Abstract
In C. elegans, unc-89 encodes a set of giant multi-domain proteins (up 8,081 residues) localized to the M-lines of muscle sarcomeres and required for normal sarcomere organization and whole-animal locomotion. Multiple UNC-89 isoforms contain two protein kinase domains. There is conservation in arrangement of domains between UNC-89 and its two mammalian homologs, obscurin and SPEG: kinase, a non-domain region of 647–742 residues, Ig domain, Fn3 domain and a second kinase domain. In all three proteins, this non-domain “interkinase region” has low sequence complexity, high proline content and lacks predicted secondary structure. We report that a major portion of this interkinase (571 residues out of 647 residues) when examined by single molecule force spectroscopy in vitro displays the properties of a random coil and acts as an entropic spring. We used CRISPR/Cas9 to create nematodes carrying an in-frame deletion of the same 571-residue portion of the interkinase. These animals display severe disorganization of all portions of the sarcomere in body wall muscle. Super-resolution microscopy reveals extra, short-A-bands lying close to the outer muscle cell membrane and between normally spaced A-bands. Nematodes with this in-frame deletion show defective locomotion and muscle force generation. We designed our CRISPR-generated in-frame deletion to contain an HA tag at the N-terminus of the large UNC-89 isoforms. This HA tag results in normal organization of body wall muscle, but approximately half the normal levels of the giant UNC-89 isoforms, dis-organization of pharyngeal muscle, small body size, and reduced muscle force, likely due to poor nutritional uptake.
Keywords: sarcomere, giant polypeptides, UNC-89, obscurin, C. elegans
Introduction
C. elegans UNC-89 is the founding member of the UNC-89/obscurin family of giant muscle proteins. Loss-of-function mutations in the C. elegans gene unc-89 result in slow moving worms with disorganized muscle sarcomeres, including a lack of M-lines [1–3]. unc-89 is a complex gene: through the use of three promoters and alternative splicing at least 16 polypeptides are generated, ranging in size from 156,000 to 900,000 Da [3–5]. Overlapping sets of isoforms are expressed in body wall muscle, pharyngeal muscle (for pumping in and grinding up bacterial food), egg laying and intestinal muscles [3], and several gonadal epithelial cells [6]. The largest of these isoforms, UNC-89-B and UNC-89-F, consist of 53 Ig domains, two Fn3 domains, a triplet of SH3, DH and PH domains near their N-termini, and two protein kinase domains (PK1 and PK2) near their C termini. Antibodies localize UNC-89 to the M-line [3,4]. To learn how UNC-89 is localized and performs its functions, our lab has systematically identified its binding partners. We have screened a yeast two hybrid library with segments representing the largest UNC-89 isoform and identified at least 7 interacting partners, and 6/7 have orthologs or homologs in humans [7–13]. Interactions have been verified by a combination of demonstrating binding in vitro using purified recombinant proteins, co-immunolocalization, and identifying muscle mutant phenotypes.
Mammalian striated muscle express three UNC-89-like proteins from separate genes, obscurin (OBSC), obscurin-like 1 (obsl1) and striated muscle preferentially expressed gene (SPEG). UNC-89 is most similar to obscurin [14,15], which also expresses many isoforms, including giant isoforms (obscurin A), and shorter isoforms with 2 protein kinase domains [16]. Although UNC-89 is located only at the M-line, various obscurin isoforms are located at either the M-line, or the Z-disk [15,17]. Mutations in the human OBSC gene result in cardiomyopathies (HCM, DCM, LVNC)[18]. In addition to serving as a platform for assembly of M-line proteins, obscurin links myofibrils to the sarcoplasmic reticulum (SR). The C-terminus of obscurin interacts with an ankyrin localized to the SR [19,20]. The obscurin KO mouse has normal myofibril organization but disorganized SR [21]. This myofibril to SR linkage is conserved in nematode UNC-89 [22]. Obsl1 is an ~200 kDa protein consisting solely of 20 tandem Ig domains, and in heart muscle is localized to M-lines, Z-disks and intercalated disks [23]. Several portions interact with titin and myomesin at the M-line [24]. However, obsl1 is ubiquitously expressed, global KO is embryonic lethal and mutations in the human Obsl1 gene result in a growth defect (3M-growth syndrome)[25]. SPEG also exists as different sized isoforms, but the largest isoform is 3,267 aa and consists of 9 Ig and 2 Fn3 domains, and 2 protein kinase domains [26]. It can be regarded as similar to the C-terminal tandem kinase domain regions of obscurin and UNC-89. SPEG is located in a complex lying between the SR and plasma membrane (JMC) in cardiomyocytes. In the mouse, cardiac-specific KO results in DCM, whereas mutations in human SPEG result in centronuclear myopathy with DCM [26]. The in vivo substrates for the kinase domains of UNC-89, obscurin and SPEG are not known. However, sequence analysis suggests that: (i) UNC-89 PK1 is catalytically inactive, whereas PK2 is an active kinase [27]; (ii) human obscurin PK1 and PK2 are both likely to be active kinases [27], and each has been shown to undergo autophosphorylation in vitro [28]. Obscurin PK2 can phosphorylate N-cadherin in vitro [28]. There is a suggestion that SPEG phosphorylates the JMC protein junctophilin 2 [29].
As shown in Fig. 1A, UNC-89, obscurin and SPEG have an overall conservation in organization around the kinase domains: PK1, a non-domain region of 647–742 residues, Ig, Fn, and PK2. There is very little sequence homology amongst the non-domain or “interkinase” (IK) regions of these proteins (Supplemental Figure 1). However, these IK regions have in common, a low sequence complexity, high proline content (11.8—13.3%), and an overall paucity of predicted α-helix and β-strands, and thus are likely to be predominantly random coils [9]. These properties of the IK regions are shared with the known highly elastic spring regions of human titin (PEVK and N2A/B)[30]. In this paper, we describe, for nematode UNC-89, the response of the interkinase to mechanical pulling and the phenotype of worms in which the interkinase has been deleted in-frame. This is the first time the function of this region has been explored for any member of the UNC-89/obscurin family.
Fig. 1. Schematic representation of domain organization around kinase domains, and secondary structure predictions for the interkinase regions of nematode UNC-89, mouse obscurin and SPEG.
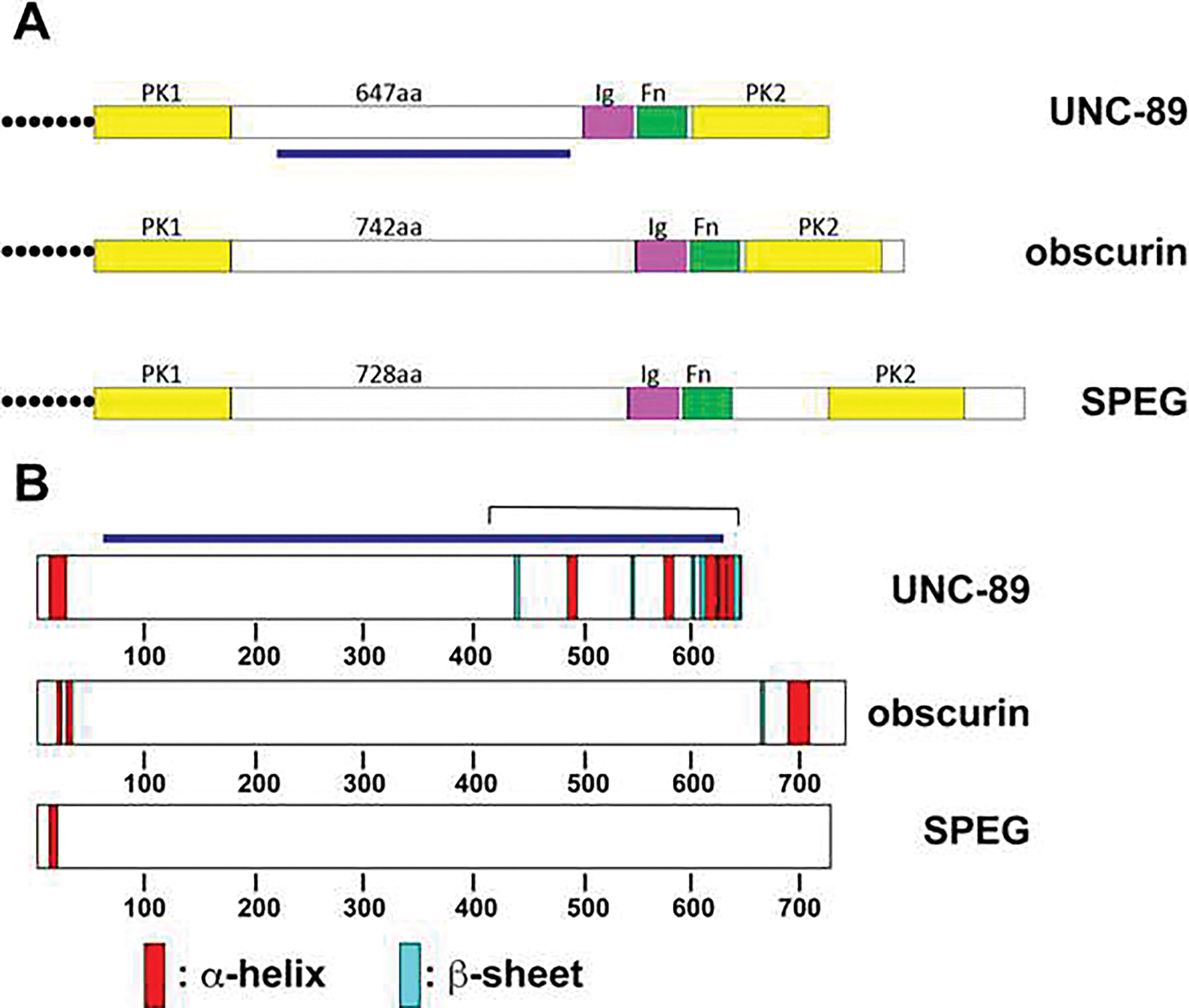
(A) Domain organization from PFAM predictions. Note that each protein, near its C-terminus consists of a protein kinase, a non-domain “interkinase” region (647–742 residues), Ig and Fn domains, and a second protein kinase domain. (B) Secondary structure predictions of the non-domain interkinase regions of UNC-89, obscurin and SPEG using JPred.
