Abstract
Respiratory syncytial virus (RSV) is one of the most common etiological agents of global acute respiratory tract infections with a disproportionate burden among infants, individuals over the age of 65, and immunocompromised populations. The two major subtypes of RSV (A and B) co-circulate with a predominance of either group during different epidemic seasons, with frequently emerging genotypes due to RSV’s high genetic variability. Global surveillance systems have improved our understanding of seasonality, disease burden, and genomic evolution of RSV through genotyping by sequencing of attachment (G) glycoprotein. However, the integration of these systems into international infrastructures is in its infancy, resulting in a relatively low number (~2200) of publicly available RSV genomes. These limitations in surveillance hinder our ability to contextualize RSV evolution past current canonical attachment glycoprotein (G)-oriented understanding, thus resulting in gaps in understanding of how genetic diversity can play a role in clinical outcome, therapeutic efficacy, and the host immune response. Furthermore, utilizing emerging RSV genotype information from surveillance and testing the impact of viral evolution using molecular techniques allows us to establish causation between the clinical and biological consequences of arising genotypes, which subsequently aids in informed vaccine design and future vaccination strategy. In this review, we aim to discuss the findings from current molecular surveillance efforts and the gaps in knowledge surrounding the consequence of RSV genetic diversity on disease severity, therapeutic efficacy, and RSV–host interactions.
Keywords: clinical outcomes, genetic diversity, genotype, molecular surveillance, respiratory syncytial virus, therapeutic design, whole-genome sequencing
Introduction
Respiratory syncytial virus (RSV) is one of the leading causes of acute respiratory tract infections (ARTIs) worldwide, with most of the disease burden occurring in pediatric,1 elderly,2 and immunocompromised3 populations. There are estimated to be over 33 million cases of RSV infection every year, with many children experiencing their first case of RSV by the age of 2.4 While most cases cause only mild to moderate symptoms, susceptible populations may experience more severe clinical manifestations, including pneumonia and respiratory failure. In addition, infants can experience serial infections over the course of their lives and into adulthood as a result of waning antibody response to RSV.5–7 With no available vaccine and a single monoclonal antibody treatment (palivizumab) that is costly and restricted to use in high-risk infants, there is an overwhelming need to identify new antibody-based and antiviral therapeutics for the treatment of severe disease.8
Despite its global significance, the disease burden of RSV has been difficult to quantify due in large part to a lack of surveillance and reporting, particularly in resource-restricted countries.9 Even less is understood about how the ongoing evolution of RSV is impacting the epidemiology of the virus, the clinical manifestation of disease, and therapeutic efficacy. There are currently two major antigenic groups that are known to circulate in the human population, RSV-A and RSV-B, but studies comparing the clinical severity of disease between these groups have failed to reach consensus.10 Variants within each antigenic group have been described through RSV genotyping and attachment glycoprotein G sequencing, but there is a lack of available sequence information to clearly define subtypes and determine their impact on disease.11–13 As a point of comparison, there are currently over 9.6 million whole-genome sequences of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) available in the Global Initiative on Sharing All Influenza Data (GISAID) database since 2020, but only about 2200 RSV whole-genome sequences are currently available despite RSV’s identification in 1955. To further complicate these assessments, the public health measures implemented to mitigate the coronavirus disease 2019 (COVID-19) pandemic have had a suppressive effect on RSV cases, decreasing overall case counts and disrupting the previously observed seasonal patterns.14,15 Predictive models suggest a resurgence of RSV once COVID-19 mitigation measures lapse, but it is unclear what new epidemiological patterns and genotypes may emerge.14 In an international longitudinal observation study, rebound RSV cases were identified across 11 countries after easing nonpharmaceutical interventions (NPIs) such as re-opening academic institutions, showcasing the need for molecular surveillance in real-time to track potential increases in potential population susceptibility.16
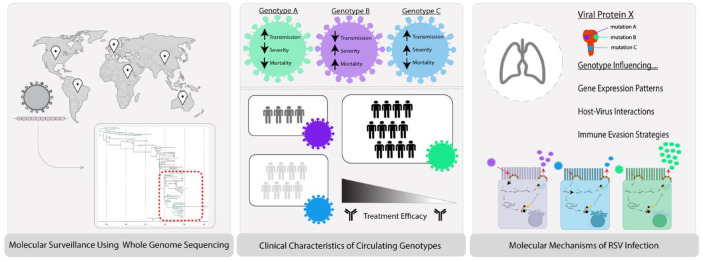
In this review, we will evaluate and discuss what is known and what remains to be discovered about the molecular epidemiology of RSV and the clinical and biological significance of RSV genetic diversity. We will address this topic in three sections: (1) RSV global surveillance and viral evolution, (2) the impact of RSV genetic diversity on clinical outcomes and therapeutics, and (3) the functional consequences of these genetic changes on RSV biology (Figure 1). Finally, we discuss new tools, techniques, and approaches that can inform future research on RSV toward development of safe, effective, and broadly applicable therapeutics.
Figure 1.
Representation of molecular surveillance pipeline. Sample collection and preparation of clinical samples from RSV positive populations from global locations in surveillance systems give rise to the ability to track RSV seasonality and rise in emerging genotypes. When performing phylogenetic analysis of the RSV whole-genome sequences, certain circulating genotypes may present differential clinical characteristics, such as clinical severity, transmission, and mortality. These genotypes (depicted in green, purple, and blue) may have varying infectivity, resulting in varying population size with a wide range of treatment efficacy. When observing the molecular underpinnings of each genotype, mutations within RSV open reading frames (ORFs) unique to defined genotypes may influence gene expression, host–pathogen interactions, and immune evasion strategies.
Methods
G and WGS sequences from GenBank
To identify the number of RSV genome sequences and G sequences publicly available and deposited, we downloaded both the accession numbers and corresponding collection_date information from NCBI GenBank during April 2020. To identify RSV G sequences, the following search terms were used:
((‘Human orthopneumovirus’[Organism] OR human respiratory syncytial virus[All Fields] AND attachment[All Fields] AND glycoprotein[All Fields]) AND ‘Human orthopneumovirus’[porgn] NOT mutant[All Fields] NOT attenuated[All Fields] NOT chimeric[All Fields] NOT recombinant[All Fields] NOT unverified [All Fields] NOT pangolin[All Fields] AND (viruses[filter] AND [PROP] AND [filter] AND [filter]).
Of 2203 of these obtained genome sequences, 2172 sequences had collection date information between 2000 and 2022; the number of sequences per year were plotted. To identify complete genome RSV sequences, the following search terms were used:
(((((((‘Respiratory syncytial virus’[Organism] OR (‘Respiratory syncytial virus’[Organism] OR Respiratory Syncytial Virus[All Fields])) OR ‘Respiratory syncytial virus’[Organism]) NOT attenuated[All Fields]) NOT mutant[All Fields]) NOT unverified[All Fields]) NOT chimeric[All Fields]) AND (‘Human orthopneumovirus’[Organism] OR (‘Human orthopneumovirus’[Organism] OR Human orthopneumovirus[All Fields])) AND complete[All Fields] AND (viruses[filter] AND biomol_genomic[PROP] AND is_nuccore[filter] AND (‘14000’[SLEN]: ‘17000’[SLEN])) NOT pangolin[All Fields] NOT unverified[All Fields]) AND ‘Human orthopneumovirus’[porgn] AND (viruses[filter] AND biomol_genomic[PROP] AND ddbj_embl_genbank[filter] AND is_nuccore[filter] AND (‘15000’[SLEN]: ‘15300’[SLEN])).
Of the 19,214 sequences obtained, 10,890 sequences had collection date information between 2000 and 2022; the number of sequences per year were plotted.
RSV lifecycle
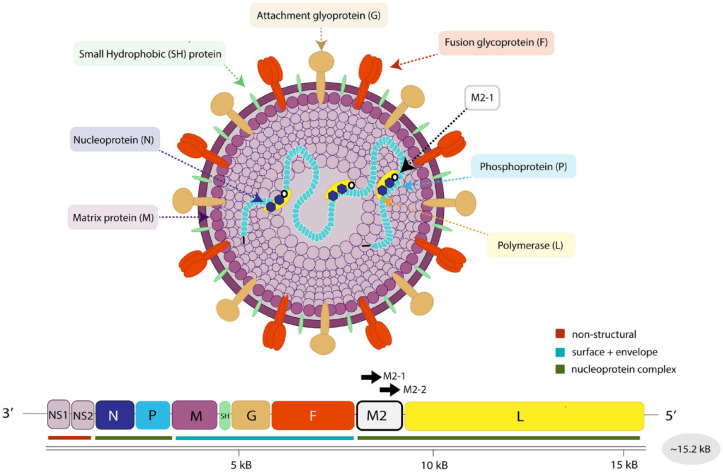
RSV was first isolated in 1957 at Johns Hopkins University in Baltimore, Maryland, from a child with bronchopneumonia and other respiratory tract infection complications.17 RSV is an enveloped virus with a single-stranded negative-sense ribonucleic acid (RNA) genome recently reclassified in 2016 from subfamily Paramyxoviridae to the Pneumoviridae family within the Orthepneumovirus genus.18 The RSV genome is approximately 15.2 kb in length and contains 10 open reading frames (ORFs) that encode nine structural and two nonstructural proteins important for pathogenicity and immune evasion (Figure 2). The RSV envelope is comprised of three surface proteins (G, F, and SH) that orchestrate binding, fusion, and entry into ciliated bronchial epithelial cells19 as well as several immune cell subsets (i.e. dendritic cells,20 regulatory B cells,21 T cells22). The heavily glycosylated G protein is responsible for host cell attachment through binding to one or more surface receptors and attachment factors. It is additionally secreted in a soluble form as a means to mediate immune evasion through antibody binding as a decoy.23 After virion attachment, the trimeric transmembrane F protein then mediates the fusion of RSV viral envelope and the host membrane following a proteolytically enabled conformational change.24 RSV entry is thought to be pH-independent with direct entry occurring at the plasma membrane, though macropinocytosis has also been postulated as an entry mechanism.25 Although not fully characterized, the small hydrophobic (SH) protein has been proposed to function as a pentameric ion channel26 viroporin with both anti-apoptotic and anti-inflammatory effects through suppression of tumor necrosis factor (TNF) signaling.27 Matrix (M) protein coats the inner envelope surface, are critical for viral assembly, and are thought to inhibit host cell transcription.28 Inside the viral envelope, phosphoprotein (P) tethers the RNA-dependent RNA polymerase (L) to the nucleoprotein (N) RNA complex, forming the core machinery necessary for viral transcription and replication. The RNA-dependent RNA polymerase (RdRp) complex is responsible for both viral mRNA production as well as genome replication. After fusion, this complex is released into the cytoplasm where replication, transcription, and viral assembly occur.29,30 Overlapping ORFs in the M2 gene encode two separate factors, M2-1 and M2-2, involved in transcription and RNA genome replication, respectively.31,32 The nonstructural proteins NS-1 and NS-2 play important roles in suppressing innate immune induction, interferon-stimulated gene (ISG) expression,33,34 and dendritic cell maturation.35 Ultimately, progeny viruses are assembled and released starting at 10–12 hours post-infection (hpi) with peak viral release around 20 hpi.36
Figure 2.
