Abstract
Proper spindle orientation is essential for cell fate determination and tissue morphogenesis. Recently, accumulating studies have elucidated several factors that regulate spindle orientation, including geometric, internal and external cues. Abnormality in these factors generally leads to defects in the physiological functions of various organs and the development of severe diseases. Herein, we first review models that are commonly used for studying spindle orientation. We then review a conservative heterotrimeric complex critically involved in spindle orientation regulation in different models. Finally, we summarize some cues that affect spindle orientation and explore whether we can establish a model that precisely elucidates the effects of spindle orientation without interfusing other spindle functions. We aim to summarize current models used in spindle orientation studies and discuss whether we can build a model that disturbs spindle orientation alone. This can substantially improve our understanding of how spindle orientation is regulated and provide insights to investigate this complex event.
Keywords: spindle, spindle orientation, spindle orientation model
1. INTRODUCTION TO SPINDLE ORIENTATION
The development of multicellular organisms begins post‐fertilization, when rapid cell division occurs in the zygote. The division process is associated with the emergence of diverse cellular functions and assembly of three‐dimensional tissue structures, which rely partially on the orientation of spindle fibres. 1 , 2 The directionality of appropriate cell division is established by the spindle orientation, which affects the precise tissue architecture of an organism. 3 , 4 , 5 Spindle misorientation results in various diseases including lissencephaly, 6 Huntington's disease 7 and some cancers. 8 , 9 hence, the study of spindle orientation will aid in understanding the connection between organismal development and human diseases.
Microtubule remodelling occurs during the formation of the specialized bipolar structure of the spindle fibres. Chromosomes are attached to the spindle microtubules at the kinetochore, which appears as a bridge between the poles. The astral microtubules interact with cortical proteins linking the spindle poles to the cell cortex. Despite research progress in regulating spindle orientation in the past decades, the lack of a suitable universal model has been a key limitation in the study of spindle orientation, suggesting a need to develop an appropriate model. This review presents several causes of spindle misorientation and discusses the possible solutions.
2. MODELS FOR STUDYING SPINDLE ORIENTATION
A correlation between the orientation of the division axis and cell fate has been discovered in Drosophila and Caenorhabditis elegans 10 , 11 , 12 and has subsequently been studied in different species. 13 First, budding yeast was used as a simple system to study asymmetrical spindle polarity 14 (Figure 1A). In budding yeast, asymmetric targeting of spindle poles to the mother and bud cell compartments orients the mitotic spindle along the mother‐bud axis. This is due to intrinsic functional asymmetry, which generates two cells with different fates. Second, the asymmetric zygotic division and differentiation during early embryogenesis have been investigated in the nervous system of flies 15 (Figure 1B). Third, mouse skin progenitors, mouse and chick neuroepithelial cells, and fish epiblast cells have been employed to explore the proliferation and differentiation of epithelial cells 16 , 17 , 18 (Figure 1C–F). These in vivo models are important in assessing new regulators and cellular processes associated with development.
FIGURE 1.
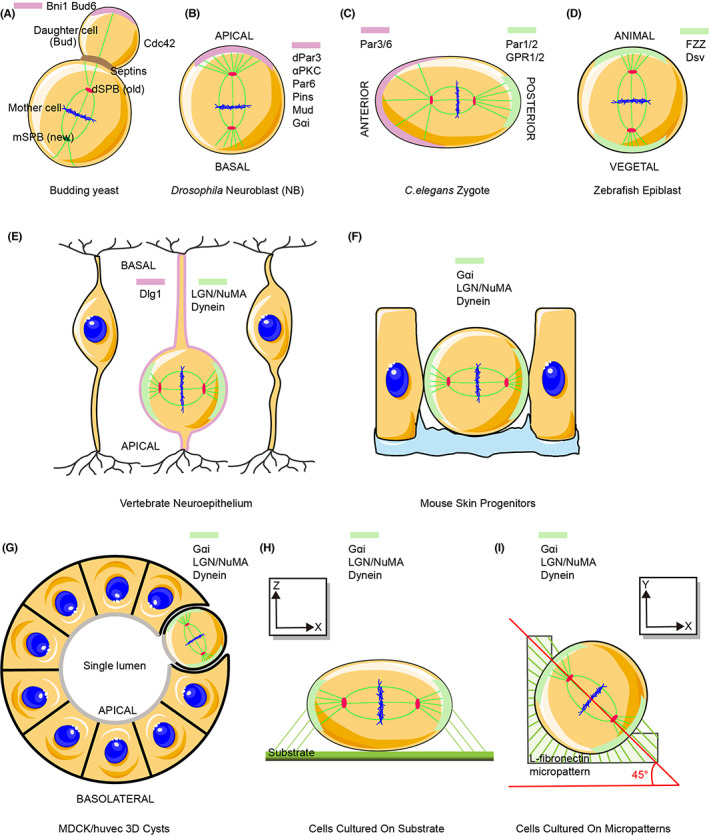