Results
To examine the nature of the IK regions of UNC-89, obscurin and SPEG in greater detail, we used JPred to predict secondary structure [30]. As shown in Supplemental Figure 2, and in Fig. 1B, there are only short stretches of α-helix (15, 9, 9, 11, 7 and 7 amino acids, from N- to C-terminus of IK), and even fewer and shorter instances of β-strands (3, 2, 3, 4 and 6 amino acids). Except for the 15-residue long region near the IK N-terminus, all the other instances of α-helices reside in the C-terminal one third of the IK sequence. These α-helices reside within the C-terminal 232 residues which is the minimal binding site determined for LIM-9 (FHL2) [9], and thus may comprise an unrecognized or novel domain. Thus, the remaining sequence of UNC-89 IK, approximately, 475 residues, is random coil. As shown in Supplemental Figure 2 and Fig. 1B, obscurin and SPEG have even fewer predicted secondary structure elements, although like UNC-89, obscurin has a fairly long α-helix of 19 residues lying within the C-terminal 52 residues.
Given the nature of the amino acid sequence of the IK region of UNC-89, we wished to determine how it might respond to a mechanical pulling force using the single molecule force spectroscopy technique. To carry this out, we first expressed in E. coli 1 571 residue segment comprising the majority of the 645 residue interkinase region (IK) flanked on the N- and C-terminus by two copies of Ig27 from human titin. The response of Ig27 to pulling force is well documented and served as an internal control. In addition, the protein had a 6XHis tag at its N-terminus (Fig. 2A). After purification (Fig. 2B), the protein was allowed to attach to a glass surface coated with Ni2+-NTA to allow binding of the N-terminus of this polyprotein via its 6XHis tag. The stylus of the single-molecule force spectrometer was allowed to bind non-specifically and at random points along the polyprotein, and then pulling force was applied by moving the piezo-electric platform downward. A representative force curve of the mechanical unfolding of the polyprotein in which the stylus had attached to the C-terminus is shown in Fig. 2C. After a long and featureless trace without energy barriers, the typical unwinding of the 4 Ig domains is found. The statistical distribution of the contour length of the long featureless peak (Fig. 2D) shows that the mean contour is approximately 185 nm. This is a good match to the expected contour length of a fully unwound polypeptide of 571 amino acids, since the average length of an amino acid is 0.34 nm, i.e. 0.34 nm/aa × 571 aa = 194 nm. Based on this force curve and contour length, we can conclude that the IK has the mechanical properties of a random coil and behaves like an entropic spring.
Fig. 2. The interkinase region (IK) of UNC-89 is a highly elastic spring.

(A) Polyprotein construct encompassing the IK flanked on either side by two titin I27 domains used in single molecule force spectroscopy (SMFS) experiments. (B) SDS-PAGE after Coomassie Blue staining of ~2 μg of the polyprotein shown in (A). (C) Representative force curve of the mechanical unfolding of the polyprotein by SMFS. The dotted line is a worm-like-chain (WLC) fit to the force-extension curve with free contour length LC and a fixed persistence length Lp = 0.36 nm. (D) Statistical distribution of the contour length of the first peak (LC = 186 ± 3 nm, n=18). The solid line represents a Gaussian fit of the histogram.
We wished to determine the in vivo consequences of removing the IK from UNC-89. To do this, we used CRISPR/Cas9 to create an in-frame deletion of the same 571 residues that we explored with the single molecule method in vitro. However, to be able to monitor the expression of UNC-89 large isoforms in which the IK had been deleted we added an HA tag to the N-terminus of the large isoforms. We thought such tagged UNC-89 could also be used in future immunoprecipitation experiments to identify proteins in a complex with UNC-89. Animals expressing HA-UNC-89 large isoforms were obtained by CRISPR/Cas9, and the strain is designated unc-89(syb797). The proper fusion of a tag containing three consecutive HA sequences to the normal N-terminus of the large UNC-89 isoforms was verified by Sanger sequencing (Supplemental Figure 3). The presence of the HA tag on the large UNC-89 isoforms was verified by western blot (Fig. 3A). Moreover, the HA-UNC-89 proteins could be detected by confocal microscopy to be normally localized to the sarcomeric M-lines of body wall muscle (Fig. 3B). The HA tag also does not affect the organization of body wall muscle sarcomeres as assessed by confocal microscopy using phalloidin for I-bands, or antibodies to myosin heavy chain A for A-bands, UNC-89 for M-lines, UNC-95 for the bases of M-lines and dense bodies, or ATN-1 (α-actinin) for the major portion of dense bodies (Fig. 4). As shown later, this HA tag also does not affect whole animal locomotion. However, HA tag does reduce the abundance of the UNC-89 giant isoforms (see Fig. 3 and Fig. 5). With an n=2, the level of expression from unc-89(syb797) is 51.2% (standard deviation of 11%) of the level from wild type.
Fig. 3. Addition of HA tags at the N-termini of the largest UNC-89 isoforms does not affect the normal M-line localization of UNC-89 in body wall muscle sarcomeres, but does moderately reduce the level of UNC-89 giant isoforms in whole animal extracts.

(A) Total Laemmli-soluble proteins from wild type and unc-89(syb797) were separated on a 5% SDS-PAGE, transferred to membrane and reacted with antibody EU30, a well-established rabbit polyclonal antibody to UNC-89. On the right is shown Ponceau S staining of the blots before reaction with antibodies. Note the presence of HA tags in unc-89(syb797) but not in wild type, and a moderate reduction in the level of UNC-89 giant isoforms in unc-89(sy797) compared with wild type. The asterisk indicates the position of the UNC-89 giant isoforms. The thin upper-most band is likely to be some UNC-89 that could not enter the separating gel. The positions of size markers are shown on the left-hand side. (B) Co-immunostaining of parts of two body wall muscle cells in unc-89(syb797) with anti-HA and MH42, a well-established monoclonal that recognizes UNC-89. Note the co-localization of each antibody to the M-lines of these muscle sarcomeres (merged image). Scale bar, 10 μm.
Fig. 4. Addition of HA tags at the N-termini of the largest UNC-89 isoforms does not affect the organization of sarcomeres in body wall muscle.

Each panel shows localization of the indicated sarcomere components using confocal microscopy. Scale bar, 10 μm.
Fig. 5. unc-89(syb797 syb1257) is an in-frame deletion of most of the IK region and results in mis-localization of UNC-89 in the sarcomeres of body wall muscle.

(A) Representation of the exon (black boxes) and intron (lines) organization of the kinase-encoding region of the unc-89 gene. Blue bar, portion deleted in syb1257 by CRISPR/Cas9. This is a 4,614 bp genomic in-frame deletion which internally deletes 571 aa of the IK beginning 66 aa C-terminal of PK1 and continuing through 10 aa N-terminal of Ig53. (B) Western blots demonstrating that the large UNC-89 isoforms are expressed in unc-89(syb797) and unc-89(syb797 syb1257). Western blots and reactions conducted as in Fig. 3. The asterisk indicates the position of the UNC-89 giant isoforms. The thin upper-most band is likely to be some UNC-89 that could not enter the separating gel. (C) Co-immunostaining of parts of several body wall muscle cells in unc-89(syb797 syb1257) with anti-HA and the anti-UNC-89 antibody MH42. Note the disorganization of M-lines in this mutant. Scale bar, 10 μm.
Next, we used unc-89(syb797) as starting point to make an in-frame deletion of 571 residues of the IK by CRISPR/Cas9, and this strain is designated unc-89(syb797 syb1257). A schematic of the genomic region deleted is shown in Fig. 5a. Sanger sequencing was used to verify that the 4,614 bp genomic deletion resulted in an in-frame deletion of the unc-89 coding sequence (Supplemental Figure 4). Before characterizing either unc-89(syb797) or unc-89(syb797syb1257) strains further, each was outcrossed 5X to wild type, to remove most possible off-target effects of CRISPR/Cas9 gene editing.
As shown in Fig. 5B, the in-frame deletion of IK (in syb797syb1257), leads to expression of UNC-89 giant isoforms that can be detected with either antibodies to the N-terminal HA tag or our standard antibodies raised against epitopes in the middle of UNC-89. We had hoped to be able to use an antibody to PK2 to prove that the deletion is in-frame, however, two attempts to make such an antibody (in rabbits or rats) failed. Nevertheless, since the levels of large isoforms did not diminish below that shown with the HA tag (in syb797), it is likely that we achieved an in-frame deletion which does not destabilize the protein. We expected to observe slightly smaller giant isoforms from syb797syb1257 compared to wild type, but there seems to be no discernible difference (Fig. 5B). Although there are at least 14 predicted giant isoforms ranging from 5,000 to 8,081 residues each, we do not know the relative abundance of each of these giant isoforms. Deletion of 571 residues out of a total of 8,081 residues for the largest isoform would be only a decrease of 7% and may not be a difference detectable by the gel system used. However, as shown in Fig. 5C, the internal deletion of the IK of UNC-89 results in disorganization of M-lines in body wall muscle. This can be seen when either an established monoclonal for UNC-89, MH42, or anti-HA is used in immunostaining. In fact, the signals from each antibody overlap.
