Abstract
Computationally identifying new targets for existing drugs has drawn much attention in drug repurposing due to its advantages over de novo drugs, including low risk, low costs, and rapid pace. To facilitate the drug repurposing computation, we constructed an automated and parameter-free virtual screening server, namely DrugRep, which performed molecular 3D structure construction, binding pocket prediction, docking, similarity comparison and binding affinity screening in a fully automatic manner. DrugRep repurposed drugs not only by receptor-based screening but also by ligand-based screening. The former automatically detected possible binding pockets of the receptor with our cavity detection approach, and then performed batch docking over drugs with a widespread docking program, AutoDock Vina. The latter explored drugs using seven well-established similarity measuring tools, including our recently developed ligand-similarity-based methods LigMate and FitDock. DrugRep utilized easy-to-use graphic interfaces for the user operation, and offered interactive predictions with state-of-the-art accuracy. We expect that this freely available online drug repurposing tool could be beneficial to the drug discovery community. The web site is http://cao.labshare.cn/drugrep/.
Keywords: computer-aided drug discovery, drug repurposing, molecular docking simulation, virtual screening
Introduction
Drug repurposing is a strategy to identify new uses or indications of existing drugs that are beyond the scope of the original medical indication [1]. In recent years, the demand for drug repurposing has dramatically increased due to the remarkable advantages over developing de novo drugs, including low risk, low costs, and rapid pace [2]. A statistics analysis showed that drug repurposing may save up to 5–7 years in average drug development time [1]. To date, numerous successful examples of drug repurposing have been derived. For example, Gleevec, a drug for chronic myeloid leukemia (CML) can also be used to treat patients with malignant gastrointestinal stromal tumors (GIST) [3]. And aspirin, an orally administered non-steroidal anti-inflammatory agent, is applied to prevent cardiovascular disease and colorectal cancer [4]. Especially, we have witnessed that a number of computational drug repurposing approaches have been applied to fight against COVID-19 [5–8].
Systematic drug repurposing can be largely divided into experimental screening approaches, and in silico approaches. The former depends on particularly designed high-throughput assays, while the latter employs existing data and evolving computational approaches that are remarkably less expensive than the former [9]. Among them, virtual screening is a promising Computer-Aided Drug Design (CADD) technology used in drug discovery to search compound libraries in a limited time [5, 10, 11]. In general, virtual screening methods are classified as receptor-based screening (also known as structure-based screening or target-based screening) and ligand-based screening. The receptor-based screening typically explores the affinity of each compound in the compound library to the target protein using molecular docking algorithms based on conformational search and scoring functions. This process consists of preparing chemical libraries, identifying binding pockets, establishing docking parameters, executing docking programs and analyzing the results. Nowadays, the tools such as Pocket [12], eFindSite [13], CavityPlus [14] and COACH [15, 16] are available for binding site detection, while GOLD [17], Glide [18], AutoDock Vina [19], DOCK 6 [20], SwissDock [21], Edock [22] and CovalentDock Cloud [23] can be used for protein-ligand docking. Unlike the receptor-based screening that indispensably depends on the 3D structure of the receptor, the ligand-based virtual screening searches for new compounds sharing the similar characteristics observed in the known active compounds. Commonly employed similarity assessment methods currently include MACSS [24], Open Babel FP2 [25], RDKit Morgan fingerprint [26], 3D similarity [27], etc. With the above principles and methods in mind, several virtual screening tools have been developed. For example, DOCK Blaster [28], VSdocker [29], iScreen [30], and MTiOpenScreen [31] are receptor-based virtual screening tools. PoLi [32], LiSiCA [33], SwissSimilarity [34], HybridSim-VS [35], and Ligity [36] are ligand-based virtual screening tools. However, most of those tools are designed for the general purpose of virtual screening but not well organized for drug repurposing. In particular, these receptor-based tools need to provide complex parameters before screening, such as the grid center coordinates and the sizes of the search space, which are difficult to obtain for non-specialists.
To address these issues with the state-of-the-art technologies, we collected three drug libraries for drug repurposing, including approved drug library, experimental drug library and traditional Chinese medicine library, and then created an automated and parameter-free tool named DrugRep, which can perform receptor-based and ligand-based virtual screening over the carefully organized drug libraries. In our tool, several innovative works developed in-house are efficiently integrated, and those include the structure-based-cavity-detection approach, CurPocket [37, 38], for receptor-based screening, the ligand-similarity-based method, LigMate [39], and the hierarchical multi-feature alignment approach, FitDock [40], for ligand-based virtual screening. Among them, CurPocket is a protein-ligand binding site prediction method that searches for cavities on the protein surface by calculating curvature factors. Our previous work has demonstrated that it is highly accurate in determining docking boxes for molecular docking [37, 41]. LigMate and FitDock both can benefit ligand-based virtual screening [39, 42]. The former combines various descriptors in different dimensions of the molecule, while the latter uses a hierarchical alignment approach to find equivalent atom pairs. Our benchmark tests showed that these two achieved more substantial enrichment power than other methods. With the state-of-the-art virtual screening methods as well as the user-friendly interface, DrugRep could serve as a convenient tool for the drug-design community at http://cao.labshare.cn/drugrep/.
