Abstract
Aim:
Lower grade gliomas [LGGs; World Health Organization (WHO) grades 2 and 3], owing to the heterogeneity of their clinical behavior, present a therapeutic challenge to neurosurgeons. The aim of this study was to explore the N6-methyladenosine (m6A) modification landscape in the LGGs and to develop an m6A-related microRNA (miRNA) risk model to provide new perspectives for the treatment and prognostic assessment of LGGs.
Methods:
Messenger RNA (mRNA) and miRNA expression data of LGGs were extracted from The Cancer Genome Atlas (TCGA) and Chinese Glioma Genome Atlas (CGGA) databases. An m6A-related miRNA risk model was constructed via least absolute shrinkage and selection operator (LASSO), univariate, and multivariate Cox regression analysis. Next, Kaplan-Meier analysis, principal-component analysis (PCA), functional enrichment analysis, immune infiltrate analysis, dynamic nomogram, and drug sensitivity prediction were used to evaluate this risk model.
Results:
Firstly, six m6A-related miRNAs with independent prognostic value were selected based on clinical information and used to construct a risk model. Subsequently, compared with low-risk group, LGGs in the high-risk group had a higher m6A writer and reader scores, but a lower eraser score. Moreover, LGGs in the high-risk group had a significantly worse clinical prognosis than those in the low-risk group. Simultaneously, this risk model outperformed other clinicopathological variables in the prognosis prediction of LGGs. Immune infiltrate analysis revealed that the proportion of M2 macrophages, regulatory T (Treg) cells, and the expression levels of exhausted immune response markers were significantly higher in the high-risk group than in the low-risk group. Finally, this study constructed an easy-to-use and free dynamic nomogram to help clinicians use this risk model to aid in diagnosis and prognosis assessment.
Conclusions:
This study developed a m6A-related risk model and uncovered two different m6A modification landscapes in LGGs. Moreover, this risk model may provide guidance and help in clinical prognosis assessment and immunotherapy response prediction for LGGs.
Keywords: Lower grade gliomas, N6-methyladenosine, microRNAs, immune infiltrate, nomogram
Introduction
Gliomas account for approximately 26% of all primary central nervous system (CNS) tumors and 81% of malignant tumors [1]. Lower grade gliomas [LGGs; World Health Organization (WHO) grades 2 and 3] often occur in the adult cerebral hemispheres and exhibit infiltrative growth, with histological types including astrocytomas and oligodendrogliomas [1, 2]. Due to the highly invasive character of the tumor, total surgical excision is very challenging, and the residual tumor will lead to recurrence and malignant progression. Some of these patients will progress to WHO grade 4 glioma and glioblastoma multiforme (GBM) within a few months, while others remain stable for many years. As a result, LGGs have a wide range of survival, from 1 year to 15 years [3]. There is no standard treatment for LGGs, and the choice of treatment varies depending on the extent of resection, histological subtype, WHO grade and molecular detection results, including surgery, chemotherapy and radiotherapy [4].
Although the histological classification of LGGs has been applied until now, it presents intra- and inter-observer heterogeneity and does not adequately predict clinical prognosis. Therefore, clinicians are increasingly depending on molecular genetic classification to guide clinical decision-making [5, 6]. Isocitrate dehydrogenase (IDH) mutations (including mutations in IDH1 or IDH2) characterize more than 70% of adult LGGs and define a subtype associated with a better prognosis [7]. Oligodendrogliomas typically have both IDH mutations and deletions in the 1p and 19q arms of chromosome (1p/19q coding deletion) and are sensitive to radiochemotherapy and have longer survival than LGGs without these alterations [8, 9]. Alpha-thalassemia/mental retardation syndrome X-linked (ATRX) mutation and loss of expression, and tumor protein p53 (TP53) mutation are more frequent in astrocytomas and telomerase reverse transcriptase (TERT) promoter mutation confers favorable prognosis in adult LGGs with IDH1/2 mutations, these are also important markers of clinical behavior [10, 11].
N6-methyladenosine (m6A) is the most common post-transcriptional modification in eukaryotic messenger RNA (mRNA) and is also engaged in regulating transfer RNA, ribosomal RNA and non-coding RNA [12, 13]. The m6A modification is primarily catalyzed by the methyltransferase (writer proteins), which is removed by the demethylase (eraser proteins) and recognized by the m6A binding protein (reader proteins) [14]. An increasing number of studies have shown that m6A modifications play an important role in both physiological and pathological processes (circadian rhythm, gene expression, cell differentiation, stress response, immune response and tumorigenesis) [15–20]. With the rapid development of RNA immunoprecipitation (RIP), next-generation sequencing (NGS) technologies, liquid chromatography (LC) and other detection technologies, the research on the mechanism of m6A in tumorigenesis and development has made rapid progress, and targeting m6A has also become a new direction for tumor clinical treatment [21].
MicroRNAs (miRNAs) are a class of non-coding RNAs with 19–22 nucleotides. Primary miRNAs produced by RNA polymerase II transcription are converted into precursor miRNAs with Drosha enzymes. The subsequent process is completed in the cytoplasm by dicerase and generates mature miRNAs.
miRNAs induce its cleavage by identifying the corresponding complementary sequence in the 3’ untranslated regions (UTRs) [22]. miRNAs have been proven to be dysfunctional in many types of tumors, and there are many mechanisms that lead to dysfunction, including genomic alterations in the genes encoding miRNAs, aberrant transcriptional regulation of miRNAs, and epigenetic alterations. Studies have also confirmed that miRNAs are involved in regulating tumor development, such as cell proliferation, apoptosis, tumor invasion, and angiogenesis [23]. Recent studies have found that they are also involved in the pathogenesis of glioma. Many studies have shown that some miRNAs are correlated with the diagnosis and prognosis of gliomas [24–26].
In this study, the transcriptional profiles of 2,157 miRNAs and 23 m6A genes were extracted from The Cancer Genome Atlas (TCGA) database. Then, we used Pearson’s correlation analysis, least absolute shrinkage and selection operator (LASSO) regression analysis, univariate and multivariate Cox regression analysis to ultimately screen six m6A-related miRNAs and construct a risk model. The model was a novel prognostic model constructed based on m6A modifications. Next, we screened drug candidates with m6A-associated miRNA signature through a public drug sensitivity database. Additionally, we estimated the relationship of this risk model with immune microenvironment and immunotherapeutic response. Finally, we developed an easy-to-use dynamic nomogram to predict the clinical prognosis of LGGs.
Materials and methods
Patients
The TCGA database was downloaded from UCSC Xena (https://xenabrowser.net/), which included 530 samples of LGG gene expression RNA sequencing (RNAseq) from the IlluminaHiSeq_RNASeqV2. 524 samples of LGG miRNA mature strand expression RNAseq from the IlluminaHiSeq_miRNASeq, relevant clinical information and 511 samples of LGG somatic mutation. The Chinese Glioma Genome Atlas (CGGA) database was downloaded from the CGGA website (http://www.cgga.org.cn/), which included 198 samples of GBMLGG (including 107 LGG and 91 GBM) gene expression miRNA from the V2.0 miRNA Expression BeadChip (Illumina). LGGs with missing overall survival (OS) values were excluded in order to reduce statistical bias during the analysis.
