Abstract
Background:
Pediatric traumatic brain injury (TBI) is common, but not all injuries require hospitalization. A computational tool for ruling-in patients who will have a clinically-relevant TBI (CRTBI) would be valuable, providing an evidence-based way to safely discharge children who are at low risk for a CRTBI. We hypothesized that an Artificial Neural Network (ANN) trained on clinical and radiologist-interpreted imaging metrics could provide a tool for identifying patients likely to suffer from a CRTBI.
Methods:
We used the prospectively-collected, publicly-available, multicenter Pediatric Emergency Care Applied Research Network (PECARN) TBI dataset. All patients with TBI under the age of 18 with admission head computed tomography (CT) imaging data were included. We constructed an ANN using clinical and radiologist-interpreted imaging metrics in order to predict CRTBI, as previously defined by PECARN: 1) Neurosurgical procedure, 2) Intubated > 24 hours as direct result of the head trauma, 3) Hospitalization ≥ 48 hours and evidence of TBI on CT, or 4) Death due to TBI.
Results:
Among 12,902 patients included in this study, 480 patients were diagnosed with CRTBI. Our ANN had a sensitivity of 99.73% with 98.19% precision, 97.98% accuracy, 91.23% negative predictive value, 0.0027% false negative rate, and 60.47% specificity for CRTBI. The area under the ROC curve was 0.9907.
Conclusions:
We are the first to utilize artificial intelligence to predict CRTBI in a clinically meaningful manner, using radiologist-interpreted CT information, in order to identify pediatric patients likely to suffer from CRTBI. This proof-of-concept study lays the groundwork for future studies incorporating iterations of this algorithm directly into the electronic medical record for real-time, data-driven predictive assistance to physicians.
Keywords: TBI, Pediatrics, Machine Learning, Artificial Intelligence
Introduction
Traumatic brain injury (TBI) affects thousands of children in the United States every year.34 Despite the large numbers of children who experience TBI, only a small percentage actually require hospitalization or prolonged surveillance.31 However, identifying which patients do require monitoring versus those that can be safety discharged from the emergency department remains an important unanswered question. Thus, creation of a tool for identifying patients at risk for clinically-relevant TBI (CRTBI) could provide an evidence-based mechanism for early safe discharge and potentially reduce unnecessary healthcare expenditures.
The Pediatric Emergency Care Applied Research Network (PECARN)1 is a consortium of 25 hospitals that developed a decision-making score based on head CT findings.16 Numerous studies have independently published on these data in an effort to develop predictive metrics to guide treatment of children with TBI, however none have used artificial neural networks (ANN).4,5,8,11,15–17,21,22,26,28,29 ANNs are a type of machine-learning (ML) algorithm that have been widely used in clinical medicine.6,7,10 ANNs are often more useful than conventional statistical methods because: 1) ANNs can take any number of input variables and predict any number of outcomes 2) ANNs are capable of improving their predictive ability over time as they are exposed to new data 3) ANNs benefit from internal validation and testing and 4) ANNs tend to have stronger discriminant ability compared to conventional statistics.2,14,32,41
Leveraging this technology, we created a model that combines clinical and radiologist-interpreted reads to predict whether or not a pediatric patient will experience a CRTBI. We quantify the accuracy and error of this algorithm and provide an open-source software package to enable prediction generation and validation. We expand on previous PECARN predictive studies by utilizing a combination of demographic, clinical, and radiologist-interpreted CT findings to investigate CRTBI in pediatric patients using ANN. We hypothesized that we could train an ANN on clinical and radiographic data to identify which pediatric TBI patients with head CT are at risk for CRTBI.
Methods
Study population
This study utilized the prospective PECARN study of children with Clinically Relevant TBI, as described previously.16,20 The PECARN TBI study enrolled patients under the age of 18 who experienced non-penetrating (i.e., blunt) head trauma that presented to the emergency department between 2004 and 2006, and had admission head CT imaging classification. All data analyzed in this study was de-identified and our study was approved by the Vanderbilt University Institutional Review Board. We included patients who had complete data available for all variables of interest, and thus did not impute any missing variables. 14,969 patients underwent head CT, of which 12,902 patients had complete imaging information.
Analysis and Variables Included
Descriptive statistics including Pearson correlation and t-test were used to evaluate the normally distributed cohort. Statistical significance was set a priori at P < 0.05. The input variables included in our ANN are as follows: 1) Mechanism of injury (e.g., motor vehicle collision, pedestrian struck by moving vehicle, bicycle rider struck by automobile, bicycle collision or fall from bicycle, other wheeled transport crash, fall to ground from standing/walking/running, walked or ran into stationary object, fall from an elevation, fall down stairs, sports, assault, objective struck head- accidental, and other etiology of injury); 2) Severity of injury mechanism [low (e.g., fall from ground level and walked/ran into stationary object), moderate (any other mechanism), high (e.g., motor vehicle collision with patient ejection, death of another passenger, or rollover, pediatric or bicyclist without helmet struck by motor vehicle, falls > 5 feet or patients 2 years and older, falls of > 3 feet < 2 years old)]; 3) Loss of consciousness; 4) Glasgow Coma Scale at presentation; 5) Age; and 6) Sex.
