Abstract
Mutants of Escherichia coli and Klebsiella aerogenes that are deficient in glutamate synthase (glutamate-oxoglutarate amidotransferase [GOGAT]) activity have difficulty growing with nitrogen sources other than ammonia. Two models have been proposed to account for this inability to grow. One model postulated an imbalance between glutamine synthesis and glutamine degradation that led to a repression of the Ntr system and the subsequent failure to activate transcription of genes required for the use of alternative nitrogen sources. The other model postulated that mutations in gltB or gltD (which encode the subunits of GOGAT) were polar on a downstream gene, gltF, which is necessary for proper activation of gene expression by the Ntr system. The data reported here show that the gltF model is incorrect for three reasons: first, a nonpolar gltB and a polar gltD mutation of K. aerogenes both show the same phenotype; second, K. aerogenes and several other enteric bacteria lack a gene homologous to gltF; and third, mutants of E. coli whose gltF gene has been deleted show no defect in nitrogen metabolism. The argument that accumulated glutamine represses the Ntr system in gltB or gltD mutants is also incorrect, because these mutants can derepress the Ntr system normally so long as sufficient glutamate is supplied. Thus, we conclude that gltB or gltD mutants grow slowly on many poor nitrogen sources because they are starved for glutamate. Much of the glutamate formed by catabolism of alternative nitrogen sources is converted to glutamine, which cannot be efficiently converted to glutamate in the absence of GOGAT activity. Finally, GOGAT-deficient E. coli cells growing with glutamine as the sole nitrogen source increase their synthesis of the other glutamate-forming enzyme, glutamate dehydrogenase, severalfold, but this is still insufficient to allow rapid growth under these conditions.
Virtually all the nitrogenous compounds in an enteric bacterium derive their nitrogen atoms from either glutamate or glutamine. It has been estimated that about 88% of the cellular nitrogen in Escherichia coli is derived from glutamate and the remaining 12% is derived from glutamine (40). Thus, glutamine and, especially, glutamate are the key intermediates in cellular nitrogen metabolism. However, neither glutamate nor glutamine is capable of supporting growth at a rate comparable to that supported by ammonium salts when provided as the sole nitrogen source, probably because of inefficient transport of both compounds. Clearly, ammonium is the preferred nitrogen source for the enteric bacteria. In the presence of ammonium, there is a strong repression of many systems that allow the cells to use alternative nitrogen sources, such as amino acids, inorganic compounds, and urea (29).
Ammonia can be fixed onto the carbon skeleton of α-ketoglutarate to form glutamate by two different pathways in the enteric bacteria. It can be fixed directly onto α-ketoglutarate in an NADPH-dependent reaction by glutamate dehydrogenase (GDH). It can also be fixed indirectly by first being attached to glutamate to form glutamine by an ATP-dependent glutamine synthetase (GS), followed by the transfer of the amide nitrogen onto α-ketoglutarate by an NADPH-dependent glutamate synthase, also called glutamate-oxoglutarate amidotransferase (GOGAT). No other pathway allows significant net assimilation of ammonia into glutamate. Mutants lacking both GDH and GOGAT are glutamate auxotrophs, requiring glutamate, a compound that can be degraded to glutamate, or a compound that can transaminate its nitrogen to α-ketoglutarate (9).
Under conditions of ammonia-limited growth, GDH plays a relatively minor role in nitrogen assimilation because of its high Km for ammonium (33). In Klebsiella spp., this role is further minimized because GDH is strongly repressed under conditions of nitrogen limitation (33). In fact, mutants lacking GDH have no altered growth phenotype in glucose minimal medium and show a competitive disadvantage relative to the wild type only during energy limitation (21). In other words, GOGAT is able to provide the cell's total glutamate supply, and during ammonia-limited growth, the indirect pathway (via glutamine and GOGAT) becomes essential (9). Under conditions of ammonia excess, either pathway is sufficient for glutamate synthesis.
The response to nitrogen excess or limitation is determined by the intracellular concentration of glutamine (23). Glutamate seems to play no role in signaling nitrogen excess or limitation. However, glutamate does play an important role in the osmoregulation of the cell, especially as the counterion that allows the accumulation of intracellular potassium (42). Glutamate and glutamine play very different regulatory roles in the cell despite their proximity in metabolic pathways. Thus, it is important that the cell have mechanisms to adjust the pools of glutamate and glutamine independent of each other and to do so during changes in nitrogen availability or osmotic challenge. The glutamate pool is filled by GDH and GOGAT and depleted by GS and general biosynthetic reactions. The glutamine pool is filled by GS and depleted largely by GOGAT and some biosynthetic reactions. There are several glutaminases reported in enteric bacteria, but it has been suggested that these are relatively inefficient in comparison to GOGAT (40). The key enzymes for maintaining the proper relative sizes of the pools of glutamate and glutamine are GS (which depletes glutamate and produces glutamine) and GOGAT (which depletes glutamine and produces glutamate).
Mutants lacking GS activity cannot accumulate glutamine (the signal of nitrogen excess) except by transport, and their nitrogen regulation is blind to exogenous ammonia (6). Such mutants mount a nitrogen starvation response (derepress the Ntr system) even in the presence of high concentrations of ammonia. This understanding was initially confused by the fact that polar mutations in glnA (the structural gene for GS) fail to express the ntrC gene product and thus mount a nitrogen excess response (no Ntr system expression) even in the absence of ammonia.
GOGAT is a heterodimeric protein whose subunits are encoded by the gltB and gltD genes. The gltB gene codes for the larger glutaminase subunit, and gltD codes for the smaller transaminase subunit analogous to GDH. Mutants that lack GOGAT cannot deplete glutamine except by biosynthetic reactions. Moreover, when ammonia is limiting (too dilute for efficient fixation by GDH), mutants that lack GOGAT cannot accumulate glutamate except by transport, transamination, the residual activity of GDH, or the weak glutaminases of the cell. Such mutants fail to grow on low concentrations of ammonia, grow poorly on glutamine, and grow poorly or not at all on a variety of nitrogen-containing compounds that can be catabolized to ammonia, glutamate, or both (9). When cells with a gltB or gltD mutation (lacking GOGAT activity) are grown with histidine as the sole nitrogen source, growth is very slow, suggesting a defect in activating the histidine utilization (hut) operons. The original explanation for this failure to grow with histidine as the sole nitrogen source was based on arguments about the size of the pool of glutamine. In the absence of GOGAT activity, the size of the glutamine pool would increase and could not be depleted, thus preventing activation of the Ntr system and consequently of histidase formation (2, 40). However, this leads to the paradoxical predictions of glutamine pools that are too high to allow derepression of the Ntr system but too low to allow growth. The discovery of a third gene downstream in the gltBD operon, gltF, led to an alternative explanation in which the GltF product was essential for nitrogen regulation and gltBD mutations might be polar on gltF expression (10, 11).
The experiments described here were designed to examine the growth defects of gltBD mutations and to determine the effects of the gltF gene on nitrogen regulation in the enteric bacteria.
MATERIALS AND METHODS
Bacterial strains.
Descriptions and genotypes of the bacterial strains used are listed in Table 1. The species Klebsiella aerogenes has been subsumed into the species Klebsiella pneumoniae; however, we have retained the older name for strains derived from strain W70 for historical reasons and to distinguish it from the nitrogen-fixing K. pneumoniae strain M5aL (which is properly Klebsiella oxytoca), from which it differs considerably. All K. aerogenes strains are derived from strain W70, all K. pneumoniae strains are derived from strain M5aL, all E. coli strains are derived from strain K-12, and all Salmonella enterica serovar Typhimurium strains are derived from strain LT-2.
TABLE 1.
List of strains
| Straina | Relevant genotype or properties | Source or reference |
|---|---|---|
| E. coli | ||
| BE3471 | W3110 gltD39::Mu d1-1734 | 15 |
| EB566 | EG47 hutC515Kb | hutC515K in EG47 by P1 transduction |
| EB3365 | YMC10 nac-28 | 35 |
| EB4504 | YMC10 recD1903::Tn10dtet | recD in YMC10 by transduction |
| EB4507 | YMC10 recD1903::Tn10dtet ΔgltF1::Ω | This work |
| EB4534 | YMC10 ΔgltF1::Ω | P1 · EB4507 × YMC10d |
| EB4539 | EB566 ΔgltF1::Ω | P1 · EB4507 × EB566 |
| EB4566 | W3110 (λRZ-5φCB540) | This work |
| EB4582 | ΔgltF1::Ω (λRZ-5φCB540) | P1 · EB4507 × EB4566 |
| EB4613 | YMC10 gltD39::Mu d1-1734 | P1 · BE3471 × YMC10 |
| EB4614 | W3110 gltD39::Mu d1-1734 | P1 · BE3471 × W3110 |
| EB4615 | Y MC10 gltD39::Mu d1-1734/pCB548 | This work |
| EB4693 | YMC10 hutCK::attλ | B. Janes and R. Bender, unpublished data |
| EB4709 | MX614 pro+ilv+ | Transduction |
| EB4713 | EB4709 hutCK::attλ | P1 · EB4693 × EB4709 |
| EB4722 | EB4713 ΔgltF1::Ω | P1 · EB4507 × EB4713 |
| EB4985 | W3110 gltD39::Mu d1-1734 | P1 · BE3471 × W3110 |
| EB5019 | YMC10 gltD39::Mu d1-1734 | P1 · BE3471 × YMC10 |
| EG47 | hsdR lac rpsL594 glk galK gal | 17 |
| MX614 | Δ[pro-lac] galE ilv-680 thi-1 | 11 |
| W3110 | Wild-type strain W3110 | R. Matthews |
| YMC10 | endA1 thi-1 hsdR17 supE44 ΔlacU169 hutC515Kb | 1 |
| K. aerogenes | ||
| MK1 | Wild-type (hutC+) | 39 |
| MK189 | gltB200 hutC | 9 |
| KB630 | gltB200 gdhA1 hutC+ | P1s of strain MK261 (9) |
| KB2560 | gltB200 gdhA1 hutC+ (Mu cts62 hP1#1) | Mu lysogen of KB630 |
| KC1043 | Wild type | 28 |
| KC1419 | KC1043/pTROY11c | This work |
| KC1467 | Mu cts63 hP1#1/pEG5005 | This laboratory |
| KC2561 | KC1043 gltB200 | This work |
| KC2668 | Δ[bla]-2 | 24 |
| KC3182 | gltB200 gdhA2::Ω | gdhA2 in KC2561 by P1 transduction |
| KC3686 | KC2668/pCB832 | This work |
| KC4540 | KC3686 gltD702::Tn5-131 | gltD702 in KC3686 by P1 transduction |
| Others | ||
| KP4387 | his+ transductant of K. pneumoniae (K. oxytoca) strain UNF122 (UNF122 is M5aL hisD2 Δlac-2002 hsdR1) | This laboratory |
| LT-2 | S. typhimurium strain MST1 | S. Maloy |
All E. coli strains are derived from E. coli K-12; all K. aerogenes strains are derived from K. aerogenes W70. Most K. aerogenes strains are derived from MK53 (39) and carry the hutC515 mutation; exceptions to this are noted as hutC+.
