Abstract
An early step in the establishment of Salmonella enterica serovar Typhimurium murine infection is the penetration of the intestinal mucosa of the small intestine. The majority of the genes responsible for the Salmonella invasive phenotype are encoded on Salmonella pathogenicity island 1, and their transcription is controlled by the hilA transcriptional activator. The expression of hilA is regulated by environmental signals including oxygen, osmolarity, pH, and growth phase such that the presence of any one suboptimal condition results in repression of hilA expression and the invasive phenotype. We have conducted a search for negative regulators of hilA by introduction of a Salmonella enterica serovar Typhimurium chromosomal DNA gene bank into a Salmonella enterica serovar Typhimurium hilA::Tn5lacZY reporter strain. This screen has identified the hha gene as a regulator that exerts a negative influence on hilA expression. Plasmid-encoded hha significantly reduces hilA::Tn5lacZY chromosomal expression, as well as expression of the invasion genes invF, prgH, and sipC. An hha null mutation results in substantial derepression of both chromosomally encoded and plasmid-encoded hilA::Tn5lacZY expression. Introduction of plasmid-encoded hha into strain SL1344 results in attenuation of invasion using in vitro and in vivo assays. Importantly, purified Hha protein was found to bind to a hilA DNA promoter fragment, suggesting that the regulatory activity of the Hha protein occurs at the hilA promoter. These data add detail to the developing model of the regulation of Salmonella invasion genes.
Pathogenic Salmonella species cause infections in humans ranging from self-limiting gastroenteritis to lethal systemic disease. After ingestion of Salmonella in contaminated food or water, the bacteria access the small intestine and invade the specialized M cells of the follicle-associated epithelium of Peyer's patches (6, 30, 47) and the absorptive enterocytes (53). Subsequently, host-adapted species move to the regional lymph nodes before spreading to the liver and spleen, where unchecked growth can result in death due to enteric fever (29, 36). Organisms that are unable to grow within the lymphatic system of the host remain localized to the intestinal epithelium, where they induce substantial inflammation that contributes to the pathology of localized gastroenteritis.
A critical step in initiation of salmonellosis is the ability to invade the intestinal cells of the host. The entry process occurs by rearrangement of the cellular membrane in the form of actin ruffles that engulf the bacteria (15). Many genetic elements responsible for the invasive phenotype of Salmonella enterica serovar Typhimurium localize to a 40-kb region of the chromosome at centisome 63, termed Salmonella pathogenicity island 1 (SPI-1) (reviewed in reference 8). Many of the SPI-1 genes encode structural components of a secretion system and are homologous to type III secretion systems found in both plant and animal pathogens including Pseudomonas, Rhizobium, Erwinia, Yersinia, Shigella, and enteropathogenic Escherichia coli (19). These systems function by translocating virulence proteins into eukaryotic cells (23). Loci within and outside SPI-1 encode proteins that are secreted through the type III secretion apparatus of SPI-1. These secreted proteins induce cellular changes in tissue culture cells and are responsible for the ability of Salmonella to invade tissue culture cells (14, 21, 22, 32, 56).
The induction of Salmonella invasion genes is tightly regulated by environmental signals that are believed to be important in the expression of the invasive phenotype in the host environment. The conditions that have been shown to repress Salmonella invasion include high oxygen, low osmolarity, low pH, and stationary-phase growth (11, 16, 34, 51). The hilA gene, located in SPI-1, encodes a transcriptional activator that modulates expression of the type III secretion apparatus proteins and the secreted effector proteins (2). Importantly, expression of hilA is modulated by the same conditions that regulate the invasive phenotype. In addition, overexpression of hilA confers a hyperinvasive phenotype and overexpression also counteracts the effects of repressing signals. Therefore, modulation of hilA expression by environmental signals appears to be a primary method of regulating the invasive phenotype of Salmonella (2, 3, 35). Results from our laboratory and from the works of others (2, 47) reveal that null mutations in hilA cause a dramatic attenuation of invasion of tissue culture cells and M cells of the Peyer's patches, in addition to attenuating virulence in mice following oral inoculation. These results establish that hilA plays a crucial role in expression of the Salmonella invasive phenotype.
Many genetic elements that exert regulatory effects on hilA have been identified including positive elements hilC/sirC/sprA (10, 48, 50), hilD (50), sirA (26), fis (55), barA, csrAB (1), and phoB, fadD, and fliZ (39) as well as negative elements such as phoPQ (5, 46). Recent work by Schechter et al. (50) provided evidence for the existence of factors that repress hilA transcription in response to environmental cues, although the putative factors were not identified. The results from a transposon mutagenesis screening conducted in our laboratory identified the hupB and ams genes, as well as two previously uncharacterized open reading frames, as negative modulators of hilA expression (12). As an alternative to the transposon mutagenesis approach, we have conducted a search for additional factors that negatively influence hilA transcription using a gene bank of Salmonella enterica serovar Typhimurium genomic DNA. We now report the identification and characterization of the role of the histone-like protein, Hha, in hilA transcription and expression of the Salmonella invasive phenotype.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The bacterial strains and plasmids used in this study are listed in Table 1. Bacterial cultures were grown in Luria-Bertani (LB) high-osmolarity medium (1% tryptone, 1% NaCl, 0.5% yeast extract) or LB low-osmolarity medium (1% tryptone, 0.5% yeast extract). Antibiotics were added to the following final concentrations (in micrograms per milliliter) where appropriate: ampicillin, 100; chloramphenicol, 25; kanamycin, 50; and tetracycline, 20. Bacterial cultures for invasion assays were grown under oxygen-limiting conditions by inoculating 3 ml of LB broth with 10 μl of a stationary-phase culture and incubating statically overnight at 37°C to a density of ≈4 × 108 to 5 × 108 CFU ml−1. High-oxygen conditions were obtained by inoculating 3 ml of LB broth with 10 μl of stationary-phase culture and shaking vigorously at 37°C to a density of ≈1.5 × 108 CFU ml−1 (31, 47).
TABLE 1.
Strains and plasmids used in the study
| Strain or plasmid | Description | Source or reference(s) |
|---|---|---|
| E. coli | ||
| DH12S | mcrA Δ(mrr-hsd RMS-mcrBC) F′ lacIqlacZΔM15 | Gibco BRL |
| YK4122H | 5K hha3::Tn5 Knr | 18, 44 |
| GS162 | K-12 lac laboratory strain | G. Stauffer |
| BJ1575 | GS162 hha3::Tn5 Knr | This work |
| BW21355 | K-12 F− ΔlacX74 | B. Wanner |
| BJ1925 | BW21355 hha3::Tn5 Knr | This work |
| S. enterica serovar Typhimurium | ||
| SL1344 | Wild-type virulent strain | 57 |
| BJ68 | SL1344 sipC::Tn5lacZY Tcr | 47 |
| BJ70 | SL1344 hilA::Tn5lacZY Tcr | 47 |
| BJ72 | SL1344 invF::Tn5lacZY Tcr | 47 |
| BJ644 | SL1344 phoP::Tn10 Tcr | 49 |
| BJ661 | SL1344 with pho-24 by P22 transduction from BJ2272 Cmr | This work |
| BJ690 | SL1344 ΔphoP hilA::Tn5lacZY Tcr | This work |
| BJ2227 | TF79 with pho-24 by P22 transduction from BJ2272 Cmr Kmr | This work |
| BJ2272 | TA2367 (pho-24) with pepT7::Mud by P22 transduction from JE2761 Cmr | This work |
| BJ2305 | BJ690 with hha::kan by P22 transduction from TF79 Kmr Tcr | This work |
| EE656 | SL1344 prgH::Tn5lacZY Tcr | 3 |
| JE2761 | LT2 pepT7::Mud Cmr | 54 |
| TA2367 | LT2 pho-24 | K. Sanderson |
| TF59 | EE251 pepT::Mud 1(x) | 12 |
| TF79 | BJ70 hha::kan Knr | This work |
| TF80 | SL1344 hha::kan Knr | This work |
| TF81 | LT2 pho-24 hha::kan Knr | This work |
| TF82 | LT2 hha::kan Knr | This work |
| Plasmids | ||
| pRW50 | Low-copy-number lacZYA reporter vector, Tcr | 37 |
| pGEM-T | High-copy-number PCR cloning vector, Apr | Promega |
| pTF120 | Cosmid carrying serovar Typhimurium DNA carrying the hha gene, Apr | This work |
| pTF137 | pGEM-T with hha from S. typhimurium, Apr | This work |
| pTF141 | pRW50 derivative carrying nt −39 to +420 of hilA fused to lacZYA Tcr | This work |
| pTF142 | pBDJ129 derivative carrying hha::kan Cmr Knr | This work |
| pLS31 | pRW50 derivative plasmid carrying −497 to +420 of hilA fused to lacZYA Tcr | 50 |
| pBDJ129 | Suicide mini-F plasmid vector, Cmr | 33 |
| pBDJ200 | repE pBR322 derivative, Apr | 33 |
| pWAM582 | Plasmid carrying the hlyCABC hemolysin genes and upstream promoter sequences | R. Welch |
Strain and plasmid construction.