The 17 variables identified by radiologists on CT imaging included the presence or absence of the following: cerebellar hemorrhage, cerebral contusion, cerebral edema, cerebral hemorrhage/intracerebral hematoma, diastasis of the skull, epidural hematoma, extra-axial hematoma, intraventricular hemorrhage, midline shift/shift of brain structures, pneumocephalus, skull fracture (and cerebral spinal fluid leak), subarachnoid hemorrhage, subdural hematoma, traumatic infarction, diffuse axonal injury, herniation and, shear injury. Head CTs were interpreted by attending radiologists at each clinical site and a blinded pediatric radiologist made definitive interpretations on scans that were difficult to interpret.20 Each site was responsible for ensuring the accuracy of their data reported to PECARN. In addition, a subsequent study detailed consistent inter-rater reliability in all data collected by the PECARN consortium.23
Our outcome of interest is “clinically-relevant TBI (CRTBI),” a composite of several variables as defined by the PECARN investigators.20 The CRTBI variables consisted any of the following: 1) Neurosurgical procedure (e.g., dura repair for cerebrospinal fluid leak, fracture elevation, hematoma drainage, intracranial pressure monitor placement, lobectomy, tissue debridement, ventriculostomy, and “other” neurosurgical procedure) 2) Intubated > 24 hours as direct result of the head trauma 3) Hospitalization ≥ 48 hours and evidence of TBI on head CT 4) Death due to TBI.
Artificial Neural Network Analysis
We trained an ANN using offline MATLAB R2016b (9.1.0.441655) on a 64-bit MacBook Pro running OS 10.11.6. We randomly partitioned patients into three groups in order to provide holdout validation on our large dataset; 70% were for training the ANN; 15% were for validating the ANN; and 15% were for subsequent final testing of the ANN. The ANN had not been exposed to any of the final test patients until after the model was finished training and validating. A two-layer, feed-forward ANN with 11 sigmoid hidden and softmax output neurons were trained using the scaled conjugate gradient back-propagation method on the dedicated partition. We tabulated confusion tables and statistics on the testing partition, as well as for the entire dataset. We assessed the predictive ability of the model rigorously with various numerical measures of accuracy, precision, and error.
Results
In this study, we included 12,902 patients of which 63% were male and the average age was 7.99 ± 5.91 years (Table 1). Of the 12,902 patients included in these analyses, 480 suffered a CRTBI. Aside from age and gender, all other clinical and imaging variables had a univariate association with CRTBI (Table 2).
Table 1: Patient Characteristics of Pediatric Traumatic Brain Injury (TBI) Patients studied using an Artificial Neural Network.
Our outcome of interest is clinically-relevant TBI, a composite of several variables as defined by the PECARN (Pediatric Emergency Care Applied Research Network) investigators. Clinically-relevant TBI included any of the following: 1) Neurosurgical procedure 2) Intubated > 24 hours as direct result of the head trauma 3) Hospitalization ≥ 48 hours, and 4) Death due to TBI.
| Variable | Non-Clinically Relevant TBI (n=12,422) | Clinically Relevant TBI (n=480) | All patients (n=12,902) | P Value* |
|---|---|---|---|---|
|
| ||||
| Age (Mean ± SD) | 8.00 ± 5.92 | 7.88 ± 5.67 | 7.00 ± 5.91 | P=0.648 |
|
| ||||
| Gender Ratio (M:F) | 1.71 | 1.89 | 1.72 | P=0.305 |
|
| ||||
| Severity of Injury | ||||
| Low | 1807 | 25 | 1832 | P <0.001 |
| Moderate | 7798 | 243 | 8041 | |
| High | 2817 | 212 | 3029 | |
|
| ||||
| Loss of Consciousness | ||||
| No | 7928 | 173 | 8101 | P <0.001 |
| Yes | 3220 | 239 | 3459 | |
| NOS | 1274 | 68 | 1342 | |
|
| ||||
| GCS Total | 15 | 15 | 15 | P <0.001 |
Univariate statistical significance examined using the t-test or Pearson’s chi-squared test.