The hut genes (with the hutC515 mutation) are from K. aerogenes.
Plasmid pTROY11 (14) confers sensitivity to bacteriophage λ.
Transduction of strain YMC10 by bacteriophage P1 grown on strain EB4507.
Growth media.
Bacterial cultures were grown either in rich (LB) medium or in W4 minimal medium (W salts [4] adjusted to an initial pH of 7.4). Minimal medium was supplemented with 0.4% (wt/vol) glucose as a carbon source and one or more of the following nitrogen sources: ammonium sulfate (0.2%); l-glutamine (0.2, 0.1, or 0.04%); l-serine, l-arginine, or potassium nitrate (each at 0.2%); or the monosodium salt of l-glutamate (0.2 or 0.4%). Thiamine HCl (10 μg/ml) was used to supplement thiamine auxotrophs where necessary. Media for plasmid and transposon selection and maintenance were supplemented with antibiotics as follows: ampicillin (75 μg/ml for E. coli strains and K. aerogenes strains carrying the Δ[bla]-2 deletion and 1,000 μg/ml for bla+ strains of K. aerogenes), spectinomycin (100 μg/ml), streptomycin (50 μg/ml), kanamycin sulfate (50 μg/ml), and tetracycline (30 μg/ml). Media for selection of strains bearing a single copy of the Ω cartridge (38) on the chromosome were supplemented with spectinomycin and streptomycin at 100 and 50 μg/ml, respectively, for some strains (W3110, YMC10, and EB4504) or at 50 and 25 μg/ml, respectively, for others (EB566, EB4566, and EB4722). Whenever glutamine was used as the sole nitrogen source, it was Calbiochem A grade.
Genetic and molecular techniques.
The hutUp-lacZ fusion was transferred from the plasmid pCB540 to the phage λRZ-5 and then to the chromosome of W3110 essentially as described by Grove and Gunsalus (20). Briefly, λRZ-5 was grown on a YMC10 transformant carrying pCB540, and the lysate thus obtained was used as a source of the double-crossover recombinant phages by transducing an attB+ E. coli strain to Apr and screening for stable lysogens. Stable lysogens were defined as those which retained Apr at a frequency of 100% after three rounds of subculture on LB medium in the absence of ampicillin. Recombinant phages spontaneously released from the stable lysogens were used to prepare a lysate of pure λRZ-5ΦCB540, which was used to transduce W3110 to Apr.
Recombinant DNA techniques were carried out essentially as described by Maniatis et al. (30). Plasmid DNA was purified using Qiaprep miniprep spin columns (Qiagen). Chromosomal DNA was purified using the Puregene DNA isolation kit (Gentra Systems). Linear DNA fragments were separated on agarose gels made in TAE buffer (30) and recovered from gel slices by using Qiaquick gel extraction spin columns (Qiagen).
Custom oligodeoxynucleotide primers were supplied by Gibco-BRL. PCR was performed with Taq DNA polymerase (Gibco-BRL). Reactions were performed in a 100-μl volume containing approximately 50 pmol of each primer, 0.1 μg of template DNA, 0.2 mM deoxyribonucleoside triphosphates, 1.5 mM MgCl2, 0.5 U of Taq DNA polymerase, and buffer conditions specified by the manufacturer of the polymerase. The PCR cycle used an initial 3-min denaturation step at 94°C followed by a 1-min primer-annealing step at 56°C, a 1-min primer extension step at 72°C, and a 1-min denaturation step at 94°C for 32 cycles, and a final extension step of 3 min at 72°C.
Cloning and sequencing of the gltBD region from K. aerogenes
The wild-type gltB gene and surrounding region was cloned from K. aerogenes strain KC1467 using the in vivo Mu lysate technique (19). Strain KB2560 (a K. aerogenes strain that lacks both GOGAT and GDH and is therefore auxotrophic for glutamate) was transduced with the lysate from KC1467 with selection for glutamate prototrophs (Glt+). Glt+ transductants were screened for growth on glucose minimal medium supplemented with serine to 0.2%, a growth phenotype which distinguishes between Gogat+ strains of K. aerogenes, which grow, and Gogat− strains, which do not (9). One of the recovered plasmids that was found to confer the Gogat+ phenotype on KB2560 (pCB507) carried approximately 20 kb of inserted DNA. pCB507 was digested with EcoRI, from which a 12-kb fragment was recovered and cloned into the EcoRI site of either pUC19 or pGB2, yielding pCB536 and its oppositely oriented subclone pCB537 (in pUC19) or pCB548 (in pGB2). Although the high-copy-number plasmids pCB536 and pCB537 could not be stably maintained in K. aerogenes, the low-copy-number plasmid pCB548 was maintained stably in K. aerogenes and was found to complement the glutamate auxotrophy of KB630. Subclones derived from pCB536 and pCB537 and maintained in E. coli (pCB593, pCB620, and pCB633) were used as templates for automated sequencing of both strands of the DNA, from which the sequence of 7,930 bp was determined.
Complementation and marker rescue analysis.
The ability of plasmids carrying portions of the wild-type K. aerogenes gltBD operon to complement the glutamate auxotrophy of KB630 (gltB200 gdhA1) or to recombine with gltB200 to yield gltB+ recombinants was tested as follows. Strain KB630 was transformed to drug resistance with the test plasmid. Drug-resistant transformants were selected on LB agar supplemented with either a mixture of streptomycin and spectinomycin (when pCB548 was used) or ampicillin (when pUC-based plasmids were used.) Fifty or more isolated colonies were then patched onto glucose ammonia minimal medium (without glutamate) with sterile toothpicks. Complementation was indicated if all patches contained solid and uniform growth after 16 to 48 h of incubation. Recombination was indicated if 30% or more of the patches contained one or more isolated colonies. If none of the patches showed uniform growth or isolated colonies, the plasmid was considered to be incapable of complementation or recombination.
The ability of pCB548 to complement the nitrogen regulatory phenotype of E. coli strains carrying the gltD39 mutation was tested by transforming EB4613 and EB4614 with pCB548 and selecting for growth on LB agar supplemented with streptomycin and spectinomycin. Purified transformants were tested for growth on minimal glucose plates with arginine as the sole nitrogen source.
Southern blot analysis.
Southern blotting and hybridization were carried out as described in the Genius nonradioactive labeling and detection kit (Boehringer Mannheim) using digoxigenin-labeled probes obtained with the random priming method. Low-stringency hybridization was carried out at 42°C with washes in 5× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) at room temperature. Moderate-stringency hybridization was carried out at 58°C with washes in 2× SSC and 0.5× SSC at 58°C. High-stringency hybridization was carried out at 65°C with washes in 2× SSC and 0.5× SSC at 65°C. The probe used to detect homologues of the gltF gene was an 830-bp PCR product corresponding to the E. coli gltF gene (as indicated in the figures). The probe used to detect homologues of the gltD gene was an 882-bp PCR product obtained by amplification of the K. aerogenes gltD gene (as indicated in the figures). Nylon filters were stripped for reblotting using the alkaline stripping method suggested by the kit's manufacturer.
Mutagenesis of E. coli gltF.
The ΔgltF1::Ω mutation was constructed in vitro and introduced into the E. coli chromosome by reverse genetics essentially as described by Horton and Pease (22). The result of this process was a fragment containing all of the DNA sequences surrounding gltF but with the coding sequence of gltF replaced by a BamHI site. This fragment was cloned into pBS SK(+), resulting in pCB1055, which carried the ΔgltF1 allele. The Ω fragment, specifying resistance to streptomycin and spectinomycin, was purified from BamHI-digested pPH45Ω (38) and ligated into the BamHI site in pCB1055, yielding pCB1063 carrying ΔgltF1::Ω.
The ΔgltF1::Ω allele was transferred to the chromosome of EB4504 (recD1903) by electroporation with the 3.7-kb Asp718-to-XbaI fragment purified from pCB1063 followed by selection for resistance to streptomycin and spectinomycin. EB4507, an ampicillin-sensitive electroporant (lacking the vector sequences from the plasmid) was screened for the presence of the ΔgltF1::Ω allele by PCR analysis. Finally, the ΔgltF1::Ω allele was transferred from EB4507 into recD+ strains using P1vir-mediated transduction with selection for the streptomycin and spectinomycin resistance encoded by the Ω fragment.
Enzyme assays.
Cultures of K. aerogenes and E. coli were grown at 30 and 37°C, respectively, in minimal medium supplemented as indicated in the tables to a density of 50 Klett units (filter 54) except where indicated. Cells were collected by centrifugation, washed once with 1% (wt/vol) KCl, and resuspended at 10-fold concentration (about 1 mg of protein/ml) in 1% KCl. The histidase, GOGAT, and β-galactosidase assays have been described previously (28). Specific activities are reported as nanomoles of product formed (or substrate degraded) per minute per milligram of protein.
Nucleotide sequence accession number.
The DNA sequence of the gltBD region from K. aerogenes was determined using the automated fluorescent dye termination method at the University of Michigan Core facility and has been deposited in GenBank under accession no. AY035435.
RESULTS
The gltBD operon of K. aerogenes.
