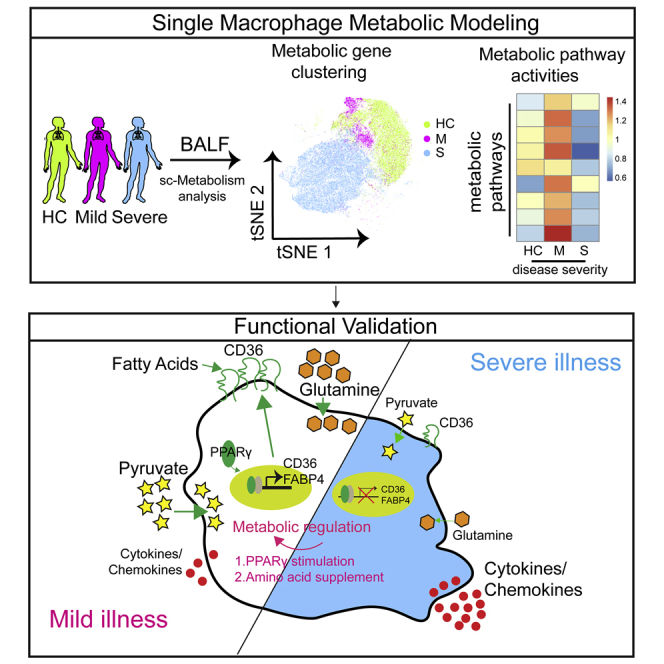
Summary
SARS-CoV-2 infection induces imbalanced immune response such as hyperinflammation in patients with severe COVID-19. Here, we studied the immunometabolic regulatory mechanisms for the pathogenesis of COVID-19. We depicted the metabolic landscape of immune cells, especially macrophages, from bronchoalveolar lavage fluid of patients with COVID-19 at single-cell level. We found that most metabolic processes were upregulated in macrophages from lungs of patients with mild COVID-19 compared to cells from healthy controls, whereas macrophages from severe COVID-19 showed downregulation of most of the core metabolic pathways including glutamate metabolism, fatty acid oxidation, citrate cycle, and oxidative phosphorylation, and upregulation of a few pathways such as glycolysis. Rewiring cellular metabolism by amino acid supplementation, glycolysis inhibition, or PPARγ stimulation reduces inflammation in macrophages stimulated with SARS-CoV-2. Altogether, this study demonstrates that metabolic imbalance of bronchoalveolar macrophages may contribute to hyperinflammation in patients with severe COVID-19 and provides insights into treating COVID-19 by immunometabolic modulation.
Subject areas: Biological sciences, Immunology, Virology, Bioinformatics
Graphical abstract
Highlights
-
•
Dysregulated metabolic activities of lung immune cells in severe COVID-19
-
•
Unbalanced metabolic reprogramming may lead to dysfunction of lung macrophages
-
•
Remodeling metabolism reduces inflammation in macrophages stimulated with SARS-CoV-2
Biological sciences; Immunology; Virology; Bioinformatics
Introduction
The coronavirus disease 2019 (COVID-19) caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has caused more than 250 million infections and more than 5 million deaths, according to the statistics of World Health Organization until December 1, 2021. It was estimated that around 80% of patients with COVID-19 experienced asymptomatic, mild, or moderate symptoms, while 20% patients progressed to severe conditions and even death. Molecular events and mechanisms underlying how SARS-CoV-2 infection leads to severe pneumonia remain largely unknown.
Accumulating studies have demonstrated that dysfunctional immune response plays a key role for severe symptoms of COVID-19 (Laing et al., 2020; Liao et al., 2020; Lucas et al., 2020; Mathew et al., 2020; Ren et al., 2021; Schulte-Schrepping et al., 2020; Wilk et al., 2020). Patients with severe COVID-19 exhibited abnormal peripheral immune activities, including lymphopenia, exhausted lymphocytes, increased immature neutrophils, and abnormal monocyte compartment and function (Arunachalam et al., 2020; Mathew et al., 2020; Schulte-Schrepping et al., 2020; Silvin et al., 2020; Wilk et al., 2020). Particularly, a cytokine storm is associated with COVID-19 severity, represented by systemic upregulation of calprotectin and other proinflammatory cytokines and chemokines (Del Valle et al., 2020; Laing et al., 2020; Ren et al., 2021; Silvin et al., 2020). We and others have discovered abundant hyperinflammatory monocyte-derived macrophages enriched in the bronchoalveolar lavage fluid (BALF) from patients with severe COVID-19, whereas highly clonal-expanded CD8+ T cells were presented in moderate patients (Liao et al., 2020; Wauters et al., 2021). Together, these studies suggest that abnormal immune response is associated with severe SARS-CoV-2 infection.
Cellular metabolism is important for the functionality of immune cells under healthy and disease conditions (Ayres, 2020; Makowski et al., 2020). Recent clinical studies have suggested an association between plasma metabolites alteration and COVID-19. Dysregulation of metabolic components and a sharp drop in blood nutrients in sera of patients with severe COVID-19 was observed in several recent studies (Lam et al., 2021; Shen et al., 2020; Song et al., 2020; Su et al., 2020; Xiao et al., 2021). It was also reported that T cells with a unique metabolic profile were expanded in acute COVID-19 (Thompson et al., 2021). However, the immunometabolic regulation of leukocytes in lungs, the major organ affected by SARS-CoV-2 infection, and its correlation with immune function and disease severity are currently unclear. To answer this question, we investigated landscape of immunometabolism of BALF cells from patients with mild-to-severe COVID-19 by using computational algorithms that quantify activities of metabolic pathways (Xiao et al., 2019) and metabolic fluxes (Damiani et al., 2019) at single-cell resolution. We discovered that COVID-19 severity was highly associated with immunometabolic dysregulation of immune cells derived from BALF. Notably, macrophages from lungs of severe COVID-19 exhibited unique metabolism with remarkably reduced of most metabolic pathways while only increased a few pathways such as glycolysis. In comparison, most of the metabolic pathways were increased in macrophages from mild cases. We proved that the antidiabetic drug rosiglitazone enhanced fatty acid metabolism and phagocytosis capacity of macrophages, and reduced proinflammatory cytokines in macrophages upon SARS-CoV-2 stimulation. Our study thus illustrated immunometabolic landscape of immune cells along progression of COVID-19, which may shed light on modulating COVID-19 by immunometabolic regulation.
Results
Study design and patient cohort
To depict the immunometabolic landscape in lungs of patients with COVID-19 and its association with disease progression, we first analyzed our single-cell transcriptomics data derived from BALF of 4 healthy controls (HCs), 3 patients with mild, and 6 patients with severe COVID-19 that we have published recently (Liao et al., 2020) and confirmed the finding with another cohort of data from BALF of 2 patients with mild and 20 patients with severe COVID-19 (Wauters et al., 2021). We determined the metabolic landscape in single BALF immune cells by using computational algorithms to quantify activities of metabolic pathways (Xiao et al., 2019) (Figure 1A). Concurrently, we performed a single-cell flux balance analysis (scFBA) to analyze differences in metabolic flux between cells from BALF of patients with mild and severe COVID-19 (Damiani et al., 2019) (Figure 1A). Leveraging our previous findings on the highly proinflammatory macrophages abundant in lungs from severe patients (Liao et al., 2020), we further investigate the functional association between metabolism and immune regulation of macrophages using correlation analysis (Figure 1A). Furthermore, we experimentally demonstrated that metabolic manipulation of human primary macrophages affects their immune response to SARS-CoV-2 or toll-like receptor agonists stimulation (Figure 1A).
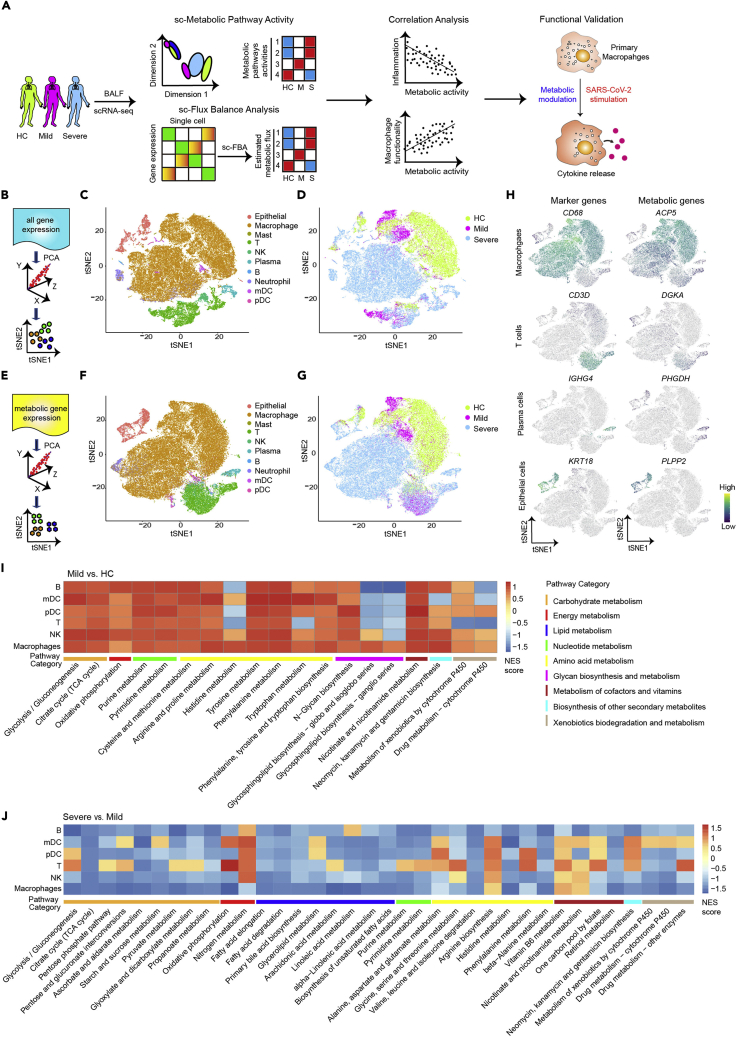
Figure 1.
Landscape of metabolic gene expression in bronchoalveolar immune cells from patients with COVID-19 at single-cell level
(A) A flowchart depicting the overall design of the study.
(B–D) A schematic diagram shows principal component analysis (PCA) (B) and t-SNE visualization of subpopulations (C and D) in BALFs of healthy controls (HCs, n = 4), mild COVID-19 (n = 3), and severe COVID-19 (n = 6) donors based on all gene expression profiles.
(E–G) A schematic diagram shows PCA (E) and t-SNE plot of metabolic genes expression profiles (F-G) of cells from BALFs based on 1,526 metabolic gene expressions. Different colors indicate distinct cell subsets (C and F) and patient groups (D and G). Cell subset identity in F was defined by using all variable genes.
(H) Feature plots show expression of cell-subset marker genes and indicated metabolic genes of macrophages, mDC, T cells, plasma cells, and epithelial cells from BALFs of HC and patients with COVID-19.
(I and J) GSEA summarizing metabolic pathway alterations of indicated subpopulations in mild versus HC (I) and patients with severe versus mild (J) COVID-19. A pathway was included in the heatmap if there was an alteration with pvalue less than 0.1 in any cell type.
Landscape of metabolic gene expression of immune cells from BALF of patients with COVID-19 at single-cell level
To investigate the metabolic profile of immune cells from BALF of patients with COVID-19, we first reproduced unsupervised clustering based on highly variable features of all genes after dimensionality reduction with t-distribution stochastic neighbor embedding (t-SNE) as we previously reported (Liao et al., 2020). Nine major cell populations including macrophages, myeloid dendritic cells (mDC), plasmacytoid dendritic cells (pDC), neutrophils, NK cells, T and B lymphocytes, plasma cells, as well as epithelial cells were found (Figures 1B–1D andTable S1). Next, we sought to determine the global structure of metabolic gene profiles of these cells. We performed unsupervised clustering analysis based on the expression levels of 1664 metabolic genes obtained from the Kyoto Encyclopedia of Genes and Genomes (KEGG) database, of which 1,526 genes have detectable values in our single-cell RNA sequencing (scRNA-seq) dataset (Figure 1E andTable S2). By doing so, we discovered that cells from BALF formed clusters matched with cell subsets as classified by all variable genes (Figures 1F,1H, and S1), indicating the metabolic heterogeneity of bronchoalveolar subpopulations. Notably, we found that cells from BALF, especially macrophages, were segregated into healthy, mild, and severe conditions based on the expression levels of 1,526 metabolic genes (Figure 1G), suggesting that SARS-CoV-2 infection potentially changed the metabolic signatures of cells in bronchoalveolar.
We next performed gene set enrichment analysis (GSEA) comparing 85 metabolic pathways (Table S2) of each major immune subset from HC and mild and severe COVID-19. We excluded neutrophils, plasma cells, and mast cells due to limited cell numbers (Table S1). The results indicated that most of the metabolic pathways such as glycolysis, citrate cycle (tricarboxylic acid [TCA] cycle), and oxidative phosphorylation were upregulated in B cells, mDC, pDC, T cells, natural killer (NK) cells, and macrophages from BALF of mild patients (Figure 1I). Strikingly, most metabolic processes including pyruvate metabolism, TCA cycle, and fatty acid elongation or degradation were downregulated in most immune subsets from severe patients compared to that from mild patients (Figure 1J). Together, these results demonstrated metabolic heterogeneity of bronchoalveolar immune subsets at transcription level and suggested that BALF cells were metabolically suppressed in patients with severe COVID-19 compared to patients with mild COVID-19.
