Abstract
Background
Current studies show that exosomal miRNAs become an important factor in cancer metastasis. Among the many miRNA studies, miR-7-5p has not been thoroughly investigated in breast cancer metastasis.
Methods
Bioinformatic screening was performed using extant data from the GEO database, and miR-7-5p expression levels in breast cancer cell lines and exosomes were further examined using real-time quantitative PCR (qRT-PCR). The extracted exosomes were characterised by transmission electron microscopy (TEM), particle size analysis and marker protein determination. Cell migration and invasion were then examined using wound healing assays and Transwell assays, respectively. Correlation between miR-7-5p and receptor-like tyrosine kinase (RYK) was analysed by luciferase reporter. The effect of miR-7-5p against RYK-related downstream factors was verified using western blot assays.
Results
In this study, we found that the expression of miR-7-5p was significantly different in exosomes secreted from breast cancer cell lines with different high and low invasiveness. Further experiments revealed that miR-7-5p has an important role in inhibiting the migration and invasion of breast cancer. In terms of mechanism of action, miR-7-5p was found to target the RYK, leading to its reduced expression, which in turn caused a reduction in the phosphorylation level of the downstream factor JNK. Reduced levels of phosphorylated JNK factors lead to reduced levels of phosphorylation of c-Jun protein, which in turn leads to increased expression of EMT transcription factors, thereby inhibiting the epithelial–mesenchymal transition (EMT) process to suppress the invasion of breast cancer.
Conclusion
Thus, we demonstrated that exosomes loaded with high levels of miR-7-5p emitted from less aggressive breast cancers can participate in the atypical WNT pathway by targeting the RYK gene and thus inhibit breast cancer metastasis.
Supplementary Information
The online version contains supplementary material available at 10.1186/s11658-022-00393-x.
Keywords: Breast cancer, Exosome, miR-7-5p, RYK, EMT, Atypical WNT pathway
Introduction
In recent years, breast cancer has been increasing in incidence each year and has become the number one killer threatening women’s health [1, 2]. Breast cancer often results in death due to the uncontrollable spread of tumour cells, causing a decline, or even failure, in the function of other organs [3, 4]. Despite the current advances in the treatment of primary breast cancer, there is as yet no effective treatment for metastatic breast cancer. Since 90% of breast cancer-related deaths are caused by metastatic breast cancer, a clearer insight into the mechanisms of breast cancer metastasis is important for the development of effective treatments for metastatic breast cancer [5].
Exosomes are extracellular vesicles approximately 40–120 nm diameter that are separated from cells and are present in body fluids (serum, urine, etc.) [6, 7]. The exosome was thought to function primarily as an intracellular waste remover when it was first discovered [8], but as research has continued, more and more studies have revealed that its function is not so simple. Exosomes can be loaded with a large number of biologically active molecules (small non-coding RNA, mRNA, DNA, proteins, etc.) released by cells into body fluids, accompanying the flow of body fluids and binding to target cells membranes to release their contents into the target cells, acting as intercellular information transfer [8–10]. MicroRNAs (miRNAs) are non-coding single-stranded RNA molecules of up to 22 nucleotides, which are essential components of the exosome. It induces protein translation inhibition in an unknown manner, primarily by complementary binding to the 3′ non-coding region (3′ UTR) portion of the target mRNA target, which in turn inhibits protein synthesis and regulates intracellular processes by modulating the translation of a set of key mRNAs [11, 12]. Since the first miRNA was discovered in 1993, a steady stream of studies has shown that human miRNAs are instrumental in a variety of cancers [13]. Exosomal miRNAs also reflect to some extent the expression pattern of dysregulated miRNAs in cancer cells. Among these, those miRNAs that are delivered by exosome loading have also been found to be up or down regulated in expression in different tumours, involved in various regulatory mechanisms of cancer growth, apoptosis and metastasis [14, 15].
In the GSE114329 dataset, by analysing the differentially expressed miRNAs of MDA231 EXO and MCF7 EXO, we found that hsa-miR-7-5p was highly expressed in exosomes secreted by low-invasive breast cancer cells. Notably, hsa-miR-7-5p has been shown to be a potential oncogenic factor involved in regulating the biology of many cancer development processes, including colorectal [16], liver [17, 18], gastric [19] and lung cancers [20]. It has also been found in some studies to have a role in drug resistance in breast cancer [21, 22]. However, research on the role and molecular mechanisms of hsa-miR-7-5p in breast cancer metastasis is limited. Therefore, we chose to further investigate the specific molecular mechanisms of hsa-miR-7-5p in breast cancer metastasis. We validated a novel target of hsa-miR-7-5p, RYK, and demonstrated the ability to further influence the JNK-mediated atypical WNT signalling pathway. This offers new ideas for the treatment of metastatic breast cancer.
Materials and methods
Bioinformatics analysis
The GSE114329 dataset was searched in the GEO database, and two subsets of the MDA231 EXO and MCF7 EXO were selected for bioinformatic analysis. After downloading the raw data from the dataset, the raw data were filtered to remove splicing and decontamination to obtain clean reads which are compared with the reference genome for subsequent information analysis. Bioinformatics analysis was carried out by Shandong Unhelix Biotechnology Corporation.
Cell culture
The two human breast cancer cell lines used in this study, MDA-MB-231 (TCHu227) and MCF7 (TCHu74), were purchased from Dalian Meilun Biotechnology Corporation. These cells were assayed for mycoplasma, and no mycoplasma contamination was found in any of them. Both cells were cultured in high-sugar DMEM medium (Solarbio, Beijing, China) comprising 10% fetal bovine serum (Biological Industries, Kibbutz Hulda, Israel) in a cell culture incubator at 5% CO2 humidity and 37 °C. To obtain cancer-derived exosomes unaffected by fetal bovine serum exosomes, cells were also cultured in high-sugar DMEM medium containing 10% exosome-free fetal bovine serum (System Biosciences, CA, USA). The cells were cultured to the logarithmic stage of growth for the experiment.
Extraction of exosomes
The cell culture supernatant was collected during the cell culture process, and the cell culture supernatant was centrifuged (Sigma, Germany) at high speed: 300g at 4 °C for 10 min, the supernatant was continued at 2000g at 4 °C for 15 min and then the supernatant was centrifuged at 10,000g at 4 °C for 30 min. Ultracentrifugation (Beckman Coulter, USA) was then performed: the supernatant was centrifuged at 110,000g for 90 min at 4 °C, then the supernatant was discarded and the precipitate was resuspended in pre-cooled PBS and continued to be centrifuged at 110,000g for 90 min at 4 °C. The precipitate was resuspended in appropriate PBS and stored in a −80 °C refrigerator.