To assess the in vivo consequences of removing the IK, we examined the organization of body wall muscle sarcomeres in greater detail, and measured whole animal locomotion and force generation. To assess sarcomere organization of the IK deletion, we took the same strategy as we did to characterize the sarcomere organization of unc-89(syb797): immunolocalization of several proteins with known sarcomere locations using confocal microscopy. Deletion of IK (in mutant syb797syb1257) results in disorganization of sarcomeres in all substructures (Fig. 6). It is important to note however that in syb797syb1257, although the pattern of UNC-89 is disorganized (i.e. not parallel straight lines as in syb797 or wild type), UNC-89 is still detectable at high levels consistent with the western blot result. Note also that in syb797syb1257, the disorganization of UNC-89 appears quite similar to the pattern of disorganization of myosin heavy chain A (MHC A). This is not unexpected as thick filaments in nematode body wall muscle have differential localization of two myosin heavy chain isoforms, with MHC A in the middle, and MHC B in the major outer portions [31]. Normally thick filaments are arranged parallel and in register, so that the M-line is the structure in the middle of the A-band, which contains MHC A, and where the thick filaments are crosslinked. Thus, when imaging C. elegans body wall muscle with light microscopy, the localization of MHC A and UNC-89 are nearly identical [4,32]. Note that upon anti-UNC-95 staining, the bases of M-lines are often branched and can overlap the bases of dense bodies (Fig. 6, right inset). Overall, these results indicate that the IK is required for the proper organization of sarcomeres.
Fig. 6. unc-89(syb797 syb1257) animals that express HA-UNC-89 isoforms with an in-frame deletion of the interkinase region have disorganized myofibrils.
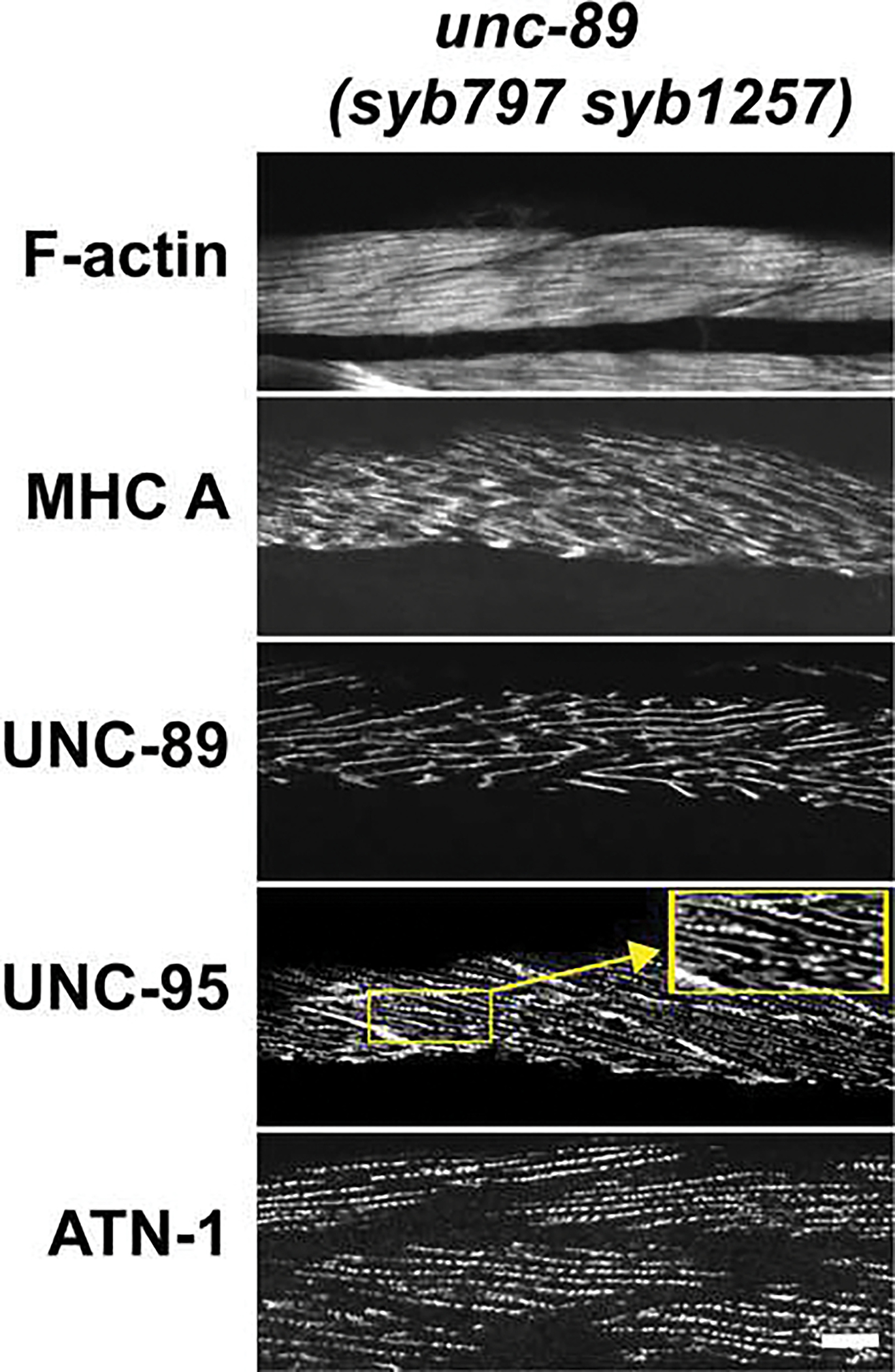
Each panel shows representative confocal images of several body wall muscle cells stained with antibodies to the indicated sarcomeric components or with rhodamine-phalloidin. Scale bar, 10 μm.
We examined the organization of MHC A and UNC-89, proxies for the middle of A-bands, at higher resolution using structured illumination microscopy (SIM). In Fig. 7 A and B, it is clear that the N-terminal HA tag on the large UNC-89 isoforms (in syb797) has no effect on the normal pattern of straight parallel A-bands. However, in-frame deletion of IK (in syb797syb1257), for both MHC A and UNC-89 immunostaining, it is evident that the A-bands are broken, and much of the parallel arrangement is missing. In addition, there appear to be extra, short A-bands (indicated by arrows in Fig. 7) that lie in the spaces between otherwise nearly normal A-bands. By capturing optical sections with an interval of 0.2 μm, we noticed that these partial A-bands lie mostly close to the outer cell membrane of the muscle cells. From top to bottom of Fig. 7A and B, images are shown from close to the outer muscle cell membrane to deeper into the cell. Note that the partial A-bands lie near the outer muscle cell but do not extend as deeply as normal A-bands.
Fig. 7. unc-89(syb797 syb1257) animals that express HA-UNC-89 isoforms with an in-frame deletion of the interkinase region show extra A-bands that are not full-depth.

After immunostaining with antibodies to MHC A (A) or antibodies to UNC-89 (B) multiple optical slices with a 0.2 μm interval were obtained by structured illumination microscopy (SIM). Yellow and blue arrows point to examples of short extra A-bands that lie between normally located A-bands and reside close to the outer muscle cell membrane. In comparison, unc-89(syb797) that expresses HA-UNC-89 shows parallel, normally spaced and full-depth A-bands. Scale bar, 5 μm.
In C. elegans, the body wall muscle is required for the animal to generate the forces required for its normal swimming, crawling and burrowing locomotion [33]. Given the disorganization of sarcomeres in the body wall muscle of nematodes expressing large isoforms of UNC-89 with an in-frame deletion of the IK, we wondered if these animals would display defects in movement or in muscle force generation. First, because our strain with the IK deletion also contains an N-terminal HA tag, we measured locomotion in the strain with the HA tag alone, syb797. As shown in Fig. 8A and B, syb797 is not slower in swimming or crawling as compared to wild type. In fact, syb797 displays a statistically faster swimming than wild type. However, clearly, syb797syb1257, containing both an HA tag and IK deletion shows a 15% reduction in swimming and a 30% reduction in crawling (Fig. 8A and B). This decrease in swimming speed is comparable to that displayed by the canonical unc-89 allele e1460, but not as severe as that of alleles su75 or tm752 (Fig. 8A). The decrease in crawling of the IK deletion strain is not as severe as 3 of 4 other alleles tested (e1460, su75, tm752). In terms of muscle force exerted, as measured by the NemaFlex assay [34], all mutant alleles (including syb797 syb1257) except for tm752 are defective compared to wild type (Fig. 8C). Interestingly, syb797, which has an HA tag at the N-terminus of the large isoforms, shows no decrease in crawling, but an increase in swimming, and decreased ability to generate force. This phenotype of increased speed of swimming, no change or even increased speed of crawling, and yet decreased force, is shared with the allele ad539. unc-89(ad539), like syb797, has normally organized sarcomeres in body wall muscle, which likely accounts for lack of any decrease in swimming or crawling. (We first showed that unc-89(ad539) has normally organized sarcomeres by EM [2], but to verify this result we have immunostained its muscle with a variety of antibodies to sarcomeric proteins, and all of these are normally localized (Supplemental Figure 5)).
Fig. 8. Whole worm locomotion and muscle force measurements of wild type and various unc-89 mutant alleles.

(A) Swimming assays show that unc-89(syb797) and unc-89(ad539) swim faster than wild type, whereas all other unc-89 mutants, including unc-89(syb797 syb1257) swim slower than wild type. (B) Crawling assays show that unc-89(ad539) crawls faster than wild type, that unc-89(syb797) crawls at the same speed as wild type, and all other unc-89 mutants crawl more slowly than wild type. C) NemaFlex force measurements show that unc-89(tm752) develops the same muscle force as wild type, whereas all other unc-89 mutants develop less force than wild type, although, unc-89(syb797 syb1257), unc-89(e1460), and unc-89(su75) are more severely affected. The actual data, including sample sizes, means, and standard errors are shown in Supplementary Table 1 for swimming and crawling and Supplementary Table 2 for NemaFlex measurements.