Different metabolic reprogramming of macrophages from BALF of patients with mild or severe COVID-19
Dysregulation of myeloid cells was a key feature of severe COVID-19 (Schulte-Schrepping et al., 2020; Silvin et al., 2020). We reported previously that highly proinflammatory macrophages were abundant in the bronchoalveolar space of patients with severe COVID-19, which may contribute to the cytokine storm (Liao et al., 2020). We speculated that the dysfunction of macrophages may be associated with their metabolic reprogramming during SARS-CoV-2 infection. Clustering analysis and t-SNE visualization based on expression profiles of 1,526 metabolic genes indicated that the global structure of metabolic gene expression in bronchoalveolar macrophages was highly different in patients with HC, mild, and severe COVID-19 (Figure 2A), suggesting potential differences in metabolic reprogramming of macrophages in mild and severe diseases. We next determined the metabolic pathway activities of macrophages using a computational algorithm that quantify activities of 85 metabolic pathways at single-cell levels (Xiao et al., 2019) (Figure 2B). The advantage of the algorithm is that instead of only considering differentially expressed genes, the algorithm defines a pathway activity score as relative gene expression value averaged over all genes in this pathway and all cells of this type. In addition, if one gene is involved in several pathways, the weighted average expression of the gene will be considered based on the number of pathways that include this gene (Figure 2B and STAR Methods). By doing so, we found that among the 85 metabolic pathways with at least 5 genes included, 71 of them were significantly highly enriched (pathway activity score >1 and permutation test p value < 0.01) in macrophages from BALF of patients with COVID-19 with mild disease compared to macrophages from HCs (Figure 2C). Intriguingly, when comparing macrophages from severe COVID-19 to macrophages from HCs, only 8 metabolic pathways were upregulated, while 65 pathways were downregulated (Figure 2D). Detailed analyses of core metabolic pathways showed that most of them such as glycolysis, citrate cycle, pyruvate metabolism, fatty acid biosynthesis, fatty acid degradation, and glutamate metabolism were upregulated in macrophages from mild disease. However, only glycolysis and fatty acid biosynthesis pathways were upregulated in macrophages from severe disease, while other pathways were downregulated (Figure 2E). Analyses of macrophages from each individual donor also confirmed that cells from severe COVID-19 showed much lower metabolic activities than cells from mild cases (Figures S2A and S2B). Expression levels of 1,526 metabolic genes with detectable value by scRNA-seq were significantly upregulated in macrophages from BALF of mild patients compared to HCs and that was downregulated in macrophage from severe patients compared to mild patients (Figure S2C and Table S3). GSEA further confirmed that the pyruvate metabolism, fatty acid degradation, oxidative phosphorylation, TCA cycle, and other metabolic pathways were downregulated in macrophages from BALF of severe COVID-19 compared to cells from mild diseases (Figure S2D). Similar results were obtained when re-analyzing metadata of another cohort (Figures S3A–S3C) including single-cell transcriptomes from BALF of 2 mild and 20 severe COVID-19 cases published recently (Wauters et al., 2021), confirming that the metabolic activity of macrophages from BALF was inhibited in patients with severe COVID-19 compared to patients with mild COVID-19.
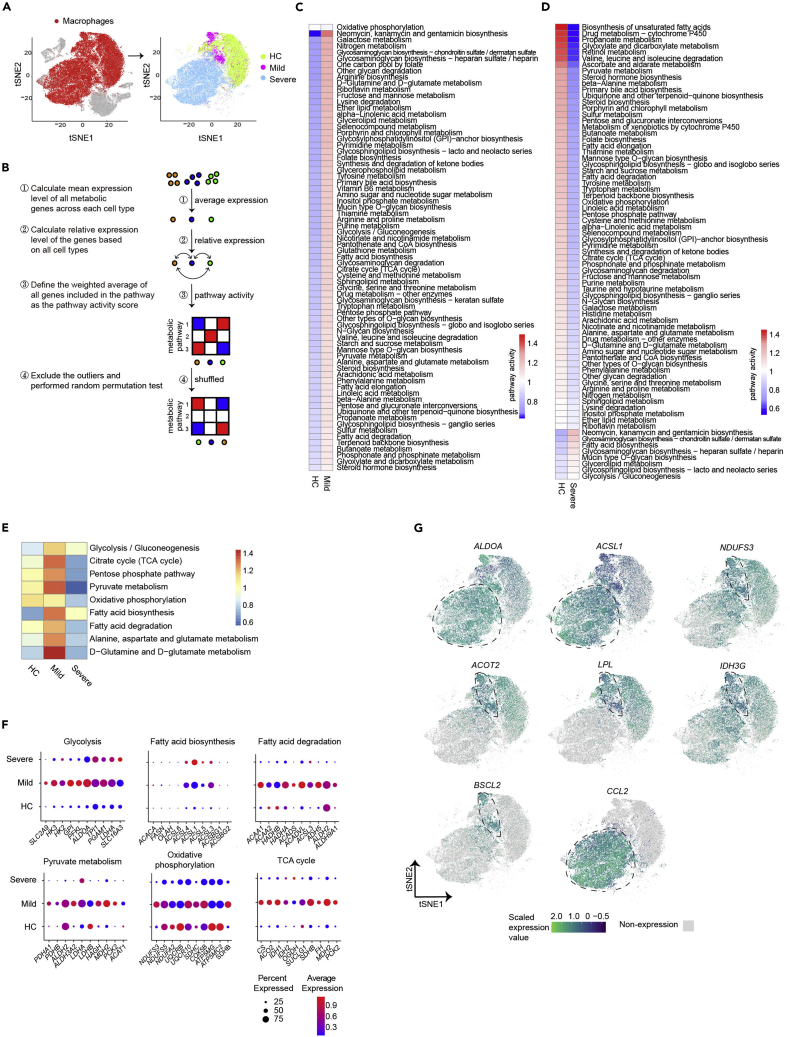
Figure 2.
Different metabolic reprogramming of macrophages in patients with mild or severe COVID-19
(A) t-SNE visualization of macrophages in BALFs of 4 HCs, 3 mild and 6 severe COVID-19 donors based on 1,526 metabolic gene-expression profiles. Macrophages were highlighted in red in left panel and color-coded for sample types in right panel.
(B) A flowchart depicting the basic principle and procedures for the calculation of metabolic pathway activity.
(C and D) Metabolic pathway activities in macrophages from HC versus patients with mild (C), or severe (D) COVID-19. Statistically non-significant values (random permutation test p > 0.01) were shown as blank.
(E) Metabolic pathway activities of core metabolic pathways in macrophages from patients with HC, mild, or severe COVID-19.
(F) Dot plots show expression of indicated metabolic genes.
(G) Feature plots show the expression level of indicated metabolic genes and chemokine CCL2 in macrophages.
Our previous results showed that SARS-CoV-2 transcripts were detected in a small percentage of BALF-derived macrophages (728 out of 30,137 cells, 2.4%) from patients with severe COVID-19, but not in macrophages from mild patients (Figure S4A). We compared the metabolic activity and scFBA of viral transcripts positive macrophages to remaining cells. Our results indicated that there was no significant difference between viral transcripts positive and negative macrophages when compared to macrophages from healthy control or patients with mild COVID-19 (Figures S4B and S4C).
Next, we compared the metabolic pathway activities and expression of specific genes involved in central metabolic pathways supporting biomass and energy synthesis, including glycolysis, fatty acid biosynthesis, TCA cycle, pyruvate metabolism, glutamine metabolism, fatty acid degradation, and oxidative phosphorylation in macrophages from 3 groups. Consistently, we found that the majority of these pathways and related genes were upregulated in macrophages from mild diseases, while only glycolysis and fatty acid biosynthesis pathways and associated genes showed increased expression in BALF macrophages from severe patients (Figures 2F and 2G). Interestingly, genes in other core metabolic pathways showed decreased expression in macrophages derived from severe patients (Figures 2F and 2G). Taken together, our results indicated a unique metabolic reprogramming profile of macrophages from lungs of patients with severe COVID-19.
Single-cell fluxomics analysis of macrophages from patients with mild and severe COVID-19
To further determine the differences of metabolism in macrophages between patients with HC, mild, and severe COVID-19, we used a mathematical approach termed flux balance analysis (FBA) to analyze the flow of metabolites through a metabolic network (Orth et al., 2010). The recently developed scFBA enables us to translate single-cell transcriptomics into single-cell fluxomics (Damiani et al., 2019). Using the multiscale stoichiometric models, we transferred the imputed metabolic gene expression matrix to predicted cell-wise metabolic flux matrix (Figure 3A). We found that among the 315 metabolic fluxes, of which 294 have detectable values, 223 of them were significantly decreased in macrophages from BALF of severe patients compared to that from mild patients at single-cell level, while only 30 fluxes were significantly increased (Figure 3B and Table S4). The decreased fluxes included lipid metabolism (e.g., acetyl-CoA to acetoacetyl-CoA), glutamine metabolism (e.g., glutamate to α-KG (α-ketoglutarate)), pyruvate metabolism (e.g., pyruvate to acetyl-CoA), TCA process (e.g., succinate to fumarate), and oxidative phosphorylation (e.g., ADP to ATP) (Figure 3C), which suggested downregulation of key substrate and energy metabolism processes in BALF macrophages from severe patients. The results were consistent with a recent report showing that blood nutrients including amino acid and lipid metabolites were significantly reduced in patients with severe COVID-19 compared to patients with mild COVID-19 (Su et al., 2020) (Figures S5A and S5B). Several metabolites that closely correlated with macrophage function, such as multiple apolipoproteins including APOA1 (Apolipoprotein AI), APOA2 (Apolipoprotein A-II), and ApoM (Apolipoprotein M), were decreased in plasma of patients with severe COVID-19 (Shen et al., 2020). Itaconate, a macrophage-specific metabolite, was also progressively reduced in plasma with COVID-19 severity (Song et al., 2020). Interestingly, metabolic fluxes associated with glucose transport were upregulated in macrophages from patients with severe COVID-19 (Figure 3C). In addition, fluxes associated with conversion of arginine to ornithine and urea were elevated in macrophages from patients with severe COVID-19 (Figure 3C), consistent with increased levels of ornithine and urea in plasma of severe patients (Su et al., 2020) (Figure S5C). Macrophages expressing arginase and producing ornithine are associated with wound healing (Albina et al., 1990), indicating lung injury in patients with severe COVID-19. The process might also lead to the consumption of arginine in the microenvironment of lungs in patients with severe COVID-19. It was reported that the decreased level of arginine and increased concentration of ornithine led to impaired antiviral or antitumor T cell immunity (Geiger et al., 2016; Lercher et al., 2019). Our results thus indicate that dysregulated metabolism in macrophages may contribute to the impairment of host immune response upon SARS-CoV-2 infection. We also compared scFBA of viral transcripts positive macrophages from severe patients to remaining cells. Our results indicated that there was no significant difference between viral transcripts positive and negative macrophages when comparing to macrophages from healthy control or patients with mild COVID-19 (Figure S4D).
Figure 3.
Single-cell fluxomics analysis of macrophages from BALF of patients with COVID-19
(A) A flowchart depicting the acquisition of an estimated single-cell flux matrix using single-cell transcriptomics.
(B) Heatmap shows significant altered metabolic fluxes (p < 0.05) in macrophages between patients with mild (n = 3) and severe (n = 3) COVID-19 at single-cell level.
(C) Dot plot shows relative levels of indicated metabolic fluxes in macrophages from patients with HCs, mild and severe COVID-19.
(D) A schematic diagram shows PCA for metabolic flux.
(E and F) t-SNE visualization of subclusters (E) and bar plot (F) show the relative contributions of macrophages from mild or severe COVID-19 in each subcluster based on 294 metabolic flux levels in each single-cell.
As an additional effort to characterize the differences in the flow of metabolites between patients with mild and severe COVID-19, we performed unsupervised clustering analysis of macrophages based on estimated metabolic flux levels of single cells (Figure 3D). We discovered 18 clusters of macrophages. Interestingly, 8 clusters (cluster 0, 1, 9, 10, 12, 14, 15, and 16) were mainly composed of macrophages from patients with mild COVID-19, while another 9 clusters (cluster 2, 4, 5, 6, 7, 8, 11, 13, and 17) mainly contained macrophages from severe patients (Figures 3D–3F). These results indicated the dramatic differences of metabolic flux between macrophages from BALF of patients with mild and severe COVID-19.
In summary, cell-wised fluxomics analysis further indicated distinct metabolism switch of macrophage from patients with severe versus mild COVID-19. The results suggested that the dysregulated metabolism of macrophage in patients with severe COVID-19 may contribute to impaired antiviral immunity.