Selection of m6A genes and m6A-related miRNAs
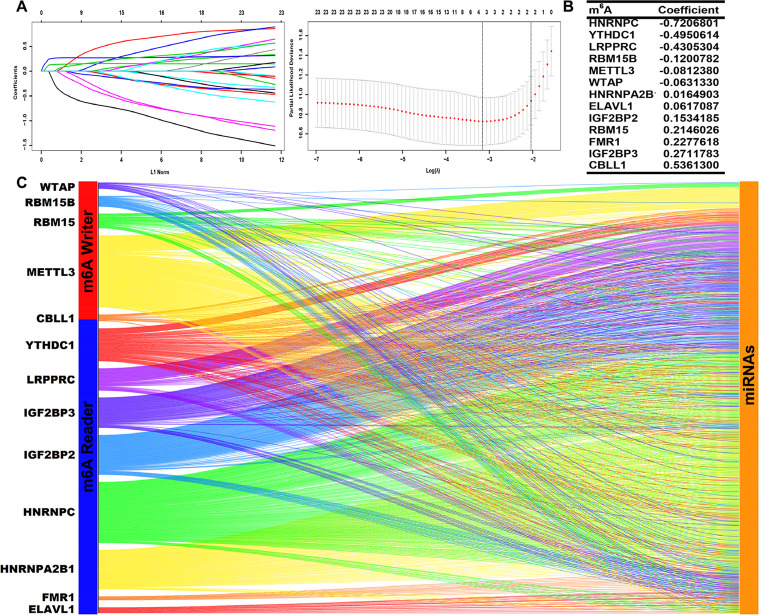
We obtained the transcriptional profiles of miRNAs and m6A genes from the TCGA database. A total of 23 m6A regulators were included for study in this research, including writers (CBLL1, KIAA1429, METTL14, METTL3, RBM15, RBM15B, WTAP and ZC3H13), erasers (ALKBH5 and FTO), and readers (ELAVL1, FMR1, HNRNPA2B1, HNRNPC, IGF2BP1, IGF2BP2, IGF2BP3, LRPPRC, YTHDC1, YTHDC2, YTHDF1, YTHDF2 and YTHDF3) [27]. LASSO regression analysis was applied using the R package glmnet, and 13 m6A genes were obtained to be significantly correlated with the prognosis of LGGs. Then, we screened m6A-related miRNAs by Pearson’s correlation analysis, and 434 m6A-related miRNAs were identified. The criteria for screening by this procedure was |Pearson R| > 0.3.
Immunofluorescence and immunohistochemistry
Immunofluorescence and immunohistochemistry of m6A-related regulatory proteins in cerebral cortex and LGGs were obtained from the Human Protein Atlas (HPA) database (http://www.proteinatlas.org/).
Risk signature construction and validation
The TCGA database was randomly divided into a training dataset and a validation dataset. Baseline characterization showed no statistically significant differences in molecular and clinical characteristics between these two cohorts (P > 0.05) (Table 1). Next, using univariate Cox regression analysis, we screened prognosis-related miRNAs from the 434 m6A-related miRNAs in the TCGA database (P < 0.05). Using the R package glmnet for LASSO regression, we screened 22 m6A-related miRNAs that were significantly associated with OS in LGGs. Then, 22 m6A-related miRNAs were identified using multivariate Cox regression analysis to identify six m6A-related miRNAs with independent prognostic value. Finally, those six m6A-related miRNAs were used to construct a risk model. The risk scores were calculated as follows. Risk score = coef(miRNA1) × expr(miRNA1) + coef(miRNA2) × expr(miRNA2) + ...... + coef(miRNAn) × expr(miRNAn), where coef(miRNAn) is the coefficient of prognosis-related miRNAs and expr(miRNAn) is the expression of miRNAs. The classification of low- and high-risk groups was defined by the mean of the risk scores.
Table 1.
Characteristics of LGGs in the training and validation cohorts
| Characteristics | Training dataset | Validation dataset | P |
|---|---|---|---|
| Age (year) | 42.54 ± 13.66 | 43.64 ± 12.53 | 0.3570 |
| KPS | 86.51 ± 12.22 | 86.8 ± 13.87 | 0.4491 |
| Gender | 0.7086 | ||
| Female | 175 (44.5%) | 61 (46.9%) | |
| Male | 218 (55.5%) | 69 (53.1%) | |
| Histological_type | 0.9071 | ||
| Astrocytoma | 146 (49.7%) | 50 (51%) | |
| Oligodendroglioma | 148 (50.3%) | 48 (49%) | |
| WHO_grade | 0.2414 | ||
| WHO grade 2 | 197 (50.1%) | 57 (44.2%) | |
| WHO grade 3 | 196 (49.9%) | 72 (55.8%) | |
| Seizure_history | 0.6963 | ||
| Yes | 230 (62.2%) | 77 (64.7%) | |
| No | 140 (37.8%) | 42 (35.3%) | |
| Sample_type | 0.5339 | ||
| Primary tumor | 381 (96.9%) | 128 (98.5%) | |
| Recurrent tumor | 12 (3.1%) | 2 (1.5%) | |
| IDH_status | 0.1918 | ||
| Wildtype | 75 (19.9%) | 18 (14.2%) | |
| Mutant | 302 (80.1%) | 109 (85.8%) | |
| Preoperative_antiseizure | 0.7701 | ||
| Yes | 201 (70%) | 63 (72.4%) | |
| No | 86 (30%) | 24 (27.6%) | |
| Preoperative_corticosteroids | 0.7680 | ||
| Yes | 162 (57%) | 53 (59.6%) | |
| No | 122 (43%) | 36 (40.4%) | |
| Headache_history | 0.4482 | ||
| Yes | 137 (38.2%) | 38 (33.6%) | |
| No | 222 (61.8%) | 75 (66.4%) | |
| Motor_changes | 0.8413 | ||
| Yes | 84 (23.9%) | 29 (25.4%) | |
| No | 267 (76.1%) | 85 (74.6%) | |
| Sensory_changes | 0.4837 | ||
| Yes | 65 (18.5%) | 17 (15%) | |
| No | 286 (81.5%) | 96 (85%) |
Bioinformatic analysis
The Database for Annotation, Visualization and Integrated Discovery (DAVID; https://david.ncifcrf.gov/) is used to carry out enrichment analysis. ESTIMATE was used to assess the percentage of stromal and immune cells in the tumor. The proportions of various immune cells in the tumor were evaluated using CIBERSORT. The R package maftools and the tumor immune dysfunction and exclusion (TIDE) algorithm were respectively used to analyze mutation data and immunotherapy response prediction. Single sample gene set enrichment analysis (GSVA; ssGSVA) was performed using GSVA to obtain a functional enrichment score for each sample [28].
Principal-component analysis and Kaplan-Meier survival analysis
High-dimensional data of 23 m6A genes and 6 m6A-associated miRNAs for risk modeling were efficiently downscaled, model identified and grouped for visualization using principal-component analysis (PCA). Kaplan-Meier survival analysis was used to assess the prognosis between the low- and high-risk groups.
Exploration of promising clinical therapeutic compounds targeting m6A-related miRNA risk model
To identify potential compounds for clinical treatment of LGGs, the half-maximal inhibitory concentration (IC50) of listed compounds were obtained from the Genomics of Drug Sensitivity in Cancer (GDSC) website. The R package oncoPredict was used to predict the IC50 of the above compounds in patients with LGG.
Independence of the m6A-related miRNA model
Univariate and multivariate Cox regression analyses were performed to identify independent variables for risk model and other clinicopathological characteristics of LGG patients. Simultaneously, we also comprehensively considered whether there is interaction and correlation between variables via R package corrgram and car.
Nomogram model construction and validation
The predictive performance of the nomogram and other variables [risk score, age, WHO grade, Karnofsky performance status (KPS), and histological type] for the 1-, 3-, and 5-year OS was established. The receiver operating characteristic (ROC) curve is used to evaluate the discrimination ability of the model. A network-based interactive dynamic nomogram application was constructed using Shiny version 1.6.0.