Transmission electron microscopy
The extracted exosomes were observed for their morphological characteristics by transmission electron microscopy (TEM). The exosome suspension was suspended as droplets on a copper grid for 15 min, then the droplets were taken up by filter paper, followed by a drop of 4% phosphotungstic acid stain for 2 min. Afterwards, the staining solution was blotted away with filter paper. The copper grids were allowed to dry overnight and then observed using TEM (JEOL, Japan).
Nanoparticle size and zeta potential analyser
The granulometry of the extracted exosomes was analysed using a nanoparticle size and zeta potential analyser (Malvern Panalytical, UK). The exosome suspension was diluted and placed in a quartz cuvette to measure the particle size.
Cellular uptake assay
Cells were inoculated in six-well plates at 2 mL per well (5 × 105 cells/mL) and incubated for 24 h. Exosomes were labelled using Dio stain (Coolaber, Beijing, China). Following the instructions, the Dio staining solution was first diluted and then mixed with the exosome suspension and incubated at 37 °C for 30 min protected from light. To remove excess dye, the stained exosomes were centrifuged at 110,000g for 90 min at 4 °C. The precipitate was then resuspended in PBS and co-incubated with cells for 3 h, and cellular uptake was observed using two-photon confocal microscope (LSM 880 NLO, Germany).
Wound healing experiment
The digested cells were inoculated in six-well plates (1 × 106 cells per well) and incubated until the cells had grown to about 80% in each well, then three monolayers of cells were scraped at medium distances in each well with the tip of a 10 μL gun. These were washed in PBS twice, and new cell culture medium was added. Then the width of the scratch was measured with an inverted microscope (Olympus, Japan). The cells were treated accordingly and incubated for a further 48 h and the width of the scratch was measured again. Cell migration was assessed by wound healing rate: wound healing rate = (0 h scratch width − 24 h scratch width)/0 h scratch width × 100%.
Transwell
Transwell chambers with an 8-μm-pore-size polycarbonate membrane at the bottom were used for detecting the migratory and invasive capacity of the cells. Transfected cells (2 × 105) were resuspended in serum-free DMEM medium and added to the upper chamber (matrix gel coated or uncoated), and then 750 μL of DMEM culture medium containing 10% serum was added to the lower chamber. After 24 h incubation at 37 °C in 5% CO2, the cells were fixed in 4% paraformaldehyde (Meilunbio, Dalian, China) for 2 min and then in methanol for 20 min. Finally, the cells were stained with 0.1% crystalline violet stain for 15 min, and then the non-migrated cells and the stromal gel coating were gently scraped off the interior of the chambers with a cotton swab and placed on a slide for imaging and counting of the migrated cells under a microscope.
Cell transfection
During cell transfection, all oligonucleotides and vectors were synthesised by GenePharma (Shanghai, China), including RYK small interfering RNA (siRYK) and its control (siNC), miR-7-5p mimic and inhibitor or its control (miR-NC mimic and miR-NC inhibitor). Cells were transfected using Lipofectamine 3000 (Mei5bio, Beijing, China) or LipoRNAiMAX (Mei5bio) according to the manufacturer’s requirements. Transfections were assayed after 48 h of normal culture. The sequences of miRNA mimics and inhibitors are shown in Additional file 1: Table S1.
RNA extraction and qRT-PCR
RNAiso (Takara Bio, Japan) for small RNA and RNAiso (Takara Bio, Japan) for plus were used to extract miRNA and total RNA from cells and exosomes. miRNA 1st Strand cDNA Synthesis Kit (Vazyme Biotech, Nanjing, China) and HiScript II 1st Strand cDNA Synthesis Kit (Vazyme Biotech) were used to perform reverse transcription of miRNA and mRNA. RNA levels were quantified using SYBR Green stain (Vazyme Biotech) on an CFX 96Touch (Bio-Rad, USA) for real-time analysis. Real-time analysis was performed using SYBR Green on an CFX 96Touch according to the following protocol. Protocol for miRNA: 95 °C for 5 min, 40 cycles including 95 °C for 15 s and 60 °C for 20 s, 72 °C for 40 s. Protocol for mRNA: 95 °C for 5 min, 40 cycles including 95 °C for 10 s and 60 °C for 30 s. The expression levels of intracellular and exosomal miRNAs were normalised to U6. The expression levels of intracellular mRNAs were normalised to GADPH. Primer sequences are listed in Additional file 1: Table S2.
Western blot
Cells or exosomes were first lysed with Western and IP Cell Lysis Solution (Coolaber) and incubated on ice for 15 min, and then the supernatant was gathered by centrifugation. Protein concentration was determined via the BCA protein assay kit (Meilunbio) for quantification. Protein samples were separated on 10% SDS–polyacrylamide gels, protein bands were transferred onto 0.45 µm PVDF membranes (Millipore, Massachusetts, USA). The PVDF films were then sealed with 5% skimmed milk powder (Bio-Rad). The closed PVDF membrane was incubated at 4 °C overnight with the corresponding primary antibody, washed with TBST buffer and incubated with secondary antibody (Proteintech, Wuhan, China) for 4–5 h at 4 °C. Washing with TBST buffer continued at the end of the secondary antibody incubation, followed by observation of the protein bands on the membrane using ECL luminescent solution (Meilunbio) and an imaging system (Bio-Rad). The primary antibodies included: TSG101 (1:20,000, cat. no. 67381-1-Ig), Alix (1:10,000, cat. no. 67715-1-Ig), CD63 (1:10,000, cat. no. 67605-1-Ig), RYK (1:1000, cat. no. 22138-1-AP), E-cadherin (1:5000, cat. no. 20874-1-AP), N-cadherin (1:6000, cat. no. 66219-1-Ig), ZEB1 (1:1000, cat. no. 21544-1-AP), Phospho-JNK (1:2000, cat. no. 80024-1-RR), Phospho-c-Jun (1:2000, cat. no. 28891-1-AP) and β-actin (1:10,000, cat. no. 66009-1-Ig) were obtained from Proteintech.
