Abstract
Flagellar number in Pseudomonas aeruginosa is controlled by FleN, a putative ATP/GTP binding protein. Disruption of fleN results in multiflagellation of the otherwise monoflagellate strains PAK and PAO1 and is associated with a chemotactic defect. We propose that flagellar number is maintained by the antiactivator FleN, which downregulates flagellar genes by binding to their transcriptional activator, FleQ, an enhancer binding protein belonging to the NifA subfamily. In this report we demonstrate direct interaction of FleN and FleQ in the yeast two-hybrid system. Mutagenesis of the putative ATP/GTP binding motif in FleN24K→Q and truncation of FleN at either the N or C terminus abrogates this interaction. FleN does not inhibit the DNA binding ability of FleQ in vitro, thus indicating that it probably utilizes another mechanism(s) to serve as a FleQ antiactivator.
Flagella play an important role in the pathogenesis of infections caused by Pseudomonas aeruginosa, Campylobacter jejuni, Helicobacter pylori, and Vibrio cholerae (16, 21). For effective motility, bacteria need to maintain the characteristic number and placement of their flagella. Studies from our laboratory indicate that in the monoflagellate P. aeruginosa, flagellar number is determined by FleN, a putative ATP/GTP binding protein (6). Disruption of fleN resulted in multiflagellation of P. aeruginosa, and overexpression of FleN from a strong plasmid promoter inhibited flagellar assembly. Analysis of other bacterial genomes including those of Pseudomonas putida, V. cholerae, H. pylori, Bacillus subtilis, and Aquifex aeolicus revealed a fleN homolog in their respective fla loci (6). Quite possibly some of these FleN homologs play a role in determining flagellar number in their respective species, as FleN does in P. aeruginosa. Except for the B. subtilis homolog Orf298, none of the other homologs have been characterized. Disruption of orf298 did not influence motility, but the effect on flagellar number, if any, was not reported (12).
Many flagellar genes and operons of P. aeruginosa are regulated by FleQ, a multidomain ς54-dependent transcriptional activator that belongs to the NifA/NtrC enhancer binding protein (EBP) family (1, 2, 23). In a fleN mutant, there is a positive correlation between multiflagellation and upregulation of FleQ-dependent flagellar promoters of various structural and regulatory genes involved in the synthesis of the flagellar motor and switch (FliM, FliN, and FliG), the basal body (FliE and FliF), the basal body rod (FlgB and FlgC), the hook (FlgD and FlgE), the cap (FliD), the filament (FliC), the regulatory proteins (FleS and FleR) (6), and the export apparatus (FlhA) (S. K. Arora, unpublished). This led us to believe that FleN exerted an antagonistic effect on FleQ-dependent transcriptional activation. FleN does not display a predictable DNA binding subsequence, making it unlikely that it is a DNA binding protein that could function as a repressor of fleQ (6) or FleQ-dependent flagellar genes. Moreover, FleQ amounts in both the wild-type PAK and the fleN mutant PAK-N (N. Dasgupta, unpublished) were similar, thus indicating that FleN inhibited FleQ posttranslationally. Therefore, the inhibition had to be mediated either through direct FleN-FleQ protein-protein interactions or indirectly through other intermediates.
EBPs of the NifA/NtrC family consist of an amino-terminal domain, a conserved central domain that catalyzes nucleoside triphosphate hydrolysis and interacts with the ς54 RNA polymerase holozyme and a C-terminal domain containing a helix-turn-helix (H-T-H) motif required for recognition of upstream activator sequences (UAS) (19). EBPs, such as NtrC of Escherichia coli, that require activation by their cognate histidine kinase (NtrB) through a phosphorylation event at their N-terminal phosphoacceptor domain (DDDK) belong to two-component systems (5, 19). Other EBPs, like NifA of Klebsiella and Azotobacter, lacking the N-terminal phosphoacceptor domain are constitutively active both in vivo and in vitro in the absence of their antiactivator (4). Under appropriate conditions, the NifA antiactivator NifL negatively modulates NifA through direct protein-protein interactions (10, 13, 14, 18, 25).
The absence of both the typical phosphoacceptor domain residues DDDK in FleQ (DDS59M instead) and a histidine kinase gene in the same operon as fleQ (1) suggests that the activation and regulation of FleQ may not involve the phosphorelay mechanism characteristic of two-component systems such as NtrB/NtrC. However, it cannot be overlooked that Ser-59 has the potential to serve as an alternative site for phosphorylation by a nonhistidine kinase (e.g., serine kinase), resulting in FleQ activation. The other alternative mechanism regulating FleQ could involve an antiactivator functioning in a similar manner to that of the NifA/NifL pair.
As disruption of fleN led to an upregulation of FleQ activity, the present study was undertaken to determine whether FleN was the putative antiactivator of FleQ. If so, we then wished to determine whether preventing FleQ from binding to DNA UAS was one of the possible mechanisms FleN employed to function as an antiactivator. Our studies indicate that FleN is the FleQ antiactivator. The entire FleN molecule, including its predicted N-terminal nucleotide binding motif, is essential for this interaction. In vitro, the interaction of FleN with FleQ did not prevent the latter's DNA binding ability.
MATERIALS AND METHODS
Strains, plasmids, and media.
The bacterial strains, yeast strains, and plasmids used in this study are listed in Table 1. Bacteria were grown in Luria-Bertani (LB) broth (24) at 37°C with shaking at 250 rpm or on LB agar plates, unless stated otherwise. The appropriate antibiotics were used to maintain the plasmids in P. aeruginosa at the following concentrations: 150 μg of carbenicillin/ml (300 μg/ml for plates) and 50 μg of tetracycline/ml (100 μg/ml for plates). In E. coli, the following concentrations were used: 200 μg of ampicillin/ml and 25 μg of tetracycline/ml. Yeast extract-peptone-dextrose and SD media (3) were used to propagate the yeast strains at 30°C.
TABLE 1.