The gltBD region from K. aerogenes was cloned as described in Materials and Methods, and the DNA sequence of 7,930 bp from the region was determined. The key features of this sequence are summarized in Fig. 1. There are two open reading frames (ORFs) which are highly similar to the gltB and gltD ORFs of E. coli (95 and 93% identity over 1,486 and 473 amino acid residues, respectively), except for a patch of 8 residues (amino acids 584 to 591) in GltB. The K. aerogenes gltB and gltD ORFs are separated by a 12-bp spacer, slightly smaller than the 15-bp spacer found between the E. coli gltB and gltD genes. An oppositely directed ORF of 324 amino acids (orfB) lies 60 bp downstream from the GltD ORF. The deduced amino acid sequence of orfB is 74% identical (but five residues longer) than the yhcJ ORF in E. coli. In E. coli, yhcJ lies more than 9 kb downstream from gltD (7).
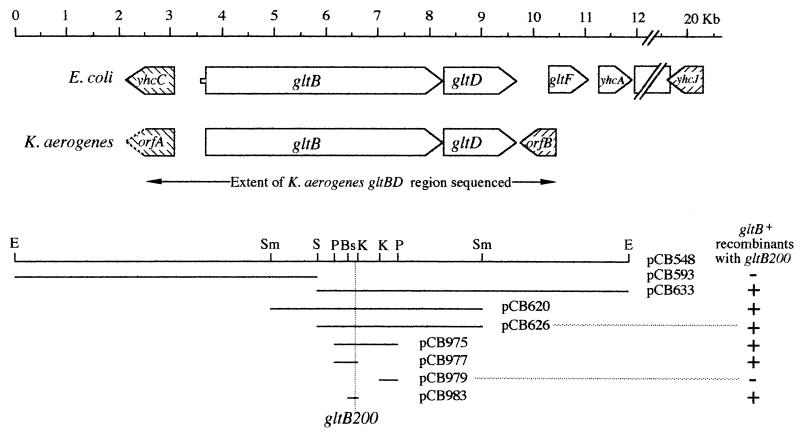
FIG. 1.
Comparison of the gltBD regions from E. coli and K. aerogenes. Homologous ORFs are indicated by similar hatching. The solid lines in the lower portion represent the DNA sequences present in the plasmids listed. E, Sm, S, P, Bs, and K represent restriction enzyme cleavage sites for EcoRI, SmaI, SalI, PstI, BssHII, and KpnI, respectively. (Note that additional sites exist in the cloned DNA for many of the indicated restriction enzymes.) Ability (+) or inability (−) to form recombinants by marker rescue with the gltB200 allele of K. aerogenes is indicated.
The 60-bp gltD-orfB intergenic region in K. aerogenes contains two 12-bp regions of inverted dyad symmetry separated by 8 bp, which might function as a bidirectional transcription terminator for gltBD and orfB operons. When a DNA fragment including these repeats was placed between the lacZ promoter and the lacZ gene, β-galactosidase expression was reduced by 95% (data not shown). Thus, this region appears to contain the termination signals for the gltBD operon of K. aerogenes. This same sequence arrangement (gltD, the 60-bp intergenic region, and orfB) was obtained from two other independent clones of the K. aerogenes gltD region. Thus, no homologue of gltF was found immediately downstream of gltD in K. aerogenes.
Upstream, 673 bp from the start of the gltB ORF, is the start of an oppositely directed ORF, orfA, which could code for a polypeptide of more than 198 amino acid residues. OrfA is highly similar (90% identity over those 198 amino acid residues) to the yhcC ORF of E. coli, which lies 674 bp upstream from the gltB ORF in E. coli. The 673 bp between orfA and gltB contain sequences that may represent divergent promoters for these ORFs. Operon fusion studies with a lacZ reporter gene suggested the presence of the gltB promoter within this region. We were unable to detect any activity from the inferred promoter of the oppositely directed ORF (orfA) under any of our conditions.
Characterization of the gltB200 mutation.
The gltB200 mutation (formerly asm-200) was originally isolated as a chemically induced mutant that was unable to use urocanate as the sole nitrogen source (9). Further studies showed that the phenotype of gltB200 was similar to those of other gltB mutations. Strains carrying gltB200 grew poorly with histidine as the sole nitrogen source, failed to grow with serine as the sole nitrogen source, lacked GOGAT activity, and caused an auxotrophic requirement for glutamate in strains carrying the gdhA1 mutation. (The gdhA1 mutation eliminates GDH activity, the only enzyme other than GOGAT capable of net glutamate synthesis from ammonia [9].) Cloned fragments containing portions of the gltBD operon were tested for the ability to give Glt+ recombinants with the gltB200 mutation. The data presented in Fig. 1 can be summarized by saying that all clones that contained the segment of gltB DNA from 2,689 to 2,881 bp downstream from the start of the gltB ORF gave rise to Glt+ recombinants. This corresponds to the BssHI and KpnI sites at positions 6.45 and 6.64 in Fig. 1. All clones tested that lacked this segment failed to yield recombinants with the gltB200 mutation. As expected, the frequency of such “marker rescue” was found to be lower for clones carrying short regions of homology than for those with more extensive regions of homology, and the introduction of a recA3011 mutation reduced the frequency of Glt+ recombinants. Thus, it appeared that the mutation responsible for the Glt− phenotype (gltB200) must lie within the region from 2,689 to 2,881 bp downstream from the start of the gltB ORF.
Recombinants were generated by such marker rescue using the smallest fragment (that in pCB983). These recombinants were able to express GOGAT activity, derepress histidase formation under conditions of nitrogen limitation, and use potassium nitrate as the sole nitrogen source just as well as a gltB+ strain generated by P1 transduction (data not shown). Thus, the gltB200 mutation, responsible for the Gogat and Ntr phenotypes, lies within the 193-bp region bounded by the BssHII and KpnI restriction sites.
This region was amplified by PCR from the chromosomes of two different gltB200 strains, KC2561 and MK189, and the DNA sequence was determined. Both strains yielded PCR products which carried a single-base-pair change at nucleotide 2,822 of the gltB ORF, a change from a G to an A in the gltB200 strains. This mutation changes a GGT codon to GAT, resulting in a G941D missense mutation in the GltB polypeptide. Of the 94 codons in the wild-type gltB ORF that code for aspartic acid, 49 are specified by the GAT triplet. Thus, the mutant GAT sequence does not generate a rare codon, and it is unlikely that the gltB200 mutation leads to polar effects on the downstream gltD gene.
Characterization of the gltD702 mutation.
The glt-702 mutation was originally isolated as a Tn5 insertion mutant of strain KC1419 (3) that was unable to use urea as the sole nitrogen source. We have shown that the phenotype of glt-702::Tn5 is similar to that of gltB200 in that strains carrying glt-702::Tn5 grew poorly with urea or histidine as the sole nitrogen source, failed to grow with serine as the sole nitrogen source, lacked GOGAT activity, and caused an auxotrophic requirement for glutamate in strains carrying the gdhA1 mutation (data not shown). Mapping studies showed that the glt-702::Tn5 mutation was about 20% linked to argG2 by P1-mediated generalized transduction, the same as the linkage of gltB200 and argG2 (16). Thus, we assumed that glt-702 was a mutation in the gltBD operon.
The Tn5 element was converted to Tn5–131 by transduction (28), and the glt-702::Tn5–131 mutation was cloned in vivo via Mu lysate, selecting for the tetracycline resistance associated with the Tn5–131 element. Tetracycline-resistant (Tcr) transductants were screened for the ability to complement the glutamate auxotrophy of KB2560 (gltB200 gdhA1). The two clones thus obtained, pCB612 and pCB613, differed in their abilities to complement the glutamate auxotrophy of a gltB200 recipient strain. The plasmid pCB613, which carried all of gltB, complemented gltB200, showing that glt-702 did not lie in gltB. As expected, pCB612, which lacked a portion of gltB, failed to complement gltB200.
Subclones derived from pCB612 were sequenced, and the point of insertion of Tn5–131 was found to be within the gltD gene, 88 codons from the 3′ end of the gltD ORF. Thus, we were able to identify the glt-702 mutation as gltD702. Moreover, the complementation of the glutamate auxotrophy of KB2560 (gltB200 gltD+) by pCB613 carrying gltB+ gltD702 proved that gltB200 was not polar on gltD. By extension, gltB200 could not be polar on gltF, a finding that is clearly inconsistent with the gltF model.
Absence of gltF in K. aerogenes.
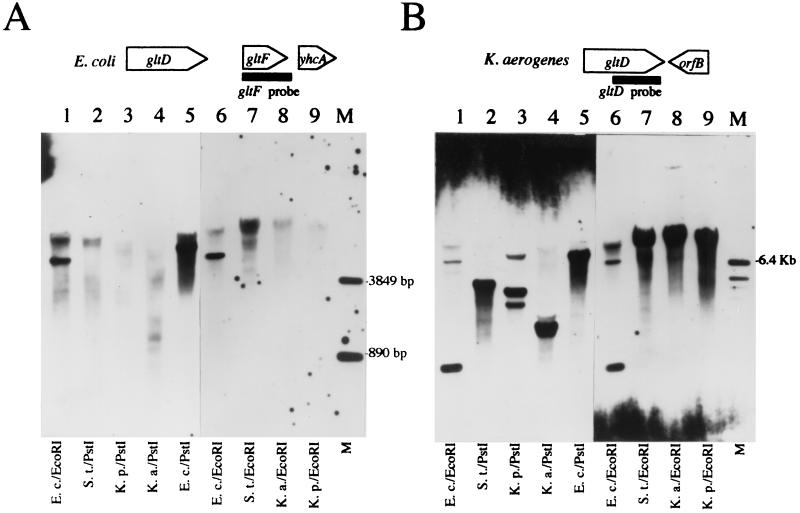
Since no gene homologous to gltF was found in the region immediately downstream of the K. aerogenes gltD gene, Southern analysis was used to see if a homologue of gltF existed anywhere in the genomes of K. aerogenes and other selected enteric bacteria. DNA from strains of K. aerogenes, K. pneumoniae (K. oxytoca), and S. enterica serovar Typhimurium was hybridized with a probe corresponding to the E. coli gltF ORF. DNA from E. coli (from which the gltF probe was derived) served as a positive control. When carried out under conditions of low hybridization stringency, a band of the size expected for E. coli gltF was observed in the E. coli lane (Fig. 2A). Less intense, smeared signals were also observed at this low stringency, which likely correspond to weaker interactions of the gltF probe with E. coli DNA other than gltF. No other DNA contained material which strongly hybridized to the gltF probe; however, as with the E. coli samples, less intense, smeared signals were obtained with the other samples. This was especially true for the S. enterica serovar Typhimurium sample and leaves open the question of whether a gene with some homology to gltF might exist somewhere in the S. enterica serovar Typhimurium genome.