The plasmid pTF137 carries the hha gene from Salmonella strain SL1344 cloned into the PCR cloning vector pGEM-T (Promega). The hha gene was amplified from the SL1344 chromosome in a standard PCR using primers hha5′ (5′-AAGCTTTGTTGTTCAGCAGCTATG-3′) and hha3′ (5′-AAGCTTCCTGCTATTGCTATGTGA-3′). The product was ligated into pGEM-T as per the instructions of the manufacturer.
The hha null mutant strain TF79 was constructed by allelic exchange of hha with a kanamycin cassette using a suicide plasmid system developed in our laboratory. This technique employs a mini-F suicide vector, pBDJ129, which relies on the RepE protein for replication, and a second high-copy-number plasmid, pBDJ200, that carries the RepE protein DNA binding sites. Since pBDJ200 is a high-copy-number plasmid and pBDJ129 is a low-copy-number plasmid, cotransformation of these plasmids into the same bacterium results in inhibition of replication of the pBDJ129 derivative due to the sequestration of RepE protein by the pBDJ200-encoded RepE DNA binding sites. Selection for an antibiotic marker on the pBDJ129 derivative allows recovery of strains in which the plasmid has integrated into the chromosome. Creation of pBDJ129 derivatives with sequences homologous to the chromosome allows integration into specific sites in the chromosome by homologous recombination. Using this system, we constructed a pBDJ129 derivative with DNA ≈900 bp upstream and ≈900 bp downstream of the hha open reading frame. This was accomplished by PCR amplification of chromosomal DNA, using primer hha3 (5′-AGATCTTTCCTAGCTATTTTCTCC-3′) with hha5 (5′-TTTTAGTTAATGGTGGATCCGACATAAATTCTAC-3′) and primer hha6 (5′-CCCCTCTTCAGGATCCAAATTCATTCGTTA-3′) with hha4 (5′-AGATCTATTTCCGCCTACCACGA-3′), to amplify the upstream and downstream sequences, respectively. The hha5 and hha6 primers incorporate a BamHI site at the 3′ end of the upstream fragment and the 5′ end of the downstream fragment, respectively. Also, hha3 and hha4 incorporate unique BglII sites into the 5′ end of the upstream and the 3′ end of the downstream PCR products, respectively. Both PCR products were digested with BamHI and ligated to pGEM-T in a triple ligation. This created a plasmid carrying ≈900 bp of DNA upstream and ≈900 bp of DNA downstream of, but not including, the hha gene joined by a unique BamHI site. A kanamycin cassette obtained from pUC4K (Pharmacia) was then ligated into the BamHI site between the cloned PCR fragments. The kanamycin cassette and flanking DNA was cut from the pGEM-T vector with BglII and ligated into the unique BglII site in the pBDJ129 suicide vector. This created a suicide plasmid, designated pTF142, with DNA homologous to sequences upstream and downstream of hha, separated by a kanamycin cassette. This plasmid was introduced into BJ70, carrying pBDJ200, and kanamycin-resistant, chloramphenicol-resistant colonies were selected. Since pTF142 is unable to replicate in the presence of pBDJ200, transformants harboring the kanamycin and chloramphenicol resistance markers have integrated pTF142 into the chromosome, which was confirmed by PCR. A screen for spontaneous kanamycin-resistant, chloramphenicol-sensitive colonies yielded the hha null mutant strain in which a second crossover had occurred, leaving a deletion of the hha gene replaced with the kanamycin cassette. Construction of the hha null mutant was verified by PCR amplification of the region and sequencing of the fragment.
Various Salmonella strains were constructed by transduction of antibiotic markers using P22 HT int−-mediated transduction (9). The E. coli hha::Tn5 mutation was moved to a lacZ background by P1-mediated transduction of Tn5 from E. coli YK4122H to E. coli GS162 to create E. coli strain BJ1575 (41).
Plasmid pTF141 was constructed in the same manner as plasmid pLS79, described in reference 50. Briefly, primers LS11 (5′-CGGATCCATGTTGAGTATGAAATCATC-3′) and LS26 (5′-GAATTCGCAGCATTTACACCCCA-3′), which incorporate BamHI or EcoRI sites into the PCR product, respectively, were used to amplify the hilA promoter from −39 bp upstream of the hilA transcriptional start site to +412 bp into the hilA transcript. The PCR product was cloned into pGEM-T (Promega) and then subcloned into pRW50 as a BamHI-EcoRI fragment to create plasmid pTF141. The hilA promoter fragment in pTF141 was sequenced prior to use to confirm that it carried the proper nucleotide sequence.
Plasmid gene bank construction.
A plasmid gene bank of S. enterica serovar Typhimurium SL1344 DNA was created by isolation of chromosomal DNA using a DNeasy Tissue kit (Qiagen). The DNA was partially digested with Sau3A1 and separated on a 0.7% agarose gel. Fragments in the range of 3 to 5 kb were cut from the gel, eluted using a QIA Quick Gel Extraction kit (Qiagen), and ligated into the BamHI site in pBluescript (Stratagene).
β-Galactosidase assays.
β-Galactosidase production from Tn5lacZY reporter constructs was quantitated using the method of Miller (41).
Hemolysin assays.
The hemolytic activity of culture supernatants was determined in a hemoglobin release assay as described by Godessart et al. (18) with minor alterations. Briefly, cultures were inoculated (100 μl into 5 ml) from an overnight culture and grown with shaking at 37°C to an optical density at 600 nm (OD600) of ≈0.6. Bacteria were removed by centrifugation, before mixing 300 μl of culture supernatant with 400 μl of phosphate-buffered saline and 300 μl of 1% washed bovine red blood cells. Samples were incubated at 40°C for 10 min, and the red blood cells were removed by centrifugation. The OD420 of each sample was measured to calculate the amount of hemoglobin released from the red blood cells. One hemolytic unit is defined as OD420/OD600 of the bacterial culture × time (in hours) × volume of culture supernatant (in milliliters).
Tissue culture conditions and invasion assays.
HEp-2 tissue culture cells (42) were maintained in RPMI 1640 (Gibco/BRL) containing 10% (vol/vol) fetal bovine serum and passaged every 2 to 3 days. Invasion assays were done as previously described (31).
Ligated loop experiments and preparation of samples for electron microscopy.
In vivo invasion of murine intestinal tissue by S. enterica serovar Typhimurium strains was assessed by the use of ligated ileal loop experiments performed as previously described (47). Briefly, ligated intestinal loops that contained ileal Peyer's patches were prepared in anesthetized BALB/c mice and injected with approximately 4 × 108 bacteria. Following incubation of the ligated loops with bacterial inocula, the mice were sacrificed and sections of the infected intestinal tissue were removed. The tissue sections were prepared for scanning electron microscopy as previously described (25) and viewed with a Hitachi S-4000 field emission scanning electron microscope.
Purification of a Hha-maltose-binding fusion protein.
The hha gene was amplified from the serovar Typhimurium chromosome with primers MBP-hha3 (5′-GTAGAAGAATTCTCTGATAAACCATTAACTAAA-3′) and MBP-hha2 (5′-TCCCAGATAACACAAGCTTGTTCTCTA-3′), which introduce an EcoRI and HindIII restriction enzyme site at the 5′ end and 3′ end of the hha gene, respectively. The hha PCR fragment was digested with HindIII and EcoRI, ligated to the pMAL-C2 vector (New England Biolabs) previously digested with HindIII and EcoRI and transformed into E. coli DH12S. Sequencing of plasmid DNA from selected colonies confirmed construction of the desired plasmid, designated TF143, encoding an Hha-MBP fusion. Hha-MBP was purified from E. coli induced for expression of the fusion protein by 0.5 mM isopropyl-β-d-thiogalactopyranoside (IPTG) according to the directions of the manufacturer. Briefly, culture extracts were passed over an amylose column and washed extensively with column buffer to remove unbound proteins. The Hha-MBP was eluted with 5 ml of column buffer containing 10% maltose. Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) analysis of the purified protein indicated that it was at least 95% pure (data not shown). The Hha portion of the fusion protein was removed from the MBP by digestion with factor Xa protease. The purified protein was quantitated using the Bradford protein quantitation kit (Bio-Rad) in preparation for gel mobility shift assays.
Gel mobility shift assay.