Schematic of RSV Virion Structure and Genome. The RSV RNA genome consists of 10 genes of 15.2 kb length that encode 11 proteins. These proteins can be categorized into ribonucleocapsid proteins [phosphoprotein (P), nucleoprotein (N), RNA polymerase (L), M2-1, and M2-2], nonstructural proteins (NS1 and NS2), three surface proteins [small hydrophobic protein (SH) attachment glycoprotein (G), and fusion protein (F)], and matrix protein (M) which surrounds the envelope and nucleocapsid.
RSV genotypes
The two antigenic groups of RSV, RSV-A and RSV-B, were originally distinguished through the use of monoclonal antibodies37,38 and complimentary deoxyribonucleic acid (cDNA) probes39 on several circulating strains in 1985. The advent of deep sequencing approaches subsequently allowed for the broader characterization of RSV diversity.40 Within these antigenic groups, there have been 37 identified genotypes documented for RSV-B41 and 13 distinct genotypes documented for RSV-A.42,43 The evolutionary rates for both subtypes differ from one another, with RSV-A proposed to have a lower rate at 1.48 × 10–3 nucleotide substitutions/site/year as opposed to RSV-B at 1.92 × 10–3 substitutions/site/year.44 However, in other studies comparing diversity within antigenic groups, RSV-A was shown to have higher consensus diversity and lower intra-host variation as opposed to RSV-B.45 Across the genome, the highest nucleotide substitution rates occur in the attachment glycoprotein (G), which is subject to substantial evolutionary pressure both as a determinant of viral transmission and as a primary target of the humoral immune response.46 These selective pressures have been thought to drive the convergent evolution of short duplication events in G, which have been observed in both RSV-A (genotype ON1) and RSV-B (genotype BA1). Given the strong diversity in this region, conventional methods to genotype RSV have relied on targeted sequencing of the G ORF.13,47 Generally, most current molecular epidemiology efforts rely on targeted sequencing of G, and specifically of the second hypervariable region, for phylogenetic analysis and classification of viral genotypes.48
Although RSV cases typically follow a seasonal pattern with the peak of infections occurring from late December to late February in the Northern Hemisphere, RSV seasonality is variable in tropical regions49 and near the equator.50 Environmental factors such as humidity and temperature were not found to be causal of RSV seasonality, and subtropical climates displayed continuous RSV cases throughout the year.51 Although there can be a heterogeneous population of genotypes co-circulating in populations during one RSV season, it has been observed that a single genotype dominates a geographic area with subtype switching occurring year to year (RSV-A followed by RSV-B and back). For example, RSV epidemics in Kilifi, Kenya, displayed sequential replacement of local circulating genotypes over 11 recurrent epidemics.52 Similar phenomena had occurred with GA2, GA5, and GA7 genotypes (RSV-A) being outcompeted by NA1, NA2, and ON1 genotypes (RSV-A), which contain a 72 nucleotide duplication in the C-terminal region of the G gene, in Malaysia,53 Canada,54 India,55 South Korea,56 and China.57 While efforts are underway to explore these trends through increased genomic surveillance, the lack of a standardized nomenclature for genotypes and limited whole-genome sequencing (WGS) data have complicated this effort.
Currently, there are approximately 13 different documented methods38 to define RSV genotypes,43 illustrating a broad lack of agreement on how to classify viral variants. This remains a major problem in the field as the adoption of standard nomenclature for genotype designations is crucial for effective surveillance, clinical risk analysis, and public health communication. For example, the rapid adoption of standard nomenclature for describing SARS-CoV-2 variants assisted scientists and epidemiologists in tracking the virus and in communicating risks with the public.58 To address this issue in the RSV field, Goya et al. worked to better define RSV clades via G-ectodomains. The authors proposed a classification system based on average genetic distance among and within RSV genotypes with a cut-off value based on the genetic distance of previously defined genotypes. Resolution of phylogenetic trees is increased when using whole-genome sequencing, but the G-ectodomain was found to be sufficient to calculate genotype designations. This method resulted in a reduction of recognized genotypes by almost threefold while additionally allowing for sub-genotypes and lineage designation.43 Likewise, efforts to standardize nomenclature when reporting RSV sequences to improve data accessibility and analysis have been proposed, akin to the standards used for influenza virus sequencing and reporting.59
While phylogenetic analysis has traditionally relied on G, several WGS strategies have since been developed to understand viral diversity in other ORFs. These same approaches are currently being explored to refine genotype and clade designation.60,61 There are other genetic ‘hotspots’ spanning the genome in addition to G that justify the need for whole-genome surveillance, including in the 5′ untranslated region (UTR) of SH, in M2, and in NS1. Both RSV-A and RSV-B have high nucleotide diversity in the 5′UTR of SH, potentially signaling selection for SH expression that may influence RSV pathogenesis.13,62 In a cohort of RSV+ infants in Vietnam, M2-2 was found to be under positive selection, with an increased ratio of nonsynonymous mutations to synonymous mutations in both RSV-A and RSV-B isolates.63 Some mutational hotspots have also been found to be subtype specific, including sites in NS1 and N in RSV-A with still uncharacterized functionality.63 Understanding the rate of viral evolution specific to these components of RSV is crucial, as some of these ORFs (i.e. M2-2, NS2, and N) contribute to RSV pathogenicity and are the primary target of some vaccine candidates and monoclonal antibodies in phase 1 clinical trials.64
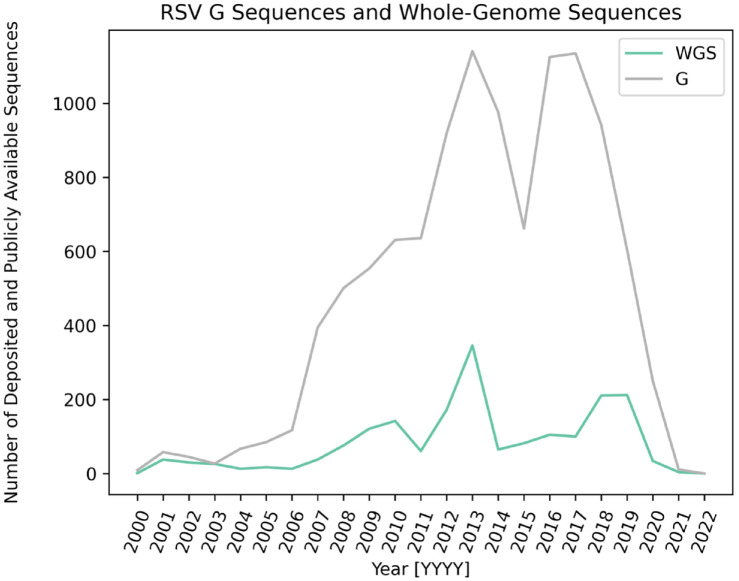
Nevertheless, WGS approaches to catalog RSV genomic diversity remain the exception. Approximately 2200 RSV whole-genome sequences have been publicly deposited in GenBank since 1956 (Figure 3), as opposed to over 9.6 million whole-genome sequences of SARS-CoV-2 since 2020. Scientific studies and public health investigations have leveraged these sequences to define the convergent evolution of G gene duplication in RSV genomes and to carry out healthcare worker outbreak investigations.65–70 All but one of these studies focus on infants less than 1 year of age, with a large subset focusing on infants with severe respiratory illness, highlighting the dearth of knowledge available on whole-genome RSV diversity and evolution in adult populations. Overall, advances in WGS creates an optimal opportunity to address ongoing questions surrounding RSV viral evolution and knowledge gaps surrounding under-served populations or under-sequenced RSV genomic regions. However, there are certain technical challenges to conduct molecular surveillance. For example, polymerase chain reaction (PCR) primer mismatches due to the increased diversity in regions of the RSV genome presents another challenge, resulting in unsuccessful amplification of certain ORF fragments of the RSV genome from one season to the next.71 Consequently, publicly available RSV sequences become critical to then aid in the design of primer schematics as a solution to primer mismatching with the goal of whole-genome coverage.72
Figure 3.
RSV sequences deposited in National Institute of Health (NIH) sequence database GenBank over time. Plot of the number of publicly available sequences deposited in GenBank from 2000 to 2022. RSV-G sequences are depicted in gray and WGS data are depicted in green.
RSV global surveillance efforts
Despite its large global disease burden, less is understood about the transmission, seasonality, and pathogenicity of RSV when compared with other pathogenic respiratory viruses. This is in part due to the lack of a widespread surveillance system to track RSV epidemiological information, in contrast to the more developed systems in place for influenza viruses and SARS-CoV-2. To resolve these issues of limited reporting, the Bill & Melinda Gates Foundation provided dedicated funding for RSV surveillance in several low- and middle-income countries (LMICs) to track mortality in infants aged < 6 months through PCR-based diagnostics between 2011 and 2013.73 The resulting data uncovered a staggering RSV disease burden and mortality in these countries, eliciting a broader call for action in Argentina, India, Pakistan, and Zambia.73 Subsequently, several countries began embedding RSV surveillance efforts into pre-existing influenza surveillance systems.74 Since then, there have been four major initiatives for increased global surveillance of RSV that have enhanced our understanding of seasonality, trends, and genomic evolution: (1) Global Influenza Surveillance and Response System (GISRS), (2) Global Epidemiology of RSV initiative (GERi), (3) Respiratory Syncytial Virus Consortium in Europe (RESCEU), and (4) International Network for Optimal Resistance Monitoring (INFORM).