FIG. 2.
Southern blot of chromosomal DNA digested with either EcoRI or PstI. (A) The probe was derived from an 830-bp fragment corresponding to the E. coli gltF ORF as depicted at the top. Hybridization conditions were low stringency (see Materials and Methods). (B) The filter was stripped and reprobed with a probe that was derived from an 882-bp fragment corresponding to the 3′ end of the K. aerogenes gltD ORF as depicted at the top. Hybridization conditions were moderate stringency. Lanes 1 through 9 contained chromosomal DNA from E. coli strain W3110 (E.c.), S. enterica serovar Typhimurium strain LT-2 (S.t.), K. aerogenes strain MK1 (K.a.), and K. pneumoniae strain KP4387 (K.p.), which is derived from strain M5aL. Lane M contained gltF standards of 3.8 and 0.89 kb and a gltD standard of 6.4 kb.
As a control, the same filter was stripped of the gltF probe and rehybridized using a probe derived from the 3′ end of the K. aerogenes gltD gene. Under conditions of low hybridization stringency, smears of hybridizing signal were seen for all DNA tested (data not shown). Under conditions of moderate hybridization stringency, distinct bands of hybridizing material were seen for E. coli DNA samples, and their sizes were consistent with those expected from digestion of the E. coli gltD region (Fig. 2B, lanes 1 and 6). A fainter signal of higher molecular weight observed with the E. coli samples may represent hybridization of the gltD probe to the E. coli aegA gene (12), which is 68% identical to the gltD probe over 872 nucleotides. All of the other samples tested displayed hybridizing material of very high molecular weight (Fig. 2B, lanes 7 to 9). The 3′ end of the E. coli gltD gene lies on the same 7.5-kb PstI fragment as the gltF gene, consistent with the position of the signal seen in Fig. 2B. Restriction maps of pCB507 show that the 3′ end of the K. aerogenes gltD gene is carried on PstI and EcoRI fragments of approximately 1.6 and 12 kb, respectively (data not shown), consistent with the positions of the signals seen in Fig. 2B, lanes 4 and 8. The sizes of gltD-bearing PstI and EcoRI fragments from the S. enterica serovar Typhimurium and K. pneumoniae samples are not known.
Since the gltD probe recognized homologues even at moderate stringency whereas the gltF probe failed to detect significant homologues even at low stringency, we concluded that the K. aerogenes, K. pneumoniae, and possibly S. enterica serovar Typhimurium strains tested lack a gene homologous to gltF.
The role of gltF in E. coli
Since homologues of gltF appear to be absent from the chromosomes of K. aerogenes and K. pneumoniae, we questioned whether the gltF gene plays a species-specific role in the function of the Ntr system in E. coli. E. coli strain EB4613 carries the psiQ39 mutation (34), in which the Mu d1–1734 phage is inserted into the gltD gene approximately one-third of the gene's length from its carboxy (3′) terminus. (Hereafter, we will refer to this mutation as gltD39::Mu d1–1734 or simply gltD39.) As expected of a gltD strain, EB4613 lacks GOGAT activity (Gogat− phenotype) and fails to grow with arginine as the sole nitrogen source (Asm− phenotype). EB4613 was transformed with pCB548, a low-copy-number plasmid carrying the gltBD operon from K. aerogenes. The resulting strain, EB4615, regained GOGAT activity and was able to use arginine as the sole nitrogen source. Thus, the presence of gltBD from K. aerogenes seemed sufficient to restore both Gogat+ and Asm+ phenotypes, even in the absence of an added gltF gene. If gltF is indeed transcribed from a promoter located upstream of the polar gltD39 mutation, then restoration of the Asm+ phenotype by a plasmid that carried only gltB and gltD (and not gltF) suggests that there is no clear role for gltF in the derepression of the Ntr system.
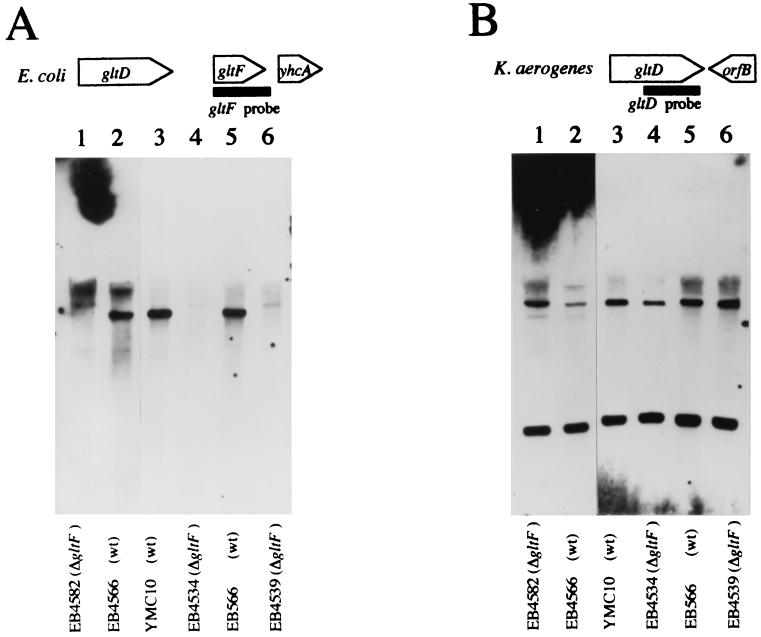
Deletion of the gltF ORF using reverse genetics allowed a more rigorous test of the role of the gltF gene in both the Ntr phenotype (ability to activate gene expression in response to nitrogen limitation) and the glutamate-dependent repression of GOGAT (another phenotype ascribed to GltF). The ΔgltF1::Ω mutation was constructed in vitro, as described in Materials and Methods, and transferred to the chromosome of the E. coli K-12 strain YMC10, where the presence of the hut genes from K. aerogenes provides a convenient reporter for the activity of the Ntr system. The resulting strain, EB4534, was checked for the loss of DNA corresponding to the gltF ORF from the chromosome by PCR analysis (data not shown), and the loss was confirmed by Southern blot analysis. Under conditions of high hybridization stringency, the EcoRI-digested wild-type chromosomal DNA sample displayed a band which hybridized to the gltF probe, while the gltF deletion mutant (EB4534) showed no such band (data not shown). Even under conditions of low stringency, similar results were obtained (Fig. 3A, lanes 3 and 4).
FIG. 3.
Southern blot analysis of EcoRI-digested chromosomal DNA from wild-type E. coli strains and derivatives carrying the ΔgltF1::Ω mutation. (A) The probe was derived from an 830-bp fragment corresponding to the gltF ORF as depicted at the top. Hybridization conditions were high stringency. (B) The filter was stripped and reprobed with a probe that was derived from an 882-bp fragment corresponding to the 3′ end of the K. aerogenes gltD ORF. Hybridization conditions were moderate stringency. Lanes 2, 3, and 5 contained DNA from three different E. coli “wild types,” EB4566 (derived from W3110), YMC10, and EB566. Lanes 1, 4, and 6 contained DNA from the ΔgltF1::Ω mutants derived from these three strains (EB4582, EB4534, and EB4539, respectively).
The specific activities of histidase and GOGAT were determined for YMC10 and its ΔgltF1 transductant, EB4534, grown under various conditions of nitrogen availability. From Table 2, it can be seen that neither the doubling time nor the levels of histidase and GOGAT were significantly altered by the presence of ΔgltF1 under any of the culture conditions tested. Several other features of Table 2 deserve comment. As the degree of nitrogen limitation increased (with a corresponding increase in doubling time), histidase levels increased and GOGAT levels decreased. The strongest repression of GOGAT activity was seen in glucose minimal medium with glutamate serving as the sole nitrogen source (Gglt), coinciding with the strongest activation of histidase expression and the slowest growth. The severe repression of GOGAT and strong activation of histidase in Gglt-grown cells were entirely dependent on an active nac gene, since the nac-28 mutant strain EB3365 failed to repress GOGAT and activate histidase expression (Table 2). Thus, the severe repression of E. coli GOGAT activity reported previously by others (11, 33) is mediated by the Ntr system acting through the NAC protein and does not appear to be a result specifically seen when glutamate serves as the sole nitrogen source.
TABLE 2.
Effect of ΔgltF1 on enzyme levels and growth rates in four E. coli backgrounds
| Origin | Strain | Genotype | Mediuma | Sp actb (nmol/min/mg)
|
Doubling time (min) | |||
|---|---|---|---|---|---|---|---|---|
| Histidase | GOGAT | GS | GDH | |||||
| YMC10 | YMC10 | WTc | GNgln0.1% | 165 | 125 | 65 | ||
| Ggln0.1% | 440 | 80 | 95 | |||||
| Ggln0.04% | 645 | 65 | 195 | |||||
| Garg0.2% | 640 | 30 | 215 | |||||
| Gglt0.2% | 855 | 15 | 720 | |||||
| EB4534 | ΔgltF1 | GNgln0.1% | 155 | 110 | 65 | |||
| Ggln0.1% | 425 | 80 | 95 | |||||
| Ggln0.04% | 655 | 70 | 180 | |||||
| Garg0.2% | 630 | 35 | 220 | |||||
| Gglt0.2% | 900 | 15 | 710 | |||||
| EB3365 | nac-28 | GNgln0.1% | 165 | 125 | 80 | |||
| Garg0.2% | 140 | 230 | 160 | |||||
| Gglt0.2% | 130 | 150 | 610 | |||||
| EG47 | EB566 | WT | GNgln0.1% | 130 | 105 | 70 | ||
| Gglt0.2% | 375 | 105 | 350 | |||||
| EB4539 | ΔgltF1 | GNgln0.1% | 120 | 100 | 70 | |||
| Gglt0.2% | 375 | 100 | 335 | |||||
| W3110 | EB4566 | WT | GNgln0.1% | 275d | 155 | 65 | ||
| Gglt0.2% | 5,460d | 10 | 1,150 | |||||
| EB4582 | ΔgltF1 | GNgln0.1% | 270d | 165 | 65 | |||
| Gglt0.2% | 5,260d | 10 | 1,130 | |||||
| MX614 | EB4713 | WT | Ggln0.1% | 345 | 175 | 1,590 | 205 | 100 |
| Gglt0.2% | 525 | 60 | 1,720 | 100 | 570 | |||
| EB4722 | ΔgltF1 | Ggln0.1% | 410 | 140 | 1,430 | 185 | 90 | |
| Gglt0.2% | 425 | 45 | 1,610 | 110 | 555 | |||
Cells were grown in minimal medium supplemented with glucose (G) at 0.4% (wt/vol) and nitrogen sources at the concentrations indicated. N, 30 mM ammonia (present as 15 mM ammonium sulfate); gln, glutamine; arg, arginine; glt, glutamate.