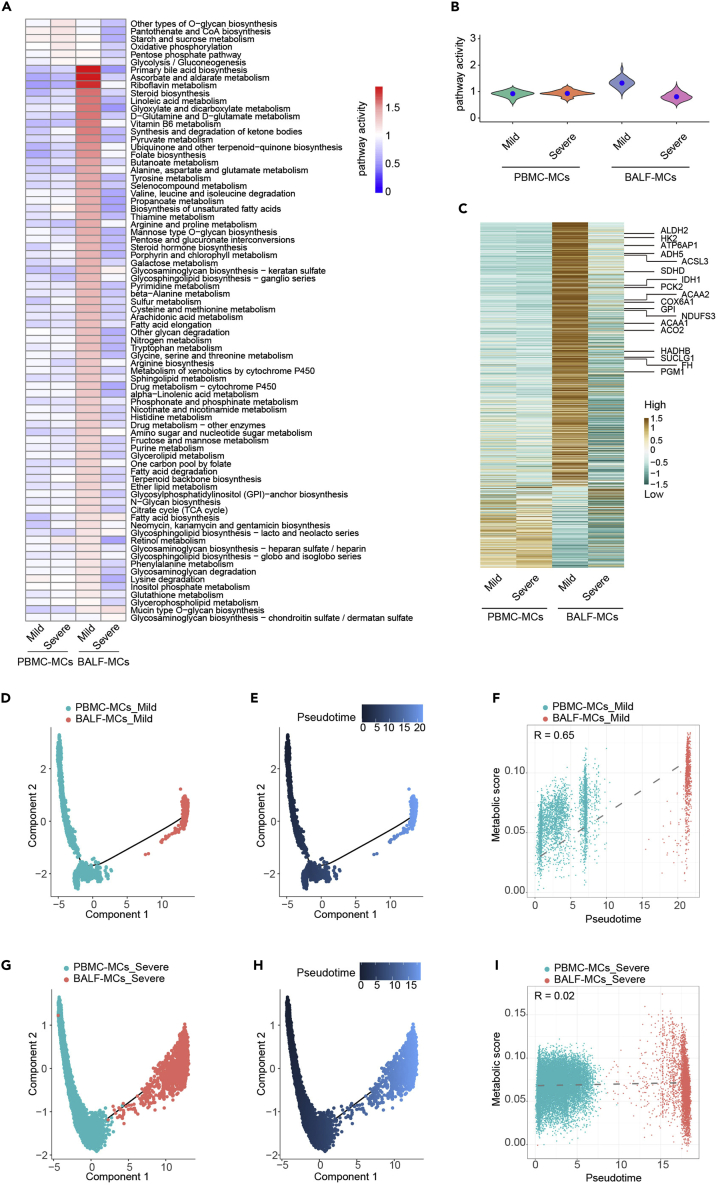
Distinct metabolic reprogramming during monocyte to macrophage transition in mild versus severe COVID-19
Macrophage differentiation from monocytes occurs in tissue during inflammation. We asked whether the metabolic transcriptome differences similarly existed in monocytes from peripheral blood of patients with mild and severe COVID-19. We have recently performed single-cell RNA-seq of paired peripheral human blood mononuclear cells (PBMCs) samples from patients with two mild and five severe COVID-19 (Xu et al., 2020), and we also analyzed BALF samples from the same patients as showed above. We annotated major cell types in PBMCs as previously reported (Figures S6A–S6C). Clustering and t-SNE visualization using metabolic gene expression profiles revealed metabolic heterogeneity among peripheral cell subsets (Figures S6D and S6E). However, we did not observe obvious segregation of monocytes among HC, mild, and severe conditions based on metabolic gene expression (Figure S6F), suggesting that immunometabolism of monocytes was not dramatically different in peripheral blood as observed in BALF.
We have recently delineated blood-toward-BALF courses of monocyte to macrophage differentiation trajectory by interrogating single-cell transcriptomics of paired blood and BALF samples from the same patients with COVID-19 (Xu et al., 2020). We next sought to determine if metabolic reprogramming occurs during monocyte-to-macrophage differentiation. CD14 expression discriminates monocyte-derived macrophages from tissue-resident macrophages (Coillard and Segura, 2019). We thus investigated metabolic changes between CD14+ monocytes from PBMC (PBMC-MCs (PBMC derived monocytes )) and CD14+ macrophage from lung (BALF-MCs) under mild and severe conditions. Most of the metabolic pathway activity and metabolic-related gene expression of PBMC-MCs remained similar between mild and severe patients, while BALF-MCs from mild patients showed significantly increased levels of metabolic activities and metabolic gene expression comparing to BALF-MCs from severe patients (Figures 4A–4C). We then asked if metabolic reprogramming occurs during differentiation of monocyte to macrophage. Unbiased pseudo time trajectory analysis using genes with high dispersion across cells revealed a route from PBMC-MCs to BALF-MCs, consistent with previous report (Xu et al., 2020) (Figures 4D, 4E,4G, and 4H). Interestingly, we observed that the metabolic activity was increased along the PBMC-MCs to BALF-MCs trajectory in mild patients (Figure 4F), whereas marginal changes of metabolic activity were observed between PBMC-MCs and BALF-MCs in severe patients (Figure 4I). Similar results were obtained when analyzing total monocytes and total macrophages from paired PBMCs and BALF samples of patients with COVID-19 (Figure S7). These results suggest that monocyte-to-macrophage transition was embedded with distinct metabolic reprogramming patterns in patients with mild and severe COVID-19, respectively.
Figure 4.
Distinct metabolic reprogramming during monocyte to macrophage transition in mild versus severe COVID-19
(A) Metabolic pathway activities of CD14+ monocyte (PBMC-MCs) or CD14+ macrophage (BALF-MCs) from PBMCs or BALF of paired patients with mild (n = 2) and severe (n = 5) COVID-19. Statistically non-significant values (random permutation test p > 0.05) were shown as blank.
(B) Distribution of pathway activities in PBMC-MCs and BALF-MCs.
(C) Heatmap shows expression of 1,526 metabolic genes in PBMC-MCs and BALF-MCs from different groups.
(D and E) Trajectory (D) and inference (E) of pseudo time ordering of PBMC-MCs and paired BALF-MCs from mild patients visualized in the DDRTree-based reduced dimensional space.
(F) Pearson correlation plot of metabolic scores and pseudo time in PBMC-MCs and paired BALF-MCs from patients with mild COVID-19.
(G–I) Trajectory (G), inference of pseudo time ordering (H) and Pearson correlation analysis (I) of PBMC-MCs and paired BALF-MCs from patients with severe COVID-19.
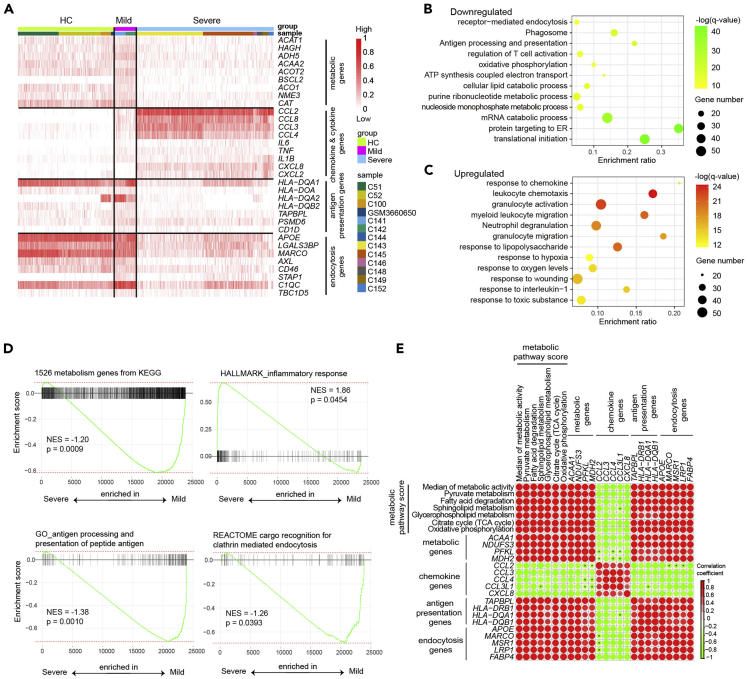
The functional impairment and hyperinflammatory phenotype of macrophages from patients with severe COVID-19 associated with metabolic dysfunction
We next determined if and how different metabolic remodeling of macrophages in mild and severe COVID-19 correlates with their functionality. We evaluated the expression of specific genes involves in metabolism and macrophage function, such as chemokines/cytokines production, antigen presentation, and endocytosis in each sample. Consistent with our above results, genes involved in most cellular metabolism processes were decreased in macrophages from patients with severe disease (Figure 5A). In comparison, genes encoding proinflammatory chemokines and cytokines such as CCL2, CCL3, CCL8, interleukin-6 (IL-6), and tumor necrosis factor-alpha (TNF-α) were significantly higher in bronchoalveolar macrophages from patients with severe disease than that from mild disease or HCs (Figure 5A), indicating a highly proinflammatory status. The result was consistent with our previous finding of elevated protein levels of proinflammatory chemokines and cytokines in BALF of patients with severe COVID-19 (Liao et al., 2020). Whereas downregulation of HLA-DR genes in macrophage from severe patients suggested the dysfunction of antigen presentation machinery (Figure 5A). Expression of MACRO (macrophage receptor with collagenous structure), AXL (AXL receptor tyrosine kinase), CD46, and other genes important for endocytosis was also downregulated in bronchoalveolar macrophage from severe patients (Figure 5A). Gene Ontology analysis also revealed that in addition to metabolic pathways, biological pathways associated with antigen processing and presentation, T cell co-stimulation, and receptor-mediated endocytosis enriched were also downregulated in BALF macrophages from severe patients compared to mild patients (Figure 5B). However, pathways related to leukocyte chemotaxis, myeloid leukocyte activation, were upregulated in bronchoalveolar macrophage from severe patients (Figure 5C). GSEA analysis revealed similar findings (Figure 5D). Similar results were obtained when analyzing another set of publicly available single-cell transcriptomes data from BALF of 2 mild and 20 severe COVID-19 cases (Wauters et al., 2021) (Figures S3D and S3E).
Figure 5.
Correlation analysis comparing metabolic activity and functionality of macrophages
(A) Heatmap shows expression of indicated genes in macrophage from BALF of patients with HC, mild, and severe COVID-19.
(B and C) Gene ontology analysis shows downregulated pathways (B) and upregulated pathways (C) in macrophages from severe versus mild COVID-19 patients.
(D) GSEA analysis of indicated gene sets in macrophages from patients with severe versus mild COVID-19.
(E) Correlation analysis comparing metabolic activity scores, or expression levels of metabolic genes with those genes related to antigen presentation, endocytosis, chemotaxis in macrophages from BALF of patients with COVID-19. Red color indicates a positive correlation, and green color indicates a negative correlation. A pvalue > 0.05 was indicated with black error within circle in the graph.
To navigate the relationship between metabolic pathways with immune response, we generated the Pearson correlation matrix using metabolic activity scores of specific pathways and expression values of genes for cytokines/chemokines, antigen presentation, and endocytosis. We found that the activity of major metabolic pathways, such as pyruvate metabolism, fatty acid degradation, TCA cycle and oxidative phosphorylation, and the expression levels of related key metabolic genes, was negatively correlated with chemokines/cytokines gene expression, but positively correlated with the expression of genes for antigen presentation and endocytosis functions (Figure 5E). These correlation analyses demonstrated the interconnected association of specific metabolism pathways with functionality of macrophages.
Amino acid supplementation or inhibition of glycolysis reduced proinflammatory cytokines production
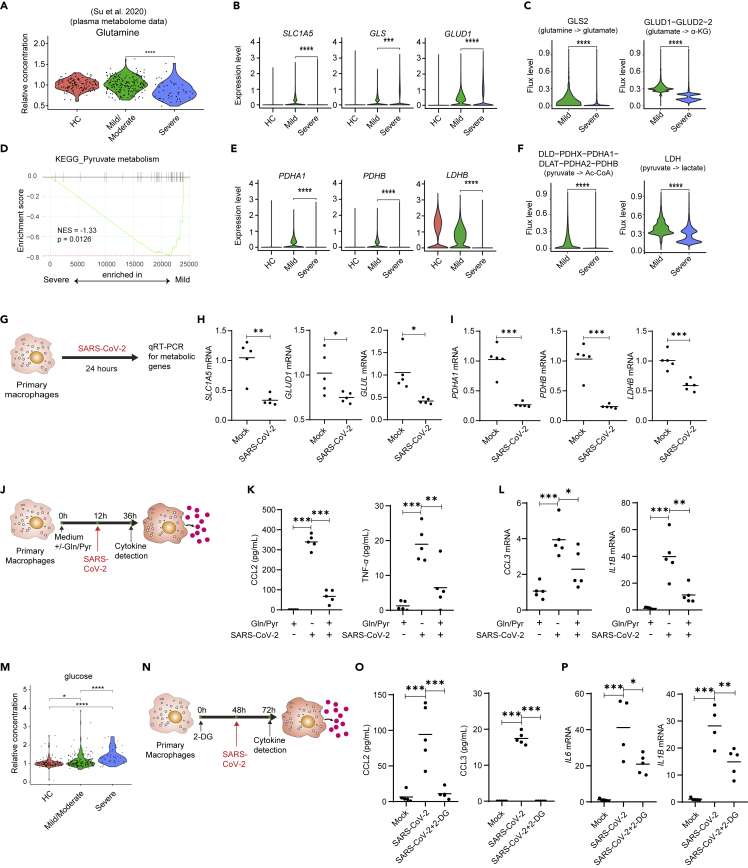
Our data above showed a unique metabolic reprogramming of macrophages from patients with severe COVID-19, as indicated by decrement of most metabolic processes such as glutamine and pyruvate metabolism but increment of glycolysis. We speculated that the unbalanced metabolic reprogramming may have contributed to dysfunction of macrophages in patients with severe COVID-19. Meta-data analysis also indicated that the levels of glutamine in plasma of patients was significantly reduced in patients with severe COVID-19 compared to HCs or patients with mild disease (Su et al., 2020) (Figure 6A). We next determine whether dysregulated glutamine and pyruvate metabolism lead to inflammatory macrophages. The expression levels of key enzymes involved in glutamine metabolism such as SCL1A5, GLS, and GLUD1 were lower in macrophages from patients with severe COVID-19 than that from patients with mild COVID-19 (Figure 6B). FBA also indicated that the flows of glutamine to glutamate, and glutamate to α-KG were reduced in macrophages from severe COVID-19 (Figure 6C). Furthermore, expression of enzymes associated with pyruvate metabolism and the levels of metabolite flux in pyruvate metabolism pathways were reduced in macrophages from BALF of patients with severe COVID-19 (Figures 6D–6F). Notably, the expression levels of key enzymes for glutamine and pyruvate metabolism were significantly reduced in human macrophages upon SARS-CoV-2 stimulation in vitro (Figures 6G–6I). We reasoned that environmental deficiency of nutrients such as glutamine and pyruvate may contribute to highly proinflammatory phenotype of BALF macrophages in patients with severe COVID-19. We thus cultured human primary macrophage in the absence or presence of glutamine and pyruvate, stimulated cells with SARS-CoV-2 (Figure 6J) or TLR7/8 agonist R848 (Figure S8A), and evaluated proinflammatory cytokines production. The results showed that glutamine and pyruvate deprivation led to high levels of proinflammatory cytokines and chemokines such as CCL2, TNF-α, IL-6, IL-1β, and CCL3, and supplementation with glutamine and pyruvate reduced inflammatory cytokines production (Figures 6K, 6L, and S8B).