Frequently used models for studying spindle orientation in vivo and in vitro. In vivo models to evaluate spindle orientation in budding yeast (A), Drosophila neuroblasts (B), C. elegans zygote (C), zebrafish epiblasts (D), vertebrate neuroepithelium (E) and mouse skin progenitors (F). In vitro spindle orientation models of cultured MDCK/HUVECs in Matrigel for real‐time observation of intracellular changes using microscopy (G), cells on fibronectin substrate (H) or micropatterns (I). Conserved polarized factors with different names of homologues in model organisms. Light pink or green represents relevant polar factors in different models. Their asymmetrical spatial distribution at the cell pole will generate two cells with different fates
However, in vitro models have more advantages in observing intracellular changes using microscopic analysis. The frequently used in vitro models include cells cultured on fibronectin‐coated plates on micropatterns, or using three‐dimensional (3D) culture methods (Figure 1H,I). Among these, cells cultured using 3D systems have been used to study epithelial morphogenesis and lumen formation (Figure 1G). For example, cell polarity with spindle orientation has been evaluated using Madin–Darby canine kidney cells 19 or human umbilical vein endothelial cells grown in Matrigel. 20 Defects in spindle orientation lead to the formation of cysts with multiple lumina and inhibit angiogenesis. 21 , 22 , 23 Furthermore, using cells cultured on fibronectin or micropatterns, changes in spindle orientation have been identified by assessing the distribution of actin retraction fibres and astral microtubules. 24 , 25 These findings are critical for the diagnosis and treatment of relative diseases.
3. CONSERVATIVE HETEROTRIMERIC COMPLEX, GɑI‐LGN‐NUMA, CONTROLS SPINDLE ORIENTATION IN DIFFERENT MODELS
A conserved heterotrimeric complex, Gɑi‐LGN‐NuMA, is reportedly involved in regulating spindle orientation both in vivo and in vitro. 26 , 27 , 28 In this complex, LGN is an adaptor molecule of Gɑi. It consists of three main domains: N‐terminal TPR domain, central ‘linker’ domain and C‐terminal GPR domain. 29 During mitosis, Gɑi is anchored to the membrane by its membrane‐anchored subunits and interacts with the GPR domain of LGN. N‐terminal TPR domain mediates interactions with multiple binding proteins such as NuMA. A functionally unknown ‘linker’ domain connects the two parts together. 30 Consequently, this complex can locate a specific region of the subcortical domain and recruit the minus‐end‐directed microtubule motor protein, dynein, directly. Dynein movement along the astral microtubule can generate a pulling force on the spindle pole, orienting the spindle at an appropriate plane and position 31 (Figure 2).
FIGURE 2.
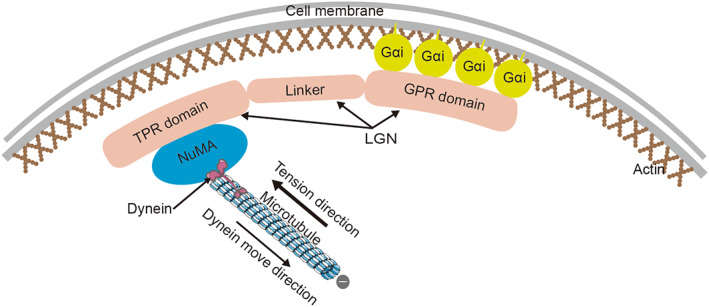
Conservative heterotrimeric complex, Gɑi‐LGN‐NuMA, in spindle orientation controlling mechanism. Gɑi is anchored to the membrane at one end and interacts with the GPR domain of LGN. The TPR domain of LGN mediates the interactions with multiple binding proteins such as NuMA. Dynein directly interacts with NuMA and moves along with the astral microtubule towards the minus end. Therefore, an appropriate pulling force is generated in the opposite direction, which is necessary for proper spindle orientation
Besides, the coiled‐coil domain of NuMA has been verified as a hairpin that can interact with LGN, dynein and microtubules simultaneously. 32 Cortical localized NuMA is also frequently observed in dividing cells, 33 which require NuMA phosphorylation. 34 Similarly, Gɑi subunits localizing to the plasma membrane are myristoylated. The modified Gɑi protein attaches to the cortex providing an anchor to the TPR domain of the LGN complex. 35 Remarkably, this conserved complex is known as Gɑi‐Pins‐Mud in Drosophila and GOA1/GPA16‐GPR1/2‐LIN5 in C. elegans (Table 1).
TABLE 1.
Genes mentioned in this review and their homologues in different model organisms
4. FACTORS REGULATING SPINDLE ORIENTATION
Specialized bipolar structures of spindle fibres are affected by several factors. They can be roughly classified as internal, external and geometric cues. They have a considerable influence on the spindle orientation. However, they eventually affect spindle orientation machinery or cell cortex interactions with astral microtubules. Here, we discuss the different factors in detail.