Results
Selection of m6A-related miRNAs in LGGs
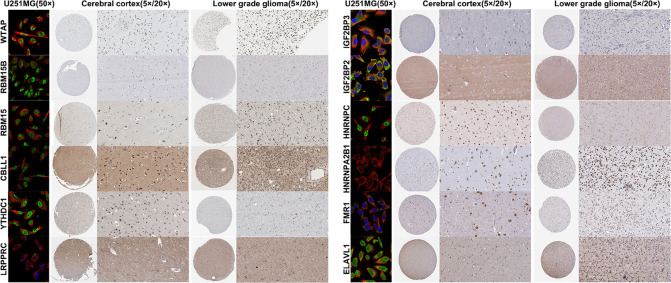
We screened prognostic m6A genes from 23 m6A genes in the TCGA database using LASSO regression analysis (Figure 1A, 1B). We defined m6A-related miRNAs as miRNAs associated with one or more of the 13 m6A genes (|Pearson R| > 0.3). Sankey plots were used to visualize the m6A-miRNA co-expression network (Figure 1C), and 434 m6A-related miRNAs were discerned as m6A-related miRNAs. Finally, immunofluorescence results revealed that m6A-related regulatory proteins were mainly expressed in the nucleus and cytoplasm of glioma cells. Immunohistochemical results demonstrated that WTAP, RBM15, IGF2BP2, HNRNPA2B1, and ELAVL1 were more expressed in LGG than in cerebral cortex, while YTHDC1 and LRPPRC were more expressed in cerebral cortex than in LGG (Figure 2, Figure S1).
Figure 1.
Identification of prognostic m6A genes in LGG patients. (A, B) LASSO regression analysis using 10-fold cross-validation; (C) Sankey plots of m6A genes and m6A-related miRNAs
Figure 2.
Immunofluorescence and immunohistochemistry of m6A-related regulatory proteins in cerebral cortex and low-grade gliomas
Risk signature construction and validation
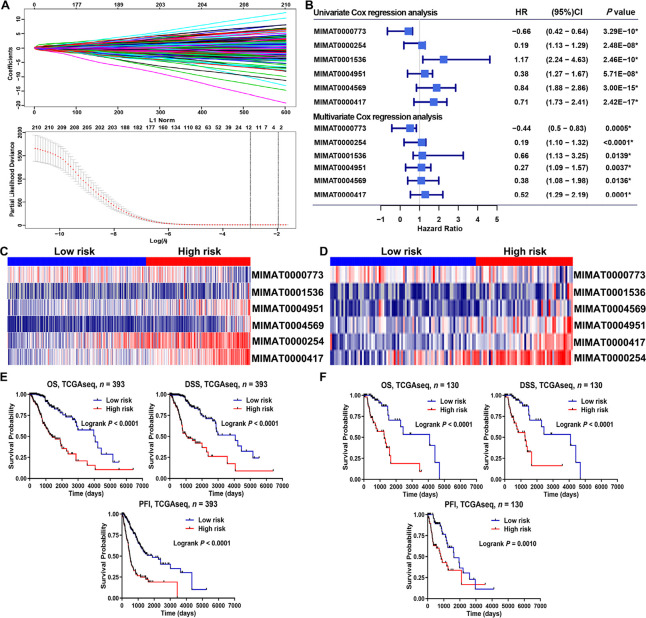
We screened m6A-related prognostic miRNAs from 434 m6A-associated miRNAs in the TCGA database using univariate Cox regression analysis. Those m6A-related miRNAs that were significantly associated with prognosis were screened in the TCGA database (P < 0.05). We further obtained 22 m6A-related miRNAs that were significantly associated with OS through the LASSO regression analysis (Figure 3A). Next, in the TCGA database, six m6A-related miRNAs with independent prognostic value were selected based on clinical information and used to construct a risk model (Figure 3B). The correlation analysis revealed that the six m6A-related miRNAs were significantly correlated with m6A regulators (Figure S2A). Then, we found that MIMAT0004951 (MIR887) has m6A modification sites (Figure S2B) based on these six miRNA transcripts through online website sequence-based RNA adenosine methylation site predictor (SRAMP; a mammalian m6A sites predictor).
Figure 3.
Risk model for LGG patients based on m6A-related miRNAs. (A) LASSO regression analysis using 10-fold cross-validation; (B) univariate and multivariate Cox regression analysis showed six independent prognostic miRNAs; (C, D) heat plots displayed the expression of six m6A-related miRNAs in the training and validation cohorts; (E, F) Kaplan-Meier analysis of the prognosis in the low-risk and high-risk groups. HR: hazard ratio; CI: confidence interval; * statistical significance
To assess the prognostic ability of the risk model, the LGG sample was divided into low- and high-risk groups based on the mean of the risk scores in the training and validation cohorts. The heatmap depicts the expression of six m6A-related miRNAs involved in the construction of the risk model in the training cohort (Figure 3C) and the validation cohort (Figure 3D). Kaplan-Meier survival analysis of both the training and validation cohorts showed that patients with LGG in the high-risk group had worse OS than those in the low-risk group (Figure 3E, 3F). To further assess the prognostic predictive ability of the risk model, we included disease-specific survival (DSS) and progression-free interval (PFI) metrics to assess the differences between low- and high-risk LGG patients. The results demonstrated that LGGs in the high-risk group had worse DSS and PFI than low-risk group. The above results suggest that the risk model of m6A-related miRNAs can effectively assess the prognosis of LGGs (Figure 3E, 3F). A total of 523 LGG patients, who were included in the study, were randomly divided into a training cohort (n = 393) and a validation cohort (n = 130). Detailed characteristics for these two cohorts showed homogeneity in these cohorts (Table 1).
PCA of m6A related miRNA risk signature model
PCA was performed to assess the differences between the low- and high-risk groups based on the transcriptional profiles of 23 m6A genes and 6 m6A-related miRNAs involved in the construction of the risk model (Figure S2C, 2D). The results of PCA indicated that 6 m6A-related miRNAs had better distinction than 23 m6A genes in terms of the distribution of the low- and high-risk groups. Next, the ssGSEA results showed compared with low-risk group, LGGs in high-risk group had a higher m6A writer and reader scores, but a lower eraser score (Figure S2E).
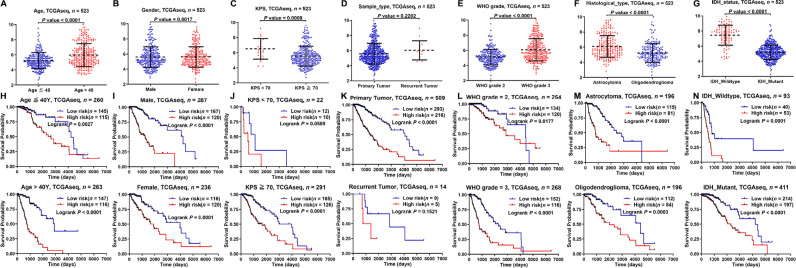
We analyzed the prognostic differences between the low- and high-risk groups in the TCGA and CGGA database stratified by clinicopathological characteristics. First of all, compared with age ≤ 40, KPS ≥ 70, primary tumor, WHO grade 2, oligodendroglioma, and IDH mutant groups, the risk score was higher in age > 40, KPS < 70, recurrent tumor, WHO grade 3, astrocytoma, and IDH wildtype groups (Figure 4A–G; Figure S3A–G). Then, according to the subgroups stratified by age, gender, KPS, sample type, WHO grade, histological type, and IDH status, the OS of the high-risk group continued to be inferior to that of the low-risk group (Figure 4H–N; Figure S3H–N).
Figure 4.