Luciferase reporter assay
Using the Biology website (http://www.targetscan.org) to predict the binding site of miR-7-5p to RYK. The pmirGLO vectors for RYK-WT and RYK-MUT were then constructed (GenePharma). Logarithmic growth phase MDA-MB-231 cells were inoculated onto 12-well plates at a density of 5 × 105 cells per well. The miR-NC mimics or miR-7-5p mimics, RYK-WT and RYK-MUT, were transfected into MDA-MB-231 cells when cell confluency reached 70%. The reporter plasmid content was 2 µg per well, while miR-7-5p mimics or miR-NC were transfected at a total of 40 pmol per well. After 48 h of transfection, cell lysis was performed and luciferase activity was measured by the Luciferase Assay Kit (Meilunbio).
Statistical analysis
Data are presented as mean ± standard deviation (SD) (n = 3). For comparisons between the two groups, Student’s t-test was used to determine the difference between the means of the normal distribution. Significance analysis was performed on all data using GraphPad Prism 5 (San Diego, CA, USA) software, and probability levels of < 0.05 were considered significant.
Results
Exosomes from highly invasive breast cancers are more likely to promote cancer cell migration than less invasive ones
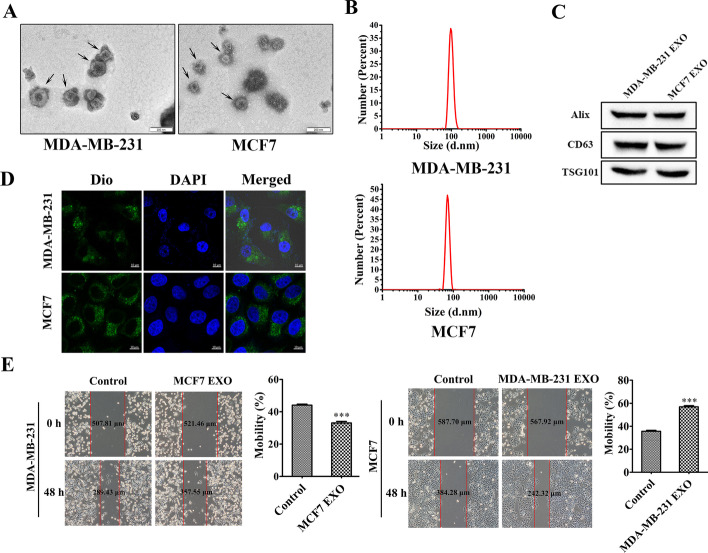
The finding that cancer cells secrete more exosomes than normal cells further suggests that exosomes secreted by cancer cells may play a central role in their metastatic mechanism. To investigate the role of breast cancer-derived exosomes in breast cancer migration and invasion, exosomes first need to be isolated. We cultured two breast cancer cell lines, MDA-MB-231 cells and MCF7 cells, using a medium containing fetal bovine serum without exosomes. After collecting the cell cultures and extracting the exosomes using differential centrifugation, they were observed using transmission electron microscopy. The classical morphology of exosomes in a homogeneous cup-like structure was observed as shown in Fig. 1A. We examined the particle size of the extracted material using a nanoparticle size and zeta potential analyser and found that most of the particles were around 100 nm in size (Fig. 1B). The presence of these three marker proteins was further demonstrated using antibodies to CD63, TSG101 and Alix (Fig. 1C). All these results indicated that the extracted vesicular structures were indeed exosomes. After isolation of the exosomes, we co-cultured MCF7-derived exosomes with MDA-MB-231 cells and MDA-MB-231-derived exosomes with MCF7 cells. The co-cultured cells were also subjected to a wound healing assay to verify whether there were some functional differences between the two different exosomes. Furthermore, to determine that exosomes from different sources could enter different cells, we first labelled the extracted exosomes with Dio staining and then observed them with a two-photon laser confocal microscope. The results showed that exosomes were able to enter cells of different origins (Fig. 1D). And the results of the wound healing assay also showed that highly invasive breast cancer-derived exosomes promoted cell migration more than less invasive ones (Fig. 1E).
Fig. 1.
Exosomes from highly invasive breast cancers are more likely to promote cancer cell migration than less invasive ones. A Transmission electron microscopy of exosomes extracted from MDA-MB-231 and MCF7 breast cancer cell lines. Scale bar, 200 nm. B Particle size distribution of isolated exosomes. C Western blot analysis of Alix, TSG101 and CD63, marker proteins of exosomes isolated from MDA-MB-231 and MCF7 cells. D Exosomes were labelled using Dio stain and co-incubated with MCF7 and MDA-MB-231 cells for 3 h. Scale bar, 10 μm. E Wound healing experiments with MCF7 and MDA-MB-231 cells treated with MDA-MB-231 exosomes and MCF7 exosomes, respectively. Data are presented as mean ± SD (n = 3). *p < 0.05, **p < 0.01, ***p < 0.001
Bioinformatic analysis to screen for differentially expressed miRNAs
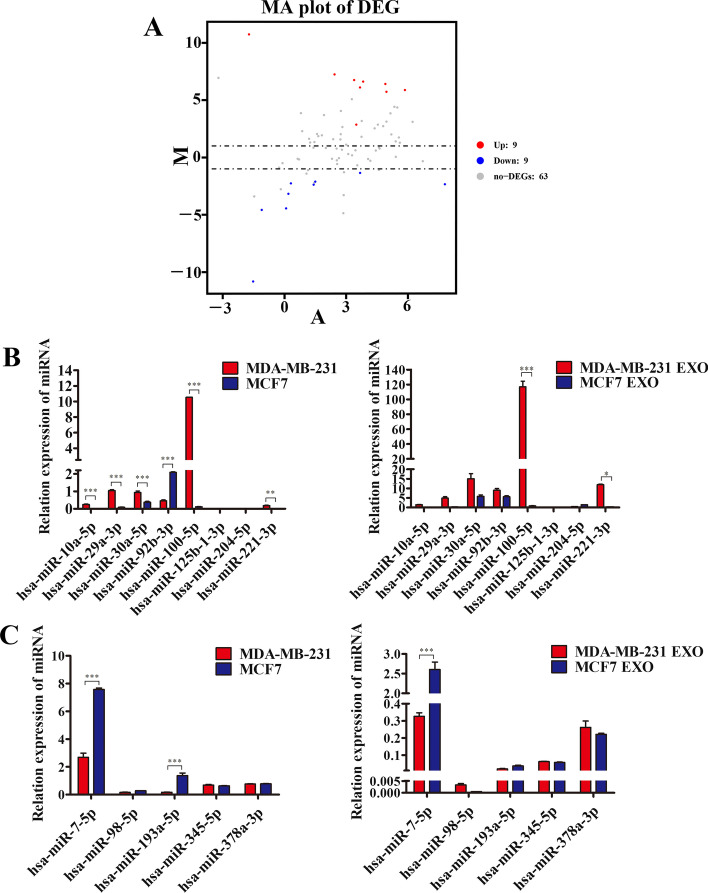
It is well known that miRNAs from cancer-derived exosomes are important regulatory molecules that mediate the link between cancer cells and host cells, so we investigated whether differentially expressed miRNAs within exosomes from different sources were responsible for mediating the functional differences between the two. We used the GSE114329 dataset from the GEO database to screen for differentially expressed miRNAs in two subsets of MDA231 EXO and MCF7 EXO by bioinformatics analysis. Nine relatively high and nine relatively low miRNAs were screened in exosomes produced by MDA-MB-231 cells (Fig. 2A). Several miRNAs with large differences in the replicate data were further excluded, several miRNAs that did not show significant differences probably owing to experimental reasons were selected, and finally eight miRNAs that were relatively highly expressed in exosomes produced by MDA-MB-231 cells and five miRNAs that were relatively lowly expressed were selected (Fig. 2A; Additional file 1: Table S3). We used qRT-PCR to validate the expression of several miRNAs in cells and exosomes. The experiments showed that hsa-miR-7-5p and hsa-miR-100-5p were most clearly different in cells and exosomes (Fig. 2B, C). We first selected hsa-miR-7-5p as our study subject. It was clear that hsa-miR-7-5p was significantly more expressed in low-invasive cells and exosomes than in highly invasive cells and exosomes.
