Abstract
Sleep occurs universally and is a biological necessity for human functioning. The consequences of diminished sleep quality impact physical and physiological systems such as neurological, cardiovascular, and metabolic processes. In fact, people impacted by common complex diseases experience a wide range of sleep disturbances. It is challenging to uncover the underlying molecular mechanisms responsible for decreased sleep quality in many disease systems owing to the lack of suitable sleep biomarkers. However, the discovery of a genetic component to sleep patterns has opened a new opportunity to examine and understand the involvement of sleep in many disease states. It is now possible to use major genomic resources and technologies to uncover genetic contributions to many common diseases. Large scale prospective studies such as the genome wide association studies (GWAS) have successfully revealed many robust genetic signals associated with sleep-related traits. With the discovery of these genetic variants, a major objective of the community has been to investigate whether sleep-related traits are associated with disease pathogenesis and other health complications. Mendelian Randomization (MR) represents an analytical method that leverages genetic loci as proxy indicators to establish causal effect between sleep traits and disease outcomes. Given such variants are randomly inherited at birth, confounding bias is eliminated with MR analysis, thus demonstrating evidence of causal relationships that can be used for drug development and to prioritize clinical trials. In this review, we outline the results of MR analyses performed to date on sleep traits in relation to a multitude of common complex diseases.
Keywords: Mendelian Randomization, GWAS, Sleep disorders, Insomnia, Sleep duration, Narcolepsy, Obstructive Sleep apnea (OSA), Restless leg syndrome (RLS), Neurodegenerative disorders, Cardiovascular disorders, obesity, cancer
Introduction
Sleep is one of the most fundamental biological processes, especially as it is so evolutionarily conserved. It is known to occur in some form in all animals ranging from jelly fish to humans [1]. It is well established that good sleep quality is essential for proper cognitive, as well as physical, performance. Architecture, continuity, timing, regularity, and satisfaction all determine the quality of sleep. Each of these measurable dimensions represents different sleep phenotypes and can be categorized as short or long sleep duration, chronotype or morningness, insomnia, hypersomnia, sleep efficiency, sleep apnea, etc [2]..
The molecular mechanisms of sleep are far from being fully understood, but clues are provided by disturbances and abnormalities in sleep that are routinely witnessed in various neurological, metabolic, and cardiovascular disorders. Principally due to a lack of reliable biomarkers, the sleep field has not been well explored. Electroencephalogram (EEG) analysis has traditionally been the only way to determine sleep abnormalities but has proven cumbersome. However, recent advances in technology involving motion tracking via video and breathing rate monitoring have enabled screening for causal factors, making it easier to explore mechanisms involved in sleep regulation. Furthermore, several molecular factors, including transcription factors, ion channels, and kinases, have been shown to modulate sleep. In addition, genetic alterations related to these factors are known to drive specific sleep abnormalities [3].
Lack of good quality sleep is associated with several diseases. Whether the lack of sleep causes health comorbidities or impaired physiological systems lead to sleep disturbances is still under investigation. Identification of genetic variants associated with various health outcomes, including sleep traits, through large scale genome wide association studies (GWAS) has greatly assisted in this investigation. The last 15 years have seen major progress in genetic discovery, largely due to the contribution of GWAS in many health settings including sleep. Sleep GWAS has enabled the detection of numerous genetic variants associated with several diseases and health traits. In turn, Mendelian Randomization (MR) analyses utilize these genetic variants identified by sleep GWAS to determine causality in the context of various health outcomes.
This review will first highlight the role of sleep GWAS in identifying critical genetic variants associated with sleep traits, and the subsequent importance of MR studies in utilizing these variants to assess causality between sleep phenotypes and health outcomes. Next, we will bring together and illustrate the many causal relations identified by MR analyses between sleep traits and various health/disease outcomes that can inform clinical or public health decision making. These findings can also empower clinical trials and drug development for disease outcomes.
GWAS
The primary goal of epidemiological studies is to identify the root cause of disease. Several such studies have identified robust associations between modifiable exposures (behavioral, pharmacological, or physiological) and disease risk. Several large-scale open-access prospective epidemiologic studies have leveraged biobanks from the United Kingdom [4], China [5], and the HUNT study [6] that have collected extensive phenotypic and genotypic information from its participants through questionnaires, physical measures, sample assays, etc., for a wide range of health-related outcomes.
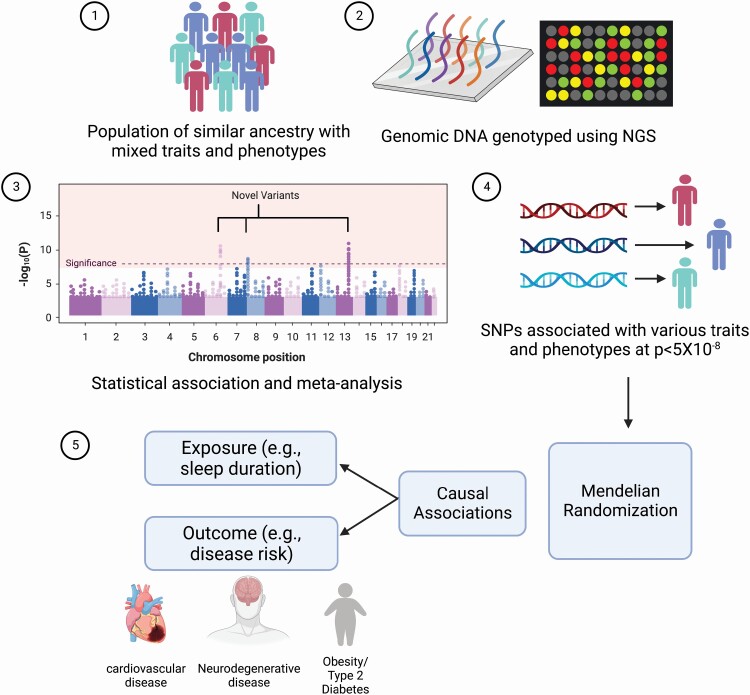
GWAS take advantage of these biobanks and other prospective studies conducted in large cohorts of individuals, recruit their participants to identify the association between a phenotype and genotype. GWAS typically scans the genome of the participants to identify genetic variants and the frequency of their occurrence between individuals who share similar ancestry but are phenotypically different [7, 8] (Figure 1). By doing so, GWAS has successfully identified many associations between genetic variation (principally single nucleotide polymorphisms [SNPs]) and a given phenotype of interest. GWAS signals are considered significantly associated with the phenotype of interest when the observed p-value is less than 5 × 10−8 [9]. Utilizing the data from all sources, GWAS has identified in excess of 300 000 SNP-trait associations to date.
Figure 1.
Overview of identification of causal associations using the Mendelian Randomization approach. 1. Population of similar ancestry but mixed phenotypes are considered for genotyping. 2. All participants are genotyped using different techniques to identify genomic regions likely to be associated with a given phenotype. 3. Statistical association tests and meta-analyses are used to identify novel causal genetic variants (single nucleotide polymorphisms – SNPs). 4. SNPs that pass the rigid thresholds for each trait are considered statistically significant and are utilized as genetic instruments for subsequent analyses. 5. By virtue of their random allocation, such genetic variants are utilized by Mendelian Randomization analyses to identify causal associations between any modifiable exposure (e.g. sleep duration) and outcome (risk for common complex disease).
In the context of sleep, GWAS efforts have typically investigated genetic variants associated with definitions of chronotype, sleep duration, and sleep efficiency, along with sleep disorders such as restless leg syndrome (RLS), narcolepsy, and insomnia. These traits and disorders were either self-reported or objectively estimated through devices such as accelerometers. Some of the notable GWAS efforts in the field of sleep are discussed below and summarized in Table 1.
Table 1.
Summary of variants identified by notable GWAS on various sleep traits
| Sleep trait | Sample Cohort | No. of cases/no. of controls | Significant variant (SNP) | Associated gene | Odds ratio | P | Reference |
|---|---|---|---|---|---|---|---|
| Restless leg syndrome | Icelandic American | 429/16 866 188/662 |
rs3923809 | BTBD9 | 1.7* 1.5 |
3 × 10−14† 0.004 |
[10] |
| European | 400/1600 | rs3923809 rs2300478 rs6494696 |
BTBD9
MEIS1 MAP2K5 and LBXCOR1 |
0.58 1.74* 1.52* |
0.024† 8.08 × 10−23† 4.74 × 10−11† |
[11] | |
| Mixed European | 15 126/95 725 | rs113851554 | MEIS1 | 1.92* | 2 × 10−280† | [12] | |
| Caucasian | 10 257/470 725 | rs10068599 rs112716420 rs10769894 |
RANBP17
MICALL2 11p15.4 |
1.09 1.25 0.9 |
6.9 × 10−10 1.5 × 10−18 9.4 × 10−14, |
[13] | |
| Asian | 264/1383 (with migraine) 1053 (normal with no migraine or RLS) |
rs79823654 rs6021854 |
CCDC141
VSTM2L |
2.1* 1,84* |
5.81 × 10–6† 4.63 × 10–7† |
[14] | |
| Periodic limb movements (PLMS) | American | 6843 | rs113851554 rs9369062 |
MEIS1
BTBD9 |
N/A N/A |
3.51 × 10−12 3.06 × 10−22 |
[15] |
| Narcolepsy | Japanese Korean European African American |
222/389 159/190 115/309 388/397 86/98 |
rs5770917 | CPT1B or CHKB | 1.74 1.97 1.4 1.33 1.86 |
1.4 × 10−4 5.2 × 10−4 0.03 0.12 0.14 |
[16] |
| Mixed European Caucasian Asian (Japanese + Koreans) African American |
807/1074 363/355 433/433 + 128/172 133/144 |
rs1154155 | TCRA | 1.87 1.8 1.54 1.31 |
1.9 × 10−13† 3.67 × 10−5 2.3 × 10−7 0.39 |
[17] | |
| European | 1886/10 421 | rs1154155 rs34593439 rs7553711 |
TCRA
CTSH TNFSF4 |
1.72 1.34 1.33 |
8.87 × 10−30 1.78 × 10−8 4.08 × 10−8 |
[18] | |
| Chinese | 1189/1997 | rs1154155 rs1551570 rs2854536 rs2834188 rs10995245 |
TCRA
P2RY11 TCRB IL10RB-INFAR1 ZNF365 |
1.64* 0.76* 0.78* 0.77* 1.23* |
5.02 × 10−49 3.77 × 10−10 3.87 × 10−8 1.95 × 10−8 1.24 × 10−11 |
[19] | |
| Mixed European | 1099 | rs12425451 | TEAD4 | N/A | 1.9 × 10−7 | [20] | |
| European Chinese African American |
807/1074 1078/1903 249/1048 |
rs3826784 | EIF3G | 0.77 0.77 0.87 |
1 × 10−4 1.47 × 10−5 0.19 |
[21] | |
| Sleep duration | UK Biobank | 127 573 | rs62158211 rs17190618 rs1380703 |
PAX8
VRK2 VRK2 |
0.94 0.96 1.06 |
4 × 10−9 0.001 0.01 |
[22] |
| UK biobank 23andMe |
111 975 | rs62158211 | PAX8 | 0.039 | 4.72 × 10−14 | [23] | |
| Excessive daytime sleepiness | UK biobank 23andMe |
111 648 | rs73536079 |
AR/OPHN1
ROBO1 ‡ TMEM132B § |
0.634 0.099 0.106 |
3.94 × 10−8 3.33 × 10−8 9.06 × 10−9 |
[23] |
| Insomnia | UK biobank 23andMe |
31 767/26 935 | rs113851554 rs145258459 rs5922858 rs13192566 rs3792900 |
MEIS1
TMEM132E CYCL1 WDR27 ‖ TGFBI ¶ |
1.26 1.23 1.12 1.14 1.10 |
9.11 × 10−19 2.13 × 10−8 1.28 × 10−8 3.17 × 10−8 2.16 × 10−8 |
[23] |
| Snoring | UK Biobank | 152 000/256 000 | rs592333 | DLEU7 | N/A | 1 × 10−17 | [24] |
| Obstructive Sleep Apnea (OSA) | FinnGen study | 16 761 | rs9937053 rs10507084 rs185932673 rs4837016 rs10928560 |
FTO
RMST/NEDD1 CAMK1D GAPVD1 CXCR4 |
1.11 1.11 1.12# 1.87 0.93 0.92 |
4.3 × 10−16 2.8 × 10−11 9.7 × 10−10Ψ 2.4 × 10−8 1.5 × 10−8 2.8 × 10−8 |
[25] |
* Odds ratio combined (different groups/stages).