Kaplan-Meier analysis of survival differences stratified by clinicopathological variables between the low- and high-risk groups in the TCGA database. (A–G) Risk score differences stratified by clinicopathological variables between the low- and high-risk groups; (H–N) Kaplan-Meier analysis of survival differences stratified by clinicopathological variables between the low- and high-risk groups
Correlation between m6A related miRNA risk signature and immune microenvironment of LGGs
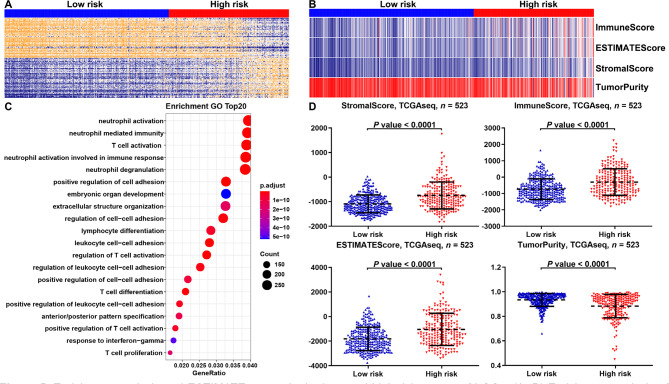
We found that the risk model of m6A-related miRNAs was mainly correlated with immune-associated biological processes by performing gene ontology (GO) enrichment analysis of differently expressed genes (DEGs) between the low- and high-risk groups (Figure 5A, 5B). Moreover, the ESTIMATE results suggested that compared with low-risk group, LGGs in high-risk group had higher immune and stromal scores, while the purity of tumors was lowered (Figure 5C, 5D).
Figure 5.
Enrichment analysis and ESTIMATE scores in the low- and high-risk groups of LGGs. (A, B) Enrichment analysis of DEGs in the low- and high-risk groups of LGGs in the TCGA database. The top 20 items were illustrated in the bubblechart; (C) the heat map displayed the distribution features of ESTIMATE scores between the low- and high-risk groups of LGGs in the TCGA database; (D) the scatter plot depicted the results of the ESTIMATE scores between the low- and high-risk groups
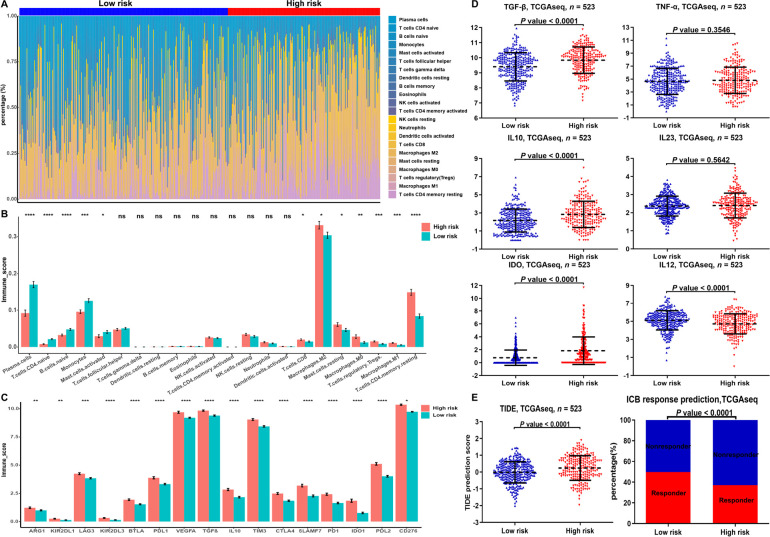
We next explored the relationship between risk models of m6A-related miRNAs and tumor-infiltrating immune cells (TICs). Immune infiltration analysis revealed higher proportion of M1 macrophage, regulatory T (Treg) cell, M0 macrophage, and M2 macrophage in the high-risk group was than that in the low-risk group (Figure 6A, 6B). We also founded that the expression of exhausted immune response markers was markedly increased in the high-risk group compared to the low-risk group (Figure 6C). Since M1 macrophages exert pro-inflammatory and anti-tumor functions, while M2 macrophages cells have anti-inflammatory and pro-tumor functions, we further analyzed the difference in secretion of immune factors between M1 and M2 macrophages in the low- and high-risk groups. The results showed that the anti-inflammatory cytokines transforming growth factor beta (TGFB), interleukin 10 (IL10) and indoleamine 2,3-dioxygenase (IDO) secreted by M2 macrophages were significantly higher in the high-risk group than in the low-risk group. The pro-inflammatory cytokines tumor necrosis factor (TNF), IL23, and IL12 secreted by M1 macrophages in the high-risk group were not different from those in the low-risk group or were lower than those in the low-risk group (Figure 6D). The above results indicate that M1 macrophages in the high-risk group occurred disability.
Figure 6.
Proportion of TICs and exhausted immune response markers in the low- and high-risk groups of LGGs. (A) Bar chart showed the percentage of various TICs in the samples from the low-risk and high-risk groups; (B, C) bar chart demonstrated the difference in the expression of various immune cells and exhausted immune response markers between the low-risk and high-risk groups; (D) expression of M1- and M2-associated genes differences between the low- and high-risk groups in the TCGA database; (E) TIDE prediction difference in the high- and low-risk patients, and accumulative bar diagram showed that the LGGs with low-risk group (49.67%, 150/302) were more likely to respond to immune checkpoint blockade (ICB) immunotherapy than the LGGs with high-risk group (37.1%, 82/221). * P < 0.05, ** P < 0.01, *** P < 0.001, **** P < 0.0001
In addition, the results of TIDE discovered that LGGs in the low-risk group had a higher response rate to immunotherapy than in the high-risk group. The above results suggest that m6A-related miRNA-based risk scores may be a potential predictor of immunotherapy response (Figure 6E).
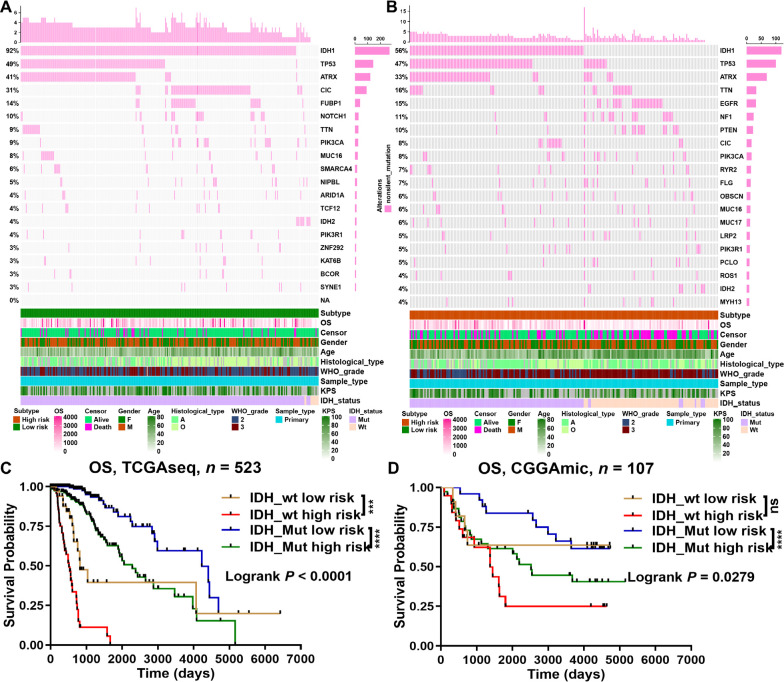
Then, we analyzed and summarized the mutation data. The waterfall plot shows the top 20 driver genes with the highest frequency of non-silent mutations between the low- and high-risk groups (Figure 7A, 7B). At the same time, the mutation frequency of IDH1 in the low-risk group (92%) was obviously higher than that in the high-risk group (56%). Therefore, we evaluated whether a risk model based on m6A-related miRNAs could better predict OS than IDH mutation status. The results showed that LGGs with IDH mutations and IDH wildtype in the high-risk group (defined as IDH_Mut high risk and IDH_wt high risk, respectively) had worse OS than those with IDH mutations and IDH wildtype in the low-risk group (defined as IDH_Mut low risk and IDH_wt low risk, respectively) (Figure 7C, 7D). Interestingly, when OS was less than 2,660 days, LGGs with IDH mutant in the high-risk group (IDH_Mut high risk) had better survival outcomes than those with IDH wildtype in the low-risk group (IDH_wt low risk). However, when OS was greater than 2,869 days (in the TCGA database) or 1,314 days (in the CGGA database), LGGs with IDH mutant in the high-risk group (IDH_Mut high risk) had worse survival outcomes than those with IDH wildtype in the low-risk group (IDH_wt low risk) (Figure 7C, 7D). Therefore, the above results suggest that risk models for m6A-related miRNAs are likely to have greater prognostic reference value than IDH mutation status.