The World Health Organization (WHO) adapted the framework of the GISRS into a pilot program for RSV surveillance from 2017 to 2019.75 GISRS tracked RSV cases in 14 countries (Argentina, Australia, Brazil, Canada, Chile, Cote d’Ivoire, Egypt, India, Mongolia, Mozambique, Russian Federation, South Africa, Thailand, and the United Kingdom) with the primary goal to establish a better understanding of RSV disease burden, seasonality, and high-risk groups. By using real-time PCR (RT-PCR) molecular diagnostic testing to detect genotype RSV, the study concluded that the seasonality of RSV was found to be dissimilar among participating surveillance locations depending on temperate and tropical climates, although there were regional overlaps with seasonal influenza virus cases. In addition, the GERi – modeled off the Global Influenza B Study (GIBS) – was founded in 2016 with the goal to determine genotype distribution, demographic factors, and environmental factors that influence RSV epidemics using national surveillance systems that provide information on RSV-subtype and clinical data.76,77 Additional regional specific efforts have emerged since that time, including RESCEU, which focuses on incidence data from a global prospective study of 10,000 infants to inform the RSV attributable economic burden and establish a national framework and biobank to support future vaccine development.78 While these efforts have focused on improved RSV surveillance to better understand disease burden, seasonality, and virological behavior, less emphasis has been placed on genomic surveillance systems and their potential long-term benefits for therapeutic design. To begin to address that gap, the INFORM–RSV study – led by the Respiratory Syncytial Virus Network (ReSViNET) Foundation – has proposed to sequence more than 4000 whole genomes from clinical isolates across 17 countries from 2017 to 2021. These data will be used to track the emergence of new genotypes and polymorphisms in RSV antigenic sites of the F protein that could impede therapeutic design.79
While traditional and molecular-based surveillance approaches are critical to improve our understanding of RSV incidence and diversity, much less is understood as to how this diversity influences clinical outcomes. These studies tend to be more regionally constrained due to the need to aggregate and compare in-depth medical record data. As one example of these efforts, the Infant Susceptibility to Pulmonary Infections and Asthma Following RSV Exposure (INSPIRE) longitudinal birth cohort of healthy infants implemented sequencing efforts in Tennessee from 2014 to 2016 to establish correlation between viral diversity and recurrent wheezing and asthma in healthy infants through WGS and phylogenetic analysis.13,80,81 Overall, these initiatives utilized similar infrastructural frameworks to increase RSV surveillance with the goal to address distinct phylogenetic, clinical, or molecular gaps in knowledge. However, all of these studies faced several challenges that highlight critical areas for improvement of RSV surveillance. For example, non-standardized platforms for testing, limited genome sequencing technologies/capabilities, and linkage to influenza surveillance systems that operate in constrained time frames limited some of the takeaways from these studies.
Efforts to perform and understand RSV surveillance were complicated even further by the COVID-19 pandemic. Widespread implementation of public health measures against respiratory viruses in 2020 brought about a stark reduction in RSV cases, ranging from a decrease of 57%82 to 85%83 in different regions of the world. This significant decrease in cases parallels that of several other respiratory viruses, such as influenza viruses, rhinoviruses, and metapneumoviruses.84–87 As public health recommendations and restrictions such as indoor masking and remote learning eased, communities around the globe began to experience a resurgence in RSV cases. Reemergence of a delayed RSV season was observed in Japan88 and Argentina89 with higher than usual cases and introductions from other countries in 2021. Likewise, in March 2021, the Centers for Disease Control and Prevention (CDC) reported increases in RSV infections through the National Respiratory and Enteric Virus Surveillance System, resulting in the issuance of a health advisory for interseasonal activity in the southern United States.90 Predictive analyses propose that the combination of implemented public health mitigation strategies and decreased exposure to RSV will increase the likelihood and intensity of an RSV epidemic reemergence with an atypical broader age range.14,15
Taken together, global surveillance efforts have substantially improved our understanding of RSV epidemiology. One major conclusion drawn from these surveillance efforts is that RSV prevalence and mortality are grossly underestimated with as many as three additional RSV-associated deaths occurring in the community for every one RSV-associated death reported in the hospital.4 These efforts additionally highlighted the absence of significant RSV surveillance in adult and older populations. In addition, the association of global RSV-subtype distributions among different age groups remain unclear.76 Finally, these global surveillance efforts have facilitated characterization of the spread of persistent global RSV subtypes, such as RSV-A ON1.91 Continued surveillance will be critical to filling in these knowledge gaps and assessing how RSV molecular epidemiology has or has not changed following the COVID-19 pandemic.
Impacts of genetic diversity on disease severity and clinical outcome
Changes in viral genotype can result in changes in viral load, cell tropism, and immune evasion, and subsequently impact disease presentation, severity, and clinical outcome. Different subtypes of influenza virus and different variants of SARS-CoV-2 have both been associated with exacerbated severe or mild disease with a similar pattern observed in patient outcomes.92,93 Several studies have examined the relationship between RSV genotype and clinical severity, although these have failed to reach consensus with studies coming to disparate conclusions.10 The reason for the contradicting conclusions is likely multifaceted and attributable to differences in inclusion criteria, cohort demographics, prior immunity, and sample size.10 These studies have largely relied on G sequencing to elucidate genotype with the assessment of clinical severity through available medical health records and severity metrics, such as proprietary severity scores, intubation, and length of hospitalization. For example, RSV-A genotype ON1 was suggested to worsen bronchiolitis clinical severity in hospitalized infants, particularly in the 2016–2017 and 2017–2018 RSV seasons.94 Likewise, another study that coupled transcriptome analysis, genotyping, and clinical metadata in a multicenter prospective study in the United States found that genotype GA5 (RSV-A) was associated with increased severity and a unique host immune response in five consecutive RSV seasons.66 Other studies, however, have found no associations between either subtype RSV-A or RSV-B and any clinical outcomes in hospitalized patients or patients with severe bronchiolitis.69,95,96 Viral load has been similarly analyzed in association to clinical severity with similarly disparate conclusions.97,98 Overall, these findings (summarized in Table 1) illustrate the complexity of measuring how circulating RSV genotypes may impact disease severity and subsequent clinical outcomes in infants, the elderly, and the immunocompromised.65–70,99–102 Furthermore, it must be reiterated that RSV genomic data used in these studies is overwhelmingly based on G sequencing; how mutations in other parts of the genome may influence these phenotypes is even less well understood. Emphasis to characterize glycoproteins and their role in viral infection has similarly been observed in other viruses, such as influenza, SARS-CoV-2, and dengue.103 However, while changes in the SARS-CoV-2 glycoprotein, Spike, have been thought to largely drive the observed changes in COVID-19 disease severity, changes outside of the influenza virus glycoproteins have been associated with more severe disease, including changes in the innate immune regulatory protein NS1.104,105
Table 1.
Representative table of studies linking RSV subtype and genotype to clinical severity and outcome.
| Location | Year | Population | Results | Reference |
|---|---|---|---|---|
| United States | 2004–2009, 2010–2011 | Infants hospitalized over 5 nonconsecutive respiratory seasons | RSV-A (GA5) was associated with increased disease severity | 66 |
| Italy | 2005–2017 | Infants hospitalized with bronchiolitis | RSV NA1 was associated with more severe bronchiolitis as opposed to ON1 or BA genotypes | 68 |
| Vietnam | 2010–2011 | Infants hospitalized over 5 nonconsecutive respiratory seasons | RSV-A virus was associated with higher clinical severity score | 65 |
| Cyprus | 2010–2013 | <12-year-old hospitalized children with acute respiratory tract infections | RSV-B BA infections was correlated with a higher risk in requiring oxygen. RSV-A ON1 was associated with a milder respiratory tract infection | 100 |
| Germany | 2011–2017 | Children with acute respiratory tract infections (ARTs) | RSV-A ON1 from children in the pediatric intensive care unit (PICU) had worsened clinical severity as opposed to non-ON1 genotypes | 70 |
| South Africa | 2012–2015 | Hospitalized patients (all ages) | No reported difference in clinical outcome between RSV-B and RSV-A. More streptococcus pneumoniae co-infections were associated with RSV-A infections | 69 |
| Japan | 2012–2015 | <5-year-old infants visiting pediatric outpatient clinics with respiratory symptoms over 3 consecutive respiratory seasons | RSV-A ON1 had a 6.9-fold increase in hospitalization risk as compared with NA1 genotype | 99 |
| Chile | 2013–2014 | Hospitalized infants with lower respiratory tract infections | No correlation in clinical outcome or severity with specific RSV-B and RSV-A genotypes | 101 |
| China | 2013–2015 | Hospitalized children 0–14 years old | RSV-A virus was associated with higher clinical severity score | 67 |
| Netherlands | 2016–2018 | Hospitalized adult patients | RSV-A strain with 8 amino acid changes unique to the region had higher clinical severity as measured by severity scores (CURB-65) | 102 |
Location: the location in which the study took place and where the samples were collected; Year: the time frame of sample collection date; Population: a description of the study population and any comorbidities or clinical symptoms of interest; Results: the conclusions of association between specified clinical severity parameters/outcomes and RSV genetic diversity.
Ultimately, there are several challenges associated with elucidating the link between RSV genotype and clinical severity. Future studies should strive to include full genome sequence data for RSV, particularly across the NS1, M2, and SH ORFs, to determine if the increased diversity across these genes correlates with any particular outcome. Furthermore, the populations included in these studies should be expanded to include other at-risk groups besides infants, such as the immunocompromised and elderly. Finally, standardization of genotype and clade designation on a global scale is crucial and needs to be achieved for conclusions to be readily shared and translated for an international audience.