Materials and methods
Workflow
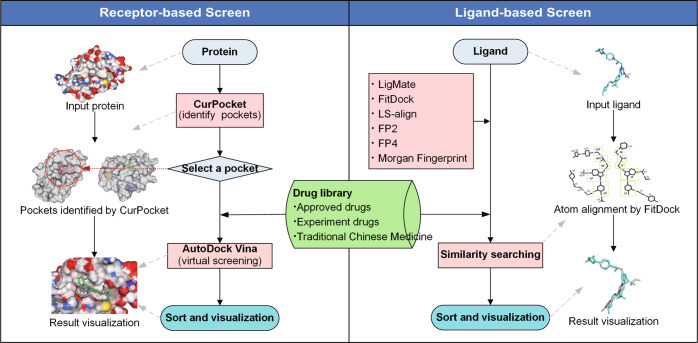
The DrugRep web interface is built by the HTML5, CSS3, PHP, and JavaScript libraries for the client-side user interface. It offers two strategies for drug repurposing, namely Receptor-based Screen (RBS), and Ligand-based Screen (LBS) (Fig. 1). For a protein receptor, RBS applies a novel cavity detection approach to automatically search the possible binding pockets and the results are shown interactively in 3D viewer for users to select. In addition to predictive pockets, RBS also utilizes the co-crystallized ligand from PDBbind [43] or user-uploaded ligand to calculate the center and sizes of the docking box using AutoDockTools [44] and eBoxSize [45]. After determining the docking parameters, RBS then performs docking for each drug with the receptor via the prevalent docking program AutoDock Vina (version 1.1.2) [19, 46], and finally gets a ranking based on the predicted affinity. For ligand-based virtual screening, LBS measures the similarity between the submitted ligand and each drug in drug libraries by LigMate, FitDock as well as five universal similarity measuring tools (i.e. Morgan fingerprint, LSalign-Rigid, LSalign-Flexi [27], FP2 and FP4 [25]). Ligand-Screen’s score ranges from 0 to 1.00, where 1.00 means totally same and 0 means not similar at all. The top-ranking compounds obtained by RBS or LBS will be displayed in tables, 2D and 3D images. The 3D visualization service is supported by NGL Viewers [47] and the superposition between the submitted ligand and drug is calculated by FitDock.
Fig. 1.
The workflow of DrugRep.
Drug libraries
DrugRep offers three drug libraries, including approved drug library (2315 drugs), experimental drug library (5935 drugs) and traditional Chinese medicine library (2390 drugs). The first two are the approved and experimental drugs from DrugBank database (version 5.1.7) [48], a state-of-the-art collection of approved, experimental, and pre-clinical drugs. Traditional Chinese medicine library which is from Topscience Company, contains 2390 monomer compounds from nearly 800 traditional Chinese medicines, and includes various structural types such as flavonoids, alkaloids, terpenoids, and glycosides. These drugs were double-checked using Open Babel [25] and assigned initial 3D conformations for drugs without 3D conformation using RDKit (http://rdkit.org) and Open Babel. Open Babel is also used to convert all drugs into the PDBQT format required for docking and other format (MOL/MOL2/SDF/SVG) required for similarity measuring tools.
Benchmark test
DUD [49], a well-established virtual screening benchmark dataset, contains 40 protein targets with 98,266 active compounds and decoys. DUD-E [50] is an enhanced DUD dataset which contains 102 targets with diverse ligands as well as decoys. The maximum unbiased validation (MUV) dataset [51] contains 17 targets, and each target has 30 actives and 15,000 decoys, which are unbiased in terms of simulation bias and artificial enrichment.
The enrichment testing is often used to evaluate virtual screening performance, and its result will be shown by the receiver operating characteristic (ROC) curve [52]. For all attainable score thresholds going from the best score to the worst, the selectivity (Se) and specificity (Sp) were calculated, as given by
Where TP is the true positive, FN is the false negative, TN is the true negative, and FP is the false positive. The ROC curve was then graphed by plotting all pairs of (1-Sp, Se). The area under the curve (AUC), which ranges from 0 to 1, is the metric used to assess a ROC curve. To be specific, AUC = 0.5 indicates that the compounds are randomly ranked, and the higher AUC means that the performance is better.
Enrichment Factor (EF), defined by the ratio of true positives in the first percent (TP1%) to all cases (N1%) divided by the ratio of actives (Na) to the total number (Nt) of test cases, is another metric used to evaluate enrichment testing.
EF greater than 1 indicates the compounds are not randomly selected, and higher EF means the enrichment effect is more significant.
Results
Performance of RBS
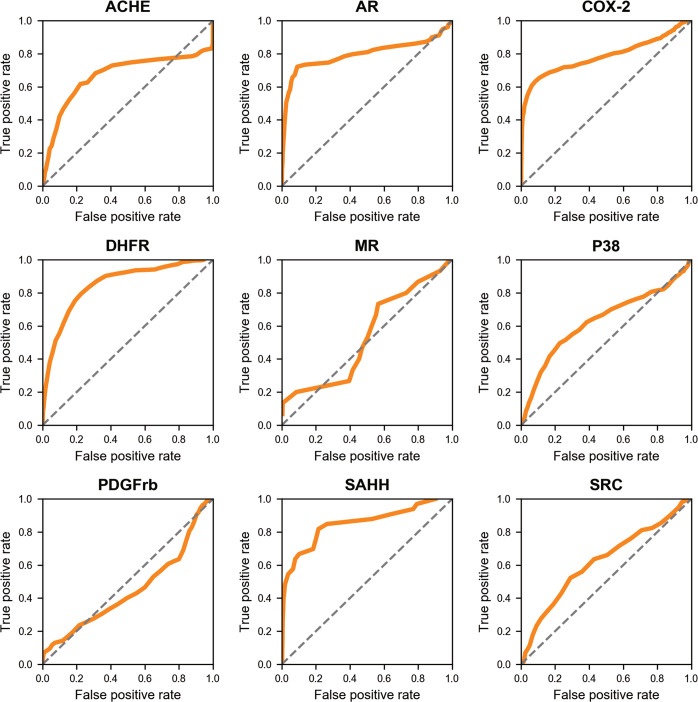
