Abstract
The min system spatially regulates division through the topological regulation of MinCD, an inhibitor of cell division. MinCD was previously shown to inhibit division by preventing assembly of the Z ring (E. Bi and J. Lutkenhaus, J. Bacteriol. 175:1118–1125, 1993); however, this was questioned in a recent report (S. S. Justice, J. Garcia-Lara, and L. I. Rothfield, Mol. Microbiol. 37:410–423, 2000) which indicated that MinCD acted after Z-ring formation and prevented the recruitment of FtsA to the Z ring. This discrepancy was due in part to alternative fixation conditions. We have therefore reinvestigated the action of MinCD and avoided fixation by using green fluorescent protein (GFP) fusions to division proteins. MinCD prevented the localization of both FtsZ-GFP and ZipA-GFP, consistent with it preventing Z-ring assembly. Consistent with a direct interaction between FtsZ and the MinCD inhibitor, we find that increased FtsZ, but not FtsA, suppresses MinCD-induced lethality. Furthermore, strains carrying various alleles of ftsZ, selected on the basis of resistance to the inhibitor SulA, displayed variable resistance to MinCD. These results are consistent with FtsZ as the target of MinCD and confirm that this inhibitor prevents Z-ring assembly.
Division in bacterial cells occurs through the concerted action of a number of division proteins localized at the division site (22, 29). These division proteins are recruited by the Z ring, which is formed through the self assembly of FtsZ, the ancestral homologue of eukaryotic tubulins (21, 25). The Z ring, along with these additional division proteins, is designated the septal ring (16), an organelle that is capable of carrying out division. The recruitment of these additional division proteins to the Z ring occurs in at least two steps. Proteins FtsA and ZipA are recruited by direct interaction with FtsZ. Many of the remaining proteins do not interact directly with FtsZ, but rather depend on FtsA (2, 14, 23, 32).
Deletion of the min locus results in the production of minicells, small anucleate cells produced by division occurring near the poles of the cell (3). These minicell divisions appear to occur at the expense of medial divisions because the nucleated mother cells have greater average cell length than wild-type cells (30). Interestingly, increased expression of essential cell division protein FtsZ suppresses this increased average cell length, suggesting that FtsZ is limiting in this mutant (5). Examination of Z rings in the min deletion mutant indicates that Z rings form at the cell poles and at interior positions (8, 34). This observation suggests that the assembly of Z rings at interior positions is not necessarily delayed by deletion of the min locus but that maturation of these Z rings into a fully functional septal ring might be hampered. It's possible that multiple Z rings within the same cell compete with each other to become fully functional.
Although the min mutant contains Z rings near the poles of the cell, polar Z rings are not observed in wild-type cells (8, 34). This fascinating ability of the min system to prevent Z rings from forming at the poles but to allow them to form at midcell has led to an intense investigation of the min system. The min system consists of three genes, minCDE, all of which are necessary to achieve topological regulation of cell division (11). Genetic evidence indicates that MinC and MinD cooperate to form an inhibitor of cell division, which is topologically regulated by MinE. Analysis of functional green fluorescent protein fusions indicates that this topological regulation by MinE is achieved by inducing MinCD to oscillate from pole to pole without allowing occupation of the midcell (17, 27, 28).
Although MinC and MinD cooperate to form an efficient inhibitor of division, several lines of evidence suggest that MinC is the inhibitor and that it is activated by MinD. First, overproduction of MinC, but not MinD, inhibits division in a Δmin strain (12). Second, MinC can combine with DicB, encoded by a cryptic phage, to efficiently inhibit division, consistent with MinC being the component of the inhibitor that contacts the division machinery (12). In all cases, the inhibition, like that caused by MinC and MinD, can be suppressed by overproduction of FtsZ, suggesting a common mechanism of inhibition. Furthermore, this suppressibility by overexpression of FtsZ suggests that FtsZ might be the target of MinC (6, 12). Immunoelectron microscopy studies revealed that Z rings were not present in filaments produced by overexpression of MinCD, suggesting that this inhibitor blocked division by preventing Z-ring formation (8). Additional support for FtsZ as the target of MinCD comes from the increased resistance of several ftsZ mutants to MinCD (6). These mutants were isolated on the basis of resistance to SulA, an inhibitor of cell division that is induced by DNA damage. Finally, a MalE-MinC fusion that is capable of blocking division and Z-ring formation in vivo binds to FtsZ and prevents accumulation of FtsZ polymers in vitro, consistent with MinC inhibiting division by preventing Z-ring assembly (18, 19).
More recently this mode of action of MinC was questioned based on several observations (20). First, fluorescence microscopy was used to observe Z rings in cells overexpressing MinCD. These rings contained ZipA but not FtsA. Second, a strain carrying ftsZ103, a sulA-resistant mutant and reported to be MinCD resistant, filamented in the presence of overexpressed MinCD. Third, increasing FtsA suppressed MinCD-induced filamentation. Last, overexpression of SulA prevented oligomerization of the endogenous FtsZ in cell extracts whereas MinCD did not. As a result it was suggested that MinCD did not prevent formation of Z rings but rather acted at a later step to prevent FtsA from localizing to the Z ring. This mode of action for MinCD seemed unlikely since it would not explain how the min system prevents Z rings from forming at the poles of cells. However, we have reanalyzed the effect of MinCD on cell division since our previous report was done using immunoelectron microscopy (8), which lacks the sensitivity of fluorescence microscopy (1). Our results are consistent with FtsZ being the target of MinC and with MinCD inhibiting division by blocking Z-ring formation.