List of Abbreviations:
GCS = Glasgow Coma Scale
M:F = Male:Female Ratio
n = number of patients
NOS = Not Otherwise Specified
Table 2: Head Computed Tomography (CT) Findings of Pediatric Traumatic Brain Injury (TBI) Patients studied using an Artificial Neural Network.
| Image Findings | Non-Clinically Important TBI (n=12,422) | Clinically Important TBI (n=480) | All patients (n=12,902) |
|---|---|---|---|
|
| |||
| Cerebellar hemorrhage | |||
| No | 12418 | 469 | 12887 |
| Yes | 4 | 11 | 15 |
|
| |||
| Cerebral contusion | |||
| No | 12365 | 365 | 12730 |
| Yes | 57 | 115 | 172 |
|
| |||
| Cerebral edema | |||
| No | 12415 | 412 | 12827 |
| Yes | 7 | 68 | 75 |
|
| |||
| Cerebral hemorrhage or Intracerebral hematoma | |||
| No | 12384 | 381 | 12765 |
| Yes | 38 | 99 | 137 |
|
| |||
| Diastasis of the skull | |||
| No | 12403 | 445 | 12848 |
| Yes | 19 | 34 | 54 |
|
| |||
| Epidural hematoma | |||
| No | 12402 | 394 | 12796 |
| Yes | 20 | 86 | 106 |
|
| |||
| Extra axial hematoma | |||
| No | 12358 | 411 | 12769 |
| Yes | 64 | 69 | 133 |
|
| |||
| Intraventricular hemorrhage | |||
| No | 12415 | 456 | 12871 |
| Yes | 7 | 24 | 31 |
|
| |||
| Midline shift of brain structures | |||
| No | 12416 | 401 | 12817 |
| Yes | 6 | 79 | 85 |
|
| |||
| Pneumocephalus | |||
| No | 12361 | 363 | 12724 |
| Yes | 61 | 117 | 178 |
|
| |||
| Skull fracture | |||
| No | 11828 | 184 | 12012 |
| Yes | 594 | 296 | 890 |
|
| |||
| Subarachnoid hemorrhage | |||
| No | 12356 | 369 | 12725 |
| Yes | 66 | 111 | 177 |
|
| |||
| Subdural hematoma | |||
| No | 12348 | 337 | 12685 |
| Yes | 74 | 143 | 217 |
|
| |||
| Traumatic infarction | |||
| No | 12422 | 476 | 12898 |
| Yes | 0 | 4 | 4 |
|
| |||
| Diffuse axonal injury | |||
| No | 12422 | 475 | 12897 |
| Yes | 0 | 5 | 5 |
|
| |||
| Herniation | |||
| No | 12422 | 475 | 12897 |
| Yes | 0 | 5 | 5 |
|
| |||
| Shear Injury | |||
| No | 12419 | 466 | 12885 |
| Yes | 3 | 14 | 17 |
Univariate statistical significance examined using the t-test or Chi-squared analysis.
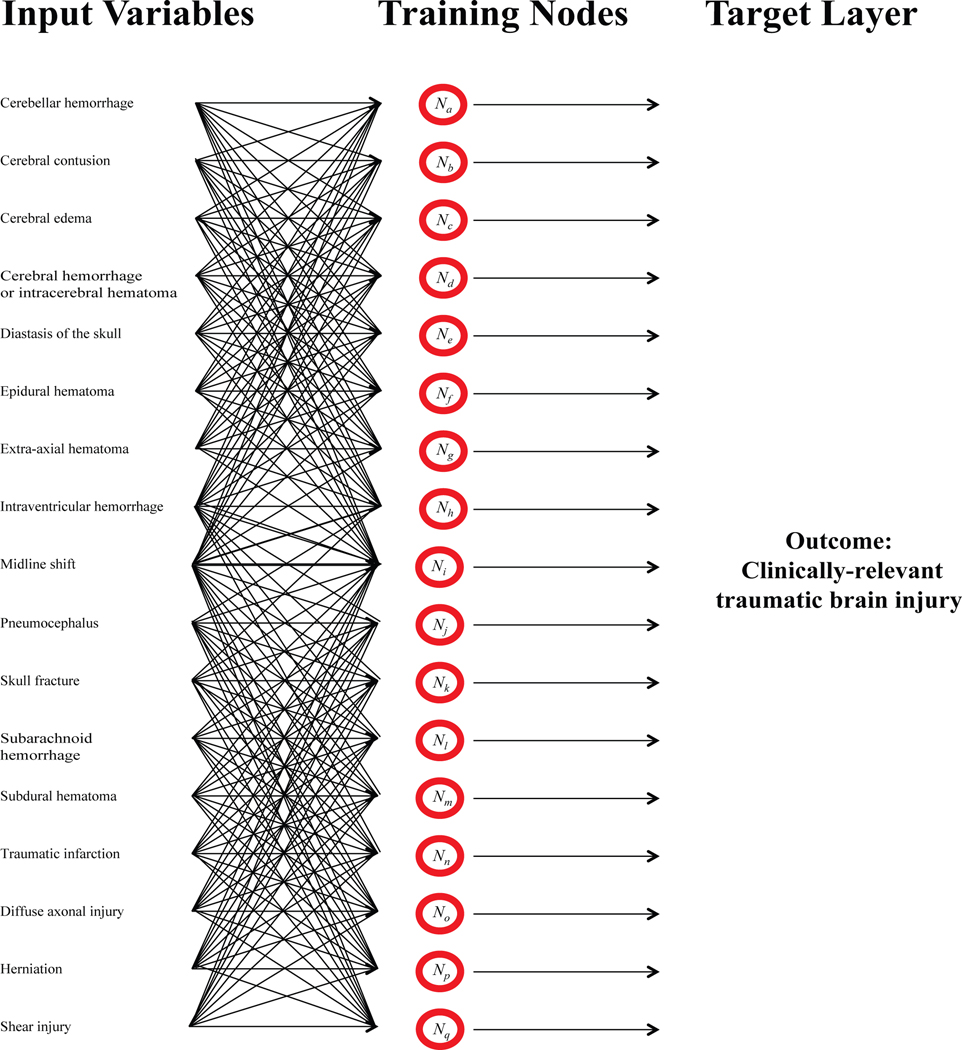