† p Value after correcting for multiple testing.
‡ Significant after adjusting for depression.
§ Significant after adjusting for BMI.
‖ Significant only in men after adjusting for sex.
¶ Significant only in women after adjusting for sex.
# Values after adjusting for BMI.
The first successful GWAS milestone was marked in sleep field when two independent studies identified genetic variants associated with RLS, a common neurological disorder causing sleep disruption which presents itself as an irresistible urge to move one’s legs in response to uncomfortable sensations [10, 11]. The first study revealed a robust association with rs3923809 embedded within the BTBD9 gene (function unknown) in Icelandic and American populations for this trait [10]. The second study performed in European ancestry identified the same association along with two others (at MEIS1 and within a region harboring both MAP2K5 and LBXCOR1) associated with RLS [11]. A recent GWAS leveraged data from four cohorts (MrOS, the Wisconsin Sleep Cohort Study, HypnoLaus, and MESA) and identified variants at the MEIS1 and the BTBD9 loci that were significantly associated with periodic leg movement syndrome (PLMS), which is a more commonly occurring sleep disorder than RLS [15]. Several meta-analyses performed on more recent GWAS efforts utilizing independent datasets confirmed the previously identified risk variants for RLS [13]. In addition, three novel variants were reported at RANBP17, MICALL2 and at a locus on 11p15.4, bringing the total to 23 risk variants [13]. Of these, MEIS1 remains to be the strongest associated locus for RLS [12]. Interestingly, MEIS1 plays an important role in brain iron metabolism. Furthermore, MEIS1 expression has been observed to inversely associate with iron levels. In fact, unpublished data from a review reported elevated levels of MEIS1 protein in patients with RLS who were reported to also present with low iron reserves [26]. There is a growing body of evidence linking deficiency in brain iron deficiency and RLS. Iron plays an important role in many neurotransmitter systems, and depleted iron reserves can alter various neurotransmitter functions, which directly or indirectly leads to the symptoms associated with RLS [27]. Two novel risk loci, at CCDC141 and VSTM2L, were found to significantly associate with RLS in people suffering from migraines. This association was indeed confirmed through functional validation efforts indicating that the pathophysiology of RLS is different in people suffering with migraines [14]. A recent reassessment of all reported associations in a large case-control GWAS dataset (17 220 individuals of European descent) revealed that 4 out of 43 variants yielded association with RLS but failed to reach the genome wide significance threshold [28]. This reassessment emphasizes the requirement of large sample sizes and stringent significance thresholds in order to identify new candidate genes.
Narcolepsy is characterized by excessive daytime sleepiness, cataplexy, and sleep paralysis. The pathogenesis of narcolepsy involves a malfunctioning hypocretin system [29]. The disorder has a strong genetic association with “Human Leukocyte Allele (HLA)” subtypes, likely suggesting an autoimmune component for the disorder [30]. The HLA association is observed in >98% of people with narcolepsy and across multiple ethnicities [30–36]. However, the presence of non-HLA positive narcoleptics implicates the role of other genes elsewhere in the human genome. A key GWAS identified a non-HLA associated variant located between CHKB and CPT1B significantly associated with narcolepsy in two independent Japanese cohorts. This association was subsequently observed in Koreans but not in Europeans or African Americans [16]. Another narcolepsy GWAS conducted in a cohort of mixed European descent identified a variant in T cell receptor alpha (TCRA) locus significantly associating with cataplexy [17]. This association went on to be replicated in both Europeans and Asians but not African Americans. The association of the variant in TCRA with narcolepsy was confirmed in two independent GWAS efforts utilizing data from European [18] and Chinese [19] participants. These studies have implicated additional risk loci (non-HLA) for narcolepsy – at CTSH, TNFSF4 [18]; P2RY11, TCRB, IL10RB-INFAR1, and ZNF365 [19]. Reassessment of the variants shortlisted by the first narcolepsy GWAS in a larger population with narcolepsy identified a variant near TEAD4 significantly associated with the age of onset of cataplexy [20]. A novel variant within EIF3G, identified through transethnic mapping, may be a better marker for narcolepsy as this variant yielded a strong association signal and was found to be in high LD with the previously identified narcolepsy variant in P2YR11 in European and Chinese cohorts, but is in low LD in African American cohorts [21].
The first GWAS on sleep duration was performed on 749 participants and identified only one association signal that did not reach genome wide significance [37]. Subsequent GWAS efforts with self-reported sleep duration have identified several suggestive variants but none reaching strict genome wide significance. The first notable GWAS on sleep duration was performed in 2016 by leveraging UK Biobank data and including >100 000 participants, identifying three significant variants across PAX8 and VRK2 [22]. The same GWAS identified 16 variants associated with chronotype or morningness; 15 of these 16 variants were previously identified in a GWAS utilizing the 23andME cohort consisting of 89 283 participants [38]. 11 of these variants reached genome wide significance. This same study also identified 9 additional variants from the 23andMe cohort associated with chronotype. More recent GWAS efforts with sleep duration have leveraged UK Biobank data derived from approximately half a million participants [23, 39, 40] and identified multiple novel genetic loci. Several of these sleep duration loci overlapped with other sleep phenotypes such as snoring, napping, excessive daytime sleepiness, and insomnia [9].
In 2017, the Saxena group performed a GWAS for several sleep traits including sleep duration, chronotype, excessive daytime sleepiness, and insomnia. This represented the first GWAS to seek loci associated with insomnia and excessive daytime sleepiness [23]. This study identified 3 variants (in MEIS1, TMEM132E, CYCL1) associated with insomnia and confirmed the previously reported association between self-reported sleep duration and the variant at the PAX8 locus. This study also reported two sex-specific insomnia signals near WDR27 (male specific) and TGFBI (female specific) that independently associated with type 1 diabetes and achieved genome wide significance through sex-stratified secondary analysis. In addition, this study identified a variant in AR/OPHN1 significantly associated with daytime sleepiness. Variants at ROBO1 and TMEM132B also achieved genome wide significance (with daytime sleepiness) through secondary analysis after adjusting for depression and BMI respectively (Table 1) [23]. The association between insomnia and the MEIS1 locus was further confirmed by yet another GWAS analysis utilizing the dataset from the UK Biobank [41]. The largest GWAS in the sleep field to date was performed by the Posthuma group on insomnia in 1 331 010 participants (approximately one quarter of participants from the UK Biobank [4] and three quarters from the biotechnology company 23andMe [42, 43]), identifying 202 genetic loci [40].
Several newer GWAS efforts have investigated variants that are significantly associated with other sleep traits such as snoring and obstructive sleep apnea (OSA). Analysis of 408 317 participants in the UK Biobank identified 41 loci for snoring of which the DLEU7 locus exhibited the strongest association [24]. To identify genetic associations with OSA, the FinnGen database was leveraged and revealed 5 variants in FTO, RMST/NEDD1, CAMK1D, GAPVD1, and CXCR4, respectively [25]. Four out of the five variants (FTO, CAMK1D, GAPVD1, and CXCR4) were previously associated with BMI indicating a BMI-dependent OSA association for these variants [44]. The association of RMST/NEDD1 locus with OSA remained significant before and after adjusting for BMI indicating a BMI-independent OSA association [25].
The public availability of GWAS datasets has accelerated evaluation of putative causal relationships between genetic loci and health outcomes [45]. The majority of loci for sleep traits other than RLS, narcolepsy, and insomnia were identified in a mixed European population participating in the UK Biobank. The data collected by the UK Biobank effort was gathered through mostly self-assessment questionnaires. Although the sample sizes of the UK Biobank are large, a lack of replication of these loci in other ethnic communities clearly represents a drawback. But the associations identified by GWAS can be used to elucidate if given sleep traits/disturbances have a causal role in disease pathogenesis.
Despite the use of strong statistical measures, linear/logistic regression analysis used in epidemiologic studies cannot justify the causal association nor estimate the directionality of the association (if present). Moreover, the factors that influence either the sleep traits/disturbances or the disease risk or both (confounding factors) make validating a causal association more complicated.
Genetic variation represents an unbiased opportunity to confirm an association between an exposure and risk of disease as they occur naturally during meiosis. MR is a powerful technique that can use these variations as unbiased instruments to define causal associations between sleep traits/disturbances and other health comorbidities.
MR
MR leverages genetic variants that occur naturally as “instruments” to elucidate if an “exposure”, such as insomnia, is responsible for a disease risk or “outcome” such as cardiovascular complications; and when widely prevalent within a population, these variants can reveal the causal nature of the association between exposure (e.g. insomnia) and the outcome (e.g. cardiovascular complications) [46, 47] (Figure 1). MR is a technique derived from Mendel’s second law of inheritance or the “law of random assortment” that addresses the inheritance of a given trait (phenotype/risk for disease) and the independence of the inheritance of additional traits and is based on these assumptions: (1) the genetic variants identified are significantly associated with exposure and outcome (p < 5 × 10−8); (2) they are not associated with any confounding factors (factors other than the exposure that have a known association with the outcome); and (3) they must be associated with the phenotype (outcome) of interest exclusively via the exposure being investigated [48].