Figure 6.
Glutamine and pyruvate supplementation reduces proinflammatory cytokines and chemokines production in macrophages
(A) Violin plot shows glutamine levels in plasma of HCs or patients with COVID-19. Each dot corresponds to an individual patient.
(B) Violin plots shows expression levels of SLC1A5, GLS, and GLUD1 in macrophages from BALF of patients with HCs, mild or COVID-19.
(C) Violin plots show relative levels of indicated metabolic fluxes in macrophages from patients with mild and severe COVID-19.
(D) GSEA analysis of pyruvate metabolism gene sets in macrophages from patients with severe versus mild COVID-19 .
(E) Expression levels of PDHA1(pyruvate dehydrogenase E1 subunit alpha 1), PDHB(pyruvate dehydrogenase E1 subunit beta), and LDHB (lactate dehydrogenase B) in macrophages from BALF of patients with HCs, mild, or severe COVID-19.
(F) Violin plots show relative levels of indicated metabolic fluxes in macrophages from patients with mild and severe COVID-19.
(G–I) Human primary monocyte-derived macrophages were stimulated with SARS-CoV-2 virus (MOI = 0.1) for 24 h (G). mRNA levels of SLC1A5, GLUD1, and GLUL (H) and PDHA1, PDHB, and LDHB (I) were detected by real-time PCR. Shown are representative data (mock, n = 5; SARS-CoV-2, n = 5) from 3 independent donors with mean values.
(J–L) Human primary macrophages cultured in complete medium or medium without glutamine and pyruvate were stimulated with SARS-CoV-2 (J). CCL2 and TFN-α levels in culture supernatant were detected by enzyme-linked immunosorbent assay(ELISA), and CCL3 and IL1B in cells were detected by real-time PCR 24 h after stimulation(K-L).
(M) Violin plot shows glucose levels in plasma of HCs or patients with COVID-19. Each dot corresponds to an individual patient.
(N–P) Human primary macrophages cultured in complete medium with 2-DG were stimulated with SARS-CoV-2 (N). CCL2 and CCL3 levels in culture supernatant were detected by ELISA, and IL6 and IL1B in cells were detected by real-time PCR 24 h after stimulation (O-P). Shown are representative data (n = 6 for each group) from 3 independent donors with mean values. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001. Unpaired, two-tailed Student’s ttest (for H and I) or one-way ANOVA and Bonferroni’s post hoc test (for K-L and O-P) were performed to compare between groups.
It was reported that diabetic individuals with high glucose levels are prone to develop the severe COVID-19 (Zhu et al., 2020). Meta-data analysis also indicated higher levels of glucose in plasma of patients with severe COVID-19 compared to HCs or patients with mild disease (Figure 6M). We cultured human primary macrophage in the presence of 2-Deoxy-D-glucose (2-DG) to inhibit glycolysis and stimulated the cells with SARS-CoV-2 or R848 (Figures 6N and S8C). The result showed that inhibition of glycolysis reduced CCL2, CCL3, IL-1β, TNF-α, and IL-6 production (Figures 6O, 6P and S8D), consistent with previous report that proinflammatory macrophages undergo metabolic reprogramming toward aerobic glycolysis (Galvan-Pena and O'Neill, 2014).
Reduced lipid metabolism and PPARγ repression in BALF macrophages from patients with severe COVID-19
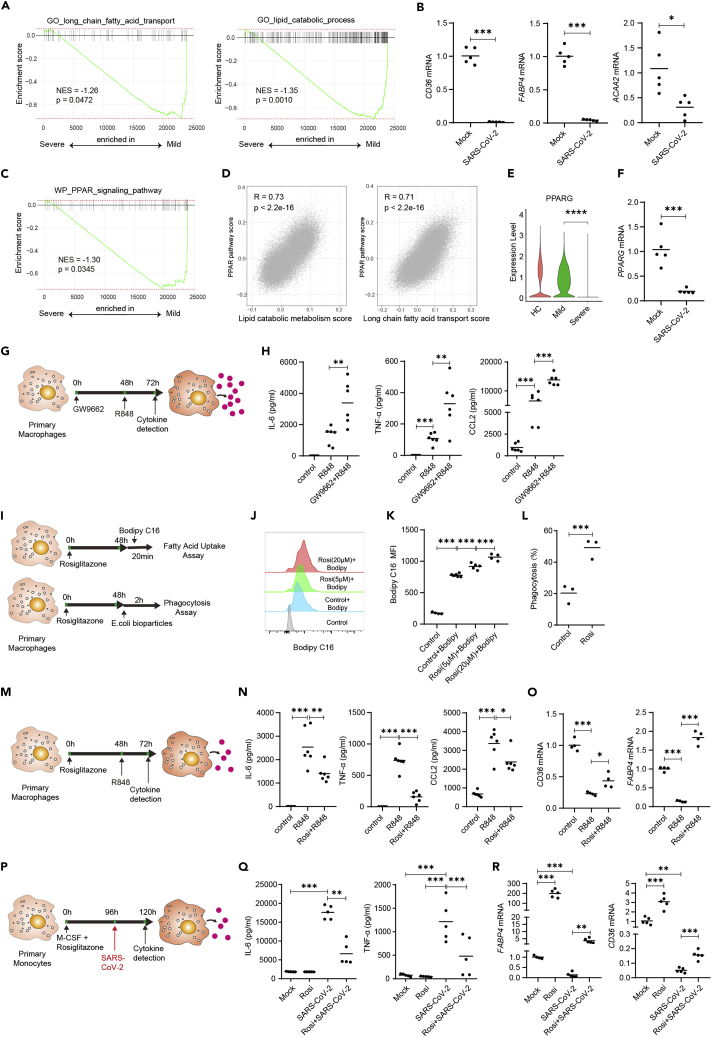
Our single-cell metabolism analysis indicated that lipid metabolism was downregulated in BALF macrophages from patients with severe COVID-19 (Figures 2 and 3). A sharp drop of lipid metabolites was also observed in patients with severe COVID-19 (Su et al., 2020). We also found that the metabolic pathway scores of several lipid metabolism pathways including fatty acid degradation, sphingolipid metabolism, and glycerophospholipid metabolism were negatively correlated with mRNA levels of proinflammatory cytokines in macrophages from BALF of patients with COVID-19 (Figure 5E). GSEA analysis also showed that the long-chain fatty acid transport and lipid catabolic process were inhibited in macrophages from BALF of patients with severe COVID-19 (Figure 7A). We stimulated human monocyte-derived macrophage with SARS-CoV-2 in vitro and observed downregulation of genes associated with lipid transport and metabolism (Hotamisligil and Bernlohr, 2015; Tontonoz et al., 1998) such as CD36, FABP4 (fatty acid binding protein 4), and ACAA2 (acetyl-CoA acyltransferase 2) (Figure 7B).
Figure 7.
The PPARγ agonist rosiglitazone reduces hyperinflammation in macrophages stimulated with SARS-CoV-2 or TLR7/8 agonist
(A) GSEA analysis of long-chain fatty acid transport and lipid metabolic process gene sets in macrophages from severe versus mild COVID-19 patients.
(B) Human primary monocyte-derived macrophages were stimulated with SARS-CoV-2 virus (MOI = 0.1) for 24 h. mRNA levels of CD36, FABP4, and ACAA2 were detected by real-time PCR.
(C) GSEA analysis of PPAR signaling pathway in macrophages from severe versus mild COVID-19 patients.
(D) Pearson correlation plot of metabolic scores versus PPAR pathway score in macrophages from BALF of HCs and patients with COVID-19.
(E) Expression levels of PPARG in macrophages from BALF of patients with HC, mild or COVID-19.
(F) Human primary monocyte-derived macrophages were stimulated with SARS-CoV-2 virus (MOI = 0.1) for 24 h. mRNA level of PPARG were detected by real-time PCR. Shown (B&F) are representative data (mock, n = 5; SARS-CoV-2, n = 5) from 3 independent donors with mean values.
(G and H) Human primary macrophages were cultured in the presence of PPARγ inhibitor GW9662 (2μM). At 48 h, the cells were stimulated with R848. IL-6, TNF-α, and CCL2 levels in culture supernatant 24 h after stimulation. Shown are representative data (mock, n = 6; R848, n = 6, Rosi + R848, n = 6) from 3 independent donors with mean values.
(I) Human primary macrophages were cultured in the presence of PPARγ activator rosiglitazone. At 48 h, the cells were incubated in medium containing 0.2μM BODIPY FL C16 for 20 min or in medium containing 0.1mg/mL pHrodo Green E. coli BioParticles Conjugate for 2 h at 37°C.
(J–L) Uptake of fatty acids (J-K) and E. coli bioparticles (L) were compared between groups using flow cytometry. Shown are representative data from 2 independent donors with mean values.
(M and N) Human primary macrophages were cultured in the presence of PPARγ activator rosiglitazone (5μM). At 48 h, the cells were stimulated with R848. IL-6, TNF-α, and CCL2 levels in culture supernatant 24 h after stimulation (N). Shown are representative data (mock, n = 6; R848, n = 6, Rosi + R848, n = 6) from 3 independent donors with mean values.
(O) Human primary macrophages were cultured as in K, CD36 and FABP4 mRNA levels in cells were detected by real-time PCR 24 h after stimulation.
(P–R) Human primary monocytes were differentiated to macrophages with M-CSF (colony stimulating factor 1) for 4 days in the presence of rosiglitazone. The differentiated cells were then stimulated with SARS-CoV-2 (MOI = 0.1) for 24 h (P). IL-6 and TNF-α levels in culture supernatant were detected by ELISA (Q). mRNA levels of CD36 and FABP4 in cells were detected by real-time PCR 24 h after stimulation (R). Shown are representative data (mock, n = 5; Rosi, n = 5, SARS-CoV-2, n = 5, Rosi + SARS-CoV-2, n = 5) from 3 independent donors with mean values. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001. Unpaired, two-tailed Student’s ttest (for B, F, and L) or one-way ANOVA and Bonferroni’s post hoc test (for H, K, N, O, Q, and R) were performed to compare between groups.
It was reported that the nuclear receptor peroxisome proliferator-activated receptor gamma (PPARγ) controls the expression of genes involved in lipid metabolism and maintains metabolic homeostasis (Ahmadian et al., 2013). PPARγ promoted the metabolic programming of anti-inflammatory macrophages, including fatty acids oxidation and mitochondrial biogenesis (Chawla et al., 2001; Kang et al., 2018), and improved their insulin resistance (Odegaard et al., 2007). GSEA analysis indicated that PPAR signaling pathway was inhibited in macrophages from patients with severe COVID-19 (Figure 7C). PPAR signaling pathway score was found positively correlated with scores of several lipid metabolism pathways in BALF macrophages at single-cell levels (Figure 7D). We examined the expression level of PPARγ gene and found that it was significantly suppressed in macrophages from BALF of severe COVID-19, as well as in monocyte-derived macrophage stimulated with SARS-CoV-2 in vitro (Figures 7E and 7F). To understand if PPARγ repression and reduced lipid metabolism contributed to dysregulated proinflammatory cytokine production, we stimulated human primary macrophages with TLR7/8 agonist resiquimod (R848) upon pharmacological inhibition of PPARγ (Figure 7G). The results showed that PPARγ repression by a small molecule GW9662 led to increased production of IL-6, TNF-α, and CCL2 (Figure 7H). Together, these results suggest that PPARγ repression may lead to reduced level of lipid metabolism, and it is closely associated with hyperinflammatory phenotype of macrophage in severe COVID-19.
The PPARγ agonist rosiglitazone enhances fatty acid uptake and phagocytosis capacity of macrophages, and reduces hyperinflammation in macrophages stimulated with SARS-CoV-2 or TLR7/8 agonist
We next determined whether activating the PPARγ signaling would enhance the function of macrophages and reduce inflammatory cytokines and chemokines production. Rosiglitazone, an FDA-approved antidiabetic drug (Campbell, 2005), was used to activate PPARγ pathway in human primary macrophage. We found that macrophages treated with rosiglitazone imported more free fatty acid, based on flow cytometry analysis of macrophages cultured with fluorescently conjugated lipid substrates (BODIPY FL C16) (Figures 7I–7K). We then detected the phagocytosis function of human primary macrophages after PPARγ signaling activation (Figure 7I). Interestingly, we found that around 50% of macrophages treated with rosiglitazone could take up Escherichiacoli bioparticles after 2 h of co-culture, while only 20% of macrophages were fluorescent positive in control group (Figure 7L). These results suggested that PPARγ agonist rosiglitazone enhanced fatty acid metabolism and phagocytosis capacity of human primary macrophages.
We next studied proinflammatory cytokines and chemokines production by macrophages after rosiglitazone treatment and toll-like receptor signaling stimulation (Figure 7M). The results showed that PPARγ activation reduced the production of proinflammatory cytokines IL-6, TNF-α, and CCL2 by macrophages stimulated with R848 (Figure 7N), and increased expression of CD36 and FABP4 (Figure 7O), two key regulators in lipid metabolism. The results suggested that lipid metabolism enhanced by rosiglitazone may contribute to the reduced inflammatory cytokines and chemokines production.