Strains, plasmids, and primers used in this study
| Strain, plasmid, or primer | Relevant information | Source or reference |
|---|---|---|
| Strains | ||
| E. coli | ||
| DH5α | hsdR recA lacZYA φ80 lacZΔM15 | GIBCO-BRL |
| XL1 blue | recA1 endA1 gyrA96 thi-1 hsdR17 supE44 relA1 lac [F′ proAB lacIqZ ΔM15 Tn10 (Tetr)] | Stratagene |
| BL21(pLysS) | F−ompT hsdSB(rB− mB−) gal dcm(DE3), T7 lysozyme gene on pLysS | Novagen Inc., Madison, Wis. |
| S. cerevisiae Y190 | MATagal4 gal80 his3 trpl-901 ade2-101 ura3-52 leu2-3 | M. S. Swanson |
| P. aeruginosa | ||
| PAK | Wild-type clinical isolate | 6 |
| PAK-Q | PAK fleQ::Gmr | 1 |
| PAK-N | PAK fleN::Gmr | 6 |
| PAK-R | PAK fleR::Gmr | 23 |
| PAO/T7(ADD1976) | PAO1 Tcr Cbs mini-D180, T7 polymerase gene in chromosome | A. Darzin |
| Plasmids | ||
| pGBT9 | Shuttle plasmid containing the GAL4 DNA binding domain | Clontech |
| pGBTQ | fleQ cloned into EcoRI-BamHI sites of pGBT9 | This study |
| pGBTN | fleN cloned into EcoRI-BamHI sites of pGBT9 | This study |
| pGBKT7-M | flgM cloned into NdeI-BamHI sites of pGADT7 | Frisk et al., submitted |
| pGBKT7-A | fliA cloned into NdeI-BamHI sites of pGADT7 | Frisk et al., submitted |
| pGAD424 | Shuttle plasmid containing the GAL4 activation domain | Clontech |
| pGADQ | fleQ cloned into EcoRI-BamHI sites of pGAD424 | This study |
| pGADN | fleN cloned into EcoRI-BamHI sites of pGAD424 | This study |
| pGADN1 | pGADNΔFleN amino acids 107–280 | This study |
| pGADN2 | pGADNΔFleN amino acids 191–280 | This study |
| pGADN3 | pGADNΔFleN amino acids 251–280 | This study |
| pGADN4 | pGADNΔFleN amino acids 261–280 | This study |
| pGADN5 | pGADNΔFleN amino acids 271–280 | This study |
| pGADN-C | pGADNΔFleN amino acids 1–106 | This study |
| pGADN24K→Q | pGADN mutated at FleN24K→Q | This study |
| pET15bVP | oriV cloned as a PstI fragment in bla of pET15b | 6 |
| pET-fleN | fleN inserted as a PCR product into the NdeI/BamHI sites of pET15bVP | 6 |
| pET-fleNΔN | fleNΔFleN amino acids 1–106 inserted as a PCR product into the NdeI/BamHI sites of pET15bVP | This study |
| pET-fleNΔC | fleNΔFleN amino acids 107–280 inserted as a PCR product into the NdeI/BamHI sites of pET15bVP | This study |
| pET-fleN24K→Q | fleN24K→Q inserted as a PCR product into the NdeI/BamHI sites of pET15bVP | This study |
| pDN19lacΩ | Promoterless lacZ oriV oriT Tetr Strr Ω fragment | 6 |
| pMSZ5 | pDN19lacΩ containing the pilA promoter region | 6 |
| placΩA | pDN19lacΩ containing the flhA promoter region | Arora, unpublished |
| pIH1119 | Expression vector to generate MBP fusion proteins | New England BioLabs |
| pIH-fleQ | fleQ cloned into EcoRI-BamHI sites of pIHI119 to express MBP-FleQ | B. Ritchings |
| pIH-fleN | fleN cloned as a PCR product into EcoRI-BamHI sites of pIHI119 to express MBP-FleN | This study |
| pMMB67EH | Broad host-range cloning vector, Ampr | S. Lory |
| pMMBQ | fleQ cloned into EcoRI-BamHI sites of pMMB67EH to express FleQ | This study |
| pMMBN | fleN cloned into EcoRI-BamHI sites of pMMB67EH to express FleN | This study |
| pMMBN4 | EcoRI-BamHI insert from pGADN4 cloned into pMMB67EH to express FleNΔ261–280 | This study |
| pMMBN5 | EcoRI-BamHI insert from pGADN5 cloned into pMMB67EH to express FleNΔ271–280 | This study |
| pMMBN24K→Q | EcoRI-BamHI insert from pGADN24K→Q cloned into pMMB67EH to express FleN24K→Q | This study |
| Primersa | ||
| fleQ5PRI | 5′ cccgaattcATGTGGCGC 3′, EcoRI site incorporated | |
| Q3Pbam | 5′ cccaaaggatccTCAATCATCCGACAG 3′, BamHI site incorporated | |
| fln5peco | 5′ cccaaagaattcATGAAGCAGATGGGGTAG 3′, EcoRI site incorporated | |
| flnBam | 5′ CCTTGCTATACgggaTCCAGAGGCCGCTG 3′, BamHI site incorporated | |
| flnN1bam | 5′ cccaaaggatcctcaCTGCATCGGCGAGAGGTG 3′, BamHI site incorporated | |
| flnN2bam | 5′ cccaaaggatcctcaCCTTCCTGCGGGCTGTGG 3′, BamHI site incorporated | |
| flnN3bam | 5′ cccaaaggatcctcaGACCTTCTGCGCGACCGCC 3′, BamHI site incorporated | |
| flnN4bam | 5′ cccaaaggatcctcaGCGCGGGTTGGCCGGC 3′, BamHI site incorporated | |
| flnN5bam | 5′ cccaaaggatcctcaCAGTCGTTCGACGAAG 3′, BamHI site incorporated | |
| flnCeco | 5′ cccaaagaattcCTCGCCGATGCAGCATGC 3′, EcoRI site incorporated | |
| flnNde | 5′ GACAACACAAcatATGAAGCAGATGGG 3′, NdeI site incorporated | |
| flnCNde | 5′ cccaaaaaacatatgTCTCGCCGATGCAGCATG 3′, NdeI site incorporated | |
| mutN1 | 5′ GGGCGGCGTCCGGCcAGACCAATGTGTCGG 3′ | |
| mutNcomp2 | 5′ CCGACACATTGGTCTgGCCGGACGCCGCCC 3′ | |
| GADf | 5′ CACTGTCACCTGGTTGGAC 3′ |
Lowercase denotes nucleotides added or modified to facilitate restriction digestion or mutagenesis at the sites marked in bold.
PCR.
PCR was performed in a DNA Thermal Cycler 480 (Perkin-Elmer Cetus, Norwalk, Conn.) using Pfu DNA polymerase (Stratagene, La Jolla, Calif.) in 100-μl reaction volumes. Briefly, the reaction consisted of 100 ng of template DNA, 1.5 mM MgCl2, 1× polymerase buffer, 0.2 mM concentrations of deoxynucleoside triphosphates, 0.5 μM each primer (Table 1) (custom synthesized at Gemini Biotech, Alachua, Fla.), 2% dimethyl sulfoxide, and 1 U of DNA polymerase. The PCR was subjected to a cycling profile with an initial denaturation of 10 min at 94°C followed by 35 cycles of the following: denaturation for 1 min at 94°C, annealing for 1 min at 45°C, and extension for 1 min/kb at 72°C. The template DNA used for PCR was either purified genomic DNA isolated using the cetyltrimethylammonium bromide procedure or a plasmid preparation using the alkaline lysis method (6). The PCR products were electrophoresed on 1% SeaPlaque GTG agarose gel (FMC Bioproducts, Rockland, Maine) and stained with ethidium bromide, and the desired bands were electroeluted for further applications.
MATCHMAKER two-hybrid system.
Sequences encoding the two functional domains of the GAL4 transcriptional activator are cloned into two different shuttle expression vectors, pGBT9 and pGAD424, which are part of the MATCHMAKER I two-hybrid system (Clontech, Palo Alto, Calif.). fleQ was amplified as a 1.5-kb PCR product using primers fleQ5PRI and Q3Pbam (Table 1), which contain engineered EcoRI and BamHI sites, respectively. The product was digested with EcoRI and BamHI and ligated into vectors pGBT9 and pGAD424 with similar cohesive ends, yielding pGBTQ and pGADQ, respectively. Similarly, fleN was amplified as an 880-bp PCR product with primers fln5peco and flnbam (Table 1) and cloned to yield pGBTN and pGADN, respectively.
To map the interacting domain of FleN, a series of C-terminal deletions in FleN was constructed by cloning PCR products generated using fln5peco as the 5′ primer and a variable 3′ primer containing a BamHI site. flnN1bam, flnN2bam, flnN3bam, flnN4bam, and flnN5bam (Table 1) served as the variable 3′ primers in the constructions of pGADN1, pGADN2, pGADN3, pGADN4, and pGADN5, respectively. An N-terminal deletion construct, pGADN-C, was constructed by cloning the PCR product of flnCeco and flnbam (Table 1). fliA and flgM were cloned in pGBKT7, yielding pGBKT7-A and pGBKT7-M, respectively (Table 1).
An appropriate combination of these recombinant plasmids was cotransformed into the yeast strain Saccharomyces cerevisiae Y190 according to the MATCHMAKER protocol and tested for interaction between FleQ and FleN, FliA and FleN, or FlgM and FleN. Transformants containing pGBT-derived plasmids were selected on SD plates lacking tryptophan, and those containing pGAD-derived plasmids were selected on SD plates lacking leucine. Cotransformants containing pGBT/pGBKT7- and pGAD-derived plasmids were selected on SD plates lacking tryptophan and leucine. For His+ selection, the cotransformants were streaked on SD plates devoid of tryptophan, leucine, and histidine and containing 50 mM 3-aminotriazole.