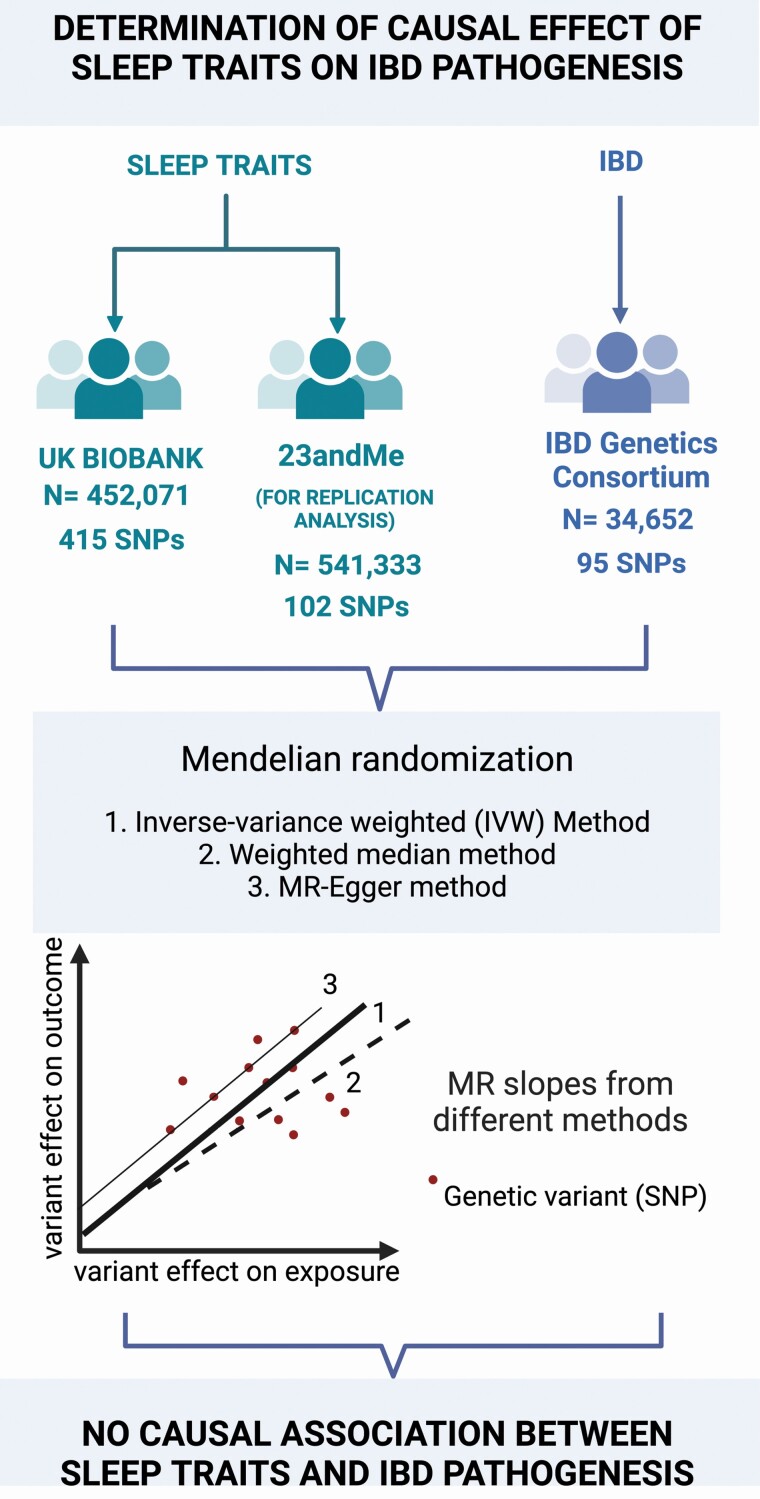
MR can reveal the impact of modifiable exposures on disease pathogenesis, representing a clear advance over what observational epidemiology could offer in the past. For example, preclinical [49], clinical [50, 51], and observational studies [52] indicate a correlation of sleep disorders with IBD. However, a causal link between these two traits has not been established. An MR analysis by Chen et al. attempted to determine the causality between differential sleep traits and IBD pathogenesis, serving as an example of how a two-sample MR can determine causality between these two trait areas [53] (Figure 2). For sleep traits, GWAS summary statistics from the UK biobank (n = 452 071) and 23andME (n = 541 333) consortia were utilized. For IBD, the data was obtained from GWAS study from the IBD Genetics Consortium (n = 34 652). The genetic variants utilized for MR were significantly associated with the sleep traits at the genome wide level (p < 5 × 10−8) (satisfying the assumption that variants should be significantly associated with exposure). Variants that are not in close linkage disequilibrium (LD) with other SNPs associated with outcome were selected for further analysis, thus eliminating confounding factors and satisfying assumption 2. The inverse-variance weighted (IVW) method was used as the primary mode of analysis to determine causality through estimating beta coefficient (β) from the SNP-outcome and SNP-exposure association estimates [54]. βis transformed into Odds Ratio (OR), which when >1.0 indicates a strong association between exposure and outcome. While estimates calculated through IVW have the highest precision, sensitivity analyses are often carried out to confirm the result. The robustness of this method was tested using weighted median (WM) method which generates a similar estimate if at least 50% of the estimated weight is through valid variants. MR-Egger analysis was employed to detect and correct for any pleiotropy [55]. If all the three methods produced a similar estimate (if slopes are similar, and significantly higher than 1 in all cases), the relationship between the two traits is most likely causal. In this study, all 3 analyses showed that none of differential sleep traits had causal effect on the pathogenesis of IBD (Figure 2).
Figure 2.
This flow chart illustrates a two sample MR analysis performed by Chen et al. to determine the causality between differential sleep traits and IBD pathogenesis. Three MR approaches were used to determine causality whose slopes are illustrated in the graph: 1. Inverse variance weighted method, 2. Weighted median method, 3. MR-Egger method. If all three methods yielded a similar estimate (with slope > 1.0), the association between the two traits maybe causal.
Initial MR analyses utilized randomly allocated genetic variants in one sample cohort to assess the impact of an exposure on disease pathogenesis (outcome) in the same sample cohort; however, it is challenging to identify a single population with both variant-exposure and variant-outcome associations [56]. Two sample MR on the other hand, utilizes the variant-exposure and variant-outcome associations from two different sample cohorts, increasing the sample size and hence power of the analysis. Public availability of GWAS data has made two sample MR a more reliable approach. Other types of MR can determine causality in more complex settings involving multiple variants and associations. Bidirectional MR analyses can aid in estimating the direction of a causal effect by using genetic variation robustly associated with exposure and outcome (from separate GWAS efforts) [57]. Factorial MR analysis can determine the combined causal effects of two or more variants responsible for a single disease [58], while multivariable MR analysis considers the variant’s pleiotropic effects on other outcomes before estimating causality.
A rapid rise in current GWAS efforts enabled a more reliable SNP heritability score and increased the reliability of the outcome of subsequent MR analyses. Given the availability of various web-based analytical platforms including MR-Base [45] and LD hub [59], it is now possible to assess all pairwise relationships to deliver leads that can be subsequently followed up in functional pursuits to provide insight into previously known/unknown potential causal relationships between exposures (sleep phenotypes) and key disease presentations.
MR of Sleep and Common Complex Disease
Over the previous decades, classical epidemiological studies determined multiple environmental and genetic factors contributing to the pathogenesis of neurodegenerative, metabolic, and cardiovascular diseases among many others. Leveraging high-dimensional molecular datasets can facilitate development of mechanistic insight into associations between environmental and genetic variants and complex disease traits. For example, a GWAS analysis in 446 118 participants from the UK Biobank identified 78 loci robustly associated with self-reported sleep duration. Genetic correlation analysis of these loci revealed associations with various traits including cardiovascular and metabolic traits [39]. Based on the polygenic risk score derived from these loci, MR analysis identified a significant association between sleep duration and multiple diseases, such as congestive heart failure, obesity, hypertension, RLS, and insomnia [60]. MR analyses can therefore provide a means to assess these associations for causality and shed light on potential protective pathways. Some of the major MR findings in the field of sleep determining the causal associations between various sleep phenotypes and health (disease) outcomes are discussed below and summarized in Table 2.
Table 2.
Summary of findings of MR analyses performed to determine causality between various sleep traits (exposures) and disease outcomes
| Disease outcome | Sleep exposure | MR model | Interpretation | Reference |
|---|---|---|---|---|
| Sleep and neurodegenerative diseases | ||||
| Cognition | Sleep duration | One and two sample MR | Positive association between short sleep duration and cognitive decline | [61] |
| Risk for Alzheimer’s disease | Sleep duration | Two sample MR | No causal association determined | [61] |
| Sleep traits | Two sample MR | No causal association determined | [62] | |
| Sleep disturbances | Bidirectional two-sample MR | AD had a causal effect on sleep disturbances, but sleep disturbances were not causal to AD | [63] | |
| Parkinson’s disease | Sleep traits | Two sample MR | No causal association determined | [64] |
| Neurodegenerative disease risk | Sleep/wake patterns | Two sample MR | Chronotype was inversely associated with Parkinson’s age of onset Sleep efficiency was associated with decreased AD risk Daytime sleepiness was associated with increased ALS risk |
[60] |
| Amyotrophic Lateral Sclerosis (ALS) | Sleep disturbances | Two sample MR | Daytime sleepiness was associated with increased ALS risk | [65] |
| Pain | Sleep disturbances | Bidirectional two-sample MR | Chronic pain had a causal effect on sleep disturbances, but sleep disturbances were not causal for pain | [66] |
| Insomnia | Bidirectional two-sample MR | Causal association between insomnia and pain was determined in both directions | [67] | |
| MR of sleep and psychiatric disorders | ||||
| Depression | Sleep duration | Two sample MR | Sleep duration was associated with decreased risk of depression | [68] |
| Daytime napping | Two sample MR | Daytime napping was associated with increased risk of depression | [68] | |
| Chronotype | Two sample MR | Chronotype was associated with decreased risk of depression | [69] | |
| Insomnia | Bidirectional multivariable MR | Causal association between insomnia and depression was determined in both directions | [70] | |
| Insomnia | Bidirectional MR | Causal association between insomnia and depression was determined in both directions | [71] | |
| Schizophrenia | Insomnia | Bidirectional MR | No causal association determined in either direction | [72] |
| Chronotype | Two sample MR | No causal association determined | [73] | |
| Sleep duration | Bidirectional two-sample MR | Long sleep duration is associated with pathogenesis of schizophrenia | [39] | |
| Bipolar disorder | Insomnia | Bidirectional MR | Insomnia had a causal effect on bipolar disorder, but bipolar disorder was not causal for insomnia | [72] |
| Autism Spectrum Disorder (ASD) | Insomnia | Bidirectional MR | Insomnia had a causal effect on ASD, but ASD was not causal for insomnia | [72] |
| MR of sleep and cardiometabolic disease | ||||
| Adiposity | Insomnia | Two sample MR | Insomnia had a causal effect on increased measure of adiposity | [74] |
| increased waist circumference | Daytime napping | Two sample MR | Daytime napping had a causal association with increased waist circumference | [74] |
| BMI (adult) | Sleep duration | Two sample MR | Long sleep duration had a causal association with increased adult BMI | [75] |
| Sleep duration | Two sample MR | Long sleep duration had a causal association with increased adult BMI | [76] | |
| Chronotype | Two sample MR | No causal association determined | [77] | |
| Daytime napping | Two sample MR | No causal association determined | [78] | |
| Snoring | Multivariable MR | Causal association between high BMI and snoring was identified | [24] | |
| BMI(Children) | Sleep duration | Two sample MR | Long sleep duration had a causal association with decreased BMI in children (2–10 years) | [79] |
| Atrial fibrillation | Obstructive sleep apnea (OSA) | Two sample MR | OSA had a causal association with increased risk of atrial fibrillation | [80] |
| Obstructive sleep apnea (OSA) | Bidirectional MR | OSA had a causal effect on atrial fibrillation, but atrial fibrillation was not causal for OSA | [81] | |
| Type 2 Diabetes (T2D) | Insomnia | Univariable/Multivariable MR | Insomnia had a causal association with T2D | [82] |
| Insomnia | Two sample MR | Insomnia had a causal association with T2D | [83] | |
| Sleep duration | Two sample MR | No causal association determined | [84] | |
| Chronotype | Two sample MR | No causal association determined | [77] | |
| Myocardial infarction (MI) | Sleep duration | Two sample MR | Short sleep duration was causally associated with MI | [85] |
| Heart failure | Sleep duration | Two sample MR | Longer sleep duration was causally associated with decreased risk for heart failure. | [86] |
| Cardiovascular diseases (CVDs) | Sleep duration | Two sample MR | Short sleep duration was causally associated with increased risk of CVDs | [87] |
| Insomnia | Two sample MR | Insomnia was causally associated with nine CVD traits | [88] | |
| Insomnia | Two sample MR | Insomnia was causally associated with overall CVD risk | [89] | |
| Stroke | Insomnia | Two sample MR | Insomnia was causally associated with increased risk for Stroke | [90] |
| Sleep duration | Two sample MR | No causal association determined | [91] | |
| Hypertension | Insomnia | Two sample MR | Insomnia was causally associated with increased risk for hypertension | [92] |
| Sleep duration | Two sample MR | Longer sleep duration was causally associated with decreased risk for hypertension | [92] | |
| MR of sleep and cancer | ||||
| Breast cancer | Chronotype | One and two sample MR | Chronotype was causally associated with decreased risk for breast cancer | [93] |
| Obstructive sleep apnea (OSA) | Two sample MR | OSA was causally associated with increased risk for breast cancer | [94] | |
| Prostate cancer | Chronotype | Univariable/Multivariable MR | Chronotype was causally associated with decreased risk for prostate cancer | [95] |
| Lung cancer | Insomnia | Two sample/Multivariable MR | Insomnia was causally associated with increased risk for lung cancer | [96] |
| Insomnia | Two sample MR | Insomnia was causally associated with increased risk for lung cancer | [97] | |
| Insomnia | Two sample MR | Insomnia was causally associated with increased risk for lung cancer | [98] | |
| MR of sleep and other health outcomes | ||||
| Peptic ulcer | Insomnia | Bidirectional MR | Insomnia was causally associated with increased risk for peptic ulcer | [99] |
| Osteoarthritis | Insomnia | Univariable/Multivariable MR | Insomnia was causally associated with increased risk for osteoarthritis | [100] |
| Sleep duration | Univariable/Multivariable MR | Short sleep duration was causally associated with increased risk for osteoarthritis | [100] | |
| Chronic kidney disease | Sleep duration | Two sample MR | Long sleep duration was causally associated with increased risk for chronic kidney disease* | [101] |
*Identified only in individuals with diabetes.