In C. elegans, the pharynx is a neuromuscular pump required for bringing bacterial food into the worm and grinding it up before its entry into the intestine. unc-89(ad539) was first isolated in a screen for mutants that show defective feeding behavior, and found to be an allele of unc-89 by complementation testing and 3 factor mapping; by polarized light microscopy the pharyngeal muscle was shown to have reduced birefringence and abnormal arcs in the terminal or posterior bulb [35]. Based on this, and the similarity of ad539 with syb797, we examined the pharyngeal muscle structure of both these alleles, and additional alleles of unc-89 using polarized light microscopy (Fig. 9). As first noted by Waterston et al. [1], in unc-89(e1460), instead of the radially oriented filaments observed in the posterior bulb of wild type, this mutant has longitudinally oriented filaments at the periphery of the bulb (indicated by yellow arrow in Fig. 9). In fact, all the unc-89 mutants that we imaged show this defect except for tm752, which shows normal pharyngeal muscle organization. Remarkably, syb797, with the HA tag, shows the same defect. However, ad539 shows not only the defect in the terminal bulb; it also shows a defect in the anterior bulb (blue arrows in Fig. 9). The only other unc-89 allele that shows both bulbs affected is su75.
Fig. 9. Polarized light images of the myofilament lattice in the pharynges of wild type and various unc-89 mutant alleles.

Yellow arrows indicate mis-localized arc-like thick filaments at the periphery of the terminal bulb, and blue arrows indicate mis-localized arc-like thick filaments at the periphery of the anterior bulb. Note that unc-89(tm752) shows pharyngeal muscle like wild type, whereas all the other unc-89 mutants show defective organization. unc-89(su75) and unc-89(ad539) are the only alleles that show defects in both the posterior and anterior bulbs. Scale bar, 50 μm.
To understand the pharyngeal muscle defect further, we examined the expression of UNC-89 protein in the pharynges of wild type, syb797 and ad539. This was done by co-immunostaining worms with antibodies to UNC-89 and to the pharyngeal myosin heavy chain MHC C [36]. We have previously demonstrated that in wild type, UNC-89 is localized to the middle of thick filaments in pharyngeal muscle [4]. As shown in Fig. 10, UNC-89 is localized to the middle of thick filaments throughout the pharynx. However, in syb797 there is no detectable expression of UNC-89 in the posterior bulb. In ad539 there is no observable expression of UNC-89 throughout the pharynx. Moreover, MHC C staining reveals that in both syb797 and ad539, there are abnormal accumulations of thick filaments at the periphery of the posterior bulb (indicated by yellow arrows in Fig. 10), corresponding to the arcs observed by polarized light. The expression defects in the mutants seem to correspond with the defects in pharyngeal muscle organization and indicate that UNC-89 is required for normal organization of sarcomeres in pharyngeal muscle.
Fig. 10. unc-89(syb797) and unc-89(ad539) have reduced levels of expression of UNC-89 in pharyngeal muscle.
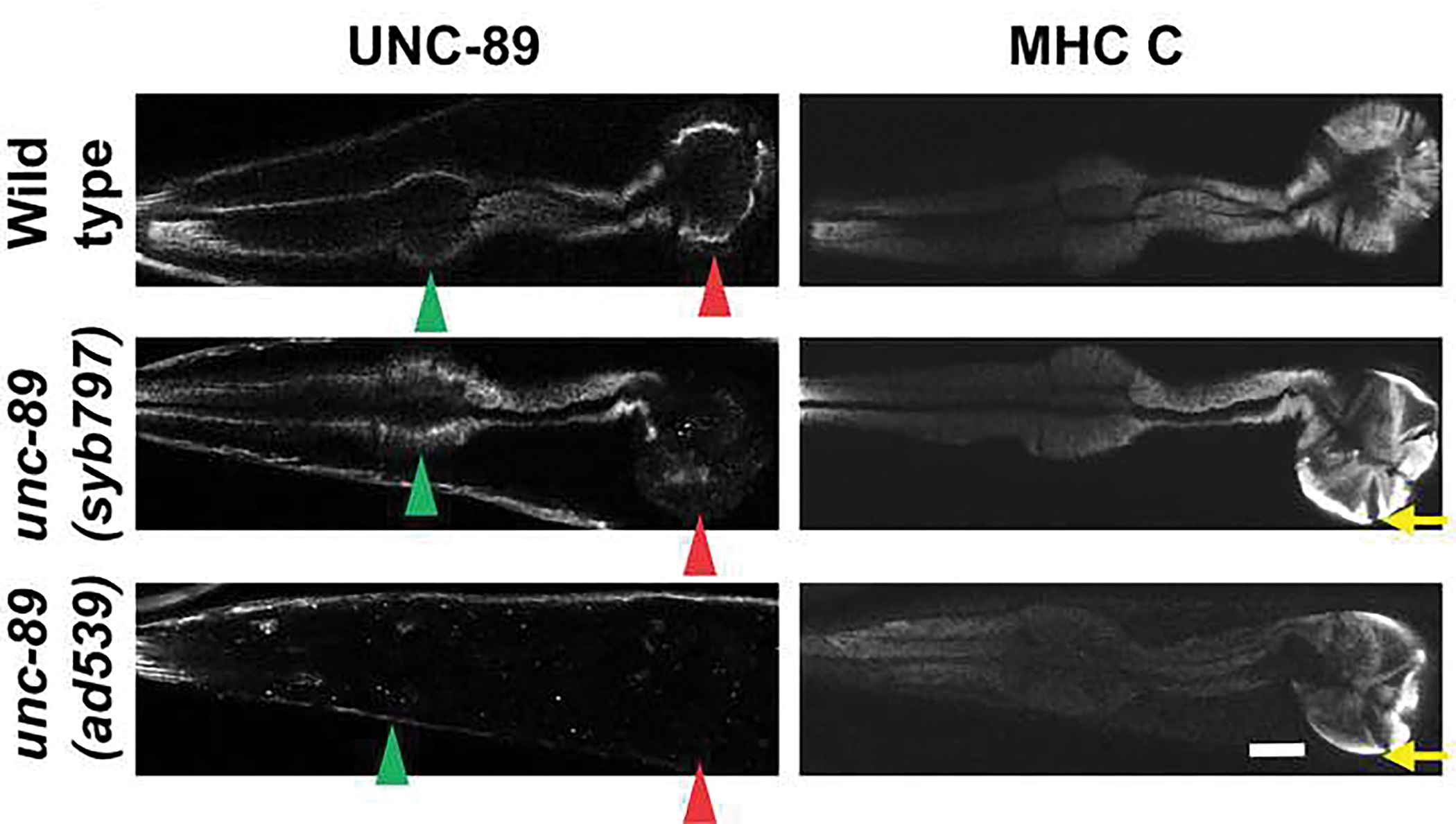
The indicated strains were immunostained with antibodies to UNC-89 and the pharyngeal-specific myosin heavy chain C (MHC C), and representative confocal images of the pharynx are shown. Note that unc-89(syb797) shows reduced UNC-89 expression in the terminal bulb, whereas unc-89(ad539) shows reduced UNC-89 expression in both the terminal and anterior bulbs. Green arrowhead, anterior bulb; red arrowhead, posterior bulb; yellow arrow, arc-like thick filaments mis-placed at the periphery of the posterior bulb. Scale bar, 10 μm.
Discussion
In this study, we explored the biophysical properties and in vivo function of an example of ~600 aa sequence that has similar features (low sequence complexity, lack of predicted domains and minimal levels of predicted secondary structure elements) and location between two protein kinase domains in the UNC-89/obscurin class of giant muscle proteins. In the case of UNC-89 IK, which was studied here, as shown in Fig. 1B, JPred predicts only short stretches of α-helices, primarily in the C-terminal third of the IK, and the majority of the sequence is predicted to be random coil. By use of single molecule force spectroscopy in vitro (Fig. 2) we showed that this IK sequence, is likely to be a random coil since it acts like an entropic spring upon application of pulling force. The nature of the IK sequence (random coil) and its physical properties (entropic spring) is shared with the highly elastic spring elements (PEVK, N2B) of another giant muscle protein, titin [37]. In the case of titin, there is good evidence to suggest that these elements unwind when muscle sarcomeres are stretched beyond their relaxed state, store potential energy and then provide a passive restoring force to allow the sarcomeres to return to their resting (relaxed) lengths [38].
To test the secondary structure prediction and have a better understanding of the response of IK to pulling force, we conducted Far-UV circular dichroism on I27-I27-IK(571)-I27-I27, and as a control protein, a polyprotein containing 9 identical repeats of the I27 domain (I279 polyprotein). As Supplemental Figure 6 shows the overall secondary structure of the I27 polyprotein (dotted line) is similar to published data on I27 polyproteins [39] showing maximum around 200 nm a minimum at ~212nm, a characteristic of a mainly β-strand structure. The overall CD spectra for the IK-I27 construct (solid line) shows a shift of the minimum to a lower wavelength (~208nm) and no clear maximum ~200 nm indicating a reduced β-strand structure and an additional random-coil component. Hence, these structural data are consistent with our AFM and JPred results and demonstrates that the interkinase region displays the properties of a random coil.
To explore the muscle function of the IK, we used CRISPR/Cas9 to create a worm strain, syb797syb1257, in which most of the IK of UNC-89 was deleted in-frame. This strain also has its giant UNC-89 isoforms N-terminally tagged with HA. syb797syb1257 also seems to show reduced expression of the UNC-89 giant isoforms similar to that shown by the HA tagged strain, syb797 (an example is shown in Fig. 5B). However, in contrast to syb797 which shows normal body wall muscle sarcomere organization, syb797syb1257, shows highly disorganized body wall muscle sarcomeres. Therefore, the body wall muscle sarcomere defect in syb797syb1257 is likely caused by the lack of the IK.