Finally, we determined whether rosiglitazone treatment could reduce inflammatory cytokines production by macrophages upon SARS-CoV-2 virus stimulation in vitro (Figure 7P). SARS-CoV-2 induced production of IL-6 and TNF-α in macrophages (Figure 7Q). Impressively, rosiglitazone inoculation significantly reduced proinflammatory cytokines production (Figure 7Q) and enhanced metabolic genes expression (Figure 7R). Taken together, our results indicated that PPARγ might serve as a target to reduce cytokine storm in patients with severe COVID-19.
Discussion
In various infectious diseases, cellular metabolism is intimately connected to the mechanisms of disease pathogenesis and the resulting pathology, as well as the host defense response. Although several recent clinical studies have suggested an association between plasma metabolites alteration and COVID-19 severity (Lam et al., 2021; Shen et al., 2020; Song et al., 2020; Su et al., 2020; Xiao et al., 2021), however, the immunometabolic regulation of leukocytes in lungs and its correlation with immune function and disease severity are currently unclear. In the present study, we depicted the immunometabolic landscape in lungs of patients with COVID-19. Our data suggested that improper metabolic reprogramming of macrophages exacerbated the imbalanced immune response such as hyperinflammatory and reduced endocytosis function. Importantly, we found that modulating lipid metabolism by PPARγ agonist rosiglitazone reduced hyperinflammation in macrophages stimulated with SARS-CoV-2. These findings shed light on treating COVID-19 by immunometabolic modulation.
Metabolism is a key regulator of immune cell phenotype and function, but it remains difficult to investigate the metabolic status of individual cells in humans. Gene expression has been used as one of the indirect means to evaluate metabolic activity (Lee et al., 2012). Recent developed computational algorithms allow for the detection of metabolic pathway activity and metabolic flux based on metabolic gene expression using single-cell transcriptomic approach (Damiani et al., 2019; Xiao et al., 2019). Here, we took advantage of the integrated single-cell metabolic profiling platforms and depicted a global picture of immunometabolic profile in lungs of patients with COVID-19 with different disease severity. Our data suggested that SARS-CoV-2 infection increased glycolysis but decreased most other core metabolic processes in BALF macrophages from severe COVID-19, which correlates with elevated inflammation and reduced antigen presentation and endocytosis functions. Our study thus established the foundation to study metabolic state and functional phenotype of individual immune cells in clinical samples from COVID-19 or other diseases.
Previous studies have reported that SARS-CoV-2 infection can induce glycolytic reprogramming of isolated monocytes in vitro (Bhatt et al., 2022; Codo et al., 2020; Cory et al., 2021). Our research uncovered dramatic metabolic shift in BALF macrophages from patients with mild to severe COVID-19. However, analysis using paired PBMCs indicated minor changes of metabolic activities of peripheral blood monocytes between patients with mild and severe COVID-19. This may be caused by the lack of direct exposure to SARS-CoV-2 virus of the circulating monocytes in vivo. In lung tissue, where direct exposure of macrophages to SARS-CoV-2 occurs, we indeed found the increased level of glycolysis pathways from patients with both mild and severe COVID-19. Interestingly, the citrate cycle, oxidative phosphorylation, and fatty acid degradation were inhibited in macrophages from patients with severe COVID-19s compared to macrophages from patients with mild diseases. The much higher levels of SARS-CoV-2 particles in lungs of severe patients may lead to the glycolytic reprogramming but inhibition of mitochondrial oxidative metabolism of macrophages. This was consistent with reports showing that SARS-CoV-2 infection suppresses Oxidative phosphorylation (OXPHOS) and alters mitochondrial function in monocytes and cell lines in vitro (Codo et al., 2020; Cortese et al., 2020; Wang et al., 2020).These results highlight the importance to study the metabolic and immunological events in SARS-CoV-2-infected lungs to further understand the mechanisms of immune dysfunction and immunopathogenesis.
Based on our single-cell metabolism study, we validated in vitro that glutamine and pyruvate supplementation or glycolysis inhibition by 2-DG reduced the levels of proinflammatory cytokines and chemokines in human primary macrophages stimulated with SARS-CoV-2 or TLR7/8 agonist. These results were consistent with the findings that proinflammatory macrophages undergo metabolic reprogramming toward aerobic glycolysis (Galvan-Pena and O'Neill, 2014). We used 2-DG as a model to inhibit glycolysis. High dose of 2-DG (10mM) was reported to suppress both glycolysis and mitochondrial metabolism, while low dose of 2-DG (<1.25mM) slightly increased oxidative phosphorylation in bone-marrow-derived macrophages (Wang et al., 2018), indicating the dose-dependent effect of 2-DG. We found here that low dose of 2-DG (1mM) can significantly suppress proinflammatory cytokines production by macrophages stimulated with either SARS-CoV-2 or R848. Consistently, 5mM of 2-DG treatment reduced SARS-CoV-2-induced proinflammatory cytokines by monocytes in vitro (Codo et al., 2020). Amino acid starvation also resulted in defective anti-inflammatory polarization in macrophages (Kimura et al., 2016). Considering that hospitalized patients with COVID-19 presented malnutrition (Clemente-Suarez et al., 2021) and patients with severe COVID-19 exhibited a sharp drop in blood nutrients such as amino acids (Su et al., 2020), we proposed that amino acid supplementation in patients with severe COVID-19 might alleviate inflammatory cytokines production by macrophages.
Lipid metabolism plays a critical role in the differentiation and function of macrophage (Chawla et al., 2001). It was reported that anti-inflammatory macrophages trigger a metabolic program of increased oxidative phosphorylation and fatty acid oxidation (Galvan-Pena and O'Neill, 2014). Our single-cell metabolism analysis showed reduced lipid metabolism in macrophages from bronchoalveolar of patients with severe COVID-19. The nuclear receptor PPARγ is known to regulate lipid metabolism in many tissues, and itis the target of antidiabetic thiazolidinedione drugs (Ahmadian et al., 2013). In macrophages, PPARγ has been shown to play important roles in inflammation and metabolism (Hevener et al., 2007; Odegaard et al., 2007). PPARγ signaling controls lipid uptake (Chawla et al., 2001; Odegaard et al., 2007) and intracellular metabolism (Odegaard et al., 2007). Impairment in PPARγ expression and lipid metabolism have been shown to be associated with blocked anti-inflammatory macrophage polarization (Kang et al., 2018). We found that the expression of PPARγ was inhibited in macrophages from patients with severe COVID-19 as reported (Desterke et al., 2020), or in human primary macrophages stimulated with SARS-CoV-2 in vitro. Most importantly, we proved that activating of PPARγ signaling in vitro by rosiglitazone, an antidiabetic drug, enhanced lipid metabolism and phagocytosis functions of macrophages, and reduced proinflammatory cytokines and chemokines production in human primary macrophages stimulated with SARS-CoV-2 or TLR7/8 agonist. The results indicated that rosiglitazone may reduce hyperinflammation in macrophages by regulating lipid metabolism. PPARγ was important for alveolar macrophages development and function (Schneider et al., 2014). It was reported that PPARγ in alveolar macrophages limits inflammation and promotes tissue recovery after respiratory viral infection (Huang et al., 2019). Depletion and altered antigen presentation of alveolar macrophages was also reported to correlate with the severe COVID-19 (Chen et al., 2022). It will be interesting to explore the role of PPARγ in alveolar macrophages function in the context of COVID-19 in the future.
Patients with insulin resistance usually have enhanced inflammation, such as increased levels of IL-6 and TNF-α (Esser et al., 2014), and other symptoms that reduced their ability to respond properly to the infection with SARS-CoV-2. Most recently, a large COVID-19-related population study showed that patients with type 2 diabetes prescribed with glucose-lowering drugs including metformin, sodium-dependent glucose transporters 2 inhibitors, and sulfonylureas had a lower risk of COVID-19-related mortality (Khunti et al., 2021). Although the study indicated that pre-infection prescription for thiazolidinediones had minor effect on reducing the risk of COVID-19-related mortality in people with type 2 diabetes, it will still be interesting to explore the anti-inflammatory properties of rosiglitazone in patients with severe COVID-19 with or without diabetes. 2-DG which targeting glycolysis has also been approved for phase II and phase III clinical trials in India (Huang et al., 2022). Other report suggested that targeting metabolism by arginine, epacadostat (a potent and selective indoleamine 2,3-dioxygenase inhibitor), and mycophenolic acid (an inosine monophosphate dehydrogenase inhibitor) reduced cytokine release in PBMCs of infected monkeys in vitro (Xiao et al., 2021). Taken together, our results and published data further support drug repurposing for targeting immunometabolism for COVID-19 treatment.
In summary, we characterized the immunometabolic landscape in lungs of patients with COVID-19 and suggested that regulating metabolism by 2-DG and rosiglitazone might reduce cytokine release syndrome in patients with severe COVID-19. Our study has identified immunometabolism dysregulation as a mechanism for COVID-19 pathogenesis and further facilitated the development of novel therapeutic for COVID-19.
Limitations of the study
We acknowledge limitations of our studies. The computational algorithms to quantify metabolic activities and fluxes used in the study were based on single-cell transcriptomes. It does not consider post-transcriptional regulation and post-translational modulation, which could be important for metabolic enzymes activity. While similar approaches have been used recently to study metabolic modeling of single cells and provided insights into targeting metabolic pathways for therapeutic interventions (Rohlenova et al., 2020; Wagner et al., 2021). We validated our major conclusion in vitro by showing that remodeling metabolism by PPARγ inhibition induced hyperinflammatory macrophages, while amino acid supplementation, glycolysis inhibition, or PPARγ stimulation reduced inflammation. In the current study, we explored the roles of PPARγ in the inflammatory potential and phagocytic capacity of macrophages by applying PPARγ-agonist rosiglitazone and antagonist GW9662. Since these chemical compounds can modify the ligand-dependent activity of PPARγ in differentiating macrophages indirectly, it will be interesting to further confirm our findings by using PPARγ genetically knockout cells. In addition, since SARS-CoV-2 transcripts were only detectable in a very small percentage of BALF-derived macrophages (728 out of 30,137 cells, 2.4%) from patients with severe COVID-19 (Bost et al., 2020), and it remains difficult to discriminate those macrophages with SARS-CoV-2 transcripts were real viral infected or that have uptake the viral RNA through phagocytosis of other virus-infected cells (Bost et al., 2020), we did not add viral biomass objective function (Nanda and Ghosh, 2021; Renz et al., 2020) of SARS-CoV-2 virus in the model to account for viral metabolic requirement from the host. To preclude the potential impact of patients’ heterogeneity and medication treatment on cellular metabolism, we analyzed the metabolic pathway activities of macrophages from BALF of individual patient and included key clinical data of each patient (Figures S2A and S2B). Our results showed no strong correlation between metabolic activity with factors such as gender or interferon atomization, ribavirin, and methylprednisolone treatment (Figures S2A and S2B). Furthermore, due to the ethical consideration and limited accessibility to samples from patients with COVID-19, especially cells from BALF of lungs, we can only use human macrophages from healthy donors to prove that targeting PPARγ with rosiglitazone reduces hyperinflammation upon SARS-CoV-2 or TLR7/8 agonist stimulation in vitro. It will be promising to test the therapeutic effect of rosiglitazone to improve the COVID-19 symptoms in patients with severe SARS-CoV-2, especially those with type 2 diabetes.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Bacterial and virus strains | ||
| SARS-CoV-2 strain SZ02 | Shenzhen Third People’s Hospital | N/A |
| Biological samples | ||
| Human spleen | Xuzhou Mine Hospital | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| Resiquimod (R848) | Sigma Aldrich | Cat# SML0196-10MG |
| GW9662 | MedChemExpress | Cat# HY-16578 |
| Rosiglitazone | MedChemExpress | Cat# HY-17386 |
| Recombinant Human M-CSF | Novoprotein | Cat# C417 |
| Glutamine | Gibco | Cat# 25030081 |
| Sodium pyruvate | Agilent | Cat# 103578-100 |
| 2-Deoxy-D-glucose (2-DG) | Sigma | Cat# D8375 |
| Critical commercial assays | ||
| Human TNF-α ELISA Kit | Mabtech | Cat# 3512-1H-6 |
| Human IL-6 ELISA Kit | Mabtech | Cat# 3460-1H-6 |
| Human MIP-1alpha (CCL3) ELISA Kit | Peprotech | Cat# 900-K35 |
| ELISA MAX Deluxe Set Human IL-1β | Biolegend | Cat# 437004 |
| ELISA MAX Deluxe Set Human MCP-1/CCL2 | Biolegend | Cat# 438804 |
| BODIPY™ FL C16 (4,4-Difluoro-5,7-Dimethyl-4-Bora-3a,4a-Diaza-s-Indacene-3-Hexadecanoic Acid) | Invitrogen | Cat# D3821 |
| pHrodo™ Green E. coli BioParticles™ Conjugate for Phagocytosis | Invitrogen | Cat# P35366 |
| SYBR qPCR Mix | Yeasen | Cat# 11198ES08 |
| RNAiso Plus | TaKaRa | Cat# 9109 |
| HiScript III RT SuperMix for qPCR | Vazyme | Cat# R222-01 |
| Oligonucleotides | ||
| qPCR primers, see Table S6 | This article | N/A |
| Software and algorithms | ||
| GraphPad Prism 8.0 | GraphPad | https://www.graphpad.com/ |
| FlowJo 10.0 | Treestar | https://www.flowjo.com/solutions/flowjo |
| R-4.0.3 | Team, 2008 | https://www.r-project.org/ |
| Seurat-3.2.2 | (Stuart et al., 2019) | https://satijalab.org/seurat/ |
| ScImpute | (Li and Li, 2018) | https://github.com/Vivianstats/scImpute |
| Pathway Activity | (Xiao et al., 2019) | https://github.com/LocasaleLab/Single-Cell-Metabolic-Landscape |
| Metascape | (Zhou et al., 2019) | https://metascape.org |
| Fgsea-1.16.0 | (Korotkevich et al., 2021) | https://github.com/ctlab/fgsea |
| Monocle-2.18.0 | (Trapnell et al., 2014) | http://cole-trapnell-lab.github.io/monocle-release/ |
| Single-cell Flux Balance Analysis | (Damiani et al., 2019) | https://github.com/BIMIB-DISCo/scFBA |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Liang Cheng (liangcheng@whu.edu.cn).
