Abstract
Hordeum californicum (H. californicum, 2n=2X=14, HcHc), one of the wild relatives of wheat (Triticum aestivum L.), harbors many desirable genes and is a potential genetic resource for wheat improvement. In this study, an elite line ND646 was selected from a BC4F5 population, which was developed using 60Co-γ irradiated wheat-H. californicum disomic addition line WJ28-1 (DA6Hc) as the donor parent and Ningchun 4 as the recurrent parent. ND646 was identified as a novel wheat-H. californicum 6HcS/6BL translocation line using genomic in situ hybridization (GISH), fluorescence in situ hybridization (FISH), and H. californicum-specific expressed sequence tag (EST) markers. Further evaluation revealed that ND646 had excellent performance in several traits, such as a higher sedimentation value (SV), higher water absorption rate (WAR), and higher hardness index (HI). More importantly, it had more kernels per spike (KPS), a higher grain yields (GY), and good resistance to powdery mildew, leaf rust, and 2,4-D butylate (2,4-D). Its excellent phenotypic performance laid the foundation for further investigation of its genetic architecture and makes ND646 a useful germplasm resource for wheat breeding.
Keywords: Wheat, H. californicum, translocation line, molecular identification, multiple desirable agronomic trait
Introduction
Wheat (Triticum aestivum L.) is one of the most important crops in the world and supplies food to at least one-third of the global population (Yang et al., 2016). However, the narrow genetic basis has resulted in a bottleneck in wheat breeding (Gupta et al., 2010). Therefore, increasing genetic variation is urgently needed for wheat improvement. Wild relatives of wheat, carrying significant genetic diversity and numerous desirable characteristics, have played an important role in wheat improvement (Mujeeb-Kazi et al., 2013). One of the most successful Chinese wheat cultivars, ‘Xiaoyan 6’ was created by the hybridization between common wheat and Elytrigia elongate (Li et al., 1990). To date, many desirable genes have been successfully introduced from wild relatives into common wheat. The wheat-rye T1RS/1BL translocations carrying disease resistance genes Pm8, Yr9, Lr26, and Sr31, as well as genes associated with superior agronomic and abiotic stress traits, have been widely used in wheat breeding (Ren et al., 2017; Howell et al., 2019; McIntosh et al., 2019; Li et al., 2020; Zhou et al., 2021). Powdery mildew resistance genes Pm21 and Pm67 from Haynaldia villosa (Xing et al., 2018; Zhang et al., 2021), and the Fusarium head blight resistance gene Fhb7 from Thinopyrum elongatum also contributed significantly to wheat genetic improvement (Wang et al., 2020).
Hordeum ssp. californicum (H. californicum), one of the most important wild relatives of wheat, has been valued as a donor of important agronomic and resistance traits, including tolerance to cold and low-nitrogen conditions, resistance to powdery mildew and leaf rust, more kernels per spike (KPS), and higher grain weight (Gupta and Fedak, 1985; Armstrong et al., 1993; Kolb et al., 2002; Kong et al., 2007; Fang et al., 2014; Mehnaz et al., 2021). To introduce favorable genes underlying the above traits into wheat, the F1 hybrid and amphiploid of intergeneric hybridization between H. californicum and commonly wheat cv. ‘Chinese Spring’ (CS) were acquired through a wide cross, (Gupta and Fedak, 1985). A series of wheat-H. californicum chromosome lines, including five disomic addition lines (DA2Hc, DA3Hc, DA4Hc, DA6Hc, and DA7Hc) and one disomic substitution line (DS6Hc), were obtained (Gupta and Fedak, 1985; Fang et al., 2014). In addition, 303 markers were assigned to chromosomes 1Hc-7Hc to identify the homoeologous groups of the alien chromosome in wheat-H. californicum derived materials and to trace the alien chromosomes (Kong et al., 2008; Fang et al., 2014). This study identified a wheat-H. californicum translocation line, ND646, by molecular cytogenetics and evaluated its phenotypic performances in multiple important traits. This research provides a novel germplasm resource for wheat breeding.
Material and Methods
Material
Wheat cv. CS-H. californicum amphiploid (2n=56, genome AABBDDHcHc) (Accession No. TA3443) was kindly provided by the Wheat Genetics and Genomics Resource Center, Kansas State University, Manhattan, Kansas, USA. Common wheat cv. CS, ‘Ningchun 4’, and ‘Hongtuzi’ were maintained by the Key Laboratory of Modern Molecular Breeding for Dominant and Special Crops in Yinchuan, Ningxia, China. WJ28-1 is a disomic addition line developed from a BC1F3 population made with CS as the recurrent parent and H. californicum amphiploid as the donor parent. WJ28-1 contains several excellent traits, such as more kernels per spike (KPS), higher grain yield (GY), and good resistance to powdery mildew.
Development of the wheat-H.californicum translocation line
The pollen of WJ28-1 plants at the booting stage were irradiated with 60Co-γ ray at a dosage of 20 Gray (Gy) and a dose rate of 0.5 Gy·min-1 (Song et al., 2013). The irradiated pollen was pollinated to an emasculated Ningchun 4. Then M1 seeds were backcrossed with Ningchun 4 or self-pollinated for several generations until stable agronomic performances were obtained. Meanwhile, their progeny were characterized by genomic in situ hybridization (GISH), fluorescence in situ hybridization (FISH), and their molecular markers.
GISH and FISH analysis
Metaphase I (MI) chromosome spreads from root-tip cells and pollen mother cells (PMCs) were prepared as described by Gill et al. (1991). Total genomic DNA was extracted following the method of Yang et al. (2006). GISH was performed according to the protocols described by Jiang and Gill (1993). FISH was performed according to the method of Jiang et al. (1994). The oligonucleotide probes pSc119.2 and pAs1 were labeled with biotin-16-dUTP and digoxigenin-11-dUTP, respectively (McIntyre et al., 1990; Nagaki et al., 1995; Du et al., 2017). Anti-digoxigenin-rhodamine Fab fragments (Roche Diagnostics GmbH, Germany) and streptavidin-fluorescein thiocyanate (FITC) (Roche Diagnostics GmbH, Germany) were used, followed by staining with 4,6-al amidine-2-phenyl indole (DAPI) to detect digoxigenin and biotin signals, respectively. Signals were visualized under an Olympus BX60 Fluorescence microscope (Olympus Optical Co. Ltd, Tokyo, Japan). Images were captured with a SPOT 32 CCD camera (SPOT Charge Coupled Device, Diagnostic Instruments, Inc., Sterling Heights, MI, USA) and analyzed using Adobe Photoshop software.