Fig. 2.
Bioinformatic analysis to screen for differentially expressed miRNAs. A Bioinformatic analysis of the volcano plot results showed that MDA-MB-231 and MCF7 source differentially expressed miRNAs in exosomes. B The expression of selected eight up-regulated miRNAs in exosomes and cells was examined using real-time quantitative PCR. C Real-time quantitative PCR was used to detect the expression of selected five down-regulated miRNAs in exosomes and cells, respectively. Data are presented as mean ± SD (n = 3). *p < 0.05, **p < 0.01, ***p < 0.001
Downregulation of exosomal miR-7-5p expression significantly facilitates the migration and invasion of breast cancer cells
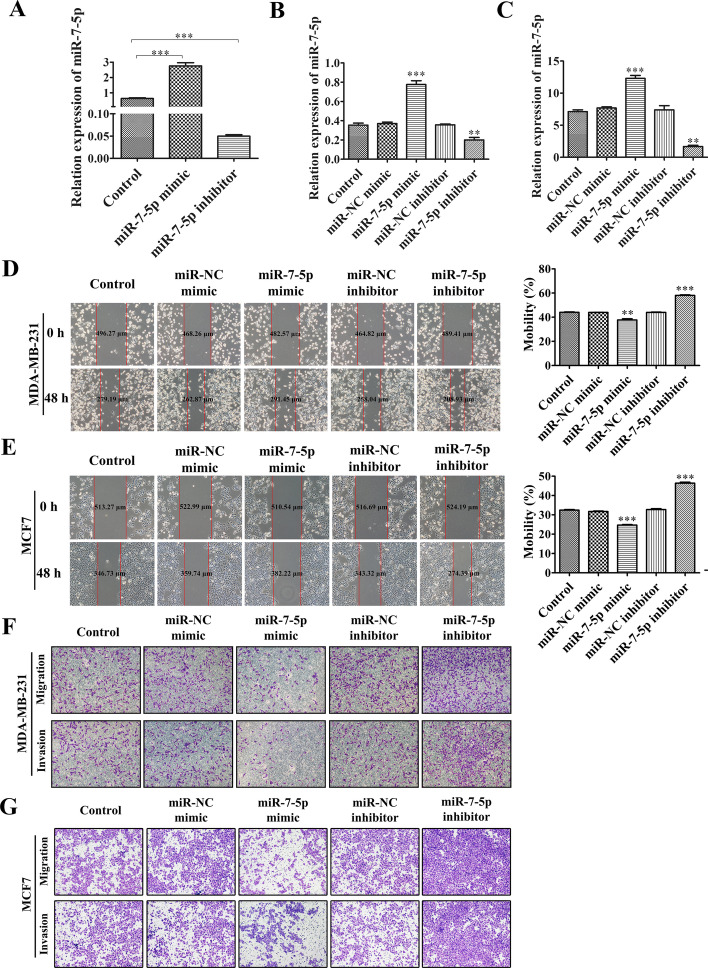
To further determine whether microRNAs function in dependence on exosomes for delivery, we used transfection experiments to increase or decrease intracellular expression of miR-7-5p. Cell cultures of transfected cells were then collected, and the exosomes released from the transfected cells were isolated. Whether changes in intracellular miR-7-5p levels cause corresponding changes in miR-7-5p within the exosomes was investigated by qRT-PCR assay. The experiments conclusively demonstrated that an increase in intracellular miR-7-5p also increased the amount of miR-7-5p in the exosomes they released, and similarly, a decrease in miR-7-5p expression led to a decrease in miR-7-5p expression in the exosomes the cells released (Fig. 3A). This suggests that intracellular miR-7-5p acts through exosomal delivery, at least to some extent.
Fig. 3.
Downregulation of exosomal miR-7-5p expression significantly facilitates the migration and invasion of breast cancer cells. A Expression of miR-7-5p in the secreted exosomes of blank control, miR-7-5p mimic and suppressor transfected MDA-MB-231 cells was measured using qRT-PCR assay. B, C The expression of miR-7-5p in MDA-MB-231 cells and MCF7 cells after transfection of blank control, miR-7-5p mimics and inhibitors and their negative controls was measured using qRT-PCR assay. D, E Wound healing assay using blank control, miR-7-5p mimic and inhibitor and its negative control after treatment of MDA-MB-231 cells and MCF7 cells. F, G The migratory invasion ability of cells after treatment of MDA-MB-231 cells and MCF7 cells with blank control, miR-7-5p mimics and inhibitors and their negative control was examined using Transwell assay. Data are presented as mean ± SD (n = 3). *p < 0.05, **p < 0.01, ***p < 0.001
We next investigate what role miR-7-5p plays in the intercellular delivery of breast cancer. We used wound healing assays and Transwell assays to investigate whether miR-7-5p has an effect on the migration invasion of breast cancer cells. We first verified that both mimics and inhibitors had been transfected into the cells within 48 h by qRT-PCR (Fig. 3B, C). The results of the wound healing and Transwell assays showed that the migration invasion ability of breast cancer cells in the miR-7-5p mimic group was significantly reduced, while the migration invasion ability of breast cancer cells in the miR-7-5p inhibitor group was significantly increased (Fig. 3D–G). This demonstrated that miR-7-5p significantly depressed breast cancer migration and invasion, which corresponds to our previous demonstration that miR-7-5p is highly expressed in low-invasive breast cancer cell lines and their exosomes, but lowly expressed in high-invasive breast cancer cell lines and their exosomes.