Materials availability
This study did not generate new unique reagents.
Experimental model and subject details
Isolation of human spleen cells
We obtained human spleen tissues from donors with ruptured spleen caused by trauma, with informed consent from the donors. Studies were approved by the Human Research Ethics Committee of Xuzhou Mine Hospital with the protocol #XZKSYY2021-002.
Human splenocytes were isolated from spleen tissue as reported (Mittag et al., 2011). Briefly, spleen tissue was disrupted with ophthalmic scissors and then digested with collagenase (2 mg/mL; Worthington Biochemical, Lakewood, NJ, USA) in the presence of DNase (0.1 mg/mL; Roche, Mannheim, Germany) at 5g tissue/20mL RPMI (Roswell Park Memorial Institute) 1640 medium (Invitrogen; Carlsbad, CA, USA) for 20 min at room temperature, followed by addition of 5mM EDTA (Sigma-Aldrich, St. Louis, MO, USA) for 5 min at room temperature. Undigested fragments were removed by filtering through a 70-mm sieve. After washing in PBS with 1% FBS and 1mM EDTA, spleen mononuclear cells were purified by Ficoll-Paque (GE Healthcare, Uppsala, Sweden) density centrifugation.
Method details
ScRNA-seq data availability and data processing
ScRNA-seq data from BALF of COVID-19 patients used in all studies except in Figure S3 can be accessed in Gene Expression Omnibus under the accession number GSE145926(Liao et al., 2020) and GSM3660650 (Morse et al., 2019). Data from paired PBMCs can be acquired in the Genome Sequence Archive (GSA) under accession number HRA000297(Xu et al., 2020). For data from BALF, we removed cells with gene number less than 200 or large than 6,000. Cells with unique molecular identifier (UMI) count less than 1,000 or mitochondrial gene percentage more than 10% (15% for PBMCs) were also removed in the study. All samples were integrated with Seurat (version 3.2.2) to avoid the influence of batch effects. We utilized first 50 dimensions of canonical correlation analysis (CCA) and PCA for further analysis.
ScRNA-seq data from BALF of another cohort of COVID-19 patients analyzed in Figure S3 can be accessed in the EGA European Genome-Phenome Archive database under the accession number EGAS00001004717 (Wauters et al., 2021). We removed cells with gene number less than 151 and large than 6,000, UMI count less than 301, and mitochondrial gene percentage more than 20% as reported (Wauters et al., 2021).
In Figure 4, integrated analysis of CD14+ monocytes and macrophages from paired BALF and PBMCs samples was performed by extracting CD14+ monocytes and macrophages and using CCA to integrated. In Figure S7, integrated analysis of total monocytes or macrophages from paired PBMCs and BALF samples was performed by extracting all monocytes and macrophages and using CCA to integrate.
Cell clustering and annotation
Cell annotation was completed as reported previously (Liao et al., 2020; Wauters et al., 2021; Xu et al., 2020). The large filtered matrix was normalized using “LogNormalize” methods in Seurat (version 3.2.2) with scale.factor = 10000 and “Percent.mt” was regressed out in the scaling step. Then, the top 2,000 variable genes were found using the “vst” method in Seurat (FindVariableFeatures function) to conduct PCA. When analyzing macrophages from BALF, we removed the doublets that were CD68+CD3D+ and CD68+IGH4+ as reported (Liao et al., 2020). To cluster cells based on a given gene list, we extracted the count matrix and annotation information to create a new cell_data_set object using monocle3 R package (version 0.2.3.0) (Cao et al., 2019). A list of metabolic genes that was obtained from the KEGG database (http://www.kegg.jp) were utilized to conduct dimensionality reduction of the whole cell population. In the meantime, a “mutual nearest neighbor” algorithm (Haghverdi et al., 2018) was used to execute alignment and subtract the effects of treatments.
Data imputation and normalization
To enable comparison of metabolic pathways activities between different cell subsets, we performed data imputation and normalization to impute missing gene expression values and minimize differences in distributions of relative gene expression levels between cell subsets and patient groups. Briefly, the scImpute algorithm first estimates the dropout probability of each gene in each cell, and it only imputes the expression of a gene if its dropout probability is higher than a threshold, which is a default value 50%, in a cell (Li and Li, 2018). In addition, scImpute predicts the expression of a dropout in a cell based on the expression of the same gene in other similar cells. Therefore, the imputed value is non-zero only when the gene is expressed in similar cells.
For normalization, the deconvolution method was selected (Lun et al., 2016). The scran R package (1.18.5) was first utilized to compute the size factor of each cell type based on the original annotation of cell type; Normalized counts value of genes with dropout rate <0.75 were then computed by dividing counts by cell type-specific size factor.
Calculation of metabolic pathway activity
To identify metabolic pathway variation among different cell subsets from different groups, we used the imputed and normalized data and utilized a computation pipeline to evaluate the pathway activity score as reported before (Xiao et al., 2019). Briefly, we first calculated the mean expression level of all metabolic genes across each cell type. Second, these absolute expression values were transferred to the relative expression level based on all cell types. We further defined the metabolic pathway activity score as the weighted average of all genes included in the corresponding pathway. Finally, we excluded the outliers (the relative expression level of genes greater than three times 75th percentile/below one-third times 25th percentile) and conducted random permutation test to select those significantly higher or lower pathway activity scores than average.
Differential gene expression analysis
Wilcoxon in Seurat (version 3.2.2) (FindAllMarkers function) was utilized to conduct differential gene expression analysis. Logfc.threshold was set to be equal to 0.25, min.pct equal to 0.1, and adjusted p-value less than 0.05. These DEGs were then used to perform enrichment analysis with clusterProfiler R package (version 3.18.1) and Metascape platform (https://metascape.org) (Zhou et al., 2019). GSEA was further used (nperm = 1000) with fgsea R package (version 1.16.0) (Korotkevich et al., 2021), which is available at Bioconductor (http://bioconductor.org/packages/fgsea) or on GitHub (https://github.com/ctlab/fgsea). The gene lists performed to GSEA were generated by presto R package (version 1.0.0) (wilcoxauc function) which is available on GitHub (https://github.com/immunogenomics/presto); Lists of genes in the metabolic pathways were obtained from the KEGG database (http://www.kegg.jp). Lists of genes in the inflammatory response, antigen processing and presentation, endocytosis, PPAR signaling pathway, long-chain fatty acid transport, lipid catabolic process, and adipogenesis pathways (Table S5) were obtained from the MSigDB database (http://www.gsea-msigdb.org/gsea/msigdb/index.jsp).
Correlation analysis
Pearson’s correlation analysis using corrplot R package (version 0.84) (https://github.com/taiyun/corrplot) was used to compute correlation coefficient and p-value.
Metabolic, inflammatory, antigen processing and presentation, endocytosis, lipid catabolic process, long-chain fatty acid transport and PPAR signaling score calculation
To define the scores of certain signaling pathways, we used a list of genes termed “GOBP_LIPID_CATABOLIC_PROCESS”, “GO_LONG_CHAIN_FATTY_ACID_TRANSPORT”, “WP_PPAR_SIGNALING_PATHWAY” from MsigDB (Bunis et al., 2020). These scores were then evaluated by using Seurat (AddModuleScore function) with default parameters. Then the scores of certain signaling pathways in single cells were utilized to perform correlation analysis.
Trajectory analysis
Monocle R package (2.18.0) was used for trajectory analysis. Briefly, we selected genes with high dispersion (adjusted p-value less than 0.05) across cells using Monocle R package (2.18.0) (dispersionTable function) to order cells, and then reduced dimension (method was set to DDRTree) and plotted trajectory with default parameters.
Gene expression visualization
The heatmaps, violin plots and bar plots of specific gene expression and correlation plots of metadata were created using pheatmap (version 1.0.12) (https://cran.r-project.org/web/packages/pheatmap) and dittoSeq R package (version 1.2.4) (Bunis et al., 2020). The feature plots of genes were generated using monocle3 R package (Cao et al., 2019). Data was auto-scaled with default parameters when using these packages.
Single-cell flux balance analysis
To obtain single-cell fluxomes of macrophages from different groups, an established computational method, namely scFBA was utilized (Damiani et al., 2019). The detailed usage of scFBA can be found on GitHub (https://github.com/BIMIB-DISCo/scFBA). Briefly, scFBA used bulk data integration approaches to set constraints on the fluxes of the individual networks, without creating context-specific models from generic ones (Jensen and Papin, 2011; Lee et al., 2012). The expression data was utilized to identify a flux distribution that maximizes the flux through highly expressed reactions, while minimizing the flux through poorly expressed reactions (Shlomi et al., 2008). As to the choice of the nutrients exchanged with biofluids, scFBA considered the main exogenous nutrients (which the overall population can uptake from 0 up to Npop⋅100 nmol/h) could be the main nutrients of cells according to the literature [1]. The imputed data of macrophages from two HCs (C100 and GSM3660650), there mild patients (C141, C142, and C144), and three severe patients (C146, C148, and C149) were used as the input of scFBA, and we chose the default objective function, the biomass exchange (Ex_biomass), which is directly related with the biomass production (biomass_synthesis). Then, a multi-scale stoichiometric model was built to estimate the cell-wise metabolic flux of macrophages based on a genome-wide metabolic network, a modified metabolic core model HMRcore, which was also available on GitHub (https://github.com/BIMIB-DISCo/scFBA), together with the scFBA suite of MATLAB functions. The flux values of each reaction were remapped in the interval [0, 1] as a way of flux rescaling. The single-cell flux matrix and normalized matrix were then transferred to seuratobject and cell_data_set object using Seurat and monocle3 R packages for further visualization. All fluxes were utilized to cluster cells and the “mutual nearest neighbor” algorithm was used to execute alignment.
Plasma Metabolome data acquisition and processing
Plasma Metabolome data from HCs and COVID-19 patients used in this study (Su et al., 2020) can be accessed on the website (https://data.mendeley.com/datasets/tzydswhhb5/5). The data table that contained metabolites info was transferred to a seuratobject using Seurat for further visualization.
Primary macrophage culture and stimulation by R848 or SARS-CoV-2
Primary human macrophages were obtained by plating fresh or frozen splenocytes obtained from donors with ruptured spleen caused by trauma with RPMI 1640 medium with 50 ng/mL M-CSF (Novoprotein, Suzhou, Jiangsu, China) and removing unattached cells after 12 hours of culturing. The attached macrophages were then cultured in complete RPMI-1640 medium or RPMI-1640 medium without glutamine and pyruvate (Seahorse Bioscience, Santa Clara, California, USA) for 12 h before simulation with R848 (Sigma, St. Louis, Missouri, USA) or SARS-CoV-2 (MOI = 1). In other experiments, macrophages were treated with 2-DG (1mM; Sigma, St. Louis, Missouri, USA), PPARγ inhibitor GW9662 (2μM; MedChemExpress, Monmouth Junction, NJ, USA), or PPARγ activator rosiglitazone (5μM; MedChemExpress, Monmouth Junction, NJ, USA) for 48 h before stimulation with R848 or SARS-CoV-2 as indicated (MOI = 1). After 24 h of stimulation, supernatants were collected for the measurement of IL-6, TNF-α, CCL2, CCL3 by ELISA and cells were harvested for the detection of IL1B, CD36, and FABP4 mRNA by real-time PCR.
Fatty acid uptake and phagocytosis assay
For measuring uptake of fatty acids, macrophages were incubated in medium containing 0.2μM BODIPY FL C16 (ThermoFisher, D3821) for 20minat 37°C. The cells were then washed and stained with LIVE/DEAD® Fixable Yellow Cell Stain Kit (ThermoFisher) for 10minat room temperature. For phagocytosis assay, macrophages were incubated in medium containing 0.1 mg/mL pHrodo™ Green E. coli BioParticles™ Conjugate (ThermoFisher, P35366) for 2 h at 37°C. The cells were then washed and stained with LIVE/DEAD® Fixable Yellow Cell Stain Kit (ThermoFisher) for 10minat room temperature.
Samples were collected on LSR-II flow cytometer (BD Biosciences) and data were analyzed with FlowJo V10 (TreeStar).
Preparation of SARS-CoV-2 virus
All studies involving SARS-CoV-2 infection were conducted in the biosafety level-3 laboratory of Shenzhen Third People’s Hospital. SARS-CoV-2 isolate SZ02 was sourced from COVID-19 patients. SARS-CoV-2 was amplified in Vero-E6 cells and viral stocks were stored at −80°C.
For detection of viral titer, Vero-E6 cells were plated in 96-well plates at 20,000 cells per well in DMEM containing 5% heat-inactivated fetal bovine serum, penicillin/streptomycin, L-glutamine, and 15mM HEPES (Life Technologies). The cells were incubated overnight in a 5% CO2 environment at 37°C, washed once with PBS and then cultured in serum-free DMEM containing penicillin/streptomycin, L-glutamine, 15mM HEPES, and 1 μg/mL TPCK-treated trypsin. A 10-fold initial dilution of samples with one freeze–thaw cycle was made in octuplicate wells of the 96-well plates followed by 6 serial 10-fold dilutions. After a 4-day incubation, the plates were observed for the presence of cytopathogenic effect using an inverted optical microscope. The endpoint titers were calculated by means of a simplified Reed & Muench method.