The ANN has a sensitivity of 99.73% and a negative predictive value (NPV) of 91.23% for CRTBI in the testing cohort (Table 3). When the data used for testing were combined with the remaining 85% of data, which the network was trained and validated on, the sensitivity remained very high at 99.54% and NPV of 84.38% (Table 3). Determination of specificity was much lower at 60.47% for testing and 64.17% for the entire dataset. We included other statistical measures of the ANN binary classifier (Table 3). A pictorial representation of the ANN constructed here is shown in Figure 1.
Table 3. Confusion Table Statistics: Testing Results* of an Artificial Neural Network on Pediatric Traumatic Brain Injury (TBI) Patients.
We randomly partitioned patients into three groups in order to provide holdout validation on our large dataset; 70% were for training the ANN; 15% were for validating the ANN; and 15% were for subsequent final testing of the ANN. Various measures of predictive ability on the test patients, as well as test patients combined with those used for training and validation are presented as proportions.
| Measure | Results (Test Group) | Results (All Patients) |
|---|---|---|
| Sensitivity | 0.9973 | 0.9954 |
| Specificity | 0.6047 | 0.6417 |
| Precision | 0.9819 | 0.9863 |
| Negative Predictive Value | 0.9123 | 0.8438 |
| False Positive Rate | 0.3953 | 0.3583 |
| False Discovery Rate | 0.0181 | 0.0137 |
| Accuracy | 0.0027 | 0.0046 |
| F1 Score | 0.9798 | 0.9823 |
| Matthews Correlation Coefficient | 0.7337 | 0.7272 |
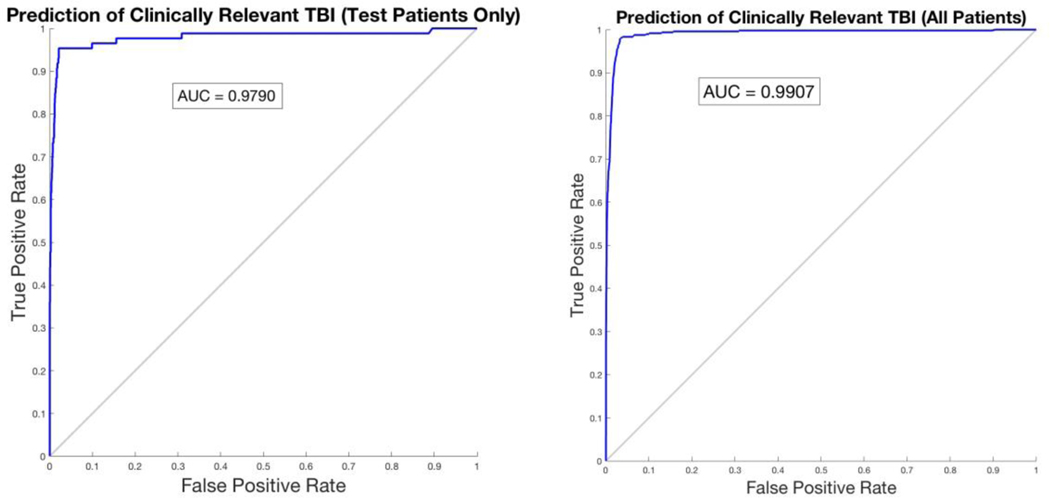
| Area under ROC Curve | 0.9907 | 0.9790 |
Figure 1: Schematic of the artificial neural network (ANN) constructed here.
Seventeen input variables were compared, converging on more than 100 training nodes (less training nodes were shown for simplicity). Each input variable connects, analogous to projections in neurons, to each training node. Arbitrary “weights” are then applied to each variable. Each training node is then used to determine the best “weights” of each variable to predict the outcome of interest (“target layer”).
Receiver Operator Characteristic (ROC) curves for both test patients and the entire dataset are calculated and provided, as seen in Figure 2. The Area Under the ROC (AUROC) was 0.9907 for prediction of CRTBI in test patients and 0.9790 for the entire data set.