Figure 7.
Genetic alteration analyses between the low- and high-risk groups of LGGs. Waterfall plot indicated gene mutations in the (A) low-risk and (B) high-risk groups; (C, D) prognostic evaluation of LGGs survival by IDH mutation status and m6A-related miRNA risk score in TCGA and CGGA databases
Exploration of promising clinical therapeutic compounds targeting m6A-related miRNA risk model
To identify potential drugs for clinical treatment of LGGs, we used the GDSC database to calculate the IC50 for each sample using the oncoPredict algorithm. A total of 104 compounds were screened for significant differences in IC50 between the low- and high-risk groups. The results of comparing the IC50 of the compounds between the two groups revealed that more compounds were more sensitive in the low- risk group compared to the high-risk group (Figure S4A). The difference between the top 5 compounds in the low- and high-risk groups of LGGs (Figure S4B–F).
Prognostic and clinical characterization of m6A-related miRNA risk model
Univariate and multivariate Cox regression analyses revealed risk score, age, WHO grade, and histological type were independent prognostic variables for LGGs (Figure S5A). Simultaneously, the results of the ROC curve analysis indicated that the area under the curve (AUC) of the risk score was always greater than other clinicopathological variables at 1, 3 and 5 years of OS. The above results suggest that the risk model based on six m6A-related miRNAs is more reliable for the prognostic evaluation of LGGs (Figure S5B). Moreover, we comprehensively considered whether there is interaction and correlation between variables (Table S1, S2), and found that the risk score and IDH mutation status have a significant correlation (Pearson r = –0.659).
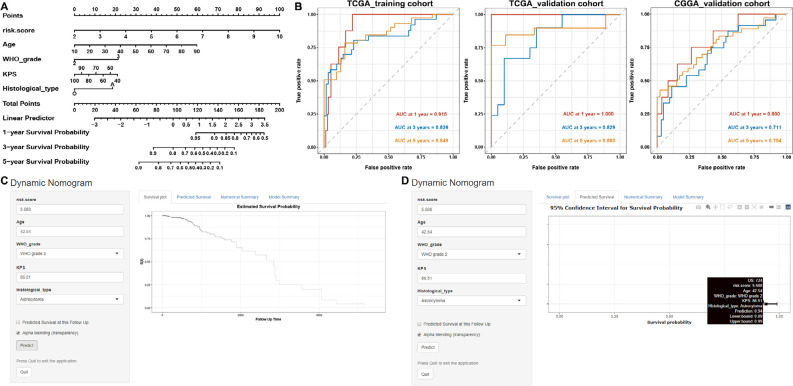
Construction and evaluation of the prognostic dynamic nomogram
The nomogram was used to predict the incidence of OS at 1, 3, and 5 years, the risk scores of the nomogram model showed dominant predictive power by comparison with other clinicopathological characteristics (Figure 8A). The ROC curve for the nomogram and its AUC displayed in Figure 8B. The final Cox model incorporated five independent predictors (risk score, age, WHO grade, KPS, and histological type) and was developed as a simple-to-use dynamic nomogram that is available online (https://fengyuan1993.shinyapps.io/LGG_m6A_Immunotherapy/) as screenshotted in Figure 8C, 8D.
Figure 8.
Construction and evaluation of a prognostic nomogram. (A) The nomogram predicts the probability of the 1-, 2-, and 3-year OS; (B) ROC curve of the nomogram predicts the probability of the 1-, 2-, and 3-year OS; (C, D) online dynamic nomogram accessible at https://fengyuan1993.shinyapps.io/LGG_m6A_Immunotherapy/
Discussion
Gliomas are the most common primary malignancies of the CNS and are correlated with poor prognosis. The identification of effective diagnostic and prognostic biomarkers is important to guide treatment [29]. An increasing number of studies have confirmed that miRNAs play essential regulatory roles in cells between normal and pathological states [30]. miRNAs have been one of the main focuses of tumor research in the past decade, and many studies have shown the importance of miRNAs in tumor biology, such as promoting tumor growth, invasion, angiogenesis, and immune evasion through the regulation of their target mRNAs [31–33]. In addition, tumor miRNA profiles can define relevant subtypes to predict patient clinical prognosis and guide treatment [34–36]. For example, exosomal miRNAs (miR-210, miR-449 and miR-5194) in plasma of gliomas serve as novel non-invasive biomarkers with high sensitivity to guide the diagnosis and prognosis of patients [37].
m6A modification is the most abundant and conserved internal transcriptional modification [18]. It was founded that miRNAs are able to repress gene expression though binding to complementary sequences in the 3’ UTRs of target genes [38]. m6A mRNA modification occurs in the region adjacent to the stop codon and close to the proximal region of the 3’ UTRs. In addition, the target sites of miRNAs are abundant in m6A peaks. Therefore, the similarity of target sites connected the potential interaction between miRNAs and m6A modifications [39]. Previous studies have demonstrated that dysfunction of miRNAs is highly involved in many important biological processes (prenatal development, immune response and tumorigenesis) [40–43]. m6A modifications are able to regulate the maturation of those miRNAs that are involved in cell proliferation and tumorigenesis. For example, during pancreatic ductal adenocarcinoma (PDAC) tumorigenesis cigarette smoke condensate (CSC) promotes the maturation of miR-25-3p through mediating METTL3 [44]. METTL3 also enhances the binding of pri-miR-221/222 to DGCR8 to regulate bladder cancer proliferation [45]. m6A could mediate arsenate-induced tumorigenesis through modification of several miRNAs (miR-106b, miR-18a/b, miR-3607, miR-423, miR-30a, and miR-320b/d/e) [46]. METTL14, another m6A regulator, is shown to suppress the invasion and metastasis of hepatocellular carcinoma (HCC) through promoting the maturation of pri-miR-126 [47].
The role of m6A modifications in the malignant progression of gliomas has been increasingly uncovered in recent years. Compared to IDH-mutant gliomas, IDH-wildtype gliomas have a significantly poorer prognosis. The fifth edition of the WHO Classification of Tumors of the CNS for GBM included only the IDH-wildtype [2]. Chang et al. [48] found that METLL3 was significantly overexpressed in IDH-wildtype GBMs, and METLL3 enhanced the stability of long non-coding RNA (lncRNA) metastasis-associated lung adenocarcinoma transcript 1 (MALAT1) through m6A modification with the help of human antigen R (HuR) and activated nuclear factor-κB (NF-κB) pathway to promote malignant progression of IDH-wildtype gliomas. Moreover, Chai et al. [49] discovered that YTHDF2 was capable of activating the NF-κB pathway in a manner that facilitated the METTL3-dependent degradation of UBX domain protein 1 (UBXN1) mRNA. Dong et al. [50] also found that under hypoxic conditions, the m6A demethylase ALKBH5 was able to promote the formation of an immunosuppressive microenvironment by promoting lncRNA NEAT1-mediated paraspeckle assembly to upregulate C-X-C motif chemokine ligand 8 (CXCL8)/IL8 expression. Both m6A and miRNAs are important regulators in the process of tumor development. However, researches on the potential mechanisms and prognostic markers of m6A-related miRNAs in LGGs are still absent. In the present study, inspired by the essential biological functions of m6A and miRNAs in malignancies, we aimed to establish a risk model based on m6A-related miRNAs.