Stimulation of human monocyte-derived macrophage by SARS-CoV-2
PBMCs were isolated by density gradient centrifugation with Ficoll-Paque Plus (GE, Chicago, Illinois, USA) of whole blood from blood donors. Monocytes were obtained by plating PBMCs with RPMI 1640 medium with 50 ng/mL M-CSF and then unattached cells were removed after 12 h of culturing. Monocyte-derived macrophage was obtained by culturing the attached monocytes for a period of 4 days in RPMI 1640 medium supplemented with 10% FBS, 50 ng/mL M-CSF in the presence or absence of 5μM PPARγ agonist Rosiglitazone (MedChemExpress, Monmouth Junction, NJ, USA) or 2μM PPARγ inhibitor GW9662 (MedChemExpress, Monmouth Junction, NJ, USA). The cells were then stimulated with SARS-CoV-2 virus with MOI = 0.1 for 24 h. Supernatants were collected for the measurement of IL-6, TNF-α, and CCL2 by ELISA, and cells were harvested for the detection of CD36 and FABP4 mRNA by real-time PCR.
RNA isolation and real-time PCR
Total RNA was isolated using RNAiso Plus (TakaRa, Kyoto, Japan) according to manufacturer’s instructions. real-time PCR was performed with the Hieff® qPCR SYBR Green Master Mix (YEASEN, Shanghai, China) on a CFX96 system (BIO RAD, CA, USA). The expression of target mRNAs was normalized with Gapdh or β-actin. The fold changes were calculated using 2−ΔΔCT method. The sequences of the primers are enlisted in Table S6.
Enzyme-linked immunosorbent assay
For the ELISA bioassay, all ELISA assays were performed using commercialized kit according to the manufacturer’s instruction. The absorbance was calculated based on a standard curve after subtracting the background.
Quantification and statistical analysis
Statistics
In sections of differential gene expression analysis, correlation analysis and of pathway activity calculation, p-value was calculated with default methods of corresponding R packages. The Wilcoxon test was used to compare metabolite levels in the plasma of COVID-19 patients in Figure 6A. Difference of specific gene expression levels in macrophages between mild versus severe COVID-19 patients was compared using ggpubr R package (version 0.4.0) (https://CRAN.R-project.org/package=ggpubr).
In all the in vitro experiments, significance levels of data were determined by using Prism8 (GraphPad Software). Experiments were analyzed by two-tailed Student’s t-test, or by one-way ANOVA and Bonferroni’s post hoc test according to the assumptions of the test, as indicated for each experiment. A p-value less than 0.05 was considered significant. The number of replicates was specified in the legend of each figure.
Acknowledgments
We thank Dr. Bo Zhong (Wuhan University) and Dr. Hudan Liu (Wuhan University) for reagents. Dr. Ziwei Dai (Southern University of Science and Technology) for suggestions on data analysis. Ms. Lian Peng for technic support. We thank the Supercomputing Center of Wuhan University for supporting on bioinformatic analysis. This study was supported by a startup fund from Wuhan University (to Liang Cheng and Li Wang), the National Science Fund for Distinguished Young Scholars (82025022 to Zheng Zhang), the National Natural Science Foundation of China (82071784 to Liang Cheng), the Non-Profit Central Research Institute Fund of Chinese Academy of Medical Sciences (2020-PT320-004 to Liang Cheng), the Key Research & Development Project of Hubei Province (2020BCA069 to Liang Cheng and Conghua Xie), the Translational Medicine and Interdisciplinary Research Joint Fund of Zhongnan Hospital of Wuhan University (ZNJC202007 to Liang Cheng and Yan Gong), and the Funding of Shenzhen Science and Technology Innovation Committee (KQTD20200909113758004 to Zheng Zhang).
Author contributions
Q.Z., L.W., L.S., Z.Z., and L.C. conceived and designed study, collected and analyzed data, and wrote the article. Q.Z., H.Y., J.C., and Y.L. performed the bioinformatic analysis. Z.Y., S.Z., X-R.S., Y.X., M.L., and F.L. performed the experiments and analyzed the data. L.Y., Q.Zhang, M.H., Y.G., C.X., and P.Z. contributed materials. L.W. and L.C. supervised the study. All authors have approved the final version of this article.
Declaration of interests
The authors declare no competing interests.
Published: November 18, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2022.105319.
Contributor Information
Li Wang, Email: liwang2020@whu.edu.cn.
Lishan Su, Email: isu@ihv.umaryland.edu.
Zheng Zhang, Email: zhangzheng1975@aliyun.com.
Liang Cheng, Email: liangcheng@whu.edu.cn.
Supplemental information
Data and code availability
Data reported in this article will be shared by the lead contact upon request. This article does not report novel RNA-seq data and original code. Any additional information required to reanalyze the data reported in this article is available from the lead contact upon request.
References
- Ahmadian M., Suh J.M., Hah N., Liddle C., Atkins A.R., Downes M., Evans R.M. PPARgamma signaling and metabolism: the good, the bad and the future. Nat. Med. 2013;19:557–566. doi: 10.1038/nm.3159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albina J.E., Mills C.D., Henry W.L., Jr., Caldwell M.D. Temporal expression of different pathways of 1-arginine metabolism in healing wounds. J. Immunol. 1990;144:3877–3880. [PubMed] [Google Scholar]
- Arunachalam P.S., Wimmers F., Mok C.K.P., Perera R.A.P.M., Scott M., Hagan T., Sigal N., Feng Y., Bristow L., Tak-Yin Tsang O., et al. Systems biological assessment of immunity to mild versus severe COVID-19 infection in humans. Science. 2020;369:1210–1220. doi: 10.1126/science.abc6261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayres J.S. A metabolic handbook for the COVID-19 pandemic. Nat. Metab. 2020;2:572–585. doi: 10.1038/s42255-020-0237-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhatt A.N., Kumar A., Rai Y., Kumari N., Vedagiri D., Harshan K.H., Chinnadurai V., Chandna S. Glycolytic inhibitor 2-deoxy-d-glucose attenuates SARS-CoV-2 multiplication in host cells and weakens the infective potential of progeny virions. Life Sci. 2022;295:120411. doi: 10.1016/j.lfs.2022.120411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bost P., Giladi A., Liu Y., Bendjelal Y., Xu G., David E., Blecher-Gonen R., Cohen M., Medaglia C., Li H., et al. Host-viral infection maps reveal signatures of severe COVID-19 patients. Cell. 2020;181:1475–1488.e12. doi: 10.1016/j.cell.2020.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunis D.G., Andrews J., Fragiadakis G.K., Burt T.D., Sirota M. dittoSeq: universal user-friendly single-cell and bulk RNA sequencing visualization toolkit. Bioinformatics. 2020;36:5535–5536. doi: 10.1093/bioinformatics/btaa1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell I.W. The clinical significance of PPAR gamma agonism. Curr. Mol. Med. 2005;5:349–363. doi: 10.2174/1566524053766068. [DOI] [PubMed] [Google Scholar]
- Cao J., Spielmann M., Qiu X., Huang X., Ibrahim D.M., Hill A.J., Zhang F., Mundlos S., Christiansen L., Steemers F.J., et al. The single-cell transcriptional landscape of mammalian organogenesis. Nature. 2019;566:496–502. doi: 10.1038/s41586-019-0969-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chawla A., Barak Y., Nagy L., Liao D., Tontonoz P., Evans R.M. PPAR-gamma dependent and independent effects on macrophage-gene expression in lipid metabolism and inflammation. Nat. Med. 2001;7:48–52. doi: 10.1038/83336. [DOI] [PubMed] [Google Scholar]
- Chen S.T., Park M.D., Del Valle D.M., Buckup M., Tabachnikova A., Thompson R.C., Simons N.W., Mouskas K., Lee B., Geanon D., et al. A shift in lung macrophage composition is associated with COVID-19 severity and recovery. Sci. Transl. Med. 2022;14:eabn5168. doi: 10.1126/scitranslmed.abn5168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clemente-Suarez V.J., Ramos-Campo D.J., Mielgo-Ayuso J., Dalamitros A.A., Nikolaidis P.A., Hormeño-Holgado A., Tornero-Aguilera J.F. Nutrition in the actual COVID-19 pandemic. Nutrients. 2021;13:1924. doi: 10.3390/nu13061924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Codo A.C., Davanzo G.G., Monteiro L.d.B., de Souza G.F., Muraro S.P., Virgilio-da-Silva J.V., Prodonoff J.S., Carregari V.C., de Biagi Junior C.A.O., Crunfli F., et al. Elevated glucose levels favor SARS-CoV-2 infection and monocyte response through a HIF-1alpha/Glycolysis-Dependent Axis. Cell Metab. 2020;32:437–446.e5. doi: 10.1016/j.cmet.2020.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coillard A., Segura E. In vivo differentiation of human monocytes. Front. Immunol. 2019;10:1907. doi: 10.3389/fimmu.2019.01907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortese M., Lee J.Y., Cerikan B., Neufeldt C.J., Oorschot V.M.J., Köhrer S., Hennies J., Schieber N.L., Ronchi P., Mizzon G., et al. Integrative imaging reveals SARS-CoV-2-induced reshaping of subcellular morphologies. Cell Host Microbe. 2020;28:853–866.e5. doi: 10.1016/j.chom.2020.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cory T.J., Emmons R.S., Yarbro J.R., Davis K.L., Pence B.D. Metformin suppresses monocyte immunometabolic activation by SARS-CoV-2 spike protein subunit 1. Front. Immunol. 2021;12:733921. doi: 10.3389/fimmu.2021.733921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damiani C., Maspero D., Di Filippo M., Colombo R., Pescini D., Graudenzi A., Westerhoff H.V., Alberghina L., Vanoni M., Mauri G. Integration of single-cell RNA-seq data into population models to characterize cancer metabolism. PLoS Comput. Biol. 2019;15:e1006733. doi: 10.1371/journal.pcbi.1006733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Del Valle D.M., Kim-Schulze S., Huang H.H., Beckmann N.D., Nirenberg S., Wang B., Lavin Y., Swartz T.H., Madduri D., Stock A., et al. An inflammatory cytokine signature predicts COVID-19 severity and survival. Nat. Med. 2020;26:1636–1643. doi: 10.1038/s41591-020-1051-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desterke C., Turhan A.G., Bennaceur-Griscelli A., Griscelli F. PPARgamma cistrome repression during activation of lung monocyte-macrophages in severe COVID-19. iScience. 2020;23:101611. doi: 10.1016/j.isci.2020.101611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esser N., Legrand-Poels S., Piette J., Scheen A.J., Paquot N. Inflammation as a link between obesity, metabolic syndrome and type 2 diabetes. Diabetes Res. Clin. Pract. 2014;105:141–150. doi: 10.1016/j.diabres.2014.04.006. [DOI] [PubMed] [Google Scholar]
- Galvan-Pena S., O'Neill L.A.J. Metabolic reprograming in macrophage polarization. Front. Immunol. 2014;5:420. doi: 10.3389/fimmu.2014.00420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiger R., Rieckmann J.C., Wolf T., Basso C., Feng Y., Fuhrer T., Kogadeeva M., Picotti P., Meissner F., Mann M., et al. L-arginine modulates T cell metabolism and enhances survival and anti-tumor activity. Cell. 2016;167:829–842.e13. doi: 10.1016/j.cell.2016.09.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haghverdi L., Lun A.T.L., Morgan M.D., Marioni J.C. Batch effects in single-cell RNA-sequencing data are corrected by matching mutual nearest neighbors. Nat. Biotechnol. 2018;36:421–427. doi: 10.1038/nbt.4091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hevener A.L., Olefsky J.M., Reichart D., Nguyen M.T.A., Bandyopadyhay G., Leung H.Y., Watt M.J., Benner C., Febbraio M.A., Nguyen A.K., et al. Macrophage PPAR gamma is required for normal skeletal muscle and hepatic insulin sensitivity and full antidiabetic effects of thiazolidinediones. J. Clin. Invest. 2007;117:1658–1669. doi: 10.1172/JCI31561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hotamisligil G.S., Bernlohr D.A. Metabolic functions of FABPs--mechanisms and therapeutic implications. Nat. Rev. Endocrinol. 2015;11:592–605. doi: 10.1038/nrendo.2015.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S., Zhu B., Cheon I.S., Goplen N.P., Jiang L., Zhang R., Peebles R.S., Mack M., Kaplan M.H., Limper A.H., Sun J. PPAR-Gamma in macrophages limits pulmonary inflammation and promotes host recovery following respiratory viral infection. J. Virol. 2019;93 doi: 10.1128/JVI.00030-19. 000300-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Z., Chavda V.P., Vora L.K., Gajjar N., Apostolopoulos V., Shah N., Chen Z.S. 2-Deoxy-D-Glucose and its derivatives for the COVID-19 treatment: an update. Front. Pharmacol. 2022;13:899633. doi: 10.3389/fphar.2022.899633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen P.A., Papin J.A. Functional integration of a metabolic network model and expression data without arbitrary thresholding. Bioinformatics. 2011;27:541–547. doi: 10.1093/bioinformatics/btq702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang S., Nakanishi Y., Kioi Y., Okuzaki D., Kimura T., Takamatsu H., Koyama S., Nojima S., Nishide M., Hayama Y., et al. Semaphorin 6D reverse signaling controls macrophage lipid metabolism and anti-inflammatory polarization. Nat. Immunol. 2018;19:561–570. doi: 10.1038/s41590-018-0108-0. [DOI] [PubMed] [Google Scholar]