Specific activities of histidase, GOGAT, GS, and GDH were measured and reported as the average of two or more assays with a standard error of the mean of less than 15%.
WT, wild type.
β-Galactosidase activity from a hut-lacZ fusion is reported, since these strains do not carry the hut operons.
The gltF deletion was also introduced into two other E. coli strains. EB4566 is a λRZ-5ΦCB540 lysogen of W3110 in which the hutUp-lacZ operon fusion directs the synthesis of β-galactosidase, making it a readily assayed reporter for Ntr activity. EB566, like YMC10, carries the K. aerogenes hut genes. Southern blots again confirmed that the ΔgltF1 strains did in fact lack gltF (Fig. 2A, lanes 1, 2, 5, and 6). In both strain backgrounds, the presence or absence of the ΔgltF1 mutation had no effect on the growth rate or enzyme expression in response to growth in Gglt medium (Table 2), confirming that gltF plays no role in nitrogen regulation.
However, comparison of the three parental strains revealed their markedly different responses to nitrogen limitation. Although YMC10, EB566, and EB4566 had similar doubling times and levels of GOGAT and, where comparisons can be made, histidase when grown under conditions of nitrogen excess (GNgln0.1%), significant background-specific differences were seen in doubling times, derepression ratios for histidase (or β-galactosidase), and repression ratios for GOGAT in Gglt medium (Table 2). These background-specific differences appeared to reflect differences in Ntr activity and the severity of nitrogen limitation imposed by the use of glutamate as the sole nitrogen source. Although these differences were most dramatic when glutamate was used as the sole nitrogen source, significant differences were also observed under less severe conditions (data not shown). Such background-specific differences reinforce the notion that observations made in one E. coli strain may not be valid in other E. coli strains, especially when multiple mutagenic treatments have intervened.
Finally, since we were unable to detect a role for GltF in any of the three E. coli strain backgrounds tested, we turned to the strain background where the original report of a GltF effect was made. Strain MX614, an ilv met mutant, was transduced to ilv+ and met+ by P1-mediated transduction, and the hut operons from strain EB4693 were also brought into the strain by P1-mediated transduction, yielding strain EB4713. An isogenic ΔgltF::Ω derivative, EB4722, was constructed by P1-mediated transduction, and the nitrogen regulation of enzyme formation in the two strains was compared. The response of strain EB4722, with gltF deleted, to nitrogen limitation was identical to that of EB4713 (Table 2). When glutamine was used as the sole nitrogen source (moderate nitrogen limitation), both strains showed derepression of GS and histidase formation. When glutamate was used as the sole nitrogen source (severe nitrogen limitation), both strains showed comparable repression of GOGAT formation as well. Even the growth rates of the two strains were similar under conditions of nitrogen limitation. Thus, we conclude that gltF plays no significant role in the nitrogen regulation of E. coli or K. aerogenes.
GOGAT mutants have difficulty deriving glutamate from glutamine.
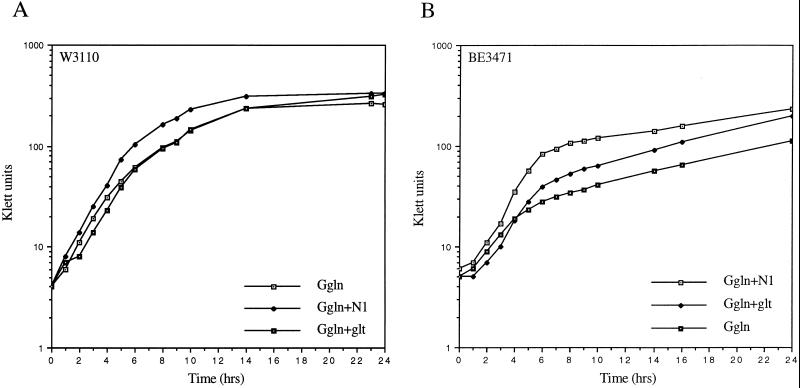
It has long been assumed that GOGAT is the principal route by which glutamine is degraded in both K. aerogenes (2) and E. coli (40). However few data have been presented to document this assumption. When wild-type E. coli strain W3110 was inoculated into glucose minimal medium with glutamine (0.1% wt/vol) as the sole nitrogen source (Ggln), it grew rapidly to high density, with a doubling time of 75 min (Fig. 4A). When the gltD mutant strain BE3471 (otherwise isogenic with W3110) was inoculated into Ggln, the growth was biphasic (Fig. 4B), with initial rapid growth (doubling time, about 85 min) followed by slow but still exponential growth (doubling time, about 658 min). Increasing the initial concentration of glutamine from 0.1 to 0.2% increased both the initial growth rate and the cell density at which the shift from fast to slow growth occurred. Conversely, lowering the glutamine from 0.1 to 0.04% decreased both the initial growth rate and the cell density at which the shift from fast to slow growth occurred (data not shown). The growth rate of the second, slow phase of growth was independent of the glutamine concentration. Such biphasic growth kinetics are reminiscent of diauxic growth, where more rapidly utilized nutrients are depleted from the growth medium before the utilization of growth rate-limiting nutrients. When a culture of strain BE3471 (gltD39) was supplemented with additional glucose or additional glutamine, the added glutamine extended the period of rapid growth whereas the glucose had no effect (data not shown). Thus, the growth rate limitation imposed on the GOGAT-deficient E. coli strain growing in Ggln was eased by the presence of additional glutamine.
FIG. 4.
Growth of E. coli strains W3110 (gltD+) (A) and BE3471 (gltD39) (B) in glucose minimal medium at 37°C with the nitrogen sources and supplements indicated. G, glucose at 0.4% (wt/vol); gln, glutamine at 0.1% (wt/vol); glt, glutamate at 0.2% (wt/vol); N1, 1 mM ammonia (added as 0.5 mM ammonium sulfate).
Glutamine undergoes spontaneous hydrolysis to ammonia and glutamate in aqueous solution, and this can affect the degree to which glutamine serves as a growth rate-limiting nitrogen source for wild-type cells (4). It is possible that the initial rapid growth of BE3471 growing in Ggln was due to the presence of ammonia or glutamate derived from the spontaneous hydrolysis of glutamine. To test this possibility, Ggln cultures of BE3471 were supplemented with sodium glutamate to 0.4% (24 mM). As expected of a glutamate-limited culture, the glutamate supplement improved the yield and growth rate of the initial rapid growth phase. The added glutamate also resulted in an increase in the growth rate of the later slow-growing phase (Fig. 4B), as well as the density obtained after 24 h of growth. The relatively small increase in growth rate is consistent with the observation that the transport of glutamate into E. coli is inefficient (31).
When Ggln cultures of BE3471 were supplemented with ammonium (1 mM), the yield and growth rate of the initial phase of growth were also improved (Fig. 4B). Although GOGAT-deficient strains of E. coli cannot assimilate ammonia through the GS/GOGAT pathway, they are able to use the GDH pathway to assimilate ammonia at concentrations above 100 μM (37). To be certain that GDH was present under these conditions, we assayed BE3471 cells grown in Ggln into the second (slow) phase of growth. It is known that the GDH of E. coli is not strongly repressed by the Ntr-dependent NAC mechanism, as is seen in K. aerogenes, but we were surprised to find that GDH levels were elevated about 10-fold above any levels seen previously in this strain (data not shown). The basis for this “hyperinduction” of GDH is unknown, but clearly there is sufficient GDH present in these cells to allow assimilation of ammonia into glutamate.
The biphasic growth, effects of glutamate and ammonia supplementation, and hyperinduction of GDH were also seen when EB4613, a gltD derivative of another wild-type strain, YMC10, was analyzed (Table 3). These effects were not unique to gltD. The gltB derivatives of both W3110 and YMC10 (strains EB4985 and EB5019, respectively) showed the same biphasic growth and supplementation patterns as the gltD derivatives (data not shown).
TABLE 3.
Effects of gltB and gltD mutations on enzyme formation and growth in E. coli and K. aerogenes
| Strain | Genotype | Mediuma | Sp act (nmol/min/mg)b
|
Doubling time (min) | |||
|---|---|---|---|---|---|---|---|
| Histidase | GOGAT | GDH | GS | ||||
| YMC10 | WTc | GNgln0.2% | 135 | 100 | 130 | 210 | 75 |
| Ggln0.2% | 250 | 120 | 165 | 765 | 85 | ||
| Ggln0.1% | 440 | 75 | 235 | 1,700 | 95 | ||
| Ggln0.04% | 645 | 65 | 270 | NDd | 175 | ||
| Gglt0.2% | 855 | 15 | 160 | ND | 680 | ||
| EB4613 | gltD39 | GNgln0.2% | 145 | 5 | 135 | 190 | 70 |
| Ggln0.2% | 130 | 5 | 425 | 665 | 100, 720e | ||
| Ggln0.1% | 165 | 0 | 1,140 | 1,260 | 140, 600e | ||
| Ggln0.04% | 230 | 8 | 1,620 | ND | 390, 1,110e | ||
| Gglt0.2% | 850 | 0 | 280 | ND | 480 | ||
| KC1043 | WT | GNgln0.2% | 35 | 170 | 370 | 260 | 55 |
| Ggln0.2% | 330 | 105 | 115 | 1,510* | 70 | ||
| Ggln0.1% | 310 | 80 | 80 | ND | 75 | ||
| Ggln0.04% | 475* | 100* | 65* | ND | 70* | ||
| Gglt0.2% | 605 | 10 | 10 | ND | 325 | ||
| KC2561 | gltB200 | GNgln0.2% | 75 | 5 | 435 | 170 | 50 |
| Ggln0.2% | 360 | 0 | 60 | 1,360 | 215 | ||
| Ggln0.1% | 335 | 0 | 45 | 1,280 | 245 | ||
| Ggln0.04% | 340 | 0 | ND | ND | 260 | ||
| Gglt0.2% | 565 | 0 | 25 | ND | 290 | ||
| KC3686 | WT | GNgln0.2% | 70 | 125 | ND | ND | 55 |
| Ggln0.04% | 355 | 70 | ND | ND | 75 | ||
| KC4540 | gltD702 | GNgln0.2% | 60 | 10 | ND | ND | 60 |
| Ggln0.04% | 305 | 0 | ND | ND | 300 | ||
Cells were grown and enzyme activities were measured as described in Table 2, note a.