DrugRep is a parameter-free virtual screening tool, which is able to automatically determine the docking box for RBS. To test the feasibility and accuracy of the receptor-based virtual screening, we performed enrichment tests using the targets (ACHE, AR, COX-2, DHFR, MR, P38, PDGFrb, SAHH, SRC) from DUD dataset, which have been benchmarked in Vina-based virtual screening by Durrant [53]. For comparison, we also performed the same test using the docking box obtained from the co-crystallized ligand (named DrugRep-xtal), since it is the near optimal parameters for screening [45]. The results show that DrugRep achieved an average AUC values of 0.69, which is superior to 0.66 by Durrant and closed to 0.74 by DrugRep-xtal (Table 1 and Fig. 2). Particularly, it outperforms the latter in 5 of the 9 targets, indicating that DrugRep is better than the virtual screening with manually-determined docking boxes, and even comparable to the virtual screening with the co-crystallized-structure-determined docking boxes. We also calculated the EF1 and EF5 (EF for top 1% and top 5%), which are more important metrics for the actual virtual screening researches. As shown in Table 1, the average EF1 and EF5 of DrugRep reach 12.25 and 6.18, which are almost the same to 11.94 and 6.38 by DrugRep-xtal. Particularly, the EF1 of AR and SAHH are 19.79 and 26.84, respectively, outperforming 13.78 and 14.92 by DrugRep-xtal significantly. Detailed analysis shows that the predicted docking boxes by DrugRep are usually slightly larger than the docking boxes of DrugRep-xtal, resulting in increased accuracy for some targets. However, in a few cases, the predicted docking boxes may shift a little such as COX-2 (Supplementary Fig. S1) and PDGFrb, and the predicted docking boxes are too big such as MR. Both result in reduced accuracy. We also benchmarked DrugRep using another popular dataset of DUD-E. The result shows that DrugRep achieves comparable performance to the state-of-the-art approach (Supplementary Table S3). Overall, these results indicate that the automatic RBS by DrugRep is feasible and accurate for drug repurposing in many targets, and its unique parameter-free manner is highly convenient for the non-expert users.
Table 1.
Enrichment tests using DUD.
| Target | AUC | EF1 | EF5 | ||||
|---|---|---|---|---|---|---|---|
| Vinaa | DrugRep-xtal | DrugRep | DrugRep-xtal | DrugRep | DrugRep-xtal | DrugRep | |
| ACHE | 0.67 | 0.65 | 0.67 | 1.86 | 5.6 | 3.17 | 4.86 |
| AR | 0.81 | 0.77 | 0.8 | 13.78 | 19.79 | 9.09 | 11.36 |
| COX-2 | 0.31 | 0.89 | 0.79 | 26.12 | 24.49 | 13.61 | 10.89 |
| DHFR | 0.76 | 0.79 | 0.85 | 9.24 | 10.7 | 4.58 | 7.66 |
| MR | 0.84 | 0.82 | 0.54 | 31 | 12.38 | 13.15 | 2.63 |
| P38 | 0.54 | 0.62 | 0.63 | 1.54 | 2.2 | 2.95 | 2.69 |
| PDGFrb | 0.53 | 0.69 | 0.45 | 5.83 | 7 | 1.17 | 2.35 |
| SAHH | 0.76 | 0.74 | 0.85 | 14.92 | 26.84 | 6.66 | 10.89 |
| SRC | 0.69 | 0.69 | 0.62 | 3.13 | 1.25 | 3.01 | 2.26 |
| AVE. | 0.66 | 0.74 | 0.69 | 11.94 | 12.25 | 6.38 | 6.18 |
DrugRep-xtal means virtual screening using calculated pockets by crystal ligand. DrugRep means virtual screening using predicted pockets by CurPocket.
aThe data were from Durrant, J.D., 2011 [53].
Fig. 2. ROC curves of virtual screening results for 9 protein targets using DrugRep.
The dashed line indicates random selection of compounds.
Performance of LBS
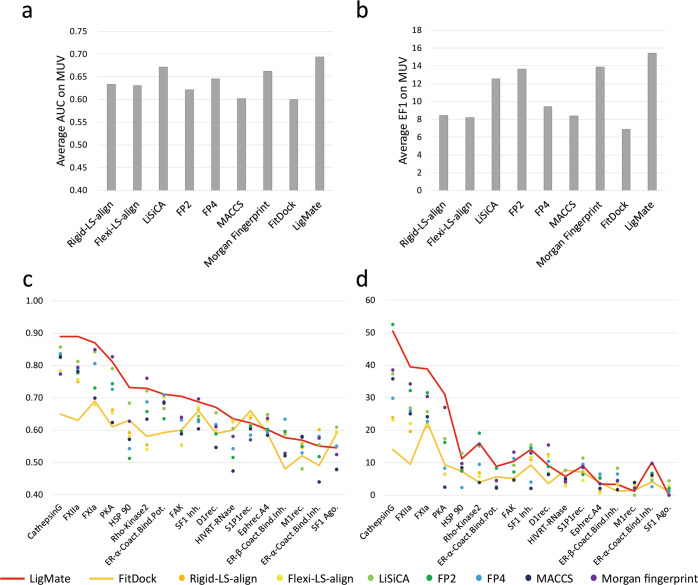
We then tested the performance of DrugRep on ligand-based virtual screening. We calculated the AUC value and the EF1 value for randomly selected actives with all corresponding decoys and the remaining actives on each target on the MUV dataset. The results of these calculations are fused using the maximum value. For each target, the above procedure was repeated 100 times to reduce the impact of randomness [39]. Figure 3a and b show the average AUC value and EF1 value of different methods for MUV dataset, respectively. The two histograms indicate LigMate surpasses all other methods. Further, we analyzed each target independently on the MUV dataset. As shown in Fig. 3c and d, LigMate achieves EF1s above 30 for targets including Cathepsin G, FXIIa, FXIa and PKA, which is significantly higher than those of most control methods. For the remaining targets, LigMate shows a comparable or even better performance. At the same time, we observed that other methods showed excellent performance for some targets (Supplementary Table S4 and S5). For example, FitDock has the highest EF1 value of 7.70 for HIVRT-RNase and the highest AUC value of 0.66 for S1P1rec. FP2 had the highest EF1 value of 19.15, 52.65, and 3.95 against Rho-Kinase2, CathepsinG, and M1rec. FP4 had the highest AUC values of 0.63, 0.71 for ER-β-Coact.Bind.Inh. and ER-α-Coact.Bind.Pot. Morgan Fingerprint achieves the highest EF1 values for FAK and D1rec., which were 13.25, 15.55; and has the highest AUC values for PKA, SF1 Inh., Rho-Kinase2, M1rec., which were 0.83, 0.70, 0.76, 0.58. Taken together, we argue that LigMate, FitDock, and other methods provided by DrugRep are complementary in enrichment test, and can contribute to ligand-based virtual screening.