MR of sleep and neurodegenerative disease
Neurodegenerative diseases consist of a varied set of conditions, with progressive degeneration of various nerve cells leading to the dysfunction of several motor and mental systems. The onset of these diseases is driven by a combination by both environmental and genetic risk factors. Neurodegenerative diseases are characterized by symptoms such as tremor, imbalance, impaired cognition and sleep disturbances. Sleep disturbances in form of sleep/wake alterations, insomnia, hypersomnia, sleep apnea, RLS etc. occur in as much as 60% of the population suffering from any neurodegenerative disease [102].
Studies have identified that sleep disturbances manifest very early for these traits, even before the main symptoms of the underlying disease [103]. Sleep disturbances can perturb circadian rhythm, which is known to regulate gene expression in several brain regions, causing impaired neural function [104, 105]. While it is important to recognize and properly manage these sleep disorders to improve symptoms observed with neurodegenerative conditions, establishing a causal relationship between sleep alterations and neurodegenerative disease has proven challenging. MR can be an effective tool to assess causality in this context, where it has gathered considerable traction in both the neurodegeneration and sleep fields. This in turn provides valuable insights into the etiology of these conditions.
Sleep and Alzheimer’s disease
Alzheimer’s disease (AD) is a severe neurodegenerative disease that impacts at least 40 million people worldwide and given the increase in aging population its prevalence is projected to dramatically increase over the next two decades. It has been widely reported that patients with AD encounter sleep disruption in the form of breathing disorders and RLS [106]. While studies report an increase in cognitive impairment associated with AD, as diagnosed through pathological markers with disturbed sleep patterns, it remains to be determined if alterations in sleep behavior are causal for AD pathogenesis. Using summary statistics from the UK Biobank (n =395 803) and the International Genomics of Alzheimer’s Project (IGAP) (n = 17 008 AD cases vs. 37 154 controls), an MR study identified a causal relationship between sleep duration and cognition, but not with the risk for AD [61]. This finding was confirmed by another independent MR effort utilizing data from the same IGAP study and essentially identified no significant causal association between disrupted sleep traits and AD risk [62]. A bidirectional 2-sample MR analysis has confirmed the lack of significant effect of sleep disturbance on AD risk; however, this study suggests that AD can causally influence sleep patterns [63]. A recent study utilized summary statistics from GWAS efforts of sleep duration [39] and insomnia [40] to investigate association with AD phenome (status of AD progression, age of AD onset, CSF levels of amyloid beta, the pathogenic form of tau and total tau, hippocampal volume, cortical surface area and thickness, neuropathological burden of neuritic plaques, neurofibrillary tangle burden, and Vascular brain injury) [107], through a combination of polygenic risk score (PRS) analysis and MR analysis. Polygenic risk score measures the individual’s chance to present with a trait (based on genotype) and reveals if possessing this trait infers susceptibility to disease risk. While the MR determined no causal association between sleep traits and AD phenome, which is consistent with our prior studies, PRS analysis identified causal association between longer sleep duration and cortical thickness and shorter sleep duration with AD pathogenesis [107].
Sleep and Parkinson’s disease
Parkinson’s disease (PD) results from the progressive loss of nigrostantial dopaminergic neurons and is characterized by tremors impairing motor coordination. Until very recently, PD was thought to be principally monogenic, but subsequently a range of genetic and environmental risk factors have been attributed to the progression of the disorder [108].
MR analyses have been undertaken to utilize GWAS meta-analysis derived summary statistics including more than 260 000 European ancestry cases to explore the causal associations of exposures on PD risk; however, this MR did not identify causal associations with any sleep related exposures risk [64].
Another MR study by Cullel et al. sought to specifically analyze causal associations between various sleep traits and various neurodegenerative diseases including PD. This study utilized genetic variants from the public domain to determine if sleep traits were causally associated with PD. Inverse causal association between the morning chronotype and age of onset of PD was identified in this MR analysis [102]. It is also noted that the same MR study identified sleep efficiency was associated with decreased AD risk and daytime sleepiness to be associated with amyotrophic lateral sclerosis (ALS).
Sleep and amyotrophic lateral sclerosis
ALS is a progressive degenerative disorder, resulting in muscle weakness [109]. ALS presents with a high degree of familial and sporadic inheritance, supporting a strong role for genetics in the development of the disease. While the exact etiology of ALS remains unknown, as with other neurodegenerative diseases the interaction between genetic and environmental needs to be characterized [110]. Although ALS predominantly presents with motor symptoms, evidence from imaging studies also indicates the presence of cognitive impairment [111]. A qualitative systematic review revealed that almost 50%–63% of patients with ALS report reduced sleep quality [112]. MR analyses make resourceful tools to assess which sleep traits are associated with ALS. Corroborating the MR findings of Cullell et al. (see above), an independent MR analysis by Zhang et al. [65] and a cross sectional study performed in a Chinese cohort [113] revealed that daytime sleepiness was causally associated with the disease.
Sleep and pain
Decrease in sleep quality increases risk for pain presentation. Studies report the presence of sleep disturbances in 67%–88% of patients with chronic pain [114]. A meta-analysis on 16 longitudinal studies involving approximately 61 000 participants assessed the impact of sleep disturbances on pain-related traits, reporting an increased risk of presenting with pain if diminished sleep quality is experienced [115]. Identification of neural and genetic pathways correlated with both sleep and pain should aid the understanding of this comorbid relationship. Indeed, twin studies report a high correlation between sleep disturbances and pain, supporting a strong genetic overlap between these two conditions [116].
A two-sample MR analysis confirmed the bidirectional causal relationship between chronic pain and sleep disturbance, with a higher degree of evidence supporting causation of sleep disruption by such pain i.e. pain leads to sleep disturbance [66]. This finding contrasts with the finding of another MR study, which also reported a bidirectional causal association between pain and insomnia, but revealing a relatively more complex relationship [67]. The discrepancy between these two MR studies is potentially explained by the differences in cohort size, and therefore statistical power of the respective studies.
MR of sleep and psychiatric disorders
Psychiatric disorders are often associated with sleep disturbances such as insomnia, hypersomnia, nightmares, etc [117]. Recent evidence suggests the dysregulation of the circadian system in several psychiatric disorders, including bipolar disorder, major depressive disorder, and schizophrenia [118] indicating the presence of complex mechanisms linking sleep disturbance phenotypes and psychiatric disorders. Genetic correlations determined through sleep GWAS efforts identified several loci not only associated with circadian genes, but also with psychiatric disorders [39, 40, 77]. Understanding this overlap could reveal novel loci that can be developed into therapeutic interventions for those presenting with sleep/psychiatric disturbances.
Sleep and depression
Depression is a major cause of mental disability and is prevalent among adults and adolescents. The trait is also a major contributor to suicide risk. Genetic vulnerability and early-life adversity are the two major unmodifiable risk factors for depression, which is now recognized as a polygenic condition [119].
A two-sample MR study leveraging UK Biobank summary statistics to assess a broad panel of modifiable factors associated with depression revealed a bidirectional association between daytime napping and depression risk [68]. Another two-sample MR analysis identified an association between morning diurnal preference and a decreased risk of depression [69].
Given that insomnia is among the most common sleep traits, a number of observational studies have assessed its association with multiple psychiatric conditions, including depression – but with mixed results. A bidirectional causal association was observed between insomnia and depression by two independent MR analyses [70, 71], while an additional independent MR analysis identified a significant causal association between insomnia and autism spectrum disorder and bipolar disorder [72].
Sleep and schizophrenia
Schizophrenia is a severe mental illness that presents with symptoms including delusions, hallucinations, thought disorders, anhedonia, social withdrawal, etc [120]. Approximately three quarters of patients with schizophrenia encounter sleep-related traits, including challenges with falling and/or staying asleep, along with daytime sleepiness [121].
MR analysis failed to identify a causal association between insomnia and schizophrenia in both directions [72]. Although genetic correlation analysis of the SNPs identified by GWAS suggests an association of schizophrenia with chronotype, MR failed to confirm the causal relationship between the two conditions [73]. However, another bidirectional two-sample MR analysis did reveal connections between longer sleep duration and the pathogenesis of schizophrenia [39].
Sleep and bipolar disorder
Bipolar disorder is characterized by several comorbidities including sleep disturbances [117]. In fact, a recent GWAS effort reported a strong association between the TRANK1 locus, which is a well-known locus associated with bipolar disorder, schizophrenia, and Kleine-Levin syndrome (KLS), a rare sleep disorder characterized by severe episodic hypersomnia, cognitive impairment, and disinhibition, providing evidence of a genetic link between bipolar disorder and sleep disturbance [122]. Despite the clinical and genetic evidence associating sleep disturbances in psychiatric disorders, a causal relationship between sleep traits and bipolar disorder has not been established [123, 124]. MR analysis was thus performed to investigate such a relationship between sleep traits and bipolar disorder. Using summary statistics for sleep disturbances from GWAS efforts and statistics from psychiatric disorders from the Psychiatric Genomics Consortium online database, two independent bidirectional MR analyses identified a causal effect of genetically predicted insomnia on bipolar disease, but not the other way around [72, 125].
MR of sleep and cardiometabolic disease
Cardiovascular disease (CVD), obesity, and diabetes collectively constitute cardiometabolic disease. These conditions share common risk factors and are often observed as comorbidities. Notably, ~70% of type 2 diabetes (T2D) related deaths are a consequence of CVD, and while patients with the related “metabolic syndrome” are at greater risk of presenting with T2D and cardiovascular complications [126]. Sleep is a vital biological process related to metabolic control. Evidence from epidemiologic studies implicates disturbed sleep increases the risk of cardiometabolic disease presentation [127]. Cardiometabolic disorders commonly have overlapping pathways of sleep-related metabolic functioning, and thus any disturbances in any such pathway can lead to elevated risk for cardiometabolic disease [127]. MR analysis can therefore assess whether suboptimal sleep is causally associated with cardiometabolic traits.
Sleep and obesity and body mass index
Obesity and body mass index (BMI) are polygenic traits that are heavily influenced by environmental factors and lifestyle choices [128]. Evidence from many epidemiological studies supports a connection between long and short sleep duration and obesity in both adults and children, the association being stronger in children and decreasing with age [129, 130]. Metabolic mediators such as leptin and ghrelin have been established as regulators of sleep and body weight suggesting shared genetic etiology between the two conditions [74]. However, genetic signals obtained from GWAS indicate mixed results. The FTO locus was the first obesity signal identified in a T2D GWAS carried out in the United Kingdom [131]. This locus confers the strongest association with BMI to date and has revealed additional associations with sleep traits including morning preference, sleep duration, and snoring. In addition, obesity loci such as SLC39A8 and HCRTR2 were identified in GWAS efforts of sleep, indicating a shared genetic link between the two conditions [74]. On the other hand, no association was identified between BMI and sleep traits as reported by a post-GWAS analysis on the polygenic risk score of 97 BMI variants derived from the UK Biobank [132]. In another UK Biobank based analysis, the obesity polygenic risk score consisting of 95 BMI variants was found to associate with daytime sleepiness but not with insomnia [23].