MATERIALS AND METHODS
Media and growth conditions.
Bacteria were grown in Luria-Bertani (LB) medium at 37°C. Spectinomycin (SPC) at 50 μg/ml, kanamycin at 25 μg/ml, and chloramphenicol at 20 μg/ml were added as needed. Glucose (always 0.2%), IPTG (isopropyl-β-d-thiogalactopyranoside), or arabinose was added at the concentrations indicated.
Bacterial strains, phages, and plasmids.
The strains and plasmids used in this study are listed in Table 1. All the JKD7.2 strains containing a pBEF plasmid were obtained by the transformation of JKD7.2/pKD3c with the desired plasmid and selecting on plates containing SPC at 37°C. Transformants were checked for sensitivity to chloramphenicol to ensure loss of pKD3c.
TABLE 1.
Strains and plasmids used in this study.
| Strain or plasmid | Description | Reference or source |
|---|---|---|
| Strains | ||
| PB114 (λDB173) | Δmin λPlac::minCD | 11 |
| JKD7.2 | ftsZ::kan recA56; FtsZ supplied by pKD3c | 10 |
| JKD7.2 (λDB173) | λDB173 lysogen of JKD7.2 | This study |
| PS269 | PB114 (λDB173/pSEB103) | This study |
| PS270 | PB114 (λDB173/pSEB104) | This study |
| PS294 | JKD7.2 (λDB173/pBEF0) | This study |
| PS293 | JKD7.2 (λDB173/pBEF2) | This study |
| PS309 | JKD7.2 (λDB173/pBEF9) | This study |
| PS310 | JKD7.2 (λDB173/pBEF100) | This study |
| PS311 | JKD7.2 (λDB173/pBEF114) | This study |
| PS299 | PB114 (λDB173/pJPB209) | This study |
| PS300 | PB114 (λDB173/pJPB222) | This study |
| PS301 | PB114 (λDB173/pJPB223) | This study |
| PS302 | PB114 (λDB173/pSEB25) | This study |
| PS298 | PB114 (λDB173/pSEB162) | This study |
| Plasmids | ||
| pBEF0 | Spcr; wild-type ftsZ on a pGB2 derivative | 4 |
| pBEF2 | Spcr; pBEF0 but contains ftsZ2 | 4 |
| pBEF9 | Spcr; pBEF0 but carries ftsZ9 | 4 |
| pBEF100 | Spcr; pBEF0 but carries ftsZ101 and ftsZ114 mutations | 4 |
| pBEF114 | Spcr; pBEF0 but carries ftsZ114 mutation (same as ftsZ103, sfiB103, or sfiB114) | 4 |
| pKD3c | Cmr; temperature-sensitive pSC101 derivative containing ftsAZ and envA | 2 |
| pSEB12 | Cmr; mini-F plasmid containing minCDE under the control of their own promoters | 26 |
| pSEB14 | Cmr; mini-F plasmid encoding minCD under the control of their own promoters | 26 |
| pJPB222 | Spcr; same as pJPB223 but ftsQAZ genes are in opposite orientation to the aadA gene; the level of expression of ftsQAZ is lower than in pJPB223 | Gift from Jean-Pierre Bouché |
| pJPB223 | Spcr; ftsQAZ cloned on pJPB209, aadA, and ftsQAZ are in the same orientation | 26 |
| PSEB25 | Spcr; pJPB223 with the PvuII fragment deleted, yielding an internal in-phase deletion of ftsA | Gift from Jean-Pierre Bouché |
| pSEB162 | Spcr; pJPB223 with the EcoRI fragment containing ftsZ deleted | This study |
| pSEB103 | Spcr; zipA-GFP under the control of PBAD on a pGB2 derivative | This study |
| pSEB104 | Spcr; ftsZ-GFP under the control of PBAD on a pGB2 derivative | This study |
| pJPB209 | Spcr; cloning vector derived from pGB2 | 26 |
| pGB2 | Spcr; cloning vector with the pSC101 origin of replication | 9 |
Plasmid pSEB14 differs from pSEB12 (minCDE) in that it expresses only the minCD genes. This plasmid was recovered in a strain that overexpressed MinE from pJPB216 (26). To test for the maintenance of pSEB14 in various strains, coextracted plasmids pSEB14 and pJPB216 were digested with EcoRI (which linearized only pJPB216) and precipitated with ethanol in order to obtain pSEB14 at about the same concentration as the pSEB12 preparation. To obtain the best transformation efficiencies, equal volumes of the pSEB12 and pSEB14 preparations were used in electroporation experiments with a Gene Pulser apparatus from Bio-Rad (0.1-cm-diameter cuvette; 1.5 kV, 25 μF, and 200 Ω). pSEB104 was generated by cloning the NgoMIV fragment carrying araC and PBAD-ftsZ-GFP from pJC104 (A. Mukherjee, C. Saez, and J. Lutkenhaus, submitted for publication) into pGB2 cut with XmaI. pSEB103 was obtained by replacing the SstI-XbaI fragment that carries ftsZ on pSEB104 by a PCR fragment containing zipA between SstI and XbaI sites. zipA was amplified using oligonucleotides 5′ZipAGFP (5′-ATGAGCTCGTTAGAACAACAGAGAAT) and 3′ZipAGFP (5′-TATCTAGAGGCGTTGGCGTCTTTGA).