Impacts of genetic diversity on therapeutic efficacy
Beyond changes in disease severity, changes in viral genotype can have massive consequences on the efficacy of antiviral therapeutics, monoclonal antibody treatments, and vaccines. The evolution of antiviral resistance has been well documented in human immunodeficiency virus type 1 (HIV-1) and influenza viruses, requiring patient-specific drug selection and the careful employment of antiviral stewardship strategies.106–108 Monoclonal antibody therapeutics are likewise highly susceptible to antigenic shift. For example, the emergence of each new variant of SARS-CoV-2 has required reassessment of monoclonal antibody efficacy.109 Finally, vaccine efficacy is strongly linked to its compatibility to the circulating variant. The ongoing antigenic drift and shift of influenza viruses have required constant surveillance and annual reformulation/administration of the preventive vaccine.110 Likewise, the emergence of the highly divergent Omicron variant of SARS-CoV-2 has resulted in an increase in breakthrough infections.111 All these represent critical tools to prevent transmission and lower disease burden, so understanding the impact of viral variation on therapeutics is essential for effective clinical care and public health response.
There are currently only two Food and Drug Administration (FDA)–approved treatments for RSV: ribavirin and palivizumab. Ribavirin is a nucleoside inhibitor with broad antiviral efficacy against RSV, hepatitis C virus, Lassa virus, and other viruses.112 Palivizumab is a monoclonal antibody that targets the F protein, though it is reserved for use as a prophylactic in high-risk infant populations.113 While there are currently limited options, there are over 14 different antiviral and immune-based therapeutics in various stages of clinical trial for the prevention and treatment of RSV infection.114 These include a number of vaccine candidates, most of which rely on either delivery of live attenuated virus or virus-like particles.115 While most of these are being formulated for use in infants, additional trials are underway for the protection of other at-risk groups, including the elderly and immunocompromised. For example, Pfizer recently commenced a phase 3 clinical trial for a RSV prefusion F vaccine candidate for individuals over the age of 60.116
RSV genetic diversity has proven to be a major challenge in the development of next-generation therapies. For example, phase 2b trials of the RSV fusion inhibitor, presatovir, employed partial fusion protein sequencing pre- and post-treatment to identify mutations associated with fusion inhibitor resistance. Of the 233 subjects administered therapy, 18 developed resistance.117 Discouragingly, certain resistance-associated mutations in F, such as V127A, were already present in a subset of subjects and had previously been documented to be circulating in South Africa,118 Korea,119 and Buenos Aires12,120 at varying frequencies. Likewise, the proposed monoclonal antibody therapeutic suptavumab failed a randomized, double-blind, placebo-controlled phase 3 trial due to the emergence of RSV-B strains with L172Q and S173 L mutations in F that decreased the binding affinity of the antibody.121 As a result, the trial failed to achieve its secondary endpoint of decreased RSV-confirmed hospitalizations or outpatient visits among enrolled patients. These mutations in F were found in the 88.6% of RSV-B genotypes in sequenced clinical samples in 2015–2016,122 demonstrating a strong need for genomic surveillance to inform therapeutic design and efficacy. Fortunately, resistance to the current monoclonal antibody therapy palivizumab has been sparse,123,124 with only a few reports emerging in Japan,125 Lebanon,126 and in a multinational cohort of patients deemed to be at high risk for respiratory virus-related illness.127 An early 2010 study identified N276S as a potential palivizumab resistance mutation in RSV-A,127 but this mutation was present in approximately 44% of RSV strains in 2008–2009 and 100% of RSV strains in 2009–2010 in Canada with no reported decrease in therapeutic efficacy.125 Overall, consideration of RSV viral evolution should be taken into account when developing next-generation therapies to minimize the potential for decreased efficacy from RSV therapeutic resistant mutations in circulating genotypes.
Improved RSV surveillance, especially longitudinal surveillance in the context of drug trials, will be critical to assessing the risk of therapeutic resistance going forward. Given the already limited options for antiviral treatment, continued surveillance for resistance to current treatment regimens will be further required to best inform clinical use and stewardship. The emphasis on therapeutic resistance has been heavily weighted toward infant populations; additional assessment of RSV evolution in response to therapeutic treatments in elderly or immunocompromised populations will be required to fully understand the risk of population-based antiviral resistance. In sum, improved molecular surveillance of RSV is sorely needed to not only inform how genetic variability impacts epidemiology and pathogenesis, but also the efficacy of current and future therapeutics.
Toward a molecular understanding of RSV biology
Understanding the mechanism by which changes in the RSV genome impact viral spread, disease pathogenesis, and therapeutic efficacy depends on an understanding of the molecular virology and selective pressures that underlie RSV replication and evolution. In general, mutations that improve a virus’ ability to replicate in a given environment (i.e. improve viral fitness) will be selected for over time and vice versa.128 For example, mutations that improve transmission, enhance immune evasion, or provide resistance to an antiviral therapeutic may enhance viral fitness in some cellular environments, providing an advantage to the viruses that carry them.129 While genomic surveillance can be remarkably effective in identifying mutations that are under selection, interpreting why they are selected requires lab-based inquiry.
The RSV glycoprotein G and fusion protein F are among the most heavily studied viral proteins given their critical role in the viral entry and their importance as therapeutic targets. F and G are expressed on the outer surface of virions and mediate binding to the target cell surface through interactions with host attachment factors and entry receptors.130 An array of entry receptors and attachment factors have been described, including CX3CR1 and heparan sulfate proteoglycans (HSPGs) that interact with G as well as nucleolin, epidermal growth factor receptor (EGFR), IGF1R, and ICAM-1 that interact with F.131 Receptor binding ultimately enables F to initiate membrane fusion and deposit the viral genetic material into the cell.25 Beyond their role in viral entry, F and G also serve critical roles in immune evasion as prominent targets for the humoral immune response, including neutralizing antibodies that block receptor binding.132 To help circumvent this response, G is heavily glycosylated to mask potential protein epitopes and is shed in a soluble form to neutralize and effectively lower antibody titers.133,134 As a result, G experiences strong selective pressure and is among the most heavily diversified ORFs in RSV. F, on the contrary, is more evolutionarily constrained and as such serves as a common therapeutic target.23 High selective pressure in the presence of fusion inhibitors, however, can lead to the emergence of resistance mutations. Understanding the impact of G and F mutations on viral entry, viral transmission, and therapeutic efficacy is therefore of critical importance.
To determine the functional impact of mutations in the RSV viral glycoproteins, a number of biochemical and cellular tools have been developed.135–137 For example, different variants of F and G can be used to pseudotype non-infectious reporter viruses in vitro to test the effect of mutations on cell entry and antibody neutralization.138 Similar systems have been developed to understand the ongoing evolution of the SARS-CoV-2 Spike protein, which faces similar selective pressures.139 Using these assays, several mutations in RSV G and F have been reported to influence their interaction with their respective host factors, influencing the efficiency of cellular attachment and membrane fusion.140 Similarly, several escape mutations reported in F following in vitro selection for fusion inhibitor resistance have been found to have secondary impacts on cell entry, resulting in a decrease in viral fitness.141 Beyond influencing viral entry and therapeutic efficacy, these mutations may also influence pathogenicity and host response. For example, mutations in the F protein of RSV-A strains 2-20 and A2 have been found to alter engagement with EGFR, which has downstream consequences on airway mucin production.142 In sum, implementation of in vitro strategies to elucidate the consequences of mutations on RSV replication will be crucial for informing the development of novel therapeutic and preventive treatments targeting these proteins.
While much is known about the function and selective pressures acting on F and G, much more needs to be done to fully understand the replicative lifecycle of RSV and the critical interactions that might drive viral evolution. Indeed, a series of genetic and biochemical studies in recent years have identified several new RSV host factors that have unique proviral or antiviral functions. For example, the role of nucleolin as a primary cellular receptor in vivo was only recently identified through transcriptomics analyses followed by siRNA knockdown and quantitative RSV plaque assays.143 Both genome-wide loss-of-function144,145 and gain-of-function38 screens in lung epithelial models have likewise identified a number of new RSV host factors. For example, a recent study identified chemokine (C-X-X Motif) Ligand 4 as a novel antiviral host factor that inhibited RSV entry and was correlated with disease severity in RSV-infected patients.146 Proteomic approaches have likewise been used to identify host proteins critical for viral replication or the immune response to infection. For instance, a study using immunoprecipitation mass spectrometry demonstrated that, in addition to perturbing immune responses, NS1 binds to mediator complexes of RNA polymerase II to regulate antiviral gene expression.147 Similar high-throughput approaches have been used to identify molecular correlates of disease that may serve as future biomarkers. For example, differentially expressed proteins involved in glycolysis (BPGM, TPI1, PRDX2, and CFL1) in nasopharyngeal secretions of children in the acute or convalescent phase of RSV infection were upregulated and identified using isobaric tags for relative and absolute quantitation (iTRAQ) analysis.148
While these studies have contributed toward our understanding of the molecular virology of RSV, most of this work fails to account for the genetic makeup of most currently circulating strains, instead relying on the use of a handful of lab-adapted strains that grow well in laboratory cultures and models. Five out of the six most commonly used strains (RSV Long, A2, Line19, Line19F, and CH-18537) were isolated before 1970 and do not recapitulate the immune response of more recent isolates.149 A more recent strain (Memphis 37) was isolated from nasal aspirates in 2001, but likewise does not contain the aforementioned duplication event in G or accurately represent currently circulating variants. Furthermore, these strains display differential disease phenotypes and severity in murine models150 and different cytopathology in primary pediatric bronchial epithelial cells when compared with clinical isolates.151 Analysis of viral gene expression between lab-adapted strains and current circulating genotypes (GA1, ON, GB1, and BA) revealed clear differences in viral lifecycle regulation between these strains that may underlie different host responses.152
Much more work needs to be done to understand what molecular determinants underlie RSV replication, how these determinants drive viral evolution, and ultimately how the resultant variants differ in their replicative lifecycle and interaction with the host. Implementation of molecular biology and systems biology strategies in tandem with molecular surveillance will be required to identify the factors driving differences in viral replication and pathogenesis in vivo. Meanwhile, the inclusion of more relevant lab models and relevant RSV isolates will be critical to refining our understanding of the proviral and antiviral host factors underlying RSV replication in different microenvironments.
