Abstract
Photoacoustic (PA) imaging has emerged as a powerful technique for the high resolution visualization of biological processes within deep tissue. Through the development and application of exogenous targeted contrast agents and activatable probes that can respond to a given cancer biomarker, researchers can image molecular events in vivo during cancer progression. This information can provide valuable details that can facilitate cancer diagnosis and therapy monitoring. In this tutorial review, we provide a step-by-step guide to select a cancer biomarker and subsequent approaches to design imaging agents for in vivo use. We envision this information will be a useful summary to those in the field, new members to the community, and graduate students taking advanced imaging coursework. We also highlight notable examples from the recent literature, with emphasis on the molecular designs and their in vivo PA imaging performance. To conclude, we provide our outlook and future perspective in this exciting field.
Graphical Abstract
1. Introduction
Photoacoustic (PA) imaging is an innovative technology that is characterized by the application of light to induce the generation of ultrasonic signals via the photoacoustic effect.1 To be more specific, the photoacoustic effect starts with the excitation of a chromophore with pulsed light, followed by the vibrational relaxation to release energy as heat, which leads to the local thermal expansion and the generation of pressure waves that is detected as sound. Owing to the unique ability of sound (at clinically relevant frequencies) to propagate through tissue with minimal attenuation, PA imaging is suitable for deep tissue applications. Moreover, by considering the speed at which sound travels through the body and the time it takes for a given signal to reach an ultrasound detector, the origin of the signal can be accurately determined to afford high resolution images. Although any light-absorbing material can be used for PA imaging, those that interact with photons in the near-infrared range (NIR) (>650 nm) are preferred for in vivo applications to limit background interference from PA-active molecules. Of note, early PA imaging studies utilized endogenous pigments such as the different forms of hemoglobin to determine the levels of oxygenation in blood. Likewise, melanin was employed to provide contrast which enabled the tracking of skin cancer cells. 2–6 However, beyond these representative examples, label-free imaging is limited by the variety of targets that can be visualized. For instance, most biomarkers (e.g., protein receptors), as well as molecular events (e.g., respiratory burst) are completely invisible to label-free PA imaging.
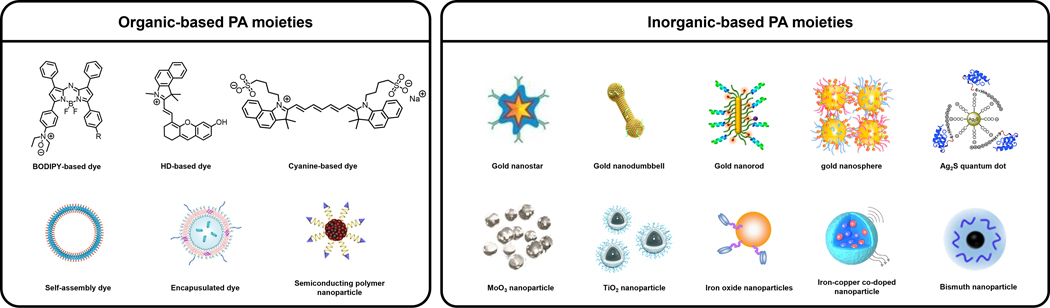
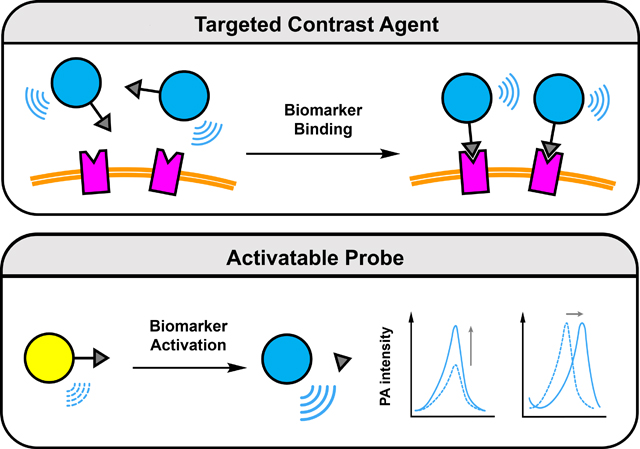
To satisfy the demand to be able to perform molecular PA imaging, a large assortment of targeted contrast agents and activatable probes (also known as acoustogenic probes) have been developed over the past decade.7–10Molecules belonging to both categories must feature a strong PA signal generating unit. Representative examples include dyes, self-assembled dye-based nanoparticles, encapsulated photoabsorbers, semiconducting polymer nanoparticles, and inorganic structures to name a few (Figure 1). In addition, targeted contrast agents are also equipped with a molecular recognition element, often referred to as a ligand, which can bind to the imaging target (e.g., cell surface protein) with high selectivity and affinity. Since this interaction does not necessarily alter the PA signal intensity, contrast is achieved through accumulation in the region of interest which is enhanced when unbound contrast agent clears from the body. On the contrary, activatable probes are often designed using the principles of activity-based sensing (ABS) which leverages the unique chemical reactivity of the imaging target to induce a change in the PA generating unit (e.g., signal enhancement, ratiometric response) upon reacting with a responsive trigger.11, 12 Regardless of the classification, imaging agents for PA have shown tremendous utility as powerful chemical biology tools that can facilitate basic scientific research. Moreover, their potential for use in human patients for diagnostic testing, real-time therapy monitoring, and surgical guidance, can have a profound impact on how various disease states are managed. For instance, in the context of cancer, the ability to visualize a primary tumor in three dimensions can inform the extent of tissue invasion prior to surgery. Likewise, non-invasively determining the presence of a biomarker or reporting on the extent of a critical cancer property such as oxygenation, is an unmet need in clinical imaging that can be addressed using PA.
Figure 1.
A summarization of representative PA moieties in this review. Reproduced from ref. 52 which is an open-access article distributed under the Creative Commons Attribution (CC BY-NC) License, 2015 Ivyspring International Publisher; from ref. 53 with permission from American Chemical Society, Copyright 2018; from ref. 55 with permission from Royal Society of Chemistry, Copyright 2021; from ref. 67 with permission from Elsevier, Copyright 2015; from ref. 70 with permission from Royal Society of Chemistry, Copyright 2018; from ref. 82 with permission from Springer Nature, Copyright 2011; from ref. 101 with permission from Springer Nature, Copyright 2019; from ref. 120 with permission from Elsevier, Copyright 2017; from ref.141 with permission from Elsevier, Copyright 2020; from ref. 145 with permission from American Chemical Society, Copyright 2017; from ref. 191 with permission from Elsevier, Copyright 2019; from ref. 287 which is an open-access article distributed under the Creative Commons Attribution (CC BY-NC) License, 2021 AAAS; from ref. 302 with permission from American Chemical Society, Copyright 2021.
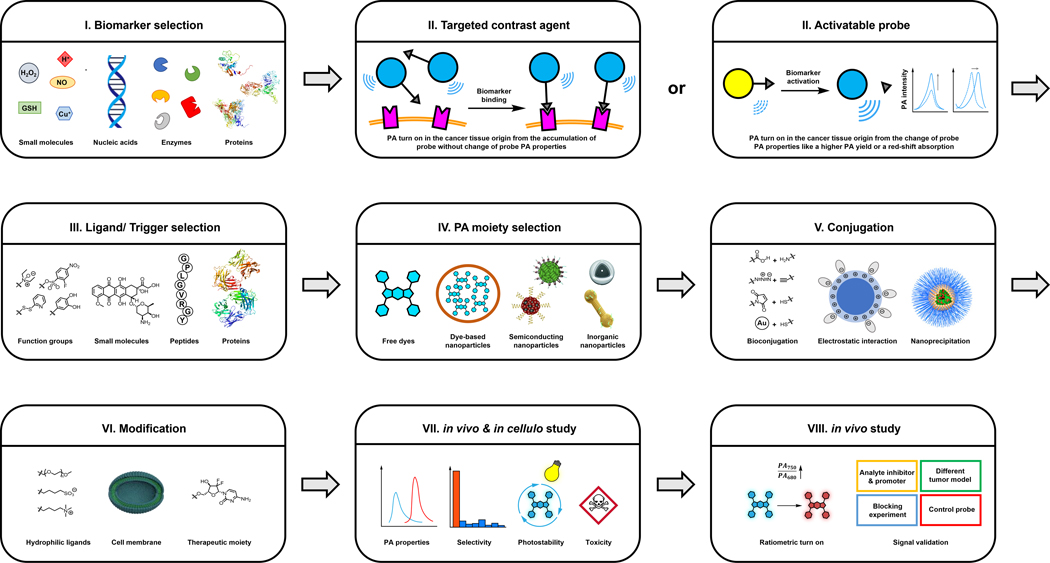
In this tutorial review, we will provide readers with a comprehensive step-by-step guide that can be used to aid in the selection of a cancer biomarker and the design of both targeted contrast agents and activatable probes for cancer. We elected to focus on cancer imaging owing to its impact on human health. Additionally, biomarkers for this group of like diseases are often not exclusive to a specific cancer type (and can be present in healthy tissue). Thus, we envision a detailed workflow describing the characteristics one must consider when selecting an imaging target and how to subsequently utilize this information to design an imaging agent would be highly beneficial. Each section will begin with a brief discussion detailing specific properties of the imaging target with respect to cancer. We will then highlight select examples for each imaging target while focusing on the key design elements that are present and discuss the corresponding in vivo PA applications. Lastly, we will provide our outlook and future perspective for the field.
2. Cancer Biomarker Selection
A cancer biomarker is defined as any analyte or cancer-associated process that when detected, may indicate the presence of cancer within a patient. On the contrary to biomarkers found in bodily fluids such as blood and urine, one that is suitable for molecular imaging must be situated close to or within the tumor region. Indeed, careful consideration is imperative when deciding whether a biomarker is an appropriate target for imaging agent development.
As a first step, one must recognize that it is rare for a biomarker to be exclusively associated with a given cancer type. Specifically, cancer cells are derived from normal cells after a sufficient number of mutations have accumulated (typically in oncogenes).13–15 Thus, it is not uncommon to see terms such as “elevated” or “overexpressed” in the literature when describing a biomarker. As such, one must approach this with caution since selecting an analyte or process that is prevalent will increase the likelihood of a false positive result. For instance, the prostate-specific antigen (PSA) screening test is notorious for drawing wrong conclusions.16 While prostate tumors do often produce high levels of PSA, so too does a urinary tract infection, taking certain medications, or even after a vigorous exercise session. Therefore, we recommend one must consider other conditions that may be associated with a potential imaging target.
If a biomarker is protein-based, an excellent starting point is to review the literature or The Human Protein Atlas (accessible via https://www.proteinatlas.org/) to compare the expression level in cancer with normal tissue. We recommend that the minimum difference should be at least 1.5-fold since it would be extremely challenging to distinguish between normal and cancerous levels if the recommended threshold was lower. Strategies will be discussed below that can improve accuracy while limiting the rate of false positives (see section 3). With regards to non-protein biomarkers such as a fleeting reactive oxygen or nitrogen species, it will be important to search for indirect clues of its levels in cancer. For instance, peroxynitrite (ONOO-) that results from the biomolecular reaction between superoxide and nitric oxide is believed to be elevated in tumors since cancer cells are often characterized by oxidative stress (leads to high superoxide) and inflammation (high nitric oxide). Since peroxynitrite can nitrosate ortho to the phenolic alcohol of a tyrosine residue found within a protein, determining the levels of nitrosated proteins can provide vital information to help decide.17 In the event one wishes to design an imaging agent to detect a novel biomarker, performing the requisite validation studies are essential. This includes rigorous in vitro testing to demonstrate target engagement, selectivity assays to determine the extent of potential off-target effects, and the development of matching control imaging agents lacking the design component that responds to or recognizes the target. Next, detection of the biomarker should be validated in live cells where a result can be confirmed using ‘blocking’ agents (e.g., small-molecule inhibitor if the target is a protein). Alternatively, one should consider experiments that increase and/or decrease the concentration of the biomarker to assess whether the signal changes accordingly. This can occur using genetic manipulation or exogenous supplementation, respectively. Lastly, after successfully transitioning into live subjects, post-imaging confirmation studies in the form of ex vivo analyses should be performed.
3. Design of PA Imaging Agents
There are many design options that one can consider after a cancer biomarker has been selected. However, it is important to first determine if the target is extracellular (accessible within the TME), localized on the cell surface, or found intracellularly. For instance, an imaging agent should ideally not be cell permeable if the cancer biomarker is extracellular since uptake into adjacent cells can result in non-specific staining. Likewise, for intracellular biomarkers, an imaging agent should be able to readily enter the cell and exhibit a propensity to be retained (at least for the duration of the imaging session). The next consideration is whether the biomarker can catalyze an enzymatic reaction or mediate a chemical transformation. In the event that this is not within the scope of the biomarker, one should consider pursuing a targeted contrast agent (TCA) design.
As mentioned in the introduction, there are many scaffolds capable of generating a strong PA signal upon irradiation. Depending on the chosen scaffold, TCAs are either small-molecule- or nanoparticle-based. For more details, we refer readers to the following review article.18 Of note, the ability to harvest light in the NIR-I (650–900 nm) and/or NIR-II (950–1300 nm) windows is an important requirement since photons in these ranges are safe and can reach deeper in tissue relative to ultraviolet and visible light. Decoration of a nanoparticle-based TCA with components such as surfactants or water solubilizing groups (e.g., PEG) may be necessary if it displays poor dispersion in water or if there is propensity to coalesce, precipitate out of solution, or form protein coronas when in vivo. In the penultimate step, one must determine if a well-annotated recognition element (ligand) is available. This may come in the form of a small molecule inhibitor (or binder), peptide, protein, affibody, or antibody. In our opinion, the corresponding binding affinity should be at least in the low micromolar range. Further, the targeting ligand should be highly selective, exhibition a minimal propensity to bind to interfering species beyond the target biomarker. For instance, cyclooxygenases are enzymes involved in the conversion of arachidonic acid to prostaglandin H2, which serves as the precursor to inflammatory molecules such as prostaglandins.19 One isoform is constitutively expressed, while a second isoform, cyclooxygenase 2, is elevated in cancer. As such, handpicking a selective small molecule inhibitor for the latter enzyme is critical if the intended goal is cancer imaging.20, 21 Finally, a strategy must be chosen to append the selected targeting ligand to the rest of the imaging agent. Conventional bioconjugation strategies such as amide coupling, maleimide chemistry, or ‘click’ reactions (e.g., Cu-catalyzed alkyne-azide coupling) may be appropriate if there is chemical compatibility with the functional groups present. An alternative strategy is to append the targeting ligand to a secondary component that can embed into the nanoparticle through hydrophobic or hydrophilic contacts. Along these lines, electrostatic interactions can also be employed to attach the targeting ligand, however, one must consider how stable this interaction will be as it encounters pH and ionic strength gradients within the body. After assembling a TCA, its PA properties, solubility, and stability should be tested in vitro. Likewise, in cellulo studies must be performed to determine cytotoxicity and validate target specificity and engagement. This should be followed by in vivo studies using at least one relevant cancer model where the biomarker is present to assess targeting, TCA accumulation, and clearance. Finally, to confirm an imaging result, we recommend the design of robust control experiments (like blocking), PA spectrum profile analysis and ex vivo assays.
With regards to the subset of biomarkers that can mediate the activation of a probe, one should utilize an ABS design strategy. As mentioned previously, ABS is different from the molecular recognition and stoichiometric binding strategies discussed above. This approach leverages the unique chemical reactivity of the biomarker to induce a change in the PA signal. For instance, proteolytic activity of enzymes such as proteases can be employed to cleave a substrate element which can unmask a latent dye or expose a key functional group that can react further to form a new photoabsorber in situ. An alternative approach that can also yield a signal change is if the enzymatic reaction changes the solubility of the corresponding product which can lead to self-assembly. Of note, if there is no product inhibition that results from turnover of the substrate, one enzyme can in theory catalyze the activation of many probe molecules, presenting an exciting opportunity to generate an enhanced signal. The same can be said for non-enzyme biomarkers including metal ions, reactive oxygen and nitrogen species, and thiols to name a few which can mediate a chemical transformation. It is noteworthy that the reactivity of the probe for these biomarkers can be adjusted to match the concentration found in cancer. For instance, if a biomarker is abundant, the reaction can be attenuated to limit false positive results from activation in healthy tissue. Alternatively, tuning can be employed to speed up a reaction for targets that are fleeting or low abundance. As a first step in selecting the appropriate reaction, we recommend that one should survey the following reviews, as well as other appropriate articles in the literature.11, 12 An alternative is draw from the extensive chemical knowledge or consider prominent organic named reactions.22 However, it is imperative to consider biocompatibility as many of these transformations are optimized for abiotic conditions (e.g., organic solvents and non-physiological temperatures). As with TCA, once an activatable probe has been developed, one should make sure the probe exhibits excellent PA properties in vitro to increase the likelihood of high-quality PA imaging in vivo. Additionally, the dose- and time-dependent response towards the target analyte should be established. This will provide vital information to predict the in vivo limit of detection and time required for probe activation, respectively. Owing to similar reactivity between certain biomarkers like members of the ROS family, careful evaluation of probe selectivity can help to avoid a false positive result. In cellulo studies are also required to obtain preliminary data like cytotoxicity that is important for related in vivo work. In the subsequent in vivo studies, beyond demonstrating probe activation in the tumor region, reliable conclusions can be achieved if one can modify the levels of an analyte through the application of inhibitor, scavenger, donor, or promoter to show that there is a correlation between the PA signal intensity and the analyte concentration. Finally, ex vivo biodistribution studies would also be necessary to determine potential off-target effects that may not be apparent during an imaging session.
4. Targeted Contrast Agents for Photoacoustic Imaging
In this section, we will focus our efforts on describing an impressive range of innovative design strategies that have been employed to develop TCAs for PA imaging. In general, a contrast agent can preferentially localize to a region of interest such as a primary tumor, sentinel lymph node, or metastatic lesion to provide a robust PA signal that can be used to distinguish between healthy adjacent tissue. Such information can then be used for diagnostic purposes or leveraged during image-guided surgery where the ability to reliably highlight an afflicted area can minimize tumor margins and ensure all cancerous tissue has been removed during a surgical debulking procedure. However, it is notable that many of the contrast agents that exist for this modality operate via passive accumulation mechanisms (e.g., the enhanced permeability and retention (EPR) effect) and thus, cannot be utilized to report on molecular details.
On the contrary, a TCA features two key elements which include a photoabsorber that can convert absorbed light into an ultrasonic readout via the PA effect and a targeting unit for biomarker recognition and binding. A photoabsorber can come in many forms since any light absorbing molecule can in theory generate a PA signal. Likewise, a variety of biomarkers have been targeted for cancer which include cell surface proteins, channels, as well as intracellular components. We have surveyed the recent literature, identified studies that apply TCAs for various cancer applications via PA imaging and highlighted select work based on targets listed below. Owing to the diversity, we have organized the various targets based on alphabetical order. References to additional biomarkers not covered in this section are also provided.23–39
4.1. B7-H3 (CD276)
Cluster of Differentiation (CD276) or B7 Homolog 3 (B7-H3) is a human protein receptor that remains at low levels in normal cells but is overexpressed in malignant tissues.40 Through a variety of mechanisms, the expression of B7-H3 is involved in tumor growth, metastasis, and evasion of the immune system.41 For these reasons, TCAs that interact with this protein have been developed to diagnose and identify cancer including those of the cervix, renal, lungs, ovaries, prostate, breast, and bladder, which have all been found to upregulate the expression of this cancer biomarker.42, 43
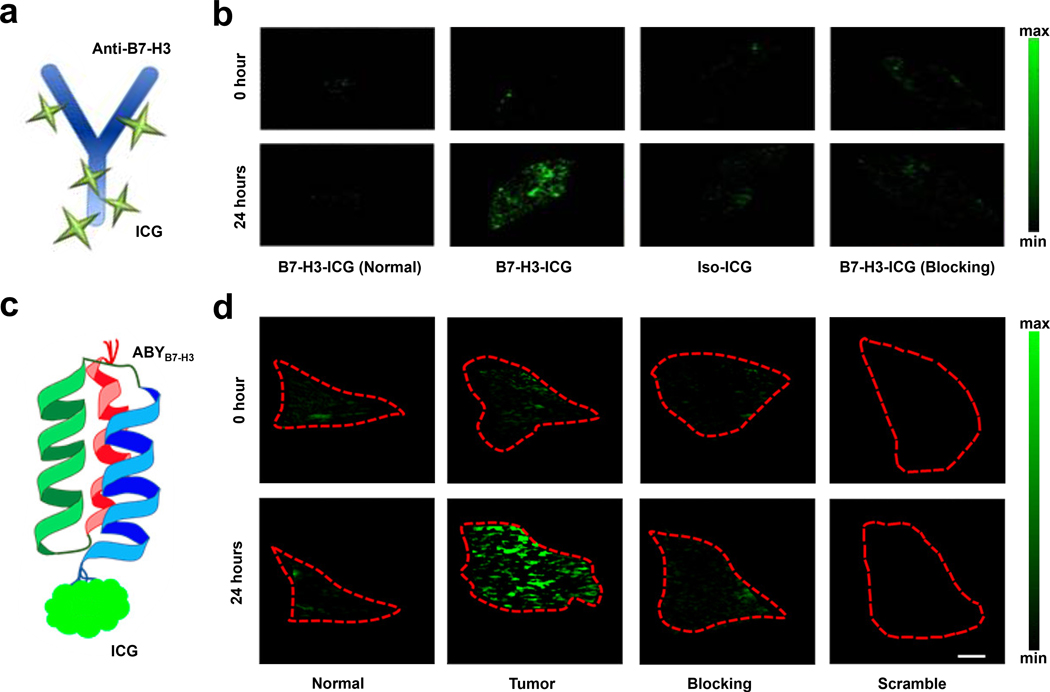
In one example, Willman and coworkers sought to develop a TCA capable of distinguishing between normal tissue and breast cancer tissue using a B7-H3-targeted antibodies conjugated to an indocyanine green (ICG) chromophore (Figure 3a).42 In vivo PA studies were conducted in transgenic breast cancer mice. Images were collected pre-injection, 24, 48, 72, and 96 hours post tail-vein injection of either the TCA (B7-H3-ICG) or control agent developed by conjugating a non-specific antibody to ICG (isotype-ICG). The PA signal in glands that contain breast cancer tissue were 3.01-fold higher relative to tissue before injection with the TCA. Moreover, none of the control conditions led to a statistically significant increase in the PA signal (Figure 3b). To confirm these results, ex vivo studies were conducted by excising breast tissue for histopathological staining which showed B7-H3 expression was elevated in the breast tumor. Researchers were also able to distinguish between their unbound TCA, non-targeted control agents, endogenous photoabsorbers, and TCA that has engaged with the receptor due to a shift in optical absorbance that occurs via endocytosis of the TCA.
Figure 3.
a) Schematic of B7-H3-ICG. b) PA images of normal mice before and after injection of B7-H3-ICG (first column) and breast cancer-bearing mice before and after injection with B7-H3-ICG, iso-ICG or B7-H3-ICG with blocking. c) Schematic of ABYB7-H3-ICG. d) PA images of normal glands before and after administration of ABYB7-H3-ICG (first column) and mammary tumors before and after administration of ABYB7-H3-ICG, ABYB7-H3-ICG with blocking, and ABYSCR-ICG scrambled control. a) and b) are reproduced from ref. 42, which is an open-access article distributed under the Creative Commons Attribution (CC BY-NC) License, 2017 Ivyspring International Publisher. c) and d) are reproduced from ref. 44 with permission from American Chemical Society, Copyright 2019.
In another report, Wilson and coworkers employed an affibody which was appended to ICG to target B7-H3 in breast cancer (ABYB7-H3-ICG) (Figure 3c).44 When ICG is conjugated to peptides/proteins, it undergoes a well-known bathochromic shift in absorbance to 850 nm relative to the free dye. Affibodies exert several benefits over antibodies as a targeting motif such as greater affinity for the protein of interest, reduced circulation time, and greater stability under a variety of physiological conditions. After successful in vitro characterization of ABYB7-H3-ICG, the authors collected PA data in tumor-bearing mice. Glands containing metastasized carcinomas were revealed upon administration of the TCA. Specifically, relative to the signal prior to injection, the PA intensity in the region of interest (ROI) was 2.1 ± 1.2-fold higher. Control mice, which was administered with ABYSCR-ICG, which is a control featuring a scrambled affibody sequence, did not yield the corresponding increase in signal, demonstrating the high selectivity of the affibody. To validate this result, the authors blocked the B7-H3 receptor in tumor-bearing mice with non-conjugated ABYB7-H3 prior to injection with the TCA and observed a significant reduction in the PA signal (Figure 3d). Lastly, they were also able to differentiate tumor tissues compared to healthy mammary tissues in breast cancer mouse models.
4.2. CD44
CD44 is a family of transmembrane glycoprotein receptors involved in cell-cell and cell-matrix interactions. Moreover, they regulate cellular processes like growth, survival, differentiation, and motility.45 Since CD44 is upregulated in many cancer types,46 it has been recognized as a potential biomarker for cancer diagnostics and therapy.47, 48 Hyaluronic acid (HA) is a non-sulfated linear glycosaminoglycan that is widely distributed in the extracellular matrix. Due to its high specificity and binding affinity towards CD44, it can be exploited as a ligand in the design of CD44-targeted molecules including TCAs for PA imaging.46, 49–55
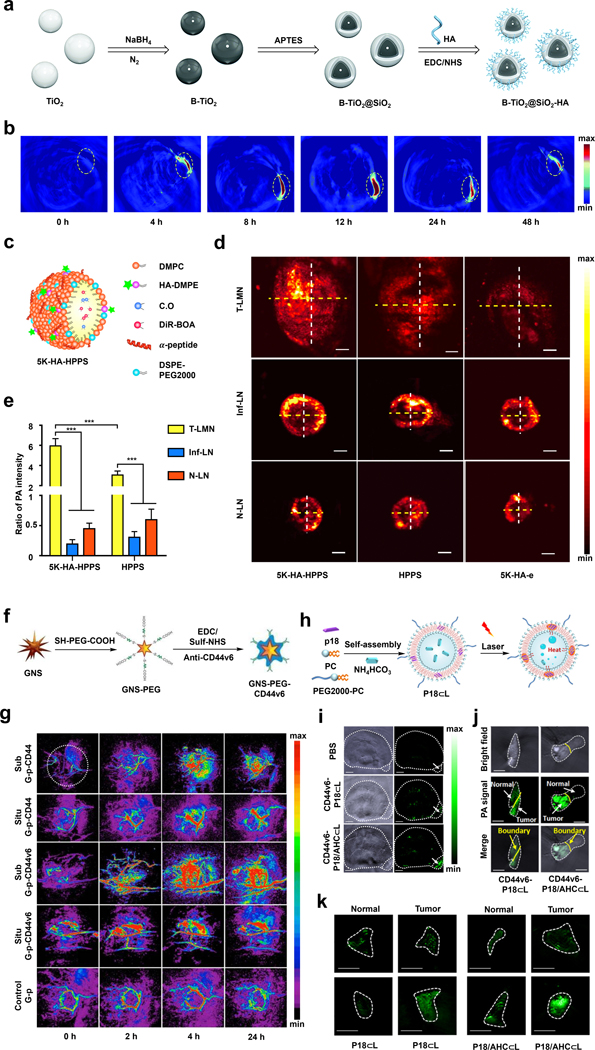
In a representative example, the laboratories of Shen and Liang have developed a TCA with NIR-II PA imaging capabilities based on B-TiO2 nanoparticles (NPs) (Figure 4a).55 Of note, TiO2 typically absorbs UV light; however, the absorption spectrum can be shifted to include NIR-II light through treatment with NaBH4, which incorporates boron atoms and introduces oxygen vacancies. After incorporating a SiO2 shell (B-TiO2-@SiO2), the authors further modified the surface with HA via amide coupling to target CD44-overexpressing tumor cells. This modification also improved the solubility of the resulting TCA (B-TiO2@SiO2-HA) in aqueous solutions which is an important consideration for in vivo applications. Since B-TiO2@SiO2-HA showed a strong photothermal effect, the authors rationalized it could also be used for PA imaging by employing a pulsed light source. PA imaging was performed in a 4T1 murine breast cancer model, in which the maximum signal enhancement in the tumor region was observed 12 hours after B-TiO2@SiO2-HA was administered (Figure 4b).
Figure 4.
a) Schematic of the synthesis of B-TiO2@SiO2-HA. b) PA images of 4T1 tumors before and at various time points after injection of B-TiO2@SiO2-HA. c) Schematic of 5K-HA-HPPS d) PA images of 4T1-related SLNs (upper), inflamed LNs (middle) and normal LNs (lower) after injection of 5K-HA-HPPS (left), HPPS (middle) and 5K-HA-e (right). e) The ratio of PA intensities at the center of the LNs compared to those at their periphery. f) Schematic of the synthesis of GNS-PEG-CD44v6. g) PA images of subcutaneous tumors (Sub) and orthotopic tumors (Situ) before and at various time points after injection of GNS-PEG-CD44v6 or GNS-PEG-CD44. h) Schematic of the synthesis of P18/AHC⊂L. i) PA images of tumors in EJ xenograft mice before and four hours after injection of PBS, CD44v6-P18 ⊂L or CD44v6-P18/AHC⊂L. j) PA images of the tumor boundary that was treated with CD44v6-P18/AHC⊂L. k) PA images of normal urothelium and tumor tissue after treatment with IgG-probes (upper) and CD44v6-probes (lower). a) and b) are reproduced from ref. 55 with permission from Royal Society of Chemistry, Copyright 2021. c), d) and e) are reproduced from ref. 54, which is an open-access article distributed under the CC BY License, 2020 Springer Nature. f) and g) are reproduced from ref. 52, which is an open-access article distributed under the Creative Commons Attribution (CC BY-NC) License, 2015 Ivyspring International Publisher. h), i), j) and k) are reproduced from ref. 53 with permission from American Chemical Society, Copyright 2018.
Likewise, the laboratories of Luo and Zhang developed a dual biomarker-targeting system (5K-HA-HPPS) based on HPPS (HDL-mimicking peptide-phospholipid scaffold) NPs for detecting sentinel lymph nodes (SLN) in breast cancer (Figure 4c).54 A critical design element of this TCA includes HA ligands which can target CD44 and the intrinsic ability of HPPS to interact with scavenger receptor class B type 1, both of which are expressed on the surface of certain breast cancer cells. To provide contrast, a NIR cyanine dye (DiR-BOA) was encapsulated within 5K-HA-HPPS since it can generate a robust PA signal upon irradiation. The authors observed that lymph nodes at various stages exhibited characteristic PA signal patterns compared with HPPS and 5K-HA emulsion (5K-HA-e), which served as control imaging agents. Specifically, for normal lymph nodes (LNs) and inflamed LNs, the signal was localized to the edge of the LNs while for 4T1-related SLNs, the PA signal was most pronounced in the central region (Figure 4d). To demonstrate the effectiveness of dual-biomarker targeting, the ratio of the PA signal between the central and the edge of the SLNs were measured using K-HA-HPPS and HPPS NPs, which was found to be 5.93 and 3.06, respectively (Figure 4e).