Photomicroscopy.
An overnight culture was diluted by a factor of 100 in LB medium plus SPC and grown to an optical density at 540 nm (OD540) of 0.05. Arabinose was added to the desired concentration, and the culture was split between two flasks. In one of these flasks 0.1 mM IPTG was added. At the time samples were taken to be observed by fluorescence microscopy, a sample was taken for sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and analyzed as indicated below. Samples were directly (not fixed) observed and photographed with a Nikon Optiphot fluorescence microscope equipped with an E Plan oil immersion lens (Nikon) with a 100× objective and a DAGE-MTIDC-330 charge-coupled device camera using Flashpoint software (Integral Technologies). Fluorescence pictures were taken using a Nikon B-2A filter block with a 450- to 490-nm exitation filter and a 520-nm barrier filter. Images were imported to Adobe Photoshop software to be assembled.
Western blot analysis.
Samples (1 ml) of cultures were centrifuged, and the cells were lysed by resuspension in 100 μl of SDS sample buffer (62.5 mM Tris-HCl) [pH 6.8], 1% SDS, 10% glycerol, 5% β-mercaptoethanol) and heating at 100°C for 5 min. Equivalent amounts of OD540 material from all of the samples were then subjected to SDS-PAGE (12.5% gel). The proteins in the gel were transferred to nitrocellulose, and FtsZ was detected by an indirect immunostaining procedure with a rabbit polyclonal antiserum against FtsZ (1:3,000) and goat anti-rabbit immunoglobulin G antibodies coupled to alkaline phosphatase (1:3,000).
RESULTS
Effect of MinCD on localization of FtsZ-GFP.
Previously, using immunoelectron microscopy we reported that overexpression of MinCD prevented Z-ring formation (8); however, this conclusion was questioned by Justice et al. (20), who used the more sensitive fluorescence microscopy. In their study various fixation conditions were utilized before immunostaining, with various results. Using our fixation conditions no Z rings were observed following overexpression of MinCD. Consistent with this, we had found that a MalE-MinC fusion blocked Z-ring assembly when assessed under these fixation conditions (19). However, using a variety of other fixation conditions they observed Z rings. Interestingly, ZipA but not FtsA was also found to localize under these alternative fixation conditions. Two of these alternate fixation conditions have in common the omission of lysozyme prior to immunostaining. We attempted to reproduce these results using these alternative fixation conditions but were unsuccessful. We observed no immunostaining if lysozyme was omitted, implying that the cells were not permeabilized. Therefore, we decided to try a different approach that avoided fixation.
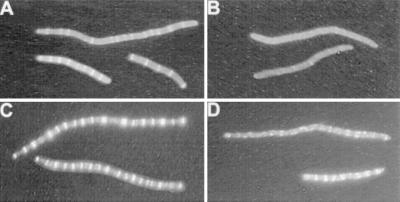
To avoid fixation, we chose to examine the localization of GFP fusions to division proteins as these fusions generally retain some function and are able to localize to the division site. We cloned ftsZ-gfp on a plasmid downstream of the arabinose promoter. This plasmid was introduced into PB114 Δmin (λDB173). The phage contains minCD downstream of the lac promoter. This combination of strain and plasmid allowed us to manipulate the levels of MinCD independently of FtsZ-GFP. In these experiments ftsZ-gfp was induced with or without the simultaneous induction of minCD. Even at low levels of induction of ftsZ-gfp the cells filamented (Fig. 1A; 0.01% arabinose) due to the increased level of FtsZ-GFP. Despite the filamentation the cells contained numerous Z rings that were well spaced. In contrast induction of minCD along with ftsZ-gfp completely prevented the formation of Z rings (Fig. 1B). No Z rings were observed in over 50 cells examined although occasional spots of fluorescence or spirals were observed. This experiment was repeated at a higher level of induction of ftsZ-gfp (0.05% arabinose). This higher level also resulted in filamentation, and the cells contained numerous Z rings that appeared brighter than at the lower arabinose concentration. The simultaneous induction of minCD at this higher level of ftsZ-gfp induction also interfered with Z-ring formation. Although Z-ring formation was not completely prevented, the filaments contained fewer rings and the rings did not have a typical appearance. FtsZ spirals were also observed.
FIG. 1.
Effect of MinCD expression on Z-ring formation. Fluorescence microscopy of PB114 Δmin (λDB173/pSEB104) (expressing FtsZ-GFP under Pbad promoter control) was performed 3 h after induction with 0.01% arabinose (A and B) or 0.05% arabinose (C and D). At the time of FtsZ-GFP induction, the culture was split in two, and no IPTG (A and C) or 1 mM IPTG (B and D) was added in order to induce MinCD from λDB173.