Fig. 3. Benchmark results of LigMate and FitDock compared to the control methods.
a, b show the average EF1 and AUC for the MUV datasets, respectively. c, d show the AUC and the EF1 for each of the 17 targets from the MUV dataset, respectively. The x-axis shows the different methods, while the y-axis shows the AUC value in (a, c), and the EF1 value in (b, d).
Case study with DrugRep
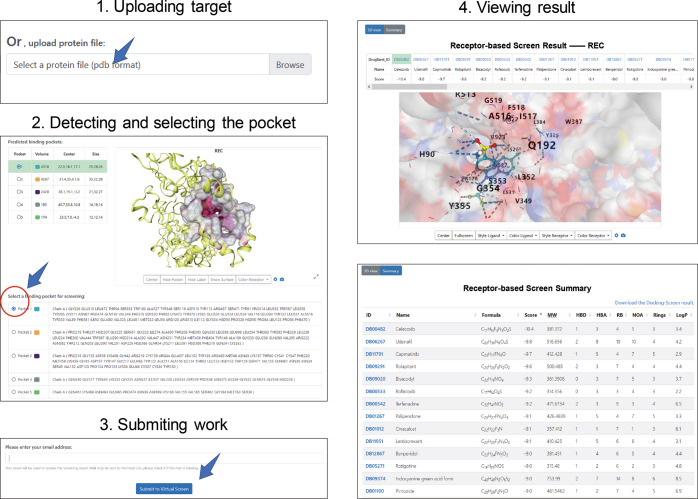
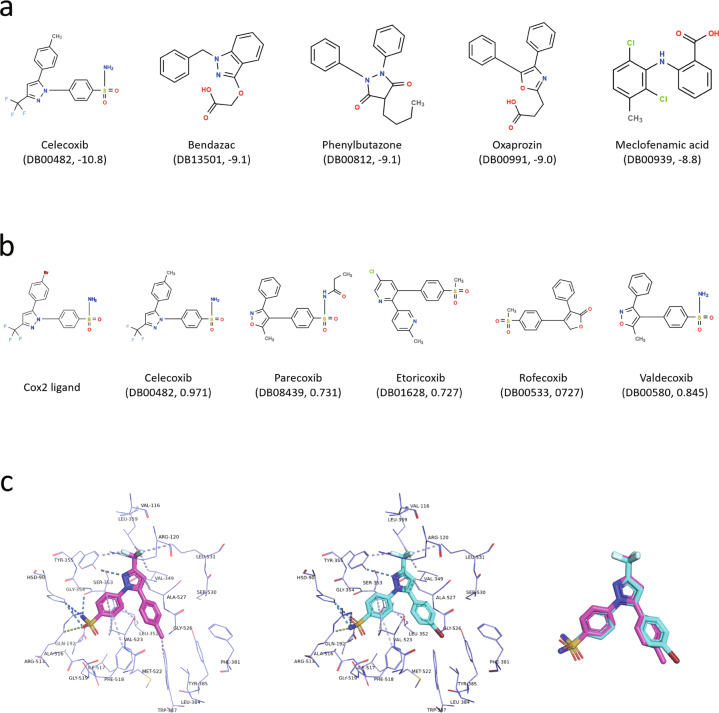
COX-2 that plays a significant role in conversion of arachidonate to prostaglandin H2 is key in the inflammatory response [54]. Targeted inhibition of COX-2’s activity can reduce the risk of peptic ulcer and is the main feature of nonsteroidal anti-inflammatory drug (NSAID) including aspirin, flurbiprofen, ibuprofen, celecoxib, flufenamic, mefenamic and tolfenamic acids [55]. Some COX-2 inhibitors may cause a significant increasing risk in heart attacks and strokes from clinical trials [56]. Therefore, screening the approved drugs which may bind with COX-2 is a general strategy in drug repurposing. Taking the screening of COX-2 with RBS as an example (Fig. 4), we only need to perform a few simple steps on the DrugRep web server, as follows: (i) uploading the target protein, (ii) detecting and selecting the pocket, (iii) submitting the task, and (iv) viewing the results of RBS. Five of the 30 best-scored drugs, namely celecoxib, oxaprozin, bendazac, meclofenamic acid and phenylbutazone, are associated with COX-2 (Fig. 5a). Of the drugs above, celecoxib, oxaprozin, meclofenamic acid and phenylbutazone are known inhibitors of COX-2. When inspecting the top 50 best-scored drugs, drug thalidomide (DrugBank ID: DB01041) that can be bioactivated by COX-2 [57] would also be found. Thalidomide is a typical example of drug repurposing. It was developed initially against morning sickness in pregnant women, but it was withdrawn years later because of its side effect of leading to severe congenital disabilities in children. Recently, researchers notice that thalidomide can also be adopted to treat some other diseases, such as leprosy, multiple myeloma, Crohn’s disease and HIV [58]. In addition, we also performed LBS against COX-2’s co-crystallized ligand using seven LBS methods embedded in DrugRep. For a drug with different scores by different LBS methods, the maximum score was adopted. Among the 50 drugs with the highest scores, five drugs, namely celecoxib, parecoxib, Etoricoxib, rofecoxib and valdecoxib (Fig. 5b and Supplementary Table S6), are known inhibitors of COX-2 and share similarities with the query ligand greater than 0.7. Among these drugs screened, celecoxib gets a similarity score of 0.971 by LBS, and gets an affinity score of −10.8 by RBS, which is greater than the co-crystallized ligand’s score of −10.2. As is shown Fig. 5c, celecoxib get an extra hydrophobic contact in a hydrophobic cavity formed by Phe381, Leu384, Tyr 385, Trp 387, Phe 513 and Ser530, because of a substitution of the bromophenyl ring to the methylbenzene ring. Example of COX-2 suggests that DrugRep can be used to screen new drugs for target, as well as to observe drug-target binding patterns.