4.1. Internal cues
The assembly of the spindle orientation machinery in the cell requires an intact actin cortex and normal astral microtubules. The cortex can effectively generate a stable force to organize the spindle at appropriate angles. 31 Latrunculin A or cytochalasin D treatments lead to spindle orientation defects, affecting cell fate. 36 Additionally, remodelling the stiff actin during mitosis can provide sufficient force to pull the spindle orientation machinery. 37 , 38 However, the precise location where this force is generated is not known. Furthermore, abnormal astral microtubules perturb the spindle dynamics and stability, or interaction with cortical proteins, leading to misorientation, irrespective of their defective nucleation/anchoring. 39 , 40 , 41 In addition, some proteins and kinases in the cell can contribute to the formation or stabilization of microtubules, thereby affecting spindle assembly or functions. For example, cylindromatosis is a deubiquitinating enzyme, which directly binds to the microtubules and regulates astral microtubule stability and dynamics via lysine 63‐linked ubiquitin hydrolysis. 42 Polycomb repressive complex 1, a minus‐end kinesin protein of the microtubule, is also important for the proper assembly, dynamics and positioning of the mitotic spindle. 43 , 44
4.2. External cues
Extracellular signals from the cell surface can control spindle orientation. The extracellular matrix (ECM), a structure composed of proteins and polysaccharides secreted by cells, is located on the cell surface or between cells and affects spindle orientation directly or indirectly. 45 , 46 One of its components, β‐integrin, can interact with focal adhesion kinase and talin to regulate spindle alignment. 47 , 48 Besides, β‐integrin knockout mice display random spindle orientation during skin stratification 49 and luminal formation. 50 β‐Integrin has been proved to be a key element in establishing apical‐basal polarity for spindle orientation and the relative molecular location of LGN, NuMA and ɑPKC. 27 Besides β‐integrin, other ECM components such as exopolysaccharides are involved in spindle orientation through sulfation and uronic acid epimerization. 51 During mitosis, JAM‐A control spindle orientation through Cdc42, further regulating cortical dynein localization. 52 Caveolin‐1 can translate the interphase adhesion geometry to mitotic spindle orientation in a RhoA‐dependent manner. 53 During kidney morphogenesis and repair, renal tubular epithelial cells lacking the transmembrane receptor Plexin‐B2 or its semaphorin ligands fail to correctly orient the mitotic spindle, leading to severe defects in epithelial architecture and function. 54 Interestingly, β‐integrin, a transmembrane protein, can interact with the ECM and cortical molecules. Therefore, we speculate that β‐integrin may act by transmitting messages from the ECM to the intracellular cortex, possibly regulating spindle orientation through microtubule‐associated proteins or actin cytoskeleton interaction. Future studies are necessary to verify the difference in the role of β‐integrin between normal cells and spindle misoriented cells.
4.3. Geometric and external force cues
In the past, cells were considered to divide along their longest axis. This is called the ‘Hertwig rule’. It has been indicated that the spindle in a mitotic cell can perceive cell shape changes to realign itself along the longest axis. 55 Continuous remodelling allows sufficient space for the formation of the spindle. In cells cultured on fibronectin or micropatterns, the distribution of actin retraction fibres dictates the orientation of the spindle. 22 Myosin 10 is considered the linker between actin and microtubules in this context. 56 Considering the relationship between myosin and dynein proteins, the formation of the classical structure of the LGN/dynein complexes is a conservative mechanism of spindle orientation. 57 Artificial altering of cell shapes causes chromosome missegregation. 58 Moreover, changes in the spindle angles have been confirmed under external magnetic field actions, 59 indicating that magnetic force can serve as a kind of external force to regulate spindle orientation. However, a recent study revealed that cell division orientation in vivo is not determined by cell shape but rather by local anisotropies in cell mechanics. 13 , 60 Studies have shown that the development of Drosophila wing is not dependent on its shape. 61 Furthermore, tissue tension and non‐interphase cell shape determine cell division, as confirmed in Drosophila follicular epithelium. 62 This tissue tension at compartmental boundaries is actomyosin‐driven tension. 63 , 64 In summary, more external forces affect spindle orientation, which need further evaluation.
5. CONCLUSIONS AND PERSPECTIVE
The findings on the molecular mechanisms that control the orientation of the mitotic spindle reveal that the conservative heterotrimeric complex Gɑi‐LGN‐NuMA regulates spindle orientation in different species. However, the molecular interplay to regulate the recruitment and maintenance of LGN to the cellular cortex is still unknown. The knockdown of LGN or NuMA results only in weak spindle orientation phenotypes, 65 , 66 suggesting an involvement of additional pathways. Additionally, different mechanisms in different tissues contribute to the process of spindle orientation regulation, highlighting the need to establish a universal spindle orientation model to study related issues.