Molecular marker analysis
Four expressed sequence tag PCR (EST-PCR) primer pairs were used to identify the origin of the H. californicum chromosome in ND646 (Fang et al., 2014). Information on 6Hc-specific markers is shown in Table 1. The EST-PCR program was one cycle at 94 °C for 3 min, followed by 32 cycles of 94 °C for 30 s, at annealing temperature at 55 °C for 45 s, and at 72 °C for 1 min, then a final extension at 72 °C for 10 min. The amplified PCR products were separated by polyacrylamide gel electrophoresis (PAGE) with an acrylamide concentration of 8% and stained with silver (Bassam and Gresshoff, 2007).
Table 1 -. Detailed information of specific molecular markers of H.californicum.
| Marker | EST ID | Chromosome location | Primer squence (5’-3’) | Arm of chromosome |
|---|---|---|---|---|
| CINAU91 | BF145253 | 6AS 0.65-1.00 6DS 0.79-0.99 | L: CCTCGTGGAGGAGAACTTCA R: GTGACCATGTCGGTGAACTG | 6HcS |
| CINAU502 | Ta#S52546774 | 6AS 0.35-0.65 6DS 0.99-1.00 | L: TTTTTCAGTGGAGGGGTCAC R: GACGGCGACTGGTTGTTAAT | 6HcS |
| CINAU506 | BE591788 | 6AL 0.90-1.00 6BL 0.36-0.40 6DL 0.74-0.80 | L: ATGGAGAGAGCGCTGTAATA R: CCCCTACATGAAATGAGAAG | 6HcL |
| CINAU511 | BE425153 | 6AL 0.90-1.00 6BL 0.40-1.00 6DL 0.47-0.68 | L: GAACATAGCCGAAGCATTAC R: CTCTACCTGGGCTACTCCTT | 6HcL |
Experimental design
Field experiments were conducted at the teaching experiment farm of Ningxia University (Regional trial Ⅰ) and the research station of the Institute of Crop Science, Ningxia Academy of Agricultural-Forestry Sciences at Yongning, Ningxia (Regional trial Ⅱ) in 2021. ND646 and Ningchun 4 were randomized at each location as a complete block design with three replications. The plot size was 12.87 m2. The plots were spaced 0.15 m apart. For each plot, approximately 3000 seeds were evenly planted. Grain yield-related traits were evaluated at both locations. All the other traits, including disease and herbicide resistance, and grain quality traits, were only evaluated at the teaching experiment farm of Ningxia University.
Disease and herbicide resistance evaluation
To evaluate powdery mildew resistance, a mixture of prevalent spore isolates of Blumeria graminis f. sp. tritici from Ningxia, China, was used to infect the susceptible wheat cultivar Hongtuzi and the experimental plants at the elongation stage. The disease grades were scored as 0 (immune, I), 0-1 (nearly immune, NI), 1-2 (highly resistant, HR), 3-4 (moderately resistant, MR), 5-6 (moderately susceptible, MS), 7-8 (highly susceptible, HS), and 9 (extremely susceptible, ES), according to Sheng and Duan (1991). For leaf rust resistance evaluation, mixed Puccinia recondita f. sp. tritici isolates was used to infect plants at the elongation stage, and the disease responses were recorded with a 0-4 rating, in which 0-2 was considered resistant and 3-4 was considered susceptible (Roelfs et al., 1992). For herbicide resistance evaluation, 0.8% 2, 4-D was sprayed onto the leaves of wheat at the elongation and heading stages.
Agronomic and grain quality traits, and grain filling characteristics
During the growing season, twenty spikes from 10 individual plants were randomly selected in each plot before 9:00 a.m. every five days from flowering to maturity. Grains were used to measure fresh grain weight (FGW), grain volume (GV), dry grain weight (DGW), and grain moisture (GM). The grain filling rate (GFR) was calculated according to Wang et al. (2016). After harvest, all materials were measured for agronomic traits, including fertile tiller number per plant (FTNPP), plant height (PH), spike length (SL), spikelets per spike (SPS), fertile spikelets per spike (FSPS), grain weight per spike (GWPS), biological yield (BY), economic yield (EY), economic coefficient (EC), kernels per spike (KPS) with ten replications and fertile spike number (FSN), thousand-kernel weight (TKW), and grain yield (GY) with three replications. A sample of 30 g of grain was used to measure grain quality traits, including moisture content (MC), crude protein content (CPC), wet gluten content (WGC), flour yield rate (FYR), sedimentation value (SV), falling number (FN), water absorption rate (WAR), hardness index (HI), stabilization time (ST), formation time (FT), volume weight (VW) and extension area (EA), with three replications by Near Infra Red Spectrum (NIRS) DA7200 (Perten, Sweden). Statistical analyses were conducted in Excel 2010 and DPS 7.05.