Fig. 4. The operation process on the web page by RBS.
The process includes (1) uploading the target protein, (2) detecting and selecting the pocket, (3) submitting the task, and (4) viewing the results of RBS.
Fig. 5. The screening result of COX-2.
a shows 2D structures of five screened drugs truly binding to the COX-2 using RBS. The score after the ID means the affinity score (kcal/mol). b shows 2D structures of COX-2 co-crystallized ligand and nine screened drugs truly binding to the COX-2 using LBS. The score after the ID means the maximum similarity score. c shows contact residues and interaction forces of COX-2’s ligand and celecoxib including hydrogen bond (color skyblue), weak hydrogen bond (color smudge), and hydrophobic contact (color lightblue).
Discussion
New drug discovery is time-consuming and costly. Computational drug repurposing provides the possibility to improve this situation, and has a promising potential in modern drug development. However, computational tasks such as preparing chemical libraries, identifying binding pockets, evaluating docking parameters, and executing docking procedures can be cumbersome for the researcher. Here, we constructed an automated and parameter-free tool called DrugRep for drug repurposing. Our efforts can be summarized into four aspects. Firstly, DrugRep is designed for the purpose of drug repurposing with well-organized drug databases including approved or experimental-stage chemical drugs, and traditional Chinese medicine drugs. Secondly, DrugRep performs large-scale RBS based on CurPocket and AutoDock Vina. CurPocket for pocket recognition achieves a success rate of 70% in predicting the top 1 docking pockets and 92.5% in top 5 on the DUD dataset (Supplementary Table S1), making it accurate and convenient to prepare docking parameters. Thirdly, compared to receptor-based screening where docking of a large number of drugs takes a certain amount of time, DrugRep provides ligand-based screening based on our latest developed LigMate and FitDock as well as other well-established similarity measuring tools, helping to discover pharmacophores and assisting molecular optimization. Finally, DrugRep offers user-friendly interface that allows even non-expert users to perform drug repurposing and visualize results with abundant information, such as 3D interactive view, 2D images, the molecular mass, the number of hydrogen bond donors, the number of hydrogen bond acceptors, and the number of rotatable bonds, the n-octanol-water partition coefficient (logP), drug targets, drug pharmacodynamics, hyperlinks to the related data [48, 59] and so on.
Despite the successful performance in the benchmark tests, DrugRep also showed some limitations, such as false positive predictions. It can be attributed to the reasons below. Firstly, proteins may adopt various binding conformations, while the RBS of DrugRep performs docking with a static structure, which is artificial and harmful to some screening. For example, a highly flexible receptor, namely HSP90 [60], showed 0 for both EF1 and EF5 in the benchmark (Supplementary Table S2). Secondly, how to rank the results of screening remains an open question, particularly for enriching low affinity actives (i.e. TK [61]). DrugRep employs Vina score and X-Score [62] to evaluate the binding modes, which shows the advantages and disadvantages of each (Supplementary Table S2). Still, there are much room for further improvement. Thirdly, the data-driven methods, such as machine learning for docking or AI-based protein-ligand binding prediction, have shown promising results [11, 63–65]. The current DrugRep has not taken advantages of those state-of-the-art methods. Fourthly, in LBS, the scores of the seven similarity measuring methods usually are inconsistent with each other due to different scoring criteria. It may be not easy for the users to select the best prediction. Take Cox-2 as an example, parecoxib (DB08439) has a score of 0.731 in Flexi-LS-align, 0.71 in Rigid-LS-align and 0.688 in FitDock, but 0.473 in FP4 and 0.297 in Morgan fingerprint (Supplementary Table S6). Finally, in terms of time cost, Flexi-LS-align and FitDock take more time (20 min) than other methods because of flexible structure comparison. And RBS is time-consuming as it takes about three hours or even more for 2000 drugs with a 500 aa-length protein on our server.
In the future, we plan to develop methods for receptor optimization and docking parameters optimization before the docking screen, improving the docking and scoring by incorporating the AI-based methods, integrating the similarity scores in LBS with a more convenient ranking system, and optimizing the algorithms and pipelines to accelerate the computation by the guidance of existing similar co-crystal structures and the introduction of GPU-based Vina docking [66]. Overall, DrugRep will be more accurate, clever, easy-to-use and rapid for drug repurposing with our continuous efforts.
Supplementary information
Acknowledgements
This work was supported by the National Natural Science Foundation of China (81973243, 31401130, and 81830108). The authors wish to thank Dr. Maximilian Grimm for his kind help in constructing the web server.
Author contributions
JHG built the DrugRep web server and wrote the manuscript. JXL built the DrugRep web server and benchmarked the program. YL and SWC tested the server. SWC revised the manuscript. WTD designed the project. ZXX guided experiments. YC designed the project and wrote the manuscript.
Competing interests
The authors declare no competing interests.