Beyond the use of HA, CD44-specific antibodies are also widely employed for CD44 targeting. It is noteworthy that antibodies produced for this application target CD44V6, an isoform of CD44 that is expressed at a higher cancer-to-healthy tissue ratio relative to CD44. The laboratories of Zhang and Cui exploited this approach to develop GNS-PEG-CD44v6, a TCA featuring a PA-active gold nano star architecture conjugated to CD44v6 monoclonal antibodies to image (and treat via the photothermal effect) gastric cancer stem cells (Figure 4f).52 In particular, the authors showed that GNS-PEG-CD44v6 had a higher PA signal (4.7-fold) than that of GNS-PEG-CD44 (2.5-fold) compared to no TCA in a subcutaneous tumor model. This difference was less obvious when orthotopic tumors were imaged. Notably, both TCAs showed higher signal than the non-targeted GNS-PEG counterpart (Figure 4g).
Similarly, the laboratories of Qiao, Xu, and Wang incorporated CD44v6 antibodies onto a liposomal nanoprobe named P18/AHC⊂L to detect bladder cancer via PA imaging (Figure 4h).53 To develop this TCA, the authors co-encapsulated purpurin-18 (P18) and NH₄HCO₃ (AHC) within liposome-based NPs. When confined, P18 is a dye that self-assembles to form aggregates which are fluorescently quenched to favor the generation of a PA signal upon irradiation. Interestingly, the local temperature increase due to the PA effect was sufficient to decompose NH₄HCO₃, leading to the local generation of CO2 bubbles. These bubbles possess higher acoustic impedance than the surrounding tissue, resulting in enhanced acoustic backscattering and an amplified signal. The increase of the PA signal due to gas generation contributed to the high sensitivity of P18/AHC⊂L that was more than three-fold higher compared to the signal of a non-amplified control known as P18⊂L (Figure 4i). Upon conjugation to CD44v6 antibodies, the resulting TCA was able to detect the tumor boundary with an 18-fold signal ratio between tumor and healthy tissue (Figure 4j), and further, was capable of imaging small bladder tumor tissue (< 5mm) with a significantly higher PA signal intensity compared to the non-amplified analog and IgG control groups (Figure 4k).
4.3. ErbB
ErbB is a family of tyrosine kinases that include EGFR (ERBB1), HER2 (ERBB2), HER3 (ERBB3), and HER4 (ERBB4). The expression levels of these transmembrane receptors are often elevated in lung, breast, gastric, and ovarian tumors, making them excellent cancer biomarkers.56, 57 Indeed, the use of ErbB for cancer detection and therapeutic delivery is prevalent in the literature.58 For example, HER2 is the molecular target for Herceptin® (trastuzumab), which is a potent antibody-based chemotherapeutic agent often prescribed to treat patients with breast cancer.59 With respect to the development of TCA for PA imaging, a number of exciting studies have been reported targeting various members of the ErbB family.60–74
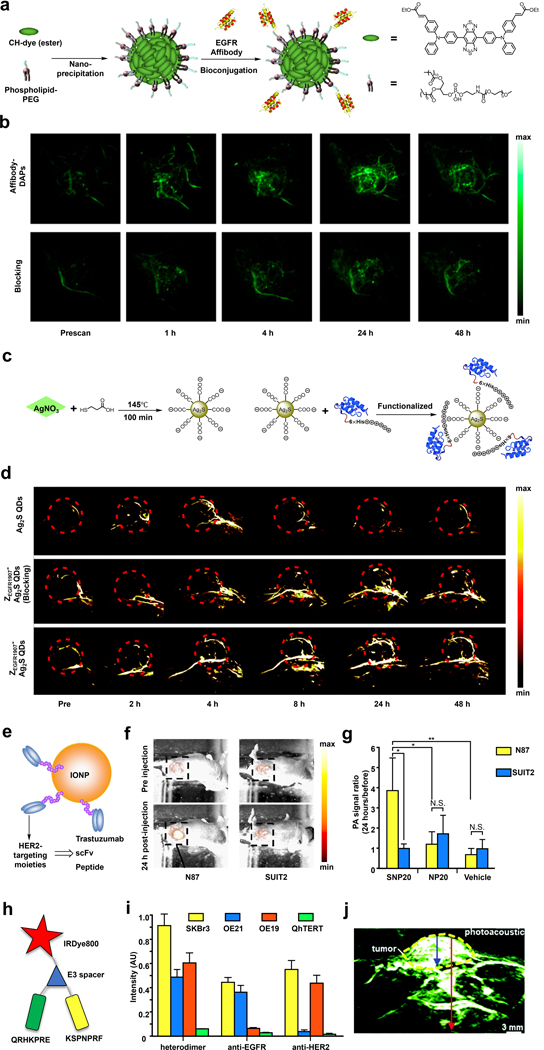
In 2017, the laboratories of Xing and Cheng employed a donor-acceptor chromophore-based nanoparticle (DAPs) design to target EGFR (epidermal growth factor receptor) which is typically responsible for mediating cell growth and division in healthy cells (Figure 5a).69 Due to the tunable energy gaps of the parent CH dye platform, the authors were able to develop congeners such as CH1000 that feature a large Stoke shift (~310 nm) and a moderate quantum yield (11.1%), as well as maximum absorbance and emission at 700 nm and 990 nm, respectively. These properties translate into a versatile multimodal imaging agent after encapsulation via nanoprecipitation using phospholipid-PEG since it can provide contrast via PA and fluorescence in the NIR-I and NIR-II regions, respectively. To augment DAPs with the ability to target EGFR, the authors utilized anti-EGFR affibodies, presumably due to their enhanced stability to pH and temperature fluctuations relative to the larger antibody counterparts. After surface modification with this targeting group and in vitro characterization, the resulting Affibody-DAPs were evaluated in vivo to map lymph nodes, visualize the vascular system, and identify EGFR-positive FTC-133 thyroid tumors. In the latter application, the authors observed a time-dependent PA signal enhancement in the tumor region that plateaued after 24 hours. However, the signal was already distinguishable four hours post-injection compared to a control group of animals that were co-treated with non-conjugated affibody which served as the blocking agent (Figure 5b).
Figure 5.
a) Schematic of the synthesis of Affibody-DAPs. b) PA images of FTC-133 tumors before and at various time points after injection of Affibody-DAPs or Affibody-DAPs with blocking. c) Schematic of the synthesis of ZEGFR:1907-Ag2S QDs. d) PA images of A431 tumor before and at various time points after injection of Ag2S QDs, ZEGFR:1907-Ag2S QDs, or ZEGFR:1907-Ag2S QDs with blocking. e) Schematic of HER2-targeting IONPs. f) PA images of N87 tumor (high expression of HER2) and SUIT2 (low expression of HER2) tumors before and 24 hours after injection of SNP20. g) The PA signal intensity ratios before and 24 hours after injection of SNP20, NP20, or vehicle (PBS). h) Schematic of QRH*-KSP*-E3-IRDye800. i) Fluorescence intensity for the heterodimer binding to SKBr3 (EGFR+/HER+) cells versus OE21 (EGFR+/HER-), OE19 (EGFR-/HER+), and QhTERT (EGFR-/HER-) cells. j) PA images of OE33 xenograft tumor at two hours after injection of QRH*-KSP*-E3-IRDye800, tumor depth was 4.8 mm (blue arrow) and total image depth (red arrow) was 1.2 cm. a) and b) are reproduced from ref. 69 with permission from American Chemical Society, Copyright 2017. c) and d) are reproduced from ref. 70 with permission from Royal Society of Chemistry, Copyright 2018. e), f) and g) are reproduced from ref. 67 with permission from Elsevier, Copyright 2015. h), i) and j) are reproduced from ref. 71 with permission from the Royal Society of Chemistry, Copyright 2018.
Similarly, the laboratories of Cheng, Zhao, and Liu developed anti-EGFR affibody (ZEGFR:1907)-modified Ag2S quantum dots (QDs) to target EGFR-expressing tumors (Figure 5c).70 Since QDs are known for their size-dependent correlation with the emission color, careful control of the particle size can yield NIR absorbing QDs suitable for PA imaging, as in the current example where the absorbance maximum is at 800 nm. A unique feature of this TCA is that the targeting moiety is not appended through covalent attachment. Instead, the authors modified their QDs using ZEGFR:1907 expressed with a His-tag to enable Coulombic interactions between the positively charged His residues and negatively charged carboxylates on the Ag2S QDs surface. With the ZEGFR:1907-Ag2S QDs in hand, in vivo targetability was evaluated in A431 tumors that are characterized by the overexpression of EGFR. When compared to non-targeted Ag2S QDs, which showed a relatively weak signal inside the tumor, the TCA gave rise to a signal enhancement four hours post-injection that persisted even after 32 hours. Localization of the TCA in the tumor was shown to be due to ZEGFR:1907-mediated interactions via a standard blocking experiment (Figure 5d).
Besides EGFR, HER2 is a member of the ErbB family that has also attracted significant interest as a drug target, as mentioned above, due to its role in cell migration, proliferation, survival, angiogenesis, and metastasis. In the context of PA imaging, Saji and coworkers have developed a series of HER2-targeted iron oxide nanoparticles (IONPs).67 This photoabsorber was selected in this study due to their low toxicity, biodegradable properties, and ability to generate a strong PA signal upon irradiation in the NIR range (Figure 5e). The first TCA developed by the authors leveraged the HER2 targeting ability of trastuzumab. However, since one gram of this drug can cost up to $10,000 USD, it is desirable to reduce the production cost by exploring other options. Thus, the authors also developed two additional TCAs using anti-HER2 scFv (single-chain fragment variable) and a 12-residue peptide. After assessing biodistribution using 111In radio tracing, it was determined that the anti-HER2 scFv-conjugated IONPs (SNP20) displayed the highest tumor-to-background ratio. As such, SNP20 was selected for subsequent PA imaging. Tumors expressing either high or low levels of HER2 were grown using N87 cells or SUIT2 cells, respectively. Results showed that SNP20 had a 3.86-fold PA signal in N87 tumors, while the PA signal in the SUIT2 tumors were unchanged (0.97-fold) 24-hour post-injection. Moreover, there was no obvious difference between the signal from N87 and SUIT2 tumors when treated with either non-targeting NP20 and a vehicle (Figure 5f and 5g).
Notably, TCAs capable of recognizing more than one type of ErbB such as EGFR and HER2 have also been reported. For instance, Wang and coworkers developed QRH*-KSP*-E3-IRDye800 which can efficiently image tumor cells which express both of these biomarkers (Figure 5h).71 Critical design elements of this TCA include: 1) a peptide targeting moiety based on heptapeptide monomers identified by the authors (QRH* for EGFR and KSP* for HER2); 2) a NIR dye (IRDye800) for harvesting light necessary to generate a PA signal; and 3) a E3 spacer to avoid steric interactions between the two heptapeptide monomers and IRDye800. To evaluate the dual-biomarker targeting capacity, QRH*-KSP*-E3-IRDye800 was tested in four cell lines, SKBr3, OE21, OE19, and QhTERT, via confocal microscopy. The results show that SKBr3 cells were the brightest since they are the only cell line of the four tested expressing both EGFR and HER2 (Figure 5i). Likewise, pharmacokinetics and biodistribution studies revealed that QRH*-KSP*-E3-IRDye800 had a PA tumor to background ratio of 2.44 ± 0.36, which was higher than 1.44 ± 0.21 for non-targeting control probe. Of note, the maximum signal was reached two hours post-injection and clearance was complete after 24 hours. Finally, a PA and ultrasound dual-modal image was captured in an OE33 xenograft tumor model to assess the imaging depth of QRH*-KSP*-E3-IRDye800. The result revealed a tumor depth of 4.8 mm and a total image depth of 1.2 cm. (Figure 5j).
4.4. Fibronectin
Fibronectin is a large glycopeptide that serves as a vital component of the extracellular matrix where it is known to bind various protein receptors including integrin and is involved in processes like embryogenesis and wound healing. While fibronectin is highly upregulated in processes like pathologic angiogenesis, its role in tumor progression is still being explored. Nevertheless, many fibronectin-targeting drug delivery and imaging studies have been reported, which highlights fibronectin’s potential for cancer applications.75–78
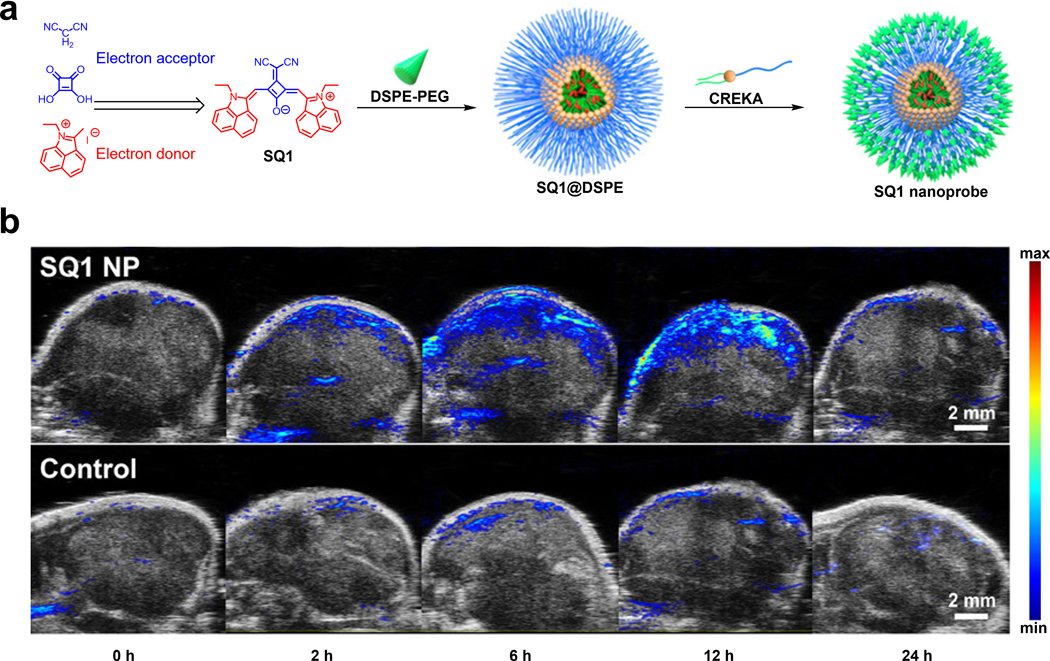
For example, the laboratories of Zhang and Wang developed a fibronectin-targeting CREKA peptide conjugated to a squaraine-based nanoprobe with NIR-II fluorescence/NIR-I PA dual imaging and photothermal therapeutic capabilities (Figure 6a).79 The squaraine dye (SQ1) used in the nanoprobe was carefully engineered based on a donor-acceptor-donor structural design. To be more specific, ethyl-grafted 1,8-naphtholactam was first introduced onto the squaric acid core as an electron donor. Subsequently, incorporation of malononitrile as an electron acceptor further lowered the band gap energy which results in a red-shifted spectrum to afford a dye with NIR-I absorption and NIR-II emission. The fibronectin targeting SQ1 NPs were synthesized through nanoprecipitation of SQ1 with DSPE-PEG2000-Mal, to provide water solubility. This was followed by decoration of the surface with a CREKA targeting peptide via maleimide-mediated conjugation. After performing in vitro experiments that showed a linear relationship between the PA signal and SQ NPs concentration, the authors conducted PA imaging in MDA-MB-231 tumor-bearing mice. A clear PA signal enhancement in the tumor region was observed in these studies which plateaued 12 hours post-injection. Conversely, the non-targeting SQ1 NPs had a much weaker signal, demonstrating the effectiveness of fibronectin-targeting (Figure 6b).
Figure 6.
a) Synthesis of SQ1 from electron acceptor and donor components and schematic of SQ1 nanoprobe assembly. b) PA imaging of MDA-MB-231 tumors at various time points after injection with SQ1 nanoprobe or SQ1@DSPE control. a) and b) are reproduced from ref. 79 with permission from American Chemical Society, Copyright 2020.
4.5. Folate Receptor
Folate (folic acid) has been implicated to play a critical role in cancer development and progression. Specifically, folate is required to produce S-adenosylmethionine (SAM), which is utilized by cells as an essential cofactor to methylate DNA and RNA.80 Since cancer cells are characterized by uncontrolled cell division and rapid growth, the resulting demand for SAM necessitates a means to obtain more folate from the local environment. Given that folate enters the cell via folate receptors (FR), targeting this cell surface protein has been employed to identify and treat numerous cancer types including ovarian and uterine tumors, as well as in lung, kidney, and breast cancer.81 In the context of PA imaging, the FR is commonly targeted to develop TCA for cancer detection.82–97
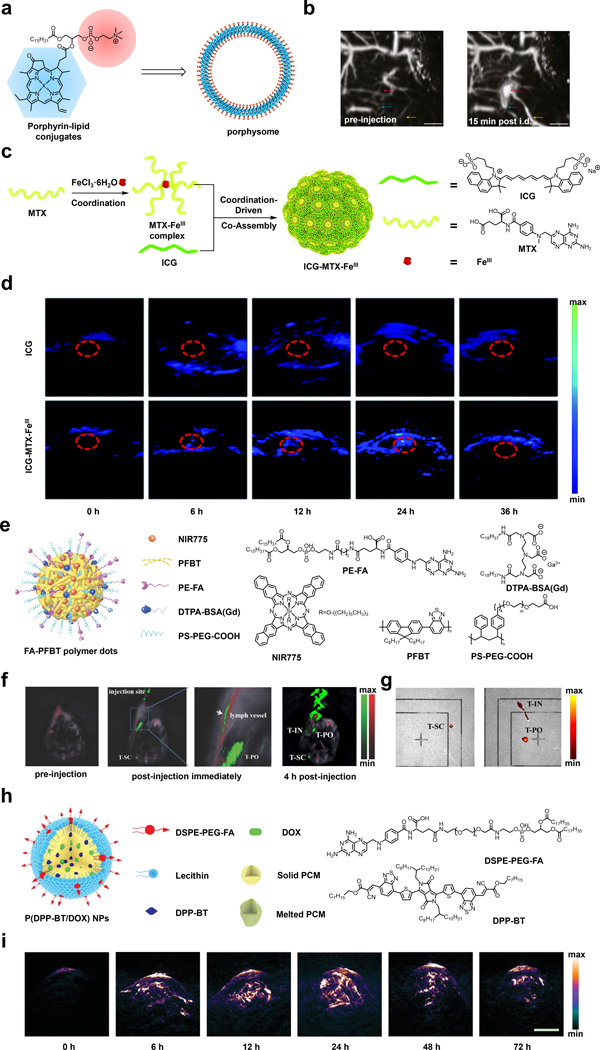
In one of the earliest examples, Zhang and coworkers developed a bilayer ‘porphysomes’ nanovesicle featuring phospholipid-porphyrin conjugates synthesized by attaching a porphyrin unit to the secondary alcohol of glycerol through an ester linkage (Figure 7a).82 A unique feature of this design is an ultrahigh extinction coefficient of 2.9 × 109 M−1cm−1 at 680 nm which can be attributed to the localization of ~8 × 104 porphyrin chromophores in the bilayer. It is important to note that this property is a strong indicator of the PA intensity of a molecule. Moreover, the high local concentration of porphyrin results in a self-quenching effect which also translates into a stronger PA signal. As noted by the authors, switching the porphyrin unit from one to another can red shift the absorption profile to expand the utility of this design. The authors also prepared folate-PEG-lipids in a similar manner, which when incorporated at one molar percent, can effectively target FRs. For instance, when mice bearing KB fibroblast tumors that overexpress the FR were treated with the porphysomes intradermally, the PA signal in the tumor region peaked 15 minutes post-injection, demonstrating efficient uptake (Figure 7b). It is noteworthy that this TCA is fully biodegradable via lipase activity which is a highly desirable property for in vivo applications.
Figure 7.
a) Schematic of porphysomes. b) PA images before and 15 mins after injection with porphysomes, showing secondary lymph vessels, lymph node, and inflowing lymph vessels. c) Schematic of synthesis of ICG-MTX-FeIII. d) PA images in HeLa tumor-bearing mice before and at various time points after injection of ICG or ICG-MTX-FeIII. e) Schematic of FA-PFBT polymer dots. f) PA images of the lymph vessel, T-SC, and T-PO in the NCI-H292 tumor-bearing mice before and immediately after injection of FA-PFBT polymer dots. g) Ex vivo fluorescence imaging of T-SC, T-IN and T-PO lymph nodes. h) Schematic of P(DPP-BT/DOX) NPs. i) PA images in HeLa tumor-bearing mice before and at various time points after injection of P(DPP-BT/DOX) NPs. a) and b) are reproduced from ref. 82 with permission from Springer Nature, Copyright 2011. c) and d) are reproduced from ref. 97 with permission from Royal Society of Chemistry, Copyright 2020. e), f) and g) are reproduced from ref. 91 with permission from John Wiley and Sons, Copyright 2018. h) and i) are reproduced from ref. 95 with permission from John Wiley and Sons, Copyright 2019.
In another example, the laboratories of Hou, Ye, and Li developed ICG-MTX-FeIII to target the FR (Figure 7c).97 The assembly of this TCA is driven by electrostatic interactions between ferric (Fe3+) ions and methotrexate (MTX). MTX serves a dual-purpose in this design by functioning as an anti-cancer drug to attenuate folate metabolism by inhibiting the enzymatic activity of dihydrofolate reductase and as a targeting moiety owing to its resemblance to folic acid. In addition, the authors selected ICG as the PA-active chromophore which can be incorporated through co-assembly with MTX since the two pendant sulfonic acid groups of ICG can also interact with Fe3+ via electrostatic contacts. It is noteworthy that iron was strategically selected because it is an MRI-active metal ion, and this property presents an opportunity for multimodal imaging along with fluorescence and PA. To assess the effectiveness of ICG-MTX-FeIII as a TCA for PA imaging, the authors generated cervical tumors in nude mice using HeLa cells. The PA signal of a group of animals treated with free ICG was barely discernible from the background, giving rise to only slight increase after six hours. However, when a second group of mice were treated with ICG-MTX-FeIII, a time-dependent PA signal increase of four-fold could be observed in the tumor region after 24 hours (Figure 7d).
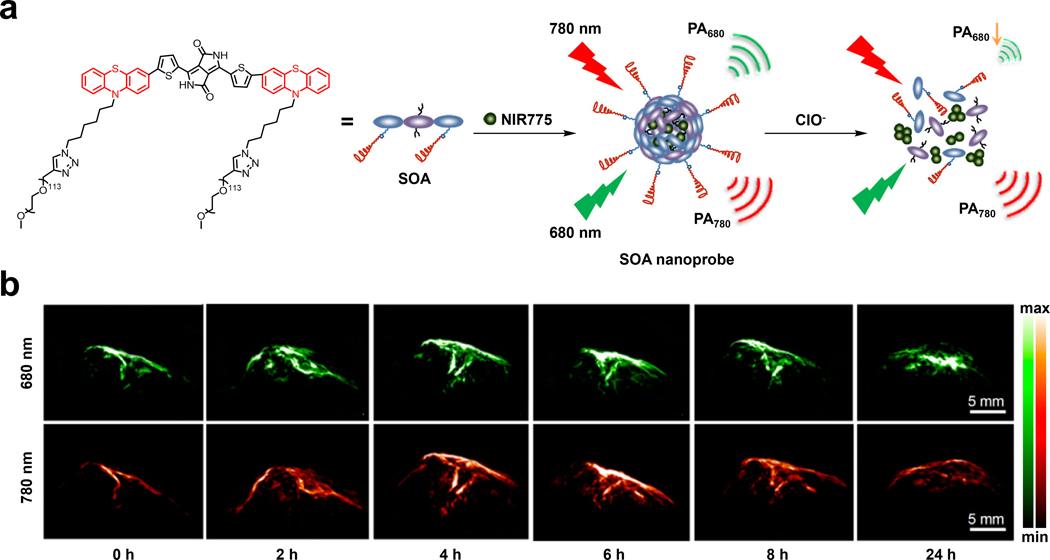
In other work, Xiong and coworkers developed a folate-functionalized polymer dot system (FA-PFBT polymer dots) that can be utilized for multimodal imaging since it features NIR fluorescence and PA imaging capabilities, amongst others, with the goal of visualizing metastatic LNs (Figure 7e).91 The polymer dot design included poly(9,9-dioctyfluorene-alt-benzothiadiazole) (PFBT) which is a fluorescent molecule as the light-absorbing component. When doped with the NIR dye, NIR775, there is a shift of the emission maximum to the NIR range. Notably, the NIR775 dye allows the system to be used for PA imaging in the NIR window along with its ability to be implemented for photodynamic therapy. In order to target cancer cells overexpressing the FR, 1,2-dipalmitoyl-sn-glycero-3-phosphoethanolamine-N-(6-((folate)amino)hexanoyl) (PE-FA) was also doped into the polymer dots. The use of the polymer dots as a TCA for PA imaging was investigated by injecting FA-PFBT polymer dots into the footbed of mucoepidermoid carcinoma NCI-H292 tumor-bearing mice. Immediately after administration, visualization of the TCA in the lymphatic vessel and T-PO lymph node was apparent. Four hours after the initial injection, strong PA signals were observed in T-IN (inguinal), T-SC (sciatic), and T-PO (popliteal) LNs (Figure 7f). Ex vivo fluorescence imaging experiments that were performed 24 hours after injection provided further support for FA-PFBT polymer dots’ utility as a PA TCA since these results were concordant with those found with in vivo PA imaging (Figure 7g). In addition, ROI evaluation of the LNs to background yielded an excellent signal-to-noise ratio of 72.24 for the T-PO LNs.
Lastly, the laboratories of Shen and Fan designed a system utilizing a diketopyrrolopyrrole (DPP)-based chromophore that exhibits its properties in the desired NIR range (Figure 7h).95 In addition to its use as a contrast agent, it also serves as a photothermal therapy / photodynamic therapy agent. The NPs referred to herein as, P(DPP-BT/DOX) NPs, also contain the chemotherapeutic drug doxorubicin (DOX). The NPs encapsulated the hydrophobic chromophore DPP-BT and DOX in an organic phase-change material composed of lauric acid and stearic acid with lecithin that is amphiphilic. The NPs were functionalized with an amphiphile (DSPE-PEG-FA) containing folic acid to target cancer cells expressing the FR. Under irradiation with 730 nm light, PA signals were generated along with photothermal and photodynamic effects, as well as the subsequent release of DOX. The authors tested the PA properties of the TCA in tumor-bearing mice and observed a 10.5-fold signal enhancement 24-hours post injection (Figure 7i).
4.6. GRPR
Gastrin-releasing peptide (GRP) is a neuropeptide that has important modulatory functions in different organs, and its role as a growth factor in tumor progression has been implicated. The corresponding gastrin-releasing peptide receptor (GRPR) is found to be overexpressed in certain types of cancers like prostate, lung and gastrinoma cancer and thus, has been recognized as a useful cancer biomarker,98–100 which has also been leveraged for PA imaging.
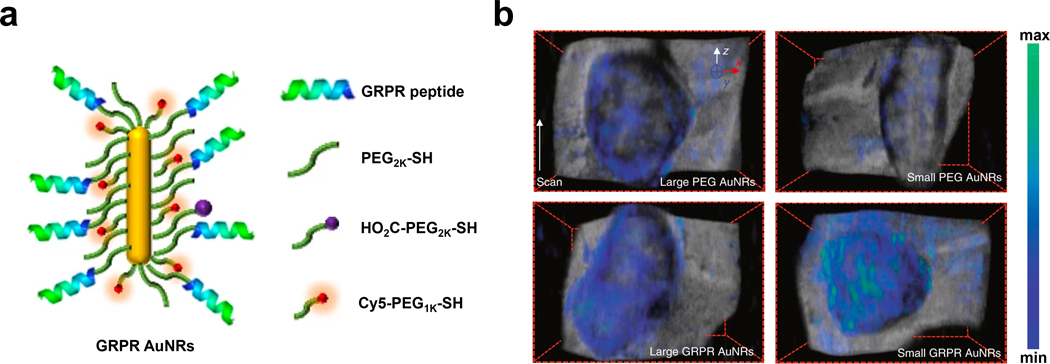
An example that targets GRPR comes from the laboratories of Gambhir and Emelianov where the authors developed miniaturized gold nanorods (AuNRs) with NIR-II absorption that could be used as a GRPR-targeting PA imaging agent (Figure 8a).101 By carefully tuning the surfactant composition, pH and reductant used in the synthesis, the authors were able to obtain AuNRs with an average size that was five to 11 times smaller than conventional AuNRs. This property is important since the smaller AuNRs were accompanied by a three-fold higher thermal stability and a 3.5-fold stronger PA signal output compared to larger AuNR samples that were optical density (OD)-matched. Further theoretical investigation revealed that the enhancement of the PA signal was quadratically proportional to the nanoparticles’ surface-to-volume ratio which rationalizes the improvement of the PA performance brought forth by miniaturizing the design. To develop TCAs, the authors leveraged the strong thiol-gold interaction that is often used to modify the surface of various Au architectures. For instance, PEG2K-SH, HO2C-PEG2K-SH, and Cy5-PEG1K-SH each contain a terminal thiol group. These components, along with GRPR-targeting peptides were conjugated onto both the small and large AuNRs. The PA imaging performance of the resulting AuNRs, as well as non-targeted analogs were tested in a murine model of prostate cancer. The results showed a PA signal increase in tumors after both small and large AuNRs were modified with the GRPR-targeting peptide. Moreover, while the difference in the PA signal within the non-targeting small and large AuNRs was insignificant, the GRPR-targeting small AuNRs showed a statistically higher PA signal than that of the OD-matched large AuNRs (Figure 8b). Of note, a decay of the PA signal for the large GRPR-targeting AuNRs was observed while there was no loss of intensity for the small AuNRs, which showcases the increase in thermal stability that results from reducing the size of the TCA.
Figure 8.
a) Schematic of GRPR AuNRs. b) PA imaging of tumor-bearing mice with non-targeted large and small PEG AuNRs and targeted large and small GRPR AuNRs. a) and b) are reproduced from ref. 101 with permission from Springer Nature, Copyright 2019.
4.7. Integrin
Integrins are a family of cell surface receptors that connect cells to the extracellular matrix (ECM) and plays a central role in processes like cell migration, invasion, proliferation, and survival, all of which are important in tumor progression.102 As such, integrins have been exploited for cancer diagnostic and therapeutic applications through the development of integrin-targeted imaging agents103 and drug-conjugates,104 respectively. A common feature of most designs is the presence of an appended RGD (arginine-glycine-aspartic acid)-based peptide unit that recognizes and binds to integrins with a half-maximal inhibitory concentration (IC50) value in the nM range.105 Relative to antibody targeting, RGD-based peptides are readily accessible, exhibit low immunogenicity, and can be conveniently conjugated to various molecular structures via conventional conjugation chemistry. Although a vast number of RGD-based TCA for PA imaging have been developed,106–137 we highlight several notable examples below.