A specialized bipolar spindle orientation machinery plays an irreplaceable role in regulating spindle angles. 67 It relies on the interaction of astral microtubules with cortical proteins to dictate spindle position and orientation. 68 , 69 Therefore, factors concerning spindle morphology and/or behaviour are likely to affect their orientation. 70 , 71 , 72 Moreover, cortical determinants and astral microtubules are not passive participants during this process. They are the core structural components of the spindle orientation machinery. The cortical proteins tune external force transmission into cells to finely regulate the spindle orientation. Understanding cellular sensing for ‘external pressure signal’ and its transmittance into cells is essential. The transmembrane protein β‐integrin is worth investigating to discover a more precise mechanism for regulating spindle orientation and establishing an extracellular controllability model. In addition, most relative proteins interacting with microtubules, especially astral microtubules, have internal cues involved in spindle orientation regulation. Indeed, several proteins perturb spindle orientation by affecting astral microtubules. For example, microtubule plus‐end protein EB1 can stabilize astral microtubules to regulate spindle orientation through phosphorylation. 73 Human microcephaly ASPM protein is a spindle pole‐focusing factor that regulates orientation by affecting the dynamics of astral microtubules. 74 Studying the differences between astral and other microtubules to control microtubule behaviour may be a new method of establishing a spindle orientation model. Meanwhile, protein kinases have been proposed to influence spindle orientation. 75
Aurora‐A kinase can regulate αPKC/Numb cortical polarity and spindle orientation to inhibit neuroblast self‐renewal in Drosophila. 76 In fission yeast, mitogen‐activated protein kinase ensures proper mitotic spindle orientation via the actin checkpoint. 77 αPKC‐mediated phosphorylation of apical Pins controls epithelial spindle orientation. 78
Adenosine‐5′‐monophosphate‐activated protein kinase has been found to regulate mitotic spindle orientation through the phosphorylation of the myosin regulatory light chain. 79 However, kinases contribute to cell signalling and complex life activities. Therefore, setting up a new spindle orientation model affected by kinases and their associated pathway will be useful.
Previous studies have compared cell division in vitro and tissue development in vivo under controlled spindle orientation. 80 , 81 , 82 , 83 , 84 The role of spindle orientation in normal and pathological development and homeostasis has been acknowledged. However, due to the lack of a suitable universal model, differential findings among the models cannot be confirmed. As spindle orientation is poorly understood, future work should aim at summarizing the similarities and differences. Overall, developing a universal spindle orientation model is necessary to study diseases and suggest possible treatments for pathologies caused by spindle misorientation.
AUTHOR CONTRIBUTIONS
Tao Zhong: Software (equal); writing – original draft (lead); writing – review and editing (lead). Xiaoxiao Gongye: Writing – original draft (equal). Minglei Wang: Software (equal); writing – original draft (equal). Jinming Yu: Conceptualization (lead); funding acquisition (supporting); project administration (lead); writing – review and editing (lead).
CONFLICT OF INTEREST
The authors confirm that there are no conflicts of interest.
ACKNOWLEDGEMENTS
This work was supported by a grant from The National Key Research and Development Projects of China (grant no. 2018YFC1312201), Radiation Oncology Innovate Unit, Chinese Academy of Medical Sciences (grant no. 2019RU071), the Academic Promotion Program of Shandong First Medical University (grant no. 2019ZL002) and the Foundation of National Natural Science Foundation of China (grant nos. 81902608, 81972863, 81627901 and 82030082).
Zhong T, Gongye X, Wang M, Yu J. Understanding the underlying mechanisms governing spindle orientation: How far are we from there? J Cell Mol Med. 2022;26:4904‐4910. doi: 10.1111/jcmm.17526
Contributor Information
Tao Zhong, Email: zhongtao@sdu.edu.cn.
Jinming Yu, Email: sdyujinming@163.com.
DATA AVAILABILITY STATEMENT
Data sharing is not applicable as no new data were generated.