- Khunti K., Knighton P., Zaccardi F., Bakhai C., Barron E., Holman N., Kar P., Meace C., Sattar N., Sharp S., et al. Prescription of glucose-lowering therapies and risk of COVID-19 mortality in people with type 2 diabetes: a nationwide observational study in England. Lancet Diabetes Endocrinol. 2021;9:293–303. doi: 10.1016/S2213-8587(21)00050-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura T., Nada S., Takegahara N., Okuno T., Nojima S., Kang S., Ito D., Morimoto K., Hosokawa T., Hayama Y., et al. Polarization of M2 macrophages requires Lamtor1 that integrates cytokine and amino-acid signals. Nat. Commun. 2016;7:13130. doi: 10.1038/ncomms13130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korotkevich G., Sukhov V., Budin N., Shpak B., Artyomov M.N., Sergushichev A. Fast gene set enrichment analysis. bioRxiv. 2021 doi: 10.1101/060012. Preprint at. [DOI] [Google Scholar]
- Lun A.T.L., Bach K., Marioni J.C., Marioni J.C. Pooling across cells to normalize single-cell RNA sequencing data with many zero counts. Genome Biol. 2016;17:75. doi: 10.1186/s13059-016-0947-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laing A.G., Lorenc A., Del Molino Del Barrio I., Das A., Fish M., Monin L., Muñoz-Ruiz M., McKenzie D.R., Hayday T.S., Francos-Quijorna I., et al. A dynamic COVID-19 immune signature includes associations with poor prognosis. Nat. Med. 2020;26:1623–1635. doi: 10.1038/s41591-020-1038-6. [DOI] [PubMed] [Google Scholar]
- Lam S.M., Zhang C., Wang Z., Ni Z., Zhang S., Yang S., Huang X., Mo L., Li J., Lee B., et al. A multi-omics investigation of the composition and function of extracellular vesicles along the temporal trajectory of COVID-19. Nat. Metab. 2021;3:909–922. doi: 10.1038/s42255-021-00425-4. [DOI] [PubMed] [Google Scholar]
- Lee D., Smallbone K., Dunn W.B., Murabito E., Winder C.L., Kell D.B., Mendes P., Swainston N. Improving metabolic flux predictions using absolute gene expression data. BMC Syst. Biol. 2012;6:73. doi: 10.1186/1752-0509-6-73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lercher A., Bhattacharya A., Popa A.M., Caldera M., Schlapansky M.F., Baazim H., Agerer B., Gürtl B., Kosack L., Májek P., et al. Type I interferon signaling disrupts the hepatic urea cycle and alters systemic metabolism to suppress T cell function. Immunity. 2019;51:1074–1087.e9. doi: 10.1016/j.immuni.2019.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W.V., Li J.J. An accurate and robust imputation method scImpute for single-cell RNA-seq data. Nat. Commun. 2018;9:997. doi: 10.1038/s41467-018-03405-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao M., Liu Y., Yuan J., Wen Y., Xu G., Zhao J., Cheng L., Li J., Wang X., Wang F., et al. Single-cell landscape of bronchoalveolar immune cells in patients with COVID-19. Nat. Med. 2020;26:842–844. doi: 10.1038/s41591-020-0901-9. [DOI] [PubMed] [Google Scholar]
- Lucas C., Wong P., Klein J., Castro T.B.R., Silva J., Sundaram M., Ellingson M.K., Mao T., Oh J.E., Israelow B., et al. Longitudinal analyses reveal immunological misfiring in severe COVID-19. Nature. 2020;584:463–469. doi: 10.1038/s41586-020-2588-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makowski L., Chaib M., Rathmell J.C. Immunometabolism: from basic mechanisms to translation. Immunol. Rev. 2020;295:5–14. doi: 10.1111/imr.12858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathew D., Giles J.R., Baxter A.E., Oldridge D.A., Greenplate A.R., Wu J.E., Alanio C., Kuri-Cervantes L., Pampena M.B., D'Andrea K., et al. Deep immune profiling of COVID-19 patients reveals distinct immunotypes with therapeutic implications. Science. 2020;369:eabc8511. doi: 10.1126/science.abc8511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittag D., Proietto A.I., Loudovaris T., Mannering S.I., Vremec D., Shortman K., Wu L., Harrison L.C. Human dendritic cell subsets from spleen and blood are similar in phenotype and function but modified by donor health status. J. Immunol. 2011;186:6207–6217. doi: 10.4049/jimmunol.1002632. [DOI] [PubMed] [Google Scholar]
- Morse C., Tabib T., Sembrat J., Buschur K.L., Bittar H.T., Valenzi E., Jiang Y., Kass D.J., Gibson K., Chen W., et al. Proliferating SPP1/MERTK-expressing macrophages in idiopathic pulmonary fibrosis. Eur. Respir. J. 2019;54:1802441. doi: 10.1183/13993003.02441-2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nanda P., Ghosh A. Genome Scale-Differential Flux Analysis reveals deregulation of lung cell metabolism on SARS-CoV-2 infection. PLoS Comput. Biol. 2021;17:e1008860. doi: 10.1371/journal.pcbi.1008860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odegaard J.I., Ricardo-Gonzalez R.R., Goforth M.H., Morel C.R., Subramanian V., Mukundan L., Red Eagle A., Vats D., Brombacher F., Ferrante A.W., Chawla A. Macrophage-specific PPARgamma controls alternative activation and improves insulin resistance. Nature. 2007;447:1116–1120. doi: 10.1038/nature05894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orth J.D., Thiele I., Palsson B.Ø. What is flux balance analysis? Nat. Biotechnol. 2010;28:245–248. doi: 10.1038/nbt.1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren X., Wen W., Fan X., Hou W., Su B., Cai P., Li J., Liu Y., Tang F., Zhang F., et al. COVID-19 immune features revealed by a large-scale single-cell transcriptome atlas. Cell. 2021;184:1895–1913.e19. doi: 10.1016/j.cell.2021.01.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renz A., Widerspick L., Dräger A. FBA reveals guanylate kinase as a potential target for antiviral therapies against SARS-CoV-2. Bioinformatics. 2020;36:i813–i821. doi: 10.1093/bioinformatics/btaa813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohlenova K., Goveia J., García-Caballero M., Subramanian A., Kalucka J., Treps L., Falkenberg K.D., de Rooij L.P.M.H., Zheng Y., Lin L., et al. Single-cell RNA sequencing maps endothelial metabolic plasticity in pathological angiogenesis. Cell Metab. 2020;31:862–877.e14. doi: 10.1016/j.cmet.2020.03.009. [DOI] [PubMed] [Google Scholar]
- Schneider C., Nobs S.P., Kurrer M., Rehrauer H., Thiele C., Kopf M. Induction of the nuclear receptor PPAR-gamma by the cytokine GM-CSF is critical for the differentiation of fetal monocytes into alveolar macrophages. Nat. Immunol. 2014;15:1026–1037. doi: 10.1038/ni.3005. [DOI] [PubMed] [Google Scholar]
- Schulte-Schrepping J., Reusch N., Paclik D., Baßler K., Schlickeiser S., Zhang B., Krämer B., Krammer T., Brumhard S., Bonaguro L., et al. Severe COVID-19 is marked by a dysregulated myeloid cell compartment. Cell. 2020;182:1419–1440.e23. doi: 10.1016/j.cell.2020.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen B., Yi X., Sun Y., Bi X., Du J., Zhang C., Quan S., Zhang F., Sun R., Qian L., et al. Proteomic and metabolomic characterization of COVID-19 patient sera. Cell. 2020;182:59–72.e15. doi: 10.1016/j.cell.2020.05.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shlomi T., Cabili M.N., Herrgård M.J., Palsson B.Ø., Ruppin E. Network-based prediction of human tissue-specific metabolism. Nat. Biotechnol. 2008;26:1003–1010. doi: 10.1038/nbt.1487. [DOI] [PubMed] [Google Scholar]
- Silvin A., Chapuis N., Dunsmore G., Goubet A.G., Dubuisson A., Derosa L., Almire C., Hénon C., Kosmider O., Droin N., et al. Elevated calprotectin and abnormal myeloid cell subsets discriminate severe from mild COVID-19. Cell. 2020;182:1401–1418.e18. doi: 10.1016/j.cell.2020.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song J.-W., Lam S.M., Fan X., Cao W.-J., Wang S.-Y., Tian H., Chua G.H., Zhang C., Meng F.-P., Xu Z., et al. Omics-driven systems interrogation of metabolic dysregulation in COVID-19 pathogenesis. Cell Metab. 2020;32:188–202.e5. doi: 10.1016/j.cmet.2020.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuart T., Butler A., Hoffman P., Hafemeister C., Papalexi E., Mauck W.M., Hao Y., Stoeckius M., Smibert P., Satija R. Comprehensive Integration of Single-Cell Data. Cell. 2019;177:1888–1902. doi: 10.1016/j.cell.2019.05.031. https://doi.org/10.1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su Y., Chen D., Yuan D., Lausted C., Choi J., Dai C.L., Voillet V., Duvvuri V.R., Scherler K., Troisch P., et al. Multi-Omics resolves a sharp disease-state shift between mild and moderate COVID-19. Cell. 2020;183:1479–1495.e20. doi: 10.1016/j.cell.2020.10.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Team R.C. R software. R Foundation for Statistical Computing. 2008;739,1 [Google Scholar]
- Thompson E.A., Cascino K., Ordonez A.A., Zhou W., Vaghasia A., Hamacher-Brady A., Brady N.R., Sun I.H., Wang R., Rosenberg A.Z., et al. Metabolic programs define dysfunctional immune responses in severe COVID-19 patients. Cell Rep. 2021;34:108863. doi: 10.1016/j.celrep.2021.108863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tontonoz P., Nagy L., Alvarez J.G., Thomazy V.A., Evans R.M. PPARgamma promotes monocyte/macrophage differentiation and uptake of oxidized LDL. Cell. 1998;93:241–252. doi: 10.1016/s0092-8674(00)81575-5. [DOI] [PubMed] [Google Scholar]
- Trapnell C., Cacchiarelli D., Grimsby J., Pokharel P., Li S., Morse M., Lennon N.J., Livak K.J., Mikkelsen T.S., Rinn J.L. The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nature biotechnology. 2014;32:381–386. doi: 10.1038/nbt.2859. https://doi.org/10.1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner A., Wang C., Fessler J., DeTomaso D., Avila-Pacheco J., Kaminski J., Zaghouani S., Christian E., Thakore P., Schellhaass B., et al. Metabolic modeling of single Th17 cells reveals regulators of autoimmunity. Cell. 2021;184:4168–4185.e21. doi: 10.1016/j.cell.2021.05.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F., Zhang S., Vuckovic I., Jeon R., Lerman A., Folmes C.D., Dzeja P.P., Herrmann J. Glycolytic stimulation is not a requirement for M2 macrophage differentiation. Cell Metab. 2018;28:463–475.e4. doi: 10.1016/j.cmet.2018.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P., Luo R., Zhang M., Wang Y., Song T., Tao T., Li Z., Jin L., Zheng H., Chen W., et al. A cross-talk between epithelium and endothelium mediates human alveolar-capillary injury during SARS-CoV-2 infection. Cell Death Dis. 2020;11:1042. doi: 10.1038/s41419-020-03252-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wauters E., Van Mol P., Garg A.D., Jansen S., Van Herck Y., Vanderbeke L., Bassez A., Boeckx B., Malengier-Devlies B., Timmerman A., et al. Discriminating mild from critical COVID-19 by innate and adaptive immune single-cell profiling of bronchoalveolar lavages. Cell Res. 2021;31:272–290. doi: 10.1038/s41422-020-00455-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilk A.J., Rustagi A., Zhao N.Q., Roque J., Martínez-Colón G.J., McKechnie J.L., Ivison G.T., Ranganath T., Vergara R., Hollis T., et al. A single-cell atlas of the peripheral immune response in patients with severe COVID-19. Nat. Med. 2020;26:1070–1076. doi: 10.1038/s41591-020-0944-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao N., Nie M., Pang H., Wang B., Hu J., Meng X., Li K., Ran X., Long Q., Deng H., et al. Integrated cytokine and metabolite analysis reveals immunometabolic reprogramming in COVID-19 patients with therapeutic implications. Nat. Commun. 2021;12:1618. doi: 10.1038/s41467-021-21907-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao Z., Dai Z., Locasale J.W. Metabolic landscape of the tumor microenvironment at single cell resolution. Nat. Commun. 2019;10:3763. doi: 10.1038/s41467-019-11738-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu G., Qi F., Li H., Yang Q., Wang H., Wang X., Liu X., Zhao J., Liao X., Liu Y., et al. The differential immune responses to COVID-19 in peripheral and lung revealed by single-cell RNA sequencing. Cell Discov. 2020;6:73. doi: 10.1038/s41421-020-00225-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y., Zhou B., Pache L., Chang M., Khodabakhshi A.H., Tanaseichuk O., Benner C., Chanda S.K. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat. Commun. 2019;10:1523. doi: 10.1038/s41467-019-09234-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu L., She Z.G., Cheng X., Qin J.J., Zhang X.J., Cai J., Lei F., Wang H., Xie J., Wang W., et al. Association of blood glucose control and outcomes in patients with COVID-19 and pre-existing type 2 diabetes. Cell Metab. 2020;31:1068–1077.e3. doi: 10.1016/j.cmet.2020.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data reported in this article will be shared by the lead contact upon request. This article does not report novel RNA-seq data and original code. Any additional information required to reanalyze the data reported in this article is available from the lead contact upon request.