Hsa-miR-7-5p targets the RYK gene
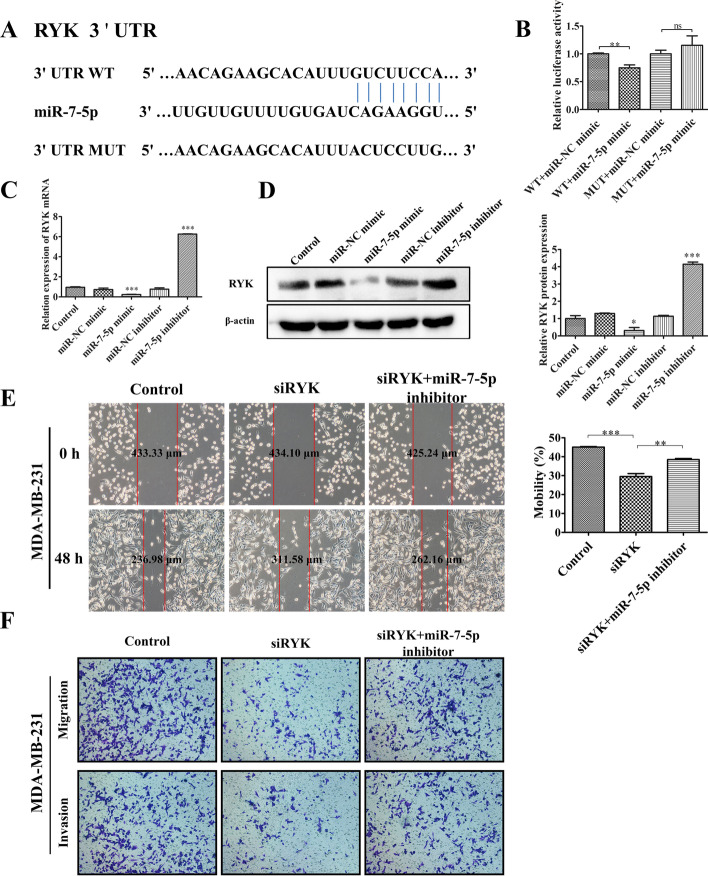
To further investigate the specific molecular mechanism by which miR-7-5p presents a significant inhibitory effect in breast cancer migration invasion, we utilised four miRNA target gene prediction software (miRTarBase, miRDB, TargetScan, miRWalk) to predict candidate target genes for miR-7-5p. We screened 41 candidate target genes (Additional file 1: Fig. S1), and then genes associated with tumour cell migration and invasion were included as further screening requirements. We finally identified the RYK genes with high scores as possible target genes for miR-7-5p. We next used only highly invasive MDA-MB-231 cells as our study subjects. We predicted the binding site for miR-7-5p in the 3′ UTR of the RYK gene (wild type) and targeted it for mutation (mutant type) and constructed plasmids for transfection (Fig. 4A). The experimental results showed that co-transfected miR-7-5p mimics significantly reduced reporter gene expression after the RYK wild type 3′ UTR, whereas no change in reporter gene expression was observed in the mutant group (Fig. 4B). Therefore, we concluded that RYK is a direct target of miR-7-5p. Consistent with the outcome of the dual luciferase reporter gene assay, the RYK gene also showed markedly reduced transcript levels and protein expression levels following transfer of miR-7-5p mimics (Fig. 4C, D). Our further in vitro experiments showed that RYK knockdown significantly inhibited the migratory and invasive ability of MDA-MB-231 cells compared with the control group. This result was identical to that obtained after treatment using miR-7-5p mimic. However, the inhibitory effect after RYK knockdown was reversed to some extent by miR-7-5p inhibitor (Fig. 4E, F). Thus, it is evident that exosomal miR-7-5p inhibits the migratory invasion of breast cancer to some extent by targeting the RYK gene.
Fig.4.
Hsa-miR-7-5p targets the RYK gene. A The wild type (WT) and mutant (MUT) of miR-7-5p at the binding site of the 3′ UTR of the RYK gene. B Validation of miR-7-5p targeting to RYK using luciferase reporter gene assays. MDA-MB-231 cells were co-transfected with negative control (NC) or miR-7-5p mimics and luciferase reporter plasmids (including RYK 3′ UTR WT or RYK 3′ UTR MUT). C Expression of RYK mRNA in MDA-MB-231 cells transfected with NC or miR-7-5p mimics and inhibitors was measured by quantitative real-time PCR. D Western blot analysis of RYK protein expression in MDA-MB-231 cells transfected with NC or miR-7-5p mimics and inhibitors. E Wound healing experiments using blank control, siRYK and siRYK + miR-7-5p inhibitor after co-treatment of MDA-MB-231 cells. F A Transwell assay was used to verify the migratory invasion ability of MDA-MB-231 cells treated with blank control, siRYK and siRYK + miR-7-5p inhibitor. Data are presented as mean ± SD (n = 3). *p < 0.05, **p < 0.01, ***p < 0.001
Hsa-miR-7-5p is involved in the JNK-mediated atypical WNT signalling pathway and significantly inhibits breast cancer metastasis
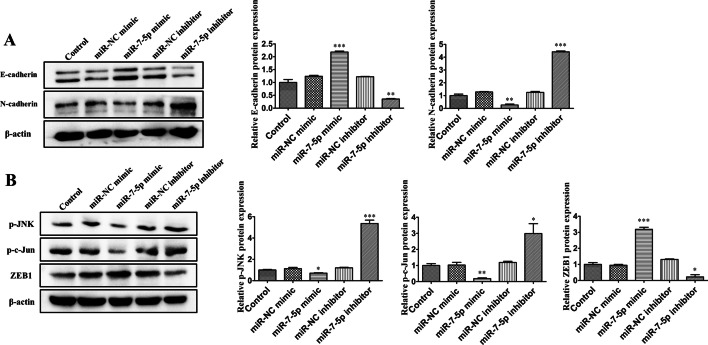
Numerous studies have demonstrated that the migratory and invasion of breast cancer is closely related to the development of EMT. Our experiments also show that transfection of miR-7-5p mimics promotes the expression of E-cadherin and inhibits the expression of N-cadherin (Fig. 5A). Thus, miR-7-5p can inhibit the development of EMT. It has also been demonstrated that JNK and c-Jun in the atypical WNT signalling pathway, in which RYK is involved, can be involved in EMT development [23], which prompted us to investigate whether JNK and c-Jun, which are downstream factors of the RYK receptor, can be affected by miR-7-5p. We found that downregulation of miR-7-5p in breast cancer cells promoted a cascade response of the atypical WNT pathway, that is, phosphorylation of JNK and c-Jun proteins (Fig. 5B). Phosphorylated c-Jun protein inhibits the expression of the EMT core transcription factor ZEB1 (Fig. 5B), thereby suppressing the expression of E-cadherin and promoting the expression of N-cadherin, thereby facilitating the EMT process and promoting breast cancer metastasis. Ultimately, we demonstrated that miR-7-5p can inhibit breast cancer migration invasion by targeting the RYK gene, which is involved in the JNK-mediated atypical WNT signalling pathway.