REFERENCES
- 1. Godard BG, Heisenberg CP. Cell division and tissue mechanics. Curr Opin Cell Biol. 2019;60:114‐120. [DOI] [PubMed] [Google Scholar]
- 2. Taneja N, Rathbun L, Hehnly H, Burnette DT. The balance between adhesion and contraction during cell division. Curr Opin Cell Biol. 2019;56:45‐52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Zhong T, Wu X, Xie W, et al. ENKD1 promotes epidermal stratification by regulating spindle orientation in basal keratinocytes. Cell Death Differ. 2022. doi: 10.1038/s41418-022-00958-5. Online ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Wu X, Zhou J, Li D. Orientation of the mitotic spindle in blood vessel development. Front Cell Dev Biol. 2020;8:583325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Lechler T, Fuchs E. Asymmetric cell divisions promote stratification and differentiation of mammalian skin. Nature. 2005;437(7056):275‐280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Li HB, Kroll T, Moll J, et al. Spindle misorientation of cerebral and cerebellar progenitors is a mechanistic cause of Megalencephaly. Stem Cell Reports. 2017;9(4):1071‐1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Molina‐Calavita M, Barnat M, Elias S, Aparicio E, Piel M, Humbert S. Mutant huntingtin affects cortical progenitor cell division and development of the mouse neocortex. J Neurosci. 2014;34(30):10034‐10040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Rothenberg ME, Cintra T, Chen EC, et al. Loss of Gpsm2 disrupts stem cell dynamics and mitotic spindle orientation in Normal colon and colon cancer. Gastroenterology. 2014;146(5):S85‐S86. [Google Scholar]
- 9. Quyn AJ, Appleton PL, Carey FA, et al. Spindle orientation bias in gut epithelial stem cell compartments is lost in precancerous tissue. Cell Stem Cell. 2010;6(2):175‐181. [DOI] [PubMed] [Google Scholar]
- 10. Chen CE, Cummings R, Mordovanakis A, et al. Cytokine receptor‐Eb1 interaction couples cell polarity and fate during asymmetric cell division. eLife. 2018;7:e33685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Gomes JE, Encalada SE, Swan KA, Shelton CA, Carter JC, Bowerman B. The maternal gene spn‐4 encodes a predicted RRM protein required for mitotic spindle orientation and cell fate patterning in early C. elegans embryos. Development. 2001;128(21):4301‐4314. [DOI] [PubMed] [Google Scholar]
- 12. Yu W, O'Brien LE, Wang F, Bourne H, Mostov KE, Zegers MM. Hepatocyte growth factor switches orientation of polarity and mode of movement during morphogenesis of multicellular epithelial structures. Mol Biol Cell. 2003;14(2):748‐763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lechler T, Mapelli M. Spindle positioning and its impact on vertebrate tissue architecture and cell fate. Nat Rev Mol Cell Biol. 2021;22(10):691‐708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Garcia‐Rodriguez LJ, Kasciukovic T, Denninger V, Tanaka TU. Aurora B‐INCENP localization at centromeres/inner kinetochores is required for chromosome bi‐orientation in budding yeast. Curr Biol. 2019;29(9):1536‐1544 e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Zhao J, Xin H, Cao L, et al. NtDRP is necessary for accurate zygotic division orientation and differentiation of basal cell lineage toward suspensor formation. New Phytol. 2016;212(3):598‐612. [DOI] [PubMed] [Google Scholar]
- 16. Bhattarai SR, Begum S, Popow R, Ezratty EJ. The ciliary GTPase Arl3 maintains tissue architecture by directing planar spindle orientation during epidermal morphogenesis. Development. 2019;146(9):dev161885. doi: 10.1242/dev.161885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Franco M, Carmena A. Eph signaling controls mitotic spindle orientation and cell proliferation in neuroepithelial cells. J Cell Biol. 2019;218(4):1200‐1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Campinho P, Behrndt M, Ranft J, Risler T, Minc N, Heisenberg CP. Tension‐oriented cell divisions limit anisotropic tissue tension in epithelial spreading during zebrafish epiboly. Nat Cell Biol. 2013;15(12):1405‐1414. [DOI] [PubMed] [Google Scholar]
- 19. Ouyang M, Yu JY, Chen Y, Deng L, Guo CL. Cell‐extracellular matrix interactions in the fluidic phase direct the topology and polarity of self‐organized epithelial structures. Cell Prolif. 2021;54(4):e13014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Montero RB, Vial X, Nguyen DT, et al. bFGF‐containing electrospun gelatin scaffolds with controlled nano‐architectural features for directed angiogenesis. Acta Biomater. 2012;8(5):1778‐1791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Gao L, Yang Z, Hiremath C, et al. Afadin orients cell division to position the tubule lumen in developing renal tubules. Development. 2017;144(19):3511‐3520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hung HF, Hehnly H, Doxsey S. The mother centriole appendage protein Cenexin modulates lumen formation through spindle orientation. Curr Biol. 2016;26(6):793‐801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Gao J, Sun L, Huo L, Liu M, Li D, Zhou J. CYLD regulates angiogenesis by mediating vascular endothelial cell migration. Blood. 2010;115(20):4130‐4137. [DOI] [PubMed] [Google Scholar]