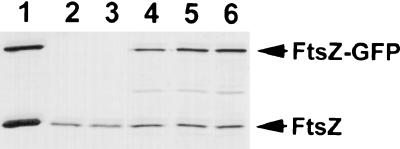
The above results indicate that expression of minCD can prevent FtsZ-GFP from localizing into Z rings. One trivial possibility is that induction of minCD interfered with induction of ftsZ-gfp. Therefore, samples were taken from the cultures with 0.05% arabinose and analyzed by immunoblotting (Fig. 2). This result shows that the level of FtsZ-GFP was not affected by minCD induction (Fig. 2, lanes 5 and 6). Furthermore, the level of FtsZ-GFP was similar to the level of the endogenous FtsZ. A sample from the culture induced with 0.01% arabinose was also analyzed. The FtsZ-GFP level was about one-third of that with the higher arabinose concentration (Fig. 2, lane 4). Since minCD induction did not interfere with ftsZ-gfp expression, we conclude that MinCD prevents FtsZ-GFP from assembling into Z rings.
FIG. 2.
Expression of GFP fusions from Pbad and induction of MinCD do not affect the level of FtsZ. Samples from the experiments in Fig. 1 and 3 were analyzed by immunoblotting using an anti-FtsZ polyclonal antibody. Lane 1, purified FtsZ and FtsZ-GFP; lanes 2 to 6, samples taken at the same time as the fluorescence pictures of Fig. 1 and 3; lane 2, sample from Fig. 3A (ZipA-GFP, no MinCD); lane 3, sample from Fig. 3B (ZipA-GFP plus MinCD); lane 4, sample from Fig. 1A (FtsZ-GFP, no MinCD); lane 5, sample from Fig. 1D (FtsZ-GFP plus MinCD); lane 6, sample from Fig. 1C (FtsZ-GFP, no MinCD).
Effect of MinCD on localization of ZipA-GFP.
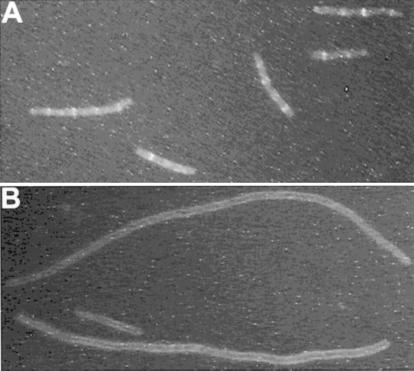
The results with FtsZ-GFP were fairly clear with respect to the effect of MinCD on Z-ring formation; however, the results are somewhat complicated since inducing FtsZ-GFP blocks cell division. We therefore analyzed the effect of MinCD on the localization of ZipA-GFP. ZipA has been shown to bind directly to FtsZ and to be a good marker for the position of Z rings (15). Also, Justice et al. (20) found that ZipA localized in the presence of MinCD. In addition, we were able to detect ZipA-GFP at a level that did not significantly inhibit division. ZipA-GFP was induced with or without induction of MinCD (Fig. 3). The heterogeneous cell length is due to using a min mutant. In the absence of MinCD induction, ZipA-GFP localized to rings, indicating that Z rings were present. With MinCD induction ZipA-GFP was not localized to rings and instead was present along the membrane. Immunoblot analysis showed that induction of ZipA-GFP did not affect the level of FtsZ (Fig. 2), eliminating this as a possible reason for the failure of Z rings to form. From these results we conclude that MinCD blocks Z-ring formation, thereby preventing ZipA from localizing to division sites.
FIG. 3.
Effect of MinCD expression on Z-ring formation using ZipA-GFP as a marker for Z rings. Shown is fluorescence microscopy of PB114 Δmin (λDB173/pSEB103) (expressing ZipA-GFP under Pbad promoter control) 3 h after induction by 0.05% arabinose. At the same time ZipA-GFP was induced, the culture was split in two and no IPTG (A) or 1 mM IPTG (B) was added to induce MinCD (under the control of lacZ promoter) from λDB173.
Effects of increased levels of FtsZ and FtsA.
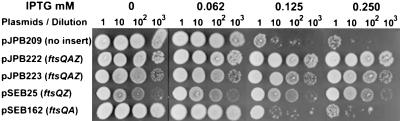
Previous results showed that increased expression of FtsZ can suppress filamentation caused by MinCD or overexpression of MinC (12, 13). However, Justice et al. (20) reported that increasing ftsA expression reduced filamentation caused by MinCD. To compare the effects of increased levels of FtsA and FtsZ on MinCD-induced lethality, we utilized a series of multicopy plasmids that contained the fts genes in various combinations. We then looked at the ability of these plasmids to suppress the lethal filamentation caused by induction of MinCD in a Δmin background. As seen from the spot tests shown in Fig. 4, IPTG at 0.125 mM reduced the plating efficiency of PB114 Δmin (λDB173) containing the vector (pJB209) by at least 3 orders of magnitude. In contrast, the presence of a plasmid containing ftsQAZ completely suppressed killing by IPTG. This suppression occurred whether the orientation of ftsQAZ was the same as or opposite to that of the aad promoter on the plasmid. The presence of just ftsQZ suppressed the sensitivity of PB114 Δmin (λDB173) to IPTG, indicating that it was the presence of ftsZ that suppresses minCD-induced killing as observed previously (6, 12). This result is supported by the results with the plasmid containing ftsQA. The presence of this plasmid offered no more protection than the vector alone. These results indicate that increased FtsZ, but not FtsA, provides protection from MinCD.