Site-directed mutagenesis.
Primer pairs mutN1 and mutNcomp2 (Table 1) were used in the QuikChange site-directed mutagenesis kit (Stratagene) to generate pGADN24K→Q according to the protocol provided in the kit. Briefly, 20 ng of column-purified (plasmid mini kit; Qiagen, Valencia, Calif.) plasmid template (pGADN) was used in a 50-μl amplification reaction mixture containing 1 μl of deoxynucleoside triphosphates, 1 μl of Pfu polymerase, 1.25 μl of each primer, and 5 μl of reaction buffer. It was subjected to a cycling profile of initial denaturation for 30 s at 95°C followed by 13 cycles of denaturation (95°C for 30 s), annealing (60°C for 1 min), and extension (68°C for 20 min). The contents were then treated with DpnI to digest the original plasmid template. One microliter of the postdigestion amplification reaction was used to transform E. coli XL1 blue cells, and transformants were selected on LB-ampicillin (100 μg/ml) plates. A clone with the desired site-specific mutagenesis, confirmed by sequencing using primer GADf (Table 1), was subsequently used for further characterization.
Construction of other recombinant plasmids.
In order to express FleQ, FleN, and FleN24K→Q in P. aeruginosa from the inducible tac promoter of the vector pMMB67EH, the fleQ- and fleN-containing inserts from pGBTQ, pGADN, and pGADN24K→Q were cloned into EcoRI-BamHI sites of the vector, yielding pMMBQ, pMMBN, and pMMBN24K→Q, respectively. Truncated versions of FleN lacking amino acids 261 to 280 and 271 to 280 were similarly expressed by cloning out the inserts from pGADN4 and pGADN5 into pMMB67EH, generating pMMBN4 and pMMBN5, respectively.
NdeI- and BamHI-digested PCR products generated by using primer pairs flnnde-flnN1bam and flnCnde-flnbam (Table 1) were cloned into pET15bVP, yielding pET-fleNΔC and pET-fleNΔN, respectively. pET-fleN24K→Q was similarly constructed by cloning the NdeI- and BamHI-digested PCR product obtained from using primer pair flnnde-flnbam (Table 1) and pGADN24K→Q as the template. pIH-fleN was constructed by cloning the EcoRI-BamHI fleN insert in pGADN into vector pIH1119. This would allow the expression of FleN as a fusion protein with the maltose binding protein (MBP) in E. coli.
Transformation of E. coli DH5α and electroporation of P. aeruginosa were performed using a standard protocol (6).
β-Galactosidase assay.
The β-galactosidase filter assay of yeast strains was performed as described in the MATCHMAKER two-hybrid system protocol. Briefly, cotransformants grown on SD plates were transferred and smeared on a piece of filter paper with a toothpick, immersed in liquid nitrogen for 30 s, and thawed on another piece of filter paper prewetted with Z buffer (15) containing X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside). The paper was incubated at 30°C until a blue color was detected. For the liquid culture assay using o-nitrophenyl-β-d-galactopyranoside as the substrate, the protocol from the yeast protocol handbook (Clontech) was followed. Briefly, overnight liquid cultures were grown in the appropriate SD selection medium and subcultured the following day in yeast extract-peptone-dextrose medium for 3 to 4 h until an optical density at 600 nm (OD600) of 0.8 to 1.0 was reached. One-milliliter culture aliquots were harvested, and cells were washed with Z buffer and resuspended in 0.2 ml of Z buffer. Aliquots (0.1 ml) were transferred to separate tubes and subjected to 4 cycles of freeze-thawing in liquid nitrogen and a 37°C water bath. Z buffer (0.7 ml) and o-nitrophenyl-β-d-galactopyranoside (0.16 ml at a concentration of 4 mg/ml) were added to each tube, and the mixtures were vortexed and incubated at 30°C until a yellow color developed. Sodium carbonate (0.4 ml at a concentration of 1 M) was added to the reaction, the tubes were centrifuged for 10 min at 12,000 rpm in a microcentrifuge (Biofuge; Heraeus), and the OD420 of the supernatant was noted. The β-galactosidase units were then calculated based on Miller's formula (15).
P. aeruginosa strains (PAK, PAK-Q, PAK-R, PAK-N) cotransformed with pMMB67EH-derived (pMMBN, pMMBN4, pMMBN5, and pMMBN24K→Q) and pDN19lacΩ-derived (placΩA and pMSZ5) plasmids were selected on LB plates containing both tetracycline and carbenicillin. The β-galactosidase activities (15) of the respective promoters were determined in liquid cultures grown in LB medium with the appropriate antibiotics under inducing (1 mM isopropyl-β-d-thiogalactopyranoside [IPTG]) and noninducing conditions at 37°C.
Expression and purification of MBP-FleQ and MBP.
To overexpress MBP-FleQ, E. coli DH5α containing pIH-fleQ was grown at 37°C with IPTG induction and MBP-FleQ purified using affinity chromatography as described in the product manual (New England BioLabs, Beverly, Mass.). The eluted protein was dialyzed at 4°C against 10 mM Tris-Cl (pH 7.9), 5% glycerol, and 1 mM dithiothreitol (DTT) prior to use. In order to purify MBP, E. coli DH5α transformed with pIH1119 was cultured and processed in a manner similar to that used for MBP-FleQ purification.
Expression and purification of MBP-FleN.
Unlike the overexpression of MBP-FleQ, overexpression of MBP-FleN from pIH-fleN in E. coli DH5α resulted in its accumulation as inclusion bodies. To purify MBP-FleN in a soluble form from the inclusion bodies, a protocol using the detergent NDSB 201 (nondetergent sulfobetaines) was adopted (28). Briefly, the bacterial pellet from a 100-ml IPTG-induced culture expressing MBP-FleN was resuspended in 2 ml of 50 mM HEPES (pH 7.5), 0.5 M NaCl, 1 mM phenylmethylsulfonyl fluoride, and 5 mM DTT containing 0.35 mg of lysozyme/ml and incubated at 20°C for 30 min. Triton X-100 was then added to a 1% concentration, and the suspension was passed through a French pressure cell at 20,000 lb/in2 to lyse the cells. The inclusion bodies were pelleted by centrifuging the lysate at 30,000 × g for 30 min at 4°C. The pellet was washed twice with phosphate-buffered saline containing 1% Triton X-100. The inclusion bodies were solubilized in 50 mM HEPES (pH 7.5), 6 M guanidine hydrochloride, and 25 mM DTT to a protein concentration of 1 mg/ml. In order to facilitate the correct folding of MBP-FleN, which would minimize precipitation, 1 ml of the protein solution was diluted quickly in 9 ml of cold folding buffer (50 mM HEPES [pH 7.5], 0.2 M NaCl, 1 mM DTT, 1 M NDSB 201) by vortexing. Some of the protein precipitated as the solution appeared turbid. After 1 h of incubation on ice, the solution was centrifuged to remove the aggregated protein fraction from the soluble protein. The supernatant was dialyzed against 10 mM Tris-Cl (pH 7.9), 5% glycerol, and 1 mM DTT and concentrated by sprinkling Sephadex G-50 on the dialysis tubing. The insoluble nature of MBP-FleN when expressed in E. coli posed a major hurdle in obtaining soluble MBP-FleN at concentrations higher than 400 to 500 ng/μl. Our attempts to purify FleN as a His-tagged fusion protein culminated in unavoidable aggregation of the protein when eluted from the Ni2+ Sepharose column. Under denaturing conditions, the protein eluted from the column efficiently, but subsequent aggregation could not be prevented in step dialysis. His-FleN purified under denaturing conditions was used as an antigen to generate polyclonal antibodies in a rabbit (Cocalico Biologicals, Reamstown, Pa.).