- 24. Malerod L, Le Borgne R, Lie‐Jensen A, et al. Centrosomal ALIX regulates mitotic spindle orientation by modulating astral microtubule dynamics. EMBO J. 2018;37(13):e97741. doi: 10.15252/embj.201797741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Nunes V, Dantas M, Castro D, et al. Centrosome‐nuclear axis repositioning drives the assembly of a bipolar spindle scaffold to ensure mitotic fidelity. Mol Biol Cell. 2020;31(16):1675‐1690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Polverino F, Naso FD, Asteriti IA, et al. The Aurora‐a/TPX2 Axis directs spindle orientation in adherent human cells by regulating NuMA and microtubule stability. Curr Biol. 2021;31(3):658‐667 e5. [DOI] [PubMed] [Google Scholar]
- 27. di Pietro F, Echard A, Morin X. Regulation of mitotic spindle orientation: an integrated view. EMBO Rep. 2016;17(8):1106‐1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Peyre E, Jaouen F, Saadaoui M, et al. A lateral belt of cortical LGN and NuMA guides mitotic spindle movements and planar division in neuroepithelial cells. J Cell Biol. 2011;193(1):141‐154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Carminati M, Gallini S, Pirovano L, Alfieri A, Bisi S, Mapelli M. Concomitant binding of Afadin to LGN and F‐Actin directs planar spindle orientation. Nat Struct Mol Biol. 2016;23(2):155‐163. [DOI] [PubMed] [Google Scholar]
- 30. Culurgioni S, Mari S, Bonetti P, et al. Insc:LGN tetramers promote asymmetric divisions of mammary stem cells. Nat Commun. 2018;9(1):1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kiyomitsu T. The cortical force‐generating machinery: how cortical spindle‐pulling forces are generated. Curr Opin Cell Biol. 2019;60:1‐8. [DOI] [PubMed] [Google Scholar]
- 32. Pirovano L, Culurgioni S, Carminati M, et al. Hexameric NuMA:LGN structures promote multivalent interactions required for planar epithelial divisions. Nat Commun. 2019;10(1):2208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Hueschen CL, Galstyan V, Amouzgar M, Phillips R, Dumont S. Microtubule end‐clustering maintains a steady‐state spindle shape. Curr Biol. 2019;29(4):700‐708 e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Sun M, Jia M, Ren H, et al. NuMA regulates mitotic spindle assembly, structural dynamics and function via phase separation. Nat Commun. 2021;12(1):7157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Bergstralh DT, Dawney NS, St JD. Spindle orientation: a question of complex positioning. Development. 2017;144(7):1137‐1145. [DOI] [PubMed] [Google Scholar]
- 36. Negishi T, Yasuo H. Distinct modes of mitotic spindle orientation align cells in the dorsal midline of ascidian embryos. Dev Biol. 2015;408(1):66‐78. [DOI] [PubMed] [Google Scholar]
- 37. Belukha UK, Plechistova LA. Immediate and remote results of treatment of patients with porphyria cutanea tarda. Vestn Dermatol Venerol. 1977;5:47‐50. [PubMed] [Google Scholar]
- 38. Schimmel L, van der Stoel M, Rianna C, et al. Stiffness‐induced endothelial DLC‐1 expression forces leukocyte spreading through stabilization of the ICAM‐1 Adhesome. Cell Rep. 2018;24(12):3115‐3124. [DOI] [PubMed] [Google Scholar]
- 39. Gai M, Bianchi FT, Vagnoni C, et al. ASPM and CITK regulate spindle orientation by affecting the dynamics of astral microtubules. EMBO Rep. 2016;17(10):1396‐1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Gambarotto D, Pennetier C, Ryniawec JM, et al. Plk4 regulates centriole asymmetry and spindle orientation in neural stem cells. Dev Cell. 2019;50(1):11‐24 e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Yi P, Goshima G. Microtubule nucleation and organization without centrosomes. Curr Opin Plant Biol. 2018;46:1‐7. [DOI] [PubMed] [Google Scholar]
- 42. Yang Y, Liu M, Li D, et al. CYLD regulates spindle orientation by stabilizing astral microtubules and promoting dishevelled‐NuMA‐dynein/dynactin complex formation. Proc Natl Acad Sci U S A. 2014;111(6):2158‐2163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Shrestha S, Wilmeth LJ, Eyer J, Shuster CB. PRC1 controls spindle polarization and recruitment of cytokinetic factors during monopolar cytokinesis. Mol Biol Cell. 2012;23(7):1196‐1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Mullen TJ, Wolff ID, Wignall SM. Minus‐end kinesins and SPD‐1(PRC1) provide complementary mechanisms to organize acentriolar C. elegans oocyte spindles. Mol Biol Cell. 2016;27:3122‐3131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Zhang JT, Nie JQ, Muhlstadt M, Gallagher H, Pullig O, Jandt KD. Stable extracellular matrix protein patterns guide the orientation of osteoblast‐like cells. Adv Funct Mater. 2011;21(21):4079‐4087. [Google Scholar]