The gel mobility shift assay was based on the methods of Fried and Crothers (13) and Garner and Revzin (17). A 457-bp hilA promoter DNA fragment, from nucleotides −497 to −39, was amplified by PCR with primers TFHilA5′ (5′-CGGAATTCGTCCAGATGACA-3′) and LS15R (5′-TGGGGTGTAAATGCTGCTT-3′). A 479-bp invF fragment, from nucleotides −156 to +323 of the invF control region, was amplified by PCR with primers InvF5′ (5′-CTGCAGAACAATAAGCCAG-3′) and InvF3′ (5′-CGGATTCAGCATATGTCG-3′). These DNA fragments were labeled by first removing the 5′ phosphate group with calf intestinal phosphatase and then end labeling the DNA with phosphonucleotide kinase and [γ-32P]-ATP. Unincorporated nucleotides and enzymes were removed with a PCR purification kit, and the specific activity of the labeled fragments was determined in a scintillation counter. The gel mobility shift assay was performed by preincubating the labeled DNA in 1× DNA binding buffer (18-μl total volume) at 37°C for 5 min and then adding 2-μl volumes of twofold serial dilutions of Hha protein in 1× DNA binding buffer. After the tubes were incubated at 37°C for 15 min, 1 μl of loading dye was added and the samples were loaded onto a nondenaturing 5% polyacrylamide–3% glycine gel.
Nucleotide sequence accession number.
The S. enterica serovar Typhimurium hha nucleotide sequence determined in this study has been deposited in GenBank under accession number AF242359.
RESULTS
Identification of a plasmid gene bank clone that represses hilA expression.
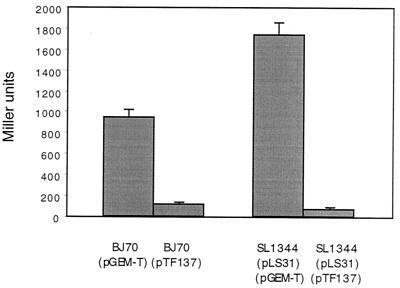
Colonies of serovar Typhimurium strain BJ70 hilA::Tn5lacZY exhibit a “fisheye” phenotype (red center with a white periphery) when grown on MacConkey lactose agar due to the oxygen regulation of the lacZY fusion (28). Expression of the hilA reporter in this strain is increased 2.5- to 3.5-fold by growth under inducing conditions (low oxygen and high osmolarity) compared to repressing conditions (high oxygen and low osmolarity). While these changes in hilA expression are relatively small, identical growth conditions are used to quantitate both β-galactosidase activity and tissue culture invasiveness. When Salmonella strains are cultured in these growth conditions to measure invasiveness, we consistently observe large changes in the ability of the strains to enter cells (≈500-fold). These reproducible findings give us a high level of confidence that small changes in hilA expression have a significant impact on downstream invasion gene transcription, invasion into mammalian cells, and host virulence. To identify factors that decrease expression of the hilA gene, we transformed strain BJ70 with a plasmid gene bank of the serovar Typhimurium SL1344 chromosome and plated ampicillin-resistant transformants on MacConkey lactose agar. White transformants that were able to significantly repress hilA::Tn5lacZY expression were isolated and retransformed into strain BJ70 to confirm the ability to repress hilA expression. One isolate, carrying a plasmid that was able to repress chromosomal hilA::Tn5lacZY expression approximately 75% compared to induced levels of expression, was obtained and selected for further study. The cloned DNA was sequenced, and the sequence obtained was used to search databases at the National Center for Biotechnology Information (NCBI) (http://www.ncbi.nlm.nih.gov) to identify the cloned DNA fragments. The partial sequence of the plasmid, pTF120, identified a gene that was highly similar to the E. coli hha gene, a gene involved in the regulation of hemolysin production (18, 43) but which has not been previously described for Salmonella species. Interestingly, a functional homolog of hha in Yersinia enterocolitica, named ymoA, has been shown to be involved in virulence gene regulation (4, 7). To determine if the hha gene was responsible for the repression of hilA::Tn5lacZY by pTF120, we amplified hha from the serovar Typhimurium chromosome by PCR, cloned the product into pGEM-T to create plasmid pTF137, and introduced the plasmid into strain BJ70. Overexpression of hha from plasmid TF137 reduced chromosomal hilA::Tn5lacZY expression more than sevenfold. Overexpression of hha also superrepresses (25-fold) hilA::Tn5lacZY expression from a plasmid reporter, which is likely due to high-level expression of hha from the high-copy-number plasmid (Fig. 1). As a control, the effect of overexpression of hha on the growth of serovar Typhimurium was assessed in growth curve experiments. No significant differences in the growth curves of Salmonella containing pGEM-T or pTF137 were observed (data not shown). Thus, overexpression of Hha causes significant repression of hilA transcription without having a measurable effect on the growth rate of Salmonella.
FIG. 1.
Effect of multicopy hha on serovar Typhimurium hilA::Tn5lacZY expression. Strains were grown statically in LB broth to an OD600 of ≈0.4 before assaying β-galactosidase activity from the hilA::Tn5lacZY reporter. Strain BJ70 carries a chromosomal hilA::Tn5lacZY reporter, while pLS31 is a low-copy-number hilA::Tn5lacZY reporter plasmid (50). Salmonella SL1344 is the wild-type virulent strain.
Overexpression of hha significantly reduces expression of invF, prgH, and sipC invasion gene reporters.
Since hha modulates hilA expression, we were interested in examining the effect of hha on invasion genes known to be regulated by hilA. To this end, we transformed strains containing chromosomal invF::Tn5lacZY, prgH::Tn5lacZY, or sipC::Tn5lacZY fusions with the vector pGEM-T or plasmid pTF137 and examined the effect of the plasmid on the expression of invF, prgH, or sipC by β-galactosidase assay. We found that, in addition to repressing hilA::Tn5lacZY expression, pTF137 caused significant decreases in chromosomal expression of invF::Tn5lacZY (159-fold), prgH::Tn5lacZY (11-fold), and sipC::Tn5lacZY (4-fold) fusions (data not shown). Thus, it seems that the repressing effect of Hha on hilA is transmitted to transcription of the invF, prgH, and sipC invasion genes. It is also possible that Hha has a repressing effect on transcription of downstream invasion genes independent of hilA transcriptional repression.
A serovar Typhimurium hha mutant is derepressed for hilA expression.
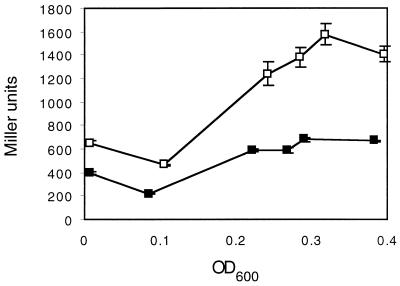
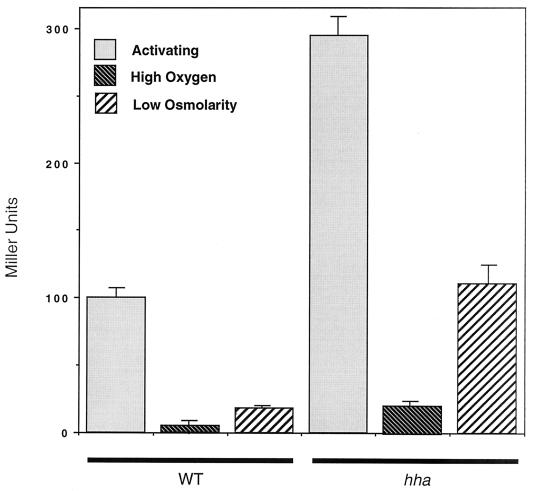
To further explore the role of Hha in the regulation of hilA, we replaced the majority of the hha gene in BJ70 with a kanamycin cassette to create the Salmonella hha null mutant TF79. We compared the expressions of the hilA::Tn5lacZY chromosomal reporter in strains BJ70 (wild type) and TF79 (hha) at various points in the growth curve of Salmonella grown in LB broth. We observed derepression of the hilA::Tn5lacZY reporter throughout the growth curve with maximal derepression (≈2.5-fold) at an OD600 of ≈0.3 (Fig. 2). Interestingly, we observed that the hilA::Tn5lacZY reporter in TF79 still seemed to be regulated by some environmental signals. Therefore, to examine the role of hha in regulating hilA expression under high-oxygen or low-osmolarity conditions, growth experiments in which oxygen was a repressing signal, but not osmolarity, or osmolarity was a repressing signal, but not oxygen, were performed. The expression of the hilA::Tn5lacZY reporter was then quantitated in the parent strain or hha mutant background. As shown in Fig. 3, hilA::Tn5lacZY is significantly repressed in both the wild-type and hha mutant strains after growth under high-oxygen (high-osmolarity) conditions, although hilA::Tn5lacZY expression is about fourfold higher in the hha mutant. Under low-osmolarity (low-oxygen) conditions, hilA::Tn5lacZY expression was increased approximately sixfold in the hha mutant compared to the wild-type strain. In fact, the levels of expression of hilA::Tn5lacZY in the hha mutant after growth under these conditions were slightly higher than those observed for the hilA::Tn5lacZY reporter in the wild-type strain when grown under fully inducing conditions. We also examined the effect of the absence of hha on expression of orgA::Tn5lacZY, invF::Tn5lacZY, and sipC::Tn5lacZY reporters under similar growth conditions and observed similar derepression of these reporters under normally repressing conditions (data not shown). Thus, deletion of hha significantly increases expression of hilA and genes (i.e., orgA, invF, and sipC) regulated by hilA. Taken together, these data indicate that hha encodes a negative regulator of the serovar Typhimurium hilA invasion transcriptional activator.