Strain-specific differences in the immune response
While limited, some studies have emphasized how RSV genotype can influence the innate and cellular immune response to infection. Much of this work has been done through comparison of different viral strains in well-characterized murine models of infection. For example, examination of strain-specific differences in the pathogenesis of RSV in BALB/c mice revealed differential levels of neutrophils and IL-13 expression in CD4+ T cells attributable to strain variation in the F protein and its fusion activity in epithelial cells.153 Similarly, when measuring phenotypic parameters in mice infected with laboratory strain A2 (RSV-A) and clinical isolate strain Line 19 (RSV-A), production of cytokines IL-10 and IL-13 was found to be higher and lower, respectively.154 Outside of murine models, some work has been done with different clinical isolates in standard cell line models to compare effects in culture. For example, a point mutation in the transcriptional termination signal of G found in 98% of circulating genotypes between 2009 and 2017 was speculated to decrease F expression relative to RSV strain A2 as a way to evade the humoral response.155
Studies of immune responses in human cohorts are more difficult given that diversity in both the hosts and the pathogen could drive meaningful changes that are hard to assign to pathogen diversity alone. For example, when assessing correlates of the host response to bronchiolitis severity, One study found that induced ISGs differed between patients infected with NA1 (RSV-A), ON1 (RSV-A), or BA (RSV-B) and that these patterns were associated with severity in bronchiolitis clinical manifestations.156 Likewise, using microarrays and high-throughput sequencing, microRNAs responsible for regulating inflammatory mediators produced by leukocytes were identified to be dysregulated in RSV-infected infants and contributes to airway hyperreactivity.157 When evaluating inflammation in these patients, elevated levels of neutrophils were identified in the bronchoalveolar lavage in children with RSV-induced bronchiolitis.158 Using paired transcriptomic analysis, specifically low-input RNA sequencing of neutrophils in sputum and blood from infants admitted in the ICU, this same study identified upregulation of neutrophil activation genes, IL-6, and NF-kB signaling.159 Although the role of neutrophilia in RSV pathophysiology is not fully understood, excessive neutrophilia has been identified in severe tissue damage and inflammation in RSV bronchiolitis.160 While these findings relate back to studies in murine models suggesting strain-specific differences in inflammation and neutrophil levels, it is still unclear if RSV genotype was a significant driver of the differential immune responses and infant outcomes. Overall, in vitro work, animal models, and measurements of immune-related gene expression patterns in clinical cohorts support the notion that RSV genetic diversity is involved in altered immune responses, though much more work needs to be done to determine how these interactions drive viral evolution.
Bridging molecular epidemiology and global systems biology
Molecular epidemiology seeks to relate the molecular mechanism of disease to environmental and genetic factors on a population-based scale to inform and predict pathogenic spread for the advancement of public health.161 The collection and dissemination of molecular surveillance data potentiates a positive feedback loop in tracking emerging variants, implementing successful public health interventions, and identifying promising molecular targets for next-generation therapeutics. Systems biology approaches seek to provide an unbiased molecular network of interactions on a genomic, metabolomic, transcriptomic, or proteomic scale to shed light on the underlying biology of a disease.162 The integration of these techniques, known as systems epidemiology, has been previously implemented in cancer biology to identify networks of host factors that contribute to translatable outcome metrics, such as tumor growth and risk of disease.163 Likewise, this multidisciplinary field has been identified as a particularly beneficial approach to rapid responses for viruses such as Zika through integration of molecular surveillance, analysis of the host response, and linkage to clinical data.164 Overall, systems epidemiology provides etiological insights and information on prevention strategies on a global scale.165 While the field of infectious disease has a long way to go before systems epidemiology can be effectively employed, the COVID-19 pandemic exemplified how molecular surveillance and systems biology approaches could be used in tandem to quickly gain insight into an evolving pathogen in near real-time. Hopefully, these tools can be effectively leveraged in the future to better inform our understanding of RSV and how viral diversity drives differences in viral replication, disease pathogenesis, patient outcome, and therapeutic efficacy.
Conclusion
The ongoing viral evolution and high global disease burden of RSV necessitate concerted efforts in molecular surveillance to understand the impact of circulating genotypes on clinical manifestations and therapeutic efficacy. Current global surveillance efforts have broadened our understanding of disease burden, seasonality, and evolution; identification of new genotypes [ON1 (RSV-A) and BA (RSV-B)] and patterns of convergent evolution have resulted from these global efforts. Continued efforts to expand these surveillance systems, standardize best practices, and implement whole-genome sequencing approaches will help advance future efforts in characterizing RSV diversity. These efforts will enable a more holistic understanding of RSV pathogenesis and pathophysiology beyond our current glycoprotein-centric understanding. The need for these efforts is especially acute in the context of viral evolution in response to existing and emerging therapeutic strategies. While enhanced surveillance will help us identify critical associations with viral genotype, improved understanding of RSV molecular virology will be critical for establishing causation and identifying the drivers of viral evolution. Expanded use of high-throughput, systems-based biochemical, cellular, and immunological assays promise rapid advancement in our understanding of underlying RSV biology. Ultimately, biomedical research and public health surveillance efforts should work to integrate molecular biology approaches within an epidemiological framework. Integration and collaboration will propel the field forward toward improved understanding of how RSV molecular diversity impacts viral replication, disease pathogenesis, patient outcome, and therapeutic efficacy.
Acknowledgments
The authors would like to thank Daphne Cornish, William J. Cisneros, Lacy M. Simons, Michael G. Ison, and Arghavan Alisotanidehkordi for their thoughtful feedback on this review. This work was supported in part through the computational resources and staff contributions provided for the Quest high performance computing facility at Northwestern University, which is jointly supported by the Office of the Provost, the Office for Research, and Northwestern University Information Technology.
Footnotes
ORCID iD: Estefany Rios Guzman  https://orcid.org/0000-0002-1399-7546
https://orcid.org/0000-0002-1399-7546
Contributor Information
Estefany Rios Guzman, Department of Medicine, Division of Infectious Diseases, Northwestern University Feinberg School of Medicine, Chicago, IL, USA; Center for Pathogen Genomics and Microbial Evolution, Institute for Global Health, Northwestern University Feinberg School of Medicine, Chicago, IL, USA.
Judd F. Hultquist, Robert H. Lurie Medical Research Center, Northwestern University, 9-141, 303 E. Superior St., Chicago, IL 60611, USA; Department of Medicine, Division of Infectious Diseases, Northwestern University Feinberg School of Medicine, Chicago, IL, USA; Center for Pathogen Genomics and Microbial Evolution, Institute for Global Health, Northwestern University Feinberg School of Medicine, Chicago, IL, USA.
Declarations
Ethics approval and consent to participate: Not applicable
Consent for publication: Not applicable
Author contributions: Estefany Rios Guzman: Conceptualization; Visualization; Writing – original draft; Writing – review & editing.
Judd F. Hultquist: Conceptualization; Funding acquisition; Project administration; Resources; Supervision; Visualization; Writing – original draft; Writing – review & editing.
Funding: The authors disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: Funding for the Center for Pathogen Genomics and Microbial Evolution was provided by a COVID-19 Supplemental Research award from the Northwestern Center for Advanced Technologies (NUCATS); the NIH-supported Third Coast CFAR P30 AI117943; NIH grant R21 AI163912; NIH grant U19 AI135964; and through a generous contribution from the Walder Foundation’s Chicago Coronavirus Assessment Network (Chicago CAN) Initiative. The funding sources had no role in the study design, data collection, analysis, interpretation, or writing of the report. Any opinions, findings, and conclusions or recommendations expressed in this material are those of the authors and do not necessarily reflect the views of the National Science Foundation and National Institutes of Health.