- 46. Tuncay H, Ebnet K. Cell adhesion molecule control of planar spindle orientation. Cell Mol Life Sci. 2016;73(6):1195‐1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Petridou NI, Skourides PA. FAK transduces extracellular forces that orient the mitotic spindle and control tissue morphogenesis. Nat Commun. 2014;5:5240. [DOI] [PubMed] [Google Scholar]
- 48. Cota CD, Davidson B. Mitotic membrane turnover coordinates differential induction of the heart progenitor lineage. Dev Cell. 2015;34(5):505‐519. [DOI] [PubMed] [Google Scholar]
- 49. Simpson CL, Patel DM, Green KJ. Deconstructing the skin: cytoarchitectural determinants of epidermal morphogenesis. Nat Rev Mol Cell Biol. 2011;12(9):565‐580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Zovein AC, Luque A, Turlo KA, et al. Beta 1 integrin establishes endothelial cell polarity and arteriolar lumen formation via a Par3‐dependent mechanism. Dev Cell. 2010;18(1):39‐51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Chung H, Multhaupt HAB, Oh ES, Couchman JR. Minireview: Syndecans and their crucial roles during tissue regeneration. FEBS Lett. 2016;590(15):2408‐2417. [DOI] [PubMed] [Google Scholar]
- 52. Tuncay H, Brinkmann BF, Steinbacher T, et al. JAM‐A regulates cortical dynein localization through Cdc42 to control planar spindle orientation during mitosis. Nat Commun. 2015;6:8128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Matsumura S, Kojidani T, Kamioka Y, et al. Interphase adhesion geometry is transmitted to an internal regulator for spindle orientation via caveolin‐1. Nat Commun. 2016;7:ncomms11858. doi: 10.1038/ncomms11858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Xia J, Swiercz JM, Banon‐Rodriguez I, et al. Semaphorin‐plexin signaling controls mitotic spindle orientation during epithelial morphogenesis and repair. Dev Cell. 2015;33(3):299‐313. [DOI] [PubMed] [Google Scholar]
- 55. Lancaster OM, Baum B. Shaping up to divide: coordinating Actin and microtubule cytoskeletal remodelling during mitosis. Semin Cell Dev Biol. 2014;34:109‐115. [DOI] [PubMed] [Google Scholar]
- 56. Lu Q, Li J, Zhang M. Cargo recognition and cargo‐mediated regulation of unconventional myosins. Acc Chem Res. 2014;47(10):3061‐3070. [DOI] [PubMed] [Google Scholar]
- 57. Kwon M, Bagonis M, Danuser G, Pellman D. Direct microtubule‐binding by Myosin‐10 orients centrosomes toward retraction fibers and subcortical Actin clouds. Dev Cell. 2015;34(3):323‐337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Wassef M, Luscan A, Aflaki S, et al. EZH1/2 function mostly within canonical PRC2 and exhibit proliferation‐dependent redundancy that shapes mutational signatures in cancer. Proc Natl Acad Sci U S A. 2019;116(13):6075‐6080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Zhang L, Hou Y, Li Z, et al. 27 T ultra‐high static magnetic field changes orientation and morphology of mitotic spindles in human cells. elife. 2017;6:e22911. doi: 10.7554/eLife.22911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Dekoninck S, Hannezo E, Sifrim A, et al. Defining the design principles of skin epidermis postnatal growth. Cell. 2020;181(3):604‐620 e22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Zhou Z, Alegot H, Irvine KD. Oriented cell divisions are not required for drosophila wing shape. Curr Biol. 2019;29(5):856‐864 e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Finegan TM, Na D, Cammarota C, et al. Tissue tension and not interphase cell shape determines cell division orientation in the drosophila follicular epithelium. EMBO J. 2019;38(3):e100072. doi: 10.15252/embj.2018100072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Scarpa E, Finet C, Blanchard GB, Sanson B. Actomyosin‐driven tension at compartmental boundaries orients cell division independently of cell geometry in vivo. Dev Cell. 2018;47(6):727‐740 e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Chanet S, Sharan R, Khan Z, Martin AC. Myosin 2‐induced mitotic rounding enables columnar epithelial cells to interpret cortical spindle positioning cues. Curr Biol. 2017;27(21):3350‐3358 e3. [DOI] [PubMed] [Google Scholar]
- 65. Zheng Z, Zhu H, Wan Q, et al. LGN regulates mitotic spindle orientation during epithelial morphogenesis. J Cell Biol. 2010;189(2):275‐288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Chu X, Chen X, Wan Q, Zheng Z, Du Q. Nuclear mitotic apparatus (NuMA) interacts with and regulates Astrin at the mitotic spindle. J Biol Chem. 2016;291(38):20055‐20067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Lew DJ, Burke DJ. The spindle assembly and spindle position checkpoints. Annu Rev Genet. 2003;37:251‐282. [DOI] [PubMed] [Google Scholar]