FIG. 4.
MinCD-induced division inhibition and the effect of multicopy ftsQAZ. Vector pJPB209 or the indicated derivatives were introduced into PB114 Δmin (λDB173), which expresses minCD under the control of Plac. One colony of each strain was resuspended in 300 μl of LB medium and serially diluted by 10. Samples (4 μl) were spotted on plates containing SPC and glucose with or without IPTG (as indicated) and incubated overnight at 37°C.
MinCD resistance due to ftsZ alleles.
Previously, it was reported that alleles of ftsZ, designated ftsZ (Rsa) for the resistance of their products to cell division inhibitor SulA, provided resistance to MinCD (6). The test compared the effects of MinCD induction on plating efficiency and filamentation for Δmin strains carrying various alleles of ftsZ on low-copy-number plasmids. The strains tested were merodiploids as each carried a copy of wild-type ftsZ on the chromosome in addition to the allele on the plasmid. In contrast, Justice et al. (20) examined a strain carrying just ftsZ103 and found that this strain filamented following induction of MinCD, suggesting that ftsZ103 provided no protection. This raised the question if any of these ftsZ alleles produced resistance to minCD in the absence of wild-type ftsZ. We used two different tests to examine the minCD resistance due to ftsZ alleles that are capable of supporting growth. The first test takes advantage of the previous observation that minCD on a single-copy vector cannot be efficiently introduced into a wild-type strain (26). The MinE expressed from the chromosome is insufficient to suppress minCD expressed from the plasmid, demonstrating that cells are sensitive to a single extra copy of minCD (26). JKD7-2 (ftsZ::kan) containing the various pBEF plasmids was transformed with either pSEB14 (minCD) or pSEB12 (minCDE). The ratio of transformation efficiencies with these two plasmids is a measure of the resistance to minCD (26). The results presented in Table 2 show that the transformation ratio is 0.5% for the control containing pBEF0 (ftsZ). This is slightly higher than that for a strain expressing ftsZ from the chromosome (26), presumably due to the slightly elevated level of FtsZ provided by pBEF0 (twofold higher). This ratio increases to 36.3% for pBEF2 (ftsZ2), the highest obtained with any of these ftsZ alleles. The other three alleles all gave intermediate levels, which were roughly equivalent, with the ratio falling in a range of 2.3 to 5.3%.
TABLE 2.
MinCD-induced division inhibition and the effect of ftsZ (Rsa) alleles
| Strain | ftsZ allele(s) | Transformation efficiency (%)a |
|---|---|---|
| JKD7.2/pBef0 | Wild type | 0.5 |
| JKD7.2/pBef2 | ftsZ2 | 36.3 |
| JKD7.2/pBef9 | ftsZ9 | 2.3 |
| JKD7.2/pBef100 | ftsZ101 + ftsZ114 | 3.7 |
| JKD7.2/pBef114 | ftsZ114 | 5.3 |
Transformation efficiencies are the ratios of transformants obtained in two parallel electroporations made with two mini-F plasmids, pSEB14, carrying minCD, and pSEB12, carrying the complete minCDE operon. The latter serves as a positive control for transformation. The values are the ratios of pSEB14/pSEB12 transformants times 100.
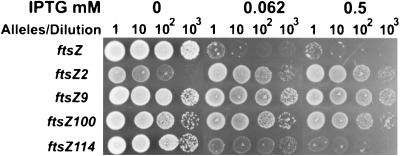
In a second test the pBEF plasmids were introduced into JKD7-2 (ftsZ::kan)(λDB173[Plac::minCD]), and the strains were examined for resistance to IPTG. The plating efficiency in the presence of pBEF0 (ftsZ+) was reduced by 3 log units at the two IPTG concentrations tested (Fig. 5). The ftsZ alleles varied in their resistance to MinCD in this test. Strains carrying ftsZ2, ftsZ9, and ftsZ100 were completely resistant to minCD, as the plating efficiency was unaffected even at the highest IPTG concentration. In contrast, a strain carrying ftsZ114 (identical to ftsZ103) was sensitive and only grew poorly at the lower IPTG concentration. However, pBEF114 (ftsZ114) supported more growth than pBEF0 (ftsZ), indicating that the strain carrying it it was more resistant. Thus, not all ftsZ alleles producing resistance to SulA produce the same level of resistance to MinCD. On the other hand, selection for alleles of ftsZ producing resistance to SulA appears to usually yield some level of resistance to MinCD.
FIG. 5.
Suppression of MinCD-induced lethality by ftsZ (Rsa) alleles. Plasmid pBEF0 (ftsZ), pBEF2 (ftsZ2), pBEF9 (ftsZ9), pBEF100 (ftsZ100), or pBEF114 (ftsZ114) was introduced into JKD7.2 (ftsZ::kan recA), lysogenic for λDB173 (Plac::minCD). In accordance with the protocol described in the legend to Fig. 4, samples were spotted on SPC and kanamycin plates with or without IPTG and incubated overnight at 37°C.