Supplementary information
The online version contains supplementary material available at 10.1038/s41401-022-00996-2.
References
- 1.Ashburn TT, Thor KB. Drug repositioning: identifying and developing new uses for existing drugs. Nat Rev Drug Discov. 2004;3:673–83. doi: 10.1038/nrd1468. [DOI] [PubMed] [Google Scholar]
- 2.Pushpakom S, Iorio F, Eyers PA, Escott KJ, Hopper S, Wells A, et al. Drug repurposing: progress, challenges and recommendations. Nat Rev Drug Discov. 2019;18:41–58. doi: 10.1038/nrd.2018.168. [DOI] [PubMed] [Google Scholar]
- 3.Druker BJ. Imatinib as a paradigm of targeted therapies. Adv Cancer Res. 2004;91:1–30. doi: 10.1016/S0065-230X(04)91001-9. [DOI] [PubMed] [Google Scholar]
- 4.Bibbins-Domingo K. Aspirin use for the primary prevention of cardiovascular disease and colorectal cancer: U.S. Preventive Services Task Force recommendation statement. Ann Intern Med. 2016;164:836–45. doi: 10.7326/M16-0577. [DOI] [PubMed] [Google Scholar]
- 5.Dotolo S, Marabotti A, Facchiano A, Tagliaferri R. A review on drug repurposing applicable to COVID-19. Brief Bioinform. 2021;22:726–41. doi: 10.1093/bib/bbaa288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fan HH, Wang LQ, Liu WL, An XP, Liu ZD, He XQ, et al. Repurposing of clinically approved drugs for treatment of coronavirus disease 2019 in a 2019-novel coronavirus-related coronavirus model. Chin Med J (Engl) 2020;133:1051–6. doi: 10.1097/CM9.0000000000000797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rothan HA, Byrareddy SN. The epidemiology and pathogenesis of coronavirus disease (COVID-19) outbreak. J Autoimmun. 2020;109:102433. doi: 10.1016/j.jaut.2020.102433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu Y, Gan J, Wang R, Yang X, Xiao Z, Cao Y. DrugDevCovid19: an atlas of anti-COVID-19 compounds derived by computer-aided drug design. Molecules. 2022;27:683. doi: 10.3390/molecules27030683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cha Y, Erez T, Reynolds IJ, Kumar D, Ross J, Koytiger G, et al. Drug repurposing from the perspective of pharmaceutical companies. Br J Pharmacol. 2018;175:168–80. doi: 10.1111/bph.13798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rester U. From virtuality to reality - Virtual screening in lead discovery and lead optimization: a medicinal chemistry perspective. Curr Opin Drug Discov Devel. 2008;11:559–68. [PubMed] [Google Scholar]
- 11.Maia EHB, Assis LC, de Oliveira TA, da Silva AM, Taranto AG. Structure-based virtual screening: from classical to artificial intelligence. Front Chem. 2020;8:343. doi: 10.3389/fchem.2020.00343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Levitt DG, Banaszak LJ. POCKET: a computer graphies method for identifying and displaying protein cavities and their surrounding amino acids. J Mol Graph. 1992;10:229–34. doi: 10.1016/0263-7855(92)80074-N. [DOI] [PubMed] [Google Scholar]
- 13.Brylinski M, Feinstein WP. eFindSite: improved prediction of ligand binding sites in protein models using meta-threading, machine learning and auxiliary ligands. J Comput Aided Mol Des. 2013;27:551–67. doi: 10.1007/s10822-013-9663-5. [DOI] [PubMed] [Google Scholar]
- 14.Xu Y, Wang S, Hu Q, Gao S, Ma X, Zhang W, et al. CavityPlus: a web server for protein cavity detection with pharmacophore modelling, allosteric site identification and covalent ligand binding ability prediction. Nucleic Acids Res. 2018;46:W374–9. doi: 10.1093/nar/gky380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wu Q, Peng Z, Zhang Y, Yang J. COACH-D: improved protein-ligand binding sites prediction with refined ligand-binding poses through molecular docking. Nucleic Acids Res. 2018;46:W438–42. doi: 10.1093/nar/gky439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yang J, Roy A, Zhang Y. Protein-ligand binding site recognition using complementary binding-specific substructure comparison and sequence profile alignment. Bioinformatics. 2013;29:2588–95. doi: 10.1093/bioinformatics/btt447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Verdonk ML, Cole JC, Hartshorn MJ, Murray CW, Taylor RD. Improved protein-ligand docking using GOLD. Proteins. 2003;52:609–23. doi: 10.1002/prot.10465. [DOI] [PubMed] [Google Scholar]
- 18.Friesner RA, Banks JL, Murphy RB, Halgren TA, Klicic JJ, Mainz DT, et al. Glide: a new approach for rapid, accurate docking and scoring. 1. Method and assessment of docking accuracy. J Med Chem. 2004;47:1739–49. doi: 10.1021/jm0306430. [DOI] [PubMed] [Google Scholar]
- 19.Trott O, Olson AJ. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem. 2010;31:455–61. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lang PT, Brozell SR, Mukherjee S, Pettersen EF, Meng EC, Thomas V, et al. DOCK 6: combining techniques to model RNA-small molecule complexes. RNA. 2009;15:1219–30. doi: 10.1261/rna.1563609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Grosdidier A, Zoete V, Michielin O. SwissDock, a protein-small molecule docking web service based on EADock DSS. Nucleic Acids Res. 2011;39:W270–7. doi: 10.1093/nar/gkr366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang W, Bell EW, Yin M, Zhang Y. EDock: blind protein-ligand docking by replica-exchange monte carlo simulation. J Cheminform. 2020;12:37. doi: 10.1186/s13321-020-00440-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ouyang X, Zhou S, Ge Z, Li R, Kwoh CK. CovalentDock Cloud: a web server for automated covalent docking. Nucleic Acids Res. 2013;41:W329–32. doi: 10.1093/nar/gkt406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Durant JL, Leland BA, Henry DR, Nourse JG. Reoptimization of MDL keys for use in drug discovery. J Chem Inf Comput Sci. 2002;42:1273–80. doi: 10.1021/ci010132r. [DOI] [PubMed] [Google Scholar]