The normal location of UNC-89 is the M-line, the structure in which thick filaments are cross linked in the middle of the A-band. Deletion of the IK, in addition to disrupting the normal continuous and parallel M-lines or A-bands, results in formation of short A-bands which are not full-depth but confined to the portion nearest the outer muscle cell membrane, and these occur between normally spaced A-bands. Sarcomere assembly is known to occur from the outside inwards, first with the deposition of ECM proteins like perlecan, followed by localization of integrins, and then localization of integrin adhesion proteins (UNC-112 (kindlin), PAT-4 (ILK), UNC-97 (PINCH), PAT-6 (α-parvin), followed by assembly of M-line or dense body (Z-disk)-specific proteins [40,41]. We know that UNC-89 interacts with CPNA-1 [11], which in turn interacts with PAT-6, a member of the 4- protein complex that interacts with the cytoplasmic tail of PAT-3 (β-integrin)[42]. It should be noted that CPNA-1 interacts with Ig1-Ig3, which is near the N-terminus of UNC-89 and remote from IK. Our results seem to indicate that these protein interactions at the membrane are not sufficient for full assembly of a M-line/A-band, and the proper spacing between A-bands. Either the length or the elasticity of UNC-89 IK is required. In future experiments, we can determine the minimum length of IK that is sufficient for full A-band assembly, and whether elasticity is crucial. We could replace the elastic IK with a sequence of similar length that is rigid. An example of a rigid sequence that could be used is the series of 4 consecutive Ig domains in human titin that are not separated by linker sequences [43]. It is tempting to speculate that the length of the interkinase is important for somehow determining the spacing between adjacent A-bands. Another possibility is that deletion of the IK in some way interferes with the activity of the active protein kinase domain of UNC-89 PK2. This is not likely because inactivation of PK2 does not result in a defect in sarcomere organization (Matsunaga et al., manuscript in preparation). So far, we have only examined the organization of body wall muscle sarcomeres in adult muscle. Therefore, we cannot distinguish a role for UNC-89 IK for sarcomere assembly vs. sarcomere stability. Indeed, it is likely that UNC-89 has more of a role in maintaining or stabilizing sarcomere structure: In strong loss of function alleles like unc-89(e1460), A-band organization is normal in early larvae [22], despite the fact that UNC-89 is expressed even in embryonic muscle [44].
In vertebrate muscle, the M-band is known to be elastic and helps titin maintain the registration of adjacent thick filaments during muscle contraction [45]. The cross-linking of thick filaments occurs by the binding of antiparallel dimers of myomesin to myosin of thick filaments. The M-band also includes complex interactions between Obsl1 and obscurin with titin, and myomesin with obscurin [46]. Although much of the elasticity and mechanical protection is provided by myomesin [47,48], our results suggest that the IK region of obscurin, which also lies in the M-band also provides some of this elasticity.
Because whole animal locomotion (swimming, crawling) and force generation are diminished upon removal of the IK, we also have learned that the level of sarcomere organization found in this mutant is not sufficient for proper contractile activity. In contrast, the better level of sarcomere organization in unc-89(tm752) is sufficient to generate normal force but is not sufficient to generate normal swimming or crawling speeds.
Our IK deletion strain also contains an HA tag appended to the N-terminus of the large UNC-89 isoforms. We designed this for future immunoprecipitation experiments to further test whether UNC-89 interacting partners that we have discovered primarily through two hybrid screens are actually associated with UNC-89: we would create in-frame deletions of segments of UNC-89 that are suspected to bind to a partner, then IP those internally deleted UNC-89 proteins, and look for absence of the association. Somewhat surprisingly, this HA tag reduces the level of expression of the giant UNC-89 isoforms to approximately half of their wild type levels in whole worm extracts. And although the HA tag does not affect the organization of sarcomeres in body wall muscle or the ability of whole animals to swim or crawl, it does reduce the ability to generate muscle force. In addition, the HA tag results in a defect in thick filaments in the terminal bulb of the pharynx. Moreover, the level of expression of UNC-89 is reduced in the terminal bulb, consistent with the western blot result. This suggests that the HA tag at the N-terminus either reduces the stability of pharyngeal-specific UNC-89 mRNA or protein isoforms, or interferes with interaction with binding partners found only in pharyngeal muscle. Although our extensive yeast two-hybrid screens did not reveal any binding partners for the N-terminus of UNC-89, these screens cannot reveal all relevant binding partners. Also, it is possible that the HA tag reduces the steady state levels of the giant UNC-89 isoforms in both pharyngeal and body wall muscle, below a critical level required for pharyngeal but not for body wall muscle sarcomere assembly and/or stability.
The reduced force generated by syb797 containing the HA tag is also puzzling. However, it should be noted that syb797 and ad539 share these properties—each has normal body wall muscle, but defective pharyngeal muscle. Then, why do these two mutants generate less force? In fact, the only mutant studied that generates normal force is tm752 that does so despite having mildly disorganized body wall muscle. The key to explaining why these two mutants generate less force appears to be their reduced body diameters. In fact, as shown in Table 1, all the mutants generating less force have smaller than wild type body diameters, and defective pharynxes. tm752 has a normal body diameter and a normal pharynx. Our speculation is that all the mutants with defective pharynxes have reduced nutrition and consequently are smaller. Although our force measurements were normalized for body diameter, it is possible that reduced nutrition results in reduced metabolism and reduced energy production, and thus less ability to generate force. It is also curious that both syb797 and ad539 display greater swimming speeds and ad539 shows greater crawling speed. Perhaps this is because these thinner animals can bend more easily.
Table 1.
The affects of unc–89 mutations on expression of UNC–89 isoforms, body wall and pharyngeal muscle structure, locomotion, body wall muscle force and body diameter.
| Strain | Mutation | Protein Expression | Body Wall Muscle Structure | Swimming | Crawling | Force by NemaFlex | Body Diameter (pm) | Pharyngeal Muscle |
|---|---|---|---|---|---|---|---|---|
| wild type | 73.5–78.3 | |||||||
| syb797 | HA tag at N–term. of giant isoforms | giant isoforms reduced by ∼50% reduced in pharynx | normal | faster | normal | reduced | 62.1 | defective posterior bulb |
| e1460 | nonsense mutation in alternatively spliced exon for Ig20–Ig26 [22] | reduced levels of giant isoforms [12] | moderate [12] | slower | slower (severe) | reduced (severe) | 57.3 | defective posterior bulb |
| syb797syb1257 | HA tag added to N–term. of giant isoforms & in–frame deletion of IK | giant isoforms reduced by ∼50% reduced in pharynx | moderate–severe | slower | slower | reduced | 55.7 | defective posterior bulb |
| su75 | 1 bp insertion in coding for Ig38 resulting in frameshift premature stop [12] | all giant isoforms missing small kinase isoforms present [3,12] | severe [12] | slower (severe) | slower (severe) | reduced (severe) | 49.9 | defective posterior & anterior bulbs |
| tm752 | out of frame deletion in IK [5] | all kinase isoforms (giant & small) missing [5] | mild [12] | slower (severe) | slower (severe) | normal | 75.9 | normal |
| ad539 | Ala to Val change in KSP region but this may not be causative | giant isoforms normal by western none detectable in pharynx | normal | faster | faster | reduced | 56.2 | defective posterior & anterior bulbs |
Finally, it should be pointed out that the C. elegans pharynx is often considered analogous to the vertebrate heart, although the two organs are likely to result from convergent evolution between two muscular pumps tasked with similar roles [49]. Thus, in the future, cardiomyopathy-causing mutations in human obscurin might be studied in C. elegans by creating the analogous mutations in unc-89, and using these mutants for genetic suppressor and drug screens.
Materials and Methods
Recombinant protein production
A cDNA encoding a 571 amino acid portion of the interkinase region of UNC-89, IYDYLRIQ…ADIEKVTP (residues 6945—7515 in UNC-89b) was synthesized using E. coli optimal codons and cloned into pUC57 by GenScript USA,. Inc. (Piscataway, New Jersey). This plasmid was digested with SpeI and SacI, and the insert was cloned into “I27 plasmid shuffled codons (003)”, in order to express a fusion protein consisting of 6xHis-I27-I27-UNC-89 IK 571 aa—I27-I27. (I27 is the human titin Ig27 domain.) The resulting plasmid was transformed into E. coli BL21 (DE3) and the fusion protein was expressed at 20° for 5 hours after induction with 750 mM Isopropyl β-D-1-thiogalactopyranoside (IPTG). The 6xHis tagged fusion protein was purified with Ni-NTA Agarose (Qiagen), as described in [13]. The buffer of the eluted protein was exchanged with PBS containing 10% glycerol.
Single-molecule force spectroscopy (SMFS) experiments
Pulling experiments using SMFS were carried out on a home-built single-molecule Atomic Force Spectrometer as previously described in the literature [50,51]. In each experiment, we deposited 10 μL of the UNC-89 IK domain polyprotein solution (0.5 mg/mL) in PBS (pH 7.4) onto a Ni2+-NTA functionalized glass coverslip [52] covered by 50 μL of PBS. We allowed the protein to adsorb onto the substrate for 10 minutes before pulling. The spring constant of each individual cantilever (MLCT, Bruker AFM Probes, Camarillo, CA) was determined experimentally by measuring the cantilever power spectrum and using the equipartition theorem [53,54] at the beginning of each experiment. All measurements were done at a constant velocity of 500 nm/s in PBS solution at room temperature. A worm-like chain (WLC) model [55] with a variable persistence length was fit to the first I27 peak to measure the contour-length (Lc) of the IK domain in the force extension data [56].
Circular Dichroism
The far UV CD spectra of the protein constructs were recorded on a Jasco J-815 Spectrometer. The protein concentration was 1 μM in 30 mM phosphate buffer pH 7.4, 100 mM KCl, 1 mM MgCl2, 1mM TCEP buffer. A 0.1 cm path length cuvette was used.