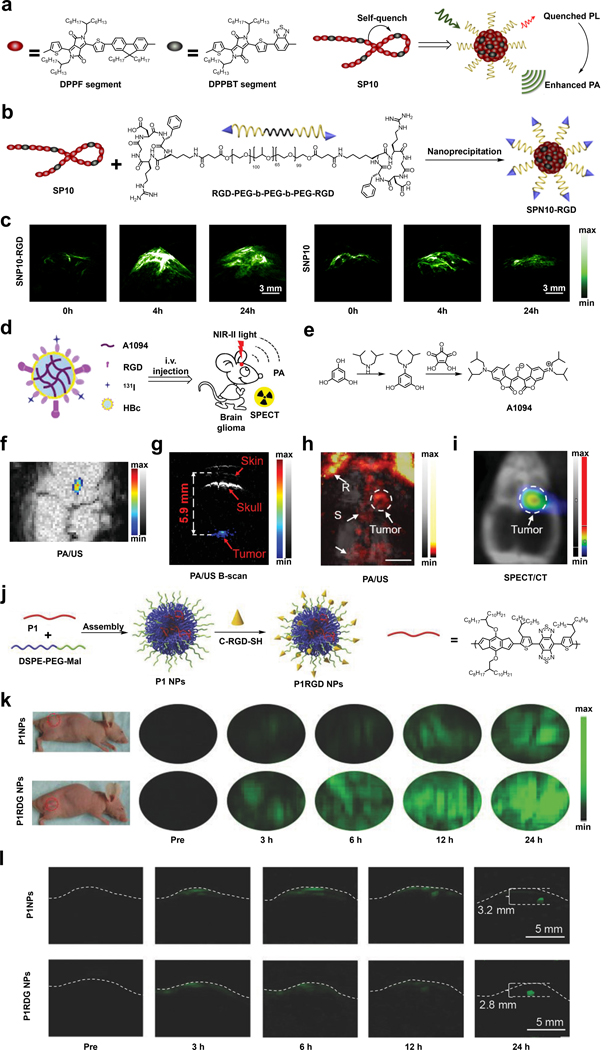
For instance, Pu and coworkers have reported an RGD-conjugated semiconducting polymer nanoparticle (SPN) to target tumors overexpressing integrin.120 The polymer that the authors developed is known as SP10 and contains two components, a DPFF segment that forms the backbone and an electronically deficient DPPBT segment that was incorporated to promote nonradiative relaxation which results in an enhancement of the PA signal (Figure 9a and 9b). Additionally, a second component known as RGD-PEG-b-PEG-b-PEG-RGD was also developed. The RGD-targeting moiety could be displayed when SP10 and the second component are nanoprecipitated to form the resulting TCA (SNP10-RGD). In a proof-of-concept study, SNP10-RGD was tested in vivo for the PA detection of murine 4T1 breast tumors. At the four-hour time point after injection, the PA signal of SPN10-RGD-treated animals reached a maximum enhancement of 4.7-fold compared to the tumor background, while the non-targeted SNP10 control only showed a 2.6-fold enhancement. Moreover, SNP10-RGD also showed a slower tumor clearance as indicated by a persistent PA signal which was still evident 24-hour post-injection (Figure 9c). This last finding can be useful if multiple imaging sessions are desirable.
Figure 9.
a) and b) Schematic of the synthesis of SNP10-RGD. c) PA images of murine 4T1 breast tumors before and at various time points after injection of SNP10-RGD or SNP10. d) Schematic of A1094@RGD-HBc. e) Synthesis of A1094 f) PA/US images of U87MG brain tumor two hours after injection of A1094@RGD-HBc. g) The corresponding PA/US B-scan to demonstrate imaging depth. h) and i) in vivo PA/US and SPECT/CT images of U87MG brain tumor two hours after injection of 131I-A1094@RGD-HBc. R: rostral rhinal vein; S: sagittal sinus. j) Schematic of the synthesis of P1RGD NPs. k) PA images of subcutaneous U87 xenograft tumor before and at various time points after administration of P1RGD NPs or P1RGDs. l) PA images of mouse brain tumor before and at various time points after administration of P1RGD NPs or P1RGDs. a), b) and c) are reproduced from ref. 120 with permission from Elsevier, Copyright 2017. d), e), f), g), h) and i) are reproduced from ref. 128, which is an open-access article distributed under the Creative Commons CC BY License, 2019 John Wiley and Sons. j), k) and l) are reproduced from ref. 124 with permission from John Wiley and Sons, Copyright 2018.
Another example comes from the laboratories of Li and Nie, who developed an integrin-targeting NIR-II TCA (A1094@RGD-HBc) which can be employed for effective in vivo PA/SPECT brain imaging.128 The A1094@RGD-HBc TCA was synthesized by encapsulating RGD-modified hepatitis B virus core protein (RGD-HBc) with A1094, a mesoionic dye exhibiting a strong aggregation-induced absorption enhancement (AIAE) effect, and 131I which was introduced for SPECT imaging (Figure 9d and 9e). The AIAE effect from A1094 is an important design element since aggregation of the dye would lead to a stronger PA signal as we have seen with other reports. The evaluation of A1094@RGD-HBc as a TCA for PA imaging was conducted in U87MG brain tumor-bearing mice model. The high efficiency of A1094@RGD-HBc to detect such tumors was supported by a strong PA signal enhancement observed in brain region even with a low imaging agent dose of 100 ug/mL (Figure 9f). Of note, the B-scans of PA and US revealed a tumor-depth of 5.9 mm, which was nearly 1.47-fold deeper than a previous study (Figure 9g).138 The targeting ability was further verified with maximum amplitude projection images of the mice brain obtained two hours post-injection, in which a tumor PA signal enhancement was observed which was consistent with the SPECT imaging results (Figure 9h and 9i).
Lastly, the laboratories of Zheng and Liu reported P1RGD NPs for brain tumor imaging via targeting of the integrin biomarker.124 The first component is P1, a conjugated polymer that was designed using a donor-acceptor structure, which features a strong NIR-II absorption that could facilitate the imaging of deep tissue via NIR-II PA imaging. The second component is DSPE-PEG-Mal. The DSPE portion of the molecule can interact with P1 during NP assembly since both are hydrophobic. On the contrary, the PEG section was included for solubility and to render the resulting NP more biocompatible. Finally, the maleimide (Mal) functional group served as a reactive handle to attach the targeting ligand (c-RGD-SH) through a Michael addition reaction with the thiol group (Figure 9j). The PA imaging performance of P1RGD NPs was evaluated in vivo with both subcutaneous and orthotopic xenograft glioma models with P1 NPs, which lacks the RGD targeting moiety, as a control. In the subcutaneous tumor model, the authors were able to show that the PA signal of P1RGD NPs was two-fold higher than that of P1 NPs three hours post-injection, and the signal-to-background of P1RGD NPs increased up to 95 at the 24-hour time point post-injection while for P1 NPs, the ratio only reached a value of 52 (Figure 9k). These results clearly demonstrate the importance of targeted delivery of a PA imaging agent. For the orthotopic tumor model, the authors were able to detect tumors up to 3.2 mm in tissue depth with a significant PA signal enhancement, and the position of the tumor was consistent with the results from MRI imaging (Figure 9l). Not surprising, in this model, the P1RGD NPs also had a signal-to-background which was higher than the P1 NPs.
4.8. Nucleolin
Nucleolin is one of the most prevalent proteins inside the nucleolus. Due to its essential role in transcription, translation, cell cycle and division, cell survival and differentiation, nucleolin dysregulation is frequently involved in tumorigenesis. Further, the overexpression of nucleolin has been reported in various cancers like pancreatic cancer, thyroid cancer, breast cancer, and colorectal cancer. Notably, while nucleolin is mostly inside the nucleolus of normal cells, the relocation and accumulation of nucleolin at the cell membrane in cancer cells have been observed, making it possible to exploit this as a potential cancer biomarker.139 140
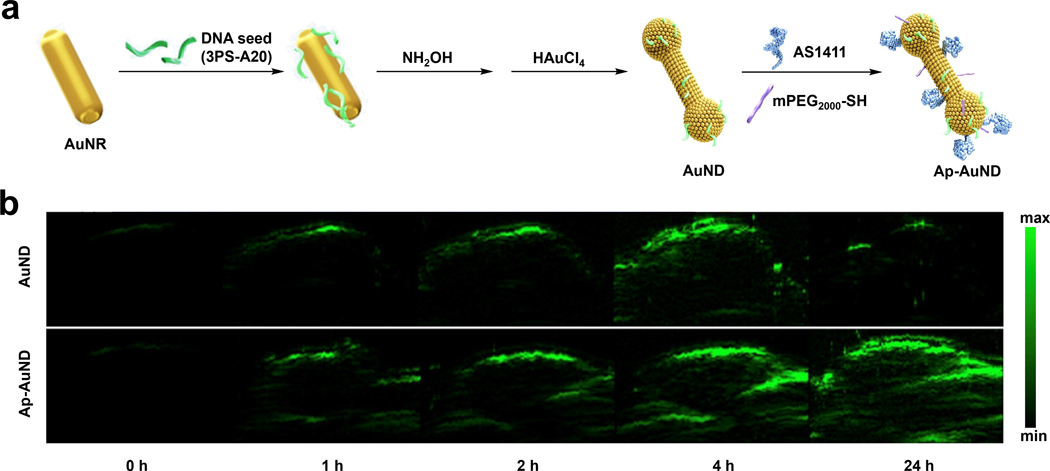
In the context of developing a TCA for PA imaging, the laboratories of Tang and Huang developed nucleolin-targeting gold nano dumbbells (AuNDs) which could additionally serve as a photothermal therapy theranostic agent (Figure 10a).141 The conventional approach to red-shift AuNPs’ absorption is by adjusting the geometry (nanorods, nanospikes, hollow gold NPs, etc.) via modification of the localized surface plasmon resonance (LSPR) properties. However, the authors applied a unique DNA-encoded seed strategy and rationalized that by varying the concentration and the sequence of the DNA seed, it would be possible to affect the morphology and dimension of AuNPs, hence allowing for fine-tuning of the corresponding absorption in a more programmable manner. Utilizing this strategy, the authors successfully shifted the AuNPs’ absorption into the NIR-II window and obtained Ap-AuND which possessed NIR-II absorption. As we have seen in the above sections with other gold-based designs, it is common to attach components such as water solubilizing PEG groups or targeting ligands via strong thiol-gold interactions. Indeed, this is the same approach the authors of this work employed. For example, their nucleolin-targeted DNA aptamer, AS1411, features a thiol group. Likewise, they decorated the surface with mPEG2000-SH through the same chemistry to increase biocompatibility. In the reported PA imaging experiments using MCF-7 breast tumor-bearing mice, the results showed that Ap-AuND could be employed for PA imaging in a highly sensitive way upon NIR-II laser irradiation, with a 97-fold tumor PA signal enhancement 24 hours post-injection compared with the pre-injection tumor background (Figure 10b). Moreover, Ap-AuND also showed better selectivity with a significantly more intense PA signal compared with the non-targeting AuND control group.
Figure 10.
a) Schematic showing synthesis of Ap-AuND from AuNR. b) PA imaging of MCF-7 tumors at various time points after injection with AuND or Ap-AuND. a) and b) are reproduced from ref. 141, which is an open-access article distributed under the Creative Commons Attribution (CC BY) License, 2021 AIP Publishing.
4.9. p32 Protein
The p32 protein is also known as gC1qR, C1qBP or HABP-1. It is a multifunctional protein involved in key biological processes such as cellular metabolism, mitochondrial quality control, and apoptosis. While it is found in nearly all healthy tissue, the expression of p32 is elevated in proliferating cells and pathological tissues like cancerous cells, suggesting its potential importance in tumor progression.142–144
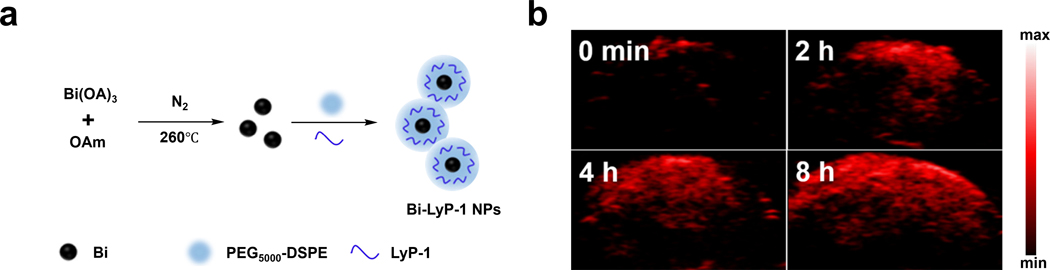
To target this biomarker, Li and coworkers developed Bi-LyP-1 NPs, which are ultra-small semimetal NPs comprised of bismuth for use as a TCA for multimodal computed tomography (CT) and PA imaging, as well as for theranostic treatment (Figure 11a).145 The bismuth NP scaffold was selected due to its strong and broad absorption profile (vide infra) and favorable optical properties such as a small carrier effective mass, a long Fermi wavelength, a small band overlap energy, and CT imaging capabilities. The authors modified the bismuth NP surface with PEG5000-DSPE to improve biocompatibility. Interestingly, ~15% of the PEG5000-DSPE molecules were modified to feature a maleimide functional group. This allowed p32-targeting LyP-1 peptides to be attached via thiol conjugation. The targeting ability of the resulting TCA was assessed in p32-overexpressing 4T1 tumors where Bi-LyP-1 NP-treated animals showed a 1.7-fold enhancement of tumor accumulation compared to a control group. This tumor-targeting ability was further confirmed via in vivo PA imaging which showed a 3.6-fold PA signal enhancement in the tumor region eight hours post intravenous injection using 700 nm laser irradiation (Figure 11b). Of note, the broad spectrum of the bismuth semimetal nanoparticle core gave similar results when irradiated at 800 and 900 nm. While having sharp absorption is beneficial and typical of many imaging agents, this example shows broad absorption can enable the use of a range of excitation wavelengths.
Figure 11.
a) Schematic of Bi-Lyp-1 NPs. b) PA images of tumors before and at various time points after injection of Bi-Lyp-1 NPs. a) and b) are reproduced from ref. 145 with permission from American Chemical Society, Copyright 2017.
4.10. Phosphatidylserine
Phosphatidylserine (PS) is a phospholipid that usually resides on the inner face of the plasma cell membrane. While in most instances, PS is not accessible when a cell is intact, it can be used to detect apoptotic or necrotic cells where the interior components are exposed or when the cell membrane becomes permeabilized. This feature, together with its prevalence in the cell membrane and consistent exposure in necrotic cells, makes PS an attractive biomarker for targeting tumor cell death upon drug treatment.146
Recently, Brindle and coworkers developed a TCA for PA imaging (C2Am-750) of early therapy-induced tumor cell death.147 The design features a fluorescent dye (VivoTag-S 750-MAL) appended to C2Am, which is a PS-targeting protein developed by the same group. Relative to its precursor, the C2A domain of Synaptotagmin-I, C2Am is smaller in size and retains nM binding affinity towards PS. Of note, a cystine residue was incorporated into C2Am at a position distal from the PS binding site which was used for conjugation to the fluorescent dye via maleimide chemistry. The authors postulated this would be essential to avoid attenuating the PS binding affinity. To assess the performance of the TCA in imaging tumor cell death, Colo205 tumors were employed as the model and MEDI3039, an anti-cancer drug, was used to trigger tumor cell death. Further, a control probe, iC2Am-680, was synthesized by conjugating IRDye 680RD to the inactive isoform of C2Am. After demonstrating the specificity of C2Am-750 to PS and its efficacy for detection of tumor cell death, PA imaging was used to visualize the retention of both C2Am-750 and iC2Am-680 in tumors. For C2Am-750, there was some PA signal without MEDI3039 treatment; however, the PA signal enhancement was substantially higher in the MEDI3039-treated tumors seven hours after drug treatment. The signal peaked three-hours post-probe injection with an apparent focus at the central region of the tumor. On the contrary, the non-targeting iC2Am-680 control showed minimal retention in the tumor with a negligible signal change before and after probe injection. Moreover, the authors were able to find that the probe was distributed in the entire tumor by reconstructing 3D images based on the axial, coronal, and sagittal views.
4.11. Protein Sulfenic Acids
Protein sulfenic acids are generated by the reversible oxidation of protein cysteine residues through the reaction with H2O2. This functionality can serve as an indicator of oxidative stress within a cell. Due to oncogenic stimulation, increased metabolic activity, and mitochondrial malfunction in cancer cells, tumors are frequently subjected to oxidative stress conditions and therefore, protein sulfenic acids can be utilized as a target for redox status-based tumor targeting.148, 149
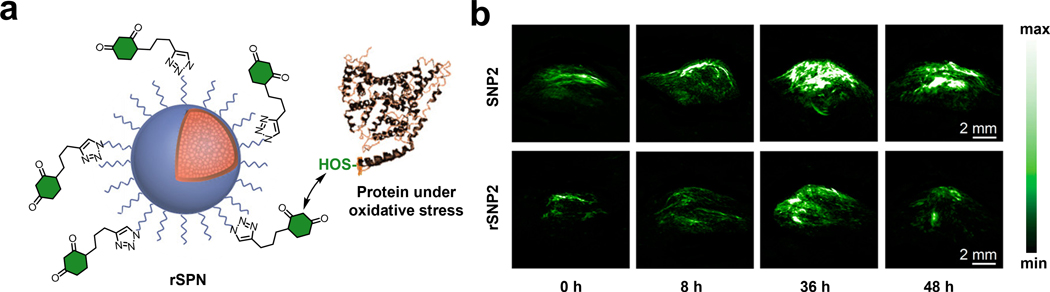
Pu and coworkers developed an activity-based TCA known as rSPN2 for in vivo PA detection of protein sulfenic acids (S-OH functional group) (Figure 12a).150 The authors based the design of rSPN2 on SPNs owing to its good biocompatibility, high PA brightness, and excellent photostability. A second key design element of the TCA is the 1,3-cyclohexanedione group, which can covalently attach to protein sulfenic acids under physiological condition through an enolization event followed by a selective SN2 reaction.151 Notably, the SPN core was shielded with a silica layer and a PEGylated shell was also installed. Overall, these modifications were necessary to prevent attenuation of the optical properties of rSPN2 from the condensation reaction and to maintain a strong absorption band at 680 nm which remained consistent regardless of the state of the SPN. The in vivo PA performance for rSPN2 in protein sulfenic acid detection was evaluated in a HeLa tumor-bearing xenograft mouse model. The PA signal of rSPN2 in the tumor was 1.3-fold higher compared with a non-reactive analog (SPN2) and was 2.2-fold higher than the tumor background 36 hours post-injection (Figure 12b).
Figure 12.
a) Schematic of rSNP. b) PA images of xenograft HeLa tumor mouse model before and at various time points after injection of rSNP2 or SNP2. a) and b) are reproduced from ref. 150 with permission from American Chemical Society, Copyright 2017.
4.12. Prostate-specific Membrane Antigen
Prostate-specific membrane antigen (PSMA) is a transmembrane protein that is present in normal tissue, but is significantly elevated in prostate tumors.152 The role of PSMA in cancer proliferation is believed to be related to glutamate release, which triggers a number of downstream effects, notably the activation of the PI3K pathway which promotes proliferation, cell growth, and angiogenesis. Since recognizing its link to cancer, PSMA has emerged as an attractive target for the development of diagnostic tools via PA imaging.153–156
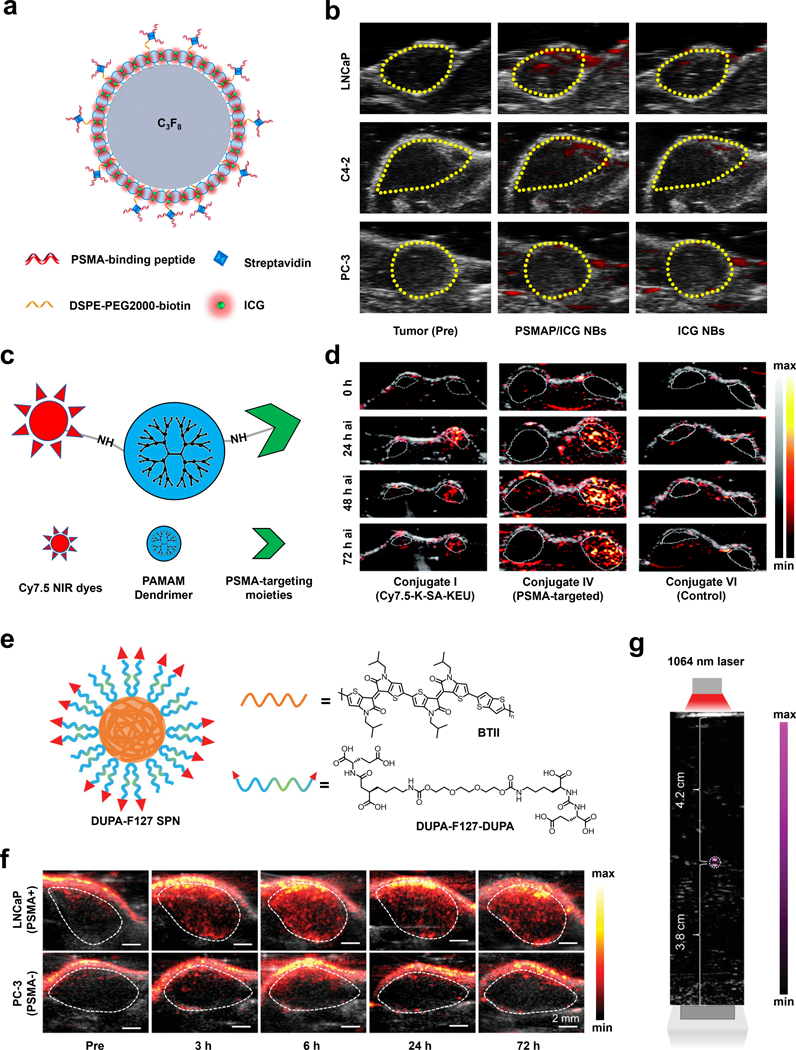
Recently, Li and coworkers utilized a nanobubble platform to develop a multimodal TCA for PSMA upregulation in prostate cancer, capable of fluorescence, ultrasound, and PA imaging (Figure 13a).154 To achieve this, authors loaded phospholipid-based bubbles with ICG and appended PSMA-binding peptide (WQPDTAHHWATL) via a biotin-streptavidin linkage. After obtaining promising in vitro results, they sought to investigate the PA enhancement of their nanobubbles, PSMAP/ICG NBs, in vivo. Using cell lines with PSMA+ expression, LNCaP and C4–2, xenograft tumors were generated in male nude mice and retro-orbital injection of their TCA was performed. Importantly, they utilized PSMA- PC-3 tumor-bearing mice as a negative control. The result showed a PA signal enhancement after PSMAP-IGB NBs injection with a much higher enhancement observed in PSMA+ mice, suggesting its usage in PSMA+ tumor imaging. The importance of the PSMA targeting moiety was further supported by a significantly enhanced PA signal in PSMAP-IGB NBs group compared with IGB NBs group in PSMA+ mice, whereas there was negligible difference between these two group in PSMA- mice. (Figure 13b)
Figure 13.
a) Schematic of PSMAP/ICG NB. C3F8 represents the perfluorocarbon gas core. b) PA images of LNCaP, C4–2, and PC-3 tumors before and after administration of PSMAP/ICG NBs or ICG NBs. c) Schematic of PAMAM dendrimer conjugated to Cy7.5 and PSMA targeting moiety. d) PA images of PSMA+ PIP (right) and PSMA− flu (left) tumors before and at various time points after injection of Conjugate I, IV, or VI. e) Schematic of DUPA-F127 SPN and chemical structures of BTII and DUPA-F127-DUPA. f) PA/US images of LNCaP and PC-3 tumors before and at various time points after injection of BTII-DUPA SPN. g) PA/US image of BTII-DUPA SPN in a polyurethane tube placed in chicken breast tissue to demonstrate imaging depth. a) and b) are reproduced from ref. 154, which is an open-access article distributed under the CC-BY-NC copyright license, 2019 Dove Medical Press. c) and d) are reproduced from ref. 156 with permission from Royal Society of Chemistry, Copyright 2021. e), f) and g) are reproduced from ref. 155 with permission from John Wiley and Sons, Copyright 2020.
In another example, the laboratories of Boctor and Pomper developed a TCA based on the poly(amidoamine) (PAMAM) dendrimer-scaffold and examined its effectiveness in prostate cancer models (Figure 13c).156 PAMAM dendrimers were selected because of their highly tunable nature that allows for multiple modalities to be accessed in one molecule. The authors designed several PSMA-targeting dendrimers, with varying numbers of PSMA-targeting motifs, chromophore moieties, and capping agents. Through a variety of in vitro experiments, the dendrimer with the best targeting ability, conjugate IV, was identified. Conjugate IV consists of nine suberic acid-lysine-glutamate-urea targeting units, six Cy7.5 signaling units, and 52 butane-1,2-diol capping groups that rendered the remaining amines of the dendrimer inactive. Overall, this TCA was able to selectively accumulate in PSMA+ PC3 PIP better than PSMA- PC3 flu cells with a ratio of 6.2 ± 1.2. Conjugate IV was also able to accumulate in PSMA+ PC3 PIP tumors in vivo five hours after tail-vein injection. The PA intensities of the decomposed dendrimers peaked 48-hour post-injection, but a strong signal persisted up to the 72-hour time-point (Figure 13d).
Finally, the laboratories of Mei and Cheng developed a SPN that can target elevated PSMA in prostate cancer (Figure 13e).155 The SPN that was employed could absorb light in the NIR-II window, and demonstrated impressive photostability. The design of the TCA known as BTII-DUPA SPN utilized a bis-isoindigo-based polymer (BTII) backbone, with its surface modified with glutamate urea targeting ligands (DUPA). In vivo experiments were performed by implanting LNCaP (PSMA+) and PC3 (PSMA-) cells on opposing flanks. Upon intravenous injection of their TCA, there was a statistically significant enhancement throughout the entire volume of the LNCaP tumor, while only a surface level enhancement was observed in the corresponding PC3 tumor, suggesting non-specific staining. The PA enhancement peaked six hours after injection in the LNCaP tumors with a 3.9-fold signal enhancement with respect to the background signal (Figure 13f). Researchers were also able to image up to a depth of 4.2 cm, which is relevant for potential clinical applications (Figure 13g). Ex vivo analysis 72 hours after injection of their TCA demonstrated accumulation of BTII-DUPA SPN in the liver and spleen, and a 2.3-fold higher concentration in PSMA+ tumors than the PSMA- tumors. In addition, BTII-DUPA SPN was found to be biocompatible as there appeared to be no damage to organs nor did the animals lose weight after injection with the TCA, which is a general indicator of health.
5. Activatable PA Probes
In this section, we will highlight various design strategies that have been employed to develop activatable probes for PA imaging. An imaging agent that is activatable must be able to interact with its intended molecular target to afford an observable signal change that may come in the form of an increase (or decrease) in the PA intensity or a shift in the wavelength of maximum absorbance. One of the unique features of an activatable probe is that it does not have to rely on stochiometric binding interactions as is the case for many TCAs highlighted in section 4. Instead, it is possible to achieve signal amplification to detect very low abundance analytes since a single equivalent of an imaging target (e.g., enzyme) can in theory activate many copies of a probe before it becomes ‘spent’. Another critical feature is the ability to target analytes that do not possess an appreciable lifetime or one where the binding interaction is transient. This is made possible by leveraging its chemical reactivity to induce a signal change (see ABS in section 3). The overall impact that ABS designs have had on the field of analyte sensing is the ability to visualize an expanded set of analytes. An activatable probe may be based on a small-molecule or nanoparticle design. Both have their merits and will be highlighted accordingly below. On this note, we will be describing activatable probes that exists for six classes of analytes including enzymes, metal ions, reactive oxygen and nitrogen species, thiols, gradients of the tumor microenvironment, and nucleic acids. For biomarkers that are not list above, we refer our readers to the following references.157–160 When appropriate, we highlight the responsive unit in red to emphasize this important design element.
5.1. Enzymes
A plethora of enzymes have evolved to ensure that biological reactions in our bodies occur within an appropriate time scale. Since they are fundamental to our health and survival, it is not surprising that the dysregulation of enzymatic activity is linked to many pathological states including cancer. In this section, we highlight various enzymes that undergo activity and/or expression changes during cancer progression, as well as the activatable probe designs that have been employed to visualize them via PA imaging.
5.1.1. Alkaline Phosphatase
Alkaline phosphatase (ALP) is a promiscuous membrane-bound enzyme capable of catalysing the removal of phosphate esters from a variety of molecular structures. Its name originates from the fact that the optimal pH is in the alkaline pH range. Although the activity of this enzyme is often used to assess liver function and bone health, irregular levels of ALP have also been linked to a variety of cancer types.161 For instance, ALP is found to be elevated in breast, prostate, and liver cancers, whereas lower levels have been observed in ovarian tumors. Despite its prevalence in the body, it has still been exploited as a potential biomarker to develop activatable probes for PA imaging if potential off-target reactivity can be avoided.162, 163
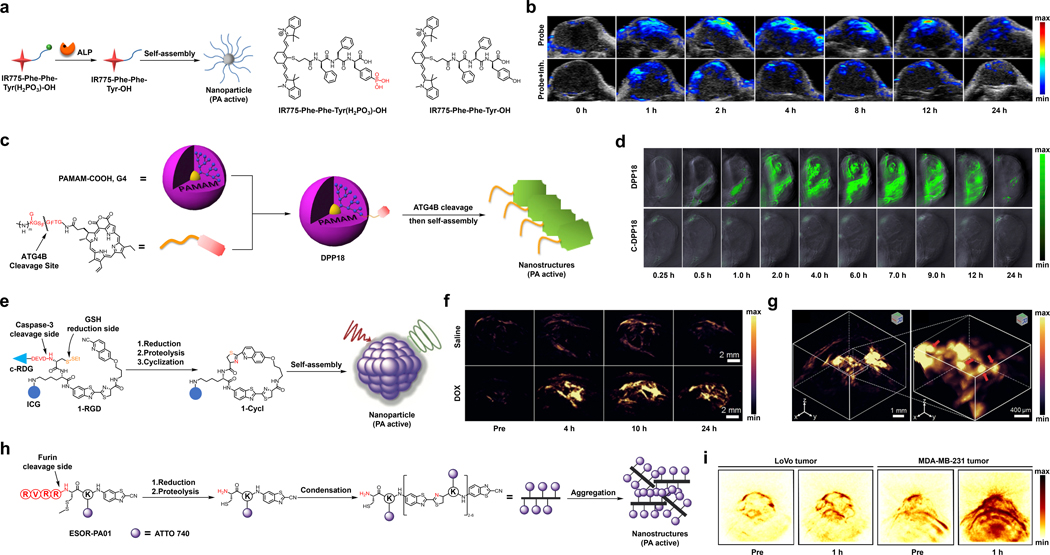
In one example, the laboratories of Cheng and Liang developed a cyanine-based activatable PA probe for ALP detection (Figure 14a).163 This probe, IR775-Phe-Phe-Tyr(H2PO3)-OH, features a IR775 dye component appended to a Phe-Phe-Tyr(H2PO3)-OH unit through substitution of the meso-Cl group of the parent dye. The design is based on the premise that once ALP catalyses the dephosphorylation reaction, the dye product becomes more hydrophobic, enters the cell, and self-assembles into a nanostructure which yields a PA signal enhancement. To test this design, the authors incubated the probe with ALP for three hours in vitro. They observed quenching of fluorescence due to formation of a non-fluorescent self-assembled NP which was confirmed using transmission electron microscopy (TEM). Since fluorescence and PA are opposing properties, the loss of fluorescence resulted in a 6.4-fold increase in the PA intensity. Additionally, the authors treated HeLa tumor-bearing mice with the probe via direct tumor injection (Figure 14b). As a control, a second group of HeLa tumor-bearing mice were administered an ALP inhibitor (l-phenylalanine) prior to injection of the probe. The maximum PA signal of the experimental mice was 2.3-fold higher than those from the control condition. In addition, ex vivo analysis revealed that the PA signal of the tumor was approximately 4.05-fold higher with respect to other major organs. It is conceivable this design could translate to other triggers if the probe is more hydrophobic than the corresponding product.