In this study, 434 m6A-associated miRNAs were identified from the TCGA database, and then six m6A-related miRNAs with independent prognostic value were screened based on clinical information and were used to construct a risk model for m6A-related miRNAs to predict the prognosis of LGGs. In particular, miR-346 is shown to inhibit glioma proliferation by targeting nuclear factor I B (NFIB) and may become a potential prognostic and therapeutic candidate [51]. miR-10b is a miRNA that is not expressed in normal brain but is significantly upregulated in glioma. Targeted inhibition of miR-10b significantly suppresses tumor growth in animal models of glioma [52]. The regulation of big mitogen-activated protein kinase 1 (BMK1) by miR-429 has an essential role in glioma invasion [53]. The expression of miR-21, miR-222 and miR-124-3p in serum exosomes was significantly higher in high-grade gliomas than in LGGs and normal controls, and the expression levels decreased rapidly after surgery. This finding implied that exosomal miR-21, miR-222 and miR-124-3p provide a minimally invasive assay to guide the clinical management of gliomas [54]. miRNAs in cerebrospinal fluid (CSF) also have the potential to serve as novel biomarkers for detection in patients with glioma. Analysis of CSF from gliomas and patients with various neurological diseases revealed that miR-15b expression levels could effectively differentiate gliomas from non-glioma patients [55]. Combined with the above studies and based on the transcripts of these six miRNAs, we discovered that only miR-887 had a potential m6A modification site through the online website SRAMP. The other five miRNAs were not predicted to contain potential m6A modification sites, but their m6A modification status should be clarified by experiments such as methylated RIP owing to their essential roles in several types of tumors, including glioma. In addition, miR-887 was first proposed by our team in glioma. We will explore the characteristics of its role in GBM and its relationship with m6A modification in future studies.
Subsequently, univariate and multivariate Cox regression analyses revealed a risk model for m6A-related miRNA as an independent prognostic risk factor for LGGs. ROC analysis indicated that this risk model outperformed other clinicopathological characteristics in the prognosis prediction of LGGs. We also developed a dynamic nomogram, which showed a highly consistent between observed and predicted rates for 1-, 3-, and 5-year OS. The risk model based on six m6A-related miRNAs is highly accurate and the predictive model will be mined for new biomarkers for subsequent studies.
The TIDE prediction score, an algorithm developed for predicting immunotherapy, has been adopted in an increasing number of studies and its predictive ability has been successfully validated [56]. In our study, the results of the TIDE algorithm analysis showed that patients in the low-risk group had a significantly better response rate to immunotherapy than the high-risk group. Immune infiltration analysis revealed higher proportion of M1 macrophage, Treg cell, M0 macrophage, and M2 macrophage in the high-risk group was than that in the low-risk group. Since M1 macrophages exert pro-inflammatory and anti-tumor functions, while M2 macrophages cells have anti-inflammatory and pro-tumor functions, we further analyzed the difference in secretion of immune factors between M1 and M2 macrophages in the low- and high-risk groups. The results showed that the immune factors TGFB, IL10, and IDO secreted by M2 macrophages in the high-risk group were obviously higher than that of the low-risk group. The immune factors TNF, IL23, and IL12 secreted by M1 macrophages in the high-risk group were not different from those in the low-risk group or were lower than those in the low-risk group. The above results indicate that M1 macrophages in the high-risk group occurred disability. We also founded that the expression of exhausted immune response markers was markedly increased in the high-risk group compared to the low-risk group. The above results suggest that this prediction model is highly promising to provide a reliable biomarker for tumor immunotherapy. Furthermore, this study provides novel perspectives on the molecular biological mechanisms of m6A-related miRNAs in LGGs.
Clinically, IDH mutation status, WHO grade and histological type are important factors influencing the prognosis of LGGs [57]. However, LGGs with the same type often present with different clinical prognosis, suggesting that current classification systems do not provide reliable prognostic predictions and accurately reflect the heterogeneity of LGG [58]. Therefore, exploring potential predictive and therapeutic biomarkers has become an important direction for LGG. The development of m6A-related miRNA risk models provides a new approach for prognosis and immunotherapy prediction of LGGs. These results also provide insights into the process and mechanism of m6A-modified miRNAs for future studies. In our study, we are also aware of some weaknesses and limitations of the present study. Such as the lack of other clinical databases for external validation and the biological mechanisms of m6A-related miRNAs have not been fully elucidated. Therefore, we will collect more LGG samples in our future work to validate the accuracy of this model and explore the functional mechanisms of miRNAs and their relationship with m6A modifications.
In conclusions, our study provides an important guidance for the prognostic assessment of LGGs and may help to shed light on the mechanisms of m6A-regulated miRNAs. Additionally, the predictive model showed sensitivity in identifying LGGs who may respond well to immunotherapy.
Abbreviations
- CGGA:
Chinese Glioma Genome Atlas
- CNS:
central nervous system
- DSS:
disease-specific survival
- GBM:
glioblastoma multiforme
- IC50:
half-maximal inhibitory concentration
- IDH:
isocitrate dehydrogenase
- IL10:
interleukin 10
- KPS:
Karnofsky performance status
- LASSO:
least absolute shrinkage and selection operator
- m6A:
N6-methyladenosine
- miRNA:
microRNA
- mRNA:
messenger RNA
- OS:
overall survival
- PCA:
principal-component analysis
- PFI:
progression-free interval
- ROC:
receiver operating characteristic
- TCGA:
The Cancer Genome Atlas
- TICs:
tumor-infiltrating immune cells
- TIDE:
tumor immune dysfunction and exclusion
- TNF:
tumor necrosis factor
- UTRs:
untranslated regions
- WHO:
World Health Organization
Supplementary materials
The supplementary material for this article is available at: https://www.explorationpub.com/uploads/Article/file/1002100_sup_1.pdf.
Declarations
Acknowledgments
We appreciate the generosity of CGGA and TCGA network for sharing the huge amount of data.
Author contributions
FY conceived the project. CM and FY supervised the project. FY, YW and XC downloaded the data and performed the statistical analysis. CM, FY, YW, XC, CD, JZ, CT and JY interpreted the results. All listed authors read and approved the final manuscript.
Conflicts of interest
The authors declare no conflicts of interest in this manuscript.
Ethical approval
Not applicable.
Consent to participate
Not applicable.
Consent to publication
Not applicable.
Availability of data and materials
Not applicable.
Funding
Not applicable.
Copyright
© The Author(s) 2022.