MR analysis can bridge this discrepancy by providing a robust approach to demonstrate causality between the GWAS variants and sleep traits. One MR analysis identified causality between insomnia and increased measures of adiposity, as well as with high daytime napping frequency and increased waist circumference [74]. Longer sleep durations (when reported as accelerometer data) were causally associated with increased adult BMI [75, 76], but other MR efforts that used self-reported sleep duration data have not identified a clear causality between sleep duration and BMI, especially in women. Interestingly, in the pediatric setting higher sleep duration has been reported to track with decreased BMI [79]. No causal association has been identified between BMI and morningness [22, 77] or daytime sleepiness [78]. A recent MR performed to determine the causal link between obstructive sleep apnea and atrial fibrillation identified five loci with significant association [80]. At least one of these variants was also associated with BMI at the genome-wide significance level, raising the possibility that OSA and BMI are causally related. MR analysis did not report a causal link between the two traits; however, a strong genetic correlation between increased BMI (plus whole body fat mass) and snoring has been established by MR when utilizing the variants identified in a European ancestry GWAS for snoring [24].
Sleep and Type 2 diabetes
The International Diabetes Federation has reported that approximately 537 million (1 in 10) adults worldwide have diabetes. T2D is characterized by the inability of the body to utilize glucose from the circulation resulting in high levels of glucose in the bloodstream i.e. hyperglycemia [133]. Poor sleep quality is associated with irregular eating patterns and an unhealthy diet both of which can lead to an increased risk of T2D.
Many observational studies indicated that insomnia and daytime napping are risk factors for T2D [134–136]. A recent study reviewed all possible observational studies of T2D and identified 97 risk factors for T2D. A multivariate MR analysis examined the causal associations between T2D and the aforementioned risk factors and identified a novel causal association between insomnia and T2D risk [82].
Another recent MR study assessed the causal relationships between key sleep-related traits and T2D and identified significant causal association with insomnia, but not other sleep related traits [83]. This latter observation is supported by other MR analyses that failed to identify causality between T2D and sleep duration [84] or morningness [77].
Sleep and cardiovascular diseases
Cardiovascular diseases (CVDs) comprise a heterogeneous group of disorders, including myocardial infarction, coronary artery disease, and hypertension [137]. CVD is in fact the leading cause of death worldwide [138].
Most observational studies that report associations between sleep traits and cardiovascular events are focused on sleep duration [139–141] and generally indicate that shorter sleep duration tracks with adverse cardiovascular outcomes.
MR has confirmed the observation that short sleep duration is causally associated with myocardial infarction [85, 87] as well as other CVDs like hypertension, ischaemic heart disease, and atrial fibrillation [87]. Corroborating this finding, another MR study revealed that longer sleep duration is causally associated with lower heart failure risk [86].
Insomnia has also been causally associated with the increased risk of several CVDs by a number of MR studies. A two-sample MR analysis using the summary statics from UK Biobank, identified a causal association between insomnia and nine cardiovascular traits [88]. These findings are in agreement with another MR study that confirmed the association between insomnia and overall CVD risk leveraging genetic variants identified from the insomnia GWAS of 1 331 010 individuals [89]. Although insomnia is a potential risk factor for stroke, it is notable that short sleep duration is not [90, 91]. A causal association between hypertension and various sleep traits, including insomnia (direct association) and sleep duration (inverse association), was identified by an MR study using data from European-descent GWAS efforts: FinnGen Study and UK biobank [92].
Leveraging the information from the FinnGen study (217 955 individuals) with 16 761 patients with OSA [25], several new MR analyses have reported an increased risk of atrial fibrillation associated with OSA [25, 80, 81, 142]. However, a bidirectional MR analysis failed to report a reverse causal association between the two traits i.e. there was no causal effect of atrial fibrillation on OSA [81].
MR of sleep and cancer
Epidemiological studies are increasingly yielding evidence that sleep impairment contributes to the pathogenesis of cancer [143]. An observational study on 23 620 participants identified that those who slept <6 h a day on average were 43% more likely to develop cancer [144]. This finding was reinforced by other studies that observed associations between short sleep duration and the pathogenesis of colorectal [145], breast [146], lung [147], and stomach [148] cancers. While these associations are highly suggestive, prospective studies, and meta-analyses have failed to report such significant findings [149, 150]. On the other hand, MR analyses can be useful to determine significance and avoid such bias associated with confounding factors.
A one sample MR analysis with UK Biobank data, and a two sample MR analysis leveraging Breast Cancer Association Consortium (BCAC) data revealed that morning preference was protective with respect to risk of developing breast cancer [93]. Through an independent MR utilizing genetic variants from the PRACTICAL consortium [151], morning preference was also shown to lower prostate cancer risk by 29% in men [95].
OSA is breathing disorder that is experienced during sleep, which presents repeated halting of breathing that can lead to a degree of hypoxia. Hypoxia is known to play an essential role in cancer progression [152]. Observational studies have revealed a causal relationship between OSA and cancer [150]. Supporting this, a two-sample MR approach identified a causal relationship between OSA and breast cancer risk in Asian populations within the BCAC cohort [94]. A recent meta-analysis including six studies and a total of 5 165 200 participants identified a two-fold increase in the risk of breast cancer in patients with OSA [153].
Three recent, independent MR analyses identified a causal relationship between insomnia and increased lung cancer risk [96–98]. Given the urgency in investigating methods to prevent lung cancer, effective intervention could involve better sleep management.
MR of sleep and other health outcomes
Over the past decade, many advances have been made in shedding light on the extent of the influence of poor sleep quality on various health outcomes. The causal associations between sleep traits and major health disease classes have been discussed in detail in the above sections. However, MR analyses have also revealed significant causal associations of sleep traits with other health outcomes.
A bidirectional MR analysis utilizing publicly available data from a large insomnia GWAS [40] implicated its role in conferring risk for peptic ulcer disease [99]. Insomnia and short sleep duration have also been shown to be causally associated with higher osteoarthritis risk [100].
Short sleep duration is widely reported to increase the risk for several cardiometabolic conditions as discussed above in turn it has been hypothesized that sleep duration impacts renal function based on evidence from two observational studies [154, 155]. Indeed, a two-sample MR analysis showed that longer sleep duration is causally associated with increased risk of presenting with chronic kidney disease, but the observations were limited to diabetic individuals [101].
While MR has been extremely useful in implicating causal associations between sleep and other health outcomes, it has also been an important tool to determine causal association between different sleep traits. Leveraging the summary statistics from a recent GWAS, MR identified a significant causal association between RLS and PLMS (periodic limb movement syndrome) [15]. Similarly, a significant causal association between snoring and sleep apnea was identified by MR utilizing summary statistics from a GWAS on snoring [24].
Limitations of MR and Next Steps
While MR is an excellent tool to assess comorbidity and causality between sleep traits and major health outcomes using genetic variants as instruments, it is not without shortcomings. Here we outline a few limitations hindering MR analyses in the identification of potential causal associations. We also outline steps that can be taken going forward to improve the outcomes through such analyses.
Pleiotropy
Pleiotropy occurs when one gene drives more than one phenotypic effect. Pleiotropism is a major limitation of MR as it violates the assumption that the genetic variant used as the instrument exclusively associates with the phenotype of interest. Genetic variants exhibit pleiotropism through multiple effects, such as modulation of intermediate gene/ protein expression and alternative splicing [46].
Many approaches are being tested to identify and remove variants exhibiting pleiotropy in order for MR analyses to yield a credible output. Heterogenous effects of variants can be visually identified by scatter plots, radial plots, or funnel plots, among others; indeed, such approaches have been traditionally used to display the effect of each variant against their association precision. Asymmetry in these plots can indicate unusual/heterogenous effects of specific variants (on the outcome), thus pinpointing pleiotropy [156]. The “leave-one-out plot” removes the identified “outlier” variants one at a time and recalculates the overall effect to select the variant with the most significant association [47]. Techniques, such as the MR-Egger Regression method, were developed for detecting such heterogeneity and bias associated with pleiotropy [55]. For example, this method has been successfully applied to account for the pleiotropy of the genetic variants associated with OSA, and to evaluate its causal association with cardiometabolic comorbidities and thus establishing a causal link between OSA and atrial fibrillation [80].
Non-linear associations
Another assumption is that the instrument leveraged in MR, principally the genetic locus, mediates association between the sleep trait and disease presentation in a linear fashion. The observations between sleep duration and various traits, however, could be non-linear and therefore, assuming a linear association can lead to a false negative output. Thus, while performing MR, it is important to consider a potential lack of linear association and to be aware of a potential different shape of association. To date, just one sleep MR study has considered both linear and non-linear associations while examining the causality between a sleep phenotype and a disease outcome, namely between sleep duration and cognitive function/dementia [61].
Diversity
In order to identify robust MR-based associations between sleep phenotypes and disease, it is important to include participants from various ethnic groups. Many of the earlier GWAS and MR efforts were performed in relatively small groups of individuals often belonging to the same ethnic background. Replication attempts in other populations have proven very underwhelming either due to small sample size or the possible lack of association in that given ethnic group. For instance, a GWAS performed for sleep duration on individuals with European ancestry identified the PAX8 and VRK2 loci [22, 157]. However, a subsequent GWAS meta-analysis and follow-up MR performed on Japanese participants failed to implicate the association of these loci with sleep duration [158].
Many individuals participating in the UK Biobank prospective study are healthy adults. Efforts to replicate any associations in the EAGLE (EArly Genetics and Lifecourse Epidemiology) cohorts consisting of children and adolescents have been relatively unsuccessful. This is an important consideration for MR studies, as some of these associations may differ with age. Only a few MR observations for sleep duration genetics coming from studies in adults have yielded modest associations for sleep duration and various disease traits in pediatric/adolescent cohorts [79, 84].
Statistical power
The precision of MR is governed by factors that include genetic variant allele frequency, magnitude of effect on health outcome, and population size. Low sample size therefore limits the rigor of MR analyses.
Within a small population, the probability of occurrence of the genetic variant responsible for the desired outcome is low. The combined risk from inheriting multiple common variants is higher than the risk from one monogenic variant, with an individual with a higher polygenic risk score being more susceptible to disease [159]. Therefore, in MR studies multiple variants are often combined to generate a polygenic risk score, which in turn increases statistical power, that can then be utilized to make more robust conclusions regarding causal relationships.
Canalization
Genetic variation co-segregates randomly during fetal formation, and some of these variants perturb normal development. However, depending on the environmental context, either through genetic redundancy (more than one gene having the same or similar function) or through the activation of alternative metabolic pathways, the same phenotypic endpoint is reached. This is called canalization, or developmental compensation [160]. Canalization poses a problem for MR analysis as this approach assumes that the genetic variation does not associate with confounding factors, but canalization introduces unanticipated confounding factors that can interfere with the analysis. Hence when relating the findings of conventional observational studies to those from MR analysis, the concept of canalization and alternate gene expression must be considered before interpreting results [46].
Conclusions
GWAS has uncovered a myriad of genetic loci associated with sleep-related traits. Use of locomotor-time measures as a way of quantifying sleep and the development of various model organisms has greatly aided in the discovery of putative genetic targets associated with sleep and enables much needed subsequent functional efforts on understanding sleep phenotypes and pathogenesis of disease.
The genetic findings have implicated association with a wide spectrum of physiological mechanisms, ranging from transcription factors to neuropeptides. All that being said, there remain key questions to be addressed in the context of sleep genetics and key processes. MR can serve as an effective tool to determine causality between various sleep phenotypes and other health outcomes utilizing the various genetic signals identified by GWAS. Characterizing the trait outcome in the context of a specific pathway can drive characterization of the underlying biology and aid in the development of new therapeutic areas.
Contributor Information
Shilpa Sonti, Center for Spatial and Functional Genomics, Children’s Hospital of Philadelphia, Philadelphia, PA, USA.
Struan F A Grant, Center for Spatial and Functional Genomics, Children’s Hospital of Philadelphia, Philadelphia, PA, USA; Department of Genetics, University of Pennsylvania, Philadelphia, PA, USA; Institute for Diabetes, Obesity and Metabolism, Perelman School of Medicine, University of Pennsylvania, Philadelphia, PA, USA; Department of Pediatrics, The University of Pennsylvania Perelman School of Medicine, Philadelphia, PA, USA; Division of Human Genetics and Endocrinology, Children’s Hospital of Philadelphia, Philadelphia, PA, USA.
