Abstract
The rpmA gene, which encodes 50S ribosomal subunit protein L27, was cloned from the extreme thermophile Aquifex aeolicus, and the protein was overexpressed and purified. Comparison of the A. aeolicus protein with its homologue from Escherichia coli by circular dichroism analysis and proton nuclear magnetic resonance spectroscopy showed that it readily adopts some structure in solution that is very stable, whereas the E. coli protein is unstructured under the same conditions. A mutant of E. coli that lacks L27 was found earlier to be impaired in the assembly and function of the 50S subunit; both defects could be corrected by expression of E. coli L27 from an extrachromosomal copy of the rpmA gene. When A. aeolicus L27 was expressed in the same mutant, an increase in the growth rate occurred and the “foreign” L27 protein was incorporated into E. coli ribosomes. However, the presence of A. aeolicus L27 did not promote 50S subunit assembly. Thus, while the A. aeolicus protein can apparently replace its E. coli homologue functionally in completed ribosomes, it does not assist in the assembly of E. coli ribosomes that otherwise lack L27. Possible explanations for this paradoxical behavior are discussed.
Protein L27 is one of the smallest and most basic polypeptides in the Escherichia coli ribosome. Previous studies of a mutant of E. coli in which the rpmA gene, which encodes L27, was deleted showed that this protein is important for both the assembly and the function of the ribosome (46). In the absence of L27, the assembly of the large or 50S ribosomal subunit is severely perturbed, resulting in the accumulation of a 40S precursor particle that is deficient also in proteins L16, L20, and L21. Although completed subunits lacking only L27 are assembled in the deletion mutant, they are impaired in peptidyltransferase activity, most likely because of a defect in the binding of aminoacyl-tRNA to the A site (46). The approximate position of protein L27 in the E. coli 50S subunit has been localized by immunoelectron microscopy to the base of the central protuberance, in the vicinity of the peptidyltransferase center (24). The exact location of L27 is still unknown, however, despite the availability of a high-resolution crystallographic structure of the 50S subunit from the archaeon Haloarcula marismortui, as this particle does not contain an L27 homologue. In the recently published 5.5-Å crystallographic structure of the Thermus thermophilus 70S ribosome, electron density has been ascribed to protein L27, but the protein has not yet been fitted to this density (51).
While the effects of L27 deletion upon peptidyltransferase and tRNA binding could be due to long-range interactions or consequences of the influence of L27 on assembly, several additional lines of evidence suggest that the protein is present at the peptidyltransferase center. For example, L27 has been affinity labeled by inhibitors of peptidyltransferase such as chloramphenicol, carbomycin, tylosin, spiramycin, and puromycin (1, 4, 27, 35, 40). Puromycin is of particular interest, as it mimics the aminoacyl moiety of the A-site tRNA and serves as a substrate for peptidyltransferase (25). This mimicry was used to advantage in the design of a transition-state analogue for peptide transfer in which puromycin represents the A-site-bound aminoacyl-tRNA (44). X-ray crystallography of this analogue complexed with the H. marismortui 50S subunit has been used to determine the location of the peptidyltransferase center (2, 28).
Further evidence for the role of L27 comes from affinity-labeling studies with a tRNAPhe derivative containing the photoreactive nucleoside 2-azidoadenosine at its 3′ terminus (39). When bound to ribosomal A or P sites, this probe cross-links predominantly to L27 (47, 49) and to nucleotides U2506 and U2585 of the 23S rRNA (48). The corresponding nucleotides in the crystallographic structure of the H. marismortui 50S subunit are located, as expected, in the vicinity of the transition-state analogue which marks the site of peptide transfer, but no proteins are seen within 20 Å of this site. These results conflict with the fact that L27 must be within 2 to 4 Å of the azido group at the 3′ terminus of the tRNA for cross-linking to occur. The presence of this protein at the peptidyltransferase center of the E. coli 50S subunit may therefore represent a significant difference between bacterial and archaeal ribosomes.
To further investigate the properties of this interesting protein, we set out to clone, overexpress, and purify protein L27 for additional structural and functional studies. Preliminary experiments with E. coli L27 indicated that it is largely unstructured in solution. We therefore decided to focus upon the homologous protein from the hyperthermophilic bacterium Aquifex aeolicus in the expectation that this protein would display greater conformational stability. This prediction was borne out by both circular dichroism (CD) and nuclear magnetic resonance (NMR) measurements. We also investigated the ability of the A. aeolicus protein to replace E. coli L27 in the E. coli ribosome in vivo. The results of this study and their possible implications for the mechanism of ribosome assembly are discussed.
MATERIALS AND METHODS
Materials.
Restriction endonucleases and T4 DNA ligase were obtained from New England Biolabs. Calf intestinal alkaline phosphatase and DNase I (grade II from bovine pancreas) were obtained from Boehringer Mannheim. Taq polymerase was obtained from Sigma-Aldrich, Inc. All enzymes were used in accordance with the manufacturer's instructions. Primers for PCR were obtained from Sigma-Genosys; those used for sequencing were the reverse sequencing and the type III or IV forward sequencing primers provided by Qiagen, Inc., for pQE-series plasmids.
Bacterial strains and plasmids.
Strains and plasmids are described in Table 1. Plasmid pSBETa was kindly provided by Hans-Henning Steinbiß of the Max-Planck-Institut für Züchtungsforschung, Cologne, Germany. A. aeolicus chromosomal DNA was a gift from Robert Huber, Universität Regensburg, Germany.
TABLE 1.
Strains and plasmids used in this work
| Strain or plasmid | Genotype | Reference or source |
|---|---|---|
| Strains | ||
| LG90 | F− Δ(lac-proAB) | 16 |
| IW312 | LG90 ΔrpmA::kan | 46 |
| XL1-Bluea | recA1 endA1 hsdR17 (F′ proAB lacIqlacZΔM15 Tn10) | 10 |
| Top10a | F− Δ(mrr-hsdRMS-mcrBC) recA1 endA1 | Invitrogen Co. |
| BL21 | F−ompT hsdS (rB− mB−) dcm gal | 38 |
| BL21(DE3) | BL21 λ(DE3) (λ lysogen encoding T7 RNA polymerase under the control of the lacUV5 promoter) | 38 |
| IW248 | BL21(DE3)(pLysS)(pETrpmA) | I. K. Wower, unpublished data |
| Plasmids | ||
| pQE70 | Ampr (LacI-regulated T5 promoter upstream of MCSb) | Qiagen |
| pREP4 | KanrlacI | Qiagen |
| pAAL27 | pQE70; A. aeolicus rpmA | This work |
| pAVM3 | pAAL27 argU | This work |
| pET3a | Ampr (T7 RNA polymerase promoter upstream of MCS) | 38 |
| pSBETa | pET3a argU | 32 |
| pETrpmA | pET3a; E. coli rpmA | Wower, unpublished |
| pLysS | Camr, T7 gene 3.5 (encoding T7 lysozyme) | 38 |
| pCR-Blunt | KanrccdB | Invitrogen |
| pEE | Ampr; pBR322 containing an E. coli chromosome fragment that includes the rplU-rpmA operon | 46 |
Only relevant markers are listed.