SDS-polyacrylamide gel electrophoresis and Western analysis.
Purified proteins were denatured by boiling in 2% sodium dodecyl sulfate (SDS)–1% β-mercaptoethanol–50 mM Tris-Cl (pH 7.5) (SSB). The samples were resolved on a 12.5% polyacrylamide gel, and the proteins were stained with Coomassie brilliant blue (24). For Western analysis (1), 3 μl of the bacterial lysate (cell pellet from 1.0 ml of bacterial culture resuspended and boiled in 100 μl of SSB) was electrophoresed on a 15% polyacrylamide gel and proteins were transferred to a polyvinylidene difluoride membrane. The blot was developed using rabbit anti-FleN antibody as the primary antibody and alkaline phosphatase-conjugated anti-rabbit immunoglobulin G as the secondary antibody. Nitroblue tetrazolium and BCIP (5-bromo-4-chloro-3-indolylphosphate) served as the substrates for the color reaction.
Gel retardation assay (GRA).
The EcoRI-BamHI insert of placΩA containing the FleQ-regulated flhA promoter was gel purified and end labeled with [α-32P] dATP and the Klenow DNA polymerase (23). PAK-Q containing either pMMB67EH or pMMBQ was grown in 10 ml of LB containing carbenicillin to an OD600 of ∼0.2, induced with 1 mM IPTG, and grown for another 2 h. The culture was harvested, and the bacterial pellet was resuspended in 1 ml of lysis buffer (50 mM Tris-Cl [pH 7.9], 1 mM EDTA, 1 mM DTT, 1 mM phenylmethylsulfonyl fluoride) and passed through a French press at 20,000 lb/in2. The lysate was cleared by centrifugation (12,000 rpm, Beckman JA-20, 4°C, 30 min) and used in the binding reaction. The reaction mixture, consisting of 1 μl of deoxyinosine-deoxyctosine (2 μg/ul), 0.5 μl of bovine serum albumin (10 mg/ml), 0.8 μl of magnesium acetate (0.1 M), 1 μl of probe (500 to 700 cpm), and 1 μl of 1:5 diluted PAK-Q lysate (vector control pMMB67EH or pMMBQ expressing FleQ), was incubated on ice for 30 min in a total volume of 10 μl unless specified otherwise.
To determine the specificity of the binding of MBP-FleQ to the flhA probe, a cold flhA promoter fragment and herring sperm DNA were included in the competition assay. Variable amounts of the purified proteins MBP, MBP-FleQ, and MBP-FleN or a combination were included in lieu of the lysate to study their respective effect(s) on the mobility of the flhA probe.
RESULTS
FleN specifically inhibits FleQ-dependent transcriptional activation.
It has been reported earlier (6) that the disruption of fleN leads to an upregulation of FleQ-dependent promoters. To ascertain whether restoring and overexpressing FleN in the fleN mutant PAK-N would specifically downregulate the activity of FleQ-dependent promoters, the flhA promoter (placΩA) was chosen to represent FleQ-dependent promoters and the PilR-dependent promoter of pilA (pMSZ5 [11]) was chosen to serve as a negative control in a β-galactosidase assay. placΩA or pMSZ5 was cotransformed with either pMMBN or pMMB67EH in PAK-N. The activities of the flhA and pilA promoters were measured in a β-galactosidase assay under noninduced and induced (1 mM IPTG) expression of FleN from pMMBN. When compared to the activities of the vector control (pMMB67EH) under the same growth conditions, the uninduced leaky expression of FleN from pMMBN downregulated the flhA promoter about 24-fold and induction (with 1 mM IPTG) of FleN from the tac promoter of the vector caused a further 4-fold reduction in the promoter activity. The pilA promoter and vector control (pDN19lacΩ) activities remained essentially unaffected by FleN (Table 2). This suggested that FleN specifically inhibited transcription of the FleQ-dependent flhA promoter in a dose-responsive manner. The same promoter exhibited downregulation in a FleR mutant background (PAK-R) when FleN expression was induced from pMMBN, whereas in a FleQ mutant background (PAK-Q), the flhA promoter showed poor baseline activity which remained unaffected when FleN was induced from pMMBN. This is consistent with the earlier observation that FleQ regulation of the flhA promoter is direct and not mediated in a cascade manner through FleR (Arora, unpublished), the other response regulator in the P. aeruginosa fla locus. The observed upregulation of the flhA promoter in PAK-N when compared to PAK-R is in accordance with the observed upregulation of FleQ dependent promoters in the fleN mutant.
TABLE 2.
Assessment of the flhA promoter activity in the fleN, fleQ, and fleR mutant strains under induced expression of FleN from pMMBN
| Strain | Plasmid (promoter) | β-Galactosidase activity (mean ± SD) (Miller units) when cotransformed with:
|
|||
|---|---|---|---|---|---|
| pMMB67EH (vector control)
|
pMMBN (expressing FleN)
|
||||
| Uninduced | Induced | Uninduced | Induced | ||
| PAK-N | placΩA (flhA) | 88,943 ± 3,939 | 87,937 ± 2,125 | 3,626 ± 229 | 854 ± 63 |
| pMSZ5 (pilA) | 12,317 ± 863 | NDa | 11,665 ± 68 | ND | |
| pDN19lacΩ | 208 ± 2 | 234 ± 14 | 210 ± 19 | 270 ± 39 | |
| PAK-Q | placΩA (flhA) | ND | 570 ± 29 | ND | 585 ± 16 |
| PAK-R | placΩA (flhA) | ND | 7,890 ± 333 | ND | 856 ± 32 |
ND, not done.
FleN and FleQ interact with one another in the yeast two-hybrid MATCHMAKER system.
FleN does not downregulate transcription of fleQ (6), and the amount of FleQ in PAK, PAK-N, and PAK-N overexpressing FleN from pMMBN as detected by anti-FleQ antibody in Western analysis remained essentially comparable (Dasgupta, unpublished). Therefore, FleN-dependent inhibition of FleQ activity is possible either through the direct interactions of FleN with FleQ or through other FleQ-dependent intermediates. To address the former possibility, we took advantage of the MATCHMAKER I two-hybrid system, which utilizes two shuttle plasmids, pGBT9 and pGAD424 (see Materials and Methods). pGBT9 was used for constructing a translational fusion with either FleQ or FleN, yielding pGBTQ and pGBTN, respectively. Similarly, pGAD424 was used for constructing a translational fusion with either FleQ or FleN, yielding pGADQ and pGADN, respectively. In the appropriate cotransformants, the direct interaction of FleN and FleQ, if any, was phenotypically tested in a β-galactosidase filter assay for LacZ activity and by growing the yeast on histidine-deficient media to examine the His+ phenotype. A positive β-galactosidase filter assay (blue color development) and a His+ phenotype were obtained with cotransformants of only pGBTN plus pGADQ and pGADN plus pGBTQ (Table 3). The combinations of pGBTN plus pGAD424, pGADN plus pGBT9, pGBTQ plus pGAD424, and pGADQ plus pGBT9 were negative for both the assays. This indicated that FleN and FleQ directly interacted with one another in the yeast strain.
TABLE 3.
Assessment of yeast cotransformants with different combinations of plasmids for a positive β-galactosidase filter assay and His+ phenotype
| Plasmid (protein expressed) | Resulta with plasmid (protein expressed):
|
||||
|---|---|---|---|---|---|
| pGBTN (FleN) | pGADN (FleN) | pGBT9 | pGAD424 | pGAD-M (FlgM) | |
| pGBTQ (FleQ) | ND | + (2.8 ± 0.28) | ND | − (not detectable) | ND |
| pGADQ (FleQ) | + (2.7 ± 0.14) | ND | − (not detectable) | ND | ND |
| pGBT9 | ND | − (not detectable) | ND | − (not detectable) | ND |
| pGAD424 | − (not detectable) | ND | − (not detectable) | ND | ND |
| pGBKT7-A (FliA) | ND | − (0.01 ± 0.0) | ND | ND | + (0.21 ± 0.06) |
| pGBKT7-M (FlgM) | ND | − (0.02 ± 0.0) | ND | ND | ND |
β-Galactosidase activity in Miller units (mean ± standard deviation), determined in a liquid culture assay, appear in parentheses. +, positive assay for both β-galactosidase and His+ phenotype; −, negative assay for both β-galactosidase and His+ phenotype; ND, assay not performed.