FIG. 2.
Effect of a hha mutation on expression of a serovar Typhimurium hilA::Tn5lacZY reporter. Cultures of strain BJ70 (hilA::Tn5lacZY) (solid squares) or TF79 (hilA::Tn5lacZY hha::kan) (open squares) were incubated statically in LB broth. Samples were taken at various time points throughout growth and assayed for β-galactosidase activity. Data are representative of three independent experiments.
FIG. 3.
Effects of an hha mutation on expression of hilA::Tn5lacZY after growth under repressing conditions. Cultures of wild-type (WT) (hilA::Tn5lacZY) or the hha mutant (hilA::Tn5lacZY hha::kan) were grown in LB medium with 1% NaCl under oxygen-limiting conditions (activating), LB medium with 1% NaCl with vigorous shaking (high oxygen), or LB medium with no NaCl under oxygen-limiting conditions (low osmolarity). β-Galactosidase of the hilA::Tn5lacZY reporter values were standardized, with the wild-type invasion after growth under inducing conditions being set to 100%.
Salmonella Hha has homology to YmoA and RmoA and is a functional homolog of E. coli Hha.
The Hha protein is a member of a family of small nucleoid-associated histone-like proteins that include H-NS, HU1, HU2, and YmoA. To perform comparative studies, the nucleotide sequence of the serovar Typhimurium hha gene was determined. A BLAST search of the NCBI database with the predicted Salmonella Hha protein identified proteins with significant homology to Hha, including Hha from hemolytic E. coli (43), RmoA from wild-type E. coli (45), and YmoA from Y. enterocolitica (7). A boxshade analysis (24, 52) revealed high conservation of amino acid residues ranging from 50% identical and 64% similar (RmoA) to 99% identical and 99% similar (E. coli Hha). The identity levels at the DNA level ranged from 54% identical (rmoA) to 86% identical (E. coli hha) (data not shown). Interestingly, a group of 10 amino acids (SAADHRLAEL) is completely conserved in the C-terminal half of each of these proteins, although no putative function for this amino acid motif has been identified.
The ymoA and hha genes have been shown to be functionally interchangeable (4, 40). We were interested in determining whether the Salmonella hha gene could function in E. coli to cause repression of hemolysin activity. The S. enterica serovar Typhimurium hha-carrying plasmid clone completely eliminated the zones of clearing around colonies on blood agar plates, indicating that the hemolytic activity of the E. coli colonies was repressed (data not shown), and quantitative analysis revealed that the hemolytic activity was repressed 34-fold; 106 ± 4 (mean ± standard deviation) hemolytic units were observed for E. coli pWAM582 pGEM-T versus 3 ± 2 units for E. coli pWAM582 pTF137. These experiments demonstrate that the hha gene in Salmonella which we have identified is a functional homolog of hha in E. coli as it is able repress E. coli hemolytic activity.
Repression of serovar Typhimurium hilA expression by Hha requires hilA URS.
The expression of hilA is known to be regulated by a variety of environmental and genetic signals with the condition that a single negative signal causes repression of hilA transcription (3). Recently, Schechter et al. (50) identified a URS between −497 bp and −39 bp of the hilA promoter that is necessary for regulation of hilA by oxygen, osmolarity, pho-24 (phoQc), sirA, and barA. To examine the role of this URS in hha repression of hilA, we measured transcription of hilA-lacZYA from a wild-type promoter (pLS31) or one lacking the −497 to −39 region of the promoter (pTF141) in strains either lacking hha or overexpressing hha. A hha mutation increased expression of hilA-lacZYA from the wild-type promoter from 2,017 ± 62 units to 4,017 ±116 units (2-fold increase) and overexpression of hha from pTF137 reduced hilA expression to 136 ± 10 units (15-fold decrease). In contrast, a hha mutation had no significant effect on hilA-lacZYA expression from the deleted URS construct carried by pTF141 (wild-type expression, 6,971 ± 314 units, versus hha mutant expression, 7,112 ± 49 units). In addition, overexpression of hha from pTF137 only slightly decreased hilA-lacZYA expression from pTF141 (5,632 ± 33 units). These data indicate that hha repression of hilA is dependent on the −497 to −39 promoter region of hilA.
Since a hha gene nearly identical to that of Salmonella is also present in E. coli, we performed experiments to determine if Hha could act as a repressor of hilA in E. coli. The hilA-lacZYA reporter plasmid pLS31 or pTF141 was introduced into the E. coli lac strain GS162 or the isogenic hha mutant BJ1575. E. coli strain GS162 had 103 ± 4 units of β-galactosidase from pLS31 compared to 250 ± 18 units from BJ1575. However, a hha mutation did not significantly increase hilA-lacZYA expression from the plasmid (pTF141) with a deletion of −497 to −39 of the hilA promoter (wild-type expression, 215 ± 13 units; hha mutant expression, 252 ± 25 units). We noted that the levels of expression of hilA-lacZYA from pTF141 were similar to those of pLS31 in an E. coli hha mutant. However, the levels of hilA-lacZYA from pTF141 were significantly below those which we observed for Salmonella and which were reported for a very similar plasmid, pLS79, by Schechter et al. in E. coli strain BW21355 (50). We examined this discrepancy further by performing an identical set of expression studies in E. coli strain BW21355. The hilA-lacZYA reporter plasmid pLS31 or pTF141 was introduced into the E. coli lac strain BW21355 or the isogenic hha mutant BJ1925. E. coli strain BW21355 expressed 69 ± 4 units of β-galactosidase from pLS31 compared to 136 ± 1 units from BJ1925. However, hilA-lacZYA expression from pTF141 was ≈100-fold higher in strain BW21355 (8,299 ± 234 units), a level similar to that observed by Schechter et al. A hha mutation did not significantly increase hilA-lacZYA expression (10,373 ± 150 units) in E. coli strain BJ1925. Our experiments with serovar Typhimurium provide additional evidence for the role of hha in hilA regulation but suggest that hilA expression can be misregulated in certain strains of E. coli, although the reason for this difference is unclear.
Hha regulates the invasive phenotype of S. enterica serovar Typhimurium.
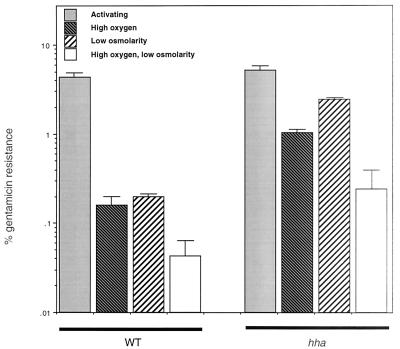
We performed experiments designed to assess the regulatory role of the Hha protein on the S. enterica serovar Typhimurium invasive phenotype. First, the effect of overexpression of hha on Salmonella invasion was examined by quantitating the invasiveness of SL1344 pGEM-T, the hha mutant TF80 pGEM-T and TF80 carrying pTF137. While the invasive phenotype of TF80 was slightly higher than that of SL1344 after growth under inducing conditions, overexpression of Hha from plasmid pTF137 in strain TF80 caused a significant reduction (40-fold) in invasion compared to SL1344 (data not shown). Invasion of SL1344 carrying pTF137 was repressed in a manner similar to that observed for TF80 pTF137 (data not shown). Next, we compared the invasiveness of SL1344 and TF80 after growth under oxygen-repressing, osmolarity-repressing, both oxygen-repressing and osmolarity-repressing, or inducing conditions. As shown in Fig. 4, a mutation in the hha gene significantly increased the invasion of Salmonella when grown under normally repressing growth conditions. Invasion was 6-fold higher (0.16 versus 1.0%) (P = 0.0005) after growth under high-oxygen conditions, 12-fold higher (0.2 versus 2.45%) (P < 0.0005) after growth under low-osmolarity conditions, and 6-fold higher (0.043 versus 0.24%) (P = 0.005) after growth in high-oxygen, low-osmolarity conditions. These results correlate well with the increased levels of β-galactosidase expression observed with the hilA-lacZY reporter in the hha mutant (Fig. 3 and 4).
FIG. 4.
Effects of an hha mutation on Salmonella SL1344 invasion after growth under repressing conditions. Cultures of SL1344 or TF80 were grown under activating low-oxygen, high-osmolarity growth conditions as the positive control or high-oxygen and high-osmolarity, low-oxygen and low-osmolarity, or high-oxygen and low-osmolarity repressing growth conditions before assaying for invasion of the bacteria into HEp-2 cells.