DISCUSSION
Our results indicate that FtsZ is the target of the MinCD inhibitor and that MinCD blocks division by preventing assembly of Z rings. This result is consistent with our previous findings (6) and disagrees with the conclusions of Justice et al. (20). They concluded that MinCD blocked division by preventing the addition of FtsA to the Z ring. However, by utilizing GFP fusions to FtsZ and ZipA, we avoided fixing cells and still observed that overexpression of MinCD blocked Z-ring formation. We also confirmed that overproduction of FtsZ suppressed MinCD-induced lethality, whereas FtsA had little effect. Finally, we show that several ftsZ (Rsa) alleles, in the absence of the wild-type ftsZ, confer various levels of resistance to MinCD.
Previously we reported that overexpression of MinCD prevented Z-ring formation (8); however, Justice et al. (20) reported that this result depended on the fixation conditions. Using our fixation conditions they did not observe Z rings; however, using a variety of other fixation conditions they observed somewhat atypical Z rings that contained ZipA but lacked FtsA. Furthermore, these rings had an abnormal appearance, so it was suggested that MinCD might interfere with the architecture of the Z ring such that it had a lower affinity for FtsA. Using GFP fusions to FtsZ and ZipA, and thereby avoiding fixation, we observed that MinCD prevented Z-ring formation. At low levels of expression of FtsZ-GFP, expression of MinCD completely prevented Z-ring formation. At higher levels of FtsZ-GFP expression, inhibition was not complete and some FtsZ structures were formed, often spirals, although a few typical Z rings were present. This ability of higher levels of FtsZ-GFP to at least partially overcome MinCD and form structures is consistent with the ability of an increased level of FtsZ to suppress MinCD. The failure of FtsZ-GFP to completely suppress MinCD is most likely due to its inability to functionally by substitute for FtsZ in division, although it can localize in vivo and polymerize in vitro (33).
The mechanism of MinCD action was confirmed using ZipA-GFP as a marker for Z rings. ZipA is known to bind tightly to the C-terminal region of FtsZ and to be a good marker for the presence of Z rings (15, 16). Expression of MinCD completely blocked the localization of ZipA-GFP, again indicating that MinCD completely blocked Z-ring assembly.
Although Justice et al. (20) reported that increased expression of ftsA showed some suppression of MinCD-induced filamentation, we observed no effect of increased FtsA on MinCD-induced lethality. We found that multicopy plasmids expressing ftsQAZ suppressed MinCD lethality. However, removing ftsZ from this plasmid eliminated suppression, whereas eliminating ftsA had no effect. Justice et al. (20) used slightly higher levels of FtsA in their studies (10-fold increase versus 5- to 7-fold in this study), which may provide some protection.
In addition to higher levels of FtsZ suppressing MinCD-induced filamentation, some mutations in ftsZ also suppress MinCD action (8). The associated mutants, designated ftsZ (Rsa), were isolated as ones that confer resistance to DNA damage-inducible inhibitor SulA (7). These mutants suppressed the lethality of overexpression of MinCD when carried on a plasmid in a strain with a wild-type copy of ftsZ on the chromosome. However, these mutants were divided into two classes based on the degree of filamentation after induction of MinCD. One class, consisting of ftsZ2 and ftsZ3, was designated very resistant, whereas another class was designated partially resistant and included ftsZ1, ftsZ9, ftsZ100, and ftsZ103 (ftsZ114). Justice et al. (20) found that ftsZ114 failed to block filamentation following induction of MinCD. This raised questions about the earlier results and interpretations. We therefore, examined those ftsZ mutants that were able to support viability for their ability to confer resistance to MinCD in the absence of wild-type ftsZ. In support of our previous report we observed that ftsZ2, ftsZ9, and ftsZ100 conferred resistance to MinCD-induced lethality, whereas ftsZ114 (ftsZ103 is identical) did not. However, ftsZ114 did support more growth in the presence of overexpressed MinCD than ftsZ, indicating that it confers some resistance. Thus, these alleles of ftsZ confer resistance to MinCD but the degree of resistance varies and depends on the test used. The most resistant strain is one carrying allele ftsZ2; this strain even appeared to grow slightly better in the presence of MinCD (Fig. 5).
Although MalE fusions to SulA and MinC block Z-ring formation and block FtsZ polymerization their modes of action are surely different. MalE-SulA blocks FtsZ GTPase, whereas MalE-MinC does not (19, 24). Thus, it was suggested that SulA prevented the interaction of FtsZ monomers that would lead to GTPase activity whereas MinC might destabilize FtsZ polymers. How might the same ftsZ mutations confer resistance to MinCD and SulA? One possibility is that they alter FtsZ such that MinC and SulA no longer interact with FtsZ. This is unlikely to be the explanation for all the mutations because they are not clustered. Another possibility is that these mutations lead to the formation of more-stable polymers, which are thus more resistant to MinC. Consistent with this we have found that FtsZ2, which is very resistant to MinC, produces stable polymers (Mukherjee et al., submitted). In contrast, FtsZ114, which is only weakly resistant, expresses nearly normal GTPase activity, implying that FtsZ114 polymers turn over rapidly. More study will be required to verify this possibility.
