Abstract
The SERINC (serine incorporator) proteins are host restriction factors that inhibit infection by HIV through their incorporation into virions. Here, we found that SERINC3 and SERINC5 exhibited additional antiviral activities by enhancing the expression of genes encoding type I interferons (IFNs) and nuclear factor κB (NF-κB) signaling. SERINC5 interacted with the outer mitochondrial membrane protein MAVS (mitochondrial antiviral signaling) and the E3 ubiquitin ligase and adaptor protein TRAF6, resulting in MAVS aggregation and polyubiquitylation of TRAF6. Knockdown of SERINC5 in target cells increased single-round HIV-1 infectivity, as well as infection by recombinant vesicular stomatitis virus (rVSV) bearing VSV-G or Ebola virus (EBOV) glycoproteins. Infection by an endemic Asian strain of Zika virus (ZIKV), FSS13025, was also enhanced by SERINC5 knockdown, suggesting that SERINC5 has direct antiviral activities in host cells in addition to the indirect inhibition mediated by its incorporation into virions. Further experiments suggested that the antiviral activity of SERINC5 was type I IFN–dependent. Together, these results highlight a previously uncharacterized function of SERINC proteins in promoting NF-κB inflammatory signaling and type I IFN production, thus contributing to its antiviral activities.
INTRODUCTION
The host immune response to viral infection is mediated by pattern recognition receptors (PRRs), including Toll-like receptors, Nod-like receptors, retinoic acid–inducible gene I (RIG-I)–like receptors (RLRs), and other cytosolic nucleic acid sensors (1, 2). Upon infection, these PRRs recognize viral DNA or RNA and trigger the activation of the transcription factors interferon (IFN) regulatory factor 3 (IRF3), IRF7, and nuclear factor κB (NF-κB), as well as inflammasome pathways, leading to the production of type I IFNs and proinflammatory cytokines, which together serve as the first line of defense to fight against pathogen invasion (3, 4). Common RLRs that recognize viral RNA include RIG-I, melanoma differentiation gene 5 (MDA5), and laboratory of genetics and physiology 2 (LGP2) (4, 5). Upon ligand binding, RIG-I and MDA5 activate the adaptor protein mitochondrial antiviral signaling (MAVS) through their caspase activation recruitment domains (CARDs) (6–9), resulting in the formation of prion-like MAVS aggregates (10), which further recruit a set of tumor necrosis factor (TNF) receptor–associated factors (TRAFs; TRAF2, TRAF5, and TRAF6) to activate the TANK- binding kinase 1 (TBK1)-IKK (IκB kinase) complex (11). These kinases subsequently activate transcription factors, such as IRF3, IRF7, and NF-κB, thereby inducing the expression of genes encoding type I IFNs and the production of proinflammatory cytokines (12).
Serine incorporator (SERINC) proteins, primarily SERINC3 and SERINC5, were identified as critical cellular restriction factors that inhibit HIV-1 infectivity when expressed in the virus-producing cell. The antiviral functions of SERINCs are counteracted by the HIV accessory protein Nef, the murine leukemia virus (MLV) protein glycoGag, and the equine infectious anemia virus S2 protein (13–16). Whereas the exact mechanism by which SERINCs restrict HIV-1 remains to be determined, incorporation of SERINC3 and SERINC5 into HIV-1 virions in virus-producing cells interferes with the infection of new target cells by HIV-1, likely at the entry step (17–21). Nef reduces the abundance of SERINC3 and SERINC5 at the plasma membrane and relocalizes SERINCs into a lysosomal compartment, leading to decreased incorporation of SERINC3 and SERINC5 into HIV-1 virions (13, 14), thus enhancing HIV-1 infectivity. Down-regulation of SERINC proteins by Nef, glycoGag, and S2 from the cell surface is mediated by direct engagement of the AP-2–dependent endocytic pathway (22–24). The cryo–electron microscopy structures of human and Drosophila SERINC5 were determined at a near-atomic resolution (25), revealing that surface-exposed regions and the interface between subdomains of SERINC5 are critical for the restriction of HIV-1 infectivity.
Many host antiviral restriction factors are produced in response to type I IFNs [and are encoded by IFN-stimulated genes (ISGs)], and in general, they are under positive selection (26–30). However, the production of SERINC proteins is not IFN inducible, nor are they under positive selection (31). Therefore, SERINCs may be considered as nonclassical restriction factors. Furthermore, many of the factors encoded by ISGs themselves regulate the type I IFN and NF-κB signaling pathways, either positively or negatively, thereby regulating antiviral immunity (32). For example, Tetherin (also known as Bst2) functions as a sensor to activate the NF-κB pathway and induce the expression of genes encoding proinflammatory cytokines (33, 34). TRIM5α, another restriction factor that targets HIV-1 uncoating, serves as a PRR to regulate cellular innate immune responses (35, 36). N4BP1, a restriction factor that targets HIV-1 RNA (37), suppresses basal NF-κB activity (38). Here, we provide evidence that SERINC3 and SERINC5 promote innate immune signaling, resulting in increased production of type I IFNs and proinflammatory cytokines, thereby inhibiting infection by HIV-1, vesicular stomatitis virus (VSV), and Zika virus (ZIKV).