- 25.O’Boyle NM, Banck M, James CA, Morley C, Vandermeersch T, Hutchison GR. Open Babel: an open chemical toolbox. J Cheminform. 2011;3:33. doi: 10.1186/1758-2946-3-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rogers D, Hahn M. Extended-connectivity fingerprints. J Chem Inf Model. 2010;50:742–54. doi: 10.1021/ci100050t. [DOI] [PubMed] [Google Scholar]
- 27.Hu J, Liu Z, Yu DJ, Zhang Y. LS-align: an atom-level, flexible ligand structural alignment algorithm for high-throughput virtual screening. Bioinformatics. 2018;34:2209–18. doi: 10.1093/bioinformatics/bty081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Irwin JJ, Shoichet BK, Mysinger MM, Huang N, Colizzi F, Wassam P, et al. Automated docking screens: a feasibility study. J Med Chem. 2009;52:5712–20. doi: 10.1021/jm9006966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Prakhov ND, Chernorudskiy AL, Gainullin MR. VSDocker: a tool for parallel high-throughput virtual screening using AutoDock on Windows-based computer clusters. Bioinformatics. 2010;26:1374–5. doi: 10.1093/bioinformatics/btq149. [DOI] [PubMed] [Google Scholar]
- 30.Tsai TY, Chang KW, Chen CY. iScreen: world’s first cloud-computing web server for virtual screening and de novo drug design based on TCM database@Taiwan. J Comput Aided Mol Des. 2011;25:525–31. doi: 10.1007/s10822-011-9438-9. [DOI] [PubMed] [Google Scholar]
- 31.Labbé CM, Rey J, Lagorce D, Vavruša M, Becot J, Sperandio O, et al. MTiOpenScreen: a web server for structure-based virtual screening. Nucleic Acids Res. 2015;43:W448–54. doi: 10.1093/nar/gkv306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Roy A, Srinivasan B, Skolnick J. PoLi: a virtual screening pipeline based on template pocket and ligand similarity. J Chem Inf Model. 2015;55:1757–70. doi: 10.1021/acs.jcim.5b00232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lešnik S, Štular T, Brus B, Knez D, Gobec S, Janežič D, et al. LiSiCA: a software for ligand-based virtual screening and its application for the discovery of butyrylcholinesterase inhibitors. J Chem Inf Model. 2015;55:1521–8. doi: 10.1021/acs.jcim.5b00136. [DOI] [PubMed] [Google Scholar]
- 34.Zoete V, Daina A, Bovigny C, Michielin O. SwissSimilarity: a web tool for low to ultra high throughput ligand-based virtual screening. J Chem Inf Model. 2016;56:1399–404. doi: 10.1021/acs.jcim.6b00174. [DOI] [PubMed] [Google Scholar]
- 35.Shang J, Dai X, Li Y, Pistolozzi M, Wang L. HybridSim-VS: a web server for large-scale ligand-based virtual screening using hybrid similarity recognition techniques. Bioinformatics. 2017;33:3480–1. doi: 10.1093/bioinformatics/btx418. [DOI] [PubMed] [Google Scholar]
- 36.Ebejer JP, Finn PW, Wong WK, Deane CM, Morris GM. Ligity: a non-superpositional, knowledge-based approach to virtual screening. J Chem Inf Model. 2019;59:2600–16. doi: 10.1021/acs.jcim.8b00779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu Y, Grimm M, Dai WT, Hou MC, Xiao ZX, Cao Y. CB-Dock: a web server for cavity detection-guided protein-ligand blind docking. Acta Pharmacol Sin. 2020;41:138–44. doi: 10.1038/s41401-019-0228-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu Y, Yang X, Gan J, Chen S, Xiao ZX, Cao Y. CB-Dock2: improved protein-ligand blind docking by integrating cavity detection, docking and homologous template fitting. Nucleic Acids Res. 2022;50:W159–64. doi: 10.1093/nar/gkac394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Grimm M, Liu Y, Yang X, Bu C, Xiao Z, Cao Y. LigMate: a multifeature integration algorithm for ligand-similarity-based virtual screening. J Chem Inf Model. 2020;60:6044–53. doi: 10.1021/acs.jcim.9b01210. [DOI] [PubMed] [Google Scholar]
- 40.Yang X, Liu Y, Gan J, Xiao ZX, Cao Y. FitDock: protein-ligand docking by template fitting. Brief Bioinform. 2022;23:bbac087. doi: 10.1093/bib/bbac087. [DOI] [PubMed] [Google Scholar]
- 41.Zhao J, Cao Y, Zhang L. Exploring the computational methods for protein-ligand binding site prediction. Comput Struct Biotechnol J. 2020;18:417–26. doi: 10.1016/j.csbj.2020.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sabe VT, Ntombela T, Jhamba LA, Maguire GEM, Govender T, Naicker T, et al. Current trends in computer aided drug design and a highlight of drugs discovered via computational techniques: a review. Eur J Med Chem. 2021;224:113705. doi: 10.1016/j.ejmech.2021.113705. [DOI] [PubMed] [Google Scholar]
- 43.Wang R, Fang X, Lu Y, Wang S. The PDBbind database: collection of binding affinities for protein-ligand complexes with known three-dimensional structures. J Med Chem. 2004;47:2977–80. doi: 10.1021/jm030580l. [DOI] [PubMed] [Google Scholar]