Funding Statement
The authors are supported by National Institutes of Health (NIH) award R01 HL143790.
Disclosure Statements
Financial Disclosure. The authors have no financial disclosures.
Non-financial Disclosure. The authors have no conflicts of interest to declare.
References
- 1. Nath RD, et al. The jellyfish Cassiopea exhibits a sleep-like state. Curr Biol. 2017;27(19): 2984–2990 e3. doi: 10.1016/j.cub.2017.08.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Buysse DJ. Sleep health: can we define it? Does it matter? Sleep. 2014;37(1):9–17. doi: 10.5665/sleep.3298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Webb JM, et al. Recent advances in sleep genetics. Curr Opin Neurobiol. 2021;69:19–24. doi: 10.1016/j.conb.2020.11.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Sudlow C, et al. UK biobank: an open access resource for identifying the causes of a wide range of complex diseases of middle and old age. PLoS Med. 2015;12(3):e1001779. doi: 10.1371/journal.pmed.1001779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Chen Z, et al. China Kadoorie Biobank of 0.5 million people: survey methods, baseline characteristics and long-term follow-up. Int J Epidemiol. 2011;40(6):1652–1666. doi: 10.1093/ije/dyr120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Krokstad S, et al. Cohort profile: the HUNT Study, Norway. Int J Epidemiol. 2013;42(4):968–977. doi: 10.1093/ije/dys095 [DOI] [PubMed] [Google Scholar]
- 7. Davey Smith G, et al. Mendelian randomization: genetic anchors for causal inference in epidemiological studies. Hum Mol Genet. 2014;23(R1):R89–R98. doi: 10.1093/hmg/ddu328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Uffelmann E, et al. Genome-wide association studies. Nat Rev Methods Primers. 2021;1:59. doi: 10.1038/s43586-021-00056-9 [DOI] [Google Scholar]
- 9. Garfield V. Sleep duration: a review of genome-wide association studies (GWAS) in adults from 2007 to 2020. Sleep Med Rev. 2021;56:101413. doi: 10.1016/j.smrv.2020.101413 [DOI] [PubMed] [Google Scholar]
- 10. Stefansson H, et al. A genetic risk factor for periodic limb movements in sleep. N Engl J Med. 2007;357(7):639–647. doi: 10.1056/NEJMoa072743 [DOI] [PubMed] [Google Scholar]
- 11. Winkelmann J, et al. Genome-wide association study of restless legs syndrome identifies common variants in three genomic regions. Nat Genet. 2007;39(8):1000–1006. doi: 10.1038/ng2099 [DOI] [PubMed] [Google Scholar]
- 12. Schormair B, et al. Identification of novel risk loci for restless legs syndrome in genome-wide association studies in individuals of European ancestry: a meta-analysis. Lancet Neurol. 2017;16(11):898–907. doi: 10.1016/S1474-4422(17)30327-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Didriksen M, et al. Large genome-wide association study identifies three novel risk variants for restless legs syndrome. Commun Biol. 2020;3(1):703. doi: 10.1038/s42003-020-01430-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Jiang YJ, et al. Genome-wide analysis identified novel susceptible genes of restless legs syndrome in migraineurs. J Headache Pain. 2022;23(1):39. doi: 10.1186/s10194-022-01409-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Edelson JL, et al. The genetic etiology of periodic leg movement in sleep. Sleep. 2022. doi: 10.1093/sleep/zsac121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Miyagawa T, et al. Variant between CPT1B and CHKB associated with susceptibility to narcolepsy. Nat Genet. 2008;40(11):1324–1328. doi: 10.1038/ng.231 [DOI] [PubMed] [Google Scholar]
- 17. Hallmayer J, et al. Narcolepsy is strongly associated with the T-cell receptor alpha locus. Nat Genet. 2009;41(6):708–711. doi: 10.1038/ng.372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Faraco J, et al. ImmunoChip study implicates antigen presentation to T cells in narcolepsy. PLoS Genet. 2013;9(2):e1003270. doi: 10.1371/journal.pgen.1003270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Han F, et al. Genome wide analysis of narcolepsy in China implicates novel immune loci and reveals changes in association prior to versus after the 2009 H1N1 influenza pandemic. PLoS Genet. 2013;9(10):e1003880. doi: 10.1371/journal.pgen.1003880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Luca G, et al. Clinical, polysomnographic and genome-wide association analyses of narcolepsy with cataplexy: a European Narcolepsy Network study. J Sleep Res. 2013;22(5):482–495. doi: 10.1111/jsr.12044 [DOI] [PubMed] [Google Scholar]
- 21. Holm A, et al. EIF3G is associated with narcolepsy across ethnicities. Eur J Hum Genet. 2015;23(11):1573–1580. doi: 10.1038/ejhg.2015.4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Jones SE, et al. Genome-wide association analyses in 128,266 individuals identifies new morningness and sleep duration loci. PLoS Genet. 2016;12(8):e1006125. doi: 10.1371/journal.pgen.1006125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lane JM, et al. Genome-wide association analyses of sleep disturbance traits identify new loci and highlight shared genetics with neuropsychiatric and metabolic traits. Nat Genet. 2017;49(2):274–281. doi: 10.1038/ng.3749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Campos AI, et al. Insights into the aetiology of snoring from observational and genetic investigations in the UK Biobank. Nat Commun. 2020;11(1):817. doi: 10.1038/s41467-020-14625-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Strausz S, et al. Genetic analysis of obstructive sleep apnoea discovers a strong association with cardiometabolic health. Eur Respir J. 2021;57(5). doi: 10.1183/13993003.03091-2020 [DOI] [PubMed] [Google Scholar]
- 26. Connor JR, et al. Iron and restless legs syndrome: treatment, genetics and pathophysiology. Sleep Med. 2017;31:61–70. doi: 10.1016/j.sleep.2016.07.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Gonzalez-Latapi P, et al. Update on restless legs syndrome: from mechanisms to treatment. Curr Neurol Neurosci Rep. 2019;19(8):54. doi: 10.1007/s11910-019-0965-4 [DOI] [PubMed] [Google Scholar]
- 28. Schormair B, et al. Reassessment of candidate gene studies for idiopathic restless legs syndrome in a large GWAS dataset of European ancestry. Sleep. 2022. doi: 10.1093/sleep/zsac098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Peyron C, et al. A mutation in a case of early onset narcolepsy and a generalized absence of hypocretin peptides in human narcoleptic brains. Nat Med. 2000;6(9):991–997. doi: 10.1038/79690 [DOI] [PubMed] [Google Scholar]
- 30. Ollila HM, et al. HLA-DPB1 and HLA class I confer risk of and protection from narcolepsy. Am J Hum Genet. 2015;96(1):136–146. doi: 10.1016/j.ajhg.2014.12.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Pelin Z, et al. HLA-DQB1*0602 homozygosity increases relative risk for narcolepsy but not disease severity in two ethnic groups. US Modafinil in Narcolepsy Multicenter Study Group. Tissue Antigens. 1998;51(1):96–100. doi: 10.1111/j.1399-0039.1998.tb02952.x [DOI] [PubMed] [Google Scholar]
- 32. Han F, et al. HLA-DQ association and allele competition in Chinese narcolepsy. Tissue Antigens. 2012;80(4):328–335. 10.1111/j.1399-0039.2012.01948.x [DOI] [PubMed] [Google Scholar]
- 33. Matsuki K, et al. Human histocompatibility leukocyte antigen (HLA) haplotype frequencies estimated from the data on HLA class I, II, and III antigens in 111 Japanese narcoleptics. J Clin Invest. 1985;76(6):2078–2083. doi: 10.1172/JCI112211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Hong SC, et al. DQB1*0301 and DQB1*0601 modulate narcolepsy susceptibility in Koreans. Hum Immunol. 2007;68(1):59–68. doi: 10.1016/j.humimm.2006.10.006 [DOI] [PubMed] [Google Scholar]
- 35. Mignot E, et al. Complex HLA-DR and -DQ interactions confer risk of narcolepsy-cataplexy in three ethnic groups. Am J Hum Genet. 2001;68(3):686–699. doi: 10.1086/318799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Alaez C, et al. Association of narcolepsy-cataplexy with HLA-DRB1 and DQB1 in Mexican patients: a relationship between HLA and gender is suggested. BMC Med Genet. 2008;9:79. doi: 10.1186/1471-2350-9-79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Gottlieb DJ, et al. Genome-wide association of sleep and circadian phenotypes. BMC Med Genet. 2007;8(Suppl 1):S9. doi: 10.1186/1471-2350-8-S1-S9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Hu Y, et al. GWAS of 89,283 individuals identifies genetic variants associated with self-reporting of being a morning person. Nat Commun. 2016;7:10448. doi: 10.1038/ncomms10448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Dashti HS, et al. Genome-wide association study identifies genetic loci for self-reported habitual sleep duration supported by accelerometer-derived estimates. Nat Commun. 2019;10(1):1100. doi: 10.1038/s41467-019-08917-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Jansen PR, et al. Genome-wide analysis of insomnia in 1,331,010 individuals identifies new risk loci and functional pathways. Nat Genet. 2019;51(3):394–403. doi: 10.1038/s41588-018-0333-3 [DOI] [PubMed] [Google Scholar]
- 41. Hammerschlag AR, et al. Genome-wide association analysis of insomnia complaints identifies risk genes and genetic overlap with psychiatric and metabolic traits. Nat Genet. 2017;49(11):1584–1592. doi: 10.1038/ng.3888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Eriksson N, et al. Web-based, participant-driven studies yield novel genetic associations for common traits. PLoS Genet. 2010;6(6):e1000993. doi: 10.1371/journal.pgen.1000993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Tung JY, et al. Efficient replication of over 180 genetic associations with self-reported medical data. PLoS One. 2011;6(8):e23473. 10.1371/journal.pone.0023473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Hoffmann TJ, et al. A large multiethnic genome-wide association study of adult body mass index identifies novel loci. Genetics. 2018;210(2):499–515. doi: 10.1534/genetics.118.301479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Hemani G, et al. The MR-Base platform supports systematic causal inference across the human phenome. Elife. 2018;7. doi: 10.7554/eLife.34408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Smith GD, et al. ‘Mendelian randomization’: can genetic epidemiology contribute to understanding environmental determinants of disease? Int J Epidemiol. 2003;32(1):1–22. doi: 10.1093/ije/dyg070 [DOI] [PubMed] [Google Scholar]
- 47. Zheng J, et al. Recent developments in Mendelian randomization studies. Curr Epidemiol Rep. 2017;4(4):330–345. doi: 10.1007/s40471-017-0128-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Emdin CA, et al. Mendelian randomization. JAMA. 2017;318(19):1925–1926. doi: 10.1001/jama.2017.17219 [DOI] [PubMed] [Google Scholar]
- 49. Tang Y, et al. Sleep deprivation worsens inflammation and delays recovery in a mouse model of colitis. Sleep Med. 2009;10(6):597–603. doi: 10.1016/j.sleep.2008.12.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Jarasvaraparn C, et al. The relationship between sleep disturbance and disease activity in pediatric patients with inflammatory bowel disease. J Pediatr Gastroenterol Nutr. 2019;68(2):237–243. doi: 10.1097/MPG.0000000000002156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Marinelli C, et al. Sleep disturbance in inflammatory bowel disease: prevalence and risk factors - a cross-sectional study. Sci Rep. 2020;10(1):507. doi: 10.1038/s41598-020-57460-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Hao G, et al. Sleep quality and disease activity in patients with inflammatory bowel disease: a systematic review and meta-analysis. Sleep Med. 2020;75:301–308. doi: 10.1016/j.sleep.2020.08.032 [DOI] [PubMed] [Google Scholar]