Specific activities were measured and reported as the average of two or more assays with a standard error of the mean of less than 15% except where indicated by an asterisk.
WT, wild type.
ND, not determined.
Growth was biphasic (Fig. 4). The first value is for the earlier phase, and the second value is for the later phase.
The hut operons from K. aerogenes are present in strain YMC10 and its gltD derivative, EB4613 (1). This allowed us to use histidase as an indicator of the activity of the Ntr system (Table 3). Growth of YMC10 on Ggln resulted in a partial derepression of histidase formation. When the glutamine concentration was reduced to 0.04%, the derepression was stronger. When glutamate was used as the sole nitrogen source (Gglt medium), the derepression was stronger still. In other words, the strength of the Ntr signal and the degree of nitrogen limitation (as indicated by growth rate) were strongly correlated. Although the derepression of histidase in strain EB4613 was essentially the same as in YMC10 under conditions of extreme nitrogen limitation (Gglt medium), the derepression of histidase in EB4613 was much less when Ggln medium was used, even when the initial glutamine concentration was reduced to 0.04%. Thus, the defect in Ntr derepression associated with GOGAT mutants of E. coli growing in Ggln appears to be the result of a physiological imbalance affecting glutamine metabolism rather than a direct genetic effect.
Growth of K. aerogenes GOGAT mutants.
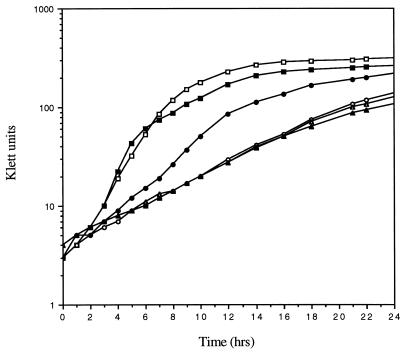
When wild-type K. aerogenes strain KC1043 was grown in Ggln medium, it grew rapidly (73-min doubling time) to high density (Fig. 5). When KC2561, a gltB derivative of KC1043, was grown in Ggln, growth was much slower (215-min doubling time) but monophasic (Fig. 5). As was true for the E. coli GOGAT-deficient strains, growth of the K. aerogenes gltB mutant KC2561 was faster when the Ggln medium was supplemented with either glutamate (not shown) or 1 mM ammonia (Fig. 5). The effectiveness of 1 mM ammonia was surprising, since GDH is strongly repressed in K. aerogenes by the Ntr system acting via NAC (Table 3). However, it is clear that GDH is required for the ammonia-mediated effect. When the GOGAT-deficient strain KC2561 was compared with strain KC3182, which lacks both GOGAT and GDH, ammonia did not increase the growth rate above that seen in Ggln medium (Fig. 5). A strain lacking GDH only (GOGAT was present) grew like the wild type in all the media tested here (data not shown). Thus, for K. aerogenes as for E. coli, growth of a GOGAT-deficient mutant on glutamine as the sole nitrogen source results in a growth rate limitation for glutamate. This argument is supported by an earlier observation (2) that the growth rate of a GOGAT-deficient mutant on Ggln medium is much lower (3- to 4-h doubling time) than that of a GOGAT-deficient mutant that also lacks GS, a glutamate-consuming enzyme (98-min doubling time).
FIG. 5.
Growth of K. aerogenes strains. Squares, strain KC1043 (wild type); circles, KC2561 (gltB200); triangles, KC3182 (gltB200 gdhA2). Growth was in minimal medium with 0.4% (wt/vol) glucose as the carbon source and 0.1% glutamine as a nitrogen source. Open symbols, no other additions; solid symbols, 1 mM ammonia was also present (as 0.5 mM ammonium sulfate).
The failure of GOGAT-deficient strains of K. aerogenes to grow with poor nitrogen sources such as histidine (Asm− phenotype) does not appear to be the result of a defect in the action of the Ntr system, either direct or indirect. The data in Table 3 show that whether Ntr action is measured by activation of histidase expression or by the more direct activation of GS expression, there is only a slight reduction in the ability of K. aerogenes GOGAT mutants grown in Ggln medium to activate the Ntr system.
DISCUSSION
Inadequacy of the GltF hypothesis.
Since the report of the original K. pneumoniae asm-1 mutation in 1971 (36), the cause of the inability of enteric bacteria lacking GOGAT activity (Gogat−) to assimilate the nitrogen from several alternative nitrogen sources (Asm− phenotype) has been open to speculation. One aspect of the problem was clear from the outset: Gogat− mutants are unable to assimilate ammonium when it is present in the medium (or generated by catabolism) at levels too low to be used by GDH, whose Km for ammonium is quite high. Thus, the growth of Gogat− mutants on low ammonia is limited by glutamate availability. But there is also one report that Gogat− mutants were unable to derepress the Ntr system and hence the Ntr-dependent operons that code for the catabolism of a variety of alternative nitrogen sources, such as histidine (9). It has been argued that GOGAT activity is required to reduce the size of the intracellular glutamine pool so that Ntr derepression can occur (2). Although a model based on glutamine pools superficially explains the Asm− and Ntr− phenotypes of Gogat− strains, this model leaves us with a paradox. GOGAT-deficient cells must have a glutamine pool that is too high to allow activation of the Ntr system but too low to allow growth. This difficulty is compounded by the observation that Gogat− mutants of K. aerogenes grow with glutamine as the sole exogenously supplied nitrogen source but apparently cannot grow on the endogenous pool of glutamine which accumulates in the presence of alternative nitrogen sources.
The discovery of a third gene in the E. coli gltBD operon, gltF, led to a simpler explanation for the inability of certain Gogat− mutants to utilize alternative nitrogen sources and derepress their Ntr systems when grown under conditions of nitrogen limitation (10, 11). According to this GltF model, mutations which interfered with the normal level of gltF expression (most likely from the promoter for the gltBD operon) prevent the derepression of the Ntr system. In particular, an insertion within the chromosomal gltF gene resulted in an Ntr− Gogat+ phenotype. Moreover, polar insertions within the gltB gene, resulting in a Gogat− Ntr− phenotype, could be complemented to a Gogat− Ntr+ phenotype if the gltF (and possibly gltD) gene were expressed at high levels. Thus, according to the GltF model, the product of the gltF gene is an essential part of the Ntr system and Ntr derepression can be independent of the Gogat phenotype, depending upon the level of gltF (and possibly gltD) expression. This GltF model had to be modified to account for the fact that growth of a gltF mutant on Gglt allowed derepression of the Ntr system whereas growth on Ggln did not. It was thus necessary to postulate that Ntr derepression occurs by a separate mechanism when glutamate is the sole nitrogen source.
As attractive as the GltF model is, the data presented here argue strongly against it. First, the GltF model fails to explain the phenotype of nonpolar mutations, like gltB200, that lead to the Ntr− phenotype. Second, it fails to explain the phenotype of gltBD mutations, polar or not, in organisms like K. aerogenes that lack a gltF gene. Third, the GltF model is inconsistent with the observation that E. coli strains lacking the entire gltF coding region were indistinguishable from the wild type with respect to Ntr regulation, growth on glucose-arginine, and GOGAT expression. Fourth, it fails to explain the Ntr+ (constitutive) phenotype of a gltB200 glnA51 (Gogat− GS−) double mutant (13). Finally, this model cannot explain how Gogat+ (but gltF mutant) cells can convert from an Ntr− to an Ntr+ phenotype when glutamate is provided as the sole nitrogen source.
Independent support for the GltF model was provided by Kuczius et al. (27), who recovered a gltF homologue from a K. pneumoniae gene library. The presence of such a K. pneumoniae gltF homologue is contradicted by our data and those of others (32). No gltF homologue was detected in K. pneumoniae strain M5aL under hybridization conditions even more permissive than those used by Kuczius et al. (27). Thus, the origin of the DNA fragment displaying homology to gltF reported by Kuczius et al. (27) is unclear. Despite the unclear origin of the DNA fragment, they found that this fragment could complement the Ntr phenotype, but not the Gogat− phenotype, of an E. coli insertion mutant. However, the complementation was tested with cells grown on 0.2% yeast extract as the nitrogen source. Under these conditions, even the wild type would fail to derepress its Ntr system. Thus, the complementation data are puzzling. The ability of the fragment to complement an E. coli Gogat− mutant for growth on arginine as the sole nitrogen source was also puzzling, since GS was not derepressed under these conditions. Derepression of the glnA ntrB ntrC operon is one of the earliest features of Ntr derepression. It is difficult to account for derepression of the systems necessary for growth on minimal medium supplemented with glucose and arginine without derepression of GS formation (40). Finally the growth of the purported Gogat− Ntr+ strain (resulting from complementation) on glycine as the sole nitrogen source is inconsistent with the fact that glycine is catabolized to ammonium, which is not readily assimilated to glutamate in a GOGAT-deficient enteric bacterium (40). Moreover, in a later report (18), this same group was unable to detect a phenotype for a gltF insertion mutant.
The glutamate starvation hypothesis.