Figure 14.
a) Reaction of IR775-Phe-Phe-Tyr(H2PO3)-OH under ALP catalysis. b) PA images in the HeLa tumor-bearing mice before and at various time points after injection of the probe with or without pre-treatment using an ALP inhibitor. c) Reaction of DPP18 under ALP catalysis d) PA images in the breast cancer xenograft autophagy-induced model at various time points after injection of DPP18 or C-DPP18 (an unreactive control probe). e) Reaction of 1-RGD under caspase-3 catalysis and GSH reduction. f) PA images of U87MG tumor (treated with saline or DOX) before and at various time points after injection of 1-RGD. g) 3D PA imaging of DOX-treated tumor at ten hours after injection of 1-RGD. h) Reaction of ESOR-PA01 under furin catalysis. g) PA images of LoVo tumor and MDA-MB-231 tumor before and one hour after injection of ESOR-PA01. a) and b) are reproduced from ref. 163 with permission from American Chemical Society, Copyright 2018. c) and d) are reproduced from ref. 166 with permission from Elsevier, Copyright 2017. e), f) and g) are reproduced from ref. 169 with permission from John Wiley and Sons, Copyright 2019. h) and i) are reproduced from ref. 172 with permission from American Chemical Society, Copyright 2013.
5.1.2. ATG4B
Autophagy related 4B cysteine peptidase (ATG4B) is a cysteine protease involved in auto phagocytosis, which is a process that eliminates all unneeded contents within a cell via a lysosome-dependent pathway.164 Damaged organelles and endogenous misfolded proteins are obliterated in this process, rendering it fundamental to maintaining cell homeostasis. ATG4B has been proposed to be used as a biomarker for autophagy efficiency. A dysregulation of ATG4B in breast, colorectal, and prostate cancers have all been reported suggesting the protease’s role in carcinoma-related invasion and proliferation.165 Due to this, a number of inhibitors and molecular tools have been developed to detect and understand this biomarker’s role in cancer progression and treatment.
For instance, Wang and co-workers developed a PA nanoprobe known as DPP18 which self-assembles in vivo in order to monitor autophagy by detecting ATG4B activity (Figure 14c).166 The authors sought to develop a chemical tool to improve the effectiveness of autophagy-induced chemotherapeutics and subsequently minimize cytotoxicity. Their probe consisted of a hydrophobic P18 dye as the PA signal generating motif, an ATG4B peptide substrate (GKGSFGFTG), and a poly(amidoamine) dendrimer which served as an agent to improve water solubility and probe biocompatibility. Once the protease cleaves the peptide, the P18 motif aggregates and results in a PA signal enhancement via formation of nanofibrous structures, along with an aggregation quenched fluorescence response. In its monomeric form, DPP18 generates a high fluorescent output, and a negligible PA signal. However, upon incubation with ATG4B for two hours, there is a dramatic 3.8-fold PA signal enhancement with a concomitant decrease in fluorescence at 735 nm. When MCF-7 breast carcinoma cells were incubated with DPP18 and then subjected to a PBS control or rapamycin to induce autophagy, the PA signal change resulting from the PBS treatment of cells was negligible, while a significant 3.7-fold PA enhancement was observed in the rapamycin- treated cells. To confirm the involvement of ATG4B, the authors performed crucial knockdown experiments and showed subsequent treatment with DPP18 does not yield the same PA signal enhancement as was previously observed for the unmodified cells. In vivo PA imaging was performed using MCF-7 tumor-bearing BALB/c mice and P-Bec1 NPs (an autophagy inducer) were utilized to generate a breast cancer xenograft autophagy-induced model system (Figure 14d). Upon intravenous administration of DPP18, a PA signal enhancement of 1.1-fold could be observed as early as 30 minutes. The intensity continued to increase in a time-dependent manner, reaching a signal maximum at six hours yielding a 3.1-fold increase. These impressive findings demonstrate the feasibility for DPP18 to be utilized as a real-time monitoring probe of autophagy in tumors.
5.1.3. Caspase-3
Caspases are a family of proteases that are known for their proapoptotic activity and play various role in tumor progression. For example, caspase-3, when it is present at elevated levels in cancerous tissue, triggers apoptosis and the release of growth and differentiation factors that result in the proliferation of neighboring cells. 167 Due to such an important role in cancer development, caspases like caspase-3, caspase-9 have been used as a biomarker for tumor detection.168
An example of an activatable PA probe for caspase-3 was developed recently by Ye and co-workers (Figure 14e).169 The design of the probe, 1-RGD, was based on a caspase-3-initiated self-assembly design strategy. 1-RGD is composed of 2-cyano-6-hydroxyquinoline (CHQ) and a d-cysteine residue (d-cys) linked to a caspase-3 substrate (peptide with a DEVD sequence), a disulfide bond that can be cleaved by GSH, and ICG which serves as the NIR light absorbing dye. To make the probe selective for cancer cells, the authors introduced a targeting ligand for the ⍺vβ3 integrin receptor which is known to be overexpressed within carcinogenic tissue (see section 4.7). The activation mechanism requires the probe to interact with caspase-3 and GSH which liberates the amino and thiol groups of d-cys. Subsequently, d-cys undergoes an intramolecular condensation reaction with CHQ to yield a rigidified and hydrophobic product, 1-cycl. The PA signal increases due to aggregation-caused quenching of ICG after the product self-assembles into a nanostructure. In vitro evaluation revealed that almost full conversion of 1-RGD to 1-Cycl occurred within five hours. This could be monitored by observing a decrease in the fluorescence intensity (22-fold) since this is associated with the proposed cyclization and self-assembly sequence. Irradiation at 780 nm yielded a three-fold PA signal enhancement after five hours. To test their probe in vivo, the authors utilized nude mice bearing subcutaneous glioblastoma (U87MG) tumors which were intravenously treated with DOX or saline (Figure 14f and 14g). As mentioned previously, DOX is a chemotherapeutic that triggers an increase in caspase-3 activity, while saline was used as a control. Upon intravenous injection of 1-RGD, the PA intensity increased and reached a maximum after 10 hours, yielding a 4.4-fold enhancement with respect to the saline-treated mice.
5.1.4. Furin
Furin is classified as a proprotein convertase. Enzymes of this sort cleave dormant proteins into their biologically active form. Furin is a Ca2+-dependent protein that operates through hydrolysing peptides at specific sites. It is involved in a variety of processes such as embryonic maturation, production of growth factors, and pathological development.170 Specifically, irregular levels of furin have been observed to enhance the development and progression of colon, neck, brain, and skin carcinomas, to name a few.171 As such, it is of utmost importance to develop chemical tools capable of visualizing this protease’s activity in such malignancies.
For instance, Gambhir and co-workers developed a furin activatable PA probe, ESOR-PA01, that works via a biorthogonal condensation to yield aggregates and nanostructures that can be detected by a PA signal enhancement (Figure 14h).172 The design of ESOR-PA01 encompasses of a furin cleavable peptide, RVRR, appended to a disulfide protected cysteine and 2-cyano-6-aminobenzothiazole (CABT), and a NIR fluorescent dye, Atto740. Researchers also generated a control probe, ESOR-PA02, differing only in the peptide sequence (RRRV) which does not act as a furin substrate. Once inside the reducing environment of the cell, the disulfide is cleaved to expose the cysteine residue. Furin then engages with its substrate and releases the amino group of the cysteine. Next, a condensation reaction between the 1,2-amino thiol moiety and the 2-cyanogroup of CABT occur to form dimers, and subsequent oligomers. Finally, these oligomeric aggregates form NPs. The aggregation quenches the fluorescence of the Atto740 dye leading to an enhancement in the PA signal. A six-fold decline in the fluorescent output was observed after incubation of ESOR-PA01 under reducing conditions for three hours with furin present. TEM studies were conducted to confirm the formation of NPs post-furin cleavage. Next, in vitro experiments were performed in which the furin-overexpressing breast cancer MDA-MB-231 cells and low-furin colorectal cancer LoVo cells were incubated with ESOR-PA01. As a result, the MDA-MB-231 cells elicited a 2.6-fold higher PA signal response of ESOR-PA01 than in the LoVo cells. In addition, in vivo studies were conducted to demonstrate the efficacy of the probe in a living organism (Figure 14i). A group of MDA-MB-231 tumor-bearing mice underwent tail-vein injections of ESOR-PA01. All mice reported an average of 2.3-fold PA signal increase 60 minutes after injection. A separate LoVo-tumor bearing group of mice similarly underwent a tail-vein injection of the probe, and a minute increase in PA signal was reported. Impressively, the MDA-MB-231 tumor-bearing mice group elicited a 7.1-fold higher PA signal than the LoVo tumor-bearing group, demonstrating its novel ability to monitor furin proteolytic activity.
5.1.5. β-Galactosidase
β-galactosidase (β-Gal) is a glycosidase that catalyses the metabolic breakdown of lactose to yield the monosaccharides galactose and glucose. This enzyme is often used as a marker of senescence since aging cells are characterized by an overexpression of β-Gal.173 Additionally, its expression is elevated in ovarian tumors, as well as in breast cancer, colon cancer, and gliomas.174 A number of activatable probes have been developed to sense β-Gal activity in cancer using PA imaging.175–179
In 2018, Pu and co-workers developed a macrotheranostic probe, CyGal-P, for sensing β-Gal activity by capping the phenolic alcohol of an hemicyanine dye (HD) with a galactose trigger (Figure 15a).176 Prior to this work, HDs were used extensively for fluorescent probe development but its potential for PA imaging had yet to be unlocked. The specific dye that was selected in this study features a long PEG chain which was included to improve the biodistribution profile of the probe. The authors developed a matching control probe (CyGal), lacking the hydrophilic PEG group. Owing to the superior solubility of CyGal-P relative to CyGal, the turn-on response was 1.8-fold higher upon incubation with β-Gal in vitro. The corresponding in vivo studies yielded similar results. After establishing ovarian cancer tumors in nude mice using SKOV3 cells, the animals were administered either CyGal-P or the control. The PA signal increased in a time-dependent matter until the maximum response was reached after one hour (Figure 15b). The low water solubility of CyGal impeded its ability to accumulate in tumor cells, which compromised probe activation. Interestingly, CyGal-P was also utilized as a photothermal therapeutic agent that was used to treat tumors upon continuous irradiation with light.
Figure 15.
a) Reaction of CyGal-P under β-Gal catalysis. b) PA images in the SKOV3-tumor bearing mice before and at various time points after injection of CyGal-P or CyGal. c) Reaction of C2X-OR2 under β-Gal catalysis. d) PA images of the abdominal cavity of the mice using C2X-OR2 at various time points after the injection of OVCAR3 cell. e) PA images of the right hind foot pad of the mice using C2X-OR2 at various time points after the injection of OVCAR3 cell. 1: Primary tumor; 2: Lymph vessel; 3: SLN. f) Schematic of SPNP. g) Ratiometric turn on (treated and untreated with BEC) of SPNP at various time after injecting into 4T1-tumor-bearing mice. h) PA images of 4T1 tumours (treated and untreated with BEC) before and at various time points after injection of SPNP under 700 or 760 nm laser irradiation. i) Reaction of Rhodol-PA under hNQO1 catalysis. j) PA images of HT-29 and MDA-MB-231 tumors before and at various time points after intratumoral injection of Rhodol-PA. k) PA images of the HT-29 and MDA-MB-231 tumors before and at various time points after tail-vein injection of Rhodol-PA. l) Schematic of the reaction of TCHM@HA under hyaluronidase catalysis. m) PA images of the bladder of 4T1 cell injected mice and healthy mice (control) using TCHM@HA at various time points after the injection of 4T1 cell. 3: artery; 4: bladder. n) PA images of the paw of 4T1 cell injected mice (LN metastasis model) and healthy mice (control) using TCHM@HA at various time points after the injection of 4T1 cell. 1: tail bone; 2: paw. o) PA images of the popliteal position of 4T1 cell injected mice (LN metastasis model) and healthy mice (control) using TCHM@HA at various time points after 4T1 cell injection. 3: artery; 4: leg. p) 3D MIP MSOT image of the 4T1 cell injected mice (lymph-node metastasis model) and healthy mice (control) using TCHM@HA at various time points after 4T1 cell injection. 1: primary tumor; 2: lymphatic vessel; 3: SLN model. For l), m), n) and 0), both PA images (up) and PA images merged with background are shown. a) and b) are reproduced from ref. 176 with permission from John Wiley and Sons, Copyright 2018. c), d) and e) are reproduced from ref. 177, which is an open-access article distributed under the Creative Commons Attribution 4.0 International License, 2018 Springer Nature. f) and h) are reproduced from ref. 181 with permission from John Wiley and Sons, Copyright 2021. i), j) and k) are reproduced from ref. 184, which is an open-access article distributed under the Creative Commons Attribution NonCommercial 3.0 Unported License, 2019 Royal Society of Chemistry. l), m), n), o) and p) are reproduced from ref. 186 with permission from John Wiley and Sons, Copyright 2019.
Around the same time, Wu and co-workers developed a series of activatable probes for PA imaging based on the HD, one of which was for β-Gal (Figure 15c).177 The major structural difference between this probe (C2X-OR2) and the one discussed above is the use of a benzoindolium acceptor to red shift the dye absorption. With C2X-OR2 in-hand, the authors sought to investigate the ability of their probe to visualize ovarian tumors generated using the OVCAR3 cell line. After intratumoral injection, an increase in the fluorescent and PA signals were observed. Furthermore, the probe was employed to image a mimic of metastatic lesions (Figure 15d). A common site of metastasis for this cancer type is the abdominal cavity. As such, the authors injected ovarian cancer cells into this part of the body to mimic the spread of cancer. After several weeks, C2X-OR2 was injected intraperitoneally to successfully detect ovarian tumors via β-Gal activity. After an additional six weeks, enhanced PA images were obtained, suggesting steady cancer proliferation over this period. Likewise, the probe also applied to detect metastases within the lymphatic system (Figure 15e). Specifically, SKOV3 cells were injected into the right hind footpad of BALB/c mice and C2X-OR2 was administered at various time. The PA imaging data showed a clear signal in both primary tumor and SLN after six weeks. Together, this shows the authors were able to map the entire metastatic route from primary tumor to SLN, suggesting C2X-OR2‘s potential in metastasis visualization.
5.1.6. Granzyme B
Granzyme B is a serine protease that is compartmentalized into granules within cytotoxic T lymphocytes (CTL) and natural killer cells (NTK). Both CTL and NK are a part of an immune response that functions by binding to virulent cells and exocytosing the contents of the granules into the malignant cell. The release of granzyme B assimilates a number of cell-killing pathways to yield a potent immune-system mediated cell death response.180 Granzyme B is a key player in immune-system activation and has been proposed to be a promising target for monitoring cancer immunotherapy.
The first granzyme B activatable PA probe was developed in the laboratories of Zhang, Pu, and Miao (Figure 15f). Researchers synthesized a dual-modal semiconducting polymer nanoprobe (SPNP) in which both the fluorescent and PA signal outputs are altered upon interaction with granzyme B in order to visualize activation of the immune system in response to immunotherapy in relation to cancer.181 The probe was comprised of an amphiphilic semiconducting cyclopentyldithiophene (CPDT) derived polymer, appended to a granzyme B cleavable peptide conjugated to IR800 dye and further decorated with PEG moieties, all denoted as SPP. SPP then self-assembles into SPNP having two distinct signal outputs, that of the semiconducting polymer (SP) and the IR800 dye. Once activated by the target, the granzyme B-specific peptide (IEFDSG) is cleaved and the IR800 dye is released. Indeed, in vitro studies revealed that SPNP possessed two absorption maxima at 700 nm and 780 nm, representing that of the SP polymer and IR800, respectively. Once incubated with granzyme B, the PA signal of SPNP at 700 nm remained constant, while the PA signal at 760 nm decreased, suggesting the anticipated cleavage of the peptide had occurred. Incubation with the enzyme resulted in a ratiometric output (PA700/PA760) enhancement of 1.4-fold compared to that of prior to incubation. After confirming activation by granzyme B, the authors sought to investigate SPNP’s response in live cells. For this purpose, granzyme B-positive cytotoxic T cells (CD8+ T cells) and granzyme B- negative 4T1 breast cancer cells were used, treated with SPNP and incubated for four hours. CD8+ T cells that were dosed with the nanoprobe revealed a 1.6-fold enhancement in the fluorescent signal, from the SP component, relative to the signal from the nanoprobe incubated with 4T1 cells. As opposed to the supernatant, which yielded a 23.5-fold fluorescent signal increase of CD8+ T cells compared to the 4T1 cells. This dramatic increase suggests that granzyme B was able to aptly cleave the peptide of the nanoprobe. Finally, researchers explored the utility of SPNP to visualize granzyme B’s response to immunotherapy in vivo. 4T1 tumors were implanted into BALB/c mice, and S‐(2‐boronoethyl)‐1‐cysteine hydrochloride (BEC) was employed as an agent to evoke an immune response in recruiting CTLs. After BEC was intravenously injected into the mouse models, SPNP then followed. The PA700 signal increased reaching a maximum after 12 hours post‐injection and the PA700/PA760 ratio reached 1.2-fold higher than that of untreated group at 36 hours post injection (Figure 15g and 15h).
5.1.7. hNQO1
Human NAD(P)H: quinone oxidoreductase isozyme 1 (hNQO1) is a multifunctional enzyme stationed within the cytosol. In normal cells, hNQO1 plays important roles in detoxification, metabolism, and protection from peroxidative damage.182 However, in cancerous cells it is proposed that the overexpression of this enzyme lessens the efficacy of chemotherapeutics by reducing their oxidative toxicity, ultimately assisting in chemoresistance. A number of cancers have been observed to have upregulated levels of hNQO1 such as colorectal, ovarian, and lung carcinomas,183 making it a promising biomarker for cancer detection.
Wang, Jiang, and co-workers developed an hNQO1 activatable dual-modal probe for PA and NIR fluorescence imaging by synthesizing a novel spirocyclic xanthene dye appended to a trimethyl locked quinone propionic acid trigger (Figure 15i).184 The design takes advantage of the equilibrium that exists between the unconjugated spirocyclic form and the conjugated red-shifted “open” conformation. This allowed researchers to develop a rhodol derivative via conjugating the rhodol backbone with a difluorophenol motif, denoted as Rhodol-NIR. The esterified dye (Rhodol-PA) adopts the closed spirocyclic conformation and upon activation by hNQO1, the phenol is uncapped, and conjugation of the dye is restored to yield Rhodol-NIR. Rhodol-PA emits a miniscule PA signal from 680 nm to 800 nm. However, after incubation with the enzyme, a 11-fold signal-to-noise ratio was obtained at 680 nm. Similarly, at 720 nm the probe yields negligible fluorescence, but exhibits a signal-to-noise ratio of approximately 59 after activation by hNQO1. After researchers were able to confirm the ability of their probe to visualize hNQO1 in living cells, they turned to investigating it in vivo. HT-29 human colon adenocarcinoma cells (with high hNQO1 levels), or MDA-MB-231 breast cancer cells (with low hNQO1 levels) were implanted in BALB/c mice to generate tumor xenografts. PA images were collected at several time points post intratumoral injection of Rhodol-PA (Figure 15j). Significant PA signals were observed in the HT-29 tumor-bearing mice after only 30 minutes and reached a maximum around three hours. As expected, MDA-MB-231 tumor-bearing mice exhibited negligible PA signals throughout the duration of the experiment. The average PA signals of the HT-29 tumors to the MD-MB-231 tumors was eight-fold higher, signifying the ability of the hNQO1 probe to obtain PA images with high contrast. Further in vivo studies were conducted as researchers administered either Rhodol-PA or PBS control through a tail-vein injection to both the HT-29 and MDA-MB-231 tumor-bearing mice (Figure 15k). Similar results were obtained as before.
5.1.8. Hyaluronidase
Hyaluronidase (HAase) is an enzyme that catalyses the depolymerization of hyaluronic acid (HA), which is a massive and composite glycosaminoglycan.185 HA increases the permeability of the cell membrane and thus, controls how penetrable tissues are. The overexpression of HAase in tumor cells increases the breakdown of HA into smaller components that allow for the cancer to metastasize. Elevated levels of HAase have been reported in cancers of the breast, prostate, and bladder. As such, it has become a promising cancer biomarker.
The laboratories of Zeng and Wu developed an HAase activatable nanoprobe to visualize and detect tumors in breast cancer xenograft and orthotopic bladder cancer models along with visualizing LN metastases (Figure 15l).186 The nanoprobe consists of a tricyanofuran-containing heptamethine dye integrated with a quaternary ammonium moiety to yield a positively charged chromophore, denoted TCHM. HA was then revamped to integrate acetate groups into the polysaccharide component to alter the hydrophobicity property to ensure the generation of NPs. By exploiting the electrostatic and hydrophobic interactions between the positively charged TCHM chromophore and negatively charged HA trigger, the nanoprobe denoted as TCHM@HA was generated. Upon interaction with HAase, HA is degraded by the enzyme which changes the charge profile. This triggers dissociation of the NP, resulting in the release of the aggregated TCHM molecules which further disaggregates. The disaggregation event results in a PA signal enhancement at approximately 882 nm which can be leveraged to image HAase activity. For instance, 4T1 cells were injected into the posterior of the mice to generate a xenograft tumor model (Figure 15m). The nanoprobe was then injected intratumorally and a significant PA signal was observed, suggesting the ability of the nanoprobes to be activated in vivo by HAase. The authors obtained similar results in an orthotopic bladder tumor model (Figure 15n). To do so, T24 cells were injected into the bladder via catheter. Administration of the probe employed the same technique. An observable PA signal in the bladder was observed five days post-cancer cell injection, suggesting the signal is indeed produced at HAase-enriched locations. Finally, visualization of cancer metastases to LNs was also successfully performed (Figure 15o and 15p).
5.1.9. Matrix Metalloproteinases
The ECM is an integral component of the tumor microenvironment (TME) that undergoes significant structural remodeling as a function of cancer progression. In this regard, the enzyme family of matrix metalloproteinases (MMPs) are responsible for the degradation of a variety of ECM proteins. This in turn releases biologically active components that can significantly alter a cell’s behavior. For instance, MMP-2 is recognized as a cancer biomarker due to its overexpression in various cancer types, including those of the colon, prostate, breast, and bladder. This has motivated the development of a variety of activatable PA probes for this enzyme.187–195
For instance, the laboratories of Ji, Shi, and Gao developed QC, a PA probe that is capable of quantitatively detecting and monitoring MMP-2 levels in vivo (Figure 16a).193 Their design encompasses an MMP-2 peptide sequence, GPLGVRGY, linked to a Cy5.5 dye and a Förster resonance energy transfer (FRET) quencher, QSY21. QC is an amphiphilic molecule that in aqueous solution self-assembles into NPs. Prior to activation, there is a large PA signal at 680 nm and a low PA signal at 730 nm. However, upon activation by MMP-2, the PA signal at 680 nm decreases, and the intensity at 730 nm remains unchanged. These properties allow for QC to be utilized as a ratiometric PA probe which is an important design feature since it can improve confidence in a given result. After demonstrating QC is non-cytotoxic, the authors showed probe activation via MMP-2 activity in 4T1 breast cancer cells. Further, they employed a 3T3 fibroblast cell line which served as the negative control owing to the low levels of MMP-2 present. The results translated well in vivo as the authors were able to successfully show that QC could detect different levels of MMP-2 as a f unction of tumor volume (Figure 16b).
Figure 16.
a) Reaction of QC under MMP-2 catalysis. b) PA images of 4T1 tumors of different sizes after injection of QC or control probe under 680 nm or 730 nm laser irradiation. c) Reaction of AlNc-pep-PEG under MMP-2 catalysis. d) PA images (with and without merged background) of the HT-1080 fibrosarcoma tumor before and after injection of AlNc-pep-PEG or AlNc-PEG under 680 nm or 760 nm laser irradiation. e) Schematic of the synthesis and reaction of PEG-pep-Dox-AuNPs under MMP-2 catalysis. f) PA images (with and without merged US signal) in SCC-7 tumor-bearing mice before and at various time points after injection of EG-Dox-AuNPs or EG-pep-Dox-AuNPs. g) Reaction of AuNS-1 and AuNS-2 with MMP-9. h) 3D PA image in a breast cancer mouse model at 24 hours after injection of saline, control probe and MMP-9 probe. Yellow: skin and surface vasculature; red: aggregated nanoparticles; light blue dashed line: tumor boundary. i) Reaction of P-Dex under uPA catalysis. j) PA images of the MDA-MB-231 tumor (with or without pre-treatment of 4-CPG) or MCF-7 tumor before and at various time after injection of P-Dex or SP. a) and b) are reproduced from ref. 193 with permission from American Chemical Society, Copyright 2019. c) and d) are reproduced from ref. 195 with with permission from American Chemical Society, Copyright 2021. e) and f) are reproduced from ref. 191 with permission from Elsevier, Copyright 2019. g) and h) are reproduced from ref. 196, which is an open-access article distributed under the Creative Commons License, 2021 Elsevier. i) and j) are reproduced from ref. 199 with permission from John Wiley and Sons, Copyright 2020.
In another example, Miki, Ohe, and coworkers also employed a self-assembly strategy to develop activatable probes for MMP-2 (Figure 16c).195 Their design is based on a photostable NIR naphthalocyanine scaffold which is conjugated to a PEG moiety through a MMP-2 substrate sequence. Cleavage of the peptide results in a naphthalocyanine (MNc) product that is poorly soluble in aqueous media and thus, spontaneously aggregates in solution. The resulting self-assembled particles exhibit an increase in the PA signal at 680 nm and a decrease at 760 nm which allows for ratiometric imaging. It is noteworthy the authors developed two versions based on the incorporation of either silicon (SiNc-pep-PEG) or aluminum (AlNc-pep-PEG). Due to the better performance of AlNc-pep-PEG in cellular studies (larger ratiometric response to MMP-2), the aluminum analog was evaluated further in vivo. In particular, the authors generated HT-1080 fibrosarcoma tumors and intravenously administered AlNc-pep-PEG or a non-activatable control probe AlNc-PEG (Figure 16d). The PA intensity of the control did not change after one hour; however, the signal ratio for the probe increased by 2.5-fold during the same time frame. This result demonstrates the ability of their probe to detect elevated MMP-2 activity in live subjects.
Moreover, Chen, Nie, and coworkers developed a AuNP-based theranostic agent known as PEG-pep-Dox-AuNPs, which is equipped with therapeutic and imaging capabilities (Figure 16e).191 The design exploits the overexpression of MMP-2 in carcinogenic cells to generate an assembly of AuNPs. Two sets of AuNPs are each decorated with complementary DNA strands on the exterior of the NPs, along with DOX appended via a heat-responsive linker. Additionally, the AuNPs surface was modified with PEG moieties that incorporate an MMP-2 substrate via a peptide linker, allowing for both enhanced circulation half-life and specificity. Once the AuNPs encounter tissue overexpressing MMP-2, the PEG component is cleaved, exposing the complementary DNA strands that allows for rapid hybridization, aggregation, and a red shift in the absorption profile into the NIR. One group of squamous cell cancer carcinoma (SCC-7) tumor-bearing mice were injected with the activatable MMP-2 probe, while another group was treated with control AuNPs (PEG-Dox-AuNPs) lacking the MMP-2-responsive peptide (Figure 16f). The average PA intensity of the probe in tumors was 1.74-fold higher than for the control particle group. Interestingly, the increase in local-temperature due to the photothermal effect releases DOX for more effective cancer killing. This design showcases the potential utility of DNA-hybridization-based strategies since other masking-responsive groups can be readily employed to block hybridization.
Besides MMP-2, there is also a PA probe reported for MMP-9. Recently, Emelianov and coworkers have developed a gold nanosphere (AuNSs)-based MMP-9 probe using a DNA aptamer as the binding moiety (Figure 16g).196 Specifically, this MMP-9 probe is based on two kinds of DNA-modified 15 nm-sized AuNSs. The first AuNSs (AuNS-1) features a surface which was modified with a double stranded aptamer for MMP-9 and its partial complementary sequence (comp-1). For the second AuNSs, AuNS-2, it was modified with a single strand of DNA (comp-2) which was complementary to comp-1. In the absence of MMP-9, the MMP-9 aptamer would bind to comp-1, preventing the interaction between comp-1 and comp-2, hence maintaining the optical properties of both AuNS-1 and AuNS-2. When MMP-9 is present, it can form a complex with the MMP-9 aptamer, opening the binding site on comp-1 and allowing the binding between comp-1 and comp-2. This binding would lead to the aggregation of AuNS-1 and AuNS-2, resulting in plasmon coupling effect that results in a red-shifted absorption profile. With this design, the authors were able to achieve the MMP-9 specific response of this probe in vitro and tested its PA image ability in vivo using a xenograft breast cancer model. At 24 hours post-injection, the MMP-9 probe showed PA signal with spectrum that matched in vitro result, while minimal signal was observed for saline control and control sensor (AuNSs modified with random DNA sequence). Moreover, the distribution of this probe was further studied volumetrically using the TriTom system together with saline and control sensor, and the results agreed with previous study that only MMP-9 probe showed the aggerated AuNS signal in the tumor, demonstrating its potential in in vivo tumor detection. (Figure 16h)
5.1.10. Urokinase-type Plasminogen Activator
Urokinase-type plasminogen activator (uPA) is another serine protease produced by kidney cells. It is fundamental for a number of processes such as embryogenesis, tissue remodeling, and wound healing.197 As a protease, it also has pathological roles like degrading the EM which facilitates cancer metastasis. Carcinomas that have been observed to overexpress uPA include colorectal, gastric, and breast cancer.198 Oftentimes, upregulated levels of uPA indicate poorer patient prognosis, demonstrating its potential as a cancer biomarker.