MCS, multiple cloning site.
Molecular biology.
Unless otherwise stated, bacteria were grown at 37°C in L broth with shaking or at 37°C on the same medium solidified with 2% (wt/vol) agar. Antibiotics were included in the media as appropriate at a concentration of 50 μg/ml (ampicillin or kanamycin) or 30 μg/ml (chloramphenicol). Plasmids for transformation or restriction analysis were prepared by the plasmid miniprep procedure (3). Those for sequencing were prepared by use of Wizard Miniprep (Promega) with modifications for automated sequencing as suggested by the manufacturer. DNA was sequenced at the University of Massachusetts DNA Sequencing Facility by automated fluorescence sequencing. Electrophoresis of DNA was performed with agarose gels in Tris-borate-EDTA buffer using standard procedures (30). PCR products and DNA fragments that had been purified on agarose gels were recovered using a Qiaex II gel extraction kit (Qiagen). Plasmids were introduced into cells made competent with calcium chloride by standard procedures (30), except for the simultaneous transformation of strain IW312 with both plasmids pAVM3 and pREP4; the latter was done using an Eppendorf model 2510 electroporator with cells made electrocompetent according to the manufacturer's instructions.
Electrophoresis of proteins.
Gel electrophoresis of proteins was carried out with an SE250 Mighty Small II gel apparatus (Hoefer Scientific Instruments). Sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) of proteins was performed as described previously (31). Extraction and electrophoresis of ribosomal proteins by two-dimensional (2D) PAGE were done by the procedure of Butler and Wild (11). For extraction of ribosomal proteins from whole cells, cell pellets were first resuspended in 1 ml of 10 mM Tris-HCl (pH 7.4)–15 mM magnesium acetate–60 mM KCl–7 mM β-mercaptoethanol. For extraction of proteins from ribonucleoprotein (RNP) particles obtained by sucrose density gradient centrifugation, the RNPs were sedimented, resuspended in the centrifugation buffer but without sucrose, and resedimented. The final pellet was dissolved in 8 M urea containing 1% β-mercaptoethanol prior to extraction. Protein concentrations were determined using the Bio-Rad Laboratories Protein Assay Reagent with bovine serum albumin as a standard.
Cloning of the A. aeolicus rpmA gene.
The A. aeolicus rpmA gene was amplified by PCR using Taq polymerase and A. aeolicus chromosomal DNA (concentration, 50 nM) as a template. The forward and reverse primers were 5′-GAGGTGTGAAACATGGCAAGTAAAGC and 5′-CGAGAAACAAGCTTAGTGAACTTTTCC, respectively; bases in bold type differ from the A. aeolicus chromosomal sequence (13), NlaIII and HindIII restriction sites are underlined, and the initiation and stop codons are in italic type. A single product of about the expected length (321 bp) was obtained. The product was gel purified, digested with both NlaIII and HindIII, and ligated into expression vector pQE70 that had been cleaved with SphI and HindIII. SphI and NlaIII produce compatible termini. The use of an NlaIII site (rather than an SphI site) in the PCR product allowed the identity of the alanine codon in the second position of the gene to be preserved. The resulting plasmids were introduced into recA mutant strain XL1-Blue. Plasmids were isolated from transformants, and the presence of the insert was confirmed by restriction digestion analysis. Both strands of one such isolate, designated pAAL27, were sequenced to confirm that the gene had been amplified and cloned without error. The A. aeolicus rpmA gene in pAAL27 is downstream of an isopropyl-β-d-galactopyranoside (IPTG)-inducible T5 promoter and a Shine-Dalgarno sequence. The T5 promoter is recognized by the E. coli host polymerase, and two lac operator sequences in pAAL27 allow repression of transcription by the lacI repressor protein encoded on a separate plasmid, pREP4.
Protein expression.
To test protein expression, strain XL1-Blue was transformed with both pAAL27 and pREP4, and the synthesis of A. aeolicus L27 was induced in early-log-phase cultures by the addition of IPTG. Although the presence of the inducer slowed growth, it did not result in significant production of the A. aeolicus protein, as judged by one-dimensional (1D) SDS-PAGE of whole-cell extracts. Inspection of the A. aeolicus rpmA coding sequence revealed that the poor expression of A. aeolicus L27 was most likely due to the presence of eight AGG or AGA arginine codons, which are rare in E. coli but common in A. aeolicus. Such codons are known to be particularly deleterious to protein expression in E. coli (7, 12, 18, 29, 37, 52). The simplest solution was to increase the copy number of the argU gene, which encodes a tRNA that, when overproduced from a plasmid, can decode both types of rare Arg codons (17, 36).
Subcloning of the argU gene.
The argU gene, together with its promoter and terminator, were excised from pSBETa with BanI and StyI to yield a 568-bp fragment, and overhangs were filled in with T4 DNA polymerase. Attempts to ligate this blunt-ended fragment into the XbaI site of pAAL27, also treated with DNA polymerase, were unsuccessful. Therefore, the fragment was cloned into the StuI site of plasmid pCR-Blunt and then excised as a 650-bp SpeI-XbaI fragment. Since SpeI and XbaI produce compatible termini, this fragment could be ligated into XbaI-cleaved pAAL27. Introduction of the argU gene into pAAL27 resulted in copious expression of A. aeolicus L27 upon induction with IPTG. One clone, pAVM3, which contained the rpmA gene in a counterclockwise orientation, was chosen for further study.
Sucrose density gradient centrifugation.