The authors declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Availability of data and materials: Not applicable
References
- 1. Dawson-Caswell M, Herbert L, Muncie J. Respiratory syncytial virus infection in children. Am Fam Physician 2011; 83: 141–146. [PubMed] [Google Scholar]
- 2. Falsey AR, Walsh EE. Respiratory syncytial virus infection in elderly adults. Drugs Aging 2005; 22: 577–587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Hynicka LM, Ensor CR. Prophylaxis and treatment of respiratory syncytial virus in adult immunocompromised patients. Ann Pharmacother 2012; 46: 558–566. [DOI] [PubMed] [Google Scholar]
- 4. Shi T. Global, regional, and national disease burden estimates of acute lower respiratory infections due to respiratory syncytial virus in young children in 2015: a systematic review and modelling study. Lancet Lond Engl 2017; 390: 946–958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. O’Shea MK, Ryan MAK, Hawksworth AW, et al. Symptomatic respiratory syncytial virus infection in previously healthy young adults living in a crowded military environment. Clin Infect Dis 2005; 41: 311–317. [DOI] [PubMed] [Google Scholar]
- 6. Kutsaya A. Prospective clinical and serological follow-up in early childhood reveals a high rate of subclinical RSV infection and a relatively high reinfection rate within the first 3 years of life. Epidemiol Infect 2016; 144: 1622–1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Nam HH, Ison MG. Respiratory syncytial virus infection in adults. BMJ 2019; 366: l5021. [DOI] [PubMed] [Google Scholar]
- 8. Smart KA, Lanctôt KL, Paes BA. The cost effectiveness of palivizumab: a systematic review of the evidence. J Med Econ 2010; 13: 453–463. [DOI] [PubMed] [Google Scholar]
- 9. Scheltema NM. Global respiratory syncytial virus-associated mortality in young children (RSV GOLD): a retrospective case series. Lancet Glob Health 2017; 5: e984–e991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Vandini S, Biagi C, Lanari M. Respiratory syncytial virus: the influence of serotype and genotype variability on clinical course of infection. Int J Mol Sci 2017; 18: E1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Mas V, Nair H, Campbell H, et al. Antigenic and sequence variability of the human respiratory syncytial virus F glycoprotein compared to related viruses in a comprehensive dataset. Vaccine 2018; 36: 6660–6673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hause AM. Sequence variability of the respiratory syncytial virus (RSV) fusion gene among contemporary and historical genotypes of RSV/A and RSV/B. PLoS ONE 2017; 12: e0175792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Schobel SA. Respiratory syncytial virus whole-genome sequencing identifies convergent evolution of sequence duplication in the C-terminus of the G gene. Sci Rep 2016; 6: 26311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Zheng Z, Pitzer VE, Shapiro ED, et al. Estimation of the timing and intensity of reemergence of respiratory syncytial virus following the COVID-19 pandemic in the US. JAMA Netw Open 2021; 4: e2141779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Di Mattia. During the COVID-19 pandemic where has respiratory syncytial virus gone? Pediatr Pulmonol 2021; 56: 3106–3109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Li Y. Understanding the potential drivers for respiratory syncytial virus rebound during the coronavirus disease 2019 pandemic. J Infect Dis 2022; 225: 957–964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Chanock R, Roizman B, Myers R. Recovery from infants with respiratory illness of a virus related to chimpanzee coryza agent (CCA). I. Isolation, properties and characterization. Am J Hyg 1957; 66: 281–290. [DOI] [PubMed] [Google Scholar]
- 18. Rima B. ICTV virus taxonomy profile: pneumoviridae. J Gen Virol 2017; 98: 2912–2913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Zhang L. Respiratory syncytial virus infection of human airway epithelial cells is polarized, specific to ciliated cells, and without obvious cytopathology. J Virol 2002; 76: 5654–5666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Tognarelli EI, Bueno SM, González PA. Immune-modulation by the human respiratory syncytial virus: focus on dendritic cells. Front Immunol 2019; 10: 0197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Zhivaki D. Respiratory syncytial virus infects regulatory B cells in human neonates via chemokine receptor CX3CR1 and promotes lung disease severity. Immunity 2017; 46: 301–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Raiden S. Respiratory syncytial virus (RSV) infects CD4+ T cells: frequency of circulating CD4+ RSV+ T cells as a marker of disease severity in young children. J Infect Dis 2017; 215: 1049–1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. McLellan JS, Ray WC, Peeples ME. Structure and function of RSV surface glycoproteins. Curr Top Microbiol Immunol 2013; 372: 83–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lamb RA, Jardetzky TS. Structural basis of viral invasion: lessons from paramyxovirus F. Curr Opin Struct Biol 2007; 17: 427–436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Krzyzaniak MA, Zumstein MT, Gerez JA, et al. Host cell entry of respiratory syncytial virus involves macropinocytosis followed by proteolytic activation of the F protein. PLoS Pathog 2013; 9: e1003309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Gan S-W. The small hydrophobic protein of the human respiratory syncytial virus forms pentameric ion channels. J Biol Chem 2012; 287: 24671–24689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Fuentes S, Tran KC, Luthra P, et al. Function of the respiratory syncytial virus small hydrophobic protein. J Virol 2007; 81: 8361–8366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ghildyal R, Ho A, Jans DA. Central role of the respiratory syncytial virus matrix protein in infection. FEMS Microbiol Rev 2006; 30: 692–705. [DOI] [PubMed] [Google Scholar]
- 29. Hacking D, Hull J. Respiratory syncytial virus – viral biology and the host response. J Infect 2002; 45: 18–24. [DOI] [PubMed] [Google Scholar]
- 30. Tawar RG. Crystal structure of a nucleocapsid-like nucleoprotein-RNA complex of respiratory syncytial virus. Science 2009; 326: 1279–1283. [DOI] [PubMed] [Google Scholar]
- 31. Collins PL, Fearns R, Graham BS. Respiratory syncytial virus: virology, reverse genetics, and pathogenesis of disease. Curr Top Microbiol Immunol 2013; 372: 3–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Tran T-L. The respiratory syncytial virus M2-1 protein forms tetramers and interacts with RNA and P in a competitive manner. J Virol 2009; 83: 6363–6374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Lo MS, Brazas RM, Holtzman MJ. Respiratory syncytial virus nonstructural proteins NS1 and NS2 mediate inhibition of Stat2 expression and alpha/beta interferon responsiveness. J Virol 2005; 79: 9315–9319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Elliott J. Respiratory syncytial virus NS1 protein degrades STAT2 by using the Elongin-Cullin E3 Ligase. J Virol 2007; 81: 3428–3436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Munir S. Respiratory syncytial virus interferon antagonist NS1 protein suppresses and skews the human T lymphocyte response. PLoS Pathog 2011; 7: e1001336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Levine S, Hamilton R. Kinetics of the respiratory syncytial virus growth cycle in HeLa cells. Arch Gesamte Virusforsch 1969; 28: 122–132. [DOI] [PubMed] [Google Scholar]
- 37. Anderson LJ. Antigenic characterization of respiratory syncytial virus strains with monoclonal antibodies. J Infect Dis 1985; 151: 626–633. [DOI] [PubMed] [Google Scholar]
- 38. Mufson MA, Orvell C, Rafnar B, et al. Two distinct subtypes of human respiratory syncytial virus. J Gen Virol 1985; 66: 2111–2124. [DOI] [PubMed] [Google Scholar]
- 39. Sullender WM, Anderson LJ, Anderson K, et al. Differentiation of respiratory syncytial virus subgroups with cDNA probes in a nucleic acid hybridization assay. J Clin Microbiol 1990; 28: 1683–1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Capobianchi MR, Giombini E, Rozera G. Next-generation sequencing technology in clinical virology. Clin Microbiol Infect Off Publ Eur Soc Clin Microbiol Infect Dis 2013; 19: 15–22. [DOI] [PubMed] [Google Scholar]
- 41. Muñoz-Escalante JC, Comas-García A, Bernal-Silva S, et al. Respiratory syncytial virus B sequence analysis reveals a novel early genotype. Sci Rep 2021; 11: 3452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Muñoz-Escalante JC, Comas-García A, Bernal-Silva S, et al. Respiratory syncytial virus A genotype classification based on systematic intergenotypic and intragenotypic sequence analysis. Sci Rep 2019; 9: 20097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Goya S. Toward unified molecular surveillance of RSV: a proposal for genotype definition. Influenza Other Respir Viruses 2020; 14: 274–285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Yu J-M, Fu Y-H, Peng X-L, et al. Genetic diversity and molecular evolution of human respiratory syncytial virus A and B. Sci Rep 2021; 11: 12941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Lin G-L. Distinct patterns of within-host virus populations between two subgroups of human respiratory syncytial virus. Nat Commun 2021; 12: 5125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Agoti CN. Local evolutionary patterns of human respiratory syncytial virus derived from whole-genome sequencing. J Virol 2015; 89: 3444–3454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Cane PA. Molecular epidemiology of respiratory syncytial virus. Rev Med Virol 2001; 11: 103–116. [DOI] [PubMed] [Google Scholar]
- 48. Wertz GW, Krieger M, Ball LA. Structure and cell surface maturation of the attachment glycoprotein of human respiratory syncytial virus in a cell line deficient in O glycosylation. J Virol 1989; 63: 4767–4776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Tang JW, Loh TP. Correlations between climate factors and incidence – a contributor to RSV seasonality. Rev Med Virol 2014; 24: 15–34. [DOI] [PubMed] [Google Scholar]
- 50. Stensballe LG, Devasundaram JK, Simoes EA. Respiratory syncytial virus epidemics: the ups and downs of a seasonal virus. Pediatr Infect Dis J 2003; 22: S21–S32. [DOI] [PubMed] [Google Scholar]
- 51. Hsu C-H. Prolonged seasonality of respiratory syncytial virus infection among preterm infants in a subtropical climate. PLoS ONE 2014; 9: e110166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Agoti CN. Successive respiratory syncytial virus epidemics in local populations arise from multiple variant introductions, providing insights into virus persistence. J Virol 2011; 89: 11630–11642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Khor C-S, Sam I-C, Hooi P-S, et al. Displacement of predominant respiratory syncytial virus genotypes in Malaysia between 1989 and 2011. Infect Genet Evol 2013; 14: 357–360. [DOI] [PubMed] [Google Scholar]
- 54. Eshaghi A. Genetic variability of human respiratory syncytial virus A strains circulating in Ontario: a novel genotype with a 72 nucleotide G gene duplication. PLoS One 2012; 7: e32807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Choudhary ML, Wadhwa BS, Jadhav SM, et al. Complete genome sequences of two human respiratory syncytial virus genotype A strains from India, RSV-A/NIV1114046/11 and RSV-A/NIV1114073/11. Genome Announc 2013; 1: e00165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Lee W-J. Complete genome sequence of human respiratory syncytial virus genotype A with a 72-nucleotide duplication in the attachment protein G gene. J Virol 2012; 86: 13810–13811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Liang X. Gradual replacement of all previously circulating respiratory syncytial virus A strain with the novel ON1 genotype in Lanzhou from 2010 to 2017. Medicine 2019; 98; e15542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Rambaut A. A dynamic nomenclature proposal for SARS-CoV-2 lineages to assist genomic epidemiology. Nat Microbiol 2020; 5: 1403–1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Salimi V. Proposal for human respiratory syncytial virus nomenclature below the species level. Emerg Infect Dis 2021; 27: e204608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Rebuffo-Scheer C. Whole genome sequencing and evolutionary analysis of human respiratory syncytial virus A and B from Milwaukee, WI 1998-2010. PLoS ONE 2011; 6: e25468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Lin G-L, Golubchik T, Drysdale S, et al. Simultaneous viral whole-genome sequencing and differential expression profiling in respiratory syncytial virus infection of infants. J Infect Dis 2020; 222: S666–S671. [DOI] [PubMed] [Google Scholar]