- 68. Chatterjee S, Som S, Varshney N, Satyadev P, Sanyal K, Paul R. Mechanics of microtubule organizing center clustering and spindle positioning in budding yeast Cryptococcus neoformans . Phys Rev E. 2021;104(3–1):034402. [DOI] [PubMed] [Google Scholar]
- 69. Cowan CR, Hyman AA. Asymmetric cell division in C. elegans: cortical polarity and spindle positioning. Annu Rev Cell Dev Biol. 2004;20:427‐453. [DOI] [PubMed] [Google Scholar]
- 70. Scepanovic G, Fernandez‐Gonzalez R. Oriented cell division: the pull of the pole. Dev Cell. 2018;47(6):686‐687. [DOI] [PubMed] [Google Scholar]
- 71. Barui A, Datta P. Biophysical factors in the regulation of asymmetric division of stem cells. Biol Rev Camb Philos Soc. 2019;94(3):810‐827. [DOI] [PubMed] [Google Scholar]
- 72. Overeem AW, Bryant DM, van Ijzendoorn SCD. Mechanisms of apical‐basal axis orientation and epithelial lumen positioning. Trends Cell Biol. 2015;25(8):476‐485. [DOI] [PubMed] [Google Scholar]
- 73. Luo YG, Ran J, Xie SB, et al. ASK1 controls spindle orientation and positioning by phosphorylating EB1 and stabilizing astral microtubules. Cell Discovery. 2016;2:16033. doi: 10.1038/celldisc.2016.33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Gai M, Bianchi FT, Vagnoni C, et al. ASPM and CITK regulate spindle orientation by affecting the dynamics of astral microtubules (vol 17, pg 1396, 2016). EMBO Rep. 2017;18(10):1870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Varshney N, Som S, Chatterjee S, et al. Spatio‐temporal regulation of nuclear division by Aurora B kinase Ipl1 in Cryptococcus neoformans . PLoS Genet. 2019;15(2):e1007959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Lee C‐Y, Andersen RO, Cabernard C, et al. Drosophila Aurora‐a kinase inhibits neuroblast self‐renewal by regulating aPKC/numb cortical polarity and spindle orientation. Genes Dev. 2006;20(24):3464‐3474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Gachet Y, Tournier S, Millar JB, Hyams JS. A MAP kinase‐dependent Actin checkpoint ensures proper spindle orientation in fission yeast. Nature. 2001;412(6844):352‐355. [DOI] [PubMed] [Google Scholar]
- 78. Hao Y, Du Q, Chen X, et al. Par3 controls epithelial spindle orientation by aPKC‐mediated phosphorylation of apical pins. Curr Biol. 2010;20(20):1809‐1818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Thaiparambil JT, Eggers CM, Marcus AI. AMPK regulates mitotic spindle orientation through phosphorylation of myosin regulatory light chain. Mol Cell Biol. 2012;32:3203‐3217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Wang X, Dong B, Zhang K, et al. E‐cadherin bridges cell polarity and spindle orientation to ensure prostate epithelial integrity and prevent carcinogenesis in vivo. PLoS Genet. 2018;14(8):e1007609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Zhong T, Zhou J. Orientation of the mitotic spindle in the development of tubular organs. J Cell Biochem. 2017;118(7):1630‐1633. [DOI] [PubMed] [Google Scholar]
- 82. Xie W, Yang Y, Gao S, et al. The tumor suppressor CYLD controls epithelial morphogenesis and homeostasis by regulating mitotic spindle behavior and adherens junction assembly. J Genet Genomics. 2017;44(7):343‐353. [DOI] [PubMed] [Google Scholar]
- 83. Tang Z, Hu Y, Wang Z, et al. Mechanical forces program the orientation of cell division during airway tube morphogenesis. Dev Cell. 2018;44(3):313‐325 e5. [DOI] [PubMed] [Google Scholar]
- 84. Sakai D, Dixon J, Dixon MJ, Trainor PA. Mammalian neurogenesis requires treacle‐Plk1 for precise control of spindle orientation, mitotic progression, and maintenance of neural progenitor cells. PLoS Genet. 2012;8(3):e1002566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Vargas E, KP MN, Cortes DB, et al. Spherical spindle shape promotes perpendicular cortical orientation by preventing isometric cortical pulling on both spindle poles during C. elegans female meiosis. Development. 2019;146(20):dev178863. doi: 10.1242/dev.178863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Jankele R, Jelier R, Gonczy P. Physically asymmetric division of the C. elegans zygote ensures invariably successful embryogenesis. eLife. 2021;10:e61714. doi: 10.7554/eLife.61714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Bergstralh DT, Lovegrove HE, Kujawiak I, et al. Pins is not required for spindle orientation in the drosophila wing disc. Development. 2016;143(14):2573‐2581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Hoffmann I. Centrosomes in mitotic spindle assembly and orientation. Curr Opin Struct Biol. 2021;66:193‐198. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data sharing is not applicable as no new data were generated.