References
- 1.Ostrom QT, Gittleman H, Truitt G, Boscia A, Kruchko C, Barnholtz-Sloan JS. CBTRUS statistical report: primary brain and other central nervous system tumors diagnosed in the United States in 2011–2015. Neuro Oncol. 2018;20:iv1–86. Erratum in: Neuro Oncol. 2018;23:508–22. 10.1093/neuonc/noy131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Louis DN, Perry A, Wesseling P, Brat DJ, Cree IA, Figarella-Branger D, et al. The 2021 WHO classification of tumors of the central nervous system: a summary. Neuro Oncol. 2021;23:1231–51. 10.1093/neuonc/noab106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.van den Bent MJ. Chemotherapy for low-grade glioma: when, for whom, which regimen? Curr Opin Neurol. 2015;28:633–938. 10.1097/WCO.0000000000000257 [DOI] [PubMed] [Google Scholar]
- 4.Schiff D, Van den Bent M, Vogelbaum MA, Wick W, Miller CR, Taphoorn M, et al. Recent developments and future directions in adult lower-grade gliomas: society for Neuro-Oncology (SNO) and European Association of Neuro-Oncology (EANO) consensus. Neuro Oncol. 2019;21:837–53. 10.1093/neuonc/noz033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.van den Bent MJ. Interobserver variation of the histopathological diagnosis in clinical trials on glioma: a clinician’s perspective. Acta Neuropathol. 2010;120:297–304. 10.1007/s00401-010-0725-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wen PY, Packer RJ. The 2021 WHO classification of tumors of the central nervous system: clinical implications. Neuro Oncol. 2021;23:1215–7. 10.1093/neuonc/noab120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yan H, Parsons DW, Jin G, McLendon R, Rasheed BA, Yuan W, et al. IDH1 and IDH2 mutations in gliomas. N Engl J Med. 2009;360:765–73. 10.1056/NEJMoa0808710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cairncross JG, Wang M, Jenkins RB, Shaw EG, Giannini C, Brachman DG, et al. Benefit from procarbazine, lomustine, and vincristine in oligodendroglial tumors is associated with mutation of IDH. J Clin Oncol. 2014;32:783–90. 10.1200/JCO.2013.49.3726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Speirs CK, Simpson JR, Robinson CG, DeWees TA, Tran DD, Linette G, et al. Impact of 1p/19q codeletion and histology on outcomes of anaplastic gliomas treated with radiation therapy and temozolomide. Int J Radiat Oncol Biol Phys. 2015;91:268–76. 10.1016/j.ijrobp.2014.10.027 [DOI] [PubMed] [Google Scholar]
- 10.Arita H, Matsushita Y, Machida R, Yamasaki K, Hata N, Ohno M, et al. TERT promoter mutation confers favorable prognosis regardless of 1p/19q status in adult diffuse gliomas with IDH1/2 mutations. Acta Neuropathol Commun. 2020;8:201. 10.1186/s40478-020-01078-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Spino M, Snuderl M. Genomic molecular classification of CNS malignancies. Adv Anat Pathol. 2020;27:44–50. 10.1097/PAP.0000000000000254 [DOI] [PubMed] [Google Scholar]
- 12.Pan T. N6-methyl-adenosine modification in messenger and long non-coding RNA. Trends Biochem Sci. 2013;38:204–9. 10.1016/j.tibs.2012.12.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ma S, Chen C, Ji X, Liu J, Zhou Q, Wang G, et al. The interplay between m6A RNA methylation and noncoding RNA in cancer. J Hematol Oncol. 2019;12:121. 10.1186/s13045-019-0805-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang Y, Gao M, Zhu F, Li X, Yang Y, Yan Q, et al. METTL3 is essential for postnatal development of brown adipose tissue and energy expenditure in mice. Nat Commun. 2020;11:1648. 10.1038/s41467-020-15488-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fustin JM, Doi M, Yamaguchi Y, Hida H, Nishimura S, Yoshida M, et al. RNA-methylation-dependent RNA processing controls the speed of the circadian clock. Cell. 2013;155:793–806. 10.1016/j.cell.2013.10.026 [DOI] [PubMed] [Google Scholar]
- 16.Zhao BS, Roundtree IA, He C. Post-transcriptional gene regulation by mRNA modifications. Nat Rev Mol Cell Biol. 2017;18:31–42. Erratum in: Nat Rev Mol Cell Biol. 2018;19:808. 10.1038/nrm.2016.132 [DOI] [PubMed] [Google Scholar]
- 17.Engel M, Eggert C, Kaplick PM, Eder M, Röh S, Tietze L, et al. The role of m6A/m-RNA methylation in stress response regulation. Neuron. 2018;99:389–403.e9. 10.1016/j.neuron.2018.07.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen XY, Zhang J, Zhu JS. The role of m6A RNA methylation in human cancer. Mol Cancer. 2019;18:103. 10.1186/s12943-019-1033-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Edens BM, Vissers C, Su J, Arumugam S, Xu Z, Shi H, et al. FMRP modulates neural differentiation through m6A-dependent mRNA nuclear export. Cell Rep. 2019;28:845–54.e5. 10.1016/j.celrep.2019.06.072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yu R, Li Q, Feng Z, Cai L, Xu Q. m6A reader YTHDF2 regulates LPS-induced inflammatory response. Int J Mol Sci. 2019;20:1323. 10.3390/ijms20061323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shen C, Xuan B, Yan T, Ma Y, Xu P, Tian X, et al. m6A-dependent glycolysis enhances colorectal cancer progression. Mol Cancer. 2020;19:72. 10.1186/s12943-020-01190-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Leung AKL. The whereabouts of microRNA actions: cytoplasm and beyond. Trends Cell Biol. 2015;25:601–10. 10.1016/j.tcb.2015.07.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Peng Y, Croce CM. The role of microRNAs in human cancer. Signal Transduct Target Ther. 2016;1:15004. 10.1038/sigtrans.2015.4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Qu S, Guan J, Liu Y. Identification of microRNAs as novel biomarkers for glioma detection: a meta-analysis based on 11 articles. J Neurol Sci. 2015;348:181–7. 10.1016/j.jns.2014.11.036 [DOI] [PubMed] [Google Scholar]
- 25.Banelli B, Forlani A, Allemanni G, Morabito A, Pistillo MP, Romani M. MicroRNA in glioblastoma: an overview. Int J Genomics. 2017;2017:7639084. 10.1155/2017/7639084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ma C, Nguyen HPT, Luwor RB, Stylli SS, Gogos A, Paradiso L, et al. A comprehensive meta-analysis of circulation miRNAs in glioma as potential diagnostic biomarker. PLoS One. 2018;13:e0189452. 10.1371/journal.pone.0189452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chong W, Shang L, Liu J, Fang Z, Du F, Wu H, et al. m6A regulator-based methylation modification patterns characterized by distinct tumor microenvironment immune profiles in colon cancer. Theranostics. 2021;11:2201–17. 10.7150/thno.52717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–50. 10.1073/pnas.0506580102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ostrom QT, Cote DJ, Ascha M, Kruchko C, Barnholtz-Sloan JS. Adult glioma incidence and survival by race or ethnicity in the United States from 2000 to 2014. JAMA Oncol. 2018;4:1254–62. 10.1001/jamaoncol.2018.1789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ebert MS, Sharp PA. Roles for microRNAs in conferring robustness to biological processes. Cell. 2012;149:515–24. 10.1016/j.cell.2012.04.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kasinski AL, Slack FJ. Epigenetics and genetics. MicroRNAs en route to the clinic: progress in validating and targeting microRNAs for cancer therapy. Nat Rev Cancer. 2011;11:849–64. 10.1038/nrc3166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stahlhut C, Slack FJ. MicroRNAs and the cancer phenotype: profiling, signatures and clinical implications. Genome Med. 2013;5:111. 10.1186/gm516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Otmani K, Lewalle P. Tumor suppressor miRNA in cancer cells and the tumor microenvironment: mechanism of deregulation and clinical implications. Front Oncol. 2021;11:708765. 10.3389/fonc.2021.708765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.He K, Li WX, Guan D, Gong M, Ye S, Fang Z, et al. Regulatory network reconstruction of five essential microRNAs for survival analysis in breast cancer by integrating miRNA and mRNA expression datasets. Funct Integr Genomics. 2019;19:645–58. 10.1007/s10142-019-00670-7 [DOI] [PubMed] [Google Scholar]