The laboratories of Miao and Pu developed a dual-modal fluorescence and photoacoustic probe that can detect malignant breast carcinoma via targeting uPA, due to its overexpression in invasive breast cancer (Figure 16i).199 The authors envisioned the probe to be highly specific and sensitive regarding uPA, and have suitable renal clearance. As such, the probe, denoted P-Dex, consisted of a dextran scaffold to facilitate renal-clearance, a hemicyanine dye (CyN3OH), a p-aminobenzyl alcohol self-immolative linker, and a uPA targeted peptide substrate (Cbz-Gly-Gly-Arg-OH). Throughout their studies, SP was utilized as a control probe, differing from P-Dex only by the absence of the dextran backbone. Prior to activation by uPA, P-Dex is essentially rendered non-fluorescent and not PA-active since the p-aminobenzyl alcohol cages the hydroxyl of the HD platform which significantly reduces its electron-donating capabilities. Once the uPA cleaves the peptide substrate, a spontaneous 1,6-elimination occurs, and a turn-on response is observed. Firstly, photophysical data was collected on both P-Dex and SP. Both probes had an absorption maximum at 610 nm, along with no PA or fluorescent signal output. However, upon incubation with uPA for only 20 minutes, both P-Dex and SP experienced a notable decrease at 610 nm concurrent with a signal increase at 695 nm corresponding to the release of the free dye, allowing for ratiometric imaging to take place. P-Dex reached a 695/610 ratio maximum after only 20 minutes, indicating the complete conversion to the free hemicyanine platform. In addition, P-Dex had a noteworthy 2.1-fold PA signal enhancement compared to that of SP. When the authors assessed the metabolism of the probe via examination of the excreted urine from mice 24 hours after intravenous injection of P-Dex or SP, they found P-Dex had a renal-clearance efficiency of 70% while SP only had a 24% efficiency. The probes were then investigated in vivo in MDA-MB-231 and MCF-7 tumor-bearing mice (Figure 16j). P-Dex was injected intravenously, and a maximum PA signal was observed just two hours after. This signal was substantially reduced when the mice were pre-treated with uPA inhibitor, 4-CPG, to block activation. SP yielded a much lower signal due to poor water solubility from the absence of the dextran component. No PA or fluorescent signals were observed in the MCF-7 tumors, highlighting the ability of P-Dex to distinguish between invasive and non-invasive breast cancers in vivo.
5.2. Metal Ions
Metal ions like copper (Cu) are indispensable to the survival of all living organisms.200 For instance, it serves as a co-factor in many enzymes (e.g., superoxide dismutase) that protect the body against oxidative stress. Additionally, the labile Cu pool, which is defined as Cu bound weakly to intracellular chelators such as GSH, has been shown to further function as a transient signaling molecule.201, 202 Owing to its general importance, the body has evolved a variety of tightly regulated mechanisms including the expression of importers, exporters, and chaperons to maintain Cu homeostasis. However, when these safeguards become dysregulated, Cu can impair cellular health and has been reported to be elevated in breast, gastrointestinal and lung cancers.203 Accordingly, the development of chemical tools that can detect and track aberrant levels of Cu with high resolution within the TME is critical to understanding its role in cancer progression.
While our group developed the first PA imaging probes for Cu (APC-1204 and CRaB-OMe-APC205), other probes (RPS1,206 LET-2207) have also been reported for Cu brain/plant imaging (Figure 17a). Each of these examples target the +2-oxidation state which is less prevalent in tumors owing to the highly reducing intracellular environment found in cancer. As such, we developed PACu-1 which features a Cu+-responsive TPA-based trigger attached to an optimized aza-BODIPY dye (Figure 17b).208 Upon binding of Cu+ to TPA, the probe undergoes an oxidative cleavage event which separates the chelator and dye moieties, restores the push-pull capabilities of the aza-BODIPY, and induces a red-shift in absorbance of 91 nm. This latter property is important since it allows for accurate ratiometric imaging of Cu+ by generating PA signals at the two wavelengths corresponding to the probe and dye product. Because the liver is a prevalent metastatic site for the cancer types mentioned above, we established an orthotopic model of liver metastasis using A549 cells to evaluate PACu-1. Prior to in vivo testing, we demonstrated that PACu-1 exhibits a robust turn-on in the presence of GSH up to 10 mM. This experiment is important since the liver and A549 cells are characterized by high levels of GSH.208 When compared to mice that received sham surgeries (1.06 ± 0.28), the PA fold turn-on was 2.31 ± 0.78 in tumor-bearing mice (Figure 17c and 17d). Our findings demonstrate that it is possible to employ imaging probes to sense aberrant metal ions like Cu+ in tumors.
Figure 17.
a) PA probes for Cu2+. b) Reaction of PACu-1 under Cu+-mediated activation. c) PA images of the A549 tumor-bearing mice and healthy mice after injection of PACu-1. d) Quantification of photoacoustic turn-on from c). c) is reproduced from ref. 208 with permission from National Academy of Sciences, Copyright 2021.
It is worth noting that in addition to the aforementioned Cu probes, a number of groups have developed PA imaging agents for detection of Li+,209 K+,210, 211 Ca2+ 212–215 and Zn2+ 216, 217. Although these have yet to be applied to track metal ions in cancer models, we anticipate this will be an exciting avenue to pursue in future studies.
5.3. Reactive Oxygen and Nitrogen Species
Reactive oxygen species (ROS) and reactive nitrogen species (RNS) are collectively known as RONS. In the context of cancer, this class of reactive molecules contribute to oxidative/nitrosative stress which is linked to cancer initiation, progression, and metastasis. In the following sections, we highlight activatable PA probes that have been developed to image these species in cancer models.
5.3.1. Hydrogen peroxide
As one of the most important ROS, hydrogen peroxide (H2O2) participates in cellular processes like hypoxic signal transduction, cell differentiation, and the immune response.218 With regards to cancer development, H2O2 is associated with many cancer hallmarks related to processes like cell proliferation, apoptosis resistance, metastasis, and angiogenesis.219–222 Due to the high H2O2 production in tumor cells,218, 223 it has been explored as a popular biomarker for cancer diagnosis and therapy.224–232 In the PA imaging field, there are also many reports related to the H2O2 imaging in cancer, some notable examples are discussed below.233–243
A popular design that has been used to generate a H2O2-specific response is to leverage the horseradish peroxidase (HPR)-catalyzed H2O2 oxidation reaction of ABTS,244 which will generate ABTS•+ (Figure 18a). This product features a strong NIR absorption profile which can be utilized for PA imaging. This design approach was first applied in PA imaging by Liu and coworkers in their H2O2-responsive liposomal nanoprobe known as Lipo@HPR&ABTS (Figure 18b).234 The use of a phospholipid-based liposome as the carrier, Lipo@HPR&ABTS was synthesized by loading the ABTS into the liposomal membrane and encapsulating the HPR enzyme into the aqueous liposomal core. Due to the low polarity of H2O2 compared with H2O, H2O2 can be transported into the liposomal core and induce the oxidation of ABTS under HPR catalysis, generating the PA signal in a highly selective and sensitive manner. To demonstrate the versatility of Lipo@HPR&ABTS, the authors employed it for inflammation detection, cancer imaging and photothermal therapy. In the cancer detection studies, Lipo@HPR&ABTS was first applied in detecting murine breast 4T1 tumors. The authors obtained a significantly higher PA signal in the tumor region compared with the control experiments where Lipo@HPR or Lipo@ABTS were applied (each control is missing one of the key components) (Figure 18c). Further, the sensitivity of Lipo@HPR&ABTS was tested by detecting tumors as small as ~2 mm in diameter, suggesting its potential use in the early-stage cancer imaging (Figure 18d). Further experiments were performed in the orthotopic brain glioma and LN metastatic models. Results from both sets of experiments show Lipo@HPR&ABTS could be employed to image oxidative stress in cancerous tissue with high PA contrast based on the elevation of H2O2 (Figure 18e and 18f).
Figure 18.
a) Reaction of ABTS under HPR-catalyzed H2O2 oxidation. b) Schematic of the reaction of Lipo@HRP&ABTS with H2O2. c) PA images of the 4T1 tumors before and at various time after injection of Lipo@ABTS, Lipo@HRP or Lipo@HRP&ABTS. d) PA images of the 4T1 tumors at different sizes before and at various time after injection of Lipo@HRP&ABTS. e) PA images of the orthotopic brain glioma tumors before and at various time after injection of Lipo@HRP&ABTS. f) PA images of SLNs with or without metastasis before and at various time points after injection of Lipo@HRP&ABTS. g) Schematic of the structure of Gd@HPRABST. h) Schematic of the synthesis of Gd@HPRABST. i) Schematic of FA-RM:GQDzyme/ABTS. j) PA images of the CNE-2 tumors before and at various time after injection of FA-RM:GQDzyme/ABTS, RM:GQDzyme/ABTS, RM-GQDzyme or RM-ABTS. k) Schematic of the synthesis of AMP NRs. l) PA images of the 4T1 tumors (treated with DMTU or FEITC, untreated as control) before and at various time after injection of AMP NRs. b), c), d), e) and f) are reproduced from ref. 234 with permission from National Academy of Sciences, Copyright 2017. g) and h) are reproduced from ref. 236 with permission from American Chemical Society, Copyright 2018. i) and j) are reproduced from ref. 239 with permission from American Chemical Society, Copyright 2019. k) and l) are reproduced from ref. 245 with permission from John Wiley and Sons, Copyright 2019.
In addition to the example above, another H2O2 probe based on HPR catalyzed oxidation of ABTS was reported by Zhang and coworkers (Figure 18g and 18h).236 Gd@HRPABTS employs the same H2O2-responsive mechanism featuring a protein-assisted biomimetic synthesis strategy. This rendered preparation of the probe easier and improved the biocompatibility of the resulting NP. The role of the HPR enzyme in Gd@HRPABTS was not only to catalyse the key oxidation reaction, but also as an incubator, stabilizer, and carrier during the synthesis. The authors found that Gd@HRPABTS was compatible for both PA and MR imaging, and its performance in vivo for oxidative stress detection in cancer was extensively studied in a following report from the same lab.242
Notably, there has also been efforts to replace HRP with an artificial enzyme to achieve the H2O2 response necessary to yield a PA signal. One such example comes from the laboratories of Nie, Yan and Fan where the researchers developed an exosome-like nanozyme known as FA-RM:GQDzyme/ABTS to image elevated H2O2 levels in nasopharyngeal carcinoma (NPC) (Figure 18i).239 The key component of this nanozyme design is GQDzyme, a graphene that is nano size which possesses intrinsic peroxidase-like activity. To construct this exosome-like structure, the authors first loaded GQDzyme with ABTS through a π−π interaction mechanism, forming the GQDzyme/ABTS NPs which was then encapsulated by folate acid conjugated natural erythrocyte membranes. The in vivo evaluation of FA-RM:GQDzyme/ABTS for NPC imaging was performed in CNE-2 tumor-bearing mice with RM-ABTS, RM-GQDzyme and RM:GQDzyme/ABTS as a control (Figure 18j). The results showed that minute signal appeared when RM-ABTS or RM-GQDzyme was employed. However, there was an enhancement in the PA signal for FA-RM:GQDzyme/ABTS compared with RM:GQDzyme/ABTS, presumably due to the tumor-targeting effect from FA ligand (section 4.5).
Similarly, Chen, Tian and coworkers discovered that MIL-100, a metal organic framework (MOF), also exhibited activity to oxidize ABTS in the presence of H2O2 (Figure 18k).245 Specifically, the authors noticed that MIL-100 had a 5.2-fold higher catalytic activity at pH 6.8 than that at pH 7.4, indicating the reactivity will be more pronounced in the acidic TME. Based on their discovery, the researchers designed a theranostic nanoreactor (AMP NRs) that would possess the PA signal turn on and the therapeutic effect in the H2O2 rich, acidic TME. The AMP NRs was synthesized by coating ABTS-preloaded MIL-100 with poly(vinylpyrrolidone) (PVP), and the feasibility of using AMP NRs in in vivo imaging was evaluated in 4T1 tumor-bearing mice (Figure 18l). The result showed that AMP NRs could image tumors at different stages of development, including sizes as small as 20 mm3. Moreover, the AMP NR’s response towards H2O2 was validated by pre-treating tumors with DMTU (a H2O2 scavenger) or FEITC (a H2O2 stimulator). The PA images showed attenuated signals in the DMTU group, whereas the strongest signal was in the FEITC treatment group.
5.3.2. Hypochlorite
Another biologically relevant ROS is hypochlorite (ClO-) which plays a role in various types of cancer-related cellular processes246–248 and thus, has attracted attention from activatable probe developers.
For instance, Pu, Fan and coworkers have developed an activatable ClO--responsive probe for ratiometric PA imaging based on a degradable SPN scaffold (Figure 19a).249 Specifically, the authors designed an optically active semiconducting oligomer amphiphile (SOA) and incorporated phenothiazine, a ROS-oxidizable unit that can react with ClO- in a manner that leads to the degradation of the polymer. Due to its amphiphilic nature, SOA can self-assemble to form NPs, which can be used to encapsulate a ROS-inert dye (NIR775) for use as an internal standard. Once SOA has been degraded by ClO-, its PA signal becomes attenuated, whereas the PA signal of NIR775 remains the same, leading to a change in the signal ratio. After demonstrating the selectivity and in vitro reactivity of the SOA nanoprobe towards ClO-, in vivo ClO- imaging was performed in the subcutaneous 4T1 xenograft tumor model (Figure 19b). The PA signals (background subtracted) at 680 nm and 780 nm were recorded, demonstrating both PA signals increased over time, presumably due to the EPR effect of nanoprobe. However, PA680 increased at a slower rate than that of PA780, leading to the increased PA780/PA680 ratio of 3.15±0.12.
Figure 19.
a) Schematic of the synthesis of SOA NPs. b) PA images of subcutaneous 4T1 xenograft tumors before and at various time points after the injection of SOA nanoprobe under 680 nm or 780 nm laser irradiation. a) and b) are reproduced from ref. 249 with permission from American Chemical Society, Copyright 2017.
5.3.3. Nitric oxide
Nitric oxide (NO) is a short-lived RNS that plays a vital role in neurological processes,250 inflammatory conditions,251 and tumor progression.252–254 In the context of cancer, it can either promote or inhibit tumor growth depending on its timing of production and local concentration.253 Therefore, methods capable of non-invasive NO detection is critical to delineating its dichotomous role in cancer biology. Until recently this was a profound challenge owing to the short biological lifetime (seconds)255 and low abundance (micromolar range) of NO in tumors. However, a number of activatable PA probes for NO have been developed.256–260
Our initial foray into NO sensing exploited a nucleophilic N-alkylaniline trigger, which reacts with NO (in the form of N2O3) to afford a N-nitrosated product.256 By installing this trigger onto an aza-BODIPY dye, we were able to detect NO in an LPS-induced inflammation model (millimolar range261); however, this first-generation probe lacked sufficient sensitivity to visualize NO in tumors. We hypothesized we could improve the limit of detection by integrating this trigger onto a new aza-BODIPY platform where two of the pendant phenyl moieties were exchanged for thiophene rings to afford SR-APNO-3, a ratiometric probe that produce a higher PA690/PA790 ratio upon reacting with NO. (Figure 20a).258 This modification was introduced because thiophenes are smaller, resulting in a red-shifted absorbance and enhanced extinction coefficient due to a more planarized structure and improved orbital overlap. Moreover, the sulfur atoms can quench fluorescence via the heavy atom effect which also favors a stronger PA signal. With this new probe, we successfully detected endogenous NO for the first time via PA imaging in a 4T1 breast cancer model. We confirmed these results in vivo by treating tumors with L-NMMA (potent NOS inhibitor) to attenuate NO biosynthesis. Relative to untreated tumors, we observed that the PA response decreased by 81% (Figure 20b and 20c).
Figure 20.
a) Reaction of SR-APNO-3 with NO via N2O3. b) Ratiometric PA turn-on of SR-APNO-3 in the 4T1 breast cancer model. c) PA imaging of the healthy tissue, 4T1 tumor and 4T1 tumor (L-NMMA treated) after SR-APNO-3 injection. Blue color bar: 690nm; red color bar: 790nm. d) Reaction of APNO-1080 with NO via N2O3. e) PA turn-on of APNO-1080 in the A549 lung cancer model. f) PA imaging of the healthy tissue and A549 tumor after APNO-1080 injection. c) is reproduced from ref. 258, which is an open-access article distributed under the Creative Commons Attribution NonCommercial 3.0 Unported License, 2019 Royal Society of Chemistry. f) is reproduced from ref. 259 with permission from American Chemical Society, Copyright 2020.
While SR-APNO-3 showed improved sensitivity, we were motivated to devise a complimentary strategy to detect NO in cancer. To this end, we developed APNO-1080, a NIR-II PA probe for NO, by employing a two-phase tuning approach.259 First, we identified a new aniline-based trigger by varying the electronics of the substituent at the para position. Second, we installed the optimized trigger onto a panel of five NIR-II absorbing cyanine dyes (Figure 20d). The resulting probe absorbs maximally in a spectral region where interference from pigments such as hemoglobin and melanin are minimized which enables more incident light to reach the probe. To test its in vivo performance, we implanted A549 lung cancer cells into a deep-seated region of the liver in Nu/J mice. It is worth noting that the liver is rich in blood and therefore, is a challenging organ to study using PA imaging. Relative to control mice, we observed a 1.65-fold turn-on response in tumor bearing animals, demonstrating the enhanced properties of APNO-1080 for deep-tissue NO imaging (Figure 20e and 20f).
5.3.4. Superoxide anion
Superoxide anion (O2•-) is known as the precursor of many other ROS and free radicals, and it can regulate processes like DNA methylation, histone methylation and histone acetylation.262, 263 Due to the central role of O2•- in the generation of other ROS, selective O2•- imaging is of interest in understanding the role of ROS in cancer development.
One report comes from Zhang and Xing where the authors have developed a BODIPY-based molecule, BDP-DOH, as an O2•- activatable PA probe (Figure 21a).264 The key feature of BDP-DOH is the ortho-phenolic hydroxyl group that can be selectively oxidized by O2•-, which leads to the enhanced PA signal at 750 nm and the attenuated PA signal at 680 nm. More importantly, the authors also demonstrated that the oxidized probe, BDP-DO, could be reduced back to BDP-DOH by GSH. They hypothesized that the redox cycle of BDP-DO could be utilized to monitor cellular redox homeostasis in cancer. After in vitro analyses which were employed to confirm BDP-DOH could be activated by O2•- and then deactivated by GSH, the authors conducted in vivo experiments in EMT6 tumor-bearing mice using the BDP-DOH/LP NP formulation, which was synthesized through self-assembly of BDP-DOH and DSPE-PEG2000-COOH to improve biocompatibility and tumor accumulation (Figure 21b). To validate their hypothesis, saline, PMA (a stimulator for O2•- generation) or PMA + LPA (a stimulator for the GSH generation) were injected intratumorally after the administration of BDP-DO (Figure 21c and 21d). The results showed an obvious enhancement in the PA750/PA680 ratio in the PMA-treated mice and a decreased PA ratiometric turn on in the PMA + LPA group, indicating O2•- could activate BDP-DO and GSH could attenuate this activation.
Figure 21.
a) Reaction of BDP-DOH with O2•- and GSH. b) Schematic of the synthesis of BDP-DOH/LP. c) PA images of the EMT6 tumor (treated with saline, PMA, or PMA + LPA) after the injection of BDP-DOH/LP under 680 or 750 nm laser irradiation. d) PA intensities of the EMT6 tumor (treated with saline, PMA, or PMA + LPA) after the injection of BDP-DOH/LP under 680 or 750 nm laser irradiation. b) and c) are reproduced from ref. 264 with permission from American Chemical Society, Copyright 2019.
5.3.5. Peroxynitrite
Peroxynitrite (ONOO-) is generated via a rapid reaction between O2•- and NO. It is characterized by its strong oxidizing ability, high reactivity, and short biological lifetime. This molecule has been linked to DNA injury, mitochondrial dysfunction, and surfactant protein damage.265, 266 It has been reported that ONOO- can nitrate amino acid residues to modulate the function and structure of cancer-related proteins like p53, CD8 and CCL2, and consequently attenuate tumor immunosuppression.267–269 With its unique role in tumor progression, activatable probes for this RONS can be beneficial in further deciphering these mechanisms in cancer.
In this regard, Pu and coworkers have made considerable advances toward PA imaging of ONOO-. For example, the authors have developed OSN-B1, an organic semiconducting nanoprobe synthesized via nanoprecipitation of a ONOO- responsive dye (BBD) and triphenylborane (TPB) under PEG-b-PPG-b-PEG assistance (Figure 22a).270 It is noteworthy that while the phenylboronic acid pinacol ester trigger in BBD was known to react with both ONOO- and H2O2,235 the researchers managed to inhibit the H2O2 reactivity through a TPB-doping strategy. To be more specific, ~4-fold of TPB compared to BBD was added during the nanoprecipitation step which served as a shield to prevent the BBD from reacting with H2O2 since only ONOO- was strong enough to oxidize TPB and reach the BBD. With this ONOO- selectivity, the authors were able to detect ONOO- in subcutaneous 4T1 xenograft tumors (Figure 22b). During this experiment, one group of mice was pre-treated with N-acetyl cysteine (NAC) (a ROS scavenger) and the ratiometric PA turn on was calculated. The data showed that, while the NAC-treated group had limited ratiometric PA turn on over 24 hours, the NAC untreated group had an obvious PA turn on that was 2.6-fold higher than the NAC treated group when it reached maximum at four hours post-injection.
Figure 22.
a) Schematic of the synthesis of OSN-B1. b) PA images of subcutaneous 4T1 tumors (with or without NAC treatment) before and 4 hours after the injection of OSN-B1 under 680 nm or 750 nm laser irradiation. c) Reaction of CySO3CF3 with ONOO-. d) PA images of subcutaneous 4T1 tumors before and at various time points after the injection of CySO3CF3. a) and b) are reproduced from ref. 270 with permission from John Wiley and Sons, Copyright 2016. d) is reproduced from ref. 271 with permission from American Chemical Society, Copyright 2018.
Beside OSN-B1, a small-molecule-based ONOO- probe (CySO3CF3) with fluorescence and PA dual imaging ability was also reported by the same group (Figure 22c).271 The design of CySO3CF3 leveraged the ONOO- selective oxidation of trifluoromethyl ketone and the following elimination reaction to uncage the HD, generating the fluorescence and PA signal. The in vivo imaging capacity of CySO3CF3 was validated in the subcutaneous 4T1 xenograft tumor model with the PA signal in the tumor region being 2.1-fold higher than the background (Figure 22d).
5.4. Glutathione and Hydrogen Sulfide
5.4.1. Glutathione
GSH is a highly abundant tripeptide thiol found in the body. It is involved in many important cellular processes including antioxidation, immune modulation and detoxification.272 For instance, in the liver where levels are as high as 10 mM, an enzyme known as glutathione S-transferase (GST) can catalyze the addition of GSH to xenobiotic molecules including drugs to aid in their removal from the body. Notably, GSH is also found to be elevated in many cancer types and plays a complex role in cancer development. In particular, GSH can protect normal cells from oncogenesis; however, the elevated levels of GSH in cancer cells can also attenuate the effect of chemo- and radiotherapy.272–274 Therefore, developing GSH probes is of interest in understanding its role in cancer development and to facilitate the application of GSH-targeted cancer therapy. Due to the deep tissue penetration of PA imaging, many activatable PA probes have been developed as tools to study the in vivo behavior of GSH,275–286 and here we introduce some recent progress in this area.
One recent example comes from Fan and coworkers where they have reported a GSH activatable PA probe with a ratiometric imaging readout (Figure 23a).287 Their reported probe (IR806-PDA) is based on the IR806 cyanine dye and features a GSH-responsive disulfide pyridine trigger. Prior to the reaction of GSH, the intact trigger is linked to the dye via the amino group which donates electron density into the conjugated π-system, leading to a blue shifted absorption profile. However, upon reaction with GSH, the disulfide trigger is cleaved to unmask a nucleophilic thiolate, which undergoes an intramolecular substitution of the amino group. The resulting thiol-link trigger is red shifted because the sulfur atom is less efficient at electron donation and thus, a change in the PA signal is accompanied by the reaction. Of note, IR806-PDA can also self-assemble to form homogenous NPs in aqueous solution due to its amphiphilic nature, which facilitates probe uptake and accumulation in tumors presumably via the EPR effect. The ratiometric change (PA820/PA620) of IR806-PDA in response to GSH was evaluated in vivo after in vitro studies demonstrated activation was mediated by GSH. The authors used a subcutaneous HeLa xenograft tumor model to show a 2.94-fold change in the ratiometric PA response (Figure 23b).
Figure 23.
a) Reaction of IR806-PDA with GSH. b) PA images of xenograft HeLa tumors before and at various time points after the injection of IR806-PDA under 680 nm or 820 nm laser irradiation. c) Schematic of the synthesis of Fe-Cu@PANI. d) PA images of normal muscle (left) and xenograft 4T1 tumor (right) before and at various time points after the injection of Fe-Cu@PANI. e) Reaction of PACDx with GSH. f) PA images of the xenograft A549 tumor before and one hour after the injection of PACDx. g) Stratification based on PA fold turn-on after the injection of PACDx. h) PA images of the lung region after the injection of vehicle and PACDx in orthotopic lung cancer model. i) PA images of the liver after the injection of vehicle and PACDx in the liver metastasis model. a) and b) are reproduced from ref. 287 with permission from John Wiley and Sons, Copyright 2018. c) and d) are reproduced from ref. 288, which is an open-access article distributed under the CC BY-NC License, 2021 AAAS. e), f), g), h) and i) are reproduced from ref. 289 with permission from Springer Nature, Copyright 2021.
In another study, Zhao, Zhang, and coworkers reported a GSH-responsive polyaniline-based NP probe (Fe-Cu@PANI) containing an iron-copper co-doped structure (Figure 23c).288 Utilizing the reductive nature of GSH, the Cu2+ in the Fe-Cu@PANI probe can undergo a redox reaction to yield NPs with a size reduction and a red-shift of the absorbance maximum. To evaluate the PA imaging performance of Fe-Cu@PANI, the probe was injected into both the normal muscle tissue (bearing no tumor) and the xenograft 4T1 tumor side of the mice. An apparent PA signal developed on the tumor side after only 30 min post-injection. On the other hand, there was only limited signal found in the normal muscle tissue (Figure 23d). This result was supported by another experiment, in which the intravenous injection of Fe-Cu@PANI showed a similar result with an obvious signal change that plateaued after four hours post-injection. Moreover, the in vivo response of Fe-Cu@PANI towards GSH was confirmed by the signal enhancement from pre-injection of GSH ethyl ester (this is a cell permeable version of GSH where the carboxylates are masked).
Finally, our group has developed a highly optimized PA imaging-based companion diagnostic test (PACDx) to selectively detect elevated GSH in numerous lung cancer models (Figure 23e).289 PACDx consists of two key elements, a dye to generate the PA signal upon irradiation and a trigger for the selective GSH response. The HD platform was chosen for this purpose because as others have shown above, it is a highly effective scaffold to construct activatable PA probes. Moreover, the lipophilic cationic character of the HD dye also favors the tumor uptake of the probe and can suppress cross-reactivity between the probe and GST. For the responsive-element, we chose the 2,4-dinitrobenzenesulfonate trigger which has been used previously as the starting point for chemical tuning.290 The reason this was not used directly was because we noticed it was too reactive and unable to discriminate GSH in healthy cells from cancerous tissues. In this regard, we leveraged the principles of physical organic chemistry to establish the structure-reactivity relationship of the probe using Hammett plots. Through this strategy, we successfully developed a highly optimized trigger featuring an ortho-fluoro group and para-nitro substituent. The resulting probe (PACDx) showed a dose-dependent turn-on up to 31.6-fold under high GSH concentrations, while negligible activation was observed with physiological GSH levels in blood or healthy cells. To validate the selectivity of PACDx towards different GSH concentrations in vivo, we first intratumorally injected PACDx into nude mice bearing A549 tumors and observed a ~1.5-fold turn-on relative to the control after one hour (Figure 23f). With this result, we further challenged PACDx by conducting a blind study, in which PACDx was used to discriminate mice bearing A549 tumors (high GSH level) and U87 tumors (low GSH level). Specifically, a group of mice were implanted with A549 or U87 cells, then their identities were concealed after they were tagged and randomized. After obtaining the diagnostic threshold using a group of healthy subjects, we successfully stratified these mice into the A549 tumor group or the U87 tumor group based on the intensity of the PA signal after the administration and imaging using PACDx (Figure 23g). The result showed that all our assignments were correct with six of seven mice being assigned with greater than 95% confidence. Finally, we applied PACDx imaging in both orthotopic lung cancer and liver metastasis models, and more than three-fold PA turn-on response was observed in both models, supporting the broad utility of PACDx (Figure 23h and 23i).
5.4.2. Hydrogen Sulfide
Hydrogen sulfide (H2S) is an endogenous gasotransmitter that plays a role in cellular processes like vasodilation, anti-inflammation, and pro-angiogenesis.291, 292 Due to the overexpression of the H2S-producing enzymes (CSE, CBS and 3MST) in many types of cancers, the H2S levels in the tumor cells is usually elevated and therefore it is recognized as a biomarker for cancer diagnosis and targeted therapy. However, like many other cancer biomarkers, the role of H2S in cancer progression is not fully studied293, 294 and developing chemical tools to visualize H2S concentrations in vivo would be important toward this end. In this regard, various H2S activatable PA probes have been developed.295–303
For instance, Yang, Tian and coworkers have published a H2S activatable probe (Au@Cu2O) based on a metal-semiconductor hybrid structure with integrated photothermal therapy capabilities (Figure 24a).299 Compared to the previously published Cu2O-based probe from the same group,304 Au@Cu2O possesses a better PA contrast and improved photothermal conversion efficiency due to the LSPR coupling effect between Au and Cu9S8, which is generated from Cu2O upon reaction with H2S. To demonstrate the ability of Au@Cu2O in detecting aberrant H2S levels in tumors, HCT tumor-bearing mice were intratumorally injected with PBS, Au@Cu2O, Au@Cu2O + AOAA (an endogenous H2S inhibitor), or Au@Cu2O + SAM (an endogenous H2S promoter). While little signal appeared in the PBS or Au@Cu2O + AOAA injection groups, an obvious signal was observed in the Au@Cu2O group, and an even stronger signal in the Au@Cu2O + SAM group (Figure 24b).
Figure 24.
a) Reaction of Au@Cu2O with H2S. b) PA images of the HCT tumors before and at various time points after the injection of PBS, Au@Cu2O, Au@Cu2O + AOAA, or Au@Cu2O + SAM. c) Schematic of the synthesis and reaction of MoO3 NPs. d) PA images of HCT116 tumors before and at various time points after the injection of PBS, MoO3 or SAM + MoO3 in the NIR-I window. e) PA images of HCT116 tumors before and at various time points after the injection of PBS, MoO3 or SAM + MoO3 in the NIR-II window. a) and b) are reproduced from ref. 299 with permission from John Wiley and Sons, Copyright 2019. c), d) and e) are reproduced from ref. 303 with permission from American Chemical Society, Copyright 2021.