To determine the specificity of the FleN-FleQ interaction, FliA, a sigma factor required for flagellin synthesis in P. aeruginosa (26), and FlgM, its antisigma factor (A. Frisk, J. Jyot, S. K. Arora, and R. Ramphal, submitted for publication), were substituted for FleQ. pGBKT7-A and pGBKT7-M carrying fliA and flgM (Frisk et al., submitted), respectively, were introduced into Y190 harboring pGADN. The pGBKT7-M plus pGAD-A cotransformant served as a positive control for the interaction between FlgM and FliA. The pGADN plus pGBKT7-A and pGADN plus pGBKT7-M cotransformants did not display positive β-galactosidase filter assays or the His+ phenotype (Table 3), indicating that FleN did not interact with either FliA or FlgM. Thus, the FleN-FleQ interaction appears to be specific. The β-galactosidase activities of the relevant cotransformants, quantitated using a liquid culture assay, are presented in Table 3.
MBP-FleQ has the ability to bind to DNA enhancer elements in vitro.
FleQ has a predicted H-T-H DNA binding motif at its C terminus. To activate transcription, an EBP like FleQ has to bind to DNA at the UAS. The antiactivator FleN could either be interfering with this process or preventing FleQ from initiating open complex formation at the ς54 holoenzyme-occupied promoters.
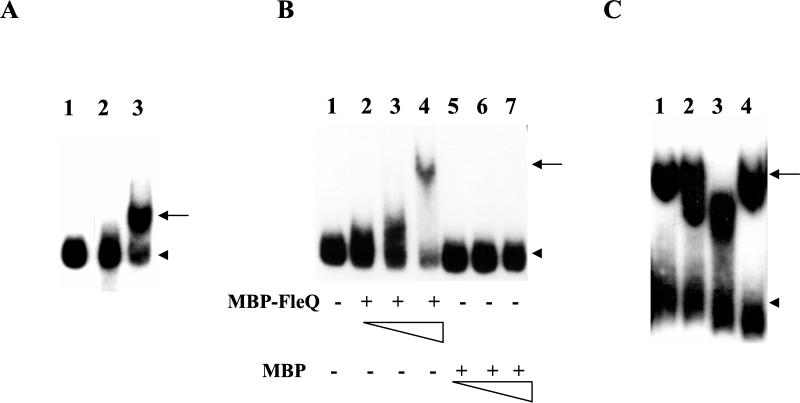
To test the former, we first established the conditions for binding FleQ to flhA UAS in vitro and then assessed the effects of FleN (if any) on it. We preferred using FleQ overexpressed from pMMBQ in the P. aeruginosa fleQ mutant PAK-Q, to ensure any posttranslational modification(s) of FleQ that might enable DNA binding. The promoter region of flhA (insert from placΩA) was end labeled (flhA probe) and examined in a GRA, using induced culture lysates of PAK-Q harboring either pMMBQ or pMMB67EH. The flhA probe exhibited a mobility shift with the FleQ-containing lysate of PAK-Q/pMMBQ but not with the vector control lysate of PAK-Q/pMMB67EH (Fig. 1A). The protein binding to the flhA probe and generating the observed mobility shift was either FleQ or some other protein that needed the FleQ activator for its synthesis. To address this, the GRA was repeated using affinity-purified MBP-FleQ and MBP proteins (Fig. 1B and 2), where FleQ was expressed as a fusion protein with MBP in E. coli. MBP-FleQ retarded the flhA promoter-containing fragment, whereas MBP alone did not generate a shift, confirming direct binding of MBP-FleQ to the flhA probe (Fig. 1B). The extent of retardation was directly proportional to the amount of MBP-FleQ included in the assay. The ability of the cold flhA fragment to compete with the probe-FleQ complex and the inability of nonspecific DNA (herring sperm DNA) to compete with the same confirmed that the binding of MBP-FleQ to the flhA probe was specific (Fig. 1C).
FIG. 1.
GRA with the flhA promoter probe. (A) Lysate from PAK-Q harboring either pMMB67EH (vector control, lane 2) or pMMBQ (expressing FleQ, lane 3). The free probe was electrophoresed in lane 1. (B) Purified MBP-FleQ and MBP proteins. Lane 1 depicts the free probe, and lanes 2 to 4 and lanes 5 to 7 contain 1.1, 2.2, and 4.4 μg of MBP-FleQ and MBP, respectively. +, present; −, absent. (C) MBP-FleQ and cold flhA promoter or herring sperm DNA. Lanes 1 to 4 contain 5.6 μg of MBP-FleQ and cold flhA promoter fragment (lanes 2 and 3, 50 and 200 ng, respectively) or herring sperm DNA (lane 4, 1 μg). The arrow and arrowhead indicate the probe-FleQ complex and the free probe, respectively.
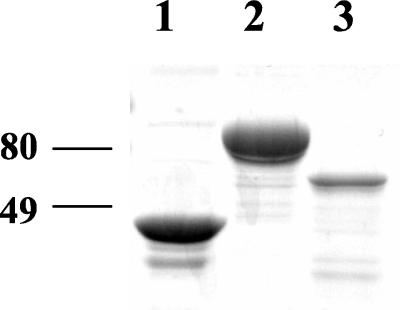
FIG. 2.
SDS-polyacrylamide gel electrophoresis profile of the purified preparations of MBP (∼3 μg), MBP-FleQ (∼5.6 μg), and MBP-FleN (∼400 ng) following Coomassie blue staining. The molecular mass (in kDa) markers are shown.
FleN does not inhibit FleQ binding to DNA.
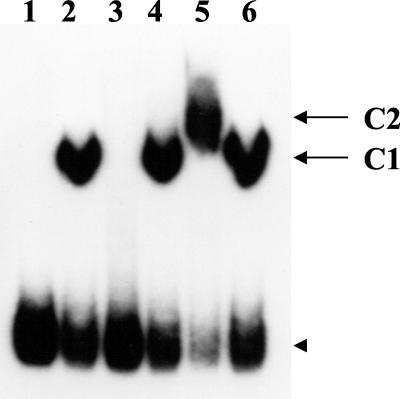
We next examined whether purified MBP-FleN inhibited the binding of purified MBP-FleQ to the flhA promoter fragment. If MBP-FleN competed with DNA to bind to MBP-FleQ, with increasing amounts of MBP-FleN there would be an accompanying increase in the availability of unbound probe. Alternatively, when interacting with MBP-FleQ, if MBP-FleN still allowed MBP-FleQ to bind to the probe, the probe–MBP-FleQ–MBP-FleN complex would be expected to be retarded in its mobility compared to the probe–MBP-FleQ complex. MBP-FleN was incubated at room temperature either with or without MBP-FleQ in the binding reaction for 30 min in a total volume of 25 μl (Fig. 3). MBP-FleN alone did not bind to the flhA probe, ruling out the possibility of FleN binding to DNA directly. The probe-protein complex in the reaction containing both MBP-FleQ and 4.2 μg of MBP-FleN (Fig. 3, lane 5) displayed a slower mobility compared to the probe–MBP-FleQ complex (Fig. 3, lane 2). One possible explanation for the observed difference in mobility is the formation of a probe–MBP-FleQ–MBP-FleN tripartite complex which is larger than the probe–MBP-FleQ bipartite complex. The mobility of the probe-protein complex in the reaction containing both MBP-FleQ and MBP (Fig. 3, lane 6) was comparable to the probe–MBP-FleQ complex, indicating that FleN, not MBP, in MBP-FleN caused the difference in the mobility (Fig. 3, lane 5). Inclusion of smaller amounts of MBP-FleN (800 ng) (Fig. 3, lane 4) did not exhibit a similar retardation, which was probably due to the insufficient amount of FleN available to complex with FleQ. Based on the in vitro results presented here, it is apparent that FleN does not inhibit FleQ from binding to DNA, thereby allowing the FleN-FleQ interacting complex to bind to DNA. The possibility of FleN modifying FleQ, and thereby influencing its transcriptional activator functions, cannot be ruled out at this stage.