Cultures were grown to an A600 of 0.5, chilled briefly on ice, and harvested by centrifugation. Cell pellets were washed in 0.5 volume of 20 mM HEPES-KOH (pH 7.8 at 0°C)–6 mM MgCl2–100 mM NaCl and resedimented. After resuspension in the same buffer containing 16% (wt/vol) sucrose, cells were lysed by the lysozyme freeze-thaw method as described by Bommer et al. (6), but with an extra freeze-thaw step. Lysates were clarified at 12,000 × g for 1 h at 4°C, and 1.5 to 3 A260 units were loaded onto 15 to 30% (wt/vol) sucrose density gradients made in 20 mM HEPES-KOH (pH 7.5 at 0°C)–10 mM MgCl2–150 mM NH4Cl–2 mM spermidine–0.05 mM spermine–4 mM β-mercaptoethanol. Centrifugation was done at 80,000 × g for 20 h in a Beckman SW41 rotor. For isolation of 70S ribosomes to be analyzed for protein content, up to 100 A260 units were loaded per gradient and centrifugation was done at 95,000 × g for 19 h in a Beckman SW28 rotor. Gradients were pumped out of the tubes with a displacing solution of 40% (wt/vol) sucrose containing uracil (40 μg/ml) to help locate the bottom of the gradients. Absorbance profiles were monitored at 254 nm using an ISCO UA-5 absorbance monitor.
Purification of protein L27 from E. coli.
Protein L27 from E. coli was isolated from cultures of strain IW248 in inclusion bodies (50), except that the cation-exchange resin was SP-Sephadex C-25 (Pharmacia Fine Chemicals, Inc) and the protein was eluted with a linear gradient of 0.2 to 0.7 M LiCl. Fractions containing pure L27, as judged by SDS-PAGE, were concentrated in an Amicon stirred cell under nitrogen pressure using a YM3 ultrafiltration membrane and then stored at −20°C.
Purification of protein L27 from A. aeolicus.
For purification of A. aeolicus L27, it was found necessary to use ompT mutant strain BL21 rather than XL1-Blue to prevent proteolytic degradation of the protein. Strain BL21 containing pAVM3 and pREP4 was grown with kanamycin and ampicillin and induced as described above for strain IW248. The procedure for purification was the same as that for the E. coli protein, but with the following modifications. The use of β-mercaptoethanol was unnecessary, since A. aeolicus L27 does not contain cysteine. Unlike E. coli L27, the A. aeolicus protein could not be solubilized from inclusion bodies with 6 M urea, but prewashing with 4 M urea removed many contaminating proteins. The A. aeolicus protein was then solubilized with 6 M guanidine HCl and dialyzed exhaustively into 6 M urea prior to cation-exchange chromatography with a 0.3 to 0.5 M linear gradient of LiCl. The resulting L27 preparation was judged greater than 98% pure by analysis of the staining pattern after SDS-PAGE by densitometry.
Renaturation of protein L27 from E. coli and A. aeolicus.
For CD and 1D NMR analyses, L27 preparations obtained by cation-exchange chromatography were renatured by exhaustive dialysis against 1 mM sodium phosphate (pH 7) (4°C) with 1 mM β-mercaptoethanol for the E. coli protein. For 2D heteronuclear NMR analysis, protein was prepared from cells grown in M9 medium supplemented with 15NH4SO4 (Cambridge Isotopes) and then refolded by the same procedure. Although several alternative treatments were tested, none altered the resulting CD spectra.
CD spectroscopy.
CD spectroscopy was performed using a J-715 spectropolarimeter (Jasco). The scans were recorded in a cell with a 0.1-cm path length at 1-nm intervals and at a rate of 10 nm/min in the range of 185 to 260 nm with a sensitivity of 10 to 20 millidegrees and a response time of 1 s. Scans were repeated 5 to 10 times, averaged, and further processed. Sample concentrations were between 0.04 and 0.25 mg/ml.
NMR spectroscopy.
NMR spectroscopy was performed using a 500-MHz Bruker NMR spectrometer; sample concentration and preparation were the same as those used for CD measurements. For Fourier-transform 1H NMR spectroscopy, a typical accumulation required at least 1,024 scans with residual water suppression by the gated irradiation method (19, 34). Tetramethylsilane phosphate was used as an internal chemical shift standard. Spectra were obtained at both 20 and 80°C. For 2D NMR spectroscopy of 15N-labeled L27, a heteronuclear single-quantum coherence experiment was performed (5).
RESULTS
Complementation.
Strain IW312 lacks E. coli L27 owing to the replacement of the gene (rpmA) that encodes it by a gene conferring kanamycin resistance (46). Plasmid pAVM3 directs the expression of A. aeolicus L27 when cells harboring the plasmid are induced with IPTG. Attempts to introduce pAVM3 into strain IW312 were unsuccessful, despite the fact that the parent plasmid, pQE70, could be easily introduced. Although the plates did not contain inducer, we suspected that leaky synthesis of A. aeolicus L27 was sufficiently high to be deleterious to growth of the mutant. In this case, the host did not contain the repressor plasmid, pREP4, since the selectable marker on pREP4 is Kanr and strain IW312 is already kanamycin resistant. However, simultaneous transformation of the mutant with both plasmids did result in viable transformants. Plasmids were isolated from three of these and subjected to restriction digestion analysis; all three were shown to contain pREP4 in addition to pAVM3.
One such isolate was grown on solid medium at a number of different IPTG concentrations (data not shown). A control strain, IW312(pQE70), grew equally well at all IPTG concentrations from 0 to 1 mM. The test strain that contained the A. aeolicus rpmA gene, IW312(pAVM3)(pREP4), grew faster than the control on plates without IPTG, suggesting that there was still some leaky expression of A. aeolicus L27. Growth increased further with 0.01 mM IPTG but progressively slowed in the range of 0.05 to 0.1 mM, with no growth at 0.5 to 1 mM, indicating again that excess expression of the A. aeolicus protein is detrimental to cell growth. Moderate expression of A. aeolicus L27, however, can compensate for the lack of the homologous E. coli protein.
These results were confirmed with liquid medium (Table 2). The strain containing the A. aeolicus rpmA gene again grew faster (doubling time, 76 min) than the control (doubling time, 105 min) in the absence of inducer. A further increase in growth rate was observed with 0.005 to 0.03 mM IPTG, while the highest concentration of IPTG tested (0.04 mM) decreased the growth rate. An optimal concentration of 0.01 mM IPTG was therefore adopted for subsequent experiments. Although A. aeolicus L27 was able to partially complement the missing E. coli L27, a greater improvement in growth was obtained when the E. coli protein was expressed from plasmid pEE (doubling time, 48 min). However, neither of the IW312 derivatives grew as fast as the wild-type strain, LG90 (doubling time, 27 min).