Results
Development of ND646
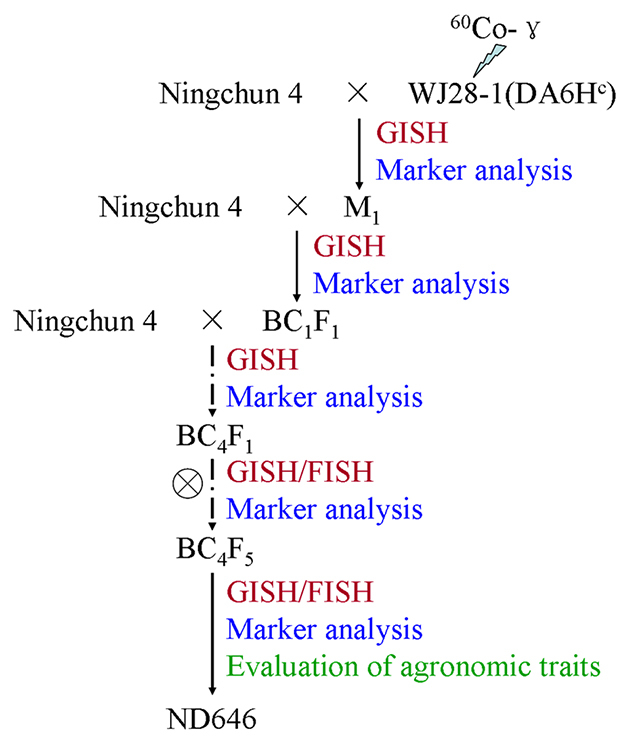
In previous work, the wheat-H. californicum disomic addition line WJ28-1 showed excellent performance for several traits, including disease resistance, more spikelets per spike, and more kernels per spike. To create wheat-H. californicum translocation lines with small alien chromosome fragments, we irradiated the pollen of WJ28-1 with 60Co-γ and pollinated them to an emasculated cultivar, ‘Ningchun 4’. The progeny were backcrossed to ‘Ningchun 4’ for three generations to produce BC4F1 seeds. BC4F1 was then crossed for four generations to produce BC4F5 seeds. For each generation of back-crossing and self-crossing, the progeny were characterized by GISH/FISH and 6Hc specific EST markers. ND646 is an elite line selected from the BC4F5 population (Figure 1).
Figure 1 -. Scheme of the development of whea-tH.californicum translocation line ND646.
Characterization of ND646 by sequential GISH/FISH and molecular markers
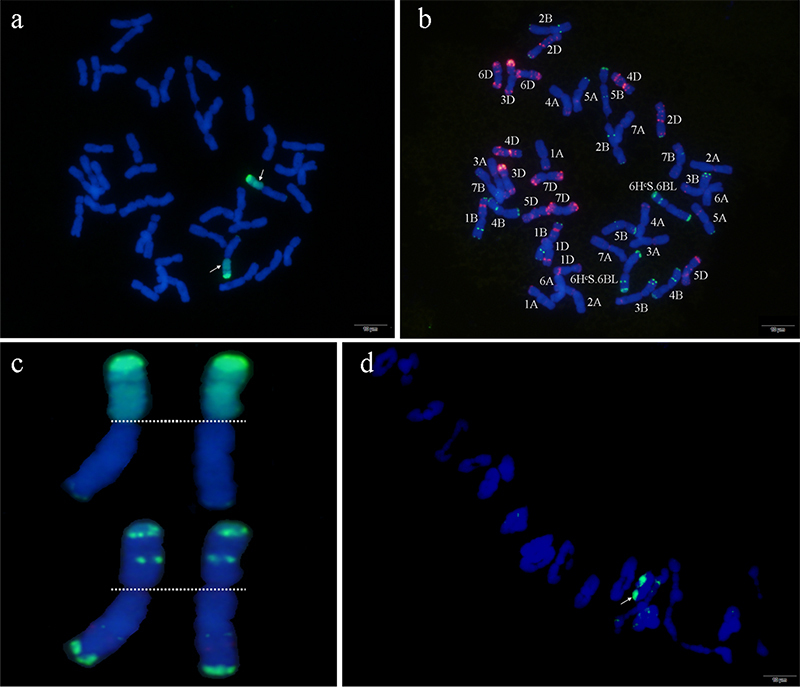
ND646, selected from the BC4F5 (Ningchun 4 × 60Co-γ irradiated WJ28-1), was analyzed using GISH/FISH. Based on the molecular karyotype of H. californicum and common wheat (Mukai et al., 1993; Fang et al., 2014; Du et al., 2017), ND646 was identified as a 6HcS/6BL translocation line (2n = 42) (Figure 2a-c). Meiosis in the PMCs of ND646 and its self-crossed progeny were investigated. Among the 100 PMCs at the metaphase I stage showing 2n = 21, all cells had one translocation bivalent (Figure 2d), and each cell contained 0.06 univalents, 3.92 rod bivalents, and 17.05 ring bivalents, on average. Thus, ND646 is a cytogenetically stable wheat-H. californicum translocation line.
Figure 2 -. Sequential GISH/FISH analysis of the wheat- H.californicum translocation line ND646. a. GISH of ND646, the genome DNA of H.californicum was visualized in bright green; b. FISH of ND646 using pSc119.2 (shown in red) and pAs1 (shown in green) repetitive DNA as probes, white arrows show the translocation chromosome; c. GISH and FISH diagram of translocation chromosomes in DN646, respectively; d. There were 21 bivalents containing one translocation bivalent (shown in green) at the metaphase I (MI) stage in pollen mother cells (PMCs) of ND646 (Scale bar = 20µm).
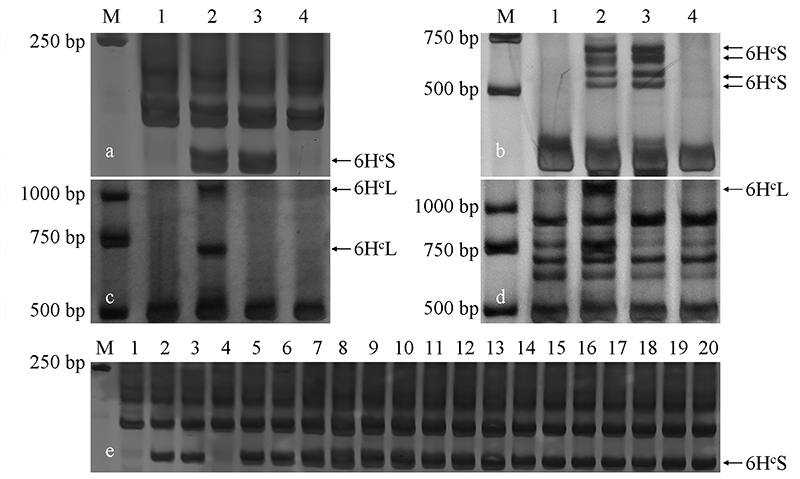
The GISH and FISH results indicated that the alien chromosome fragment of ND646 is 6Hc. To further verify the origin of alien chromosome fragments, 6Hc-specific markers were used to amplify the total genomic DNA of CS, WJ28-1, ND646, and Ningchun 4. All the ND646 plants could be amplified by the 6Hc short-arm specific markers but not by the 6Hc long-arm specific markers (Figure 3). Thus, the alien chromosome fragments of ND646 were confirmed as the short arm of chromosome 6Hc.