FIG. 3.
Assessment of the influence of FleN on the FleQ-flhA promoter complex by GRA. The mobility profiles of the flhA probe alone (lane 1) and when incubated with either MBP-FleQ (5.1 μg, lane 2), MBP-FleN (4.2 μg, lane 3), MBP-FleQ (5.1 μg) plus MBP-FleN (800 ng) (lane 4), MBP-FleQ (5.1 μg) plus MBP-FleN (4.2 μg) (lane 5), or MBP-FleQ (5.1 μg) plus MBP (4.2 μg) (lane 6) are shown. C1 and C2 indicate the probe–MBP-FleQ and probe–MBP-FleQ–MBP-FleN complexes, respectively. The arrowhead indicates the mobility of the free probe in all of the lanes.
Conformation of FleN is central to its interaction with FleQ.
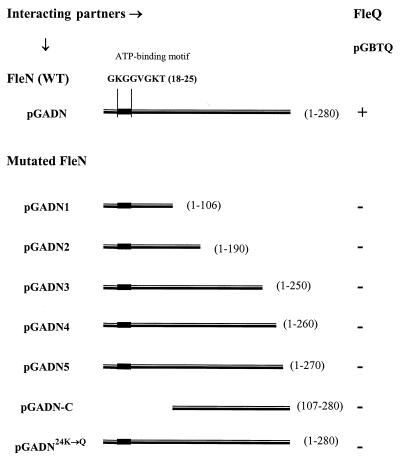
The deduced amino acid sequence of FleN has 280 residues. In order to map the interacting domain of FleN, a series of FleN C-terminal deletion constructs, namely, pGADN1, pGADN2, pGADN3, pGADN4, and pGADN5, were tested in interaction studies with pGBTQ (GAL4BD-FleQ) (Fig. 4). As none of them exhibited a positive β-galactosidase filter assay or His+ phenotype, we deduced that none of them interacted with FleQ. As pGADN5 (FleN containing amino acids 1 to 270), which contained the smallest deletion (the last 10 amino acids), did not interact, it indicated that the interacting domain probably mapped in that region. To test this hypothesis, an N-terminally truncated construct (pGADN-C) with an intact C terminus (FleN containing amino acids 107 to 280) was tested. The absence of an interaction (Fig. 4) disproved the hypothesis, suggesting that in the yeast two-hybrid system, the overall conformation of FleN, rather than a domain structure that could function in isolation, probably determined its interaction with FleQ.
FIG. 4.
Two-hybrid assay to determine the interaction between mutated FleN constructs and FleQ. Symbols: +, positive β-galactosidase filter assay and His+ phenotype; −, negative β-galactosidase filter assay and His− phenotype.
FleN has a putative ATP/GTP binding motif at its N terminus (Fig. 4), referred to as the A consensus sequence or the P loop (17, 29). An appreciable proportion of proteins that bind to ATP or GTP share this glycine-rich conserved-sequence motif. It typically forms a flexible loop between a beta strand and an alpha helix, which interacts with one of the phosphate groups of the nucleotide. In FleN, this motif maps between a predicted beta strand and alpha helix, indicating possible ATP/GTP binding. Mutagenesis of this motif in MinD (K→Q), an ATPase involved in the inhibition of cell division in E. coli, abrogated MinD activity (7). To examine the effects of a similar mutagenesis in FleN on its interaction with FleQ, pGADN24K→Q (AAG→CAG) was generated and cotransformed with pGBTQ into the yeast strain Y190. Absence of a positive β-galactosidase filter assay and His+ phenotype indicated that the FleN24K→G fusion did not interact with the FleQ fusion (Fig. 4), thus indicating that the ATP/GTP binding motif of FleN was essential for interacting with FleQ in vivo in yeast. We speculate that binding of ATP/GTP probably facilitates the correct folding of FleN into a conformation that promotes interaction with FleQ.
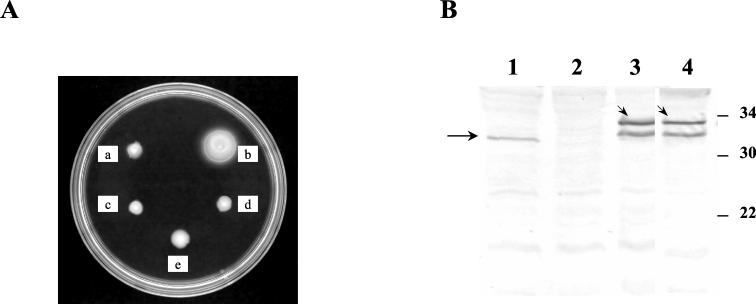
To assess whether the overall conformation of FleN and its nucleotide binding motif were essential for the functioning of FleN in the P. aeruginosa milieu, three FleN mutants with either an N-terminal truncation, a C-terminal truncation, or a mutated ATP binding motif were studied in PAK-N. From earlier studies (6), we were aware that the restoration of motility was sensitive to the amounts of FleN. Excessive FleN was detrimental to flagellar biogenesis. The expression of FleN (His-FleN) from the T7 promoter of pET-fleN without the T7 RNA polymerase in PAK-N restores motility and restricts the number of flagella to a single polar flagellum. Therefore, the same vector background (pET15BVP) was chosen to insert fleN regions present in pGADN1 (amino acids 1 to 106), pGADN-C (amino acids 107 to 280), and pGADN24K→Q as PCR products, yielding pET-fleNΔC (C terminus deleted), pET-fleNΔN (N terminus deleted), and pET-fleN24K→Q (nucleotide binding motif mutated), respectively. These plasmids were electroporated into PAK-N and assessed for the ability to complement the fleN mutation. None of them restored motility (Fig. 5A). To examine the stable expression of the FleN mutant proteins from the plasmid, Western analysis using polyclonal anti-FleN antibodies was performed (Fig. 5B). The amount of expression of FleN24K→Q from pET-fleN24K→Q was comparable to that of FleN from pET-fleN, denoting that FleN24K→Q was unable to complement the motility defect in the FleN mutant probably due to its inability to functionally interact with FleQ. The expression of truncated FleN from pET-fleNΔC and pET-fleNΔN was not detectable under the same conditions. The same plasmids directed the detectable expression of the truncated FleN proteins when introduced into P. aeruginosa (PAO1/T7) and E. coli (BL21 pLysS) strains carrying the T7 RNA polymerase gene (data not shown), indicating that the plasmid constructs used were correct. These data suggest that the truncated FleN proteins FleNΔ107–280 and FleNΔ1–106 either are too poorly expressed in the absence of T7 RNA polymerase in PAK-N to allow immunodetection or are unstable.
FIG. 5.
(A) Comparison of the motility phenotypes of PAK-N containing pET15bVP (a, vector control), pET-fleN (b, wild-type FleN), pET-fleNΔC (c, FleN containing amino acids 1 to 106), pET-fleNΔN (d, FleN containing amino acids 107 to 280), and pET-fleN24K→Q (e, FleN24K→Q). (B) Western analysis of wild-type PAK and PAK-N strains with anti-FleN polyclonal antibody (1:2,000). Lane 1, wild-type PAK; lane 2, PAK-N (pET15bVP); lane3, PAK-N (pET-fleN) and lane 4, PAK-N (pET-fleN24K→Q). The arrow indicates FleN in wild-type PAK (lane 1), and the arrowheads indicate His-FleN (lane 3) and His-FleN24K→Q (lane 4) expressed from the respective plasmids in PAK-N. The molecular mass (in kDa) markers are indicated.