Another recent example is reported by Zhao and Zhang, where the researchers have developed water soluble MoO3 NPs as an activatable NIR-II PA probe for H2S detection and for photothermal therapy (Figure 24c).303 The MoO3 NPs were prepared by ultrasonication and oxidation of Mo2C. Initially, there was no light absorption in the NIR region, however a broad absorption band ranging from 400 nm to 1350 nm was observed after its reaction with sodium sulfide. This significant change in the absorption was proposed to be due to the redox reaction between H2S and the MoO3 NPs, which generated Mo(V) and formed the highly stable polyoxometalate (POM) cluster. Due to the wide absorption of the activated MoO3 NPs, its H2S imaging performance was evaluated in both the NIR-I and NIR-II regions using HCT116 tumor-bearing mice (Figure 24d and 24e). Specifically, PBS, MoO3, or MoO3 + SAM were injected separately into the mice, and their PA signals were recorded at both 760 nm (NIR-I) and 1080 nm (NIR-II) wavelengths. For both the NIR-I and NIR-II imaging windows, the PA signal increased in the MoO3 and MoO3 + SAM groups, with a significantly more intense signal enhancement in the latter group, demonstrating MoO3 NPs’ in vivo response towards H2S. More importantly, this result also showed that changing the imaging window from the NIR-I to NIR-II region could significantly reduce the background signal, which highlights the advantage of developing NIR-II PA probes.
5.5. Gradients of the TME
As mentioned previously, the TME is defined as the complex surrounding in which a tumor resides. Several of the most fascinating features of the TME that imaging scientists, cancer biologists, and drug developers are interested in sensing is the extent of oxygenation deficiency (also known as hypoxia) and the local pH of cancer tissue. Owing to the heterogeneous nature of the TME and the unpredictable nature in which blood vessels form when a tumor grows, natural gradients are established. For instance, the oxygen content decreases in cancer cells that are further away from blood vessels since the diffusion of oxygen is limited by distance. Similarly, the pH also decreases in regions far removed from blood flow, resulting in a condition known as acidosis.
5.5.1. Tumor Hypoxia
Tissue hypoxia occurs when the demand for oxygen outweighs the available supply. Although this condition is associated with a variety of pathological states including coronary and peripheral artery disease,305 alcoholic liver injury,306 and gastrointestinal inflammatory conditions,307, 308 to name a few, it is a hallmark of many solid tumors owing to two critical factors. First, the diffusion of oxygen from blood vessels to distal cancer cells limits the amount of oxygen accessible to these regions. Second, cancer cells grow at a rapid pace and thus, have a high metabolic demand that consumes copious levels of oxygen. Several design strategies have been employed to develop PA probes to visualize tumor hypoxia.179, 205, 309–323
One approach, introduced by our group in 2017, is based on the cytochrome P450 enzyme (CYP)-mediated bio-reduction of a new N-oxide-based trigger (Figure 25a) which we installed onto an aza-BODIPY dye platform to yield HyP-1.309 One of the advantages of this design is that CYPs are ubiquitously found throughout the body,324–326 since overexpression is not necessary it is possible to use HyP-1 to detect both acute and chronic hypoxia. Beyond this first-generation probe, we also developed other congeners including rHyP-1 which features improved photophysical properties such as a red-shifted absorbance profile to facilitate ratiometric imaging.311 Using rHyP-1, we were able to reveal the extent and location of hypoxic domains in 4T1 breast tumors via 3D reconstruction (Figure 25b).
Figure 25.
a) Reaction of HyP-1/rHyP-1 via CYP activity under hypoxic conditions. b) 3D PA imaging of 4T1 tumor using rHyP-1 to reveal hypoxic domains in green. c) Reaction of NR-azo via CYP activity under hypoxic conditions. d) PA images of tumor bearing mice and heathy mice at 0.5 hour after intratumoral injection of PBS, NR-CLB or NR-azo. e) PA images of tumor bearing mice before and at various time points after injection of Lipo-NR-azo. f) Reaction of IR1048-MZ via NTR activity under hypoxic conditions. g) PA images in A549 tumor-bearing BALB/c nude mice before and at various time points after injection of IR1048-MZ. h) Reaction of PA-HD-NTR via NTR activity under hypoxic conditions. b) is reproduced from ref. 311 with permission from American Chemical Society, Copyright 2018. d) and e) are reproduced from ref. 317, which is an open-access article distributed under the Creative Commons Attribution (CC BY-NC) License, 2019 Ivyspring International Publisher. g) is reproduced from ref. 329 with permission from John Wiley and Sons, Copyright 2021.
Besides N-oxide, azo compounds can also be reduced under hypoxic conditions by P450 enzymes.327 Based on this reactivity, Zeng, Wu, and coworkers have developed an activatable hypoxia-responsive probe (NR-azo) featuring an azo linkage used to append a drug (Figure 25c).317 Upon conversion of the azo group into the amine under hypoxia, a red shift in the PA signal resulted due to the formation of the activated product (NR-NH2). Notably, the reduction of the azo linkage would also lead to the release of the masked nitrogen mustard (an anti-cancer drug). To assess the PA imaging performance of NR-azo for the detection of tumor hypoxia, NR-azo, NR-CLB (an inactive control probe) and a vehicle control (PBS buffer), were injected intratumorally into tumor-bearing mice while healthy mice were also used as controls (Figure 25d). The result showed that a strong PA signal was only evident in the tumor-bearing mice treated with NR-azo. Moreover, to enhance the tumor accumulation of NR-azo through the EPR effect, NR-azo was encapsulated into liposomes using a phospholipid formulation. The resulting Lipo-NR-azo agent was injected intravenously into tumor-bearing mice (Figure 25e). A PA signal was apparent in the tumor region at one-hour post-injection indicating successful delivery. Further, the intensity reached its peak after 6 hours. Taken together, these results show the potential of this probe to detect hypoxia in tumors.
The final strategy involves leveraging the ability of nitroreductases (NTRs) which are enzymes elevated in chronic hypoxic tissues to reduce aryl nitro groups to the corresponding hydroxyl amine or amino products.328 For instance, Cai and coworkers developed IR1048-MZ by substituting the meso-chloro group of IR-1048 with a nitro imidazole trigger (Figure 25f).312 NTR-mediated reduction of the nitro moiety induces an increase in absorbance at 980 nm resulting in an enhancement of the PA signal. The authors were able to successfully employ IR1048-MZ in A549 lung cancer xenografts to perform 3D PA imaging of hypoxia (Figure 25g). Recently, in proof-of-concept studies, our group developed PA-HD-NTR which features a 4-nitrobenzyl trigger appended to a dye platform optimized for PA imaging through a cleavable self-immolative linker (Figure 25h).329 When PA-HD-NTR was applied in vivo to detect hypoxia in 4T1 tumors, we observed a 1.4-fold turn-on relative to the non-cancerous tissue where the signal change was negligible. It is noteworthy that one limitation of NTR-based probes is that they are prone to potential off-target activation by bacterial NTR, which are oxygen-independent, to give false positive results.330–332
5.5.2. Tumor pH
The dysregulation of pH in the TME is a known hallmark of cancer333 which contributes to poor vascular perfusion, regional hypoxia and an increased flux of carbon building blocks through fermentative glycolysis of tumor cells.334 It is noteworthy that this is different from normal cells in which the intracellular pH (~7.2) is lower than the extracellular pH (~7.4). On the other hand, cancer cells exhibit the opposite pH gradient where the extracellular pH (~6.7–7.1) is lower than the intracellular pH (≥7.4).335–338 This unique pH gradient can promote proliferation, the evasion of apoptosis, metabolic adaptation, migration, and invasion of cancer cells.333 Due to its central role in cancer development, the tumor pH is an important biomarker in cancer that has been exploited for targeted drug delivery and imaging,339–341 as well as the development of many activatable PA probes.342–371 Here we highlight some recent examples.
In one example, Liu, Sessler and coworkers have successfully developed an expanded porphyrin-based probe, octaphyrin, for in vivo imaging of tumor pH.366 Under acidic conditions, octaphyrin can undergo a series of proton transfer electron transfer (PTET) or concerted proton electron transfer (CPET) processes, generating molecules that possess longer absorption wavelengths, up to 1200 nm (which is within the NIR-II region) (Figure 26a). This is an important property for PA imaging as shown in previous examples, since operating in this window allows for visualization of deeper tissue. After encapsulating octaphyrin using DSPE-PEG2000, the PA imaging performance of the resulting probe (OctaNPs) was assessed in a HepG2 tumor model. Notably, the authors demonstrated that while tumor pH might not be low enough to induce the PCET process, their probe could also leverage the reducing nature of tumor tissue to facilitate probe activation. In this in vivo experiment, the authors observed a statistically significant PA signal in the tumor region within only one minute. On the contrary, there was negligible signal change in healthy tissue. When comparing the overall PA signal intensity between the tumor and healthy tissue, the authors found a difference of ~47-fold which demonstrates the utility of OctaNPs as a reliable PA probe for tumor pH determination (Figure 26b).
Figure 26.
a) Reaction of octaphyrin under acidic conditions. b) PA/US B-scan image of BALB/c nude mice and HepG2-tumor bearing mice before and at various time points after injection of OctaNPs. c) Schematic of the synthesis of LET-5. d) PA images in 4T1 tumor-bearing mice before and at various time points after injection of Croc-PEG5K or LET-5. e) PA images of 4T1 tumor before and after injection of LET-5 with pre-treatment using saline, citrate buffer or a solution of NaHCO3. f) Reaction of CypHRGD-C under acidic conditions. g) PA images in A549 tumor-bearing mice before and at various time points after injection of CypHRGD-C. a) and b) are reproduced from ref. 366, which is an open-access article distributed under the Creative Commons Attribution (CC BY-NC) License, 2019 Ivyspring International Publisher. c), d) and e) are reproduced from ref. 367 with permission from Elsevier, Copyright 2021. g) is reproduced from ref. 370 with permission from American Chemical Society, Copyright 2021.
In another report, Lin and coworkers developed a pH-responsive probe (LET-5) based on a croconium dye platform which features a 1,2-dicarbonyl motif (Figure 26c).367 The chemical structure of LET-5 contains two key elements. The first component is an amphiphilic Croc-PEG5K molecule where the Croc dye is the pH-sensing moiety capable of generating a PA signal change under acidic conditions, and a PEG5K chain to render the molecule hydrophilic. The second part is a red blood cell membrane (RBCm)-based vesicle, which was introduced to improve the tumor accumulation of Croc-PEG5K by enhancing its stability and prolonging blood circulation. Finally, these two elements were combined through hydrophobic interactions between Croc and the RBCm vesicles. The in vivo evaluation of LET-5 was conducted in 4T1 tumor-bearing mice. First, the authors injected Croc-5K and LET-5 to perform PA imaging which allowed a direct comparison of the signal intensities within the tumor region. An apparent PA signal was observed for the LET-5-treated tumors which was absent in the Croc-5K-treated tumors. This result demonstrates the successful targeting ability of the RBCm vesicle (Figure 26d). However, to confirm the signal change was due to pH-mediated probe activation, the authors pre-treated tumors with saline (control), citrate buffer (pH = 5.0), and a solution of NaHCO3 (pH = 9.0) prior to LET-5 administration. Compared to the saline and alkaline groups, the PA intensity was strongest for the citrate buffer group, providing compelling evidence that LET-5 was sensing tumor pH (Figure 26e).
A final notable example comes from Miki, Ohe, and coworkers, where the authors reported a pH activatable probe (CypHRGD-C) with PA and fluorescence dual-imaging abilities.370 This probe exploits an ICG-based pH-sensing platform previously published by the same group.372 In this design, the cyclization event that results from the mercapto group attacking the central carbon leads to disruption of π-conjugation which attenuates the PA and fluorescent signals. Under acidic conditions, the reversible ring-opening process is driven by the protonation of sulfur atom which restores conjugation resulting in a turn on response in both modes (Figure 26f). Additionally, to modify this platform for in vivo tumor imaging, a sulfonate chain was introduced to increase water solubility, the methyl group was replaced by a bulky phenyl group to prevent aggregation, and a cRGD peptide was introduced for enhanced tumor recognition (see section 5.7). Together, these modifications led to an optimized probe which was tested in the A549 tumor-bearing mice for its tumor pH sensing capabilities. A clear PA signal appeared within the tumor region within three hours and this intensity reached the maximum after nine hours (Figure 26g).
5.6. Nucleic Acids
Tumor related nucleic acids (DNA, mRNA, miRNA) plays an important role in tumor progression and they are upregulated in many types of tumors373–376. Therefore, tumor related nucleic acids have become a useful cancer biomarker for activatable PA probe development.
One example comes from Tang, Li, and coworkers where the authors reported a TK1 mRNA targeting PA/FL dual-modal imaging probe based on the carboxylated graphene oxide (GO) scaffold.377 Specifically, the synthesis of this GO probe starts with the covalent conjugation of the amino-modified DNA onto the GO, then the Cy5.5 dye conjugated recognition sequence, which could bind with TK1 mRNA, was hybridized with the DNA on the GO. The hybridization of surface DNA and recognition DNA would bring the Cy5.5 dye close to the GO surface, which led to quenching of fluorescence signal and enhancement of the PA signal (Figure 27a). When TK1 was present, the recognition DNA would separate from the GO surface due to the stronger binding with TK1 than the surface DNA, which would result in an increase in the fluorescent signal and reduction of the PA signal from Cy5.5. The TK1 mRNA detection capacity of the GO probe was first evaluated in live cells using confocal microscopy. The TK1 mRNA overexpression in MCF-7 and HepG2 cells were selected as model cell lines, and MCF-10A cells together with HL-7702 cells were chosen as control as the expression of TK1 mRNA is negligible. The results from the imaging studies showed a clear fluorescent turn-on response in the MCF-7 and HepG2 cells, providing in cellulo evidence that TK1 mRNA detection is possible with the GO probe (Figure 27b). Further, in vivo TK1 mRNA detection by this GO nanoprobe was performed in subcutaneous xenograft mouse models with MCF-7 and HepG2 cells. In both models a clear turn-off of the PA signal was observed compared to the control probe due to the TK1 mRNA expression in these tumors. Of note, in vivo fluorescence imaging in the same model was not achieved due to limited penetration depth, demonstrating the value of PA imaging (Figure 27c).
Figure 27.
a) Schematic of GO probe. b) Confocal microscopy images of MCF-10A, HL-7702, MCF-7 and HepG2 after treatment of GO probe. c) PA images of tumors with MCF-7 or HepG2 cells after injection of control probe and GO probe. d) Schematic of the synthesis of DMSN-DP@CM. e) Schematic of how DNA-PA probe detect miRNA-21 and how fuel DNA amplify signal. f) PA image in BALB/c mice with subcutaneous tumor before and at various time points after injection of DMSN-DP@CM. a), b) and c) are reproduced from ref. 377 with permission from Royal Society of Chemistry, Copyright 2018. d), e) and f) are reproduced from ref. 378 with permission from John Wiley and Sons, Copyright 2019.
Besides mRNA, miRNA also attracted significant interest 375. For instance, Zhang and co-workers have developed a miRNA-21 ratiometric PA probe (DMSN-DP@CM) based on MCF-7 cancer cell membrane-encapsulated dendritic mesoporous silica nanoparticles (DMSN) (Figure 27d).378 The miRNA activation of DMSN-DP@CM was achieved by two key components on the DMSN surface. The first is the DNA-PA probe, which consisted of three types of DNA strands. The first DNA strand is conjugated to IRDye 800CW (F strand) or IRDye QC-1 as quencher (Q strand), and the final type of strand served as the linker (linker strand) to connect the previous two strands together and induce contact-mediated quenching. Of note, this linker strand could also be recognized by miRNA-21 which can lead to the separation of the F and Q strands. Separation of these components result in an increase of the PA780/PA725 ratio. The other component was the DNA fuel strands, which was designed to be released by GSH. The DNA fuel strand could displace the miRNA-21 on the linker strand through a toe-hole strand displacement reaction,379 the released miRNA-21 could further react with other linker strands and therefore, a strong PA signal could be achieved with a low concentration of miRNA-21 (Figure 27e). The in vivo PA imaging by DMSN-DP@CM was conducted in BALB/c mice with subcutaneous tumors. The result showed an increase in the PA780/PA725 ratio which reached the maximum intensity six hours post-injection (Figure 27f). To further investigate the origin of the PA signal, PBS buffer (control), DMSN-Linker/F/Q@CM (lacking the DNA fuel strand), DMSN-DP@CM and DMSN-DP@CM+miRNA-21 were injected separately. The PA780/PA725 ratio of DMSN-Linker/F/Q@CM was lower than that of DMSN-DP@CM, suggesting the PA signal amplification ability of the DNA fuel strands. Moreover, the miRNA-21 imaging ability was further validated by an increase of the PA780/PA725 ratio when miRNA-21 was co-injected with DMSN-DP@CM into the tumor.
6. Conclusion and Future Outlook
In this tutorial review, we have provided a simple to use, step-by-step guide that aims to assist developers select an appropriate cancer biomarker. We also outline important design criteria to consider when developing an imaging agent for PA applications. As demonstrated by the vast number of innovative design strategies, it is evident that PA imaging is one of the most vibrant and exciting fields to have recently emerged. The ability to sense a molecular event within a native in vivo context while achieving exquisite spatial resolution is no longer confined to the imaging of cell cultures, thin tissue slices, or shallow regions of the body. Indeed, extending the detection of a cancer biomarker or the visualization of a cancer-related process into deep tissue has profound implications for fundamental scientific research. Particularly, PA imaging agents are primed to serve as powerful chemical biology tools to help uncover the complex and often confounding mechanistic underpinnings of tumorigenesis, cancer progression, and metastasis using live animal models. Likewise, many of the examples discussed above have shown tremendous potential in a preclinical setting for the early diagnosis of cancer, treatment monitoring, and image-guided surgery. To translate these impressive results into human patients, it will be critical for developers to exploit existing design strategies or consider new approaches that focus on increasing the PA signal output to reduce the overall dosing requirement which can greatly improve safety. Likewise, as can be seen in the various examples highlighted throughout this review, more efficient cancer targeting and a greater turn-on response after biomarker engagement yields more reliable data. However, many PA-active photoabsorbers found within the body (e.g., hemoglobin and lipids) can lower imaging agent performance, both in regard to high background or attenuation of incident light which restricts the penetration depth. An emerging area within PA imaging that is primed to address these limitations is the shift of the wavelength of maximum absorbance into the NIR-II regime where there is less interference from the aforementioned endogenous pigments. We anticipate these advances will have a significant impact on both the small-molecule and nanoparticle imaging moiety fronts. Another approach to increase reliability of an imaging result is the development of imaging agents that can target two or more cancer biomarkers which will be critical in offsetting potential false positive results. For instance, beyond the development of activatable probes that simply accumulate via passive uptake mechanisms and subsequently ‘turn-on’ in tumors, the attachment of a cancer-specific targeting ligand can improve reliability since multiple conditions must be met prior to probe activation. Indeed, in an example we highlighted, the attachment of an RGD-targeting moiety to an activatable sensor for tumor acidity has allowed for tumor-specific pH monitoring. By substituting the targeting group, as well as the responsive trigger, additional cancer types and biomarkers can be detected, respectively. Lastly, we envision the development of PA imaging agents with dual- or multi-modal capabilities can aid in clinical translation. If the additional imaging mode is used clinically, existing hospital instrumentation can be employed to validate PA imaging results. Along these lines, although the availability of suitable PA equipment is a potential barrier, we can take comfort that recent developments on the instrumentation front have yielded convenient hand-held scanners and reliable endoscopy setups designed for human use. Together, we envision that progress in these two areas (imaging agent design and engineering of new equipment), will help to bring PA imaging to the forefront of biomedical imaging techniques.
Figure 2.
Step-by-step roadmap for cancer biomarker selection and subsequent photoacoustic imaging agent design. Reproduced from ref. 120 with permission from Elsevier, Copyright 2017; from ref. 69 with permission of American Chemical Society, Copyright 2017; from ref. 141 with permission from Elsevier, Copyright 2020; from ref. 55 with permission from Royal Society of Chemistry, Copyright 2021; from ref. 79 with permission from American Chemical Society, Copyright 2020; from ref. 377 with permission from John Wiley and Sons, Copyright 2019.
Key learning points:
Advantages of photoacoustic imaging that make it an ideal modality for preclinical and clinical applications in cancer imaging.
Criteria for selecting an appropriate cancer biomarker for imaging agent development.
Design strategies that are available to develop targeted contrast agents or activatable probes for this imaging modality.
Notable examples from the literature to highlight the various cancer biomarker targets, imaging designs, validation experiments and applications.
Perspective in the cancer PA imaging field.
Acknowledgements
The authors thank the National Science Foundation (1752879) and the National Institutes of Health (R35GM133581) for supporting this work.
Biographies

Zhenxiang Zhao graduated from Nankai University in 2020 with a BSc in Chemistry. During this time, he conducted undergraduate research in the laboratory of Prof. Gong Chen, mainly focusing on the synthesis of macrocycle peptides via C-H activation. He is currently pursuing a PhD in Chemistry with Prof. Jeff Chan at UIUC. His research interests include the development of probes to detect metal ions and aggregated proteins in human diseases.

Chelsea Swartchick graduated from the University of Portland in 2019, where she synthesized naphthoquinones with antimalarial properties under Prof. Warren Wood. In the same year, she began her PhD studies at UIUC with Jeff Chan where her research focuses on the synthesis of chemical tools to elucidate the role of calcium in neurodegeneration and aging. Chelsea has since earned the Illinois Chemistry Enta Fellowship, as well as recognition for excellence in teaching.

Prof. Jeff Chan began his independent career in 2014 and was promoted to the rank of Associate Professor in 2021. He is a member of the Beckman Institute for Advanced Science and Technology where he is the current leader of the Photoacoustic Imaging work group. His research interests include the development of photoacoustic probes, design of tools to study the chemical biology of aging, and the application of new reactions for targeted drug delivery.
Footnotes
Conflicts of interest
There are no conflicts to declare.
References
- 1.Wang LV and Hu S, Science, 2012, 335, 1458–1462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhang HF, Maslov K, Stoica G. and Wang LHV, Nat. Biotechnol, 2006, 24, 848–851. [DOI] [PubMed] [Google Scholar]
- 3.Zhang HF, Maslov K, Sivaramakrishnan M, Stoica G. and Wang LV, Appl. Phys. Lett, 2007, 90, 053901. [Google Scholar]
- 4.Beard P, Interface Focus, 2011, 1, 602–631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Xia J, Yao J. and Wang LHV, Progress In Electromagnetics Research, 2014, 147, 1–22.25642127 [Google Scholar]
- 6.Attia ABE, Balasundaram G, Moothanchery M, Dinish US, Bi R, Ntziachristos V. and Olivo M, Photoacoustics, 2019, 16, 100144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Miao Q. and Pu K, Bioconjugate Chem., 2016, 27, 2808–2823. [DOI] [PubMed] [Google Scholar]
- 8.Huang J. and Pu K, Angew. Chem. Int. Ed, 2020, 59, 11717–11731. [DOI] [PubMed] [Google Scholar]
- 9.Cheng P. and Pu K, Nature Reviews Materials, 2021, DOI: 10.1038/s41578-021-00328-6. [DOI] [Google Scholar]
- 10.Zeng Z, Liew SS, Wei X. and Pu K, Angew. Chem. Int. Ed, 2021, DOI: 10.1002/anie.202107877. [DOI] [PubMed] [Google Scholar]
- 11.Chan J, Dodani SC and Chang CJ, Nat. Chem, 2012, 4, 973–984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bruemmer KJ, Crossley SWM and Chang CJ, Angew. Chem. Int. Ed, 2020, 59, 13734–13762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Loeb LA, Loeb KR and Anderson JP, Proceedings of the National Academy of Sciences, 2003, 100, 776–781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bozic I, Antal T, Ohtsuki H, Carter H, Kim D, Chen S, Karchin R, Kinzler KW, Vogelstein B. and Nowak MA, Proceedings of the National Academy of Sciences, 2010, 107, 18545–18550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Takeshima H. and Ushijima T, npj Precision Oncology, 2019, 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Martin RM, Donovan JL, Turner EL, Metcalfe C, Young GJ, Walsh EI, Lane JA, Noble S, Oliver SE, Evans S, Sterne JAC, Holding P, Ben-Shlomo Y, Brindle P, Williams NJ, Hill EM, Ng SY, Toole J, Tazewell MK, Hughes LJ, Davies CF, Thorn JC, Down E, Davey Smith G, Neal DE, Hamdy FC and f. t. C. T. Group, JAMA, 2018, 319, 883–895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Radi R, Proceedings of the National Academy of Sciences, 2004, 101, 4003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Weber J, Beard PC and Bohndiek SE, Nat Methods, 2016, 13, 639–650. [DOI] [PubMed] [Google Scholar]