TABLE 2.
Effect of expression of the L27 protein on the rate of growth of strain IW312
| Strain | IPTG (mM) | L27 protein expressed from the plasmid | Doubling time (min) |
|---|---|---|---|
| IW312(pQE70) | 0 | 105 | |
| IW312(pAVM3)(pREP4) | 0 | A. aeolicus L27 | 76 |
| 0.005 | A. aeolicus L27 | 72 | |
| 0.01 | A. aeolicus L27 | 70 | |
| 0.02 | A. aeolicus L27 | 70 | |
| 0.03 | A. aeolicus L27 | 72 | |
| 0.04 | A. aeolicus L27 | 78 | |
| IW312(pEE) | 0 | E. coli L27 | 48 |
| LG90 (wild type) | 0 | 27 |
Protein expression.
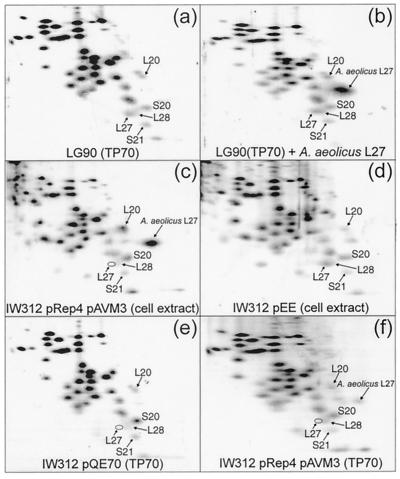
Protein expression was analyzed by 2D PAGE. Proteins extracted from the 70S ribosomes of wild-type strain LG90 (Fig. 1a) yielded the expected pattern of spots, including that of L27. When purified A. aeolicus L27 was mixed with this extract, a prominent additional spot corresponding to heterologous L27 was observed in the gel pattern (Fig. 1b). Figure 1c shows the pattern of ribosomal proteins in whole-cell extracts of strain IW312(pAVM3)(pREP4) grown in the presence of inducer. As anticipated, E. coli L27 is absent, while a spot corresponding to A. aeolicus L27 is clearly visible. When IW312 contains instead plasmid pEE, which expresses E. coli L27, the spot corresponding to E. coli L27 is restored (Fig. 1d). The contents of proteins in 70S ribosomes from strains IW312(pQE70) and IW312(pAVM3)(pREP4) grown in the presence of inducer are shown in Fig. 1e and f, respectively. As expected, the 70S ribosomes of both strains lack E. coli L27, but the latter now contain A. aeolicus L27. These results demonstrate that the heterologous protein can be assembled into E. coli ribosomes.
FIG. 1.
2D electrophoresis of ribosomal proteins. Total proteins were extracted from 70S ribosomes of strain LG90 (TP70) (a), 70S ribosomes of strain LG90 mixed with purified A. aeolicus L27 (b), whole cells of strain IW312(pAVM3)(pREP4) grown with IPTG (c), whole cells of strain IW312(pEE) (d), 70S ribosomes of strain IW312(pQE70) (e), and 70S ribosomes of strain IW312(pAVM3)(pREP4) grown with IPTG (f). The positions of both L27 proteins are indicated, along with the positions of some adjacent proteins for orientation.
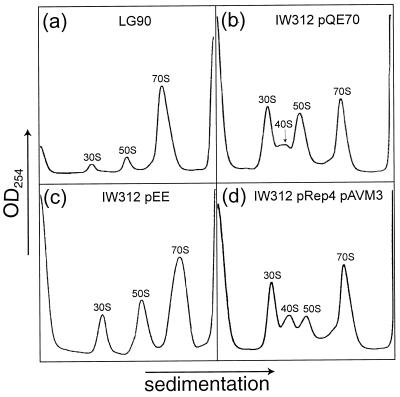
Analysis of ribosome assembly in the presence of A. aeolicus L27.
Cell extracts from different strains were centrifuged through sucrose density gradients, and the resulting absorbance profiles were compared. The wild-type parent strain, LG90, shows a normal profile, with large amounts of 70S ribosomes and small peaks corresponding to 30S and 50S subunits (Fig. 2a). Owing to the absence of L27 in strain IW312, a 40S precursor to the 50S ribosomal subunits accumulates, so that the relative amount of completed 70S is reduced (46). This finding is shown for strain IW312 containing control plasmid pQE70 (Fig. 2b; profiles from a culture without the plasmid were identical). Resupply of the E. coli L27 protein from plasmid pEE largely removes this defect (Fig. 2c). However, when the A. aeolicus protein is supplied from plasmid pAVM3 (Fig. 2d), assembly seems even more perturbed than with no L27 at all, with a smaller peak of the 50S subunits relative to the 40S precursor. This finding could, in theory, reflect a greater propensity of 50S subunits to associate with 30S subunits to form 70S couples. The result would be a corresponding increase in the 70S material and a decrease in the 30S peak. However, such a redistribution is not apparent, suggesting that the shortage of free 50S subunits in strain IW312(pAVM3)(pREP4) is due to a decrease in their net synthesis rather than to increased sequestration into 70S couples.
FIG. 2.
Sucrose density gradient absorbance profiles. Ribosomal particles were from strains grown with IPTG. The positions of the 70S ribosomes, the 30S and 50S subunits, and the 40S precursor are indicated.
Since this comparison is complicated by poor resolution of the 30S, 40S, and 50S peaks, a second set of gradients were centrifuged for longer times (data not shown). Analysis of the peak areas confirmed that the ratio of 50S material to 30S material in strain IW312(pQE70) was one-third lower than that in LG90 and that the ratio in IW312(pAVM3)(pREP4) was two-thirds lower. This result can be explained by the presence of a slowly sedimenting precursor to the 50S subunits in the 30S peak or by the breakdown of a precursor to the 50S subunits. Either way, expression of the A. aeolicus protein does not improve 50S subunit assembly in the mutant. Thus, the expression of A. aeolicus L27 presumably increases the rate of growth of the mutant by improving the function of the otherwise L27-deficient 50S subunits rather than by aiding their assembly.
Structural comparison of L27 from E. coli and A. aeolicus.