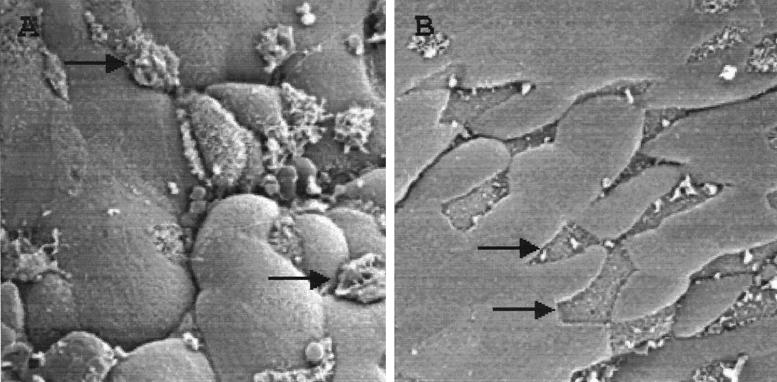
Wild-type S. enterica serovar Typhimurium invades and destroys M cells of the follicle-associated epithelium of Peyer's patches in a ligated loop model of infection (27, 30). Because hilA and many of the genes that it regulates are essential for M cell invasion, the murine ligated loop model was used to assess the effect of overexpression of Hha on invasion of M cells. Cultures of wild-type SL1344 transformed with the control vector pGEM-T or pTF137 were grown under conditions that induce invasiveness before the inocula were introduced into ligated intestinal loops of mice. Following processing of the tissue for microscopy, examination of the intestinal tissue by scanning electron microscopy revealed that more than 90% of M cells in murine ileal follicle-associated epithelium infected with wild-type bacteria had large apical membrane ruffles or were destroyed. By comparison, very few (<10%) of M cells infected with the strain overexpressing Hha displayed any membrane alterations (Fig. 5). These data establish that the presence of multicopy hha in wild-type Salmonella greatly reduces the ability of the strain to invade and destroy M cells in vivo.
FIG. 5.
Effect of multicopy hha on in vivo invasion. Intestinal ligated loops of BALB/c mice were prepared and injected with ≈4 × 108 CFU of SL1344 (wild type) containing the vector (pGEM-T) or hha (pTF137) plasmid. Following a 1-h incubation, infected intestinal tissue was harvested and prepared for viewing by scanning electron microscopy. (A) Murine Peyer's patch epithelium infected with SL1344 (pGEM-T). (B) Murine Peyer's patch epithelium infected with SL1344 (pTF137). Arrows, M cells.
Relationship of the regulatory activity of hha and phoPQ on hilA expression.
A point mutation (pho-24) in the phoQ gene results in constitutive activation of phoP (20) and is known to cause significant repression of hilA transcription (3). Our work indicates that hha also plays a negative regulatory role in hilA expression. We were interested in determining whether hha negative effects were mediated by the phoPQ two-component system, whether constitutive phoP activity was mediated by the hha gene, or whether the two regulators exert separate effects on hilA expression. To this end, Salmonella strains that carry mutations in either phoP, hha or both the phoP and hha genes were constructed in a BJ70 (hilA::Tn5lacZY) background. Other strains that carry the constitutive phoP mutation (pho-24), overexpress hha, or both carry pho-24 and overexpress hha were constructed. In addition, we constructed and examined a strain that carries the pho-24 mutation but has a mutation in hha as well as a strain that overexpresses hha but has a mutation in the phoP gene. β-Galactosidase experiments were performed to measure the effects of the different mutations on expression of the hilA::Tn5lacZY reporter (Table 2). The Salmonella strain carrying a mutation in hha (TF79) had 2.7-fold higher levels of expression of β-Galactosidase than did the parent strain BJ70. The strain carrying the phoP mutation, BJ690, had slightly increased levels of hilA::Tn5lacZY expression compared to BJ70. The hha phoP double-mutant strain had hilA levels of expression comparable to those of the hha single mutant (1,531 ± 33 units and 1,528 ± 20 units, respectively). Overexpression of hha from a plasmid resulted in 5.0-fold repression of the hilA::Tn5lacZY reporter, while the pho-24 constitutive mutation decreased the lacZY reporter levels >100-fold. The strain overexpressing hha and carrying the pho-24 mutation had β-galactosidase levels comparable to those of the strain carrying only the pho-24 mutation. Interestingly, overexpression of hha still significantly repressed hilA::Tn5lacZY expression (3.7-fold) in a strain lacking a functional phoP gene, although not quite as well as when the phoP gene was intact. In addition, the pho-24 constitutive mutation still induced efficient repression of hilA::Tn5lacZY expression (32-fold) in the absence of the hha gene, although not as efficiently as when a functional hha gene was present. These results confirm that both phoPQ and hha exert negative regulatory influences on hilA expression and indicate that each can significantly repress hilA expression in the absence of the other regulator.
TABLE 2.
Effects of altered phoPQ and hha levels on hilA::Tn5lacZY expression
| Strain (plasmid) | Strain genotype | Plasmid genotype | Units of β-galactosidase activitya |
|---|---|---|---|
| BJ70 | hilA::Tn5lacZY | None | 563 ± 3 |
| TF79 | hilA::Tn5lacZY hha::kan | None | 1,528 ± 20 |
| BJ690 | hilA::Tn5lacZY ΔphoP | None | 702 ± 12 |
| BJ2305 | hilA::Tn5lacZY hha::kan ΔphoP | None | 1,531 ± 33 |
| BJ70 (pGEM-T) | hilA::Tn5lacZY | Vector | 575 ± 4 |
| BJ70 (pTF137) | hilA::Tn5lacZY | hha+ | 116 ± 4 |
| BJ661 (pGEM-T) | hilA::Tn5lacZY pho-24 | Vector | 5 ± 1 |
| BJ661 (pTF137) | hilA::Tn5lacZY pho-24 | hha+ | 2 ± 1 |
| BJ690 (pTF137) | hilA::Tn5lacZY ΔphoP | hha+ | 155 ± 2 |
| BJ2227 (pGEM-T) | hilA::Tn5lacZY hha::kan pho-24 | Vector | 18 ± 8 |
β-Galactosidase activity was determined after incubation in growth-inducing conditions.
The Hha protein binds to hilA upstream regulatory sequences.
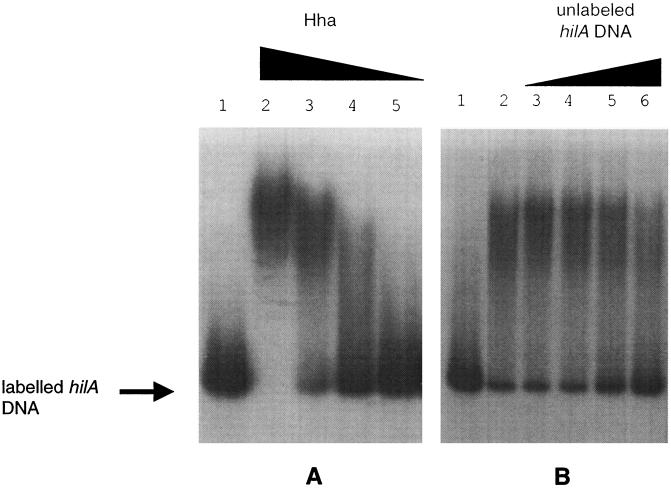
Based on the evidence presented here that hha is a negative modulator of hilA transcription and invasion, we investigated whether the Hha protein could bind directly to the hilA promoter. First, plasmid pTF143, which encodes Hha-MBP, was tested and found to possess the ability to repress hilA::Tn5lacZY expression 3.6-fold compared to the parent strain (data not shown). The result from this control indicated that Hha purified using the MBP system still possessed the ability to repress hilA expression. The ability of twofold dilutions of purified Hha protein to bind to and alter the gel mobility of a 32P-labeled hilA promoter fragment (−497 to −39) was tested. As seen in Fig. 6A, 1.0 μmol of Hha retarded the mobility of virtually all of the radiolabeled hilA promoter fragment, 0.5 μmol of Hha shifted the majority of the DNA, and 0.25 μmol of Hha still shifted a substantial portion of the radiolabeled hilA promoter fragment. We next examined the ability of unlabeled hilA promoter DNA to compete for Hha binding to a 32P-labeled hilA promoter fragment. Twofold dilutions of the unlabeled hilA promoter fragment (12.5 to 100 ng) were added to 100 ng of radiolabeled hilA promoter incubated with 1.0 μmol of Hha. As observed in Fig. 6B, the unlabeled hilA promoter DNA effectively competed for binding of the Hha protein, as the concentration of the unlabeled fragment approached that of the radiolabeled hilA fragment. Importantly, unlabeled nonspecific Salmonella chromosomal DNA did not compete for Hha binding to the hilA promoter (data not shown). As another specificity control, the invF promoter and portions of the open reading frame (−156 to +323) were amplified for use in a gel mobility shift assay. The invF promoter fragment was labeled and incubated with twofold dilutions of Hha protein (0.25 to 1.0 μmol of protein) or Fis protein (0.25 to 1.0 μmol of protein) under conditions similar to those described for the experiments illustrated in Fig. 6A. No shifting of the radiolabeled invF DNA was observed with the highest amounts of Hha protein used (1.0 μmol of protein), although the Fis protein was able to shift the invF promoter at 0.5 μmol of protein, providing additional evidence that Hha binding to the hilA promoter is specific under the conditions tested (data not shown).