- 19.Malkowski MG, in Encyclopedia of Inorganic and Bioinorganic Chemistry, DOI: 10.1002/9781119951438.eibc0547.pub2, pp. 1–18. [DOI] [Google Scholar]
- 20.Gardner SH, Reinhardt CJ and Chan J, Angewandte Chemie International Edition, 2021, 60, 5000–5009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yadav AK, Reinhardt CJ, Arango AS, Huff HC, Dong L, Malkowski MG, Das A, Tajkhorshid E. and Chan J, Angewandte Chemie International Edition, 2020, 59, 3307–3314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bearrood TE and Chan J, Aldrichimica Acta, 2017, 50, 31–42. [Google Scholar]
- 23.Kim JW, Galanzha EI, Shashkov EV, Moon HM and Zharov VP, Nature Nanotechnology, 2009, 4, 688–694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Joshi PP, Yoon SJ, Hardin WG, Emelianov S. and Sokolov KV, Bioconjugate Chem., 2013, 24, 878–888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xi L, Grobmyer SR, Zhou GY, Qian WP, Yang L. and Jiang HB, Journal of Biophotonics, 2014, 7, 401–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lozano N, Al-Ahmady ZS, Beziere NS, Ntziachristos V. and Kostarelos K, Int. J. Pharm, 2015, 482, 2–10. [DOI] [PubMed] [Google Scholar]
- 27.Yang M, Fan QL, Zhang RP, Cheng K, Yan JJ, Pan DH, Ma XW, Lu A. and Cheng Z, Biomaterials, 2015, 69, 30–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hu WB, Ma HH, Hou B, Zhao H, Ji Y, Jiang RC, Hu XM, Lu XM, Zhang L, Tang YF, Fan QL and Huang W, Acs Applied Materials & Interfaces, 2016, 8, 12039–12047. [DOI] [PubMed] [Google Scholar]
- 29.Xie BW, Tomaszewski MR, Neves AA, Ros S, Hu DE, McGuire S, Mullins SR, Tice D, Sainson RCA, Bohndiek SE, Wilkinson RW and Brindle KM, Clinical Cancer Research, 2017, 23, 6893–6903. [DOI] [PubMed] [Google Scholar]
- 30.Liu CB, Chen JQ, Zhu Y, Gong XJ, Zheng RQ, Chen NB, Chen D, Yan HX, Zhang P, Zheng HR, Sheng ZH and Song L, Nano-Micro Lett., 2018, 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cai K, Zhang WY, Foda MF, Li XY, Zhang J, Zhong YT, Liang HG, Li HQ, Han HY and Zhai TY, Small, 2020, 16. [DOI] [PubMed] [Google Scholar]
- 32.Jiang Y, Duan XC, Bai JH, Tian HK, Ding D. and Geng YH, Biomaterials, 2020, 255. [DOI] [PubMed] [Google Scholar]
- 33.Wu FS, Yue LL, Cheng K, Chen J, Wong KL, Wong WK and Zhu XJ, Acs Biomaterials Science & Engineering, 2020, 6, 5230–5239. [DOI] [PubMed] [Google Scholar]
- 34.Cheng Y, Bao DD, Chen XJ, Wu YJ, Wei YH, Wu ZB, Li FZ and Piao JG, Int. J. Pharm, 2021, 593. [DOI] [PubMed] [Google Scholar]
- 35.Li L, Patil D, Petruncio G, Harnden KK, Somasekharan JV, Paige M, Wang LV and Salvador-Morales C, Acs Nano, 2021, 15, 2413–2427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu SL, Zhang W, Chen QQ, Hou JX, Wang JR, Zhong YX, Wang XY, Jiang WX, Ran HT and Guo DJ, Nanoscale, 2021, 13, 14049–14066. [DOI] [PubMed] [Google Scholar]
- 37.Qi S, Zhang YC, Liu GY, Chen JB, Li XZ, Zhu Q, Yang YQ, Wang F, Shi JH, Lee CS, Zhu GY, Lai PX, Wang LD and Fang CH, Acta Biomater., 2021, 129, 245–257. [DOI] [PubMed] [Google Scholar]
- 38.Roberts S, Khera E, Choi C, Navaratna T, Grimm J, Thurber GM and Reiner T, Journal of Nuclear Medicine, 2021, 62, 839–848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhou HC, Ren J, Lin YJ, Gao DY, Hu DH, Yin TH, Qiu C, Miao XY, Liu CB, Liu X, Zheng HR, Zheng RQ and Sheng ZH, Journal of Materials Chemistry B, 2021, 9, 3005–3014. [DOI] [PubMed] [Google Scholar]
- 40.Yang S, Wei W. and Zhao Q, International Journal of Biological Sciences, 2020, 16, 1767–1773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liu S, Liang J, Liu Z, Zhang C, Wang Y, Watson AH, Zhou C, Zhang F, Wu K, Zhang F, Lu Y. and Wang X, Front Oncol, 2021, 11, 654684–654684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wilson KE, Bachawal SV, Abou-Elkacem L, Jensen K, Machtaler S, Tian L. and Willmann JK, Theranostics, 2017, 7, 1463–1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bam R, Laffey M, Nottberg K, Lown PS, Hackel BJ and Wilson KE, Bioconjug Chem, 2019, 30, 1677–1689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bam R, Laffey M, Nottberg K, Lown PS, Hackel BJ and Wilson KE, Bioconjugate Chem., 2019, 30, 1677–1689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ponta H, Sherman L. and Herrlich PA, Nature Reviews Molecular Cell Biology, 2003, 4, 33–45. [DOI] [PubMed] [Google Scholar]
- 46.Chen C, Zhao S, Karnad A. and Freeman JW, J Hematol Oncol, 2018, 11, 64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Basakran NS, Saudi Med J, 2015, 36, 273–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Orian-Rousseau V. and Ponta H, Arch Toxicol, 2015, 89, 3–14. [DOI] [PubMed] [Google Scholar]
- 49.Galanzha EI, Kim J-W and Zharov VP, Journal of Biophotonics, 2009, 2, 725–735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Swierczewska M, Choi KY, Mertz EL, Huang X, Zhang F, Zhu L, Yoon HY, Park JH, Bhirde A, Lee S. and Chen X, Nano Letters, 2012, 12, 3613–3620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Galanzha EI, Nedosekin DA, Sarimollaoglu M, Orza AI, Biris AS, Verkhusha VV and Zharov VP, Journal of Biophotonics, 2015, 8, 81–93. [DOI] [PubMed] [Google Scholar]
- 52.Liang S, Li C, Zhang C, Chen Y, Xu L, Bao C, Wang X, liu G, zhang F. and Cui D, Theranostics, 2015, 5, 970–984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhang D, Wang Z, Wang L, Wang Z, Wang H, Li G, Qiao Z-Y, Xu W. and Wang H, ACS Applied Materials & Interfaces, 2018, 10, 28331–28339. [DOI] [PubMed] [Google Scholar]
- 54.Dai Y, Yu X, Wei J, Zeng F, Li Y, Yang X, Luo Q. and Zhang Z, Light: Science & Applications, 2020, 9, 164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Guo X, Wen C, Xu Q, Ruan C, Shen X-C and Liang H, Journal of Materials Chemistry B, 2021, 9, 2042–2053. [DOI] [PubMed] [Google Scholar]
- 56.Wang Z, Methods Mol Biol, 2017, 1652, 3–35. [DOI] [PubMed] [Google Scholar]
- 57.Appert-Collin A, Hubert P, Crémel G. and Bennasroune A, Frontiers in Pharmacology, 2015, 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chen W, Shen B. and Sun X, Molecular Imaging, 2019, 18, 1536012118823473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Plosker GL and Keam SJ, Drugs, 2006, 66, 449–475. [DOI] [PubMed] [Google Scholar]
- 60.Copland JA, Eghtedari M, Popov VL, Kotov N, Mamedova N, Motamedi M. and Oraevsky AA, Mol Imaging Biol, 2004, 6, 341–349. [DOI] [PubMed] [Google Scholar]
- 61.Kim G, Huang SW, Day KC, O’Donnell M, Agayan RR, Day MA, Kopelman R. and Ashkenazi S, J Biomed Opt, 2007, 12, 044020. [DOI] [PubMed] [Google Scholar]
- 62.Bhattacharyya S, Wang S, Reinecke D, Kiser W Jr., Kruger RA and DeGrado TR, Bioconjug Chem, 2008, 19, 1186–1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Li PC, Wang CR, Shieh DB, Wei CW, Liao CK, Poe C, Jhan S, Ding AA and Wu YN, Opt Express, 2008, 16, 18605–18615. [DOI] [PubMed] [Google Scholar]
- 64.Zhang T, Cui H, Fang C-Y, Jo J, Yang X, Chang H-C and Forrest ML, Proc SPIE Int Soc Opt Eng, 2013, 8815, 881504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Xi L, Satpathy M, Zhao Q, Qian W, Yang L. and Jiang H, Nanomedicine, 2014, 10, 669–677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Maeda A, Bu J, Chen J, Zheng G. and DaCosta RS, Mol Imaging, 2014, 13. [DOI] [PubMed] [Google Scholar]
- 67.Kanazaki K, Sano K, Makino A, Shimizu Y, Yamauchi F, Ogawa S, Ding N, Yano T, Temma T, Ono M. and Saji H, Nanomedicine, 2015, 11, 2051–2060. [DOI] [PubMed] [Google Scholar]
- 68.Zhang T, Cui H, Fang C-Y, Cheng K, Yang X, Chang H-C and Forrest ML, Nanomedicine, 2015, 10, 573–587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Cheng K, Chen H, Jenkins CH, Zhang G, Zhao W, Zhang Z, Han F, Fung J, Yang M, Jiang Y, Xing L. and Cheng Z, ACS Nano, 2017, 11, 12276–12291. [DOI] [PubMed] [Google Scholar]
- 70.Zhang Y, Zhao N, Qin Y, Wu F, Xu Z, Lan T, Cheng Z, Zhao P. and Liu H, Nanoscale, 2018, 10, 16581–16590. [DOI] [PubMed] [Google Scholar]
- 71.Chen J, Gao Z, Li G. and Wang TD, Chemical Communications, 2018, 54, 13196–13199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ye Z, Srivastava PK, Xu Y, Wang W, Jing L, Chen S-L and Tu C-C, ACS Appl. Nano Mater, 2019, 2, 7577–7584. [Google Scholar]
- 73.Wang Y, Pasternak M, Sathiyamoorthy K. and Kolios MC, Biomed Opt Express, 2021, 12, 2171–2185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wang Y, Fadhel MN, Hysi E, Pastenak M, Sathiyamoorthy K. and Kolios MC, Journal of Biophotonics, n/a, e202100099. [DOI] [PubMed] [Google Scholar]
- 75.Kumra H. and Reinhardt DP, Adv Drug Deliv Rev, 2016, 97, 101–110. [DOI] [PubMed] [Google Scholar]
- 76.Lin T-C, Yang C-H, Cheng L-H, Chang W-T, Lin Y-R and Cheng H-C, Cells, 2020, 9. [Google Scholar]
- 77.Rick JW, Chandra A, Dalle Ore C, Nguyen AT, Yagnik G. and Aghi MK, Semin Oncol, 2019, 46, 284–290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wang J, Li R, Li M. and Wang C, Journal of Receptors and Signal Transduction, 2021, 41, 313–320.32900261 [Google Scholar]
- 79.Yao D, Wang Y, Zou R, Bian K, Liu P, Shen S, Yang W, Zhang B. and Wang D, ACS Applied Materials & Interfaces, 2020, 12, 4276–4284. [DOI] [PubMed] [Google Scholar]
- 80.Frigerio B, Bizzoni C, Jansen G, Leamon CP, Peters GJ, Low PS, Matherly LH and Figini M, Journal of Experimental & Clinical Cancer Research, 2019, 38, 125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ledermann JA, Canevari S. and Thigpen T, Ann Oncol, 2015, 26, 2034–2043. [DOI] [PubMed] [Google Scholar]
- 82.Lovell JF, Jin CS, Huynh E, Jin H, Kim C, Rubinstein JL, Chan WCW, Cao W, Wang LV and Zheng G, Nature Materials, 2011, 10, 324–332. [DOI] [PubMed] [Google Scholar]
- 83.Wang Y-W, Fu Y-Y, Peng Q, Guo S-S, Liu G, Li J, Yang H-H and Chen G-N, Journal of Materials Chemistry B, 2013, 1, 5762–5767. [DOI] [PubMed] [Google Scholar]
- 84.Zhong J, Yang S, Zheng X, Zhou T. and Xing D, Nanomedicine, 2013, 8, 903–919. [DOI] [PubMed] [Google Scholar]
- 85.Wang H, Liu C, Gong X, Hu D, Lin R, Sheng Z, Zheng C, Yan M, Chen J, Cai L. and Song L, Nanoscale, 2014, 6, 14270–14279. [DOI] [PubMed] [Google Scholar]
- 86.Zhong J, Wen L, Yang S, Xiang L, Chen Q. and Xing D, Nanomedicine: Nanotechnology, Biology and Medicine, 2015, 11, 1499–1509. [DOI] [PubMed] [Google Scholar]
- 87.Bai LY, Yang XQ, An J, Zhang L, Zhao K, Qin MY, Fang BY, Li C, Xuan Y, Zhang XS, Zhao YD and Ma ZY, Nanotechnology, 2015, 26, 315701. [DOI] [PubMed] [Google Scholar]
- 88.Balasundaram G, Ho CJ, Li K, Driessen W, Dinish US, Wong CL, Ntziachristos V, Liu B. and Olivo M, Int J Nanomedicine, 2015, 10, 387–397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.An J, Yang XQ, Cheng K, Song XL, Zhang L, Li C, Zhang XS, Xuan Y, Song YY, Fang BY, Hou XL, Zhao YD and Liu B, ACS Appl Mater Interfaces, 2017, 9, 41748–41759. [DOI] [PubMed] [Google Scholar]
- 90.Li Y, Song Y, Chen W, Xu Q. and Liu Z, Materials Chemistry Frontiers, 2017, 1, 916–921. [Google Scholar]
- 91.Cao F, Guo Y, Li Y, Tang S, Yang Y, Yang H. and Xiong L, Advanced Functional Materials, 2018, 28, 1707174. [Google Scholar]
- 92.Zhang RY, Wang ZY, Yang XQ, Xuan Y, Cheng K, Li C, Song XL, An J, Hou XL and Zhao YD, Nanotechnology, 2018, 29, 055101. [DOI] [PubMed] [Google Scholar]
- 93.Yang Q, Li P, Ran H, Wan J, Chen H, Chen H, Wang Z. and Zhang L, Acta Biomaterialia, 2019, 90, 337–349. [DOI] [PubMed] [Google Scholar]
- 94.Nima ZA, Watanabe F, Jamshidi-Parsian A, Sarimollaoglu M, Nedosekin DA, Han M, Watts JA, Biris AS, Zharov VP and Galanzha EI, Scientific Reports, 2019, 9, 887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Wang Q, Dai Y, Xu J, Cai J, Niu X, Zhang L, Chen R, Shen Q, Huang W. and Fan Q, Advanced Functional Materials, 2019, 29, 1901480. [Google Scholar]
- 96.Yang C, Zhang Y, Luo Y, Qiao B, Wang X, Zhang L, Chen Q, Cao Y, Wang Z. and Ran H, Journal of Materials Chemistry B, 2020, 8, 380–390. [DOI] [PubMed] [Google Scholar]
- 97.Tu L, Fan Z, Zhu F, Zhang Q, Zeng S, Chen Z, Ren L, Hou Z, Ye S. and Li Y, Journal of Materials Chemistry B, 2020, 8, 5667–5681. [DOI] [PubMed] [Google Scholar]
- 98.Cornelio DB, Roesler R. and Schwartsmann G, Ann Oncol, 2007, 18, 1457–1466. [DOI] [PubMed] [Google Scholar]
- 99.Ischia J, Patel O, Bolton D, Shulkes A. and Baldwin GS, BJU International, 2014, 113, 40–47. [DOI] [PubMed] [Google Scholar]
- 100.Baratto L, Laudicella R, Picchio M, Baldari S. and Iagaru A, Clinical and Translational Imaging, 2019, 7, 39–44. [Google Scholar]
- 101.Chen Y-S, Zhao Y, Yoon SJ, Gambhir SS and Emelianov S, Nature Nanotechnology, 2019, 14, 465–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Aksorn N. and Chanvorachote P, Anticancer Res., 2019, 39, 541–548. [DOI] [PubMed] [Google Scholar]
- 103.Ye Y. and Chen X, Theranostics, 2011, 1, 102–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Arosio D. and Casagrande C, Advanced Drug Delivery Reviews, 2016, 97, 111–143. [DOI] [PubMed] [Google Scholar]
- 105.Kapp TG, Rechenmacher F, Neubauer S, Maltsev OV, Cavalcanti-Adam EA, Zarka R, Reuning U, Notni J, Wester H-J, Mas-Moruno C, Spatz J, Geiger B. and Kessler H, Scientific Reports, 2017, 7, 39805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Wang L, Xie X, Oh JT, Li ML, Ku G, Ke S, Similache S, Li C. and Stoica G, Conf Proc IEEE Eng Med Biol Soc, 2005, 2006, 190–192. [DOI] [PubMed] [Google Scholar]
- 107.De La Zerda A, Zavaleta C, Keren S, Vaithilingam S, Bodapati S, Liu Z, Levi J, Smith BR, Ma T-J, Oralkan O, Cheng Z, Chen X, Dai H, Khuri-Yakub BT and Gambhir SS, Nature Nanotechnology, 2008, 3, 557–562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Xiang L, Yuan Y, Xing D, Ou Z, Yang S. and Zhou F, J Biomed Opt, 2009, 14, 021008. [DOI] [PubMed] [Google Scholar]
- 109.Zerda A. d. l, Liu Z, Bodapati S, Teed R, Vaithilingam S, Khuri-Yakub BT, Chen X, Dai H and Gambhir SS, Nano Letters, 2010, 10, 2168–2172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Pan D, Pramanik M, Senpan A, Allen JS, Zhang H, Wickline SA, Wang LV and Lanza GM, FASEB J, 2011, 25, 875–882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.de la Zerda A, Bodapati S, Teed R, May SY, Tabakman SM, Liu Z, Khuri-Yakub BT, Chen X, Dai H. and Gambhir SS, ACS Nano, 2012, 6, 4694–4701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Zhou T, Wu B. and Xing D, Journal of Materials Chemistry, 2012, 22, 470–477. [Google Scholar]
- 113.Fan Q, Cheng K, Hu X, Ma X, Zhang R, Yang M, Lu X, Xing L, Huang W, Gambhir SS and Cheng Z, Journal of the American Chemical Society, 2014, 136, 15185–15194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Wang C, Bao C, Liang S, Fu H, Wang K, Deng M, Liao Q. and Cui D, Nanoscale Research Letters, 2014, 9, 264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Cheng K, Kothapalli S-R, Liu H, Koh AL, Jokerst JV, Jiang H, Yang M, Li J, Levi J, Wu JC, Gambhir SS and Cheng Z, Journal of the American Chemical Society, 2014, 136, 3560–3571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Zhang D, Qi GB, Zhao YX, Qiao SL, Yang C. and Wang H, Adv Mater, 2015, 27, 6125–6130. [DOI] [PubMed] [Google Scholar]
- 117.Qin H, Zhou T, Yang S. and Xing D, Small, 2015, 11, 2675–2686. [DOI] [PubMed] [Google Scholar]
- 118.Cai X, Liu X, Liao L-D, Bandla A, Ling JM, Liu Y-H, Thakor N, Bazan GC and Liu B, Small, 2016, 12, 4873–4880. [DOI] [PubMed] [Google Scholar]
- 119.Li Y, Jiang C, Zhang D, Wang Y, Ren X, Ai K, Chen X. and Lu L, Acta Biomater., 2017, 47, 124–134. [DOI] [PubMed] [Google Scholar]
- 120.Xie C, Upputuri PK, Zhen X, Pramanik M. and Pu K, Biomaterials, 2017, 119, 1–8. [DOI] [PubMed] [Google Scholar]
- 121.Zhen X, Feng X, Xie C, Zheng Y. and Pu K, Biomaterials, 2017, 127, 97–106. [DOI] [PubMed] [Google Scholar]
- 122.Deng X, Li K, Cai X, Liu B, Wei Y, Deng K, Xie Z, Wu Z, P a. Ma, Z Hou, Z Cheng and J Lin, Advanced Materials, 2017, 29, 1701266. [DOI] [PubMed] [Google Scholar]
- 123.Haedicke K, Brand C, Omar M, Ntziachristos V, Reiner T. and Grimm J, Photoacoustics, 2017, 6, 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Guo B, Sheng Z, Hu D, Liu C, Zheng H. and Liu B, Advanced Materials, 2018, 30, 1802591. [DOI] [PubMed] [Google Scholar]
- 125.Sheng Z, Guo B, Hu D, Xu S, Wu W, Liew WH, Yao K, Jiang J, Liu C, Zheng H. and Liu B, Adv Mater, 2018, DOI: 10.1002/adma.201800766,e1800766. [DOI] [PubMed] [Google Scholar]
- 126.Yu G, Yang Z, Fu X, Yung BC, Yang J, Mao Z, Shao L, Hua B, Liu Y, Zhang F, Fan Q, Wang S, Jacobson O, Jin A, Gao C, Tang X, Huang F. and Chen X, Nature Communications, 2018, 9, 766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Capozza M, Blasi F, Valbusa G, Oliva P, Cabella C, Buonsanti F, Cordaro A, Pizzuto L, Maiocchi A. and Poggi L, Photoacoustics, 2018, 11, 36–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Liu Y, Liu H, Yan H, Liu Y, Zhang J, Shan W, Lai P, Li H, Ren L, Li Z. and Nie L, Advanced Science, 2019, 6, 1801615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Li Z, Guo T, Hu Y, Qiu Y, Liu Y, Wang H, Li Y, Chen X, Song J. and Yang H, ACS Appl Mater Interfaces, 2019, 11, 9860–9871. [DOI] [PubMed] [Google Scholar]
- 130.Cao Y, Wu T, Zhang K, Meng X, Dai W, Wang D, Dong H. and Zhang X, Acs Nano, 2019, 13, 1499–1510. [DOI] [PubMed] [Google Scholar]
- 131.Zhang Y, Lv J, Liu P, Zhao X, Chen K, Li Q, Nie L. and Fang C, Acs Biomaterials Science & Engineering, 2020, 6, 5874–5885. [DOI] [PubMed] [Google Scholar]
- 132.Park YD, Park J-E, Kim HS, Choi S-H, Park JE, Jeon J. and Park S-H, Bioconjugate Chem., 2020, 31, 2607–2617. [DOI] [PubMed] [Google Scholar]
- 133.Fang X, Lui KH, Li S, Lo WS, Li X, Gu Y. and Wong WT, Int J Nanomedicine, 2020, 15, 10271–10284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Liu N, Gujrati V, Malekzadeh-Najafabadi J, Werner JPF, Klemm U, Tang L, Chen Z, Prakash J, Huang Y, Stiel A, Mettenleiter G, Aichler M, Blutke A, Walch A, Kleigrewe K, Razansky D, Sattler M. and Ntziachristos V, Photoacoustics, 2021, 22, 100263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Liu N, O’Connor P, Gujrati V, Gorpas D, Glasl S, Blutke A, Walch A, Kleigrewe K, Sattler M, Plettenburg O. and Ntziachristos V, Advanced Healthcare Materials, 2021, 10, 2002115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Liu R, Xu F, Wang L, Liu M, Cao X, Shi X. and Guo R, Nanomaterials, 2021, 11, 394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Ouyang Z, Li D, Xiong Z, Song C, Gao Y, Liu R, Shen M. and Shi X, ACS Appl Mater Interfaces, 2021, 13, 6069–6080. [DOI] [PubMed] [Google Scholar]
- 138.Guo B, Sheng Z, Kenry D. HuX Lin, Xu s, Liu C, Zheng H and Liu B., Materials Horizons, 2017, 4, 1151–1156. [Google Scholar]
- 139.Carvalho LS, Gonçalves N, Fonseca NA and Moreira JN, Pharmaceuticals, 2021, 14, 60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Ferrara B, Belbekhouche S, Habert D, Houppe C, Vallée B, Bourgoin-Voillard S, Cohen JL, Cascone I. and Courty J, Nanotechnology, 2021, 32. [DOI] [PubMed] [Google Scholar]
- 141.Zhang Y, Song T, Feng T, Wan Y, Blum NT, Liu C, Zheng C, Zhao Z, Jiang T, Wang J, Li Q, Lin J, Tang L. and Huang P, Nano Today, 2020, 35, 100987. [Google Scholar]
- 142.Saha P, Kaul R. and Datta K, The Nucleus, 2017, 60, 221–226. [Google Scholar]
- 143.Ghebrehiwet B, Geisbrecht BV, Xu X, Savitt AG and Peerschke EIB, Semin Immunol, 2019, 45, 101338. [DOI] [PubMed] [Google Scholar]
- 144.Matsumoto K. and Bay BH, Curr Med Chem, 2021, 28, 5048–5065. [DOI] [PubMed] [Google Scholar]
- 145.Yu X, Li A, Zhao C, Yang K, Chen X. and Li W, ACS Nano, 2017, 11, 3990–4001. [DOI] [PubMed] [Google Scholar]
- 146.Sharma B. and Kanwar SS, Seminars in Cancer Biology, 2018, 52, 17–25. [DOI] [PubMed] [Google Scholar]
- 147.Xie B, Tomaszewski MR, Neves AA, Ros S, Hu D-E, McGuire S, Mullins SR, Tice D, Sainson RCA, Bohndiek SE, Wilkinson RW and Brindle KM, Clinical Cancer Research, 2017, DOI: 10.1158/1078-0432.Ccr-17-1029. [DOI] [PubMed] [Google Scholar]
- 148.Gupta V. and Carroll KS, Biochimica et Biophysica Acta (BBA) - General Subjects, 2014, 1840, 847–875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Seo YH and Carroll KS, Proceedings of the National Academy of Sciences, 2009, DOI: 10.1073/pnas.0903015106,pnas.0903015106. [DOI] [Google Scholar]
- 150.Lyu Y, Zhen X, Miao Y. and Pu K, ACS Nano, 2017, 11, 358–367. [DOI] [PubMed] [Google Scholar]
- 151.Paulsen CE, Truong TH, Garcia FJ, Homann A, Gupta V, Leonard SE and Carroll KS, Nature Chemical Biology, 2012, 8, 57–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Chang SS, Rev Urol, 2004, 6 Suppl 10, S13–S18. [PMC free article] [PubMed] [Google Scholar]
- 153.Behnoosh T, Ying C, Xiaoyu G, Hyun Jae K, Martin P. and Emad MB, 2015. [Google Scholar]
- 154.Wang Y, Lan M, Shen D, Fang K, Zhu L, Liu Y, Hao L. and Li P, Int J Nanomedicine, 2020, 15, 4289–4309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Wu J, Lee HJ, You L, Luo X, Hasegawa T, Huang K-C, Lin P, Ratliff T, Ashizawa M, Mei J. and Cheng J-X, Small, 2020, 16, 2001215. [DOI] [PubMed] [Google Scholar]
- 156.Lesniak WG, Wu Y, Kang J, Boinapally S, Ray Banerjee S, Lisok A, Jablonska A, Boctor EM and Pomper MG, Nanoscale, 2021, 13, 9217–9228. [DOI] [PubMed] [Google Scholar]
- 157.Yao JJ, Kaberniuk AA, Li L, Shcherbakova DM, Zhang RY, Wang LD, Li G, Verkhusha VV and Wang LHV, Nature Methods, 2016, 13, 67-+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Xiao W, Ruan SB, Yu WQ, Wang RR, Hu C, Liu R. and Gao HL, Molecular Pharmaceutics, 2017, 14, 3489–3498. [DOI] [PubMed] [Google Scholar]
- 159.Kim JJ, Lee YA, Su D, Lee J, Park SJ, Kim B, Lee JHJ, Liu X, Kim SS, Bae MA, Lee JS, Hong SC, Wang L, Samanta A, Kwon HY, Choi SY, Kim JY, Yu YH, Ha HH, Wang ZX, Tam WL, Lim B, Kang NY and Chang YT, Journal of the American Chemical Society, 2019, 141, 14673–14686. [DOI] [PubMed] [Google Scholar]
- 160.Moore C, Borum RM, Mantri Y, Xu M, Fajtova P, O’Donoghue AJ and Jokerst JV, Acs Sensors, 2021, 6, 2356–2365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Rao SR, Snaith AE, Marino D, Cheng X, Lwin ST, Orriss IR, Hamdy FC and Edwards CM, British Journal of Cancer, 2017, 116, 227–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Gao X, Ma G, Jiang C, Zeng L, Jiang S, Huang P. and Lin J, Analytical Chemistry, 2019, 91, 7112–7117. [DOI] [PubMed] [Google Scholar]
- 163.Wu C, Zhang R, Du W, Cheng L. and Liang G, Nano Lett., 2018, 18, 7749–7754. [DOI] [PubMed] [Google Scholar]
- 164.Agrotis and Ketteler, Cells, 2019, 9, 53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Bosc D, Vezenkov L, Bortnik S, An J, Xu J, Choutka C, Hannigan AM, Kovacic S, Loo S, Clark PGK, Chen G, Guay-Ross RN, Yang K, Dragowska WH, Zhang F, Go NE, Leung A, Honson NS, Pfeifer TA, Gleave M, Bally M, Jones SJ, Gorski SM and Young RN, Scientific Reports, 2018, 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Lin Y-X, Wang Y, Qiao S-L, An H-W, Wang J, Ma Y, Wang L. and Wang H, Biomaterials, 2017, 141, 199–209. [DOI] [PubMed] [Google Scholar]
- 167.Shalini S, Dorstyn L, Dawar S. and Kumar S, Cell Death Differ, 2015, 22, 526–539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Yang Q, Cai S, Forrest ML, Cui H. and Yang X, Journal of Biomedical Optics, 2011, 16, 116026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Wang Y, Hu X, Weng J, Li J, Fan Q, Zhang Y. and Ye D, Angewandte Chemie International Edition, 2019, 58, 4886–4890. [DOI] [PubMed] [Google Scholar]
- 170.Thomas G, Nature Reviews Molecular Cell Biology, 2002, 3, 753–766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Braun E. and Sauter D, Clinical & Translational Immunology, 2019, 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Dragulescu-Andrasi A, Kothapalli S-R, Tikhomirov GA, Rao J. and Gambhir SS, Journal of the American Chemical Society, 2013, 135, 11015–11022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Debacq-Chainiaux F, Erusalimsky JD, Campisi J. and Toussaint O, Nature Protocols, 2009, 4, 1798–1806. [DOI] [PubMed] [Google Scholar]
- 174.Asanuma D, Sakabe M, Kamiya M, Yamamoto K, Hiratake J, Ogawa M, Kosaka N, Choyke PL, Nagano T, Kobayashi H. and Urano Y, Nature Communications, 2015, 6, 6463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Cai X, Li L, Krumholz A, Guo Z, Erpelding TN, Zhang C, Zhang Y, Xia Y. and Wang LV, PLoS One, 2012, 7, e43999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Zhen X, Zhang J, Huang J, Xie C, Miao Q. and Pu K, Angewandte Chemie International Edition, 2018, 57, 7804–7808. [DOI] [PubMed] [Google Scholar]
- 177.Wu Y, Huang S, Wang J, Sun L, Zeng F. and Wu S, Nature Communications, 2018, 9, 3983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Zeng Q, Wu Y. and Zhang T, Journal of Innovative Optical Health Sciences, 2019, 12, 1941001. [Google Scholar]
- 179.Gardner SH, Brady CJ, Keeton C, Yadav AK, Mallojjala SC, Lucero MY, Su S, Yu Z, Hirschi JS, Mirica LM and Chan J, Angewandte Chemie International Edition, 2021, 60, 18860–18866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Shield MA and Mirkes PE, in Handbook of Developmental Neurotoxicology, eds. Slikker W and Chang LW, Academic Press, San Diego, 1998, DOI: 10.1016/B978-012648860-9.50012-1, pp. 159–188. [DOI] [Google Scholar]
- 181.Zhang Y, He S, Chen W, Liu Y, Zhang X, Miao Q. and Pu K, Angew. Chem. Int. Ed, 2021, 60, 5921–5927. [DOI] [PubMed] [Google Scholar]
- 182.Ross D. and Siegel D, Redox Biology, 2021, 41, 101950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Srijiwangsa P. and Na-Bangchang K, Journal of Clinical & Experimental Oncology, 2017, 2017, 1–6. [Google Scholar]
- 184.Liu F, Shi X, Liu X, Wang F, Yi H-B and Jiang J-H, Chemical Science, 2019, 10, 9257–9264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Biol Cancer. Ther., 2005, 4, 917–917. [Google Scholar]
- 186.Wu Y, Chen J, Sun L, Zeng F. and Wu S, Adv. Funct. Mater, 2019, 29, n/a. [Google Scholar]
- 187.Levi J, Kothapalli SR, Ma T-J, Hartman K, Khuri-Yakub BT and Gambhir SS, Journal of the American Chemical Society, 2010, 132, 11264–11269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Levi J, Kothapalli S-R, Bohndiek S, Yoon J-K, Dragulescu-Andrasi A, Nielsen C, Tisma A, Bodapati S, Gowrishankar G, Yan X, Chan C, Starcevic D. and Gambhir SS, Clinical Cancer Research, 2013, 19, 1494–1502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Yang K, Zhu L, Nie L, Sun X, Cheng L, Wu C, Niu G, Chen X. and Liu Z, Theranostics, 2014, 4, 134–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Liu C, Li S, Gu Y, Xiong H, Wong WT and Sun L, Mol Imaging Biol, 2018, 20, 919–929. [DOI] [PubMed] [Google Scholar]
- 191.Yang K, Liu Y, Wang Y, Ren Q, Guo H, Matson JB, Chen X. and Nie Z, Biomaterials, 2019, 223, 119460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Xia F, Niu J, Hong Y, Li C, Cao W, Wang L, Hou W, Liu Y. and Cui D, Acta Biomater, 2019, 89, 289–299. [DOI] [PubMed] [Google Scholar]