If the GltF model must be discarded, that leaves the glutamine imbalance model with its glutamine paradox to explain the Ntr− phenotype of Gogat− mutants. Our data provide a clarification of this apparent paradox by suggesting that Gogat− mutants fail to grow on alternative nitrogen sources because they are starved for glutamate. In other words, the inability to derepress the Ntr system of E. coli is not a cause of the phenotype but rather an effect of it. If growth is limited by the supply of glutamate, then any supply of glutamine will, by definition, be excess. Thus, Gogat− mutants fail to grow on Ntr-dependent nitrogen sources for the same reason they fail to grow on low concentrations of ammonia: they are glutamate starved.
The hypothesis of glutamate starvation also explains the fact that the E. coli gltD mutant grew very slowly (with doubling times of greater than 10 h) with glutamine as the sole nitrogen source. Several glutaminases have been identified in E. coli, so one might expect that E. coli gltD mutants would use them to derive glutamate from glutamine. It is possible that these glutaminases lack the affinity or capacity to derive sufficient glutamate from the glutamine that E. coli can transport. It is also possible that the intracellular hydrolysis of glutamine to equimolar amounts of glutamate and ammonia by glutaminases is quickly reversed by the ATP-dependent synthesis of glutamine by GS, again depriving the cell of glutamate. If the ammonia concentration rose to levels comparable to the Km of GDH (about 1 mM), then the efficient GDH could yield a net synthesis of glutamate that could balance the glutamine synthesis in a way that the weaker glutaminases could not.
K. aerogenes gltB and gltD mutants grew much faster with glutamine as the sole nitrogen source (4-h doubling time) than their E. coli counterparts. This might be explained if K. aerogenes has stronger glutaminase activity than E. coli. The catabolic asparaginase (41) has a Km for glutamine of about 1 mM, more than sufficient to allow it to function in Ggln0.1%, where glutamine is present at about 7 mM. Apparently, this asparaginase can supply glutamate at a rate sufficient to support a moderate growth rate. Another feature of this enzyme that may be important is its location in the periplasm rather than the cytoplasm. The hydrolysis of glutamine in the periplasm would leave the glutamate available for active transport via the glutamate transport system, which is very weakly expressed (31), but would leave the ammonia available only for passive diffusion through the ammonia facilitator. Thus, glutamate might not be stoichiometrically reconverted to glutamine if some ammonia is lost to the medium. In fact, a K. aerogenes gltB mutant growing on glutamine as the sole nitrogen source accumulates ammonia in the culture medium at levels below 1 mM (9).
One final note about the effectiveness of the catabolic asparaginase in the hydrolysis of glutamine should be mentioned here. Although the Km for glutamine is more than adequate to carry out this function, the Vmax of the glutaminase activity of this enzyme is low. Thus, although the catabolic asparaginase may allow K. aerogenes gltB mutants to use glutamine better than their E. coli counterparts (which lack the catabolic asparaginase), even the K. aerogenes mutants are growth rate limited by the amount of glutamate that they can produce (via catabolic asparaginase) and maintain (in the face of depletion by GS). The argument that GS traps the glutamate generated from glutaminase is supported by the fact that the K. aerogenes glnA gltB double mutants grow much faster with glutamine as the sole nitrogen source than do gltB single mutants.
Perhaps the simplest way to understand these effects is to consider the opposing effects of the two key enzymes of nitrogen assimilation, GS and GOGAT. GS forms glutamine at the expense of glutamate, and GOGAT forms glutamate at the expense of glutamine. In a wild-type strain growing on Gglt, GOGAT depletes glutamine from an already-limited supply and converts it back to glutamate, further exacerbating the glutamine limitation. However, in a Gogat− strain which lacks the cell's primary glutaminase activity, the limited glutamine supply is spared for use in biosynthesis. Thus, under these conditions, the Gogat− strain actually grows more rapidly than the wild type (Table 3). During growth on Ggln, the exogenous glutamine supply shifts the growth limitation from glutamine to glutamate in Gogat− strains only, as Gogat+ strains readily convert glutamine to glutamate. GS depletes this much-needed glutamate from an already-limited supply. Thus, K. aerogenes glnA gltB mutants grow faster than glnA+ gltB mutants on Ggln, though still more slowly than a glnA+ gltBD+ wild type (2).
Ninfa and colleagues have reported a role for α-ketoglutarate in the control of the Ntr system in E. coli. According to their findings (26), when α-ketoglutarate levels are extremely low, the PII protein does not signal inactivation of the Ntr system, whatever the glutamine levels in the cell. However, these levels are far below anything expected under physiological conditions. At moderate levels of α-ketoglutarate, up to 100 μM, the ability of glutamine to signal uridylylation or deuridylylation of PII is the dominant signal, and the Ntr system responds to the glutamine signal alone. If α-ketoglutarate levels rise high enough to saturate PII, then there would be a reduced affinity of unmodified PII for the phosphatase activity of NtrB, leading to a higher level of phosphorylated NtrC than might be predicted on the basis of glutamine pools alone. One would expect that the absence of GOGAT would lead to an accumulation of α-ketoglutarate (as well as glutamine) and thus that the Ntr system might appear more active than in a wild-type strain. Since the effort here has been to explain why the Ntr system sometimes seems to be less active than expected in GOGAT mutants, it appears that the effect of α-ketoglutarate on the interactions of PII with NtrB cannot explain the regulatory phenotype of the gltBD mutants.
In summary, the data presented here resolve the paradox provided by the first Ntr mutations (gltB). Their failure to derepress Ntr systems under some conditions that allow derepression of Ntr in the wild type is a simple reflection of their specific starvation for glutamate rather than a general starvation for nitrogen. These data also provide another confirmation that the enteric bacteria treat their two sources of organic nitrogen (glutamate and glutamine) differently. Glutamine is used to signal the nitrogen status of the cell, and glutamate is ignored in this role. Glutamate is used as a counterion to maintain cellular potassium levels for osmotic balance and not for nitrogen signaling.
Open questions about glutamate synthesis.
The data presented here deal directly with effects of aberrant regulation of glutamate metabolism, but they also raise a number of questions about the normal regulation of glutamate formation. It is clear that gltBD expression requires activation by the leucine-responsive regulatory protein Lrp (15). The data presented here show clearly that NAC is capable of severely repressing gltBD expression (Table 2). We and others have noted a strong repression of both gltBD and gdhA expression in cells grown in broth (8). For gltBD, this repression can be explained by the lack of Lrp under such conditions (15), but for gdhA (which codes for GDH), Lrp is not necessary (unpublished observations). The severe repression of gdhA expression by broth is independent of the severe NAC-mediated repression of gdhA, but the nature of the effector responsible for broth repression of gdhA is unknown. The data in Table 3 contain one further unexpected finding: when E. coli GOGAT mutants were starved for glutamate (by growing them with glutamine as the sole nitrogen source), GDH levels increased dramatically to as much as 10-fold that seen in cells grown in high ammonia. The mechanism of this increase is unknown, but the increase has important consequences for cells limited for glutamate. GOGAT mutants are dependent on GDH for the assimilation of ammonia into glutamate. At concentrations of ammonium lower than about 1 mM, K. aerogenes NAC causes severe repression of GDH, and the poor Km of GDH for ammonia means that the residual GDH is ineffective. In E. coli and S. enterica serovar Typhimurium, GDH is repressed either only slightly or not at all by NAC, allowing GOGAT mutants to grow with ammonia concentrations as low as 100 μM. It has long been known that mutations (or high-copy-number clones) that increase the production of GDH allow growth at even lower concentrations of ammonia (5). However, except for one class of mutation that creates a new, stronger promoter for gdhA in K. aerogenes (25), the nature of these GDH overexpression mutations is unknown. Given the central role of glutamate in biosynthesis and osmoregulation, it is not surprising that its synthesis is subject to multiple forms of regulation. What is surprising is how little is known about that regulation.
A reinterpretation of the data that led to the GltF hypothesis.
Finally, the data presented here suggest an explanation for some of the irreproducible results that have been reported regarding regulatory effects that are or are not seen in cells grown with 0.2% (and perhaps 0.1%) glutamine as the sole nitrogen source. The effect is clearest in the E. coli gltD mutant growing with glutamine as the sole nitrogen source. When the cell density is sufficiently low, the spontaneous decomposition of the glutamine in the medium provides enough glutamate (and ammonia) to allow rapid growth. When the cell density reaches a critical value, dependent on the concentration of glutamine, the supply of glutamate and ammonia becomes insufficient and the cells are truly dependent on glutamine alone as the nitrogen source. If we extrapolate that finding to wild-type cells, we can assume that, at low cell densities, cells are growing on a mixture of glutamine and small amounts of glutamate and ammonia; at higher densities, the nitrogen source is glutamine alone. Thus, early in the growth of the culture, Ntr-activated enzymes (histidase) are not formed and Ntr-repressed enzymes (GDH) are formed and accumulate. At a cell density of about 108 cells/ml, Ntr-activated enzymes (histidase) are formed rapidly and accumulate while Ntr-repressed enzymes (GDH) stop accumulating and begin to be diluted. Thus, if cells are harvested 1 or 2 generations after the transition occurs, they will still have substantial quantities of GDH and will have already accumulated substantial quantities of histidase. Moreover, the exact point at which the transition occurs will depend on the size of the inoculum, the growth rate of the cells, and the concentration and quality of the glutamine in the medium. In short, small changes in the growth conditions can have the result that 0.2% glutamine is no longer barely derepressing and may make 0.2% glutamine either a strongly derepressing or nonderepressing medium. This may explain why effects that actually result from slight variations in growth rates and conditions have been attributed to gltF.
ACKNOWLEDGMENT
This work was supported by Public Health Service grant GM47156 from the National Institutes of Health to R.A.B.