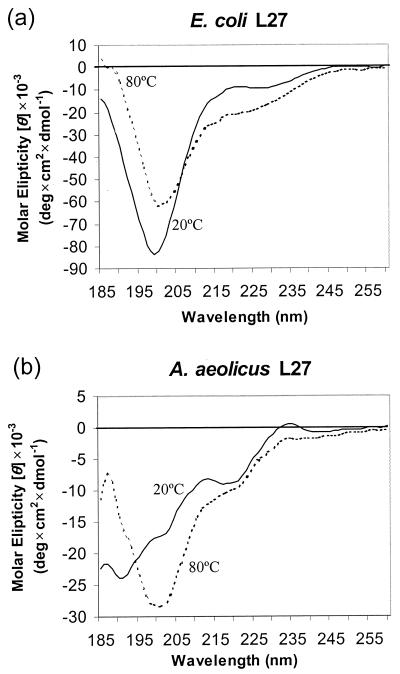
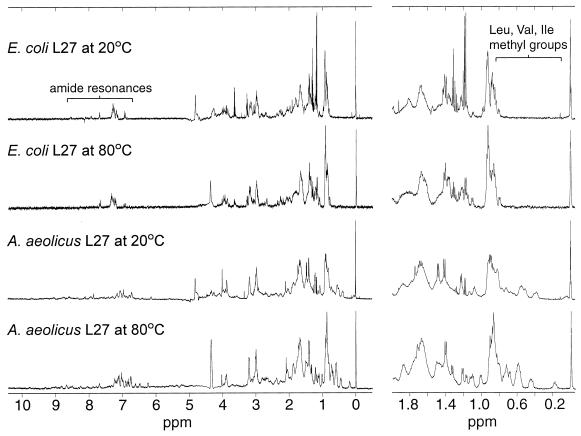
E. coli and A. aeolicus L27 proteins were purified and renatured as detailed in Materials and Methods and subjected to CD and NMR analyses. The CD spectrum of E. coli L27 (Fig. 3a) indicates that the protein is unstructured. This inference was reinforced by 2D heteronuclear NMR analysis of a 15N-labeled sample of the E. coli protein, which demonstrated that there was no dispersion of the backbone amide or side-chain amine resonances (data not shown). The CD spectrum of A. aeolicus L27 is clearly different from that of E. coli L27 at 20°C but resembles that of the E. coli protein at 80°C. Together, these observations suggest that A. aeolicus L27 may be at least partially structured, a conclusion that was confirmed by 1D proton NMR analysis. Figure 4 shows that there are several striking differences between the NMR spectra of the E. coli and A. aeolicus proteins. In particular, the methyl groups of the isoleucine, valine, and leucine side chains, at 0 to 1.0 ppm, and the amide resonances, at 6.8 to 9.8 ppm, are much more dispersed in A. aeolicus L27 than in the E. coli protein. The NMR spectrum of the E. coli protein at 80°C differed very little from that at 20°C. In contrast, the dispersion of nonpolar side chain resonances in A. aeolicus L27 was reduced but nevertheless still showed evidence of structure.
FIG. 3.
CD spectra of E. coli L27 and A. aeolicus L27.
FIG. 4.
Proton NMR spectra of E. coli L27 and A. aeolicus L27. Resonances that show clear differences in structure between the two proteins are indicated by brackets.
DISCUSSION
The effects of heterologous protein expression on ribosome assembly in an E. coli ribosomal protein deletion mutant have not previously been reported. In two earlier studies, heterologous ribosomal proteins were expressed in the presence of a truncated (33) or undersynthesized (22) E. coli ribosomal protein, but the consequences for ribosome assembly were not investigated. Most of the data on the ability of heterologous proteins to assemble into E. coli ribosomes derives from studies in which the heterologous protein is expressed in the presence of its E. coli counterpart and so must compete for assembly. Although there have been many such studies, most of the proteins studied so far, from all three phylogenetic domains, seem to compete efficiently with their E. coli homologues even when expressed at low levels. Some striking examples are found among the chloroplast ribosomal proteins that have been expressed in E. coli. For instance, overexpression in E. coli of a chloroplast S18 protein, which has N- and C-terminal extensions that more than double the size of this protein compared to that of E. coli, resulted in incorporation of the protein into 30 to 40% of the ribosomes (43). Another chloroplast protein (L23), which has only 26% identity in amino acid sequence to its E. coli homologue, was also shown to assemble (8). A more extreme example is provided by the chloroplast protein Psrp-1, which has no E. coli counterpart. When expressed at a low level in E. coli, about 84% of this protein was assembled, with the result that about half of the ribosomes contained the foreign protein (9). Only a few examples exist so far of heterologous proteins that fail to assemble into E. coli ribosomes (21, 45) or that assemble to form inactive ribosomes (43). Thus, many ribosomal proteins have proven to be interchangeable across wide phylogenetic distances, indicating a degree of structural and functional conservation similar to that of rRNA.
In many cases, the incorporation of heterologous proteins into polysomes has been demonstrated, suggesting that the ribosomes which contain them are functional. In one study (42), it was shown not only that the human and H. marismortui homologues of E. coli ribosomal protein L2 are incorporated into E. coli polysomes but also that the resulting hybrid ribosomes are as active in in vitro protein synthesis as wild-type E. coli ribosomes. This is a surprising result, given that protein L2 is one of the most important for full peptidyltransferase activity (14, 20) and that the two homologous proteins exhibit only 30 and 36% amino acid identity with the E. coli protein.