- 53. Chen M, et al. Differential sleep traits have no causal effect on inflammatory bowel diseases: a Mendelian randomization study. Front Pharmacol. 2021;12:763649. doi: 10.3389/fphar.2021.763649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Burgess S, et al. Mendelian randomization analysis with multiple genetic variants using summarized data. Genet Epidemiol. 2013;37(7):658–665. doi: 10.1002/gepi.21758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Bowden J, et al. Mendelian randomization with invalid instruments: effect estimation and bias detection through Egger regression. Int J Epidemiol. 2015;44(2):512–525. doi: 10.1093/ije/dyv080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Burgess S, et al. Guidelines for performing Mendelian randomization investigations. Wellcome Open Res. 2019;4:186. doi: 10.12688/wellcomeopenres.15555.2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Haycock PC, et al. Best (but oft-forgotten) practices: the design, analysis, and interpretation of Mendelian randomization studies. Am J Clin Nutr. 2016;103(4):965–978. doi: 10.3945/ajcn.115.118216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Montgomery AA, et al. Design, analysis and presentation of factorial randomised controlled trials. BMC Med Res Methodol. 2003;3:26. doi: 10.1186/1471-2288-3-26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Zheng J, et al. LD Hub: a centralized database and web interface to perform LD score regression that maximizes the potential of summary level GWAS data for SNP heritability and genetic correlation analysis. Bioinformatics. 2017;33(2):272–279. doi: 10.1093/bioinformatics/btw613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Dashti HS, et al. Polygenic risk score identifies associations between sleep duration and diseases determined from an electronic medical record biobank. Sleep. 2019;42(3). doi: 10.1093/sleep/zsy247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Henry A, et al. The relationship between sleep duration, cognition and dementia: a Mendelian randomization study. Int J Epidemiol. 2019;48(3):849–860. doi: 10.1093/ije/dyz071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Anderson EL, et al. Is disrupted sleep a risk factor for Alzheimer’s disease? Evidence from a two-sample Mendelian randomization analysis. Int J Epidemiol. 2021;50(3):817–828. doi: 10.1093/ije/dyaa183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Huang J, et al. Sleep, major depressive disorder, and Alzheimer disease: a Mendelian randomization study. Neurology. 2020;95(14):e1963–e1970. doi: 10.1212/WNL.0000000000010463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Noyce AJ, et al. The Parkinson’s disease Mendelian randomization research portal. Mov Disord. 2019;34(12):1864–1872. doi: 10.1002/mds.27873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Zhang G, et al. Daytime sleepiness might increase the risk of ALS: a 2-sample Mendelian randomization study. J Neurol. 2021;268(11):4332–4339. doi: 10.1007/s00415-021-10564-z [DOI] [PubMed] [Google Scholar]
- 66. Sun J, et al. Polygenic evidence and overlapped brain functional connectivities for the association between chronic pain and sleep disturbance. Transl Psychiatry. 2020;10(1):252. doi: 10.1038/s41398-020-00941-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Broberg M, et al. Mendelian randomization highlights insomnia as a risk factor for pain diagnoses. Sleep. 2021;44(7). doi: 10.1093/sleep/zsab025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Choi KW, et al. An exposure-wide and Mendelian randomization approach to identifying modifiable factors for the prevention of depression. Am J Psychiatry. 2020;177(10):944–954. doi: 10.1176/appi.ajp.2020.19111158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Daghlas I, et al. Genetically proxied diurnal preference, sleep timing, and risk of major depressive disorder. JAMA Psychiatry. 2021;78(8):903–910. doi: 10.1001/jamapsychiatry.2021.0959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Cai L, et al. Causal links between major depressive disorder and insomnia: a Mendelian randomisation study. Gene. 2021;768:145271. doi: 10.1016/j.gene.2020.145271 [DOI] [PubMed] [Google Scholar]
- 71. Zhou F, et al. Assessing the causal associations of insomnia with depressive symptoms and subjective well-being: a bidirectional Mendelian randomization study. Sleep Med. 2021;87:85–91. doi: 10.1016/j.sleep.2021.08.025 [DOI] [PubMed] [Google Scholar]
- 72. Gao X, et al. The bidirectional causal relationships of insomnia with five major psychiatric disorders: a Mendelian randomization study. Eur Psychiatry. 2019;60:79–85. doi: 10.1016/j.eurpsy.2019.05.004 [DOI] [PubMed] [Google Scholar]
- 73. Lane JM, et al. Genome-wide association analysis identifies novel loci for chronotype in 100,420 individuals from the UK Biobank. Nat Commun. 2016;7:10889. doi: 10.1038/ncomms10889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Dashti HS, et al. Genetics of sleep and insights into its relationship with obesity. Annu Rev Nutr. 2021;41:223–252. doi: 10.1146/annurev-nutr-082018-124258 [DOI] [PubMed] [Google Scholar]
- 75. Doherty A, et al. GWAS identifies 14 loci for device-measured physical activity and sleep duration. Nat Commun. 2018;9(1):5257. doi: 10.1038/s41467-018-07743-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Jones SE, et al. Genetic studies of accelerometer-based sleep measures yield new insights into human sleep behaviour. Nat Commun. 2019;10(1):1585. doi: 10.1038/s41467-019-09576-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Jones SE, et al. Genome-wide association analyses of chronotype in 697,828 individuals provides insights into circadian rhythms. Nat Commun. 2019;10(1):343. doi: 10.1038/s41467-018-08259-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Wang H, et al. Genome-wide association analysis of self-reported daytime sleepiness identifies 42 loci that suggest biological subtypes. Nat Commun. 2019;10(1):3503. doi: 10.1038/s41467-019-11456-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Wang J, et al. Sleep duration and adiposity in children and adults: observational and Mendelian randomization studies. Obesity (Silver Spring). 2019;27(6):1013–1022. doi: 10.1002/oby.22469 [DOI] [PubMed] [Google Scholar]
- 80. Chen W, et al. Causal effect of obstructive sleep apnea on atrial fibrillation: a Mendelian randomization study. J Am Heart Assoc. 2021;10(23):e022560. doi: 10.1161/JAHA.121.022560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Chen L, et al. Obstructive sleep apnea and atrial fibrillation: insights from a bidirectional Mendelian randomization study. BMC Med Genomics. 2022;15(1):28. doi: 10.1186/s12920-022-01180-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Yuan S, et al. An atlas on risk factors for type 2 diabetes: a wide-angled Mendelian randomisation study. Diabetologia. 2020;63(11):2359–2371. doi: 10.1007/s00125-020-05253-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Gao X, et al. Investigating causal relations between sleep-related traits and risk of type 2 diabetes mellitus: a Mendelian randomization study. Front Genet. 2020;11:607865. doi: 10.3389/fgene.2020.607865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Wang J, et al. Sleep duration and risk of diabetes: observational and Mendelian randomization studies. Prev Med. 2019;119:24–30. doi: 10.1016/j.ypmed.2018.11.019 [DOI] [PubMed] [Google Scholar]
- 85. Daghlas I, et al. Sleep duration and myocardial infarction. J Am Coll Cardiol. 2019;74(10):1304–1314. doi: 10.1016/j.jacc.2019.07.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. van Oort S, et al. Modifiable lifestyle factors and heart failure: a Mendelian randomization study. Am Heart J. 2020;227:64–73. doi: 10.1016/j.ahj.2020.06.007 [DOI] [PubMed] [Google Scholar]
- 87. Ai S, et al. Causal associations of short and long sleep durations with 12 cardiovascular diseases: linear and nonlinear Mendelian randomization analyses in UK Biobank. Eur Heart J. 2021;42(34):3349–3357. doi: 10.1093/eurheartj/ehab170 [DOI] [PubMed] [Google Scholar]
- 88. Liu X, et al. Genetically predicted insomnia in relation to 14 cardiovascular conditions and 17 cardiometabolic risk factors: a Mendelian randomization study. J Am Heart Assoc. 2021;10(15):e020187. doi: 10.1161/JAHA.120.020187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Larsson SC, et al. Genetic liability to insomnia and cardiovascular disease risk. Circulation. 2019;140(9):796–798. doi: 10.1161/CIRCULATIONAHA.119.041830 [DOI] [PubMed] [Google Scholar]
- 90. Titova OE, et al. Sleep duration and stroke: prospective cohort study and Mendelian randomization analysis. Stroke. 2020;51(11):3279–3285. doi: 10.1161/STROKEAHA.120.029902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Lu H, et al. Sleep duration and stroke: a Mendelian randomization study. Front Neurol. 2020;11:976. doi: 10.3389/fneur.2020.00976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. van Oort S, et al. Association of cardiovascular risk factors and lifestyle behaviors with hypertension: a Mendelian randomization study. Hypertension. 2020;76(6):1971–1979. doi: 10.1161/HYPERTENSIONAHA.120.15761 [DOI] [PubMed] [Google Scholar]
- 93. Richmond RC, et al. Investigating causal relations between sleep traits and risk of breast cancer in women: Mendelian randomisation study. BMJ. 2019;365:l2327. doi: 10.1136/bmj.l2327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Gao XL, et al. Obstructive sleep apnea syndrome and causal relationship with female breast cancer: a Mendelian randomization study. Aging (Albany NY). 2020;12(5):4082–4092. doi: 10.18632/aging.102725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Sun X, et al. Genetically proxied morning chronotype was associated with a reduced risk of prostate cancer. Sleep. 2021;44(10). doi: 10.1093/sleep/zsab104 [DOI] [PubMed] [Google Scholar]
- 96. Wang J, et al. Association between sleep traits and lung cancer: a Mendelian randomization study. J Immunol Res. 2021;2021:1893882. doi: 10.1155/2021/1893882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Shen J, et al. Genetic liability to insomnia and lung cancer risk: a Mendelian randomization analysis. Front Genet. 2021;12:756908. doi: 10.3389/fgene.2021.756908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Huo Z, et al. Genetically predicted insomnia and lung cancer risk: a Mendelian randomization study. Sleep Med. 2021;87:183–190. doi: 10.1016/j.sleep.2021.06.044 [DOI] [PubMed] [Google Scholar]
- 99. Wu Y, et al. GWAS of peptic ulcer disease implicates Helicobacter pylori infection, other gastrointestinal disorders and depression. Nat Commun. 2021;12(1):1146. doi: 10.1038/s41467-021-21280-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Ni J, et al. Evidence for causal effects of sleep disturbances on risk for osteoarthritis: a univariable and multivariable Mendelian randomization study. Osteoarthritis Cartilage. 2021;30(3):443–450. doi: 10.1016/j.joca.2021.11.021 [DOI] [PubMed] [Google Scholar]
- 101. Mazidi M, et al. Longer sleep duration may negatively affect renal function. Int Urol Nephrol. 2021;53(2):325–332. doi: 10.1007/s11255-020-02624-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Cullell N, et al. Sleep/wake cycle alterations as a cause of neurodegenerative diseases: a Mendelian randomization study. Neurobiol Aging. 2021;106:320 e1–320 e12. doi: 10.1016/j.neurobiolaging.2021.05.008 [DOI] [PubMed] [Google Scholar]
- 103. Iranzo A. Sleep in neurodegenerative diseases. Sleep Med Clin. 2016;11(1):1–18. doi: 10.1016/j.jsmc.2015.10.011 [DOI] [PubMed] [Google Scholar]
- 104. Schmidt C, et al. Age-related changes in sleep and circadian rhythms: impact on cognitive performance and underlying neuroanatomical networks. Front Neurol. 2012;3:118. doi: 10.3389/fneur.2012.00118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Abbott SM, et al. Chronic sleep disturbance and neural injury: links to neurodegenerative disease. Nat Sci Sleep. 2016;8:55–61. doi: 10.2147/NSS.S78947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Brzecka A, et al. Sleep disorders associated with Alzheimer’s disease: a perspective. Front Neurosci. 2018;12:330. doi: 10.3389/fnins.2018.00330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Andrews SJ, et al. Causal associations between modifiable risk factors and the Alzheimer’s phenome. Ann Neurol. 2021;89(1):54–65. doi: 10.1002/ana.25918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Billingsley KJ, et al. Genetic risk factors in Parkinson’s disease. Cell Tissue Res. 2018;373(1):9–20. doi: 10.1007/s00441-018-2817-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Saberi S, et al. Neuropathology of amyotrophic lateral sclerosis and its variants. Neurol Clin. 2015;33(4):855–876. doi: 10.1016/j.ncl.2015.07.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Al-Chalabi A, et al. The epidemiology of ALS: a conspiracy of genes, environment and time. Nat Rev Neurol. 2013;9(11):617–628. doi: 10.1038/nrneurol.2013.203 [DOI] [PubMed] [Google Scholar]