- 62. Ivancic-Jelecki J, Slovic A, Ljubin-Sternak S, et al. Variability analysis and inter-genotype comparison of human respiratory syncytial virus small hydrophobic gene. Virol J 2018; 15: 109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Do LAH, Wilm A, van Doorn HR, et al. Direct whole-genome deep-sequencing of human respiratory syncytial virus A and B from Vietnamese children identifies distinct patterns of inter- and intra-host evolution. J Gen Virol 2015; 96: 3470–3483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Mazur NI. The respiratory syncytial virus vaccine landscape: lessons from the graveyard and promising candidates. Lancet Infect Dis 2018; 18: e295–e311. [DOI] [PubMed] [Google Scholar]
- 65. Tran DN. Molecular epidemiology and disease severity of human respiratory syncytial virus in Vietnam. PLoS ONE 2013; 8: e45436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Rodriguez-Fernandez R. Respiratory syncytial virus genotypes, host immune profiles, and disease severity in young children hospitalized with bronchiolitis. J Infect Dis 2018; 217: 24–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Liu W. Epidemiology and clinical presentations of respiratory syncytial virus subgroups A and B detected with multiplex real-time PCR. PLoS One 2016; 11: e0165108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Midulla F. How respiratory syncytial virus genotypes influence the clinical course in infants hospitalized for Bronchiolitis. J Infect Dis 2019; 219: 526–534. [DOI] [PubMed] [Google Scholar]
- 69. Valley-Omar Z. Human respiratory syncytial virus diversity and epidemiology among patients hospitalized with severe respiratory illness in South Africa, 2012–2015. Influenza Other Respir Viruses 2022; 16: 222–235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Streng A. Spread and clinical severity of respiratory syncytial virus A genotype ON1 in Germany, 2011–2017. BMC Infect Dis 2019; 19: 613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Beerenwinkel N, Günthard H, Roth V, et al. Challenges and opportunities in estimating viral genetic diversity from next-generation sequencing data. Front Microbiol 2012; 3: 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Kellam P. Molecular identification of novel viruses. Trends Microbiol 1998; 6: 160–166. [DOI] [PubMed] [Google Scholar]
- 73. Srikantiah P, Vora P, Klugman KP. Assessing the full burden of respiratory syncytial virus in young infants in low- and middle-income countries: the importance of community mortality studies. Clin Infect Dis 2021; 73: S177–S179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Obando-Pacheco P. Respiratory syncytial virus seasonality: a global overview. J Infect Dis 2018; 217: 1356–1364. [DOI] [PubMed] [Google Scholar]
- 75. Chadha M, Hirve S, Bancej C, et al. Human respiratory syncytial virus and influenza seasonality patterns – Early findings from the WHO global respiratory syncytial virus surveillance. Influenza Other Respir Viruses 2020; 14: 638–646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Staadegaard L. The global epidemiology of RSV in community and hospitalized care: findings from 15 countries. Open Forum Infect Dis 2021; 8: ofab159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Staadegaard L. Temporal variations in respiratory syncytial virus epidemics, by virus subtype, 4 countries. Emerg Infect Dis 2021; 27: 1537–1540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Wildenbeest JG. Respiratory Syncytial Virus Consortium in Europe (RESCEU) Birth Cohort Study: defining the burden of infant respiratory syncytial virus disease in Europe. J Infect Dis 2020; 222: S606–S612. [DOI] [PubMed] [Google Scholar]
- 79. Langedijk AC. Global molecular diversity of RSV – the ‘INFORM RSV’ study. BMC Infect Dis 2020; 20: 450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Larkin EK. Objectives, design and enrollment results from the Infant Susceptibility to Pulmonary Infections and Asthma Following RSV Exposure Study (INSPIRE). BMC Pulm Med 2015; 15: 45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Achten NB. Interference between respiratory syncytial virus and human rhinovirus infection in infancy. J Infect Dis 2017; 215: 1102–1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Zhu Y. Epidemiological and virological characteristics of respiratory tract infections in children during COVID-19 outbreak. BMC Pediatr 2021; 21: 195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Wagatsuma K, Koolhof IS, Shobugawa Y, et al. Decreased human respiratory syncytial virus activity during the COVID-19 pandemic in Japan: an ecological time-series analysis. BMC Infect Dis 2021; 21: 734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Poole S, Brendish NJ, Clark TW. SARS-CoV-2 has displaced other seasonal respiratory viruses: results from a prospective cohort study. J Infect 2020; 81: 966–972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Olsen SJ. Decreased influenza activity during the COVID-19 pandemic – United States, Australia, Chile, and South Africa, 2020. Am J Transplant 2020; 20: 3681–3685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Hu C, Tang YW, Su QM, et al. Public health measures during the COVID-19 pandemic reduce the spread of other respiratory infectious diseases. Front Public Health 2021; 9: 771638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Grochowska M, Ambrożej D, Wachnik A, et al. The impact of the COVID-19 pandemic lockdown on pediatric infections – a single-center retrospective study. Microorganisms 2022; 10: 178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Ujiie M, Tsuzuki S, Nakamoto T, et al. Resurgence of respiratory syncytial virus infections during COVID-19 pandemic, Tokyo, Japan. Emerg Infect Dis 2021; 27: 2969–2970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Dolores A. RSV reemergence in Argentina since the SARS-CoV-2 pandemic. J Clin Virol 2022; 149: 105126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. CDC Health Alert Network. Increased interseasonal respiratory syncytial virus (RSV) activity in parts of the Southern United States. HAN Archive, CDC Health Alert Network. Increased interseasonal respiratory syncytial virus (RSV) activity in parts of the Southern United States [Google Scholar]
- 91. Duvvuri VR. Genetic diversity and evolutionary insights of respiratory syncytial virus A ON1 genotype: global and local transmission dynamics. Sci Rep 2015; 5: 14268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Avni T. Comparison of clinical outcomes of influenza A and B at the 2017–2018 influenza season: a cohort study. Eur J Clin Microbiol Infect Dis 2020; 39: 1109–1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Giles B, Meredith P, Robson S, et al. The SARS-CoV-2 B.1.1.7 variant and increased clinical severity – the jury is out. Lancet Infect Dis 2021; 21: 1213–1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Midulla F, Mattia GD, Nenna R, et al. Novel variants of respiratory syncytial virus A ON1 associated with increased clinical severity of bronchiolitis. J Infect Dis 2020; 222: 102–110. [DOI] [PubMed] [Google Scholar]
- 95. Laham FR, Mansbach JM, Piedra PA, et al. Clinical profiles of respiratory syncytial virus subtypes A and B among children hospitalized with bronchiolitis. Pediatr Infect Dis J 2017; 36: 808–810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Papadopoulos NG, Gourgiotis D, Javadyan A, et al. Does respiratory syncytial virus subtype influences the severity of acute bronchiolitis in hospitalized infants? Respir Med 2004; 98: 879–882. [DOI] [PubMed] [Google Scholar]
- 97. Garcia-Mauriño C, Moore-Clingenpeel M, Thomas J, et al. Viral load dynamics and clinical disease severity in infants with respiratory syncytial virus infection. J Infect Dis 2019; 219: 1207–1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. El Saleeby CM, Bush AJ, Harrison LM, et al. Respiratory syncytial virus load, viral dynamics, and disease severity in previously healthy naturally infected children. J Infect Dis 2011; 204: 996–1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Hibino A, Saito R, Taniguchi K, et al. Molecular epidemiology of human respiratory syncytial virus among children in Japan during three seasons and hospitalization risk of genotype ON1. PLoS ONE 2018; 13: e0192085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Panayiotou C, Richter J, Koliou M, et al. Epidemiology of respiratory syncytial virus in children in Cyprus during three consecutive winter seasons (2010–2013): age distribution, seasonality and association between prevalent genotypes and disease severity. Epidemiol Infect 2014; 142: 2406–2411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Espinosa Y, Martín CS, Torres AA, et al. Genomic loads and genotypes of respiratory syncytial virus: viral factors during lower respiratory tract infection in Chilean hospitalized infants. Int J Mol Sci 2017; 18: 654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Efstathiou C, Abidi SH, Harker J, et al. Revisiting respiratory syncytial virus’s interaction with host immunity, towards novel therapeutics. Cell Mol Life Sci 2020; 77: 5045–5058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Banerjee N, Mukhopadhyay S. Viral glycoproteins: biological role and application in diagnosis. VirusDisease 2016; 27: 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Harvey WT, Carabelli AM, Jackson B, et al. SARS-CoV-2 variants, spike mutations and immune escape. Nat Rev Microbiol 2021; 19: 409–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Wang S, Zhang L, Zhang R, et al. Identification of two residues within the NS1 of H7N9 influenza A virus that critically affect the protein stability and function. Vet Res 2018; 49: 98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Kantor R, Shafer RW, Follansbee S, et al. Evolution of resistance to drugs in HIV-1-infected patients failing antiretroviral therapy. AIDS Lond Engl 2004; 18: 1503–1511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Monto AS. Implications of antiviral resistance of influenza viruses. Clin Infect Dis 2009; 48: 397–399. [DOI] [PubMed] [Google Scholar]
- 108. Van der Vries E, Ison MG. Antiviral resistance in influenza viruses: clinical and epidemiological aspects. Antimicrob Drug Resist 2016; 2: 1165–1183. [Google Scholar]
- 109. Murano K, Guo Y, Siomi H. The emergence of SARS-CoV-2 variants threatens to decrease the efficacy of neutralizing antibodies and vaccines. Biochem Soc Trans 2021; 49: 2879–2890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Houser K, Subbarao K. Influenza vaccines: challenges and solutions. Cell Host Microbe 2015; 17: 295–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Christensen PA, Oslen RA, Long SW, et al. Signals of significantly increased vaccine breakthrough, decreased hospitalization rates, and less severe disease in patients with coronavirus disease 2019 caused by the omicron variant of severe acute respiratory syndrome coronavirus 2 in Houston, Texas. Am J Pathol 2022; 192: 642–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Beaucourt S, Vignuzzi M. Ribavirin: a drug active against many viruses with multiple effects on virus replication and propagation. Molecular basis of ribavirin resistance. Curr Opin Virol 2014; 8: 10–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Empey KM, Peebles RS, Kolls JK. Pharmacologic advances in the treatment and prevention of respiratory syncytial virus. Clin Infect Dis Off Publ Infect Dis Soc Am 2010; 50: 1258–1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Behzadi MA, Leyva-Grado VH. Overview of current therapeutics and novel candidates against influenza, respiratory syncytial virus, and Middle East respiratory syndrome coronavirus infections. Front Microbiol 2019; 10: 1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Rey-Jurado E, Kalergis AM. Immunological features of respiratory syncytial virus-caused pneumonia – implications for vaccine design. Int J Mol Sci 2017; 18: 556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Pfizer. Study to evaluate the efficacy, immunogenicity, and safety of RSVpreF in adults. (RENOIR). Washington, DC: NIH U.S. National Library of Medicine, 2022. [Google Scholar]