Figure 2: Receiver Operating Characteristic curve for Artificial Neural Network (ANN) predictions of clinically-relevant traumatic brain injury.
We randomly partitioned patients into three groups in order to provide holdout validation on our large dataset; 70% were for training the ANN; 15% were for validating the ANN; and 15% were for subsequent final testing of the ANN. The ANN had not been exposed to any of the test patients until after the model was finished training. The ROC for the testing set of patients (left) and the ROC for the entire dataset (right).
Discussion
We constructed and validated an artificial neural network (ANN), a machine-learning computational algorithm, to predict CRTBI in children using clinical and imaging data. This platform has apparent clinical utility for the inexperienced pediatric emergency care provider to assign admission for children with TBI given its very high sensitivity for CRTBI, which has small prevalence (<5%) but serious consequence, such as future intracranial procedure(s), respiratory failure, prolonged hospitalization, and/or mortality.34 This would be the first study to our knowledge aiming at predicting TBI of any type in any patient population.
Predictive outcome and prognostication models are becoming increasingly important across medicine and surgery (e.g., CHA2DS2-VASc score,12,13 Acute Physiology and Chronic Health Evaluation [APACHE],18 Sequential Organ Failure Assessment [SOFA]40), and modeling techniques have evolved over the years. These models have classically relied on logistic regression or conventional statistics to generate predictions, and often use fewer input variables that are manually entered. More recently, artificial neural networks (ANN) have been shown to robustly predict complications, outcomes and prognosis among numerous fields,6,14,32,35–37 including TBI.9,19,27,33,38,39 Thus, an ANN tool could yield predictive information for CRTBI would helpful and provide an evidence-based mechanism for treating these patients.
ANNs are computational constructs used to interpret the maximum number of combinations of data in complex systems, like making medical diagnoses, where many competing factors influence the outcome.3 During training of the ANN, random “weights” are assigned to each input variable, compared against every variable in the model, and then used to predict the strength of correlation with the outcome of interest (Figure 1). While there is no maximum number of variables that can be included in an ANN, addition of irrelevant variables will not make the data prediction any stronger.42 Thus, we chose to rationally design the ANN described here by only including variables which had previously been shown by univariate statistics to be significantly associated with CRTBI.
We trained an ANN on data collected from PECARN and successfully developed a very sensitive (Sensitivity= 99.73% AUC= 0.9907) tool for identifying CRTBI (NPV = 91%) in children. We optimized the ANN for sensitivity over specificity to conservatively identify patients likely to be diagnosed with CRTBI. Future iterations of this ANN with additional variables and data not available through PECARN, could be similarly leveraged to optimize specificity, thereby safely ruling out disease. However, since PECARN is a group consisting of 25 hospitals and collected data prospectively, these data most accurately reflect the epidemiological and treatment diversity seen across North America for pediatric TBI. Importantly, the number of variables included in the predictive ANN algorithm can be greatly increased compared to prior risk-calculation tools due to the overwhelmingly computational superiority of machine-learning compared to conventional statistical approaches, which are limited by degrees of freedom.30,32,38,42 Our intent was to provide software allowing for real-world incorporation of data into a standalone application or an EMR. Future applications could self-collect this clinical data (i.e., our published algorithm depends on collected and interpreted data), and therefore would integrate all necessary input data from electronic medical record systems, and provide results for the clinician on-the-ground. Another strength of this study is its external generalizability, as we did not further divide our cohort based on any further head-injury patterns (e.g., subdural, epidural, intraparenchymal hematoma, shear) often seen in the literature, as we wanted to reflect the full-spectrum of pediatric patients with all severities and pathoanatomic types of TBI presenting to any emergency department.
In the authors’ opinion, future iterations of ANN-based predictive modeling should be center around three guiding principles: 1) prospective data collection leading to real-time updates and refinement of the algorithm, 2) directly linking ANN models to the electronic health record and 3) increase in the granularity of data available for training the ANN, for instance, using image-based processing. First, compared to traditional statistical approaches which require new analyses to be performed each time new data is added, ANNs can be constantly updated, providing real-time, up-to-date information and quantitative evidence. ANNs could be designed to be using national, regional, or even provider-specific data. Second, directly linking ANNs to the electronic health record would provide streamlined data collection and up-to-date predictive capabilities based on the most current evidence. Lastly, ANNs could be trained directly on the CT images themselves, leading to quicker diagnosis, prognosis and better utilization of hospital resources.