- 193.Yin L, Sun H, Zhang H, He L, Qiu L, Lin J, Xia H, Zhang Y, Ji S, Shi H. and Gao M, Journal of the American Chemical Society, 2019, 141, 3265–3273. [DOI] [PubMed] [Google Scholar]
- 194.Meng T, Fan B, Li Q, Peng X, Xu J. and Zhang R, Journal of Materials Chemistry B, 2020, 8, 9888–9898. [DOI] [PubMed] [Google Scholar]
- 195.Miki K, Imaizumi N, Nogita K, Oe M, Mu H, Huo W, Harada H. and Ohe K, Bioconjugate Chem., 2021, 32, 1773–1781. [DOI] [PubMed] [Google Scholar]
- 196.Kim J, Yu AM, Kubelick KP and Emelianov SY, Photoacoustics, 2022, 25, 100307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Mahmood N, Mihalcioiu C. and Rabbani SA, Frontiers in Oncology, 2018, 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Dass K, Ahmad A, Azmi AS, Sarkar SH and Sarkar FH, Cancer Treatment Reviews, 2008, 34, 122–136. [DOI] [PubMed] [Google Scholar]
- 199.Li Q, Li S, He S, Chen W, Cheng P, Zhang Y, Miao Q. and Pu K, Angew. Chem. Int. Ed, 2020, 59, 7018–7023. [DOI] [PubMed] [Google Scholar]
- 200.Que EL, Domaille DW and Chang CJ, Chem Rev, 2008, 108, 1517–1549. [DOI] [PubMed] [Google Scholar]
- 201.Linder MC, International Journal of Molecular Sciences, 2020, 21, 4932. [Google Scholar]
- 202.Ackerman CM and Chang CJ, J. Biol. Chem, 2018, 293, 4628–4635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Shanbhag VC, Gudekar N, Jasmer K, Papageorgiou C, Singh K. and Petris MJ, Biochimica et Biophysica Acta (BBA) - Molecular Cell Research, 2021, 1868, 118893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Li H, Zhang P, Smaga LP, Hoffman RA and Chan J, J. Am. Chem. Soc, 2015, 137, 15628–15631. [DOI] [PubMed] [Google Scholar]
- 205.Zhou EY, Knox HJ, Liu C, Zhao W. and Chan J, J Am Chem Soc, 2019, 141, 17601–17609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Wang S, Sheng Z, Yang Z, Hu D, Long X, Feng G, Liu Y, Yuan Z, Zhang J, Zheng H. and Zhang X, Angew. Chem. Int. Ed, 2019, 58, 12415–12419. [DOI] [PubMed] [Google Scholar]
- 207.Zeng L, Ma G, Xu H, Mu J, Li F, Gao X, Deng Z, Qu J, Huang P. and Lin J, Small, 2019, 15, 1803866. [DOI] [PubMed] [Google Scholar]
- 208.Lucero MY, Tang Y, Zhang CJ, Su S, Forzano JA, Garcia V, Huang X, Moreno D. and Chan J, Proceedings of the National Academy of Sciences, 2021, 118, e2106943118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Cash KJ, Li C, Xia J, Wang LV and Clark HA, ACS Nano, 2015, 9, 1692–1698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Lee CH, Folz J, Zhang W, Jo J, Tan JWY, Wang X. and Kopelman R, Anal. Chem, 2017, 89, 7943–7949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Ning J, Lin X, Su F, Sun A, Liu H, Luo J, Wang L. and Tian Y, Analytical and Bioanalytical Chemistry, 2020, 412, 6947–6957. [DOI] [PubMed] [Google Scholar]
- 212.Mishra A, Jiang YY, Roberts S, Ntziachristos V. and Westmeyer GG, Anal. Chem, 2016, 88, 10785–10789. [DOI] [PubMed] [Google Scholar]
- 213.Roberts S, Seeger M, Jiang YY, Mishra A, Sigmund F, Stelzl A, Lauri A, Symvoulidis P, Rolbieski H, Preller M, Dean-Ben XL, Razansky D, Orschmann T, Desbordes SC, Vetschera P, Bach T, Ntziachristos V. and Westmeyer GG, Journal of the American Chemical Society, 2018, 140, 2718–2721. [DOI] [PubMed] [Google Scholar]
- 214.Slikboer S, Naperstkow Z, Janzen N, Faraday A, Soenjaya Y, Le Floc’h J, Al-Karmi S, Swann R, Wyszatko K, Demore CEM, Foster S. and Valliant JF, Molecular Pharmaceutics, 2020, 17, 3369–3377. [DOI] [PubMed] [Google Scholar]
- 215.Shemetov AA, Monakhov MV, Zhang QR, Canton-Josh JE, Kumar M, Chen MM, Matlashov ME, Li X, Yang W, Nie LM, Shcherbakova DM, Kozorovitskiy Y, Yao JJ, Ji N. and Verkhusha VV, Nat. Biotechnol, 2021, 39, 368-+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Zhang C, Gao R, Zhang L, Liu C, Yang Z. and Zhao S, Anal. Chem, 2020, 92, 6382–6390. [DOI] [PubMed] [Google Scholar]
- 217.Fang HB, Wang CJ, Chen YC, Chen ZY, Yao SK, Yang SP, Dong L, Guo ZJ and He WJ, Inorganic Chemistry Frontiers, 2021, 8, 3402–3410. [Google Scholar]
- 218.Lennicke C, Rahn J, Lichtenfels R, Wessjohann LA and Seliger B, Cell Communication and Signaling, 2015, 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.López-Lázaro M, Cancer Letters, 2007, 252, 1–8. [DOI] [PubMed] [Google Scholar]
- 220.Winterbourn CC, Nature Chemical Biology, 2008, 4, 278–286. [DOI] [PubMed] [Google Scholar]
- 221.Lisanti MP, Martinez-Outschoorn UE, Lin Z, Pavlides S, Whitaker-Menezes D, Pestell RG, Howell A. and Sotgia F, Cell Cycle, 2011, 10, 2440–2449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Zhou Y, Wang L, Wang C, Wu Y, Chen D. and Lee TH, Arch. Pharmacal Res, 2020, 43, 187–203. [DOI] [PubMed] [Google Scholar]
- 223.Szatrowski TP and Nathan CF, Cancer Research, 1991, 51, 794. [PubMed] [Google Scholar]
- 224.Schaeferling M, Groegel DBM and Schreml S, Microchim. Acta, 2011, 174, 1–18. [Google Scholar]
- 225.Guo H, Aleyasin H, Dickinson BC, Haskew-Layton RE and Ratan RR, Cell and Bioscience, 2014, 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Bilan DS and Belousov VV, Antioxidants & Redox Signaling, 2018, 29, 569–584. [DOI] [PubMed] [Google Scholar]
- 227.Rezende F, Brandes RP and Schroeder K, Antioxidants & Redox Signaling, 2018, 29, 585–602. [DOI] [PubMed] [Google Scholar]
- 228.Zheng D-J, Yang Y-S and Zhu H-L, Trac-Trends in Analytical Chemistry, 2019, 118, 625–651. [Google Scholar]
- 229.Wei Y, Liu Y, He Y. and Wang Y, Journal of Materials Chemistry B, 2021, 9, 908–920. [DOI] [PubMed] [Google Scholar]
- 230.Kim KS, Lee D, Song CG and Kang PM, Nanomedicine, 2015, 10, 2709–2723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Yu LD, Chen Y. and Chen HR, Chin. Chem. Lett, 2017, 28, 1841–1850. [Google Scholar]
- 232.Yang N, Xiao WY, Song XJ, Wang WJ and Dong XC, Nano-Micro Lett., 2020, 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Pu K, Shuhendler AJ, Jokerst JV, Mei J, Gambhir SS, Bao Z. and Rao J, Nature Nanotechnology, 2014, 9, 233–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Chen Q, Liang C, Sun X, Chen J, Yang Z, Zhao H, Feng L. and Liu Z, Proceedings of the National Academy of Sciences of the United States of America, 2017, 114, 5343–5348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Xie C, Zhen X, Lyu Y. and Pu K, Adv. Mater, 2017, 29. [DOI] [PubMed] [Google Scholar]
- 236.Yang W, Shi X, Shi Y, Yao D, Chen S, Zhou X. and Zhang B, ACS Nano, 2018, 12, 12169–12180. [DOI] [PubMed] [Google Scholar]
- 237.Yang Z, Dai Y, Yin C, Fan Q, Zhang W, Song J, Yu G, Tang W, Fan W, Yung BC, Li J, Li X, Li X, Tang Y, Huang W, Song J. and Chen X, Adv. Mater, 2018, 30. [DOI] [PubMed] [Google Scholar]
- 238.Bouche M, Puehringer M, Iturmendi A, Arnirshaghaghi A, Tsourkas A, Teasdale I. and Cormode DP, Acs Applied Materials & Interfaces, 2019, 11, 28648–28656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Ding H, Cai Y, Gao L, Liang M, Miao B, Wu H, Liu Y, Xie N, Tang A, Fan K, Yan X. and Nie G, Nano Lett., 2019, 19, 203–209. [DOI] [PubMed] [Google Scholar]
- 240.Li C, Lin W, Liu S, Zhang W. and Xie Z, Journal of Materials Chemistry B, 2019, 7, 4655–4660. [DOI] [PubMed] [Google Scholar]
- 241.Liu F, Lin L, Zhang Y, Wang Y, Sheng S, Xu C, Tian H. and Chen X, Adv. Mater, 2019, 31. [DOI] [PubMed] [Google Scholar]
- 242.Wang P, Yang W, Shen S, Wu C, Wen L, Cheng Q, Zhang B. and Wang X, ACS Nano, 2019, 13, 11168–11180. [DOI] [PubMed] [Google Scholar]
- 243.Weber J, Bollepalli L, Belenguer AM, Antonio MD, De Mitri N, Joseph J, Balasubramanian S, Hunter CA and Bohndiek SE, Cancer Research, 2019, 79, 5407–5417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Matsuda H, Tanaka H, Blas BL, Nosenas JS, Tokawa T. and Ohsawa S, Japanese Journal of Experimental Medicine, 1984, 54, 131–138. [PubMed] [Google Scholar]
- 245.Liu F, Lin L, Zhang Y, Wang Y, Sheng S, Xu C, Tian H. and Chen X, Adv. Mater, 2019, 31, 1902885. [DOI] [PubMed] [Google Scholar]
- 246.Klebanoff SJ, Journal of Leukocyte Biology, 2005, 77, 598–625. [DOI] [PubMed] [Google Scholar]
- 247.Gungor N, Knaapen AM, Munnia A, Peluso M, Haenen GR, Chiu RK, Godschalk RWL and Van Schooten FJ, Mutagenesis, 2010, 25, 149–154. [DOI] [PubMed] [Google Scholar]
- 248.Pan B, Ren H, Lv X, Zhao Y, Yu B, He Y, Ma Y, Niu C, Kong J, Yu F, Sun W-B, Zhang Y, Willard B. and Zheng L, J. Transl. Med, 2012, 10, 65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Yin C, Zhen X, Fan Q, Huang W. and Pu K, Acs Nano, 2017, 11, 4174–4182. [DOI] [PubMed] [Google Scholar]
- 250.Snyder SH, Science, 1992, 257, 494–496. [DOI] [PubMed] [Google Scholar]
- 251.Wink DA, Hines HB, Cheng RYS, Switzer CH, Flores-Santana W, Vitek MP, Ridnour LA and Colton CA, Journal of Leukocyte Biology, 2011, 89, 873–891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Fukumura D, Kashiwagi S. and Jain RK, Nature Reviews Cancer, 2006, 6, 521–534. [DOI] [PubMed] [Google Scholar]
- 253.Vannini F, Kashfi K. and Nath N, Redox Biology, 2015, 6, 334–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Heinecke JL, Ridnour LA, Cheng RYS, Switzer CH, Lizardo MM, Khanna C, Glynn SA, Hussain SP, Young HA, Ambs S. and Wink DA, Proceedings of the National Academy of Sciences, 2014, 111, 6323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Thomas DD, Liu ZP, Kantrow SP and Lancaster JR, Proceedings of the National Academy of Sciences of the United States of America, 2001, 98, 355–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Reinhardt CJ, Zhou EY, Jorgensen MD, Partipilo G. and Chan J, Journal of the American Chemical Society, 2018, 140, 1011–1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Wang S, Li Z, Liu Y, Feng G, Zheng J, Yuan Z. and Zhang X, Sensors and Actuators B: Chemical, 2018, 267, 403–411. [Google Scholar]
- 258.Reinhardt CJ, Xu R. and Chan J, Chemical Science, 2020, 11, 1587–1592. [Google Scholar]
- 259.Lucero MY, East AK, Reinhardt CJ, Sedgwick AC, Su S, Lee MC and Chan J, Journal of the American Chemical Society, 2021, 143, 7196–7202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Qi J, Feng L, Zhang X, Zhang H, Huang L, Zhou Y, Zhao Z, Duan X, Xu F, Kwok RTK, Lam JWY, Ding D, Xue X. and Tang BZ, Nature Communications, 2021, 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.Reinhardt CJ, Zhou EY, Jorgensen MD, Partipilo G. and Chan J, J. Am. Chem. Soc, 2018, 140, 1011–1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Afanas’ev I, Aging Dis, 2015, 6, 216–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Xiao H, Zhang W, Li P, Zhang W, Wang X. and Tang B, Angew. Chem. Int. Ed, 2020, 59, 4216–4230. [DOI] [PubMed] [Google Scholar]
- 264.Zheng J, Zeng Q, Zhang R, Xing D. and Zhang T, Journal of the American Chemical Society, 2019, 141, 19226–19230. [DOI] [PubMed] [Google Scholar]
- 265.Alvarez B. and Radi R, Amino Acids, 2003, 25, 295–311. [DOI] [PubMed] [Google Scholar]
- 266.Szabó C, Ischiropoulos H. and Radi R, Nature Reviews Drug Discovery, 2007, 6, 662–680. [DOI] [PubMed] [Google Scholar]
- 267.Pardo FS, Hsu DW, Zeheb R, Efird JT, Okunieff PG and Malkin DM, British Journal of Cancer, 2004, 91, 1678–1686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.Nagaraj S, Gupta K, Pisarev V, Kinarsky L, Sherman S, Kang L, Herber DL, Schneck J. and Gabrilovich DI, Nature Medicine, 2007, 13, 828–835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269.Gabrilovich DI and Nagaraj S, Nature Reviews Immunology, 2009, 9, 162–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270.Zhang J, Zhen X, Upputuri PK, Pramanik M, Chen P. and Pu K, Adv. Mater, 2017, 29. [DOI] [PubMed] [Google Scholar]
- 271.Zhang J, Zhen X, Zeng J. and Pu K, Anal. Chem, 2018, 90, 9301–9307. [DOI] [PubMed] [Google Scholar]
- 272.Balendiran GK, Dabur R. and Fraser D, Cell Biochem. Funct, 2004, 22, 343–352. [DOI] [PubMed] [Google Scholar]
- 273.Nunes S. and Serpa J, International Journal of Molecular Sciences, 2018, 19, 1882. [Google Scholar]
- 274.Gupta S. and Sahu AN, International Journal of Pharmaceutical Sciences and Research, 2020, 11, 3608–3618. [Google Scholar]
- 275.Ni D, Jiang D, Valdovinos HF, Ehlerding EB, Yu B, Barnhart TE, Huang P. and Cai W, Nano Letters, 2017, 17, 3282–3289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 276.Chu C, Lin H, Liu H, Wang X, Wang J, Zhang P, Gao H, Huang C, Zeng Y, Tan Y, Liu G. and Chen X, Advanced Materials, 2017, 29, 1605928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 277.Gong F, Cheng L, Yang N, Jin Q, Tian L, Wang M, Li Y. and Liu Z, Nano Letters, 2018, 18, 6037–6044. [DOI] [PubMed] [Google Scholar]
- 278.Liu Y, Yang Z, Huang X, Yu G, Wang S, Zhou Z, Shen Z, Fan W, Liu Y, Davisson M, Kalish H, Niu G, Nie Z. and Chen X, ACS Nano, 2018, 12, 8129–8137. [DOI] [PubMed] [Google Scholar]
- 279.Yin C, Tang Y, Li X, Yang Z, Li J, Li X, Huang W. and Fan Q, Small, 2018, 14, e1703400. [DOI] [PubMed] [Google Scholar]
- 280.Tang L, Yu F, Tang B, Yang Z, Fan W, Zhang M, Wang Z, Jacobson O, Zhou Z, Li L, Liu Y, Kiesewetter DO, Tang W, He L, Ma Y, Niu G, Zhang X. and Chen X, ACS Applied Materials & Interfaces, 2019, 11, 27558–27567. [DOI] [PubMed] [Google Scholar]
- 281.Lin X, Liu S, Zhang X, Zhu R, Chen S, Chen X, Song J. and Yang H, Angewandte Chemie International Edition, 2020, 59, 1682–1688. [DOI] [PubMed] [Google Scholar]
- 282.Zhu Q, Fan Z, Zuo W, Chen Y, Hou Z. and Zhu X, ACS Applied Materials & Interfaces, 2020, 12, 51314–51328. [DOI] [PubMed] [Google Scholar]
- 283.An L, Cao M, Zhang X, Lin J, Tian Q. and Yang S, ACS Applied Materials & Interfaces, 2020, 12, 8050–8061. [DOI] [PubMed] [Google Scholar]
- 284.Lin X, Zhu R, Hong Z, Zhang X, Chen S, Song J. and Yang H, Advanced Functional Materials, 2021, 31, 2101278. [Google Scholar]
- 285.Wang S, Zhang L, Zhao J, He M, Huang Y. and Zhao S, Sci Adv, 2021, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 286.Yan C, Xu H, Wu M, Zhao Z, Zhao W, Tang J. and Guo Z, Science China Chemistry, 2021, DOI: 10.1007/s11426-021-9970-4. [DOI] [Google Scholar]
- 287.Yin C, Tang Y, Li X, Yang Z, Li J, Li X, Huang W. and Fan Q, Small, 2018, 14, 1703400. [DOI] [PubMed] [Google Scholar]
- 288.Wang S, Zhang L, Zhao J, He M, Huang Y. and Zhao S, Science Advances, 2021, 7, eabe3588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 289.Lucero MY and Chan J, Nature Chemistry, 2021, 13, 1248–1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 290.Maeda H, Matsuno H, Ushida M, Katayama K, Saeki K. and Itoh N, Angew. Chem. Int. Ed, 2005, 44, 2922–2925. [DOI] [PubMed] [Google Scholar]
- 291.Cao X, Ding L, Xie Z-Z, Yang Y, Whiteman M, Moore PK and Bian J-S, Antioxidants & Redox Signaling, 2019, 31, 1–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 292.Wang R-H, Chu Y-H and Lin K-T, International Journal of Molecular Sciences, 2021, 22, 6562.34207284 [Google Scholar]
- 293.Wu D, Si W, Wang M, Lv S, Ji A. and Li Y, Nitric Oxide, 2015, 50, 38–45. [DOI] [PubMed] [Google Scholar]
- 294.Kashfi K, Biochem. Pharmacol, 2018, 149, 205–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 295.Shi B, Gu X, Fei Q. and Zhao C, Chemical Science, 2017, 8, 2150–2155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 296.An L, Wang X, Rui X, Lin J, Yang H, Tian Q, Tao C. and Yang S, Angewandte Chemie International Edition, 2018, 57, 15782–15786. [DOI] [PubMed] [Google Scholar]
- 297.Ma T, Zheng J, Zhang T. and Xing D, Nanoscale, 2018, 10, 13462–13470. [DOI] [PubMed] [Google Scholar]
- 298.Sun L, Wu Y, Chen J, Zhong J, Zeng F. and Wu S, Theranostics, 2019, 9, 77–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 299.Tao C, An L, Lin J, Tian Q. and Yang S, Small, 2019, 15, 1903473. [DOI] [PubMed] [Google Scholar]
- 300.Yan C, Liu D, An L, Wang Y, Tian Q, Lin J. and Yang S, Anal Chem, 2020, 92, 8254–8261. [DOI] [PubMed] [Google Scholar]
- 301.Wang R, Gu X, Li Q, Gao J, Shi B, Xu G, Zhu T, Tian H. and Zhao C, Journal of the American Chemical Society, 2020, 142, 15084–15090. [DOI] [PubMed] [Google Scholar]
- 302.Zhang W, Wang J, Su L, Chen H, Zhang L, Lin L, Chen X, Song J. and Yang H, Science China Chemistry, 2020, 63, 1315–1322. [Google Scholar]
- 303.Wang S, Zhang L, Zhao J, Hou L, Wen C, Liang H. and Zhao S, Acs Appl Bio Mater, 2021, 4, 974–983. [Google Scholar]
- 304.An L, Wang X, Rui X, Lin J, Yang H, Tian Q, Tao C. and Yang S, Angew. Chem. Int. Ed, 2018, 57, 15782–15786. [DOI] [PubMed] [Google Scholar]
- 305.Weitz JI, Byrne J, Clagett GP, Farkouh ME, Porter JM, Sackett DL, Strandness DE and Taylor LM, Circulation, 1996, 94, 3026–3049. [DOI] [PubMed] [Google Scholar]
- 306.Nath B. and Szabo G, Hepatology, 2012, 55, 622–633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 307.Semenza Gregg L., Cell, 2012, 148, 399–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 308.Taylor CT and Colgan SP, Journal of Molecular Medicine, 2007, 85, 1295–1300. [DOI] [PubMed] [Google Scholar]
- 309.Knox HJ, Hedhli J, Kim TW, Khalili K, Dobrucki LW and Chan J, Nat Commun, 2017, 8, 1794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 310.Feng L, Cheng L, Dong Z, Tao D, Barnhart TE, Cai W, Chen M. and Liu Z, ACS Nano, 2017, 11, 927–937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 311.Knox HJ, Kim TW, Zhu Z. and Chan J, ACS Chem Biol, 2018, 13, 1838–1843. [DOI] [PubMed] [Google Scholar]
- 312.Meng X, Zhang J, Sun Z, Zhou L, Deng G, Li S, Li W, Gong P. and Cai L, Theranostics, 2018, 8, 6025–6034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 313.An L, Wang Y, Lin J, Tian Q, Xie Y, Hu J. and Yang S, ACS Applied Materials & Interfaces, 2019, 11, 15251–15261. [DOI] [PubMed] [Google Scholar]
- 314.Umehara Y, Kageyama T, Son A, Kimura Y, Kondo T. and Tanabe K, RSC Advances, 2019, 9, 16863–16868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 315.Lin Y, Sun L, Zeng F. and Wu S, Chemistry – A European Journal, 2019, 25, 16740–16747. [DOI] [PubMed] [Google Scholar]
- 316.Chen M, Knox HJ, Tang Y, Liu W, Nie L, Chan J. and Yao J, Opt. Lett, 2019, 44, 3773–3776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 317.Huang J, Wu Y, Zeng F. and Wu S, Theranostics, 2019, 9, 7313–7324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 318.Zeng Z, Ouyang J, Sun L, Zeng C, Zeng F. and Wu S, Analytical Chemistry, 2020, 92, 9257–9264. [DOI] [PubMed] [Google Scholar]
- 319.Qu J, Teng D, Sui G, Guan S, Wang Y, Wang Q, Lin Y, Ran H, Wang Z. and Wang H, Biomaterials Science, 2020, 8, 3116–3129. [DOI] [PubMed] [Google Scholar]
- 320.Ouyang J, Sun L, Zeng Z, Zeng C, Zeng F. and Wu S, Angewandte Chemie International Edition, 2020, 59, 10111–10121. [DOI] [PubMed] [Google Scholar]
- 321.Li M, Li H, Wu Q, Niu N, Huang J, Zhang L, Li Y, Wang D. and Tang BZ, iScience, 2021, 24, 102261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 322.Wang J, Huang J, Zhou W, Zhao J, Peng Q, Zhang L, Wang Z, Li P. and Li R, Journal of Nanobiotechnology, 2021, 19, 87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 323.Zhang S, Chen H, Wang L, Qin X, Jiang B-P, Ji S-C, Shen X-C and Liang H, Angewandte Chemie International Edition, n/a. [Google Scholar]
- 324.Nishida CR, Lee M. and de Montellano PRO, Mol. Pharmacol, 2010, 78, 497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 325.Nishida CR and Ortiz de Montellano PR, J. Med. Chem, 2008, 51, 5118–5120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 326.Sugiura M. and Kato R, J. Pharmacol. Exp. Ther, 1977, 200, 25. [PubMed] [Google Scholar]
- 327.Zbaida S, Drug Metabolism Reviews, 1995, 27, 497–516. [DOI] [PubMed] [Google Scholar]
- 328.Brown JM and Wilson WR, Nature Reviews Cancer, 2004, 4, 437–447. [DOI] [PubMed] [Google Scholar]
- 329.Gardner SH, Brady CJ, Keeton C, Yadav AK, Mallojjala SC, Lucero MY, Su S, Yu Z, Hirschi JS, Mirica LM and Chan J, Angew. Chem. Int. Ed. Engl, 2021, n/a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 330.Kizaka-Kondoh S. and Konse-Nagasawa H, Cancer Sci., 2009, 100, 1366–1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 331.Jane VS, Richard JK and Terence CJ, Mini-Reviews in Medicinal Chemistry, 2001, 1, 293–306. [DOI] [PubMed] [Google Scholar]
- 332.Chevalier A, Zhang Y, Khdour OM, Kaye JB and Hecht SM, Journal of the American Chemical Society, 2016, 138, 12009–12012. [DOI] [PubMed] [Google Scholar]
- 333.Webb BA, Chimenti M, Jacobson MP and Barber DL, Nature Reviews Cancer, 2011, 11, 671–677. [DOI] [PubMed] [Google Scholar]
- 334.Damaghi M, Wojtkowiak J. and Gillies R, Front. Physiol, 2013, 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 335.Gillies RJ, Raghunand N, Karczmar GS and Bhujwalla ZM, Journal of Magnetic Resonance Imaging, 2002, 16, 430–450. [DOI] [PubMed] [Google Scholar]
- 336.Stüwe L, Müller M, Fabian A, Waning J, Mally S, Noël J, Schwab A. and Stock C, The Journal of Physiology, 2007, 585, 351–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 337.Gallagher FA, Kettunen MI, Day SE, Hu D-E, Ardenkjær-Larsen JH, Zandt RIT, Jensen PR, Karlsson M, Golman K, Lerche MH and Brindle KM, Nature, 2008, 453, 940–943. [DOI] [PubMed] [Google Scholar]
- 338.Busco G, Cardone RA, Greco MR, Bellizzi A, Colella M, Antelmi E, Mancini MT, Dell’Aquila ME, Casavola V, Paradiso A. and Reshkin SJ, The FASEB Journal, 2010, 24, 3903–3915. [DOI] [PubMed] [Google Scholar]
- 339.Kanamala M, Wilson WR, Yang M, Palmer BD and Wu Z, Biomaterials, 2016, 85, 152–167. [DOI] [PubMed] [Google Scholar]
- 340.Tang H, Zhao W, Yu J, Li Y. and Zhao C, Molecules, 2018, 24, 4. [Google Scholar]
- 341.Chen Y, Anal. Biochem, 2021, 612, 113900. [DOI] [PubMed] [Google Scholar]
- 342.Duan Z, Gao Y-J, Qiao Z-Y, Fan G, Liu Y, Zhang D. and Wang H, Journal of Materials Chemistry B, 2014, 2, 6271–6282. [DOI] [PubMed] [Google Scholar]
- 343.Lin L-S, Cong Z-X, Li J, Ke K-M, Guo S-S, Yang H-H and Chen G-N, Journal of Materials Chemistry B, 2014, 2, 1031–1037. [DOI] [PubMed] [Google Scholar]
- 344.Kimbrough CW, Khanal A, Zeiderman M, Khanal BR, Burton NC, McMasters KM, Vickers SM, Grizzle WE and McNally LR, Clin Cancer Res, 2015, 21, 4576–4585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 345.Chen Q, Liu X, Chen J, Zeng J, Cheng Z. and Liu Z, Advanced Materials, 2015, 27, 6820–6827. [DOI] [PubMed] [Google Scholar]
- 346.Wu X, Lin B, Yu M, Yang L, Han J. and Han S, Chemical Science, 2015, 6, 2002–2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 347.Chen Q, Liu X, Zeng J, Cheng Z. and Liu Z, Biomaterials, 2016, 98, 23–30. [DOI] [PubMed] [Google Scholar]
- 348.Gurka MK, Pender D, Chuong P, Fouts BL, Sobelov A, McNally MW, Mezera M, Woo SY and McNally LR, J Control Release, 2016, 231, 60–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 349.Zeiderman MR, Morgan DE, Christein JD, Grizzle WE, McMasters KM and McNally LR, ACS Biomaterials Science & Engineering, 2016, 2, 1108–1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 350.Miao Q, Lyu Y, Ding D. and Pu K, Adv. Mater, 2016, 28, 3662–3668. [DOI] [PubMed] [Google Scholar]
- 351.Song J, Kim J, Hwang S, Jeon M, Jeong S, Kim C. and Kim S, Chemical Communications, 2016, 52, 8287–8290. [DOI] [PubMed] [Google Scholar]
- 352.Ju K-Y, Kang J, Pyo J, Lim J, Chang JH and Lee J-K, Nanoscale, 2016, 8, 14448–14456. [DOI] [PubMed] [Google Scholar]
- 353.Shi S, Liu Y, Chen Y, Zhang Z, Ding Y, Wu Z, Yin J. and Nie L, Theranostics, 2016, 6, 2170–2182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 354.Tang Q, Xiao W, Huang C, Si W, Shao J, Huang W, Chen P, Zhang Q. and Dong X, Chemistry of Materials, 2017, 29, 5216–5224. [Google Scholar]
- 355.Dai C, Lin H, Xu G, Liu Z, Wu R. and Chen Y, Chemistry of Materials, 2017, 29, 8637–8652. [Google Scholar]
- 356.Jo J, Lee CH, Kopelman R. and Wang X, Nature Communications, 2017, 8, 471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 357.Yu Z, Wang M, Pan W, Wang H, Li N. and Tang B, Chemical Science, 2017, 8, 4896–4903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 358.Samykutty A, Grizzle WE, Fouts BL, McNally MW, Chuong P, Thomas A, Chiba A, Otali D, Woloszynska A, Said N, Frederick PJ, Jasinski J, Liu J. and McNally LR, Biomaterials, 2018, 182, 114–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 359.Lee S, Lee H, Han H, Yoo H, Kim H, Chang JH and Kim H, Science of Advanced Materials, 2018, 10, 665–673. [Google Scholar]
- 360.Zhang Y, Chang J, Huang F, Yang L, Ren C, Ma L, Zhang W, Dong H, Liu J. and Liu J, ACS Biomaterials Science & Engineering, 2019, 5, 1589–1601. [DOI] [PubMed] [Google Scholar]
- 361.Wang Z, Upputuri PK, Zhen X, Zhang R, Jiang Y, Ai X, Zhang Z, Hu M, Meng Z, Lu Y, Zheng Y, Pu K, Pramanik M. and Xing B, Nano Research, 2019, 12, 49–55. [Google Scholar]
- 362.Roberts S, Strome A, Choi C, Andreou C, Kossatz S, Brand C, Williams T, Bradbury M, Kircher MF, Reshetnyak YK, Grimm J, Lewis JS and Reiner T, Scientific Reports, 2019, 9, 8550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 363.Yang Z, Dai Y, Shan L, Shen Z, Wang Z, Yung BC, Jacobson O, Liu Y, Tang W, Wang S, Lin L, Niu G, Huang P. and Chen X, Nanoscale Horizons, 2019, 4, 426–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 364.Zhu W, Chen M, Liu Y, Tian Y, Song Z, Song G. and Zhang X, Nanoscale, 2019, 11, 20630–20637. [DOI] [PubMed] [Google Scholar]
- 365.Li S, Lui K-H, Tsoi T-H, Lo W-S, Li X, Hu X, Chi-Shing Tai W, Hiu-Ling Hung C, Gu Y-J and Wong W-T, Nanoscale Advances, 2019, 1, 554–564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 366.Yang Z, Song J, Tang W, Fan W, Dai Y, Shen Z, Lin L, Cheng S, Liu Y, Niu G, Rong P, Wang W. and Chen X, Theranostics, 2019, 9, 526–536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 367.Gao X, Jiang S, Li C, Chen Y, Zhang Y, Huang P. and Lin J, Biomaterials, 2021, 267, 120454. [DOI] [PubMed] [Google Scholar]
- 368.Yang Z, Gu B, Jiang C, Zhang L, Liu Q. and Song S, Nanomedicine: Nanotechnology, Biology and Medicine, 2021, 33, 102356. [DOI] [PubMed] [Google Scholar]
- 369.Li S, Lui K-H, Li X, Fang X, Lo W-S, Gu Y-J and Wong W-T, ACS Applied Bio Materials, 2021, 4, 4152–4164. [DOI] [PubMed] [Google Scholar]
- 370.Mu H, Miki K, Harada H, Tanaka K, Nogita K. and Ohe K, ACS Sensors, 2021, 6, 123–129. [DOI] [PubMed] [Google Scholar]
- 371.Chen J, Sedgwick AC, Sen S, Ren Y, Sun Q, Chau C, Arambula JF, Sarma T, Song L, Sessler JL and Liu C, Chemical Science, 2021, 12, 9916–9921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 372.Miki K, Kojima K, Oride K, Harada H, Morinibu A. and Ohe K, Chem. Commun, 2017, 53, 7792–7795. [DOI] [PubMed] [Google Scholar]
- 373.Sawyers CL, Nature, 2008, 452, 548–552. [DOI] [PubMed] [Google Scholar]
- 374.Schwarzenbach H, Hoon DSB and Pantel K, Nature Reviews Cancer, 2011, 11, 426–437. [DOI] [PubMed] [Google Scholar]
- 375.Peng Y. and Croce CM, Signal Transduction and Targeted Therapy, 2016, 1, 15004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 376.Zheng Y, Luo Y, Chen X, Li H, Huang B, Zhou B, Zhu L, Kang X. and Geng W, Molecular Cancer, 2021, 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 377.Yang L, Li J, Pan W, Wang H, Li N. and Tang B, Chem. Commun, 2018, 54, 3656–3659. [DOI] [PubMed] [Google Scholar]
- 378.Zhang K, Meng X, Yang Z, Cao Y, Cheng Y, Wang D, Lu H, Shi Z, Dong H. and Zhang X, Adv. Mater, 2019, 31, 1807888. [DOI] [PubMed] [Google Scholar]
- 379.Yijun Guo BW, Shiyan Xiao, Dongbao Yao, Hui Li, Huaguo Xu, Tingjie Song, Xiang Li, Haojun Liang, Quant. Biol, 2017, 5, 25–41. [Google Scholar]