Generation of CRISPR/Cas9 edited nematodes
Strains PHX797 and PHX1257 were generated by SunyBiotech (http://www.sunybiotech.com) using CRISPR/Cas9 technology in the following ways. PHX797, unc-89(syb797), was created from wild type strain N2 to contain an HA tag at the N-terminus of the large UNC-89 isoforms, using sgRNAs with the following sequences: sg1: ccatcATGGCTAGTCGACGCCAAAAG, and sg2: cccaccttacccttaccatcATGG. PHX1257 unc-89(syb797syb1257) was created from PHX797 to contain a 4,614 bp in-frame deletion of the interkinase region, using sgRNAs with the following sequences: Sg1:CCTGTAGCACCGGAAGGTCGACG, Sg2:CCACCACCGACTGTGGAATACGT, Sg3:AACAGCAATTGAGAGAATTAAGG and Sg4:CCACAAAGAAGAATGATGATGGA. Sanger sequencing was used to verify the sequence alterations of each strain (Supplemental Figures 3 and 4). Both strains were outcrossed 5 times to wild type to remove most of the possible off target effects of CRISPR/Cas9; PHX797 to GB312, and PHX1257 to GB313.
Antibodies and western blots
We used the procedure of Hannak et al. [57] to prepare total protein lysates from wild type, unc-89(syb797) and unc-89(syb797syb1257) animals. Equal amounts of total protein from these strains were separated by a 5% separating and 3% stacking Laemmli SDS-PAGE gel, transferred to nitrocellulose membrane for 2 hours and reacted with anti-UNC-89 EU30 affinity-purified rabbit polyclonal antibodies [4] at 1:100 dilution, or with anti-HA antibodies (rabbit monoclonal antibody C29F4 from Cell Signaling Technology) at 1:100 dilution, followed by reaction with anti-mouse Ig-horseradish peroxidase, and ECL. For the EU30 antibody we used Pierce ECL Western Blotting Substrate (cat. no. 32106 from Thermo Scientific), but for the anti-HA antibody we needed the increased sensitivity of SuperSignal West Pico Chemiluminescent Substrate (cat. no. 34080, from Thermo Scientific). We found that using freshly prepared or freshly purchased acrylamide solutions (e.g. ProtoGel, cat. No. EC-890 from National Diagnostics) was absolutely essential to prevent horrible streaking of the protein bands.
Immunofluorescence microscopy
Adult worms were fixed as described previously [58,59]. For body wall muscle staining, these primary antibodies were used at the following dilutions: anti-myosin heavy chain A (MHC A; mouse monoclonal 5–6; [31]) at 1:200; anti-UNC-89 (mouse monoclonal MH42 [4, 44]) at 1:100; anti-UNC-95 (rabbit polyclonal Benian-13; [60]) at 1:200; and anti-ATN-1 (α-actinin; MH35; [61]) at 1:200. For pharyngeal muscle staining, the fixed worms were incubated with anti-UNC-89 (rabbit polyclonal EU30; [4]) at 1:100, and anti-myosin heavy chain C (MHC C; mouse monoclonal 9.2.1; [36]), at 1:50. MH35 antibodies were kindly provided by Dr. Pamela Hoppe (Western Michigan University). 5–6 and 9.2.1 antibodies were obtained from the Developmental Studies Hybridoma Bank, created by the NICHD of the NIH and maintained at The University of Iowa, Department of Biology, Iowa City, IA 52242. The secondary antibodies included anti-mouse IgG-Alexa 594 (Invitrogen A11032), and anti-rabbit IgG Alexa 488 (Invitrogen A21206), each at 1:200 dilution. Images shown in Figures 3–6, and the Supplemental Figure 5 were captured at room temperature with a Zeiss confocal system (LSM510) equipped with an Axiovert 100M microscope and an Apochromat ×63/1.4 numerical aperture oil objective. The color balances of the images were adjusted by using Adobe Photoshop. We examined the staining of at least 3 worms in each strain.
Super-resolution microscopy was performed with a Nikon N-SIM system in 3D structured illumination mode on an Eclipse Ti-E microscope equipped with a 100×/1.49 NA oil immersion objective, 488- and 561-nm solid-state lasers, and an EM-CCD camera (DU-897, Andor Technology). The color balances of the images were adjusted by using Adobe Photoshop (Adobe, San Jose, CA). A Z-series at 0.2 μm intervals was taken from the outer muscle cell membrane deeper into the muscle cell, as shown in Fig. 7.
Nematode locomotion assays
Swimming Assay
Day 2 adults from two, 6 cm NGM OP50 seeded plates were washed off the plates, washed free from bacteria and collected into M9 buffer such that the ratio of worms to buffer was 1:1. Two ml of M9 buffer was added to an unseeded 6 cm NGM plate where upon 5 μl of worm suspension was added to the center of the plate. Worms were allowed 5 minutes to adapt before a video recording of their swimming motions was made using a dissecting stereoscopic microscope fitted with a CMOS camera (Thorlabs). Ten, 10 second videos were recorded for each nematode strain from different sections of the plate, each video tracking the motion of ~10 worms. The video data was analyzed by Image J FIJI WrmTracker software [62] to obtain body bends per second (BBPS) from a total of 48–83 individual worms for each strain. The resulting worm tracks for each animal were observed, with immobilized animals in addition to upper and lower range outliers being removed from BBPS values for each strain prior to the statistical analysis. The final values for each strain were compared to wild type and tested for statistical significance using a Student T-test.
Crawling Assay
Day 2 adults were harvested as above, except that all washing steps used M9 buffer containing 0.2 g/L gelatin. 5 μl of worm suspension was added to the center of a 6 cm unseeded NGM plate, and the excess liquid was removed. After 5 minutes for adaptation, worm crawling was recorded using the above-mentioned strategy for extraction of body bends per second (BBPS) from a total of 66–90 individual worms for each strain. The resulting values for each strain were compared to wild type and tested for statistical significance using a Student T-test.
Measurement of C. elegans muscle force using NemaFlex
The maximum exertable muscle force by C. elegans strains was measured using the NemaFlex technique as previously described [34]. The technique is based on deflection of soft micropillars as the animals are crawling through the micropillar arena. The micropillar devices had pillars arranged in a square lattice with a pillar diameter of 44 μm and height of 87 μm. The gap between the pillars is 71 μm. Age synchronized day 1 adult animals were loaded individually in each chamber [63], and a 1-minute video was captured for each animal in a food-free environment at a temperature of 20 ± 1 °C. Imaging was performed in brightfield using a Nikon Ti-E microscope and Andor Zyla sCMOS 5.5 camera. Images were acquired at 5 frames per second with a 4x objective at a pixel resolution of 1.63 μm per pixel. Movies were then processed and analyzed for force values using our in-house-built image processing software (MATLAB, R2016a). Animal force measurements were calculated by identifying the maximal force exerted in each frame for an individual animal. We define the maximum exertable force f95 corresponding to the 95th percentile of these maximal forces. To account for differences in animal body diameter, we normalize f95 by the cube of animal body diameter [34]. To compare muscle forces of mutants with wild-type animals, we take the ratio of this normalized maximum exertable force and denote it as fold change in muscle force. Statistical analysis was performed using Wilcoxon rank-sum test.
Polarized light microscopy
Polarized light images were captured with a Zeiss Axioskop microscope equipped with 20X/0.40 Pol strain-free objective and an AxioCam MRm digital camera and software (Carl Zeiss, Jena, Germany).
Supplementary Material
Supplemental Fig. 1. Sequence alignment of IK regions of UNC-89, obscurin and SPEG using ClustalW. (https://www.genome.jp/tools-bin/clustalw).
Supplemental Fig. 2. Secondary structure predictions of IK regions of UNC-89, obscurin and SPEG using JPred. (http://www.compbio.dundee.ac.uk/jpred/index.html)
Supplemental Fig. 3. Results of Sanger sequencing and conceptual translation of PHX797, unc-89(syb797).
Supplemental Fig. 4. Results of Sanger sequencing and conceptual translation across genomic deletion in PHX1257, unc-89(syb797syb1257).
Supplementary Fig. 5. The body wall muscle of unc-89(ad539) has normally organized sarcomeres. Each panel shows representative confocal images of several body wall muscle cells stained with antibodies to the indicated sarcomeric components or with rhodamine-phalloidin for F-actin. Scale bar, 10 μm.
Supplementary Figure 6. Secondary structure of the I272-IK-I272 protein and a control I279 polyprotein determined by circular dichroism. (A) The overall secondary structure determined by Far-UV circular dichroism of the I272-IK-I272 used in AFM experiments (solid line) and a control protein containing 9 identical repeats of the I27 domain (dotted line). The protein concentration was 1 μM in 30 mM phosphate buffer pH 7.4, 100 mM KCl, 1 mM MgCl2, 1mM TCEP buffer. A 0.1 cm path length cuvette was used. (B) The % of β-strand, α-helical and random-coil content was estimated by using the BeStSel tool [64] and these are: 16% β-strand, 29% α-helix, 55% random for the IK construct (box on the right) and 40% β-strand, 20% α-helix, 36% random-coil for the I27 polyprotein (box on the left).
Supplementary Table 1. Detailed statistics for swimming and crawling assays on the different strains used in this study and summarized in Fig. 6A and B.
Supplementary Table 2: Muscle force and body diameter data for the different strains used in this study and summarized in Fig. 6C.
Highlights.