Against this background, the role of A. aeolicus L27 in E. coli appears to be somewhat paradoxical in that its incorporation appears to improve the function of 50S subunits that lack E. coli L27 but, at the same time, impairs their assembly even in the absence of a competing homologue. There are several possible explanations for the poor incorporation of A. aeolicus L27 into 50S subunits. Although its amino acid sequence is 60% identical to that of its E. coli homologue, it is possible that differences occur in portions of the polypeptide that are critical for assembly of this protein in E. coli. To assess the extent of phylogenetic variation in L27, sequences related to that of E. coli L27 were obtained from GenBank. Thirty-five bacterial and 22 eukaryotic sequences were obtained. No archaeal homologues were found, and the majority of the eukaryotic sequences are known to be from organelles. An alignment of the E. coli and A. aeolicus L27 sequences (Fig. 5) revealed that 16 amino acids in the A. aeolicus sequence are neither identical nor similar to those in the E. coli sequence. An alignment of all bacterial sequences showed that only three of these are unique to A. aeolicus. These are Ala 5, Tyr 67, and Pro 83. While 9 and 12 different amino acids are seen at the latter two positions, respectively, Ala 5 is unique to A. aeolicus; all other bacterial sequences have lysine, barring two arginines. All eukaryotic sequences also have lysine or arginine at this position, with the exceptions of one histidine and one alanine. The presence of a positively charged side group at this position might be required for assembly in E. coli (and other organisms). Alternatively, it is possible that the 15-amino-acid C-terminal extension of the A. aeolicus protein (relative to E. coli L27) impedes its assembly into E. coli ribosomes. A third possibility is that A. aeolicus L27 may be too structured in its unassembled state to be efficiently incorporated into the subunit structure. The E. coli protein is one of the least structured of the E. coli ribosomal proteins in solution (15), and it is possible that its assembly depends upon such flexibility. The A. aeolicus protein, on the other hand, readily adopts structure in solution that is very robust. As such, it may lack the plasticity necessary for assembly.
FIG. 5.
Comparison of the amino acid sequences of L27 from E. coli and A. aeolicus. Amino acids that are identical in the two sequences are indicated by asterisks, very similar ones are indicated by colons, similar ones are indicated by periods, and dissimilar ones are left unmarked. Ala 5, Tyr 67, and Pro 83 in the A. aeolicus sequence are highlighted in bold and underlined. Conservation in 35 bacterial sequences is represented below the sequences by a histogram, where the height of the bars reflects the degree of conservation. Positions in the A. aeolicus sequence that are dissimilar from those in the E. coli sequence are also indicated by a caret below the corresponding histogram bar. Alignments were performed with ClustalX, version 1.81 (41).
The L27 proteins from E. coli and A. aeolicus had very different CD spectra, and each one was essentially invariant, despite the use of several different renaturation protocols. For E. coli L27, this appears to be the case because the isolated protein is not structured at all, while A. aeolicus L27 seems to readily adopt some structure that is highly stable and independent of the renaturation procedure. For the E. coli protein, the lack of structure was confirmed by 2D heteronuclear NMR analysis. However, there remained a possibility that suitable renaturation conditions had not been found, since the protein was purified under denaturing conditions. Dijk and coworkers extracted E. coli L27 from ribosomes using mild “nondenaturing” conditions, so that the resulting protein should have been as close to its native folded state as possible. The CD spectrum that they obtained for their protein (15) was very similar to ours, although they predicted that it contained 50% β-sheet structure. Prediction of secondary structure from the CD spectrum was not attempted in the present study for two reasons. First, such predictions can be difficult to perform with confidence. Second, protein concentrations in the present study were estimated by the Bradford assay, while more accurate quantitation would be required to allow structural prediction. Nonetheless, CD spectroscopy is quite useful for comparing the degrees of structure in different preparations of related proteins, as was done here. The proton NMR spectra for the proteins measured by Littlechild and collaborators (23) and Morrison et al. (26) were also similar to ours, although they concluded that this technique could not entirely discount the presence of structure. However, given the similarity of the CD and 1D proton NMR spectra, together with the 2D NMR spectrum, we conclude that E. coli L27 is unstructured in solution regardless of the method of isolation.
The fact that the A. aeolicus protein gives a CD spectrum that is different from that of E. coli L27 suggests that it is structured in solution to at least some extent. This idea is reinforced by the observation that after thermal denaturation at 80°C, its spectrum resembles that of E. coli L27 at 20°C. Moreover, proton NMR analysis shows clear evidence of structure, some of which persists even at 80°C. As such, the A. aeolicus protein is a much better candidate for physical and structural studies, such as X-ray crystallography, than its E. coli counterpart. Efforts are also under way to identify the amino acids within L27 that are cross-linked from the acceptor terminus of tRNA. This information will define more precisely the proposed juxtaposition of the tRNA with protein L27 at the peptidyltransferase center.
ACKNOWLEDGMENTS
We are grateful to Ken Rotondi for assistance with one-dimensional NMR and to Muppulla Sukumar for performing two-dimensional NMR.
This work was funded by NIH grant GM22807, NSF grant MCB-9818051, and PRF award 34868-AC4 from the American Chemical Society. A.V.M. was supported by National Research Service award T32 GM08515 from the NIH.
REFERENCES