Figure 3 -. Amplification of EST-PCR primer pairs in the parents and ND646. a-d. Markers CINAU 91, CINAU 502, CINAU 506 and CINAU 511, respectively. 1-4 were CS, WJ28-1, ND646 and Ningchun 4, respectively; 5-20 were the amplification of marker CINAU 91 in 16 individual plants of ND646; M was a DNA marker with a molecular weight of 100, 250, 500, 750, 1000 and 2000 bp. Arrows showed specific banding of 6HcS and 6HcL chromosomes.
Agronomic, grain yield, and grain quality traits of ND646
The agronomic traits, including SL (11.83 cm vs. 10.92 cm), SPS (21.50 vs. 20.20), and GWPS (2.78 g vs. 2.04 g) in regional trial I. KPS (61.10 vs. 44.50 in regional trial I and 49.51 vs. 43.78 in regional trial 2), and GY (8614 kg.ha-1 vs. 7974 kg.ha-1 in regional trial I and 11093 kg.ha-1 vs. 10417 kg.ha-1 in regional trial II) were significantly (p < 0.05) higher in ND646 than in Ningchun 4 (Table 2). The grain quality traits, including SV (30.17 mL vs. 26.94 mL), WAR (59.94% vs. 58.81%) and HI (69.27 vs. 63.69), were also significantly (p < 0.05) higher in ND646 than in Ningchun 4 in regional trial I (Table 2).
Table 2 -. Performances on agronomic, grain yield and grain quality traits of Ningchun 4 and ND646.
| Traits | Index | Ningchun 4 | ND646 |
|---|---|---|---|
| Agronomic | FTNPP (No.) | 2.30±0.675 | 2.60±0.843 |
| PH (cm) | 91.07±2.948 | 93.25±0.674 | |
| SL (cm) | 10.92±0.585 | 11.83±0.897 * | |
| SPS (No.) | 20.20±1.549 | 21.50±0.707 * | |
| FSPS (No.) | 18.50±1.581 | 19.30±1.636 | |
| GWPS (g) | 2.04±0.433 | 2.78±0.539 * | |
| EC | 0.45±0.051 | 0.46±0.041 | |
| Yield-related | FSN (million myriad·ha-1) | 450.56±91.320 | 506.48±60.266 |
| KPS (No.) | 44.50±10.233 | 61.10±12.096 * | |
| TKW (g) | 41.56±0.439 | 42.70±0.453 | |
| TY (kg·ha-1) | 9307.22±84.374 | 12372.19±833.186 | |
| GY(kg·ha-1) | 7974.70±625.512 | 8614.75±641.201 * | |
| FSN (million myriad·ha-1) | 448.89±134.220 | 462.22±94.595 | |
| KPS (No.) | 43.78±6.641 | 49.51±5.706 * | |
| TKW (g) | 43.48±2.898 | 46.54±3.850 | |
| TY (kg·ha-1) | 8597.30±3257.577 | 10533.02±1625.845 | |
| GY(kg·ha-1) | 10417.97±93.869 | 11093.65±18.894 * | |
| Grain Quality | CPC (%) | 14.29±0.053 | 13.97±0.225 |
| WGC (%) | 29.96±0.265 | 30.14±0.572 | |
| FYR (%) | 67.94±0.414 * | 65.09±0.203 | |
| SV (mL) | 26.94±0.532 | 30.17±1.259 * | |
| FN (s) | 448.10±3.779 | 429.81±6.542 | |
| WAR (%) | 58.81±0.555 | 59.94±0.303 * | |
| HI | 63.69±1.529 | 69.27±0.451 * | |
| ST (min) | 3.53±0.365 | 5.03±0.820 | |
| FT (min) | 3.76±0.302 | 4.15±0.078 | |
| VW (g·L-1) | 818.32±0.894 * | 793.09±1.203 | |
| EA (cm2) | 103.04±6.904 | 109.18±16.620 |
PTNPP=Fertile tiller number per plant, PH=Plant height, SL=Spike length, SPS=Spikelets per spike, FSPS=Fertile spikelets per spike, GWPS=Grain weight per spike, EC=Economic coefficient, FSN=Fertile spike number, KPS=Kernels per spike, TKW=Thousand kernel weight, TY=Theoretical yield, GY=Grain yield, CPC=Crude protein content, WGC=Wet gluten content, FYR=Flour yield rate, SV=Sedimentation value, FN=Falling number, WAR=Water absorption rate, HI=Hardness index, ST=Stabilization time, FT=Formation time, VW=Volume weight, EA=Extension area. * represented significant differences within the same trait between the two materials (p<0.05).
Resistance of ND646 to disease and herbicide
Resistance of the adult ND646 plants to powdery mildew, leaf rust, and herbicide were evaluated in the field. ND646 was resistant to powdery mildew, whereas its common wheat parent, Ningchun 4, was susceptible when infected with a mixture of Blumeria graminis f. sp. tritici isolates (Figure 4a). Furthermore, ND646 had a higher level of resistance to leaf rust than Ningchun 4, with disease ratings of 3 and 2, respectively (Figure 4b). When treated with 2,4-D, the whole plant and spikes of Ningchun 4 were seriously distorted. However, ND646 showed high resistance to 2,4-D and showed robustly growth (Figure 4c). Therefore, ND646 is a novel gene source for improving the disease and herbicide resistance in wheat in Ningxia.