- 111. Kew JJ, et al. The relationship between abnormalities of cognitive function and cerebral activation in amyotrophic lateral sclerosis. A neuropsychological and positron emission tomography study. Brain. 1993;116(Pt 6):1399–1423. doi: 10.1093/brain/116.6.1399 [DOI] [PubMed] [Google Scholar]
- 112. Lucia D, et al. Disorders of sleep and wakefulness in amyotrophic lateral sclerosis (ALS): a systematic review. Amyotroph Lateral Scler Frontotemporal Degener. 2021;22(3–4):161–169. doi: 10.1080/21678421.2020.1844755 [DOI] [PubMed] [Google Scholar]
- 113. Liu S, et al. Excessive daytime sleepiness in Chinese patients with sporadic amyotrophic lateral sclerosis and its association with cognitive and behavioural impairments. J Neurol Neurosurg Psychiatry. 2018;89(10):1038–1043. doi: 10.1136/jnnp-2018-318810 [DOI] [PubMed] [Google Scholar]
- 114. Finan PH, et al. The association of sleep and pain: an update and a path forward. J Pain. 2013;14(12):1539–1552. doi: 10.1016/j.jpain.2013.08.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Afolalu EF, et al. Effects of sleep changes on pain-related health outcomes in the general population: a systematic review of longitudinal studies with exploratory meta-analysis. Sleep Med Rev. 2018;39:82–97. doi: 10.1016/j.smrv.2017.08.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Gasperi M, et al. Genetic and environmental influences on sleep, pain, and depression symptoms in a community sample of twins. Psychosom Med. 2017;79(6):646–654. doi: 10.1097/PSY.0000000000000456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Harvey AG. A transdiagnostic approach to treating sleep disturbance in psychiatric disorders. Cogn Behav Ther. 2009;38(Suppl 1):35–42. doi: 10.1080/16506070903033825 [DOI] [PubMed] [Google Scholar]
- 118. Walker WH II, et al. Circadian rhythm disruption and mental health. Transl Psychiatry. 2020;10(1):28. doi: 10.1038/s41398-020-0694-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Wray NR, et al. Genome-wide association analyses identify 44 risk variants and refine the genetic architecture of major depression. Nat Genet. 2018;50(5):668–681. doi: 10.1038/s41588-018-0090-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Winship IR, et al. An overview of animal models related to schizophrenia. Can J Psychiatry. 2019;64(1):5–17. doi: 10.1177/0706743718773728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Cohrs S. Sleep disturbances in patients with schizophrenia: impact and effect of antipsychotics. CNS Drugs. 2008;22(11):939–962. doi: 10.2165/00023210-200822110-00004 [DOI] [PubMed] [Google Scholar]
- 122. Ambati A, et al. Kleine-Levin syndrome is associated with birth difficulties and genetic variants in the TRANK1 gene loci. Proc Natl Acad Sci USA. 2021;118(12). doi: 10.1073/pnas.2005753118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Dolsen MR, et al. Insomnia as a transdiagnostic process in psychiatric disorders. Curr Psychiatry Rep. 2014;16(9):471. doi: 10.1007/s11920-014-0471-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Gregory AM, et al. Sleep, emotional and behavioral difficulties in children and adolescents. Sleep Med Rev. 2012;16(2):129–136. doi: 10.1016/j.smrv.2011.03.007 [DOI] [PubMed] [Google Scholar]
- 125. Nassan M, et al. Genetic evidence for a potential causal relationship between insomnia symptoms and suicidal behavior: a Mendelian randomization study. Neuropsychopharmacology. 2022;47(9):1672–1679. doi: 10.1038/s41386-022-01319-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Vasudevan AR, et al. Cardiometabolic risk assessment: an approach to the prevention of cardiovascular disease and diabetes mellitus. Clin Cornerstone. 2005;7(2–3):7–16. doi: 10.1016/s1098-3597(05)80063-8 [DOI] [PubMed] [Google Scholar]
- 127. Rangaraj VR, et al. Association between sleep deficiency and cardiometabolic disease: implications for health disparities. Sleep Med. 2016;18:19–35. doi: 10.1016/j.sleep.2015.02.535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Bouchard C. Gene-environment interactions in the etiology of obesity: defining the fundamentals. Obesity (Silver Spring). 2008;16(Suppl 3):S5–S10. doi: 10.1038/oby.2008.528 [DOI] [PubMed] [Google Scholar]
- 129. Cappuccio FP, et al. Meta-analysis of short sleep duration and obesity in children and adults. Sleep. 2008;31(5):619–626. doi: 10.1093/sleep/31.5.619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Ogilvie RP, et al. The epidemiology of sleep and obesity. Sleep Health. 2017;3(5):383–388. doi: 10.1016/j.sleh.2017.07.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Frayling TM, et al. A common variant in the FTO gene is associated with body mass index and predisposes to childhood and adult obesity. Science. 2007;316(5826):889–894. doi: 10.1126/science.1141634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. Celis-Morales C, et al. Sleep characteristics modify the association of genetic predisposition with obesity and anthropometric measurements in 119,679 UK Biobank participants. Am J Clin Nutr. 2017;105(4):980–990. doi: 10.3945/ajcn.116.147231 [DOI] [PubMed] [Google Scholar]
- 133. Cappuccio FP, et al. Sleep and cardio-metabolic disease. Curr Cardiol Rep. 2017;19(11):110. doi: 10.1007/s11886-017-0916-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Vgontzas AN, et al. Insomnia with objective short sleep duration is associated with type 2 diabetes: a population-based study. Diabetes Care. 2009;32(11):1980–1985. doi: 10.2337/dc09-0284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. Hein M, et al. Prevalence and risk factors of type 2 diabetes in insomnia sufferers: a study on 1311 individuals referred for sleep examinations. Sleep Med. 2018;46:37–45. doi: 10.1016/j.sleep.2018.02.006 [DOI] [PubMed] [Google Scholar]
- 136. Chen GC, et al. Daytime napping and risk of type 2 diabetes: a meta-analysis of prospective studies. Sleep Breath. 2018;22(3):815–824. doi: 10.1007/s11325-017-1528-z [DOI] [PubMed] [Google Scholar]
- 137. Andersson C, et al. Epidemiology of cardiovascular disease in young individuals. Nat Rev Cardiol. 2018;15(4):230–240. doi: 10.1038/nrcardio.2017.154 [DOI] [PubMed] [Google Scholar]
- 138. Roth GA, et al. Global and regional patterns in cardiovascular mortality from 1990 to 2013. Circulation. 2015;132(17):1667–1678. doi: 10.1161/CIRCULATIONAHA.114.008720 [DOI] [PubMed] [Google Scholar]
- 139. Yin J, et al. Relationship of sleep duration with all-cause mortality and cardiovascular events: a systematic review and dose-response meta-analysis of prospective cohort studies. J Am Heart Assoc. 2017;6(9):e005947. doi: 10.1161/JAHA.117.005947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140. Wang C, et al. Association of estimated sleep duration and naps with mortality and cardiovascular events: a study of 116 632 people from 21 countries. Eur Heart J. 2019;40(20):1620–1629. doi: 10.1093/eurheartj/ehy695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141. Park S, et al. Short or long sleep duration and CKD: a Mendelian randomization study. J Am Soc Nephrol. 2020;31(12):2937–2947. doi: 10.1681/ASN.2020050666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142. Li Y, et al. Assessment of the causal effects of obstructive sleep apnea on atrial fibrillation: a Mendelian randomization study. Front Cardiovasc Med. 2022;9:843681. doi: 10.3389/fcvm.2022.843681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143. Chen Y, et al. Sleep duration and the risk of cancer: a systematic review and meta-analysis including dose-response relationship. BMC Cancer. 2018;18(1):1149. doi: 10.1186/s12885-018-5025-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144. von Ruesten A, et al. Association of sleep duration with chronic diseases in the European Prospective Investigation into Cancer and Nutrition (EPIC)-Potsdam study. PLoS One. 2012;7(1):e30972. doi: 10.1371/journal.pone.0030972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145. Jiao L, et al. Sleep duration and incidence of colorectal cancer in postmenopausal women. Br J Cancer. 2013;108(1):213–221. doi: 10.1038/bjc.2012.561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146. Wang P, et al. Night-shift work, sleep duration, daytime napping, and breast cancer risk. Sleep Med. 2015;16(4):462–468. doi: 10.1016/j.sleep.2014.11.017 [DOI] [PubMed] [Google Scholar]
- 147. Luojus MK, et al. Sleep duration and incidence of lung cancer in ageing men. BMC Public Health. 2014;14:295. doi: 10.1186/1471-2458-14-295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148. Gu F, et al. Sleep duration and cancer in the NIH-AARP Diet and Health Study Cohort. PLoS One. 2016;11(9):e0161561. doi: 10.1371/journal.pone.0161561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149. Markt SC, et al. Sleep duration and disruption and prostate cancer risk: a 23-year prospective study. Cancer Epidemiol Biomarkers Prev. 2016;25(2):302–308. doi: 10.1158/1055-9965.EPI-14-1274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150. Lu Y, et al. Association between sleep duration and cancer risk: a meta-analysis of prospective cohort studies. PLoS One. 2013;8(9):e74723. doi: 10.1371/journal.pone.0074723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151. Schumacher FR, et al. Association analyses of more than 140,000 men identify 63 new prostate cancer susceptibility loci. Nat Genet. 2018;50(7):928–936. doi: 10.1038/s41588-018-0142-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152. Kukwa W, et al. Obstructive sleep apnea and cancer: effects of intermittent hypoxia? Future Oncol. 2015;11(24):3285–3298. doi: 10.2217/fon.15.216 [DOI] [PubMed] [Google Scholar]
- 153. Yap DWT, et al. The association of obstructive sleep apnea with breast cancer incidence and mortality: a systematic review and meta-analysis. J Breast Cancer. 2022;25(3):149–163. doi: 10.4048/jbc.2022.25.e11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154. Turek NF, et al. Sleep disturbances as nontraditional risk factors for development and progression of CKD: review of the evidence. Am J Kidney Dis. 2012;60(5):823–833. doi: 10.1053/j.ajkd.2012.04.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155. Cheungpasitporn W, et al. The effects of short sleep duration on proteinuria and chronic kidney disease: a systematic review and meta-analysis. Nephrol Dial Transplant. 2017;32(6):991–996. doi: 10.1093/ndt/gfw072 [DOI] [PubMed] [Google Scholar]
- 156. Bowden J, et al. Improving the visualization, interpretation and analysis of two-sample summary data Mendelian randomization via the Radial plot and Radial regression. Int J Epidemiol. 2018;47(4):1264–1278. doi: 10.1093/ije/dyy101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157. Gottlieb DJ, et al. Novel loci associated with usual sleep duration: the CHARGE Consortium Genome-Wide Association Study. Mol Psychiatry. 2015;20(10):1232–1239. doi: 10.1038/mp.2014.133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158. Nishiyama T, et al. Genome-wide association meta-analysis and Mendelian randomization analysis confirm the influence of ALDH2 on sleep durationin the Japanese population. Sleep. 2019;42(6):zsz046. doi: 10.1093/sleep/zsz046 [DOI] [PubMed] [Google Scholar]
- 159. Khera AV, et al. Genome-wide polygenic scores for common diseases identify individuals with risk equivalent to monogenic mutations. Nat Genet. 2018;50(9):1219–1224. doi: 10.1038/s41588-018-0183-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160. Wilkins AS. Canalization:a molecular genetic perspective. Bioessays. 1997;19(3):257–262. doi: 10.1002/bies.950190312 [DOI] [PubMed] [Google Scholar]