- 1.Arevalo M A, Tejedor F, Polo F, Ballesta J P. Synthesis and biological activity of photoactive derivatives of erythromycin. J Med Chem. 1989;32:2200–2204. doi: 10.1021/jm00129a027. [DOI] [PubMed] [Google Scholar]
- 2.Ban N, Nissen P, Hansen J, Moore P B, Steitz T A. The complete atomic structure of the large ribosomal subunit at 2.4 Å resolution. Science. 2000;289:905–920. doi: 10.1126/science.289.5481.905. [DOI] [PubMed] [Google Scholar]
- 3.Birnboim H C, Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979;7:1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bischof O, Urlaub H, Kruft V, Wittman-Liebold B. Peptide environment of the peptidyl transferase center from Escherichia coli 70 S ribosomes as determined by thermoaffinity labeling with dihydrospiramycin. J Biol Chem. 1995;270:23060–23064. doi: 10.1074/jbc.270.39.23060. [DOI] [PubMed] [Google Scholar]
- 5.Bodenhausen G, Ruben D J. Natural abundance nitrogen-15 NMR by enhanced heteronuclear spectroscopy. Chem Phys Lett. 1980;69:185–189. [Google Scholar]
- 6.Bommer U, Burkhardt N, Jünemann R, Spahn C, Triana-Alonso F, Nierhaus K. Ribosomes and polysomes. In: Graham J, Richwood D, editors. Subcellular fractionation—a practical approach. Washington, D.C.: IRL Press; 1997. pp. 271–301. [Google Scholar]
- 7.Bonekamp F, Jensen K F. The AGG codon is translated slowly in E. coli even at very low expression levels. Nucleic Acids Res. 1988;16:3013–3024. doi: 10.1093/nar/16.7.3013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bubunenko M G, Schmidt J, Subramanian A R. Protein substitution in chloroplast ribosome evolution. A eukaryotic cytosolic protein has replaced its organelle homologue (L23) in spinach. J Mol Biol. 1994;240:28–41. doi: 10.1006/jmbi.1994.1415. [DOI] [PubMed] [Google Scholar]
- 9.Bubunenko M G, Subramanian A R. Recognition of novel and divergent higher plant chloroplast ribosomal proteins by Escherichia coli ribosome during in vivo assembly. J Biol Chem. 1994;269:18223–18231. [PubMed] [Google Scholar]
- 10.Bullock W O, Fernandez J M, Short J M. A high efficiency plasmid transforming recA Escherichia coli strain with beta-galactosidase selection. BioTechniques. 1987;5:376. [Google Scholar]
- 11.Butler P D, Wild D G. Ribosomal protein synthesis by a mutant of Escherichia coli. Eur J Biochem. 1984;144:649–654. doi: 10.1111/j.1432-1033.1984.tb08514.x. [DOI] [PubMed] [Google Scholar]
- 12.Day A J, Aplin R T, Willis A C. Overexpression, purification, and refolding of link module from human TSG-6 in Escherichia coli: effect of temperature, media, and mutagenesis on lysine misincorporation at arginine AGA codons. Protein Expr Purif. 1996;8:1–16. doi: 10.1006/prep.1996.0068. [DOI] [PubMed] [Google Scholar]
- 13.Deckert G, Warren P V, Gaasterland T, Young W G, Lenox A L, Graham D E, Overbeek R, Snead M A, Keller M, Aujay M, Huber R, Feldman R A, Short J M, Olsen G J, Swanson R V. The complete genome of the hyperthermophilic bacterium Aquifex aeolicus. Nature. 1998;392:353–358. doi: 10.1038/32831. [DOI] [PubMed] [Google Scholar]
- 14.Diedrich G, Spahn C M, Stelzl U, Schäfer M A, Wooten T, Bochkariov D E, Cooperman B S, Traut R R, Nierhaus K H. Ribosomal protein L2 is involved in the association of the ribosomal subunits, tRNA binding to A and P sites and peptidyl transfer. EMBO J. 2000;19:5241–5250. doi: 10.1093/emboj/19.19.5241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dijk J, Littlechild J A, Freund A M, Pouyet J, Daune M, Provencher S W. The secondary structure of salt-extracted ribosomal proteins from Escherichia coli as studied by circular dichroic spectroscopy. Biochim Biophys Acta. 1986;874:227–234. doi: 10.1016/0167-4838(86)90122-6. [DOI] [PubMed] [Google Scholar]
- 16.Guarente L, Ptashne M. Fusion of Escherichia coli lacZ to the cytochrome c gene of Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1981;78:2199–2203. doi: 10.1073/pnas.78.4.2199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gurskii Y, Marimont N, Bibilashvili R. The effect of intracellular concentrations of tRNAs corresponding to rare arginine codons AGG and AGA on gene expression in Escherichia coli. Mol Biol. 1992;26:719–723. [PubMed] [Google Scholar]
- 18.Hu X, Shi Q, Yang T, Jackowski G. Specific replacement of consecutive AGG codons results in high-level expression of human cardiac troponin T in Escherichia coli. Protein Expr Purif. 1996;7:289–293. doi: 10.1006/prep.1996.0041. [DOI] [PubMed] [Google Scholar]
- 19.Hwang T L, Shaka A J. Water suppression that works. Excitation sculpting using arbitrary wave-forms and pulsed-field gradients. J Magn Reson Ser A. 1995;112:275–279. [Google Scholar]
- 20.Khaitovich P, Mankin A S, Green R, Lancaster L, Noller H F. Characterization of functionally active subribosomal particles from Thermus aquaticus. Proc Natl Acad Sci USA. 1999;96:85–90. doi: 10.1073/pnas.96.1.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Köpke A K, Paulke C, Gewitz H S. Overexpression of the methanococcal ribosomal protein L12 in Escherichia coli and its incorporation into halobacterial 50 S subunits yielding active ribosomes. J Biol Chem. 1990;265:6436–6440. [PubMed] [Google Scholar]
- 22.Li Y, Huff M O, Hanic-Joyce P J, Ellis S R. Derivatives of the yeast mitochondrial ribosomal protein MrpS28 replace ribosomal protein S15 as functional components of the Escherichia coli ribosome. J Mol Biol. 1993;233:606–614. doi: 10.1006/jmbi.1993.1539. [DOI] [PubMed] [Google Scholar]
- 23.Littlechild J, Malcolm A, Paterakis K, Ackermann I, Dijk J. The tertiary structure of salt-extracted ribosomal proteins from Escherichia coli as studied by proton magnetic resonance spectroscopy and limited proteolysis experiments. Biochim Biophys Acta. 1987;913:245–255. doi: 10.1016/0167-4838(87)90336-0. [DOI] [PubMed] [Google Scholar]
- 24.Lotti M, Stöffler-Meilicke M, Stöffler G. Localization of ribosomal protein L27 at the peptidyl transferase centre of the 50 S subunit, as determined by immuno-electron microscopy. Mol Gen Genet. 1987;210:498–503. doi: 10.1007/BF00327203. [DOI] [PubMed] [Google Scholar]
- 25.Monroe R E, Marcker K A. Ribosome-catalysed reaction of puromycin with formylmethionine-containing oligonucleotide. J Mol Biol. 1967;25:347–350. doi: 10.1016/0022-2836(67)90146-5. [DOI] [PubMed] [Google Scholar]