Figure 4 -. Responses to diseases and herbicides of Ningchun 4 and ND646. 1 and 2 represent Ningchun 4 and ND646, respectively. a. Powdery mildew on flag leaves; b. Leaf rust on flag leaves; c. Effects of 2,4-D on spikes and whole plants, respectively.

Grain filling characteristic of ND646
In Ningxia, the grain filling of spring wheat occurs from late May to early July. During the late period of grain filling, the temperature could be higher than 25 °C. The grain filling of many wheat cultivars in Ningxia is severely impacted by this high temperature. To investigate the grain filling characteristics of ND646, the dynamics of different morphology traits, including the GV, FGW, DGW, GM, and GFR of ND646 and Ningchun 4, were monitored during grain filling stage. The dynamics of GV, FGW, DGW, and GM of Ningchun 4 and ND646 both showed “S” curves with an increasing trend in the first phase after anthesis followed by a plateau and then a decreasing trend (Figure 5).
Figure 5 -. Dynamics of grain volume (GV), fresh grain weight (FGW), dry grain weight (DGW), and grain moisture (GM) of Ningchun 4 and ND646 during grain filling.
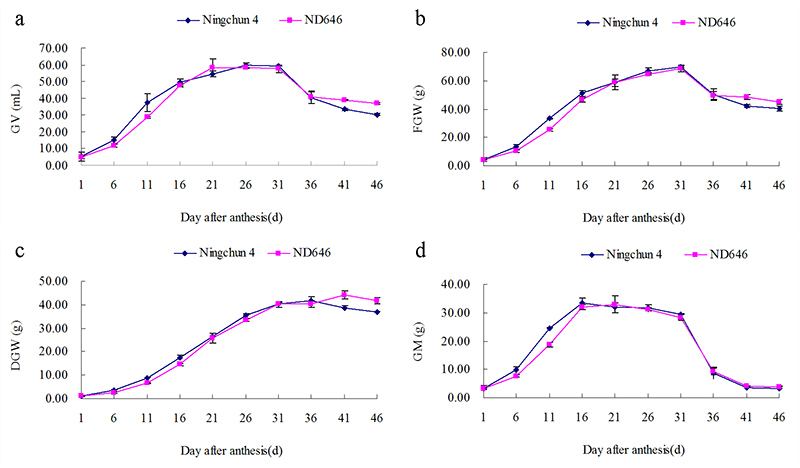
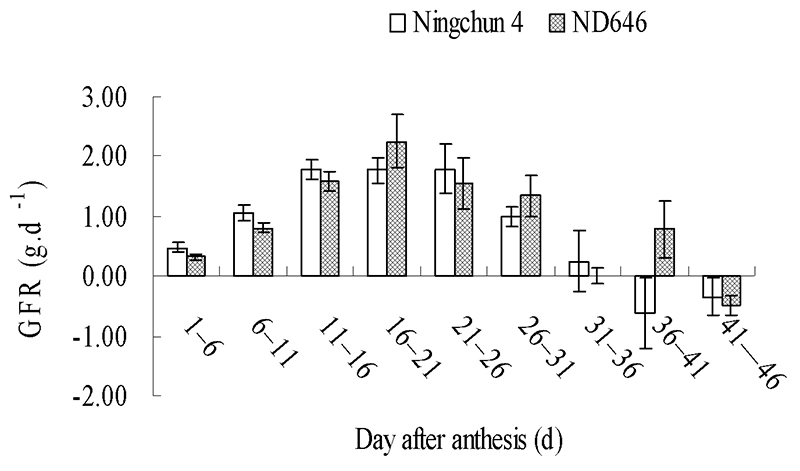
The increases in GV, FGW, and GM during the first 21 days and DGW during the first 36 days after anthesis were higher in ND646 than in Ningchun 4 (Table 3). The GV of Ningchun 4 and ND646 reached a maximum of 59.581 mL on the 26th day and 58.433 mL on the 21st day after anthesis, respectively (Table 3; Figure 5a). The FGW of Ningchun 4 and ND646 reached a maximum of 69.544 g and 68.544 g, respectively, both on the 31st day after anthesis (Table 3; Figure 5b). The DGW of Ningchun 4 and ND646 reached a maximum of 41.513 g on the 36th day and 44.207 g on the 41st day after anthesis, respectively (Table 3; Figure 5c). The GM of Ningchun 4 and ND646 reached a maximum of 33.458 g on the 16th day and 32.824 g on the 21st day after anthesis, respectively (Table 3; Figure 5d). The GFR of Ningchun 4 and ND646 reached the peaks of 1.792 g.d-1 on the 21st-26th day and 2.255 g.d-1 on the 16th-21st day, respectively (Table 4; Figure 6). Grain filling of Ningchun4 was completed on the 36th-41st day after anthesis, while the grain filling of ND646 was fully completed on the 41st-46th day after anthesis. By monitoring the dynamics of grain morphology traits during grain filling, we found that ND646 had a longer grain filling period and a higher maximum GFR at the critical filling stage than Ningchun 4.
Table 3 -. Grain morphological characteristics of Ningchun 4 and ND646 at the grain filling stage.
| Days after anthesis (d) | GV (mL) | FGW (g) | DGW (g) | GM (g) | ||||
|---|---|---|---|---|---|---|---|---|
| Ningchun 4 | ND646 | Ningchun 4 | ND646 | Ningchun 4 | ND646 | Ningchun 4 | ND646 | |
| 1 | 5.281 | 4.824 | 4.194 | 4.137 | 0.919 | 0.871 | 3.275 | 3.266 |
| 6 | 15.214 * | 11.736 | 13.091 * | 10.096 | 3.320 * | 2.476 | 9.771 * | 7.620 |
| 11 | 37.261 | 28.507 | 33.103 ** | 25.127 | 8.671 ** | 6.533 | 24.433 ** | 18.594 |
| 16 | 49.685 | 47.462 | 50.981 | 46.409 | 17.523 * | 14.444 | 33.458 | 31.966 |
| 21 | 54.618 | 58.433 | 58.441 | 58.544 | 26.365 | 25.721 | 32.076 | 32.824 |
| 26 | 59.581 | 58.103 | 66.944 | 64.557 | 35.323 | 33.490 | 31.621 | 31.067 |
| 31 | 59.483 | 57.784 | 69.544 | 68.544 | 40.275 | 40.213 | 29.269 | 28.331 |
| 36 | 40.359 | 40.597 | 50.251 | 49.520 | 41.513 | 40.287 | 8.738 | 9.233 |
| 41 | 33.304 | 39.085 ** | 41.966 | 48.252 ** | 38.443 | 44.207 * | 3.523 | 4.045 * |
| 46 | 30.353 | 36.991 ** | 40.322 | 44.845 * | 36.702 | 41.702 * | 3.143 | 3.814 * |
GV = Grain volume, FGW = Fresh grain weight; DGW = Dry grain weight, GM = Grain moisture. * and ** represent significant (p < 0.05) and highly significant (p < 0.01) differences, respectively, in the corresponding traits between Ningchun 4 and ND646.