Justice et al. (20) induced SulA and MinCD to block cell division and then examined the ability of these inhibitors to block FtsZ oligomerization in the extracts upon raising the temperature. They found that SulA prevented oligomerization, as expected from previous studies (19, 31), but found that MinCD did not. Although this is consistent with MinC acting after assembly, possibly destabilizing FtsZ polymers as reported earlier (19), it may not actually be supportive due to limitations of this approach. In vivo the activity of MinC is concentrated at the membrane by MinD (17, 27). In the absence of MinD, MinC has to be overexpressed 25- to 50-fold to block division. The concentrating effect of MinD is lost once the cells are broken. Thus, Justice et al. (20) overexpressed MinCD sufficiently to inhibit division in vivo but were unlikely to have overexpressed MinC sufficiently to affect FtsZ polymerization in vitro (a 1:1 stoichiometry [19], which is unlikely to be achieved by the single-copy vector used to express minC, is required). In contrast to MinC, SulA does not appear to be localized to the membrane and presumably binds FtsZ in the cytoplasm, preventing its assembly into polymers.
Finally, it is unlikely that MinCD acts by preventing recruitment of FtsA to the Z ring, as it would not explain the ability of the min system to prevent Z-ring formation at the cell poles. The present results using GFP fusions, whose use avoids any possible artifacts due to fixation, confirm that MinCD inhibits cell division by preventing formation of Z rings.
ACKNOWLEDGMENTS
This work was supported by grant GM 29764 from the National Institutes of Health.
ADDENDUM IN PROOF
It was recently reported that overexpression of minCD in Bacillus subtilis also inhibits division by blocking Z-ring formation (P. A. Levin, R. L. Schwartz, and A. D. Grossman, J. Bacteriol. 183:5449–5452, 2001).
REFERENCES
- 1.Addinall S G, Bi E, Lutkenhaus J. FtsZ ring formation in fts mutants. J Bacteriol. 1996;178:3877–3884. doi: 10.1128/jb.178.13.3877-3884.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Addinall S G, Lutkenhaus J. FtsA is localized to the septum in an FtsZ-dependent manner. J Bacteriol. 1996;178:7167–7172. doi: 10.1128/jb.178.24.7167-7172.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Adler H I, Fisher W D, Cohen A, Hardigree A I. Miniature Escherichia coli cells deficient in DNA. Proc Natl Acad Sci USA. 1967;57:321–326. doi: 10.1073/pnas.57.2.321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bi E, Lutkenhaus J. Analysis of ftsZ mutations that confer resistance to the cell division inhibitor SulA (SfiA) J Bacteriol. 1990;172:5602–5609. doi: 10.1128/jb.172.10.5602-5609.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bi E, Lutkenhaus J. FtsZ regulates frequency of cell division in Escherichia coli. J Bacteriol. 1990;172:2765–2768. doi: 10.1128/jb.172.5.2765-2768.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bi E, Lutkenhaus J. Interaction between the min locus and ftsZ. J Bacteriol. 1990;172:5610–5616. doi: 10.1128/jb.172.10.5610-5616.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bi E, Lutkenhaus J. Isolation and characterization of ftsZ alleles that affect septal morphology. J Bacteriol. 1992;174:5414–5423. doi: 10.1128/jb.174.16.5414-5423.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bi E, Lutkenhaus J. Cell division inhibitors SulA and MinCD prevent formation of the FtsZ ring. J Bacteriol. 1993;175:1118–1125. doi: 10.1128/jb.175.4.1118-1125.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Churchward G, Belin D, Nagamine Y. A pSC101-derived plasmid which shows no sequence homology to other commonly used cloning vectors. Gene. 1984;31:165–171. doi: 10.1016/0378-1119(84)90207-5. [DOI] [PubMed] [Google Scholar]
- 10.Dai K, Lutkenhaus J. ftsZ is an essential cell division gene in Escherichia coli. J Bacteriol. 1991;173:3500–3506. doi: 10.1128/jb.173.11.3500-3506.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.de Boer P A, Crossley R E, Rothfield L I. A division inhibitor and a topological specificity factor coded for by the minicell locus determine proper placement of the division septum in E. coli. Cell. 1989;56:641–649. doi: 10.1016/0092-8674(89)90586-2. [DOI] [PubMed] [Google Scholar]
- 12.de Boer P A, Crossley R E, Rothfield L I. Central role for the Escherichia coli minC gene product in two different cell division-inhibition systems. Proc Natl Acad Sci USA. 1990;87:1129–1133. doi: 10.1073/pnas.87.3.1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.de Boer P A J, Crossley R E, Rothfield L I. Roles of MinC and MinD in the site-specific septation block mediated by the MinCDE system of Escherichia coli. J Bacteriol. 1992;174:63–70. doi: 10.1128/jb.174.1.63-70.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guzman L, Belin D, Carson M J, Beckwith J. Tight regulation, modulation, and high-level expression by vectors containing the arabinose Pbad promoter. J Bacteriol. 1995;177:4121–4130. doi: 10.1128/jb.177.14.4121-4130.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hale C A, de Boer P A. Direct binding of FtsZ to ZipA, an essential component of the septal ring structure that mediates cell division in E. coli. Cell. 1997;88:175–185. doi: 10.1016/s0092-8674(00)81838-3. [DOI] [PubMed] [Google Scholar]