- 35.Lizarte Neto FS, Rodrigues AR, Trevisan FA, de Assis Cirino ML, Matias CCMS, Pereira-da-Silva G, et al. microRNA-181d associated with the methylation status of the MGMT gene in Glioblastoma multiforme cancer stem cells submitted to treatments with ionizing radiation and temozolomide. Brain Res. 2019;1720:146302. 10.1016/j.brainres.2019.146302 [DOI] [PubMed] [Google Scholar]
- 36.Pyman B, Sedghi A, Azizi S, Tyryshkin K, Renwick N, Mousavi P. Exploring microRNA regulation of cancer with context-aware deep cancer classifier. Pac Symp Biocomput. 2019;24:160–71. 10.1142/9789813279827_0015 [DOI] [PubMed] [Google Scholar]
- 37.Tabibkhooei A, Izadpanahi M, Arab A, Zare-Mirzaei A, Minaeian S, Rostami A, et al. Profiling of novel circulating microRNAs as a non-invasive biomarker in diagnosis and follow-up of high and low-grade gliomas. Clin Neurol Neurosurg. 2020;190:105652. 10.1016/j.clineuro.2019.105652 [DOI] [PubMed] [Google Scholar]
- 38.Sekar D, Lakshmanan G, Mani P, Biruntha M. Methylation-dependent circulating microRNA 510 in preeclampsia patients. Hypertens Res. 2019;42:1647–8. 10.1038/s41440-019-0269-8 [DOI] [PubMed] [Google Scholar]
- 39.Alarcón CR, Lee H, Goodarzi H, Halberg N, Tavazoie SF. N6-methyladenosine marks primary microRNAs for processing. Nature. 2015;519:482–5. 10.1038/nature14281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cho YK, Son Y, Kim SN, Song HD, Kim M, Park JH, et al. MicroRNA-10a-5p regulates macrophage polarization and promotes therapeutic adipose tissue remodeling. Mol Metab. 2019;29:86–98. 10.1016/j.molmet.2019.08.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rahmanian S, Murad R, Breschi A, Zeng W, Mackiewicz M, Williams B, et al. Dynamics of microRNA expression during mouse prenatal development. Genome Res. 2019;29:1900–9. 10.1101/gr.248997.119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wu CJ, Cho S, Huang HY, Lu CH, Russ J, Cruz LO, et al. MiR-23~27~24-mediated control of humoral immunity reveals a TOX-driven regulatory circuit in follicular helper T cell differentiation. Sci Adv. 2019;5:eaaw1715. 10.1126/sciadv.aaw1715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cen B, Lang JD, Du Y, Wei J, Xiong Y, Bradley N, et al. Prostaglandin E2 induces miR675-5p to promote colorectal tumor metastasis via modulation of p53 expression. Gastroenterology. 2020;158:971–84.e10. 10.1053/j.gastro.2019.11.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang J, Bai R, Li M, Ye H, Wu C, Wang C, et al. Excessive miR-25-3p maturation via N6-methyladenosine stimulated by cigarette smoke promotes pancreatic cancer progression. Nat Commun. 2019;10:1858. 10.1038/s41467-019-09712-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Han J, Wang JZ, Yang X, Yu H, Zhou R, Lu HC, et al. METTL3 promote tumor proliferation of bladder cancer by accelerating pri-miR221/222 maturation in m6A-dependent manner. Mol Cancer. 2019;18:110. 10.1186/s12943-019-1036-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gu S, Sun D, Dai H, Zhang Z. N6-methyladenosine mediates the cellular proliferation and apoptosis via microRNAs in arsenite-transformed cells. Toxicol Lett. 2018;292:1–11. 10.1016/j.toxlet.2018.04.018 [DOI] [PubMed] [Google Scholar]
- 47.Ma JZ, Yang F, Zhou CC, Liu F, Yuan JH, Wang F, et al. METTL14 suppresses the metastatic potential of hepatocellular carcinoma by modulating N6-methyladenosine-dependent primary microRNA processing. Hepatology. 2017;65:529–43. 10.1002/hep.28885 [DOI] [PubMed] [Google Scholar]
- 48.Chang YZ, Chai RC, Pang B, Chang X, An SY, Zhang KN, et al. METTL3 enhances the stability of MALAT1 with the assistance of HuR via m6A modification and activates NF-κB to promote the malignant progression of IDH-wildtype glioma. Cancer Lett. 2021;511:36–46. 10.1016/j.canlet.2021.04.020 [DOI] [PubMed] [Google Scholar]
- 49.Chai RC, Chang YZ, Chang X, Pang B, An SY, Zhang KN, et al. YTHDF2 facilitates UBXN1 mRNA decay by recognizing METTL3-mediated m6A modification to activate NF-κB and promote the malignant progression of glioma. J Hematol Oncol. 2021;14:109. 10.1186/s13045-021-01124-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dong F, Qin X, Wang B, Li Q, Hu J, Cheng X, et al. ALKBH5 facilitates hypoxia-induced paraspeckle assembly and IL8 secretion to generate an immunosuppressive tumor microenvironment. Cancer Res. 2021;81:5876–88. 10.1158/0008-5472.CAN-21-1456 [DOI] [PubMed] [Google Scholar]
- 51.Li Y, Xu J, Zhang J, Zhang J, Zhang J, Lu X. MicroRNA-346 inhibits the growth of glioma by directly targeting NFIB. Cancer Cell Int. 2019;19:294. 10.1186/s12935-019-1017-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gabriely G, Yi M, Narayan RS, Niers JM, Wurdinger T, Imitola J, et al. Human glioma growth is controlled by microRNA-10b. Cancer Res. 2011;71:3563–72. 10.1158/0008-5472.CAN-10-3568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chen W, Zhang B, Guo W, Gao L, Shi L, Li H, et al. miR-429 inhibits glioma invasion through BMK1 suppression. J Neurooncol. 2015;125:43–54. 10.1007/s11060-015-1887-x [DOI] [PubMed] [Google Scholar]
- 54.Santangelo A, Imbrucè P, Gardenghi B, Belli L, Agushi R, Tamanini A, et al. A microRNA signature from serum exosomes of patients with glioma as complementary diagnostic biomarker. J Neurooncol. 2018;136:51–62. 10.1007/s11060-017-2639-x [DOI] [PubMed] [Google Scholar]
- 55.Baraniskin A, Kuhnhenn J, Schlegel U, Maghnouj A, Zöllner H, Schmiegel W, et al. Identification of microRNAs in the cerebrospinal fluid as biomarker for the diagnosis of glioma. Neuro Oncol. 2012;14:29–33. 10.1093/neuonc/nor169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jiang P, Gu S, Pan D, Fu J, Sahu A, Hu X, et al. Signatures of T cell dysfunction and exclusion predict cancer immunotherapy response. Nat Med. 2018;24:1550–8. 10.1038/s41591-018-0136-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gittleman H, Sloan AE, Barnholtz-Sloan JS. An independently validated survival nomogram for lower-grade glioma. Neuro Oncol. 2020;22:665–74. 10.1093/neuonc/noz191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cancer Genome Atlas Research Network. Brat DJ, Verhaak RG, Aldape KD, Yung WK, Salama SR, Cooper LA, et al. Comprehensive, integrative genomic analysis of diffuse lower-grade gliomas. N Engl J Med. 2015;372:2481–98. 10.1056/NEJMoa1402121 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Not applicable.