Although we lay the ground-work for incorporating machine-learning into evaluation of children with TBI, this study is not without limitations. First, because machine-learning algorithms are computational constructs that are not familiar to most physicians, these models can be seen as foreign and/or unproven entities.7 However, as the importance of utilizing “big data” increases, utilizing AI and ML will inevitably be tools used going forward.7,10,25 Second, despite the very large number of total patients, the number of patients in each individual subset of CRTBI was low. However, with additional data, we believe we can create more sophisticated models with higher specificity in the future providing even better data on who can be safely discharged without risk for readmission. Furthermore, we were not able to incorporate standardized metrics observed during the patient’s physical exam, details that are difficult to quantify and capture. Thus, while algorithm-based decision tools can be useful in guiding the physician’s decision, these constructs absolutely do not replace the information that can only be obtained by a trained physician. Third, each patient in our study obtained head CT imaging, an assumption in itself that our model is heavily dependent on, a decision that is not standardized across institutions and likely changes over time. There is an extensive literature on the utility and safety of head CT for mild TBI in children since these data’s collection.24 These imaging data have been dichotomized without providing further quantification per covariate (e.g. degree of midline shift, quantification of hemorrhage). In reality, these CT images are interpreted by a combination of emergency medicine and/or night-hawk radiologists, such that decisions would be made way before a complex research-level interpretation could be accomplished. Lastly, we used a single-data source (PECARN) that is publicly-available and has undergone rigorous quality-improvement. However, we are limited by the clinical-practice standards of those years (2004–2006), including the rationale and threshold to obtain head-CT imaging in children. Further computational restructuring of our ANN model may also provide additional metrics for future studies that analyze the CT data directly instead of the radiologist interpretation.
We posit that in-hospital use of the model may actually increase the power of the algorithm as ANNs can be trained on new data, and has potential to be incorporated as a future online tool or packaged into the electronic medical records system (available for download in the Supplemental Material), but would require much of the heavy research-level classification to be performed immediately for this to be time-sensitive and clinically relevant. Currently, much of the trauma registry classifications, clinical documentation, and final imaging reads are done well-after clinical decisions are made, and often times only fully complete well after patient discharge or death.
Conclusions
Training an ANN model using data from PECARN, we have constructed a highly sensitive tool to diagnose CRTBI. Further iterations of this ANN may bring real-time, data-driven updates to the hands of pediatric emergency providers in order to provide the most accurate evidence-based care, and particularly aid mid-level and/or inexperience practitioners in small outlying or austere facilities. Immediate identification of pediatric TBI patients who are likely to require additional hospital resources allows clinical teams and hospital administrators to work synergistically to provide the best clinical care. We believe that approaches like our ANN can offer more robust and accurate predictions that can be updated prospectively in real-time.
Acknowledgements
A.T.H. receives salary and tuition support through the Vanderbilt University School of Medicine Medical Scientist Training Program (5T32GM007347). This project is supported by the Vanderbilt Clinical and Translational Science Award (UL1RR024975, NIH/NCATS, M.B.P.). M.B.P. is funded by R01 (GM120484, NIH/NIGMS), and a Vanderbilt Faculty Scholars Award. We would also like to thank the Surgical Outcomes Center for Kids at Monroe Carell Jr. Children’s Hospital of Vanderbilt University for administrative support. No portions of these data have been published or presented elsewhere. The authors declare no conflicts of interest. A.T.H. and D.P.S. had full access to all the data in the study and take full responsibility for the integrity of the data and the accuracy of the data analysis. A.T.H. lead the study and was responsible for its conception. A.T.H and D.P.S. designed the study, drafted the manuscript, and are responsible for data integrity. D.P.S. performed the computational modeling and software packaging. J.L. collected data, analyzed data, and critically revised the manuscript. O.D.G., C.N.S., M.B.P, supervised the study, aided with interpretation of analysis, provided feedback, and critically revised the manuscript.