- 26.Morrison C A, Bradbury E M, Littlechild J, Dijk J. Proton magnetic resonance studies to compare Escherichia coli ribosomal proteins prepared by two different methods. FEBS Lett. 1977;83:348–352. doi: 10.1016/0014-5793(77)81038-7. [DOI] [PubMed] [Google Scholar]
- 27.Nicholson A W, Cooperman B S. Photoaffinity labeling of Escherichia coli ribosomes with an aryl azide analogue of puromycin. FEBS Lett. 1978;90:203–208. doi: 10.1016/0014-5793(78)80369-x. [DOI] [PubMed] [Google Scholar]
- 28.Nissen P, Hansen J, Ban N, Moore P B, Steitz T A. The structural basis of ribosome activity in peptide bond synthesis. Science. 2000;289:920–930. doi: 10.1126/science.289.5481.920. [DOI] [PubMed] [Google Scholar]
- 29.Rosenberg A H, Goldman E, Dunn J J, Studier F W, Zubay G. Effects of consecutive AGG codons on translation in Escherichia coli, demonstrated with a versatile codon test system. J Bacteriol. 1993;175:716–722. doi: 10.1128/jb.175.3.716-722.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 31.Schägger H, von Jagow G. Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal Biochem. 1987;166:368–379. doi: 10.1016/0003-2697(87)90587-2. [DOI] [PubMed] [Google Scholar]
- 32.Schenk P M, Baumann S, Mattes R, Steinbiss H H. Improved high-level expression system for eukaryotic genes in Escherichia coli using T7 RNA polymerase and rare Arg tRNAs. BioTechniques. 1995;19:196–198. , 200. [PubMed] [Google Scholar]
- 33.Schnier J, Thamm S, Lurz R, Hussain A, Faist G, Dobrinski B. Cloning and characterization of a gene from Rhizobium meliloti 2011 coding for ribosomal protein S1. Nucleic Acids Res. 1988;16:3075–3089. doi: 10.1093/nar/16.7.3075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sklenar V, Piotto M, Leppik R, Saudek V. Gradient-tailored water suppression for 1H-15N HSQC experiments optimized to retain full sensitivity. J Magn Reson Ser A. 1993;102:241–245. [Google Scholar]
- 35.Sonenberg N, Wilchek M, Zamir A. Mapping of Escherichia coli ribosomal components involved in peptidyl transferase activity. Proc Natl Acad Sci USA. 1973;70:1423–1426. doi: 10.1073/pnas.70.5.1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Spanjaard R A, Chen K, Walker J R, van Duin J. Frameshift suppression at tandem AGA and AGG codons by cloned tRNA genes: assigning a codon to argU tRNA and T4 tRNA(Arg) Nucleic Acids Res. 1990;18:5031–5036. doi: 10.1093/nar/18.17.5031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Spanjaard R A, van Duin J. Translation of the sequence AGG-AGG yields 50% ribosomal frameshift. Proc Natl Acad Sci USA. 1988;85:7967–7971. doi: 10.1073/pnas.85.21.7967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Studier F W, Rosenberg A H, Dunn J J, Dubendorff J W. Use of T7 RNA polymerase to direct expression of cloned genes. Methods Enzymol. 1990;185:60–89. doi: 10.1016/0076-6879(90)85008-c. [DOI] [PubMed] [Google Scholar]
- 39.Sylvers L A, Wower J, Hixson S S, Zimmermann R A. Preparation of 2-azidoadenosine 3′,5′-[5′-32P]bisphosphate for incorporation into transfer RNA. Photoaffinity labeling of Escherichia coli ribosomes. FEBS Lett. 1989;245:9–13. doi: 10.1016/0014-5793(89)80180-2. [DOI] [PubMed] [Google Scholar]
- 40.Tejedor F, Ballesta J P. Ribosome structure: binding site of macrolides studied by photoaffinity labeling. Biochemistry. 1985;24:467–472. doi: 10.1021/bi00323a033. [DOI] [PubMed] [Google Scholar]
- 41.Thompson J D, Gibson T J, Plewniak F, Jeanmougin F, Higgins D G. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ühlein M, Weglöhner W, Urlaub H, Wittmann-Liebold B. Functional implications of ribosomal protein L2 in protein biosynthesis as shown by in vivo replacement studies. Biochem J. 1998;331:423–430. doi: 10.1042/bj3310423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Weglöhner W, Jünemann R, von Knoblauch K, Subramanian A R. Different consequences of incorporating chloroplast ribosomal proteins L12 and S18 into the bacterial ribosomes of Escherichia coli. Eur J Biochem. 1997;249:383–392. doi: 10.1111/j.1432-1033.1997.00383.x. [DOI] [PubMed] [Google Scholar]
- 44.Welch M, Chastang J, Yarus M. An inhibitor of ribosomal peptidyl transferase using transition-state analogy. Biochemistry. 1995;34:385–390. doi: 10.1021/bi00002a001. [DOI] [PubMed] [Google Scholar]
- 45.Wittmann-Liebold B, Ühlein M, Urlaub H, Müller E C, Otto A, Bischof O. Structural and functional implications in the eubacterial ribosome as revealed by protein-rRNA and antibiotic contact sites. Biochem Cell Biol. 1995;73:1187–1197. doi: 10.1139/o95-128. [DOI] [PubMed] [Google Scholar]
- 46.Wower I K, Wower J, Zimmermann R A. Ribosomal protein L27 participates in both 50 S subunit assembly and the peptidyl transferase reaction. J Biol Chem. 1998;273:19847–19852. doi: 10.1074/jbc.273.31.19847. [DOI] [PubMed] [Google Scholar]
- 47.Wower J, Hixson S S, Zimmermann R A. Labeling the peptidyltransferase center of the Escherichia coli ribosome with photoreactive tRNA(Phe) derivatives containing azidoadenosine at the 3′ end of the acceptor arm: a model of the tRNA-ribosome complex. Proc Natl Acad Sci USA. 1989;86:5232–5236. doi: 10.1073/pnas.86.14.5232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wower J, Kirillov S V, Wower I K, Guven S, Hixson S S, Zimmermann R A. Transit of tRNA through the Escherichia coli ribosome. Cross-linking of the 3′ end of tRNA to specific nucleotides of the 23 S ribosomal RNA at the A, P, and E sites. J Biol Chem. 2000;275:37887–37894. doi: 10.1074/jbc.M005031200. [DOI] [PubMed] [Google Scholar]
- 49.Wower J, Wower I K, Kirillov S V, Rosen K V, Hixson S S, Zimmermann R A. Peptidyl transferase and beyond. Biochem Cell Biol. 1995;73:1041–1047. doi: 10.1139/o95-111. [DOI] [PubMed] [Google Scholar]
- 50.Wu H, Wower I, Zimmermann R A. Mutagenesis of ribosomal protein S8 from Escherichia coli: expression, stability, and RNA-binding properties of S8 mutants. Biochemistry. 1993;32:4761–4768. doi: 10.1021/bi00069a010. [DOI] [PubMed] [Google Scholar]
- 51.Yusupov M M, Yusupova G Z, Baucom A, Lieberman K, Earnest T N, Cate J H, Noller H F. Crystal structure of the ribosome at 5.5 Å resolution. Science. 2001;292:883–896. doi: 10.1126/science.1060089. [DOI] [PubMed] [Google Scholar]
- 52.Zahn K. Overexpression of an mRNA dependent on rare codons inhibits protein synthesis and cell growth. J Bacteriol. 1996;178:2926–2933. doi: 10.1128/jb.178.10.2926-2933.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]