Table 4 -. Dynamic changes in the grain filling rate of Ningchun 4 and ND646.
| Days after anthesis (d) | Ningchun 4 | ND646 |
|---|---|---|
| 1-6 | 0.480±0.092 * | 0.321±0.056 |
| 6-11 | 1.070±0.132 | 0.811±0.082 |
| 11-16 | 1.771±0.162 | 1.582±0.159 |
| 16-21 | 1.768±0.208 | 2.255±0.439 |
| 21-26 | 1.792±0.418 | 1.554±0.440 |
| 26-31 | 0.990±0.165 | 1.345±0.345 |
| 31-36 | 0.247±0.504 | 0.015±0.128 |
| 36−41 | −0.614±0.593 | 0.784±0.469 |
| 41−46 | −0.348±0.311 | −0.501±0.163 |
* represents a significant difference (p<0.05) of corresponding traits between Ningchun 4 and ND646.
Figure 6 -. Dynamics of grain filling rate of Ningchun 4 and ND646.
Discussion
Transferring desirable genes from wheat wild relatives could be an efficient approach to broadening genetic diversity and boosting wheat genetic improvement (Qi et al., 2010). The creation of wheat-alien translocation lines is an efficient method to transfer desirable genes from wild relatives to common wheat compared with genetically unstable addition and substitution lines (Danilova et al., 2014). H. californicum has many potentially valuable traits, so it is vital to produce wheat-H. californicum translocation lines to transfer its useful genes to wheat for wheat improvement. A series of wheat-H. californicum addition lines was created. WJ28-1 is one of these lines and has been identified as DA6Hc by molecular markers (Fang et al., 2014). WJ28-1 showed several excellent traits, including disease resistance, more spikelets per spike, and more kernels per spike. Wheat translocation lines can be induced by spontaneous translocation, tissue culture, gametocidal chromosomes, Ph-systems, and ionization irradiation (Li et al., 2016). Therefore, WJ28-1 was irradiated by 60Co-γ to create translocation lines with small alien chromosome fragments. 60Co-γ irradiated WJ28-1 was backcrossed to Ningchun 4 for several generations to transfer the excellent alien genes from WJ28-1 into the Ningxia wheat background. A stable translocation line ND646 was developed during this process.
Determining chromosome constitutions is a crucial step in the introgression of elite genes into wheat. Sequential C-banding and GISH are extremely useful in identifying wheat-alien introgressions (Jiang and Gill, 1993). After the introduction of alien chromosomes into the wheat genome, sequential C-banding and GISH can be used to detect alien chromosomal fragments. However, repeated DNA probes are inefficient in identifying homoeologous chromosomes, as the abundance and distribution of repetitive elements vary among homoeologous chromosomes within species and chromosomes of closely related species (Badaeva et al., 1992; Friebe and Gill, 1993; Dedkova et al., 2007). PCR-based markers have been used extensively as effective tools for alien chromosome segment identification under wheat background (Wang et al., 2010; Zhuang et al., 2011; Zhao et al., 2014). In this study, ND646 was characterized by FISH and EST-PCR markers as a wheat-H. californicum 6HcS/6BL translocation line.
Wild relatives of wheat are valuable resources for expanding the gene pools of wheat breeding (Wu et al., 2006; Mullan et al., 2009). Although a large number of wheat-alien translocation lines carrying excellent alien genes have been produced, only a few have been successfully incorporated into wheat breeding programs. The development of translocation lines with lower linkage drag and regular meiotic behavior will increase the efficiency of using wild relatives in wheat breeding. In this study, ND646 carrying the short arm of H. californicum chromosome 6Hc exhibited many desirable traits, such as more kernels per spike, higher grain yields, higher sedimentation value, higher water absorption rate, higher hardness index, and resistance to powdery mildew, leaf rust, and 2,4-D. As a valuable germplasm for wheat breeding, ND646 will be continuously evaluated in regional trials of Ningxia and regional trials of the spring wheat region in northern China. Although the wheat 6BS chromosome was replaced by the 6HcS chromosome, it had a good compensatory effect, and ND646 showed excellent comprehensive agronomic traits. Due to whole 6Hc short arm translocation in ND646, the loss of alien chromosome fragments will inevitably occur in the process of inheritance, resulting in instability of the translocation line. To eliminate the genetic burden of large chromosome fragments, we will further improve ND646 using homoeologous recombination induced by the Ph mutant.
Acknowledgments
We are grateful to Prof. Dr. Xiue Wang (State Key Laboratory of Crop Genetics and Germplasm Enhancement, Cytogenetics Institute, Nanjing Agricultural University) for her useful advice. We thank LetPub (https://www.letpub.com/) for its linguistic assistance during the preparation of this manuscript. This research was funded by the National Natural Science Foundation of China, grant number 32160452; The Ningxia Key Research and Development Program (Special Talents), grant number 2021BEB04070; The Natural Science Foundation of Ningxia Province, grant number 2021AAC03069; The Ningxia Hui Autonomous Region Agricultural Breeding Special Project, grant number 2018NYYZ02.