Since the expression of the truncated FleN proteins from the pET constructs in PAK-N was not optimal for detection, in order to allow assessment of the respective truncations in complementing the fleN mutant based on the motility assay, we resorted to measuring the activity of the FleQ-dependent flhA promoter, which is sensitive to the levels of functional FleN. We tested the ability of the smallest truncations in FleN (FleNΔ261–280 and FleNΔ271–280) and FleN24K→Q to inhibit FleQ-dependent transcriptional activation of the flhA promoter. pMMBN4, pMMBN5, and pMMBN24K→Q, expressing FleNΔ261–280, FleNΔ271–280, and FleN24K→Q, respectively, were introduced into PAK-N/placΩA. The flhA promoter activities of these strains were examined in a β-galactosidase assay (Table 4). PAK-N cotransformed with placΩA and pMMBN served as the positive control for downregulation of the flhA promoter by FleN. There was no appreciable downregulation of the promoter activity by either of the truncated FleN constructs (pMMBN4 and pMMBN5) or FleN24K→Q (pMMBN24K→Q), whereas the wild-type FleN-expressing construct of pMMBN downregulated the activity of the same promoter. The level of expression of each of the truncated FleN proteins (FleNΔ261–280 and FleNΔ271–280) and FleN24K→Q from the respective plasmids was comparable to that of wild-type FleN from pMMBN (data not shown). Thus, the truncated FleN proteins and FleN24K→Q tested here are not competent to inhibit FleQ dependent transcriptional activation. This incompetence could be attributed to their inabilities to interact with FleQ, as observed earlier in the yeast two-hybrid system.
TABLE 4.
Comparison of wild-type and mutated FleN proteins in their ability to downregulate the flhA promoter (placΩA) activity in a β-galactosidase reporter assay
| Plasmid | (protein expressed) | β-galactosidase activity (mean ± SD) (Miller units) |
|---|---|---|
| pMMBa | 8,422 ± 107 | |
| pMMBN | (FleN) | 1,123 ± 84 |
| pMMBN4 | (FleNΔ261–280) | 8,590 ± 56 |
| pMMBN5 | (FleNΔ271–280) | 8,256 ± 67 |
| pMMBN24K→Q | (FleN24K→Q) | 7,198 ± 92 |
Vector control, no protein expressed.
DISCUSSION
The general mechanism of activation for EBPs involves a modification in their N termini through phosphorylation at their phosphoacceptor domains (17). On the other hand, certain EBPs (activator), e.g., NifA, in the γ subdivision of the Proteobacteria, which lack the N-terminal domain for modification, remain constitutively active and are modulated through inhibition of their activities when bound to another protein, e.g., the antiactivator NifL (4, 9, 18). The absence of both a typical phosphoacceptor domain in FleQ and an accompanying sensor kinase gene favors the modulation of FleQ activity by an antiactivator rather than by phosphorylation and dephosphorylation. FleN appeared to be the ideal candidate for an antiactivator of FleQ because the absence of FleN resulted in the upregulation of FleQ-dependent promoters (7) including the flhA promoter (this study), and the overexpression of FleN downregulated the FleQ-dependent flhA promoter as observed in this study (Table 2).
A positive assay for interaction between FleN and FleQ in the yeast two-hybrid system suggested direct protein-protein interactions between FleN and FleQ in regulating the flagellar number of P. aeruginosa. In order to map the interacting domain of FleN, interaction studies of yeast with six truncated constructs (FleNΔ107–280, FleNΔ191–280, FleNΔ251–280, FleNΔ261–280, FleNΔ271–280, and FleNΔ1–106) and one mutated construct (ATP/GTP binding motif, FleN24K→Q) of FleN were conducted. Inability of the truncated and the mutated constructs to interact with FleQ in the yeast two-hybrid system (Fig. 4) suggested that the interacting domain in FleN was not restricted to the N or C terminus.
In P. aeruginosa, three of the above described mutated FleN proteins (FleNΔ261–280, FleNΔ271–280, and FleN24K→Q), when tested in β-galactosidase assays (Table 4), failed to downregulate FleQ-dependent transcriptional activation (flhA promoter), whereas wild-type FleN succeeded. Thus, interaction of FleN and FleQ correlates with FleN-dependent inhibition of FleQ activity. It is likely that the conformation of FleN and its ability to bind to ATP/GTP are important for interacting with FleQ in vivo. This is unlike NifL, where the C-terminal domain is sufficient to interact with NifA and inhibit its activity (20). Recently, NifL of Azotobacter vinelandii was reported to be competent in inhibiting NifA via a concerted mechanism in which DNA binding, catalytic activity, and potential interaction with the RNA polymerase were controlled by NifL in order to prevent transcriptional activation under detrimental environmental conditions (4).
FleQ is predicted to bind to UAS of FleQ-dependent promoters by virtue of its C-terminal H-T-H motif. Purified MBP-FleQ expressed in E. coli was competent to bind to the flhA UAS in vitro, indicating that a posttranslational modification (e.g., phosphorylation) restricted to a P. aeruginosa host was not essential for enabling MBP-FleQ to bind to DNA. To function as an antiactivator of FleQ, FleN could either be inhibiting open complex formation at FleQ-dependent promoters or preventing efficient DNA binding at UAS. Alternatively, it could be a concerted inhibition of both the processes leading to a cumulative synergistic effect as seen in the NifA/NifL pair (4). We chose to examine the effect of FleN (if any) on the ability of FleQ to bind to DNA utilizing the flhA promoter probe as a representative. Inclusion of MBP-FleN in a GRA binding reaction with MBP-FleQ indicated that the presence of MBP-FleN apparently did not abrogate or reduce MBP-FleQ-DNA complex formation (Fig. 3). It instead resulted in the formation of a larger complex presumably consisting of MBP-FleQ, MBP-FleN, and the flhA promoter probe owing to the interaction of FleN with FleQ. These results suggest that FleN does not inhibit FleQ from binding to the DNA enhancer elements in vitro. The exact mechanism of FleN-imposed downregulation of FleQ through FleN-FleQ interaction remains to be uncovered.
As FleN is an antiactivator of FleQ, one would expect its expression to be regulated, rather than constitutive, to allow balanced synthesis of the two proteins. Unlike the coordinated synthesis of NifA and NifL from one operon in Klebsiella pneumoniae (8, 9), FleQ and FleN do not belong to the same operon in P. aeruginosa (1, 6, 27). Regulation of fleN transcription could serve as an alternative mechanism for achieving a balanced level of FleN synthesis. As analysis of the immediate upstream sequences of fleN did not reveal a putative promoter (6), we believe fleN is part of the flhF fleN operon which may be cotranscribed from the promoter upstream of flhF (Arora, unpublished; Dasgupta, unpublished), a gene responsible for the polar placement of flagella (22). In P. aeruginosa, the flhF fleN promoter is positively regulated by RpoN (ς54) and FleQ (Arora, unpublished), indicating that FleQ apparently drives the synthesis of its own antiactivator, FleN. This suggests that under physiological conditions, FleN and FleQ could be envisaged to utilize a feedback mechanism to regulate the activities of one another and thereby control (i) flagellar number and (ii) the unnecessary continuing synthesis of early flagellar components in P. aeruginosa. The interaction of FleN with FleQ thus serves as a major regulatory checkpoint in controlling the synthesis of the flagellar components.
ACKNOWLEDGMENTS
We thank M. S. Swanson for providing the yeast strain and plasmids used in the yeast two-hybrid system and S. K. Arora for helpful discussion during preparation of the manuscript.