FIG. 6.
Gel retardation assay of hilA promoter DNA with purified Hha protein. (A) 32P-labeled hilA promoter DNA (100 ng) was incubated with various concentrations of Hha and then run on a nondenaturing 5% polyacrylamide-3% glycine gel. The concentrations of Hha used were as follows: lane 1, no protein; lane 2, 1 μmol; lane 3, 0.5 μmol; lane 4, 0.25 μmol; lane 5, 0.13 μmol. Bands were visualized by autoradiography. The arrow indicates the unshifted radiolabeled hilA band. (B) The binding of Hha to labeled hilA promoter DNA was competed with unlabeled hilA promoter DNA. Labeled hilA promoter DNA (100 ng) was mixed with 1 μmol of Hha and various amounts of unlabeled hilA promoter competitor DNA as indicated: lane 1, labeled hilA DNA only (control); lane 2, labeled hilA DNA with 1 μmol of Hha; lane 3, labeled hilA DNA with 1 μmol of Hha and 12.5 ng of unlabeled hilA promoter DNA; lane 4, labeled hilA DNA with 1 μmol of Hha and 25.0 ng of unlabeled hilA promoter DNA; lane 5, labeled hilA DNA with 1 μmol of Hha and 50.0 ng of unlabeled hilA promoter DNA; lane 6, labeled hilA DNA with 1 μmol of Hha and 100.0 ng of unlabeled hilA promoter DNA. Bands were visualized by autoradiography.
DISCUSSION
We have used a plasmid gene bank approach to search for factors that down-regulate expression of S. enterica serovar Typhimurium hilA expression. A plasmid clone that has the ability to repress a hilA::Tn5lacZY reporter was identified, and subsequent work revealed that the plasmid carried the hha gene which was responsible for the hilA::Tn5lacZY repressing activity. An S. enterica serovar Typhimurium hha knockout mutant was constructed by allelic exchange, and the mutation was found to significantly derepress hilA expression. Hha-mediated derepression of hilA also resulted in significantly increased levels of tissue culture invasion after growth of the bacteria under high-osmolarity repressing conditions, as well as high-oxygen repressing conditions. Overexpression of hha repressed transcription of invasion genes known to be regulated by HilA (i.e., invF, prgH, and sipC). Finally, overexpression of hha repressed the invasiveness of S. enterica serovar Typhimurium for HEp-2 tissue culture cells and for murine M cells in an intestinal ligated loop model. These data lead us to conclude that Hha is a negative regulator of the S. enterica serovar Typhimurium invasion gene transcriptional activator hilA.
Interestingly, we found that the invF::lacZY reporter was derepressed significantly more (159-fold) than the sipC::lacZY reporter (4-fold). Since invF encodes a transcriptional activator of the sipBCDA operon, this result is somewhat surprising. One possible explanation for this difference is that basal levels of invF and sipC differed substantially, with sipC expression levels under repressing conditions being significantly higher than those of invF. Thus, the apparent effect of the hha mutation on sipC expression was minimized due to the high basal levels of expression. Another possible explanation is that other regulatory factors are important for invF transcription, but not sipC transcription, and while the absence of Hha results in a significant derepression of sipC the additional factors still exert a significant effect on invF transcription. Future experiments will be performed to address these questions.
Work characterizing positive and negative regulation of hilA is in progress in many laboratories. It is now clear that there are multiple genes that contribute to the activation of hilA, including hilC/sirC/sprA (10, 48, 50), hilD (50), sirA, barA (1), csrAB (1), fis (55), and phoB, fadD, and fliZ (39). Schechter et al. (50) have demonstrated that hilA expression is repressed in the absence of hilD. However, deletion of the URS promoter sequence of hilA led to unregulated expression of hilA, even in the absence of hilD. More recently, it was found that PhoP/PhoQ, FadD, FliZ, PhoB, SirA, and EnvZ require the URS for modulation of hilA expression, but these factors are not responsible for regulating hilA in response to oxygen and osmolarity (38). It was hypothesized that these factors may function by altering the expression of hilD or the unidentified repressor(s). Our present findings indicate that the absence of hha leads to derepression of hilA under low-osmolarity conditions and partial derepression under high-oxygen conditions. Importantly, the derepression of hilA transcription under these conditions, in the hha mutant, leads to a corresponding increase in tissue culture invasion. Our present findings do not allow us to determine whether positive regulatory factors (i.e., fadD, fliZ, sirA, barA, csrAB, phoB, and fis) alter the expression or activity of hha or whether they modulate HilD activity posttranscriptionally. However, the activities of these regulators have been shown not to entail modulation of hilD transcription, although mutations in pstS and sirA were found to have a mild effect on hilD transcription (38). Interestingly, since a hha mutation did not totally derepress hilA expression under all repressing growth conditions, it is likely that another repressor (hilE, ams, pag, and/or another unidentified repressor) also plays an important role in modulating hilA expression in response to repressing growth conditions.
Despite the fact that a variety of environmental conditions are known to downregulate invasion gene expression, until recently the only gene known to repress hilA expression was the phoQ constitutive allele (5, 46). We have performed genetic experiments to determine if mutations in phoPQ and hha have similar effects on hilA transcription. The results suggest that the mechanisms of regulation of hha and phoPQ on hilA transcription differ. A mutation in hha increases hilA transcription approximately threefold, and overexpression of hha decreases hilA transcription approximately fivefold. These data seem to be consistent with the idea that Hha binds at the hilA promoter to exert a regulatory effect on transcription of the gene. In contrast, deletion of phoP increases hilA transcription only slightly (≈20%) but the pho-24 phoQ constitutive mutation decreases hilA expression 115-fold. These results suggest that the high levels of phosphorylation activity present in strains with the pho-24 mutation are required for phoPQ regulation of hilA. From the data it seems possible that phosphorylated PhoP is not responsible for the hilA repression caused by the pho-24 mutation since a phoP mutation itself does not substantially derepress hilA transcription, although the data do not eliminate this possibility. Alternatively, hyperphosphorylation resulting from the pho-24 mutation may downregulate the transcription or protein activity of critical positive regulatory factors such as hilD, fis, or any of the other identified factors to significantly reduce hilA transcription. It is also possible that hyperphosphorylation resulting from the pho-24 mutation upregulates or increases the binding activity of negative regulatory factors. Answers to these detailed questions await further work.
Our group recently identified transposon insertions within ams, the gene encoding RNase E; hupB, a subunit of the nucleoid-associated protein HU; and an unidentified pag gene and a newly described hilE gene that cause upregulation of a hilA::Tn5lacZY reporter (12). While the mechanism by which these genes alter hilA expression is unknown, it is possible that some of these genes encode hilA repressors that may be the targets of the derepressors HilC and HilD. In the present work we have identified another gene, hha, that also appears to have the ability to repress hilA expression. Thus, the number of hilA negative regulators is approaching that of the identified positive regulators of hilA, suggesting that a mutation in any single repressor protein will not result in constitutive expression of hilA. Interestingly, Schechter et al. (50) recently postulated that small nucleoid-associated proteins are good candidates for hilA repressors because this family of proteins modulates expression of genes in response to conditions such as temperature, osmolarity, pH, and oxygen tension. Our recent findings appear to confirm this speculation.
We have provided evidence indicating that at least one activity of Hha is at the hilA promoter. In a model of hilA regulation proposed by Schechter et al. (50), a repressor protein present in both E. coli and S. enterica serovar Typhimurium acts at the −497 to −39 hilA promoter region to modulate hilA expression. Essentially identical Hha proteins (1-amino-acid difference) in E. coli and S. enterica serovar Typhimurium are functionally homologous, as S. enterica serovar Typhimurium Hha can repress expression of E. coli hemolysin. We have demonstrated that the S. enterica serovar Typhimurium Hha protein is able to repress expression of hilA::Tn5lacZY from a reporter plasmid. Further, the repressing activity of Hha requires the −497 to −39 region of the hilA promoter, since a reporter plasmid lacking this sequence was not repressed by Hha in S. enterica serovar Typhimurium. Finally, we have shown using a gel shift assay that purified Hha protein binds to hilA promoter DNA. This binding could be inhibited in competitive-binding studies with unlabeled hilA sequences but not by unlabeled nonspecific Salmonella chromosomal DNA sequences. Hha was also unable to bind to and shift a DNA fragment carrying the invF promoter. Collectively, these data provide evidence that Hha binds specifically to the hilA promoter, which we believe is the likely mechanism by which it represses hilA transcription. The specific sites of binding of the Hha protein await future experiments.