References
- Armstrong K, Fedak G, Molnar S, Pandeya R, Robert L. Hoisington D and McNab A (eds) Progress in genome mapping of wheat and related species: Proceedings of the 3rd public workshop of the International Triticeae Mapping lnitiative. CIMMYT; Mexico: 1993. Genome analysis in the Triticeae; pp. 62–63. [Google Scholar]
- Badaeva ED, Badaev NS, Gill BS, Filatenko AA. Intraspecific karyotype divergence in Triticum araraticum (Poaceae) Plant Syst Evo. 1992;192:117–145. [Google Scholar]
- Bassam BJ, Gresshoff PM. Silver staining DNA in polyacrylamide gels. Nat Protoc. 2007;2:2649–2654. doi: 10.1038/nprot.2007.330. [DOI] [PubMed] [Google Scholar]
- Danilova TV, Friebe B, Gill BS. Development of a wheat single gene FISH map for analyzing homoeologous relationship and chromosomal rearrangements within the Triticeae. Theor Appl Genet. 2014;127:715–730. doi: 10.1007/s00122-013-2253-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dedkova OS, Badaeva ED, Mitrofanova EN, Bilinskaya EN, Pukhalskiy VA. Analysis of intraspecific diversity of cultivated emmer Triticum dicoccum (Schrank.) Schuebl. using C-banding technique. Russ J Genet. 2007;43:1271–1285. [PubMed] [Google Scholar]
- Du P, Zhuang L, Wang Y, Yuan L, Wang Q, Wang D, Tan L, Shen J, Xu H, Zhao H, et al. Development of oligonucleotides and multiplex probes for quick and accurate identification of wheat and Thinopyrum bessarabicum chromosomes. Genome. 2017;60:93–103. doi: 10.1139/gen-2016-0095. [DOI] [PubMed] [Google Scholar]
- Fang Y, Yuan J, Wang Z, Wang H, Xiao J, Yang Z, Zhang R, Qi Z, Xu W, Hu L, et al. Development of T. aestivum L.-H. californicum alien chromosome lines and assignment of homoeologous groups of Hordeum californicum chromosomes. J Genet Genomics. 2014;41:439–447. doi: 10.1016/j.jgg.2014.06.004. [DOI] [PubMed] [Google Scholar]
- Friebe B, Gill BS. C-band polymorphism and structural rearrangements detected in common wheat (Triticum aestivum) Euphytica. 1993;78:1–5. [Google Scholar]
- Gill BS, Friebe B, Endo TR. Standard karyotype and nomenclature system for description of chromosome bands and structural aberrations in wheat (Triticum aestivum L.) Genome. 1991;34:830–839. [Google Scholar]
- Gupta PK, Fedak G. Intergeneric hybrids between Hordeum californicum and Triticum aestivum. J Hered. 1985;76:365–368. [Google Scholar]
- Gupta PK, Langridge P, Mir RR. Marker-assisted wheat breeding: Present status and future possibilities. Mol Breed. 2010;261:145–161. [Google Scholar]
- Howell T, Moriconi JI, Zhao X, Hegarty J, Fahima T, Santa-Maria GE, Dubcovsky J. A wheat/rye polymorphism affects seminal root length and yield across different irrigation regimes. J Exp Bot. 2019;70:4027–4037. doi: 10.1093/jxb/erz169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang J, Gill BS. Sequential chromosome banding and in situ hybridization analysis. Genome. 1993;36:792–795. doi: 10.1139/g93-104. [DOI] [PubMed] [Google Scholar]
- Jiang JM, Friebe B, Gill BS. Recent advances in alien gene transfer in wheat. Euphytica. 1994;73:199–212. [Google Scholar]
- Kolb A, Alpert P, Enters D, Holzapfel C. Pattern of invasion within a grassland community. J Ecol. 2002;9:871–881. [Google Scholar]
- Kong F, Wang H, Cao A, Qin B, Ji J, Wang S, Wang XE. Characterization of T. aestivum-H. californicum chromosome addition lines DA2H and MA5H. J Genet Genomics. 2008;35:673–678. doi: 10.1016/S1673-8527(08)60089-2. [DOI] [PubMed] [Google Scholar]
- Kong F, Wang HY, Zhao Y, Ji JH, Cao AZ, Wang SL. Giemsa C-banding and karyotype analysis of California barley chromosome. Acta Agrestia Sin. 2007;15:103–108. [Google Scholar]
- Li H, Lv M, Song L, Zhang J, Gao A, Li L, Liu W. Production and identification of wheat-Agropyron cristatum 2P translocation lines. PLoS One. 2016;11:e0145928. doi: 10.1371/journal.pone.0145928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li SQ, Tang HP, Zhang H, Mu Y, Lan XJ, Ma J. A 1BL/1RS translocation contributing to kernel length increase in three wheat recombinant inbred line populations. Czech J Genet Plant Breed. 2020;56:43–51. [Google Scholar]
- Li WL, Li ZS, Mu SM. A cytological study of chromosomal structure changes in a common wheat variety, Xiaoyan No.6. J Genet Genomics. 1990;6:430–437. [Google Scholar]
- McIntosh RA, Dubcovsky J, Rogers WJ, Xia XC, Raupp WJ. Catalogue of gene symbols for wheat: 2019 supplement. In: Raupp WJ (ed) Annual wheat newsletter, the wheat genetic and genomic resources at Kansas State University. Manhattan. 2019:98–113. [Google Scholar]
- McIntyre CL, Pereira S, Moran LB, Appels R. New Secale cereal (rye) DNA derivatives for the detection of rye chromosome segments in wheat. Genome. 1990;33:635–640. doi: 10.1139/g90-094. [DOI] [PubMed] [Google Scholar]
- Mehnaz M, Dracatos P, Pham A, March T, Maurer A, Pillen K, Forrest K, Kulkarni T, Pourkheirandish M, Park RF, et al. Discovery and fine mapping of Rph28: A new gene conferring resistance to Puccinia hordei from wild barley. Theor Appl Genet. 2021;134:2167–2179. doi: 10.1007/s00122-021-03814-1. [DOI] [PubMed] [Google Scholar]