- 16.Hale C A, de Boer P A. Recruitment of ZipA to the septal ring of Escherichia coli is dependent on FtsZ and independent of FtsA. J Bacteriol. 1999;181:167–176. doi: 10.1128/jb.181.1.167-176.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hu Z, Lutkenhaus J. Topological regulation of cell division in Escherichia coli involves rapid pole to pole oscillation of the division inhibitor MinC under the control of MinD and MinE. Mol Microbiol. 1999;34:82–90. doi: 10.1046/j.1365-2958.1999.01575.x. [DOI] [PubMed] [Google Scholar]
- 18.Hu Z, Lutkenhaus J. Analysis of MinC reveals two independent domains involved in interaction with MinD and FtsZ. J Bacteriol. 2000;182:3965–3971. doi: 10.1128/jb.182.14.3965-3971.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hu Z, Mukherjee A, Pichoff S, Lutkenhaus J. The MinC component of the division site selection system in Escherichia coli interacts with FtsZ to prevent polymerization. Proc Natl Acad Sci USA. 1999;96:14819–14824. doi: 10.1073/pnas.96.26.14819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Justice S, Garcia-Lara J, Rothfield L I. Cell division inhibitors SulA and MinC/MinD block septum formation at different steps in the assembly of the Escherichia coli division machinery. Mol Microbiol. 2000;37:410–423. doi: 10.1046/j.1365-2958.2000.02007.x. [DOI] [PubMed] [Google Scholar]
- 21.Lowe, J., and L. A. Amos. 1998. Crystal structure of the bacterial cell-division protein FtsZ. Nature 203–206. [DOI] [PubMed]
- 22.Lutkenhaus J, Addinall S G. Bacterial cell division and the Z ring. Annu Rev Biochem. 1997;66:93–116. doi: 10.1146/annurev.biochem.66.1.93. [DOI] [PubMed] [Google Scholar]
- 23.Ma X, Ehrhardt D W, Margolin W. Colocalization of cell division proteins FtsZ and FtsA to cytoskeletal structures in living Escherichia coli cells by using green fluorescent protein. Proc Natl Acad Sci USA. 1996;93:12998–13003. doi: 10.1073/pnas.93.23.12998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mukherjee A, Cao C, Lutkenhaus J. Inhibition of FtsZ polymerization by SulA, an inhibitor of septation in Escherichia coli. Proc Natl Acad Sci USA. 1998;95:2885–2890. doi: 10.1073/pnas.95.6.2885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mukherjee A, Lutkenhaus J. Guanine nucleotide-dependent assembly of FtsZ into filaments. J Bacteriol. 1994;176:2754–2758. doi: 10.1128/jb.176.9.2754-2758.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pichoff S, Vollrath B, Touriol C, Bouche J P. Deletion analysis of gene minE which encodes the topological specificity factor of cell division in Escherichia coli. Mol Microbiol. 1995;18:321–329. doi: 10.1111/j.1365-2958.1995.mmi_18020321.x. [DOI] [PubMed] [Google Scholar]
- 27.Raskin D M, de Boer P A. MinDE-dependent pole-to-pole oscillation of division inhibitor MinC in Escherichia coli. J Bacteriol. 1999;181:6419–6424. doi: 10.1128/jb.181.20.6419-6424.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Raskin D M, de Boer P A. Rapid pole-to-pole oscillation of a protein required for directing division to the middle of Escherichia coli. Proc Natl Acad Sci USA. 1999;96:4971–4976. doi: 10.1073/pnas.96.9.4971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rothfield L, Justice S, Garcia-Lara J. Bacterial cell division. Annu Rev Genet. 1999;33:423–438. doi: 10.1146/annurev.genet.33.1.423. [DOI] [PubMed] [Google Scholar]
- 30.Teather R M, Collins J F, Donachie W D. Quantal behavior of a diffusible factor which initiates septum formation at potential division sites in Escherichia coli. J Bacteriol. 1974;118:407–413. doi: 10.1128/jb.118.2.407-413.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Trusca D, Scott S, Thompson C, Bramhill D. Bacterial SOS checkpoint protein SulA inhibits polymerization of purified FtsZ cell division protein. J Bacteriol. 1998;180:3946–3953. doi: 10.1128/jb.180.15.3946-3953.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Weiss D S, Chen J C, Ghigo J M, Beckwith J. Localization of FtsI (PBP3) to the septal ring requires its membrane anchor, the Z ring, FtsA, FtsQ, and FtsL. J Bacteriol. 1999;181:508–520. doi: 10.1128/jb.181.2.508-520.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yu X C, Margolin W. Ca2+-mediated GTP-dependent dynamic assembly of bacterial cell division protein FtsZ into asters and polymer networks in vitro. EMBO J. 1997;16:5455–5463. doi: 10.1093/emboj/16.17.5455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yu X C, Margolin W. FtsZ ring clusters in min and partition mutants: role of both the Min system and the nucleoid in regulating FtsZ ring localization. Mol Microbiol. 1999;32:315–326. doi: 10.1046/j.1365-2958.1999.01351.x. [DOI] [PubMed] [Google Scholar]