In summary, we report that the S. enterica serovar Typhimurium Hha protein is a repressor of hilA transcription. However, it seems clear that the Hha protein is only one of many repressors of hilA. Previous work by our group has identified several other genes that may encode proteins with hilA-repressing activity (12). In addition, phoPQ repression appears to be independent of the hha pathway. The identification of Hha as a regulator of hilA suggests that hilA may be regulated by alterations in DNA topology, which may be the common link between the effects of environmental signals on the invasive phenotype and the activity of the Hha repressor protein. Future work will be directed at more carefully defining the sites of interaction of regulator proteins at the hilA promoter as well as determining how repressors and activators interact with each other.
ACKNOWLEDGMENTS
We thank the Central Microscopy Research Facility of the University of Iowa College of Medicine for technical assistance with electron microscopy. We gratefully acknowledge Antonio Juárez, Cathy Lee, Lisa Schechter, George Stauffer, Barry Wanner, Rod Welch, and members of the Jones lab for providing information, strains, or plasmids used in this work.
T.F.F. was supported by a predoctoral fellowship on NIH Training Grant 5T32 AI07511. R.L.W. was supported by NIH postdoctoral training grant HL07638. This work was supported by NIH grant AI28268 awarded to B.D.J.
REFERENCES
- 1.Altier C, Suyemoto M, Ruiz A I, Burnham K D, Maurer R. Characterization of two novel regulatory genes affecting Salmonella invasion gene expression. Mol Microbiol. 2000;35:1872–1882. doi: 10.1046/j.1365-2958.2000.01734.x. [DOI] [PubMed] [Google Scholar]
- 2.Bajaj V, Hwang C, Lee C A. hilA is a novel ompR/toxR family member that activates the expression of Salmonella typhimurium invasion genes. Mol Microbiol. 1995;18:715–727. doi: 10.1111/j.1365-2958.1995.mmi_18040715.x. [DOI] [PubMed] [Google Scholar]
- 3.Bajaj V, Lucas R L, Hwang C, Lee C A. Co-ordinate regulation of Salmonella typhimurium invasion genes by environmental and regulatory factors is mediated by control of hilA expression. Mol Microbiol. 1996;22:703–714. doi: 10.1046/j.1365-2958.1996.d01-1718.x. [DOI] [PubMed] [Google Scholar]
- 4.Balsalobre C, Juarez A, Madrid C, Mourino M, Prenafeta A, Munoa F J. Complementation of the hha mutation in Escherichia coli by the ymoA gene from Yersinia enterocolitica: dependence on the gene dosage. Microbiology. 1996;142:1841–1846. doi: 10.1099/13500872-142-7-1841. [DOI] [PubMed] [Google Scholar]
- 5.Behlau I, Miller S I. A PhoP-repressed gene promotes Salmonella typhimurium invasion of epithelial cells. J Bacteriol. 1993;175:4475–4484. doi: 10.1128/jb.175.14.4475-4484.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Clark M A, Jepson M A, Simmons N L, Hirst B H. Preferential interaction of Salmonella typhimurium with mouse Peyer's patch M cells. Res Microbiol. 1994;145:543–552. doi: 10.1016/0923-2508(94)90031-0. [DOI] [PubMed] [Google Scholar]
- 7.Cornelis G R, Sluiters C, Delor I, Geib D, Kaniga K, Lambert de Rouvroit C, Sory M P, Vanooteghem J C, Michiels T. ymoA, a Yersinia enterocolitica chromosomal gene modulating the expression of virulence functions. Mol Microbiol. 1991;5:242–246. doi: 10.1111/j.1365-2958.1991.tb01875.x. [DOI] [PubMed] [Google Scholar]
- 8.Darwin K H, Miller V L. Molecular basis of the interaction of Salmonella with the intestinal mucosa. Clin Microbiol Rev. 1999;12:405–428. doi: 10.1128/cmr.12.3.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Davis R W, Botstein D, Roth J R. Advanced bacterial genetics: a manual for genetic engineering. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1980. [Google Scholar]
- 10.Eichelberg K, Hardt W D, Galán J E. Characterization of SprA, an AraC-like transcriptional regulator encoded within the Salmonella typhimurium pathogenicity island 1. Mol Microbiol. 1999;33:139–152. doi: 10.1046/j.1365-2958.1999.01458.x. [DOI] [PubMed] [Google Scholar]
- 11.Ernst R K, Dombroski D M, Merrick J M. Anaerobiosis, type 1 fimbriae, and growth phase are factors that affect invasion of HEp-2 cells by Salmonella typhimurium. Infect Immun. 1990;58:2014–2016. doi: 10.1128/iai.58.6.2014-2016.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fahlen T F, Mathur N, Jones B D. Identification and characterization of mutants with increased expression of hilA, the invasion gene transcriptional activator of Salmonella typhimurium. FEMS Immunol Med Microbiol. 2000;28:25–35. doi: 10.1111/j.1574-695X.2000.tb01453.x. [DOI] [PubMed] [Google Scholar]
- 13.Fried M, Crothers D M. Equilibria and kinetics of lac repressor-operator interactions by polyacrylamide gel electrophoresis. Nucleic Acids Res. 1981;9:6505–6525. doi: 10.1093/nar/9.23.6505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fu Y, Galán J E. The Salmonella typhimurium tyrosine phosphatase SptP is translocated into host cells and disrupts the actin cytoskeleton. Mol Microbiol. 1998;27:359–368. doi: 10.1046/j.1365-2958.1998.00684.x. [DOI] [PubMed] [Google Scholar]
- 15.Galán J E. Interactions of bacteria with non-phagocytic cells. Curr Opin Immunol. 1994;6:590–595. doi: 10.1016/0952-7915(94)90146-5. [DOI] [PubMed] [Google Scholar]
- 16.Galán J E, Curtiss R., III Expression of Salmonella typhimurium genes required for invasion is regulated by changes in DNA supercoiling. Infect Immun. 1990;58:1879–1885. doi: 10.1128/iai.58.6.1879-1885.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Garner M M, Revzin A. A gel electrophoresis method for quantifying the binding of proteins to specific DNA regions: application to components of the Escherichia coli lactose operon regulatory system. Nucleic Acids Res. 1981;9:3047–3060. doi: 10.1093/nar/9.13.3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Godessart N, Munoa F J, Regue M, Juarez A. Chromosomal mutations that increase the production of a plasmid-encoded haemolysin in Escherichia coli. J Gen Microbiol. 1988;134:2779–2787. doi: 10.1099/00221287-134-10-2779. [DOI] [PubMed] [Google Scholar]
- 19.Groisman E A, Ochman H. Cognate gene clusters govern invasion of host epithelial cells by Salmonella typhimurium and Shigella flexneri. EMBO J. 1993;12:3779–3787. doi: 10.1002/j.1460-2075.1993.tb06056.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gunn J S, Hohmann E L, Miller S I. Transcriptional regulation of Salmonella virulence: a PhoQ periplasmic domain mutation results in increased net phosphotransfer to PhoP. J Bacteriol. 1996;178:6369–6373. doi: 10.1128/jb.178.21.6369-6373.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hardt W D, Chen L M, Schuebel K E, Bustelo X R, Galán J E. S. typhimurium encodes an activator of Rho GTPases that induces membrane ruffling and nuclear responses in host cells. Cell. 1998;93:815–826. doi: 10.1016/s0092-8674(00)81442-7. [DOI] [PubMed] [Google Scholar]
- 22.Hong K H, Miller V L. Identification of a novel Salmonella invasion locus homologous to Shigella ipgDE. J Bacteriol. 1998;180:1793–1802. doi: 10.1128/jb.180.7.1793-1802.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hueck C J. Type III protein secretion systems in bacterial pathogens of animals and plants. Microbiol Mol Biol Rev. 1998;62:379–433. doi: 10.1128/mmbr.62.2.379-433.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jeanmougin F, Thompson J D, Gouy M, Higgins D G, Gibson T J. Multiple sequence alignment with Clustal X. Trends Biochem Sci. 1998;23:403–405. doi: 10.1016/s0968-0004(98)01285-7. [DOI] [PubMed] [Google Scholar]
- 25.Jensen V B, Harty J T, Jones B D. Interactions of the invasive pathogens Salmonella typhimurium, Listeria monocytogenes, and Shigella flexneri with M cells and murine Peyer's patches. Infect Immun. 1998;66:3758–3766. doi: 10.1128/iai.66.8.3758-3766.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Johnston C, Pegues D A, Hueck C J, Lee A, Miller S I. Transcriptional activation of Salmonella typhimurium invasion genes by a member of the phosphorylated response-regulator superfamily. Mol Microbiol. 1996;22:715–727. doi: 10.1046/j.1365-2958.1996.d01-1719.x. [DOI] [PubMed] [Google Scholar]