Fig. 5.
Hsa-miR-7-5p is involved in the JNK-mediated atypical WNT signalling pathway and significantly inhibits breast cancer metastasis. A Western blot analysis of E-cadherin and N-cadherin protein expression levels after treatment of MDA-MB-231 cells with blank controls or miR-7-5p mimics, inhibitors and their negative controls. B Western blot analysis of p-JNK, p-c-Jun and ZEB1 protein expression levels after treatment of MDA-MB-231 cells with blank control or miR-7-5p mimics, inhibitors and their negative controls. Data are presented as mean ± SD (n = 3). *p < 0.05, **p < 0.01, ***p < 0.001
Discussion
Breast cancer is currently the leading malignancy threatening women’s mortality worldwide [24]. Metastatic malignant breast cancer accounts for 20–30% of breast cancer cases and the survival rate of the disease is only 22%, making it the leading cause of death for patients [5, 25, 26]. Metastatic spread of breast cancer involves many steps, including EMT, migration, invasion, extravasation, mesenchymal–epithelial transformation (MET) and colonisation [27]. An increasing range of research has demonstrated the importance of exosomes for all metastatic processes such as migration invasion of cancer [28, 29]. It has been shown that cancer cells secrete much higher levels of exosomes than those released by normal cells, and therefore the exosomes secreted by cancer cells themselves are critical for the exchange of genetic information and the reprogramming of metabolism between cells, as well as the microenvironment in which the cells live [30, 31]. We screened through validation that miR-7-5p was relatively highly expressed in low-invasive breast cancer cell lines and less expressed in highly invasive cells. Further investigation of the mechanism of miR-7-5p action revealed that the RYK gene may act as a new target gene for miR-7-5p.
Among these, the RYK gene is a receptor-like tyrosine kinase, characterised by impaired kinase activity, which can exert regulatory effects by binding WNT ligands through its structural region [32]. In fact, RYK has been studied extensively in human development and more initially in cancer [33]. However, in recent years, RYK has been investigated as a receptor in atypical signalling pathways in many cancers [32, 34]. One study found that inhibition of β-catenin, a part of the typical WNT pathway, did not antagonise the RYK-induced metastatic phenotype in gastric cancer, demonstrating that the pro-metastatic effects of RYK in gastric cancer are mediated, at least in part, through the atypical Wnt signalling pathway [34]. It has been demonstrated that in prostate cancer the WNT5A/RYK signalling pathway is involved in the apoptotic and proliferative processes of cancer [35]. It has also been demonstrated that, as a response to WNT5A, RYK promotes the proliferation of breast stem and progenitor cells, and further studies have found that RYK plays a role as a receptor for the atypical WNT signalling pathway in the promotion of brain metastasis in breast cancer [36]. And in glioblastoma, RYK knockdown inhibited the invasiveness of wnt5a-induced U-105MG and U251MG glioma cells [37]. Various studies have demonstrated that abnormal WNT/RYK signalling may promote the oncogenic properties of cancer, particularly in tissues where RYK development is important. It has been reported that the atypical WNT pathway is often not dependent on β-catenin, but instead involves small GTPases of the Rho family, such as Rho or Rac, and that signalling enhances cell motility through the JNK/c-JUN cascade signalling response [36]. However, phosphorylated c-Jun protein is often able to affect the expression of EMT transcription factors (e.g. ZEB1) and thereby influence the EMT process. EMT is widely known to be an important process affecting the migration and invasion of breast cancer [23, 38, 39]. Interestingly, we demonstrated experimentally that miR-7-5p expression was significantly positively correlated with E-cadherin expression and significantly negatively correlated with N-cadherin expression. This indicates that miR-7-5p can inhibit the EMT process. We further experimentally verified that miR-7-5p was also able to affect the phosphorylation of JNK and c-Jun protein and ZEB1 protein.
Therefore, we determined that miR-7-5p targets the RYK gene, causing deletion of the RYK receptor and affecting the JNK-mediated cascade of the atypical WNT pathway, leading to effects on the phosphorylation of JNK and c-Jun proteins, resulting in increased expression of EMT transcription factors such as ZEB1. This in turn affects the expression of E-cadherin and suppresses EMT, ultimately inhibiting the migration and invasion of breast cancer (Additional file 1: Fig. S2).
There are of course limitations to our study. (1) We were not able to analyse the clinical significance of exosomal miR-7-5p in breast cancer by collecting case samples of tissue. (2) Given the limited experimental conditions, we should have used more breast cancer cell lines to validate our results. (3) We should further validate the effect of miR-7-5p in vivo by nude mouse tumorigenesis assay (Additional file 2).
Conclusions
In conclusion, our experimental data show that miR-7-5p in exosomes is closely associated with breast cancer migration and invasion. We identified a new target factor for miR-7-5p, RYK. We linked the oncogenic effect of miR-7-5p to the atypical WNT signalling pathway via the RYK gene. Our findings provide new targets for the treatment of advanced breast cancer and new ideas for the study of the molecular mechanisms of miRNAs.
Supplementary Information
Additional file 1: Table S1. Sequences of oligonucleotide fragment. Table S2 Sequences of primers required for the experiment. Table S3 Relative expression of several miRNAs screened from the GSE114329 dataset.
Acknowledgements
No applicable.