By single molecule force spectroscopy, the ~645 residue non-domain region lying between the two protein kinase domains of UNC-89 behaves as an elastic random coil
Nematodes lacking this non-domain region have disorganized sarcomeres and reduced whole animal locomotion and force generation
UNC-89 non-domain region is required for proper assembly or stability of A-bands from thick filaments
Acknowledgements
This study was supported in-part by a grant from the National Institutes of Health (R01 AR064307) to G.M.B. J.C.M. was supported in-part by the National Science Foundation-Graduate Research Fellowship Program (Grant #DGE 1444932). T.M. was partially supported by a grant from the National Institutes of Health (R01GM118534) to A.F.O. and G.M.B. L.L. and S.A.V. were partially supported by the National Aeronautics and Space Administration (Grant # NNX15AL16G).
Abbreviations:
- Ig domain
immunoglobulin domain
- Fn3 domain
fibronectin type 3 domain
- SPEG
striated muscle preferentially expressed gene
- HA
hemagglutinin
Footnotes
Credit Author Statement
Hiroshi Qadota conceptualized the project, obtained most of the data, analyzed most of the data, supervised other authors, wrote the original draft and reviewed and edited the manuscript. Jasmine Moody obtained some data, analyzed some data, wrote and edited several sections of the manuscript, and provided resources through her NSF fellowship. Leila Lesanpezeshki obtained some data, analyzed some data, and wrote and edited several sections of the manuscript. Taylor Moncrief obtained some data, analyzed some data, and wrote and edited several sections of the manuscript. Deborah Kitzler obtained some data, analyzed some data, and wrote and edited several sections of the manuscript. Purnima Devaki Bhat obtained some data, analyzed some data, and wrote and edited several sections of the manuscript. Siva A. Vanapalli helped with the conceptualization and data analysis, reviewed and edited the manuscript and provided some resources through existing grants. Andres F. Oberhauser obtained some data, analyzed some data, wrote and edited several sections of the manuscript and also performed overall editing of the manuscript and provided some resources through existing grants. Guy M. Benian was the corresponding author and was involved in the overall conceptualization of the project, obtained some data, was involved in data curation, data analysis, project administration, wrote multiple versions (editing) of the manuscript, and provided most of the resources through existing grants.
References
- [1].Waterston RH, Thomson JN, Brenner S, Mutants with altered muscle structure in C. elegans, Dev. Biol. 77 (1980) 271–302. [DOI] [PubMed] [Google Scholar]
- [2].Benian GM, Ayme-Southgate A, Tinley TL, The genetics and molecular biology of the titin/connectin-like proteins of invertebrates, Rev. Physio. Biochem. Pharm 138 (1999) 235–268. [DOI] [PubMed] [Google Scholar]
- [3].Small TM, Gernert KM, Flaherty DB, Mercer KB, Borodovsky M, Benian GM, Three new isoforms of C. elegans UNC-89 containing MLCK-like protein kinase domains, J. Mol. Biol. 342 (2004) 91–108. [DOI] [PubMed] [Google Scholar]
- [4].Benian GM, Tinley TL, Tang X, Borodovsky M, The Caenorhabditis elegans gene unc-89, required for muscle M-line assembly, encodes a giant modular protein composed of Ig and signal transduction domains, J. Cell Biol. 132 (1996) 835–848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Ferrara TM, Flaherty DB, Benian GM, Titin / connectin-related proteins in C. elegans: a review and new findings, J. Musc. Res. Cell Motil. 26 (2005) 435–447. [DOI] [PubMed] [Google Scholar]
- [6].Ono K, Yu R, Ono S, Structural components of the nonstriated contractile apparatuses in the C. elegans gonadal myoepithelial sheath and their essential roles for ovulation, Dev. Dyn. (2007) 236, 1093–1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Qadota H, McGaha LA, Mercer KB, Stark TJ, Ferrara TM, Benian GM, A novel protein phosphatase is a binding partner for the protein kinase domains of UNC-89 (obscurin) in C. elegans, Mol. Biol. Cell 19 (2008a) 2424–2432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Qadota H, Blangy A, Xiong G, Benian GM, The DH-PH region of the giant protein UNC-89 activates RHO-1 GTPase in C. elegans body wall muscle, J. Mol. Biol. 383 (2008b) 747–752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Xiong G, Qadota H, Mercer KB, McGaha LA, Oberhauser AF, Benian GM, A LIM-9 (FHL) / SCPL-1 (SCP) complex interacts with the C-terminal protein kinase regions of UNC-89 (obscurin) in C. elegans muscle, J. Mol. Biol. 386 (2009) 976–988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Wilson KJ, Qadota H, Mains PE, Benian GM, UNC-89 (obscurin) binds to MEL-26, a BTB-domain protein, and affects the function of MEI-1 (katanin) in striated muscle of C. elegans, Mol. Biol. Cell 23 (2012) 2623–2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Warner A, Xiong G, Qadota H, Rogalski T, Vogl AW, Moerman DG, Benian GM, CPNA-1, a copine domain protein, is located at integrin adhesion sites and is required for myofilament stability in Caenorhabditis elegans, Mol. Biol. Cell 24 (2013) 601–616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Qadota H, Mayans O, Matsunaga Y, McMurry JL, Wilson KJ, Kwon GE, Stanford R, Deehan K, Tinley TL, Ngwa VM, Benian GM, The SH3 domain of UNC-89 (obscurin) interacts with paramyosin, a coiled-coil protein, in Caenorhabditis elegans muscle, Mol. Biol. Cell 27 (2016) 1606–1620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Qadota H, Matsunaga Y, Bagchi P, Lange KI, Carrier KJ, Pols WV, Swartzbaugh E, Wilson KJ, Srayko M, Pallas DC, Benian GM, Protein phosphatase 2A is crucial for sarcomere organization in Caenorhabditis elegans striated muscle, Mol. Biol. Cell 29 (2018) 2084–2097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Bang ML, Centner T, Fornoff F, Geach AJ, Gotthardt M, McNabb M, Witt CC, Labeit D, Gregorio CC, Granzier H, Labeit S, The complete gene sequence of titin, expression of an unusual approximately 700-kDa titin isoform, and its interaction with obscurin identify a novel Z-line to I-band linking system, Circ. Res. 89 (2001) 1065–1072. [DOI] [PubMed] [Google Scholar]
- [15].Young P, Ehler E, Gautel M, Obscurin, a giant sarcomeric Rho guanine nucleotide exchange factor protein involved in sarcomere assembly, J. Cell Biol. 154 (2001) 123–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Perry NA, Ackermann MA, Shriver M, Hu LY, Kontrogianni-Konstantopoulos A, Obscurins: unassuming giants enter the spotlight, IUBMB Life 65 (2013) 479–486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Bowman AL, Kontrogianni-Konstantopoulos A, Hirsch SS, Geisler SB, Gonzalez-Serratos H, Russell MW, Bloch RJ, Different obscurin isoforms localize to distinct sites at sarcomeres, FEBS Lett. 581 (2007) 1549–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Grogan A, Kontrogianni-Konstantopoulos A, Unraveling obscurins in heart disease, Pflugers Arch. 471 (2018) 735–743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Kontrogianni-Konstantopoulos A, Jones EM, Van Rossum DB, Bloch RJ RJ., Obscurin is a ligand for small ankyrin 1 in skeletal muscle, Mol Biol Cell. 14 (2003) 1138–1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Bagnato P, Barone V, Giacomello E, Rossi D, Sorrentino V, Binding of an ankyrin-1 isoform to obscurin suggests a molecular link between the sarcoplasmic reticulum and myofibrils in striated muscles, J Cell Biol. 160 (2003) 245–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Lange S, Ouyang K, Meyer G, Cui L, Cheng H, Lieber RL, Chen J, Obscurin determines the architecture of the longitudinal sarcoplasmic reticulum, J Cell Sci. 122 (2009) 2640–2650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Spooner PM, Bonner J, Maricq AV, Benian GM, Norman KR, Large isoforms of UNC-89 (obscurin) are required for muscle cell architecture and optimal calcium release in Caenorhabditis elegans, PLoS ONE 7(7)(2012) e40182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Geisler SB, Robinson D, Hauringa M, Raeker MO, Borisov AB, Westfall MV, Russell MW, Obscurin-like 1, OBSL1, is a novel cytoskeletal protein related to obscurin, Genomics 89 (2007) 521–531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Fukuzawa A, Lange S, Holt M, Vihola A, Carmignac V, Ferreiro A, Udd B, Gautel M M, Interactions with titin and myomesin target obscurin and obscurin-like 1 to the M-band: implications for hereditary myopathies, J. Cell Sci. 121 (2008) 1841–1851. [DOI] [PubMed] [Google Scholar]
- [25].Hanson D, Murray PG, Sud A, Temtamy SA, Aglan M, Superti-Furga A, Holder SE, Urquhart J, Hilton E, Manson FD, Scambler P, Black GC, Clayton PE, The primordial growth disorder 3-M syndrome connects ubiquitination to the cytoskeletal adaptor OBSL1, Am. J. Hum. Genet. 84 (2009) 801–806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Agrawal PB, Pierson CR, Joshi M, Liu X, Ravenscroft G, Moghadaszadeh B, Talabere T, Viola M, Swanson LC, Haliloğlu G, Talim B, Yau KS, Allcock RJ, Laing NG, Perrella MA, Beggs AH, SPEG interacts with myotubularin, and its deficiency causes centronuclear myopathy with dilated cardiomyopathy, Am. J. Hum. Genet. 95 (2014) 218–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Mayans O, Benian GM, Simkovic F, Rigden DJ, Mechanistic and functional diversity in the mechanosensory kinases of the titin-like family, Biochem. Soc. Trans. 41 (2013) 1066–1071. [DOI] [PubMed] [Google Scholar]