REFERENCES
- 1.Backman K, Chen Y-M, Magasanik B. Physical and genetic characterization of the glnA-glnG region of the E. coli chromosome. Proc Natl Acad Sci USA. 1981;78:3742–3747. doi: 10.1073/pnas.78.6.3743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bender R A. Ph.D. thesis. Cambridge, Mass: Massachusetts Institute of Technology; 1976. [Google Scholar]
- 3.Bender R A, Friedrich B. Regulation of assimilatory nitrate reductase formation in Klebsiella aerogenes W70. J Bacteriol. 1990;172:7256–7259. doi: 10.1128/jb.172.12.7256-7259.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bender R A, Janssen K A, Resnick A D, Blumenberg M, Foor F, Magasanik B. Biochemical parameters of glutamine synthetase from Klebsiella aerogenes. J Bacteriol. 1977;129:1001–1009. doi: 10.1128/jb.129.2.1001-1009.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bender R A, Macaluso A, Magasanik B. Glutamate dehydrogenase: genetic mapping and isolation of regulatory mutants of Klebsiella aerogenes. J Bacteriol. 1976;127:141–148. doi: 10.1128/jb.127.1.141-148.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bender R A, Magasanik B. Regulatory mutations in the Klebsiella aerogenes structural gene for glutamine synthetase. J Bacteriol. 1977;132:100–105. doi: 10.1128/jb.132.1.100-105.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Blattner F R, Plunkett G, Bloch C A, Perna N T, Burland V, Riley M, Collado-Vides J, Glasner J D, Rode C K, Mayhew G F, Gregor J, Davis N W, Kirkpatrick H A, Goeden M A, Rose D J, Mau B, Shao Y. The complete genome sequence of Escherichia coli K-12. Science. 1997;277:1452–1474. doi: 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
- 8.Brenchley J E, Baker C A, Patil L G. Regulation of the ammonia assimilatory enzymes in Salmonella typhimurium. J Bacteriol. 1975;124:182–189. doi: 10.1128/jb.124.1.182-189.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brenchley J E, Prival M J, Magasanik B. Regulation of the synthesis of enzymes responsible for glutamate formation in Klebsiella aerogenes. J Biol Chem. 1973;248:6122–6128. [PubMed] [Google Scholar]
- 10.Castano I, Bastarrachea F, Covarrubias A A. gltBDF operon of Escherichia coli. J Bacteriol. 1988;170:821–827. doi: 10.1128/jb.170.2.821-827.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Castano I, Flores N, Valle F, Covarrubias A A, Bolivar F. gltF, a member of the gltBDF operon of Escherichia coli, is involved in nitrogen-regulated gene expression. Mol Microbiol. 1992;6:2733–2741. doi: 10.1111/j.1365-2958.1992.tb01450.x. [DOI] [PubMed] [Google Scholar]
- 12.Cavicchioli R, Kolesnikow T, Chiang R C, Gunsalus R P. Characterization of the aegA locus of Escherichia coli: control of gene expression in response to anaerobiosis and nitrate. J Bacteriol. 1996;178:6968–6974. doi: 10.1128/jb.178.23.6968-6974.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.DeLeo A B, Magasanik B. Identification of the structural gene for glutamine synthetase in Klebsiella aerogenes. J Bacteriol. 1975;121:313–319. doi: 10.1128/jb.121.1.313-319.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.de Vries G E, Raymond C K, Ludwig R A. Extension of bacteriophage λ host range: selection, cloning, and characterization of a constitutive λ receptor gene. Proc Natl Acad Sci USA. 1984;81:6080–6084. doi: 10.1073/pnas.81.19.6080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ernsting B R, Denninger J W, Blumenthal R M, Matthews R G. Regulation of the gltBDF operon of Escherichia coli: how is a leucine-insensitive operon regulated by the leucine-responsive regulatory protein? J Bacteriol. 1993;175:7160–7169. doi: 10.1128/jb.175.22.7160-7169.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gaillardin C, Magasanik B. Involvement of the product of the glnF gene in the autogenous regulation of glutamine synthetase formation in Klebsiella aerogenes. J Bacteriol. 1978;133:1329–1338. doi: 10.1128/jb.133.3.1329-1338.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Goldberg R B, Bender R A, Streicher S L. Direct selection for P1-sensitive mutants of enteric bacteria. J Bacteriol. 1974;118:810–814. doi: 10.1128/jb.118.3.810-814.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Grassl G, Bufe B, Muller B, Rosel M, Kleiner D. Characterization of the gltF gene product of Escherichia coli. FEMS Microbiol Lett. 1999;179:79–84. doi: 10.1111/j.1574-6968.1999.tb08711.x. [DOI] [PubMed] [Google Scholar]
- 19.Groisman E A, Casadaban M J. Mini-Mu bacteriophage with plasmid replicons for in vivo cloning and lac gene fusing. J Bacteriol. 1986;168:357–364. doi: 10.1128/jb.168.1.357-364.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Grove C L, Gunsalus R P. Regulation of the aroH operon of Escherichia coli K-12 by the tryptophan repressor. J Bacteriol. 1987;169:2158–2164. doi: 10.1128/jb.169.5.2158-2164.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Helling R B. Why does Escherichia coli have two primary pathways for synthesis of glutamate? J Bacteriol. 1994;176:4664–4668. doi: 10.1128/jb.176.15.4664-4668.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Horton R M, Pease L R. Recombination and mutagenesis of DNA sequences using PCR. In: McPherson M J, editor. Directed mutagenesis: a practical approach. Oxford, England: IRL Press; 1991. pp. 217–247. [Google Scholar]
- 23.Ikeda T P, Shauger A E, Kustu S. Salmonella typhimurium apparently perceives external nitrogen limitation as internal glutamine limitation. J Mol Biol. 1996;259:589–607. doi: 10.1006/jmbi.1996.0342. [DOI] [PubMed] [Google Scholar]
- 24.Janes B K, Bender R A. Alanine catabolism in Klebsiella aerogenes: molecular characterization of the dadAB operon and its regulation by the nitrogen assimilation control protein. J Bacteriol. 1998;180:563–570. doi: 10.1128/jb.180.3.563-570.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Janes B K, Pomposiello P J, Perez-Matos A, Najarian D J, Goss T J, Bender R A. Growth inhibition caused by overexpression of the structural gene for glutamate dehydrogenase (gdhA) from Klebsiella aerogenes. J Bacteriol. 2001;183:2709–2714. doi: 10.1128/JB.183.8.2709-2714.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jiang P, Peliska J A, Ninfa A J. Reconstitution of the signal-transduction bicyclic cascade responsible for the regulation of Ntr gene transcription in Escherichia coli. Biochemistry. 1998;37:12795–12801. doi: 10.1021/bi9802420. [DOI] [PubMed] [Google Scholar]
- 27.Kuczius T, Eitinger T, D'Ari R, Castorph H, Kleiner D. The gltF gene of Klebsiella pneumoniae: cloning and initial characterization. Mol Gen Genet. 1991;229:479–482. doi: 10.1007/BF00267472. [DOI] [PubMed] [Google Scholar]
- 28.Macaluso A, Best E A, Bender R A. Role of the nac gene product in the nitrogen regulation of some NTR-regulated operons of Klebsiella aerogenes. J Bacteriol. 1990;172:7249–7255. doi: 10.1128/jb.172.12.7249-7255.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Magasanik B. Regulation of nitrogen utilization. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C.: American Society for Microbiology; 1996. pp. 1344–1356. [Google Scholar]
- 30.Maniatis T, Fritsch E F, Sambrook J. Molecular cloning: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1982. [Google Scholar]
- 31.Marcus M, Halpern Y S. Genetic analysis of the glutamate permease in Escherichia coli K-12. J Bacteriol. 1969;97:1118–1128. doi: 10.1128/jb.97.3.1118-1128.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McClelland M, Florea L, Sanderson K, Clifton S W, Parkhill J, Churcher C, Cougan G, Wilson R K, Miller W. Comparison of the Escherichia coli K-12 genome with sampled genomes of a Klebsiella pneumoniae and three Salmonella enterica serovars, Typhimurium, Typhi, and Paratyphi. Nucleic Acids Res. 2000;28:4974–4986. doi: 10.1093/nar/28.24.4974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Meers J L, Tempest D W, Brown C M. Glutamine (amide):2-oxoglutarate amino transferase oxido-reductase (NADP), an enzyme involved in the synthesis of glutamate by some bacteria. J Gen Microbiol. 1970;64:187–194. doi: 10.1099/00221287-64-2-187. [DOI] [PubMed] [Google Scholar]
- 34.Metcalf W W, Steed P M, Wanner B L. Identification of phosphate starvation-inducible genes in Escherichia coli K-12 by DNA sequence analysis of psi::lacZ (Mu d1) transcriptional fusions. J Bacteriol. 1990;172:3191–3200. doi: 10.1128/jb.172.6.3191-3200.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Muse W B, Bender R A. The nac (nitrogen assimilation control) gene from Escherichia coli. J Bacteriol. 1998;180:1166–1173. doi: 10.1128/jb.180.5.1166-1173.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nagatani H, Shimizu M, Valentine R C. The mechanism of ammonia assimilation in nitrogen fixing bacteria. Arch Mikrobiol. 1971;79:164–175. doi: 10.1007/BF00424923. [DOI] [PubMed] [Google Scholar]
- 37.Pahel G, Zelenetz A D, Tyler B M. gltB gene and regulation of nitrogen metabolism by glutamine synthetase in Escherichia coli. J Bacteriol. 1978;133:139–148. doi: 10.1128/jb.133.1.139-148.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Prentki P, Krisch H M. In vitro insertional mutagenesis with a stable DNA fragment. Gene. 1984;29:303–313. doi: 10.1016/0378-1119(84)90059-3. [DOI] [PubMed] [Google Scholar]
- 39.Prival M J, Magasanik B. Resistance to catabolite repression of histidase and proline oxidase during nitrogen-limited growth of Klebsiella aerogenes. J Biol Chem. 1971;246:6288–6296. [PubMed] [Google Scholar]
- 40.Reitzer L J. Ammonia assimilation and the biosynthesis of glutamine, glutamate, aspartate, asparagine, l-alanine, and d-alanine. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C.: American Society for Microbiology; 1996. pp. 391–407. [Google Scholar]
- 41.Resnick A D, Magasanik B. l-Asparaginase of Klebsiella aerogenes. J Biol Chem. 1976;251:2722–2728. [PubMed] [Google Scholar]
- 42.Yan D, Ikeda T P, Shauger A E, Kustu S. Glutamate is required to maintain the steady-state potassium pool in Salmonella typhimurium. Proc Natl Acad Sci USA. 1996;93:6527–6531. doi: 10.1073/pnas.93.13.6527. [DOI] [PMC free article] [PubMed] [Google Scholar]