RESULTS
Transient expression of SERINC5 enhances the type I IFN and NF-κB signaling pathways
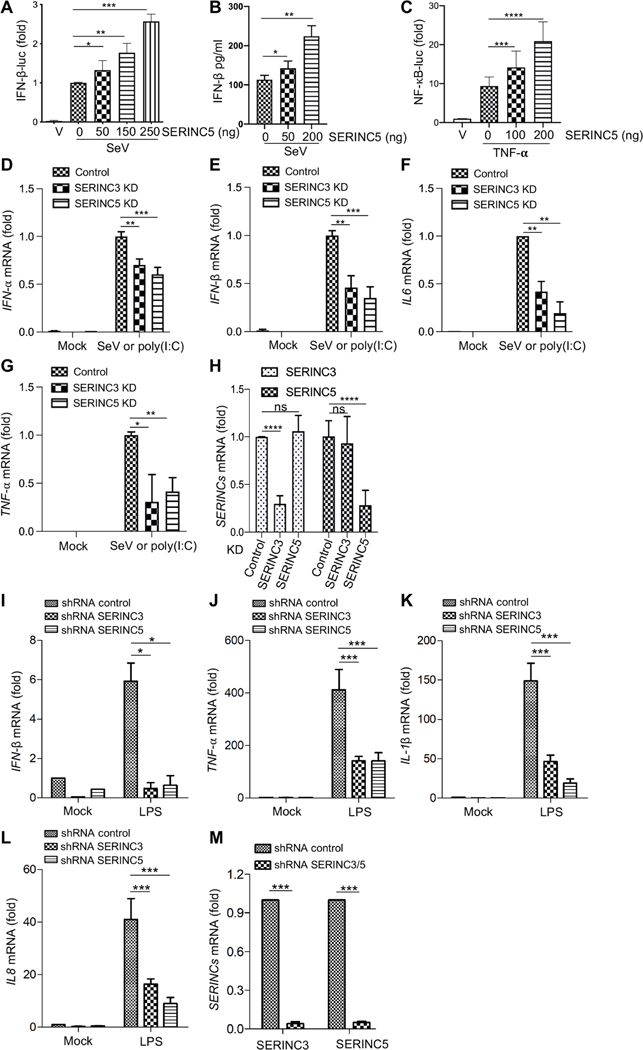
We cotransfected human human embryonic kidney (HEK) 293T cells with increasing amounts (0, 50, 150, and 250 ng) of a SERINC5 expression plasmid, either pBJ-SERINC5-HA (hemagglutinin) or pQCXIP-SERINC5-FLAG, together with an IFN-β–luciferase reporter vector. After infection of the transfected cells with Sendai virus (SeV), we observed increased IFN-β–driven luc activities and IFN-β production in a manner that correlated with the increased amount of plasmid used (Fig. 1, A and B). We treated similarly transfected HEK293T cells with TNF-α (10 ng/ml) for 12 hours and found that NF-kB–driven luc activities were also increased with increased amount of plasmid (Fig. 1C). To confirm these results in other cell types, we applied doxycycline (0.5 μg/ml) to a phorbol 12-myristate 13-acetate (PMA)–treated THP-1 monocytic cell line (to induce their maturation into macrophages) inducibly expressing SERINC5, and we observed increased amounts of mRNA encoding IFN-β, IFN-α, ISG15, TNF-α, and IL-6 (interleukin-6) 8 hours after SeV infection (fig. S1, A to F). Consistent with the overexpression data, knockdown of SERINC5 in PMA-treated THP-1 cells led to decreased amounts of mRNAs encoding IFN-β and TNF-α (fig. S1, G and I). Together, these data support the hypothesis that SERINC5 promotes the expression of type I IFNs and the NF-κB signaling pathway in human HEK293T cells and PMA-treated THP-1 cells.
Fig. 1. SERINC5 enhances the type I IFN and NF-κB signaling pathways.