References
- 1.The Pediatric Emergency Care Applied Research Network (PECARN): rationale, development, and first steps. Pediatr Emerg Care 19:185–193, 2003 [DOI] [PubMed] [Google Scholar]
- 2.Ahmed FE: Artificial neural networks for diagnosis and survival prediction in colon cancer. Mol Cancer 4:29, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Amato F, López A, Peña-Méndez EM, Vaňhara P, Hampl A, Havel J: Artificial neural networks in medical diagnosis. Journal of Applied Biomedicine 11:47–58, 2013 [Google Scholar]
- 4.Babl FE, Borland ML, Phillips N, Kochar A, Dalton S, McCaskill M, et al. : Accuracy of PECARN, CATCH, and CHALICE head injury decision rules in children: a prospective cohort study. Lancet 389:2393–2402, 2017 [DOI] [PubMed] [Google Scholar]
- 5.Badawy MK, Dayan PS, Tunik MG, Nadel FM, Lillis KA, Miskin M, et al. : Prevalence of Brain Injuries and Recurrence of Seizures in Children With Posttraumatic Seizures. Acad Emerg Med 24:595–605, 2017 [DOI] [PubMed] [Google Scholar]
- 6.Baxt WG: Application of artificial neural networks to clinical medicine. Lancet 346:1135–1138, 1995 [DOI] [PubMed] [Google Scholar]
- 7.Beam AL, Kohane IS: Translating Artificial Intelligence Into Clinical Care. Jama 316:2368–2369, 2016 [DOI] [PubMed] [Google Scholar]
- 8.Borgialli DA, Mahajan P, Hoyle JD Jr., Powell EC, Nadel FM, Tunik MG, et al. : Performance of the Pediatric Glasgow Coma Scale Score in the Evaluation of Children With Blunt Head Trauma. Acad Emerg Med 23:878–884, 2016 [DOI] [PubMed] [Google Scholar]
- 9.Chong SL, Liu N, Barbier S, Ong ME: Predictive modeling in pediatric traumatic brain injury using machine learning. BMC Med Res Methodol 15:22, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Darcy AM, Louie AK, Roberts LW: Machine Learning and the Profession of Medicine. Jama 315:551–552, 2016 [DOI] [PubMed] [Google Scholar]
- 11.Dayan PS, Holmes JF, Hoyle J Jr., Atabaki S, Tunik MG, Lichenstein, et al. : Headache in traumatic brain injuries from blunt head trauma. Pediatrics 135:504–512, 2015 [DOI] [PubMed] [Google Scholar]
- 12.Gage BF, van Walraven C, Pearce L, Hart RG, Koudstaal PJ, Boode BS, et al. : Selecting patients with atrial fibrillation for anticoagulation: stroke risk stratification in patients taking aspirin. Circulation 110:2287–2292, 2004 [DOI] [PubMed] [Google Scholar]
- 13.Gage BF, Waterman AD, Shannon W, Boechler M, Rich MW, Radford MJ: Validation of clinical classification schemes for predicting stroke: results from the National Registry of Atrial Fibrillation. Jama 285:2864–2870, 2001 [DOI] [PubMed] [Google Scholar]
- 14.Gholipour C, Rahim F, Fakhree A, Ziapour B: Using an Artificial Neural Networks (ANNs) Model for Prediction of Intensive Care Unit (ICU) Outcome and Length of Stay at Hospital in Traumatic Patients. J Clin Diagn Res 9:OC19–23, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Glass T, Ruddy RM, Alpern ER, Gorelick M, Callahan J, Lee L, et al. : Traumatic brain injuries and computed tomography use in pediatric sports participants. Am J Emerg Med 33:1458–1464, 2015 [DOI] [PubMed] [Google Scholar]
- 16.Greenberg JK, Yan Y, Carpenter CR, Lumba-Brown A, Keller MS, Pineda JA, et al. : Development and Internal Validation of a Clinical Risk Score for Treating Children With Mild Head Trauma and Intracranial Injury. JAMA Pediatr 171:342–349, 2017 [DOI] [PubMed] [Google Scholar]
- 17.Ide K, Uematsu S, Tetsuhara K, Yoshimura S, Kato T, Kobayashi T: External Validation of the PECARN Head Trauma Prediction Rules in Japan. Acad Emerg Med 24:308–314, 2017 [DOI] [PubMed] [Google Scholar]
- 18.Knaus WA, Draper EA, Wagner DP, Zimmerman JE: APACHE II: a severity of disease classification system. Crit Care Med 13:818–829, 1985 [PubMed] [Google Scholar]
- 19.Kreif N, Grieve R, Diaz I, Harrison D: Evaluation of the Effect of a Continuous Treatment: A Machine Learning Approach with an Application to Treatment for Traumatic Brain Injury. Health Econ 24:1213–1228, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kuppermann N, Holmes JF, Dayan PS, Hoyle JD, Jr., Atabaki SM, Holubkov R, et al. : Identification of children at very low risk of clinically-important brain injuries after head trauma: a prospective cohort study. Lancet 374:1160–1170, 2009 [DOI] [PubMed] [Google Scholar]
- 21.Lee LK, Monroe D, Bachman MC, Glass TF, Mahajan PV, Cooper A, et al. : Isolated loss of consciousness in children with minor blunt head trauma. JAMA Pediatr 168:837–843, 2014 [DOI] [PubMed] [Google Scholar]
- 22.Magana JN, Kuppermann N: The PECARN TBI Rules Do Not Apply to Abusive Head Trauma. Acad Emerg Med 24:382–384, 2017 [DOI] [PubMed] [Google Scholar]