- 44.Morris GM, Huey R, Lindstrom W, Sanner MF, Belew RK, Goodsell DS, et al. AutoDock4 and AutoDockTools4: automated docking with selective receptor flexibility. J Comput Chem. 2009;30:2785–91. doi: 10.1002/jcc.21256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Feinstein WP, Brylinski M. Calculating an optimal box size for ligand docking and virtual screening against experimental and predicted binding pockets. J Cheminform. 2015;7:18. doi: 10.1186/s13321-015-0067-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Eberhardt J, Santos-Martins D, Tillack AF, Forli S. AutoDock Vina 1.2.0: new docking methods, expanded force field, and python bindings. J Chem Inf Model. 2021;61:3891–8. doi: 10.1021/acs.jcim.1c00203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rose AS, Bradley AR, Valasatava Y, Duarte JM, Prlic A, Rose PW. NGL viewer: web-based molecular graphics for large complexes. Bioinformatics. 2018;34:3755–8. doi: 10.1093/bioinformatics/bty419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wishart DS, Feunang YD, Guo AC, Lo EJ, Marcu A, Grant JR, et al. DrugBank 5.0: a major update to the DrugBank database for 2018. Nucleic Acids Res. 2018;46:D1074–82. doi: 10.1093/nar/gkx1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Huang N, Shoichet BK, Irwin JJ. Benchmarking sets for molecular docking. J Med Chem. 2006;49:6789–801. doi: 10.1021/jm0608356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mysinger MM, Carchia M, Irwin JJ, Shoichet BK. Directory of useful decoys, enhanced (DUD-E): better ligands and decoys for better benchmarking. J Med Chem. 2012;55:6582–94. doi: 10.1021/jm300687e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rohrer SG, Baumann K. Maximum unbiased validation (MUV) data sets for virtual screening based on PubChem bioactivity data. J Chem Inf Model. 2009;49:169–84. doi: 10.1021/ci8002649. [DOI] [PubMed] [Google Scholar]
- 52.Triballeau N, Acher F, Brabet I, Pin JP, Bertrand HO. Virtual screening workflow development guided by the “receiver operating characteristic” curve approach. Application to high-throughput docking on metabotropic glutamate receptor subtype 4. J Med Chem. 2005;48:2534–47. doi: 10.1021/jm049092j. [DOI] [PubMed] [Google Scholar]
- 53.Durrant JD, McCammon JA. NNScore 2.0: a neural-network receptor-ligand scoring function. J Chem Inf Model. 2011;51:2897–903. doi: 10.1021/ci2003889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lucido MJ, Orlando BJ, Vecchio AJ, Malkowski MG. Crystal structure of aspirin-acetylated human cyclooxygenase-2: insight into the formation of products with reversed stereochemistry. Biochemistry. 2016;55:1226–38. doi: 10.1021/acs.biochem.5b01378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rao P, Knaus EE. Evolution of nonsteroidal anti-inflammatory drugs (NSAIDs): cyclooxygenase (COX) inhibition and beyond. J Pharm Pharm Sci. 2008;11:81s–110s. doi: 10.18433/J3T886. [DOI] [PubMed] [Google Scholar]
- 56.Sharma JN, Jawad NM. Adverse effects of COX-2 inhibitors. ScientificWorldJournal. 2005;5:629–45. doi: 10.1100/tsw.2005.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhou SF, Zhou ZW, Yang LP, Cai JP. Substrates, inducers, inhibitors and structure-activity relationships of human Cytochrome P450 2C9 and implications in drug development. Curr Med Chem. 2009;16:3480–675. doi: 10.2174/092986709789057635. [DOI] [PubMed] [Google Scholar]
- 58.Vargesson N. Thalidomide-induced teratogenesis: history and mechanisms. Birth Defects Res C Embryo Today. 2015;105:140–56. doi: 10.1002/bdrc.21096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhou Y, Zhang Y, Lian X, Li F, Wang C, Zhu F, et al. Therapeutic target database update 2022: facilitating drug discovery with enriched comparative data of targeted agents. Nucleic Acids Res. 2022;50:D1398–407. doi: 10.1093/nar/gkab953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Biamonte MA, Van de Water R, Arndt JW, Scannevin RH, Perret D, Lee WC. Heat shock protein 90: inhibitors in clinical trials. J Med Chem. 2010;53:3–17. doi: 10.1021/jm9004708. [DOI] [PubMed] [Google Scholar]
- 61.Bissantz C, Folkers G, Rognan D. Protein-based virtual screening of chemical databases. 1. Evaluation of different docking/scoring combinations. J Med Chem. 2000;43:4759–67. doi: 10.1021/jm001044l. [DOI] [PubMed] [Google Scholar]
- 62.Wang R, Lai L, Wang S. Further development and validation of empirical scoring functions for structure-based binding affinity prediction. J Comput Aided Mol Des. 2002;16:11–26. doi: 10.1023/A:1016357811882. [DOI] [PubMed] [Google Scholar]
- 63.Li H, Sze KH, Lu G, Ballester PJ. Machine-learning scoring functions for structure-based virtual screening. Wiley Interdiscip Rev Comput Mol Sci. 2021;11:e1478. doi: 10.1002/wcms.1478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Luo Y, Zhao X, Zhou J, Yang J, Zhang Y, Kuang W, et al. A network integration approach for drug-target interaction prediction and computational drug repositioning from heterogeneous information. Nat Commun. 2017;8:573. doi: 10.1038/s41467-017-00680-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zeng X, Zhu S, Lu W, Liu Z, Huang J, Zhou Y, et al. Target identification among known drugs by deep learning from heterogeneous networks. Chem Sci. 2020;11:1775–97. doi: 10.1039/C9SC04336E. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tang S, Chen R, Lin M, Lin Q, Zhu Y, Ding J, et al. Accelerating AutoDock Vina with GPUs. Molecules. 2022;27:3041. doi: 10.3390/molecules27093041. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.