Abbreviations
- qRT-PCR
Real-time quantitative PCR
- EXO
Exosome
- TEM
Transmission electron microscopy
- RYK
Receptor-like tyrosine kinase
- EMT
Epithelial–mesenchymal transition
- miRNAs
MicroRNAs
- 3′ UTR
3′ non-coding region
- WT
Wild type
- MUT
Mutant type
- MET
Mesenchymal–epithelial transformation
- p-JNK
Phosphorylated JNK protein
- p-c-Jun
Phosphorylated c-Jun protein
Author contributions
Z.L. analysed and studied the experiment; Z.L. and L.L. collated the data and wrote the manuscript; R.G. and C.C. assisted with the experiment. Y.G. designed and directed the study. All authors have read and approved the final manuscript.
Funding
This work was supported by Natural Science Foundation of Shandong Province (ZR2020MC064).
Availability of data and materials
Please contact the corresponding author for data requests.
Declarations
Ethics approval and consent to participate
No applicable.
Consent for publication
All authors have read and approved the final manuscript.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71(3):209–249. doi: 10.3322/caac.21660. [DOI] [PubMed] [Google Scholar]
- 2.Liu M, Gong C, Xu RY, Chen Y, Wang XD. MicroRNA-5195-3p enhances the chemosensitivity of triple-negative breast cancer to paclitaxel by downregulating EIF4A2. Cell Mol Biol Lett. 2019;24(1):1–11. doi: 10.1186/s11658-019-0168-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Abdulrahman GO, Jr Rahman, Adebisi G. Epidemiology of breast cancer in Europe and Africa. J Cancer Epidemiol. 2012;2012:915610. [DOI] [PMC free article] [PubMed]
- 4.Ding JJ, Xu Z, Zhang YY, Tan CL, Hu WZ, Wang M, et al. Exosome-mediated miR-222 transferring: an insight into NF-kappa-mediated breast cancer metastasis. Exp Cell Res. 2018;369(1):129–138. doi: 10.1016/j.yexcr.2018.05.014. [DOI] [PubMed] [Google Scholar]
- 5.Wu Q, Li JJ, Zhu S, Wu J, Chen C, Liu Q, et al. Breast cancer subtypes predict the preferential site of distant metastases: a SEER based study. Oncotarget. 2017;8(17):27990–27996. doi: 10.18632/oncotarget.15856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gallo A, Tandon M, Alevizos I, Illei GG, et al. The majority of microRNAs detectable in serum and saliva is concentrated in exosomes. PLoS ONE. 2012;7(3):e30679. doi: 10.1371/journal.pone.0030679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zomer A, Vendrig T, HopmansErik S, van Eijndhoven M, Middeldorp Jaap M, Michiel PD, et al. Exosomes: fit to deliver small RNA. Commun Integr Biol. 2010;3(5):447–450. doi: 10.4161/cib.3.5.12339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Record M, Carayon K, Poirot M, Silvente-Poirot S. Exosomes as new vesicular lipid transporters involved in cell–cell communication and various pathophysiologies. BBA Mol Cell Bio L. 2014;1841(1):108–120. doi: 10.1016/j.bbalip.2013.10.004. [DOI] [PubMed] [Google Scholar]
- 9.Akers JC, Gonda D, Kim R, Carter BS, Chen CC. Biogenesis of extracellular vesicles (EV): exosomes, microvesicles, retrovirus-like vesicles, and apoptotic bodies. J Neuro Oncol. 2013;113(1):1–11. doi: 10.1007/s11060-013-1084-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lu L, Huang JJ, Mo JT, Da X, Li QX, Fan M, et al. Exosomal lncRNA TUG1 from cancer-associated fibroblasts promotes liver cancer cell migration, invasion, and glycolysis by regulating the miR-524-5p/SIX1 axis. Cell Mol Biol Lett. 2022;27(1):1–17. doi: 10.1186/s11658-022-00309-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Png KJ, Halberg N, Yoshida M, Tavazoie SF. A microRNA regulon that mediates endothelial recruitment and metastasis by cancer cells. Nature. 2012;481(7380):190–194. doi: 10.1038/nature10661. [DOI] [PubMed] [Google Scholar]
- 12.Rabinowits G, Gercel-Taylor C, Day JM, Taylor DD, Kloecker GH. Exosomal microRNA: a diagnostic marker for lung cancer. Clin Lung Cancer. 2009;10(1):42–46. doi: 10.3816/CLC.2009.n.006. [DOI] [PubMed] [Google Scholar]
- 13.Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75(5):843–854. doi: 10.1016/0092-8674(93)90529-Y. [DOI] [PubMed] [Google Scholar]
- 14.Zhang GX, Zhang W, Li BJ, Stringer-Reasor E, Chu CJ, Sun LY, et al. MicroRNA-200c and microRNA-141 are regulated by a FOXP3-KAT2B axis and associated with tumor metastasis in breast cancer. Breast Cancer Res. 2017;19(1):1–13. doi: 10.1186/s13058-017-0858-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hong BS, Ryu HS, Kim N, Kim J, Lee E, Moon H, et al. Tumor suppressor miRNA-204-5p regulates growth, metastasis, and immune microenvironment remodeling in breast cancer. Cancer Res. 2019;79(7):1520–1534. doi: 10.1158/0008-5472.CAN-18-0891. [DOI] [PubMed] [Google Scholar]
- 16.Ling YT, Cao CX, Li SJ, Qiu M, Shen GX, Chen ZW, et al. TRIP6, as a target of miR-7, regulates the proliferation and metastasis of colorectal cancer cells. Biochem Biophys Res Commun. 2019;514(1):231–238. doi: 10.1016/j.bbrc.2019.04.092. [DOI] [PubMed] [Google Scholar]
- 17.Wang Y, Yang HT, Zhang GY, Luo CJ, Zhang SZ, Luo RY, et al. Hsa-miR-7-5p suppresses proliferation, migration and promotes apoptosis in hepatocellular carcinoma cell lines by inhibiting SPC24 expression. Biochem Biophys Res Commun. 2021;561:80–87. doi: 10.1016/j.bbrc.2021.05.020. [DOI] [PubMed] [Google Scholar]