- 23.Marcin JP, Romano PS, Dharmar M, Chamberlain JM, Dudley N, Macias CG, et al. : Implicit Review Instrument to Evaluate Quality of Care Delivered by Physicians to Children in Emergency Departments. Health Serv Res, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Miglioretti DL, Johnson E, Williams A, Greenlee RT, Weinmann S, Solberg LI, et al. : The use of computed tomography in pediatrics and the associated radiation exposure and estimated cancer risk. JAMA Pediatr 167:700–707, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Murdoch TB, Detsky AS: The inevitable application of big data to health care. Jama 309:1351–1352, 2013 [DOI] [PubMed] [Google Scholar]
- 26.Nakhjavan-Shahraki B, Yousefifard M, Hajighanbari MJ, Oraii A, Safari S, Hosseini M: Pediatric Emergency Care Applied Research Network (PECARN) prediction rules in identifying high risk children with mild traumatic brain injury. Eur J Trauma Emerg Surg 43:755–762, 2017 [DOI] [PubMed] [Google Scholar]
- 27.Nielson JL, Cooper SR, Yue JK, Sorani MD, Inoue T, Yuh EL, et al. : Uncovering precision phenotype-biomarker associations in traumatic brain injury using topological data analysis. PLoS One 12:e0169490, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nigrovic LE, Lee LK, Hoyle J, Stanley RM, Gorelick MH, Miskin M, et al. : Prevalence of clinically important traumatic brain injuries in children with minor blunt head trauma and isolated severe injury mechanisms. Arch Pediatr Adolesc Med 166:356–361, 2012 [DOI] [PubMed] [Google Scholar]
- 29.Nigrovic LE, Lillis K, Atabaki SM, Dayan PS, Hoyle J, Tunik MG, et al. : The prevalence of traumatic brain injuries after minor blunt head trauma in children with ventricular shunts. Ann Emerg Med 61:389–393, 2013 [DOI] [PubMed] [Google Scholar]
- 30.Obermeyer Z, Emanuel EJ: Predicting the Future - Big Data, Machine Learning, and Clinical Medicine. N Engl J Med 375:1216–1219, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Palchak MJ, Holmes JF, Vance CW, Gelber RE, Schauer BA, Harrison MJ, et al. : Does an isolated history of loss of consciousness or amnesia predict brain injuries in children after blunt head trauma? Pediatrics 113:e507–513, 2004 [DOI] [PubMed] [Google Scholar]
- 32.Penny W, Frost D: Neural networks in clinical medicine. Med Decis Making 16:386–398, 1996 [DOI] [PubMed] [Google Scholar]
- 33.Pourahmad S, Hafizi-Rastani I, Khalili H, Paydar S: Identifying Important Attributes for Prognostic Prediction in Traumatic Brain Injury Patients. A Hybrid Method of Decision Tree and Neural Network. Methods Inf Med 55:440–449, 2016 [DOI] [PubMed] [Google Scholar]
- 34.Quayle KS, Powell EC, Mahajan P, Hoyle JD Jr., Nadel FM, Badawy MK, et al. : Epidemiology of blunt head trauma in children in U.S. emergency departments. N Engl J Med 371:1945–1947, 2014 [DOI] [PubMed] [Google Scholar]
- 35.Rughani AI, Dumont TM, Lu Z, Bongard J, Horgan MA, Penar PL, et al. : Use of an artificial neural network to predict head injury outcome. J Neurosurg 113:585–590, 2010 [DOI] [PubMed] [Google Scholar]
- 36.Segal ME, Goodman PH, Goldstein R, Hauck W, Whyte J, Graham JW, et al. : The accuracy of artificial neural networks in predicting long-term outcome after traumatic brain injury. J Head Trauma Rehabil 21:298–314, 2006 [DOI] [PubMed] [Google Scholar]
- 37.Senders JT, Arnaout O, Karhade AV, Dasenbrock HH, Gormley WB, Broekman ML, et al. : Natural and Artificial Intelligence in Neurosurgery: A Systematic Review. Neurosurgery, 2017 [DOI] [PubMed] [Google Scholar]
- 38.Senders JT, Staples PC, Karhade AV, Zaki MM, Gormley WB, Broekman MLD, et al. : Machine Learning and Neurosurgical Outcome Prediction: A Systematic Review. World Neurosurg 109:476–486.e471, 2018 [DOI] [PubMed] [Google Scholar]
- 39.Senders JT, Zaki MM, Karhade AV, Chang B, Gormley WB, Broekman ML, et al. : An introduction and overview of machine learning in neurosurgical care. Acta Neurochir (Wien), 2017 [DOI] [PubMed] [Google Scholar]
- 40.Vincent JL, Moreno R, Takala J, Willatts S, De Mendonca A, Bruining H, et al. : The SOFA (Sepsis-related Organ Failure Assessment) score to describe organ dysfunction/failure. On behalf of the Working Group on Sepsis-Related Problems of the European Society of Intensive Care Medicine. Intensive Care Med 22:707–710, 1996 [DOI] [PubMed] [Google Scholar]
- 41.Walczak S: Artificial neural network medical decision support tool: predicting transfusion requirements of ER patients. IEEE Trans Inf Technol Biomed 9:468–474, 2005 [DOI] [PubMed] [Google Scholar]
- 42.Zou J, Han Y, So SS: Overview of artificial neural networks. Methods Mol Biol 458:15–23, 2008 [DOI] [PubMed] [Google Scholar]