- 27.Jones B, Pascopella L, Falkow S. Entry of microbes into the host: using M cells to break the mucosal barrier. Curr Opin Immunol. 1995;7:474–478. doi: 10.1016/0952-7915(95)80091-3. [DOI] [PubMed] [Google Scholar]
- 28.Jones B D, Falkow S. Identification and characterization of a Salmonella typhimurium oxygen-regulated gene required for bacterial internalization. Infect Immun. 1994;62:3745–3752. doi: 10.1128/iai.62.9.3745-3752.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jones B D, Falkow S. Typhoid fever: host immune response and Salmonella virulence determinants. Annu Rev Immunol. 1996;14:533–561. doi: 10.1146/annurev.immunol.14.1.533. [DOI] [PubMed] [Google Scholar]
- 30.Jones B D, Ghori N, Falkow S. Salmonella typhimurium initiates murine infection by penetrating and destroying the specialized epithelial M cells of the Peyer's patches. J Exp Med. 1994;180:15–23. doi: 10.1084/jem.180.1.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jones B D, Lee C A, Falkow S. Invasion by Salmonella typhimurium is affected by the direction of flagellar rotation. Infect Immun. 1992;60:2475–2480. doi: 10.1128/iai.60.6.2475-2480.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kaniga K, Uralil J, Bliska J B, Galán J E. A secreted protein tyrosine phosphatase with modular effector domains in the bacterial pathogen Salmonella typhimurium. Mol Microbiol. 1996;21:633–641. doi: 10.1111/j.1365-2958.1996.tb02571.x. [DOI] [PubMed] [Google Scholar]
- 33.Klein J R, Jones B D. Transcriptional organization and function of invasion genes within Salmonella pathogenicity island 1, including the prgH, prgI, prgJ, prgK, orgA, orgB, and orgC genes. Infect Immun. 2000;68:3368–3376. doi: 10.1128/iai.68.6.3368-3376.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lee C A, Falkow S. The ability of Salmonella to enter mammalian cells is affected by bacterial growth state. Proc Natl Acad Sci USA. 1990;87:4304–4308. doi: 10.1073/pnas.87.11.4304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lee C A, Jones B D, Falkow S. Identification of a Salmonella typhimurium invasion locus by selection for hyperinvasive mutants. Proc Natl Acad Sci USA. 1992;89:1847–1851. doi: 10.1073/pnas.89.5.1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Levine M M, Galen J, Barry E, Noriega F, Chatfield S, Sztein M, Dougan G, Tacket C. Attenuated Salmonella as live oral vaccines against typhoid fever and as live vectors. J Biotechnol. 1996;44:193–196. doi: 10.1016/0168-1656(95)00094-1. [DOI] [PubMed] [Google Scholar]
- 37.Lodge J, Fear J, Busby S, Gunasekaran P, Kamini N R. Broad host range plasmids carrying the Escherichia coli lactose and galactose operons. FEMS Microbiol Lett. 1992;74:271–276. doi: 10.1016/0378-1097(92)90441-p. [DOI] [PubMed] [Google Scholar]
- 38.Lucas R L, Lee C A. Roles of hilC and hilD in regulation of hilA expression in Salmonella enterica serovar Typhimurium. J Bacteriol. 2001;183:2733–2745. doi: 10.1128/JB.183.9.2733-2745.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lucas R L, Lostroh C P, DiRusso C C, Spector M P, Wanner B L, Lee C A. Multiple factors independently regulate hilA and invasion gene expression in Salmonella enterica serovar Typhimurium. J Bacteriol. 2000;182:1872–1882. doi: 10.1128/jb.182.7.1872-1882.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mikulskis A V, Delor I, Thi V H, Cornelis G R. Regulation of the Yersinia enterocolitica enterotoxin yst gene. Influence of growth phase, temperature, osmolarity, pH and bacterial host factors. Mol Microbiol. 1994;14:905–915. doi: 10.1111/j.1365-2958.1994.tb01326.x. [DOI] [PubMed] [Google Scholar]
- 41.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1972. [Google Scholar]
- 42.Moore A E, Sabachewsky L, Toolan H W. Culture characteristics of four permanent lines of human cancer cells. Cancer Res. 1955;15:598. [PubMed] [Google Scholar]
- 43.Nieto J M, Carmona M, Bolland S, Jubete Y, de la Cruz F, Juarez A. The hha gene modulates haemolysin expression in Escherichia coli. Mol Microbiol. 1991;5:1285–1293. doi: 10.1111/j.1365-2958.1991.tb01902.x. [DOI] [PubMed] [Google Scholar]
- 44.Nieto J M, Mourino M, Balsalobre C, Madrid C, Prenafeta A, Munoa F J, Juarez A. Construction of a double hha hns mutant of Escherichia coli: effect on DNA supercoiling and alpha-haemolysin production. FEMS Microbiol Lett. 1997;155:39–44. doi: 10.1111/j.1574-6968.1997.tb12683.x. [DOI] [PubMed] [Google Scholar]
- 45.Nieto J M, Prenafeta A, Miquelay E, Torrades S, Juarez A. Sequence, identification and effect on conjugation of the rmoA gene of plasmid R100-1. FEMS Microbiol Lett. 1998;169:59–66. doi: 10.1111/j.1574-6968.1998.tb13299.x. [DOI] [PubMed] [Google Scholar]
- 46.Pegues D A, Hantman M J, Behlau I, Miller S I. PhoP/PhoQ transcriptional repression of Salmonella typhimurium invasion genes: evidence for a role in protein secretion. Mol Microbiol. 1995;17:169–181. doi: 10.1111/j.1365-2958.1995.mmi_17010169.x. [DOI] [PubMed] [Google Scholar]
- 47.Penheiter K L, Mathur N, Giles D, Fahlen T, Jones B D. Non-invasive Salmonella typhimurium mutants are avirulent because of an inability to enter and destroy M cells of ileal Peyer's patches. Mol Microbiol. 1997;24:697–709. doi: 10.1046/j.1365-2958.1997.3741745.x. [DOI] [PubMed] [Google Scholar]
- 48.Rakeman J L, Bonifield H R, Miller S I. A HilA-independent pathway to Salmonella typhimurium invasion gene transcription. J Bacteriol. 1999;181:3096–3104. doi: 10.1128/jb.181.10.3096-3104.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rathman M, Sjaastad M D, Falkow S. Acidification of phagosomes containing Salmonella typhimurium in murine macrophages. Infect Immun. 1996;64:2765–2773. doi: 10.1128/iai.64.7.2765-2773.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schechter L M, Damrauer S M, Lee C A. Two AraC/XylS family members can independently counteract the effect of repressing sequences upstream of the hilA promoter. Mol Microbiol. 1999;32:629–642. doi: 10.1046/j.1365-2958.1999.01381.x. [DOI] [PubMed] [Google Scholar]
- 51.Schiemann D A, Shope S R. Anaerobic growth of Salmonella typhimurium results in increased uptake by Henle 407 epithelial and mouse peritoneal cells in vitro and repression of a major outer membrane protein. Infect Immun. 1991;59:437–440. doi: 10.1128/iai.59.1.437-440.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Smith R F, Wiese B A, Wojzynski M K, Davison D B, Worley K C. BCM Search Launcher–an integrated interface to molecular biology data base seach and analysis services available on the World Wide Web. Genome Res. 1996;6:454–462. doi: 10.1101/gr.6.5.454. [DOI] [PubMed] [Google Scholar]
- 53.Takeuchi A. Electron microscope studies of experimental Salmonella infection. I. Penetration into the intestinal epithelium by Salmonella typhimurium. Am J Pathol. 1967;50:109–136. [PMC free article] [PubMed] [Google Scholar]
- 54.Tsang A W, Escalante S J. CobB, a new member of the SIR2 family of eucaryotic regulatory proteins, is required to compensate for the lack of nicotinate mononucleotide:5,6-dimethylbenzimidazole phosphoribosyltransferase activity in cobT mutants during cobalamin biosynthesis in Salmonella typhimurium LT2. J Biol Chem. 1998;273:31788–31794. doi: 10.1074/jbc.273.48.31788. [DOI] [PubMed] [Google Scholar]
- 55.Wilson R L, Libby S J, Freet A M, Boddicker J D, Fahlen T F, Jones B D. Fis, a DNA nucleoid-associated protein, is involved in Salmonella typhimurium SPI-1 invasion gene expression. Mol Microbiol. 2001;39:79–88. doi: 10.1046/j.1365-2958.2001.02192.x. [DOI] [PubMed] [Google Scholar]
- 56.Wood M W, Rosqvist R, Mullan P B, Edwards M H, Galyov E E. SopE, a secreted protein of Salmonella dublin, is translocated into the target eukaryotic cell via a sip-dependent mechanism and promotes bacterial entry. Mol Microbiol. 1996;22:327–338. doi: 10.1046/j.1365-2958.1996.00116.x. [DOI] [PubMed] [Google Scholar]
- 57.Wray C, Sojka W J. Experimental Salmonella typhimurium in calves. Res Vet Sci. 1978;25:139–143. [PubMed] [Google Scholar]