This work was supported by NIH grant AI45014 to R.R.
REFERENCES
- 1.Arora S K, Ritchings B W, Almira E C, Lory S, Ramphal R. A transcriptional activator, FleQ, regulates mucin adhesion and flagellar gene expression in Pseudomonas aeruginosa in a cascade manner. J Bacteriol. 1997;179:5574–5581. doi: 10.1128/jb.179.17.5574-5581.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arora S K, Ritchings B W, Almira E C, Lory S, Ramphal R. The Pseudomonas aeruginosa flagellar cap protein, FliD, is responsible for mucin adhesion. Infect Immun. 1998;66:1000–1007. doi: 10.1128/iai.66.3.1000-1007.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons, Inc.; 1995. [Google Scholar]
- 4.Barrett J, Ray P, Sobczyk A, Little R, Dixon R. Concerted inhibition of the transcriptional activation functions of the enhancer-binding protein NIFA by the anti-activator NIFL. Mol Microbiol. 2001;39:480–494. doi: 10.1046/j.1365-2958.2001.02243.x. [DOI] [PubMed] [Google Scholar]
- 5.Bourret R B, Borkovich K A, Simon M I. Signal transduction pathways involving protein phosphorylation in prokaryotes. Annu Rev Biochem. 1991;60:401–411. doi: 10.1146/annurev.bi.60.070191.002153. [DOI] [PubMed] [Google Scholar]
- 6.Dasgupta N, Arora S K, Ramphal R. fleN, a gene that regulates flagellar number in Pseudomonas aeruginosa. J Bacteriol. 2000;182:357–364. doi: 10.1128/jb.182.2.357-364.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.de Boer P A J, Crossley R E, Hand A R, Rothfield L I. The MinD protein is a membrane ATPase required for the correct placement of the Escherichia coli division site. EMBO J. 1991;10:4371–4380. doi: 10.1002/j.1460-2075.1991.tb05015.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Govantes F, Andujar E, Santero E. Mechanism of translational coupling in the nifLA operon of Klebsiella pneumoniae. EMBO J. 1998;17:2368–2377. doi: 10.1093/emboj/17.8.2368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Govantes F, Molina-Lopez J A, Santero E. Mechanism of coordinated synthesis of the antagonistic regulatory proteins NifL and NifA of Klebsiella pneumoniae. J Bacteriol. 1996;178:6817–6823. doi: 10.1128/jb.178.23.6817-6823.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Henderson N, Austin S A, Dixon R A. Role of metal ions in negative regulation of nitrogen fixation by the nifL gene product from Klebsiella pneumoniae. Mol Gen Genet. 1989;216:484–491. [Google Scholar]
- 11.Ishimoto K S, Lory S. Formation of pilin in Pseudomonas aeruginosa requires the alternative sigma factor (RpoN) of RNA polymerase. Proc Natl Acad Sci USA. 1989;86:1954–1957. doi: 10.1073/pnas.86.6.1954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kirsch M L, Carpenter P B, Ordal G W. A putative ATP-binding protein from the che/fla locus of Bacillus subtilis. DNA Sequence. 1994;4:217–275. doi: 10.3109/10425179409020852. [DOI] [PubMed] [Google Scholar]
- 13.Lee H-S, Narberhaus F, Kustu S. In vitro activity of NifL, a signal transduction protein for biological nitrogen fixation. J Bacteriol. 1993;175:7683–7688. doi: 10.1128/jb.175.23.7683-7688.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lei S, Pulakat L, Gavini N. Genetic analysis of nif regulatory genes by utilizing the yeast two-hybrid system detected formation of a NifL-NifA complex that is implicated in regulated expression of nif genes. J Bacteriol. 1999;181:6535–6539. doi: 10.1128/jb.181.20.6535-6539.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1972. pp. 352–355. [Google Scholar]
- 16.Moens S, Vanderleyden J. Functions of bacterial flagella. Crit Rev Microbiol. 1996;22:67–100. doi: 10.3109/10408419609106456. [DOI] [PubMed] [Google Scholar]
- 17.Moller W, Amons R. Phosphate-binding sequences in nucleotide-binding proteins. FEBS Lett. 1985;186:1–7. doi: 10.1016/0014-5793(85)81326-0. [DOI] [PubMed] [Google Scholar]
- 18.Money T, Jones T, Dixon R, Austin S. Isolation and properties of the complex between the enhancer binding protein NIFA and the sensor NIFL. J Bacteriol. 1999;181:4461–4468. doi: 10.1128/jb.181.15.4461-4468.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Morett E, Segovia L. The sigma 54 bacterial enhancer-binding protein family: mechanism of action and phylogenetic relationship of their functional domains. J Bacteriol. 1993;175:6067–6074. doi: 10.1128/jb.175.19.6067-6074.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Narberhaus F, Lee H-S, Schmitz R A, He L, Kustu S. The C-terminal domain of NifL is sufficient to inhibit NifA activity. J Bacteriol. 1995;177:5078–5087. doi: 10.1128/jb.177.17.5078-5087.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ottemann K M, Miller J F. Roles of motility in bacterial-host interactions. Mol Microbiol. 1997;24:1109–1117. doi: 10.1046/j.1365-2958.1997.4281787.x. [DOI] [PubMed] [Google Scholar]
- 22.Pandza S, Baetens M, Park C H, Au T, Keyman M, Matin A. The G-protein FlhF has a role in polar flagellar placement and general stress response induction in Pseudomonas putida. Mol Microbiol. 2000;36:414–423. doi: 10.1046/j.1365-2958.2000.01859.x. [DOI] [PubMed] [Google Scholar]
- 23.Ritchings B W, Almira E C, Lory S, Ramphal R. Cloning and phenotypic characterization of fleS and fleR, new response regulators of Pseudomonas aeruginosa which regulate motility and adhesion to mucin. Infect Immun. 1995;63:4868–4876. doi: 10.1128/iai.63.12.4868-4876.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 25.Schmitz R, He L, Kustu S. Iron is required to relieve inhibitory effects of NifL on transcriptional activation by NifA in Klebsiella pneumoniae. J Bacteriol. 1996;178:4679–4687. doi: 10.1128/jb.178.15.4679-4687.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Starnbach M N, Lory S. The fliA (rpoF) gene of Pseudomonas aeruginosa encodes an alternative sigma factor required for flagellin synthesis. Mol Microbiol. 1992;6:459–469. doi: 10.1111/j.1365-2958.1992.tb01490.x. [DOI] [PubMed] [Google Scholar]
- 27.Stover C K, Pham X Q, Erwin A L, Mizoguchi S D, Warrener P, Hickey M J, Brinkman F S, Hufnagle W O, Kowalik D J, Lagrou M, Garber R L, Goltry L, Tolentino E, Westbrock-Wadman S, Yuan Y, Brody L L, Coulter S N, Folger K R, Kas A, Larbig K, Lim R, Smith K, Spencer D, Wong G K, Wu Z, Paulsen I T. Complete genome sequence of Pseudomonas aeruginosa PA01, an opportunistic pathogen. Nature. 2000;406:959–964. doi: 10.1038/35023079. [DOI] [PubMed] [Google Scholar]
- 28.Vuillard L, Rabilloud T, Goldberg M E. Interactions of non-detergent sulfobetaines with early folding intermediates facilitate in vitro protein renaturation. Eur J Biochem. 1998;256:128–135. doi: 10.1046/j.1432-1327.1998.2560128.x. [DOI] [PubMed] [Google Scholar]
- 29.Walker J E, Saraste M, Runswick M J, Gay N J. Distantly related sequences in the alpha- and beta-subunits of ATP synthase, myosin, kinases and other ATP-requiring enzymes and a common nucleotide binding fold. EMBO J. 1982;1:945–951. doi: 10.1002/j.1460-2075.1982.tb01276.x. [DOI] [PMC free article] [PubMed] [Google Scholar]