- Mujeeb-Kazi A, Kazi AG, Dundas I, Rasheed A, Ogbonnaya F, Kishii M. Genetic diversity for wheat improvement as a conduit for food security. Adv Agron. 2013;122:179–257. [Google Scholar]
- Mukai Y, Nakahara Y, Yamamoto M. Simultaneous discrimination of the three genomes in hexaploid wheat by multicolor fluorescence in situ hybridization using total genomic and highly repeated DNA probes. Genome. 1993;36:489–494. doi: 10.1139/g93-067. [DOI] [PubMed] [Google Scholar]
- Mullan DJ, Mirzaghaderi G, Walker E, Colmer TD, Francki MG. Development of wheat-Lophopyrum elongatum recombinant lines for enhanced sodium ‘exclusion’ during salinity stress. Theor Appl Genet. 2009;119:1313–1323. doi: 10.1007/s00122-009-1136-9. [DOI] [PubMed] [Google Scholar]
- Nagaki K, Tsujimoto H, Isono K, Sasakuma T. Molecular characterization of a tandem repeat, Afa family, and its distribution among Triticeae. Genome. 1995;38:479–486. doi: 10.1139/g95-063. [DOI] [PubMed] [Google Scholar]
- Qi Z, Du P, Qian B, Zhuang L, Chen H, Chen T, Shen J, Guo J, Feng Y, Pei Z. Characterization of a wheat-Thinopyrum bessarabicum (T2JS-2BS.2BL) translocation line. Theor Appl Genet. 2010;121:589–597. doi: 10.1007/s00122-010-1332-7. [DOI] [PubMed] [Google Scholar]
- Ren TH, Tang ZX, Fu SL, Yan BJ, Tan FQ, Ren ZL, Li Z. Molecular cytogenetic characterization of novel wheat-rye T1RS.1BL translocation lines with high resistance to diseases and great agronomic traits. Front Plant Sci. 2017;8:799. doi: 10.3389/fpls.2017.00799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roelfs AP, Singh RP, Saari EE. Rust diseases of wheat: Concepts and methods of disease management. CIMMYT; México: 1992. 80 [Google Scholar]
- Sheng BQ, Duan XY. Improvement of scale 0-9 method for scoring adult plant resistance to powdery mildew of wheat. Beijing Agricult Sci. 1991;1:38–39. [Google Scholar]
- Song L, Jiang L, Han H, Gao A, Yang X, Li L, Liu W. Efficient induction of wheat-Agropyron cristatum 6P translocation lines and GISH detection. PLoS One. 2013;8:e69501. doi: 10.1371/journal.pone.0069501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Sun S, Ge W, Zhao L, Hou B, Wang K, Lyu Z, Chen L, Xu S, Guo J, et al. Horizontal gene transfer of Fhb7 from fungus underlies Fusarium head blight resistance in wheat. Science. 2020;368:eaba5435. doi: 10.1126/science.aba5435. [DOI] [PubMed] [Google Scholar]
- Wang MJ, Zhang Y, Lin ZS, Ye XG, Yuan YP, Ma W, Xin ZY. Development of EST-PCR markers for Thinopyrum intermedium chromosome 1Ai#2 and their application in characterization of novel wheat-grass recombinants. Theor Appl Genet. 2010;121:1369–1380. doi: 10.1007/s00122-010-1394-6. [DOI] [PubMed] [Google Scholar]
- Wang ZJ, Zhang WJ, Wan JY, Bai LJ, Jia YY, Liu Y. Identification of translocation line of Triticum aestivum-Lolium perenne L. and analysis on grain filling and yield traits of its offsprings. J Triticum Crops. 2016;36:841–848. [Google Scholar]
- Wu J, Yang X, Wang H, Li H, Li L, Li X, Liu W. The introgression of chromosome 6P specifying for increased numbers of florets and kernels from Agropyron cristatum into wheat. Theor Appl Genet. 2006;114:13–20. doi: 10.1007/s00122-006-0405-0. [DOI] [PubMed] [Google Scholar]
- Xing L, Hu P, Liu J, Witek K, Zhou S, Xu J, Zhou W, Gao L, Huang Z, Zhang R, et al. Pm21 from Haynaldia villosa encodes a CC-NBS-LRR protein conferring powdery mildew resistance in wheat. Mol Plant. 2018;11:874–878. doi: 10.1016/j.molp.2018.02.013. [DOI] [PubMed] [Google Scholar]
- Yang MJ, Huang KY, Han QD. Research progresses on wheat powdery mildew and its resistance. Mol Plant Breed. 2016;14:1244–1254. [Google Scholar]
- Yang ZJ, Li GR, Chang ZJ, Zhou JP, Ren ZL. Characterization of a partial amphiploid between Triticum aestivum cv Chinese Spring and Thinopyrum intermedium ssp. trichophorum. Euphytica. 2006;49:11–17. [Google Scholar]
- Zhang RQ, Xiong CX, Mu HQ, Yao RN, Meng XR, Kong LN. Pm67, a new powdery mildew resistance gene transferred from Dasypyrum villosum chromosome 1V to common wheat (Triticum aestivum L.) Crop J. 2021;9:882–888. [Google Scholar]
- Zhao RH, Wang HY, Jia Q, Xiao J, Yuan CX, Zhang YJ. Development of EST-PCR markers for the chromosome 4V of Haynaldia villosa and their application in identification of 4V chromosome structural aberrants. China Agi Sci. 2014;13:282–289. [Google Scholar]
- Zhou S, Zhang CM, Zhao H, Lu MY, Dong FS, Liu YW. RNA interference targeting ω-secalin genes differentially affects the processing quality in a wheat T1BL·1RS translocation line. Crop J. 2021;9:456–464. [Google Scholar]
- Zhuang LF, Sun L, Li AX, Chen TT, Qi ZJ. Identification and development of diagnostic markers for a powdery mildew resistance gene in chromosome 2R of Chinese rye cultivar Jingzhouheimai. Mol Breed. 2011;27:455–465. [Google Scholar]