- [28].Hu LY, Kontrogianni-Konstantopoulos A, The kinase domains of obscurin interact with intercellular adhesion proteins, FASEB J. 27 (2013) 2001–2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Quick AP, Wang Q, Philippen LE, Barreto-Torres G, Chiang DY, Beavers D, Wang G, Khalid M, Reynolds JO, Campbell HM, Showell J, McCauley MD, Scholten A, Wehrens XH, SPEG (Striated Muscle Preferentially Expressed Protein Kinase) Is Essential for Cardiac Function by Regulating Junctional Membrane Complex Activity, Circ. Res. 120 (2016) 110–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Drozdetskiy A, Cole C, Procter J, Barton GJ, JPred4: a protein secondary structure prediction server, Nucl. Acids Res. 43 (2015) W389–W394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Miller DM, Ortiz I, Berliner GC, Epstein HF, Differential localization of two myosins within nematode thick filaments, Cell 34 (1983) 477–490. [DOI] [PubMed] [Google Scholar]
- [32].Qadota H, Matsunaga Y, Nguyen KCQ, Mattheyses A, Hall DH, Benian GM, High-resolution imaging of muscle attachment structures in C. elegans, Cytoskeleton (2017) 1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Waterston RH, Muscle, In The Nematode Caenorhabditis elegans (1988) (Wood WB, ed), pp. 281–335, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y. [Google Scholar]
- [34].Rahman M, Hewitt JE, Van-Bussel F, Edwards H, Blawzdziewicz J, Szewczyk NJ, Driscoll M, Vanapalli SA, NemaFlex: a microfluidics-based technology for standardized measurement of muscular strength of C. elegans, Lab on a Chip 18.15 (2018) 2187–2201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Avery L, The genetics of feeding in C. elegans, Genetics 133 (1993) 897–917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Miller DM, Stockdale FE, Karn J, Immunological identification of the genes encoding the four myosin heavy chain isoforms of C. elegans, Proc. Natl. Acad. Sci. 83 (1986) 2305–2309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Li H, Linke WA, Oberhauser AF, Carrion-Vazquez M, Kerkvliet JG, Lu H, Marszalek PE, Fernandez JM, Reverse engineering of the giant muscle protein titin, Nature 418 (2002) 998–1002. [DOI] [PubMed] [Google Scholar]
- [38].Granzier H, Labeit S, Cardiac titin: an adjustable multi-functional spring, J. Physiol. 541(2002) 335–342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Oroz J, Bruix M, Laurents DV, Galera-Prat A, Schonfelder J, Canada FJ, Carrion-Vasquez M, The Y9P variant of the titin I27 module: structural determinants of its revisited nanomechanics, Structure 24 (2016) 606–616. [DOI] [PubMed] [Google Scholar]
- [40].Moerman DG, Williams BD, Sarcomere assembly in C. elegans muscle, WormBook; (2006) 1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Gieseler K, Qadota H, Benian GM, Development, structure, and maintenance of C. elegans body wall muscle, WormBook; (2017) 1–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Lin X, Qadota H, Moerman DG, Williams BD, elegans PAT-C 6/actopaxin plays a critical role in the assembly of integrin adhesion complexes in vivo, Curr. Biol. 13(2003) 922–932. [DOI] [PubMed] [Google Scholar]
- [43].von Castelmur E, Marino M, Svergun DI, Kreplak L, Ucurum-Fotiadis Z, Konarev PV, Urzhumtsev A, Labeit D, Labeit S, Mayans O, A regular pattern of Ig super-motifs defines segmental flexibility as the elastic mechanism of the titin chain, Proc. Natl. Acad. Sci. U S A. 105 (2008) 1186–1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Hresko MC, Williams BD, Waterston RH, Assembly of body wall muscle and muscle cell attachment structures in Caenorhabditis elegans, J. Cell Biol. 124 (1994) 491–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Agarkova I, Perriard J-C, The M-band: an elastic web that crosslinks thick filaments in the center of the sarcomere, Trends Cell Biol. 15 (2005) 477–485. [DOI] [PubMed] [Google Scholar]
- [46].Lange S, Pinotsis N, Agarkova I, Ehler E, The M-band: The underestimated part of the sarcomere, BBA-Mol. Cell Res. 1867 (2020) 118440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Berkemeier F, Bertz M, Xiao S, Pinotsis N, Wilmanns M, Grater F, Rief M, Fast-folding alpha-helices as reversible strain absorbers in the muscle protein myomesin, Proc. Natl. Acad. Sci. USA 108 (2011) 14139–14144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Pernigo S, Fukuzawa A, Beedle AEM, Holt M, Round A, Pandini A, Garcia-Manyes S, Gautel M, Steiner RA, Binding of myomesin to obscurin-like-1 at the muscle M-band provides a strategy for isoform-specific mechanical protection, Structure 25 (2017) 107–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Mango S, The C. elegans pharynx: a model for organogenesis. WormBook, ed. The C. elegans Research Community, WormBook, 10.1895/wormbook.1.129.1 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Bujalowski PJ, Oberhauser AF, Tracking unfolding and refolding reactions of single proteins using atomic force microscopy methods, Methods 60 (2013) 151–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Rabbi M, Marszalek PE, Measuring Protein Mechanics by Atomic Force Microscopy [DOI] [PubMed] [Google Scholar]
- [52].Hossain MD, Furuike S, Kinosita K Jr, The rotor tip inside a bearing of a thermophilic F1-ATPase is dispensable for torque generation, Biophys. J. 90 (2006) 4195–4203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Florin E-L, Rief M, Lehmann H, Ludwig M, Dornmair C, Moy VT, Gaub HE, Sensing specific molecular interactions with the atomic force microscope. Biosens. and Bioelectr. 10 (1995) 895–901. [Google Scholar]
- [54].Kim M, Abdi K, Marszalek PE, Fast and forceful refolding of stretched alpha-helical solenoid proteins, Biophys. J. 98 (2010) 3086–3092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Bustamante C, Marko JF, Smith S S, Entropic elasticity of lambda-phage DNA. Science 265 (1994) 1599–1600. [DOI] [PubMed] [Google Scholar]
- [56].Marko JF, Siggia ED, Stretching DNA, Macromolecules 28 (1995) 8759–8770. [Google Scholar]
- [57].Hannak E, Oegema K, Kirkham M, Gonczy P, Habermann B, Hyman AA, The kinetically dominant assembly pathway for centrosomal asters in Caenorhabditis elegans is gamma-tubulin dependent, J. Cell Biol. 157 (2002) 591–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Nonet ML, Grundahl K, Meyer BJ, Rand JB, Synaptic function is impaired but not eliminated in C. elegans mutants lacking synaptotagmin, Cell 73 (1993) 1291–1305. [DOI] [PubMed] [Google Scholar]
- [59].Wilson KJ, Qadota H, Benian GM, Immunofluorescent localization of proteins in Caenorhabditis elegans muscle, Methods Mol. Biol. 798 (2012) 171–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Qadota H, Mercer KB, Miller RK, Kaibuchi K, Benian GM, Two LIM domain proteins and UNC-96 link UNC-97/pinch to myosin thick filaments in Caenorhabditis elegans muscle, Mol. Biol. Cell 18 (2007) 4317–4326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Francis R, Waterston RH, Muscle cell attachment in Caenorhabditis elegans, J. Cell Biol. 114 (1991) 465–479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Nussbaum-Krammer C, Neto MF, Brielmann RM, Pedersen JS, Morimoto RI, Investigating the spreading of toxicity of prion-like proteins using the metazoan model organism C. elegans, J. Vis. Exp. 95 (2015) 52321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Hewitt JE, Pollard AK, Lesanpezeshki L, Deane CS, Gaffney CJ, Etheridge T, Szewczyk NJ, Vanapalli SA, Muscle strength deficiency and mitochondrial dysfunction in a muscular dystrophy model of Caenorhabditis elegans and its functional response to drugs, Disease Mod. & Mechan. 11.12 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Micsonai A, Wien F, Bulyaki E, Kun J, Moussong E, Lee Y-H, Goto Y, Refregiers M, Kardos J, BeStSel: a web server for accurate protein secondary structure prediction and fold recognition from the circular dichroism spectra, Nucl. Acids Res. 46 (2018) W315–W322. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Fig. 1. Sequence alignment of IK regions of UNC-89, obscurin and SPEG using ClustalW. (https://www.genome.jp/tools-bin/clustalw).
Supplemental Fig. 2. Secondary structure predictions of IK regions of UNC-89, obscurin and SPEG using JPred. (http://www.compbio.dundee.ac.uk/jpred/index.html)
Supplemental Fig. 3. Results of Sanger sequencing and conceptual translation of PHX797, unc-89(syb797).
Supplemental Fig. 4. Results of Sanger sequencing and conceptual translation across genomic deletion in PHX1257, unc-89(syb797syb1257).
Supplementary Fig. 5. The body wall muscle of unc-89(ad539) has normally organized sarcomeres. Each panel shows representative confocal images of several body wall muscle cells stained with antibodies to the indicated sarcomeric components or with rhodamine-phalloidin for F-actin. Scale bar, 10 μm.
Supplementary Figure 6. Secondary structure of the I272-IK-I272 protein and a control I279 polyprotein determined by circular dichroism. (A) The overall secondary structure determined by Far-UV circular dichroism of the I272-IK-I272 used in AFM experiments (solid line) and a control protein containing 9 identical repeats of the I27 domain (dotted line). The protein concentration was 1 μM in 30 mM phosphate buffer pH 7.4, 100 mM KCl, 1 mM MgCl2, 1mM TCEP buffer. A 0.1 cm path length cuvette was used. (B) The % of β-strand, α-helical and random-coil content was estimated by using the BeStSel tool [64] and these are: 16% β-strand, 29% α-helix, 55% random for the IK construct (box on the right) and 40% β-strand, 20% α-helix, 36% random-coil for the I27 polyprotein (box on the left).
Supplementary Table 1. Detailed statistics for swimming and crawling assays on the different strains used in this study and summarized in Fig. 6A and B.
Supplementary Table 2: Muscle force and body diameter data for the different strains used in this study and summarized in Fig. 6C.


