin C from Streptomyces lavendulaeNRRL 2564. Chem Biol. 1999;6:251–263. doi: 10.1016/S1074-5521(99)80040-4. [DOI] [PubMed] [Google Scholar]
- 26.Meulien P, Downing R G, Broda P. Excision of the 40kb segment of the TOL plasmid from Pseudomonas putidamt-2 involves direct repeats. Mol Gen Genet. 1981;184:97–101. doi: 10.1007/BF00271202. [DOI] [PubMed] [Google Scholar]
- 27.Mo K, Lora C O, Wanken A E, Javanmardian M, Yang X, Kulpa C F. Biodegradation of methyl t-butyl ether by pure bacterial cultures. Appl Microbiol Biotechnol. 1997;47:69–72. doi: 10.1007/s002530050890. [DOI] [PubMed] [Google Scholar]
- 28.Nagy I, Schoofs G, Compernolle F, Proost P, Vanderleyden J, De Mot R. Degradation of the thiocarbamate herbicide EPTC (S-ethyl dipropylcarbamothioate) and biosafing by Rhodococcussp. strain NI86/21 involve an inducible cytochrome P-450 system and aldehyde dehydrogenase. J Bacteriol. 1995;177:676–687. doi: 10.1128/jb.177.3.676-687.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Peterson J A, Lu J-Y, Geisselsoder J, Graham-Lorence S, Carmona C, Witney F, Lorence M C. Cytochrome P-450terp. Isolation and purification of the protein and cloning and sequencing of its operon. J Biol Chem. 1992;267:14193–14203. [PubMed] [Google Scholar]
- 30.Picardeau M, Varnerot A, Rauzier J, Gicquel B, Vincent V. Mycobacterium xenopi IS1395, a novel insertion sequence expanding the IS256family. Microbiology. 1996;142:2453–2461. doi: 10.1099/00221287-142-9-2453. [DOI] [PubMed] [Google Scholar]
- 31.Prince R C. Biodegradation of methyl tertiary-butyl ether (MTBE) and other fuel oxygenates. Crit Rev Microbiol. 2000;26:163–178. doi: 10.1080/10408410008984175. [DOI] [PubMed] [Google Scholar]
- 32.Reddy B R, Shaw L E, Sayers J R, Williams P A. Two identical copies of IS1246, a 1275 base pair sequence related to other bacterial insertion sequences, enclose the xylgenes on TOL plasmid pWW0. Microbiology. 1994;140:2305–2307. doi: 10.1099/13500872-140-9-2305. [DOI] [PubMed] [Google Scholar]
- 33.Reisch M S. Top 50 chemicals production rose modestly last year. Chem Eng News. 1994;72:12–16. [Google Scholar]
- 34.Rhee S, Martin R G, Rosner J L, Davies D R. A novel DNA-binding motif in MarA: the first structure for an AraC family transcriptional activator. Proc Natl Acad Sci USA. 1998;95:10413–10418. doi: 10.1073/pnas.95.18.10413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schagger H, von Jagow G. Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal Biochem. 1987;166:368–379. doi: 10.1016/0003-2697(87)90587-2. [DOI] [PubMed] [Google Scholar]
- 36.Seimieniak D R, Slightom J L, Chung S-T. Nucleotide sequence of Streptomyces fradiae transposable element Tn4556: a class-II transposon related to Tn3. Gene. 1990;86:1–9. doi: 10.1016/0378-1119(90)90107-3. [DOI] [PubMed] [Google Scholar]
- 37.Steffan R J, McKlay K, Vainberg S, Condee C W, Zhang D. Biodegradation of the gasoline oxygenates methyl tert-butyl ether, ethyl tert-butyl ether, and tert-amyl ether by propane-oxidizing bacteria. Appl Environ Microbiol. 1997;63:4216–4222. doi: 10.1128/aem.63.11.4216-4222.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Weirenga R K, Terpstra P, Hol W G J. Prediction of the occurrence of the ADP-binding βαβ-fold in proteins, using an amino-acid sequence fingerprint. J Mol Biol. 1986;187:101–107. doi: 10.1016/0022-2836(86)90409-2. [DOI] [PubMed] [Google Scholar]
- 39.Zheng H, Tkachuk-Saad O, Prescott J F. Development of a Rhodococcus equi-Escherichia coliplasmid shuttle vector. Plasmid. 1997;38:180–187. doi: 10.1006/plas.1997.1311. [DOI] [PubMed] [Google Scholar]