- 18.Tang Y, Tang ZY, Yang JR, Liu TQ, Tang YT. MicroRNA-7-5p inhibits migration, invasion and metastasis of intrahepatic cholangiocarcinoma by inhibiting MyD88. J Clin Transl Hepato. 2021;9(6):809–817. doi: 10.14218/JCTH.2021.00021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ye TB, Yang MH, Huang DC, Wang X, Xue BQ, Tian N, et al. MicroRNA-7 as a potential therapeutic target for aberrant NF-kappa B-driven distant metastasis of gastric cancer. J Exp Clin Cancer Res. 2019;38(1):1–18. doi: 10.1186/s13046-019-1074-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xiao HP. MiR-7-5p suppresses tumor metastasis of non-small cell lung cancer by targeting NOVA2. Cell Mol Biol Lett. 2019;24(1):1–13. doi: 10.1186/s11658-019-0188-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huang Q, Wu YY, Xing SJ, Yu ZW. Effect of miR-7 on resistance of breast cancer cells to adriamycin via regulating EGFR/PI3K signaling pathway. Eur Rev Med Pharmaco. 2019;23(12):5285–5292. doi: 10.26355/eurrev_201906_18195. [DOI] [PubMed] [Google Scholar]
- 22.Hong TZ, Ding J, Li WL. MiR-7 reverses breast cancer resistance to chemotherapy by targeting MRP1 and BCL2. Oncotargets Ther. 2019;12:11097–11105. doi: 10.2147/OTT.S213780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xu XF, Zhang MF, Xu FY, Jiang SJ. Wnt signaling in breast cancer: biological mechanisms, challenges and opportunities. Mol Cancer. 2020;19(1):1–35. doi: 10.1186/s12943-020-01276-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lv MH, Mao QX, Li JT, Qiao JH, Chen XC, Luo SX. Knockdown of LINC00665 inhibits proliferation and invasion of breast cancer via competitive binding of miR-3619-5p and inhibition of catenin beta 1. Cell Mol Biol Lett. 2020;25(1):1–13. doi: 10.1186/s11658-020-00235-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kia V, Paryan M, Mortazavi Y, Biglari A, Mohammadi-Yeganeh S. Evaluation of exosomal miR-9 and miR-155 targeting PTEN and DUSP14 in highly metastatic breast cancer and their effect on low metastatic cells. J Cell Biochem. 2019;120(4):5666–5676. doi: 10.1002/jcb.27850. [DOI] [PubMed] [Google Scholar]
- 26.Giordano C, La Camera G, Gelsomino L, Barone I, Bonofiglio D, Ando S, et al. The biology of exosomes in breast cancer progression: dissemination, immune evasion and metastatic colonization. Cancers. 2020;12(8):2179. doi: 10.3390/cancers12082179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Danac JMC, Uy AGG, Garcia RL. Exosomal microRNAs in colorectal cancer: overcoming barriers of the metastatic cascade (review) Int J Mol Med. 2021;47(6):1–16. doi: 10.3892/ijmm.2021.4945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang Y, Liu YF, Liu HY, Tang WH. Exosomes: biogenesis, biologic function and clinical potential. Cell Biosci. 2019;9(1):1–18. doi: 10.1186/s13578-019-0282-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Huang HH, Zheng XC, Cai CQ, Yao ZC, Lu ST, Meng XJ, et al. Exosomes derived from breast cancer lung metastasis subpopulations promote tumor self-seeding. Biochem Biophys Res Commun. 2018;503(1):242–248. doi: 10.1016/j.bbrc.2018.06.009. [DOI] [PubMed] [Google Scholar]
- 30.Harris DA, Patel SH, Gucek M, Hendrix A, Westbroek W, Taraska JW. Exosomes released from breast cancer carcinomas stimulate cell movement. PLoS ONE. 2015;10(3):e0117495. doi: 10.1371/journal.pone.0117495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li KY, Chen YH, Li A, Tan CL, Liu XB. Exosomes play roles in sequential processes of tumor metastasis. Int J Cancer. 2019;144(7):1486–1495. doi: 10.1002/ijc.31774. [DOI] [PubMed] [Google Scholar]
- 32.Roy JP, Halford MM, Stacker SA. The biochemistry, signalling and disease relevance of RYK and other WNT-binding receptor tyrosine kinases. Growth Factors. 2018;36(1–2):15–40. doi: 10.1080/08977194.2018.1472089. [DOI] [PubMed] [Google Scholar]
- 33.Kessenbrock K, Smith P, Steenbeek SC, Pervolarakis N, Kumar R, Minami Y, et al. Diverse regulation of mammary epithelial growth and branching morphogenesis through noncanonical Wnt signaling. Proc Natl Acad Sci USA. 2017;114(12):3121–3126. doi: 10.1073/pnas.1701464114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fu YG, Chen YL, Huang JH, Cai ZD, Wang YQ. RYK, a receptor of noncanonical Wnt ligand Wnt5a, is positively correlated with gastric cancer tumorigenesis and potential of liver metastasis. AM J Physiol-gastr L. 2020;318(2):G352–G360. doi: 10.1152/ajpgi.00228.2019. [DOI] [PubMed] [Google Scholar]
- 35.Thiele S, Zimmer A, Gobel A, Rachner TD, Rother S, Fuessel S, et al. Role of WNT5A receptors FZD5 and RYK in prostate cancer cells. Oncotarget. 2018;9(43):27293–27304. doi: 10.18632/oncotarget.25551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Klemm F, Bleckmann A, Siam L, Chuang HN, Rietkotter E, Behme D, et al. Beta-catenin-independent WNT signaling in basal-like breast cancer and brain metastasis. Carcinogenesis. 2011;32(3):434–442. doi: 10.1093/carcin/bgq269. [DOI] [PubMed] [Google Scholar]
- 37.Adamo A, Fiore D, De Martino F, Roscigno G, Affinito A, Donnarumma E, et al. RYK promotes the stemness of glioblastoma cells via the WNT/beta-catenin pathway. Oncotarget. 2017;8(8):13476–13487. doi: 10.18632/oncotarget.14564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lamouille S, Xu J, Derynck R. Molecular mechanisms of epithelial–mesenchymal transition. Nat Rev Mol Cell Biol. 2014;15(3):178–196. doi: 10.1038/nrm3758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Byun Y, Choi YC, Jeong Y, Lee G, Yoon S, Jeong Y, et al. MiR-200c downregulates HIF-1 and inhibits migration of lung cancer cells. Cell Mol Biol Lett. 2019;24(1):1–14. doi: 10.1186/s11658-019-0152-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Table S1. Sequences of oligonucleotide fragment. Table S2 Sequences of primers required for the experiment. Table S3 Relative expression of several miRNAs screened from the GSE114329 dataset.
Data Availability Statement
Please contact the corresponding author for data requests.