- 117. Porter DP, Guo Y, Perry J, et al. Assessment of drug resistance during phase 2b clinical trials of presatovir in adults naturally infected with respiratory syncytial virus. Antimicrob Agents Chemother 2020; 64: e02312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Liu H, Lu B, Tabor DE, et al. Characterization of human respiratory syncytial virus (RSV) isolated from HIV-exposed-uninfected and HIV-unexposed infants in South Africa during 2015-2017. Influenza Other Respir Viruses 2020; 14: 403–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Choi S-H, Park KS, Kim Y-J. Analysis of respiratory syncytial virus fusion protein from clinical isolates of Korean children in palivizumab era, 2009–2015. J Infect Chemother 2019; 25: 514–519. [DOI] [PubMed] [Google Scholar]
- 120. Trento A, Casas I, Calderón A, et al. Ten years of global evolution of the human respiratory syncytial virus BA genotype with a 60-nucleotide duplication in the G protein gene. J Virol. 84, 7500–7512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Simões EAF, Forleo-Neto E, Geba GP, et al. Suptavumab for the prevention of medically attended respiratory syncytial virus infection in preterm infants. Clin Infect Dis 2021; 73: e4400–e4408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Lu B, Liu H, Tabor DE, et al. Emergence of new antigenic epitopes in the glycoproteins of human respiratory syncytial virus collected from a US surveillance study, 2015–17. Sci Rep 2019; 9: 3898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Zhu Q, Patel NK, McAuliffe JM, et al. Natural polymorphisms and resistance-associated mutations in the fusion protein of respiratory syncytial virus (RSV): effects on RSV susceptibility to palivizumab. J Infect Dis 2012; 205: 635–638. [DOI] [PubMed] [Google Scholar]
- 124. DeVincenzo JP, Hall CB, Kimberlin DW, et al. Surveillance of clinical isolates of respiratory syncytial virus for palivizumab (synagis)–resistant mutants. J Infect Dis 2004; 190: 975–978. [DOI] [PubMed] [Google Scholar]
- 125. Hashimoto K, Hosoya M. Neutralizing epitopes of RSV and palivizumab resistance in Japan. Fukushima J Med Sci 2017; 63: 127–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Abou-El-Hassan H, Massaad E, Soudani N, et al. Detection of ON1 and novel genotypes of human respiratory syncytial virus and emergence of palivizumab resistance in Lebanon. PLoS ONE 2019; 14: e0212687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Adams O, Bonzel L, Kovacevic A, et al. Palivizumab-resistant human respiratory syncytial virus infection in infancy. Clin Infect Dis 2010; 51: 185–188. [DOI] [PubMed] [Google Scholar]
- 128. Wargo AR, Kurath G. Viral fitness: definitions, measurement, and current insights. Curr Opin Virol 2012; 2: 538–545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Irwin KK, Renzette N, Kowalik TF, et al. Antiviral drug resistance as an adaptive process. Virus Evol 2016; 2: vew014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Battles MB, McLellan JS. Respiratory syncytial virus entry and how to block it. Nat Rev Microbiol 2019; 17: 233–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Feng Z, Xu L, Xie Z. Receptors for respiratory syncytial virus infection and host factors regulating the life cycle of respiratory syncytial virus. Front Cell Infect Microbiol 2022; 12: 858629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. Murin CD, Wilson IA, Ward AB. Antibody responses to viral infections: a structural perspective across three different enveloped viruses. Nat Microbiol 2019; 4: 734–747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. García-beato R, Martínez I, Francí C, et al. Host cell effect upon glycosylation and antigenicity of human respiratory syncytial virus G glycoprotein. Virology 1996; 221: 301–309. [DOI] [PubMed] [Google Scholar]
- 134. Bukreyev A, Yang L, Fricke J, et al. The secreted form of respiratory syncytial virus g glycoprotein helps the virus evade antibody-mediated restriction of replication by acting as an antigen decoy and through effects on fc receptor-bearing leukocytes. J Virol 2008; 82: 12191–12204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. Hamza A, Shafat Z, Parray ZA, et al. Structural characterization and binding studies of the ectodomain g protein of respiratory syncytial virus reveal the crucial role of ph with possible implications in host–pathogen interactions. ACS Omega 2021; 6: 10403–10414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136. Karron RA, Buonagurio DA, Georgiu AF, et al. Respiratory syncytial virus (RSV) SH and G proteins are not essential for viral replication in vitro: clinical evaluation and molecular characterization of a cold-passaged, attenuated RSV subgroup B mutant. Proc Natl Acad Sci 1997; 94: 13961–13966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. Vanover D, Smith DV, Blanchard EL, et al. RSV glycoprotein and genomic RNA dynamics reveal filament assembly prior to the plasma membrane. Nat Commun 2017; 8: 667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138. Haid S, Grethe C, Bankwitz D, et al. Identification of a human respiratory syncytial virus cell entry inhibitor by using a novel lentiviral pseudotype system. J Virol 2016; 90: 3065–3073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139. Duan L, Zheng Q, Zhang H, et al. The SARS-CoV-2 spike glycoprotein biosynthesis, structure, function, and antigenicity: implications for the design of spike-based vaccine immunogens. Front Immunol 2020; 11: 576622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140. Yan D, Lee S, Thakkar VD, et al. Cross-resistance mechanism of respiratory syncytial virus against structurally diverse entry inhibitors. Proc Natl Acad Sci 2014; 111: E3441–E3449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141. Battles MB, Langedijk JP, Furmanova-Hollenstein P, et al. Molecular mechanism of respiratory syncytial virus fusion inhibitors. Nat Chem Biol 2016; 12: 87–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142. Currier MG, Lee S, Stobart CC, et al. EGFR interacts with the fusion protein of respiratory syncytial virus strain 2-20 and mediates infection and mucin expression. PLoS Pathog 2016; 12: e1005622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143. Mastrangelo P, Chin AA, Tan S, et al. Identification of RSV Fusion Protein Interaction Domains on the Virus Receptor, Nucleolin. Viruses 2021; 13: 261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144. Kolokoltsov A, Deniger D, Fleming EH, et al. Small interfering RNA profiling reveals key role of clathrin-mediated endocytosis and early endosome formation for infection by respiratory syncytial virus. J Virol 2007; 81: 7786–7800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145. Anderson DE, Pfeffermann K, Kim SY, et al. Comparative loss-of-function screens reveal ABCE1 as an essential cellular host factor for efficient translation of paramyxoviridae and pneumoviridae. mBio 2019; 10: e00826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146. Han Z, Rao J, Xie Z, et al. Chemokine (C-X-C Motif) Ligand 4 is a restrictor of respiratory syncytial virus infection and an indicator of clinical severity. Am J Respir Crit Care Med 2020; 202: 717–729. [DOI] [PubMed] [Google Scholar]
- 147. Pei J, Beri NR, Zou AJ, et al. Nuclear-localized human respiratory syncytial virus NS1 protein modulates host gene transcription. Cell Rep 2021; 37: 109803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148. Yin G-Q, Zeng HX, Li ZL, et al. Differential proteomic analysis of children infected with respiratory syncytial virus. Braz J Med Biol Res 2021; 54: e9850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149. Pandya MC, Callahan SM, Savchenko KG, et al. A contemporary view of respiratory syncytial virus (RSV) biology and strain-specific differences. Pathogens 2019; 8: 67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150. Stokes KL, Chi MH, Sakamoto K, et al. Differential pathogenesis of respiratory syncytial virus clinical isolates in BALB/c mice. J Virol 2011; 85: 5782–5793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151. Villenave R, O’Donoghue D, Thavagnanam S, et al. Differential cytopathogenesis of respiratory syncytial virus prototypic and clinical isolates in primary pediatric bronchial epithelial cells. Virol J 2011; 8: 43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152. Rajan A, Piedra F-A, Aideyan L, et al. Multiple respiratory syncytial virus (RSV) strains infecting HEp-2 and A549 cells reveal cell line-dependent differences in resistance to RSV infection. J Virol 2022; 96: e01904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153. Stokes K, Currier MG, Sakamoto K, et al. The respiratory syncytial virus fusion protein and neutrophils mediate the airway mucin response to pathogenic respiratory syncytial virus infection. J Virol 2013; 87: 10070–10082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154. Lukacs NW, Moore ML, Rudd BD, et al. Differential immune responses and pulmonary pathophysiology are induced by two different strains of respiratory syncytial virus. Am J Pathol 2006; 169: 977–986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155. Human S, Hotard AL, Rostad CA, et al. A respiratory syncytial virus attachment gene variant associated with more severe disease in infants decreases fusion protein expression, which may facilitate immune evasion. J Virol 2020; 95: e01201–e01220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156. Pierangeli A, Viscido A, Bitossi C, et al. Differential interferon gene expression in bronchiolitis caused by respiratory syncytial virus-A genotype ON1. Med Microbiol Immunol (Berl.) 2020; 209: 23–28. [DOI] [PubMed] [Google Scholar]
- 157. Liu Z, Fan P, Chen M, et al. miRNAs and leukotrienes in respiratory syncytial virus infection. Front Pediatr 2021; 9: 602195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158. Smith P, Wang S-Z, Dowling K, et al. Leucocyte populations in respiratory syncytial virus-induced bronchiolitis. J Paediatr Child Health 2001; 37: 146–151. [DOI] [PubMed] [Google Scholar]
- 159. Besteman SB, Callaghan A, Langedijk AC, et al. Transcriptome of airway neutrophils reveals an interferon response in life-threatening respiratory syncytial virus infection. Clin Immunol 2020; 220: 108593. [DOI] [PubMed] [Google Scholar]
- 160. Yasui K, Baba A, Iwasaki Y, et al. Neutrophil-mediated inflammation in respiratory syncytial viral bronchiolitis. Pediatr Int 2005; 47: 190–195. [DOI] [PubMed] [Google Scholar]
- 161. Honardoost M, Rajabpour A, Vakili L. Molecular epidemiology; new but impressive. Med J Islam Repub Iran 2018; 32: 53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162. Aderem A. Systems biology: its practice and challenges. Cell 2005; 121: 511–513. [DOI] [PubMed] [Google Scholar]
- 163. Lund E, Dumeaux V. Systems epidemiology in cancer. Cancer Epidemiol Biomarkers Prev 2008; 17: 2954–2957. [DOI] [PubMed] [Google Scholar]
- 164. Rasmussen AL, Katze MG. Genomic signatures of emerging viruses: a new era of systems epidemiology. Cell Host Microbe 2016; 19: 611–618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165. Smith MT, Hainaut P, Perera F, et al. Future perspectives on molecular epidemiology. IARC Sci Publ 2011; 163: 493–500. [PubMed] [Google Scholar]