(A to C) HEK293T cells were transfected with the indicated amounts of SERINC5-expressing plasmids together with that of IFN-β (A) or NF-κB luc (C) and pRLTK expression vectors. Twenty-four hours later, the cells were infected with SeV at an MOI of 2 (A and B) or treated with TNF-α (10 ng/ml) (C) for 8 hours. Forty- eight hours after transfection, the cells were lysed, and firefly and renilla luc activities were measured. IFN-β production was measured by ELISA (B). All data are plotted as relative values from five (A and C) or three (B) independent experiments by setting the values in cells not transfected with SERINC5-expressing plasmid to zero. (D to H) Human MDMs from three different donors (n = 3) were transduced with lentiviral shRNAs targeting SERINC3 or SERINC5 or with a scrambled control. The cells were incubated with SeV or poly(I:C), and the relative amounts of mRNAs for IFN-α, IFN-β, IL-6, and TNF-α were measured by qPCR analysis. (H) Analysis of the relative KD efficiencies of SERINC3 and SERINC5. (I to L) Human MDMs from four different donors (n = 4) were transduced with lentiviral shRNAs targeting SERINC3 or SERINC5 or with scrambled control shRNA. The cells were stimulated with LPS (100 ng/ml) for 8 hours, and then the relative amounts of mRNAs for IFN-β, TNF-α, IL-1β, and IL-8 were measured by qPCR analysis. (M) The average KD efficiencies of SERINC3 and SERINC5 mRNA in all four donors were determined. Note that in all cases, the values of cells transduced with scrambled control shRNA and then incubated with SeV, poly(I:C), or LPS were set to 1.00 for comparison. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001. ns, not significant.
KD of SERINC3 or SERINC5 in human MDMs diminishes type I IFN and NF-κB signaling
To determine whether endogenous SERINC proteins regulate the expression of genes encoding type I IFNs and NF-κB signaling, we measured the amounts of mRNA encoding IFN-α, IFN-β, IL-6, TNF-α, and several other cytokines in primary human monocyte-derived macrophages (MDMs) that were transduced with lentiviruses expressing short hairpin RNAs (shRNAs) targeting SERINC3 or SERINC5 and then were infected with SeV or were treated with polyinosinic:polycytidylic acid [poly(I:C)] or lipopolysaccharide (LPS). KD of SERINC3 or SERINC5 reduced the amounts of mRNAs encoding IFN-α, IFN-β, IL-6, and TNF-α in cells infected with SeV or treated with poly(I:C) by about two- to threefold (Fig. 1, D to G, and fig. S2, A to C), reflecting the KD efficiency of SERINC3 and SERINC5 in these cells (Fig. 1H and fig. S2, A to C). We also stimulated MDMs with LPS (100 ng/ml) for 8 hours to activate the NF-κB pathway, and we observed decreased cytokine mRNA abundance upon SERINC KD. The amount of mRNA encoding IFN-β was decreased ~10-fold after KD of SERINC3 or SERINC5 in LPS-stimulated MDMs (Fig. 1I and fig. S3, A to D). The amounts of mRNA encoding TNF-α, IL-1β, and IL-8 were also greatly decreased in SERINC3- and SERINC5-KD cells upon stimulation with LPS, albeit to different extents (Fig. 1, J to L, and fig. S3, A to D). The KD efficiency of SERINC3 and SERINC5 in LPS-stimulated MDMs from four donors was about ~90% based on analysis of mRNA abundance (Fig. 1M and fig. S3E). Together, these results suggest that endogenous SERINC3 and SERINC5 proteins in human MDMs positively regulate the expression of type I IFNs and the NF-κB signaling pathway.
SERINC5 promotes IRF3 and IκBα phosphorylation and enhances IRF3 nuclear translocation
The hallmark of the activation of type I IFN production and NF-κB signaling pathways is the phosphorylation of key cellular molecules, leading to expression of select target genes in the nucleus (39). We observed that upon stimulation of cells expressing exogenous SERINC5 with poly(I:C), the extent of phosphorylation of IRF3 and IκBα was increased in 293T cells (fig. S4, A and C). Note that the total abundance of IκBα was decreased in cells expressing high amounts of SERINC5, which inversely correlated with increased amounts of phosphorylated IκBα (fig. S4, A and C). We next infected HEK293 cells stably transfected with an empty retroviral vector (pQCXIP) or a retroviral vector expressing SERINC5 (pQCXIP-SERINC5) with SeV for different times and observed that the amounts of p52 and p100, which are a proxy for assessment of the activation of the noncanonical NF-κB pathway (40), were also increased in SERINC5-expressing cells, in particular at 24 hours after SeV infection (fig. S4, B and C). Together, these results suggest that the expression of SERINC5 in HEK293T cells potentiates signaling by the type I IFN production and NF-κB pathways, including the noncanonical NF-κB pathway.
We next performed immunofluorescence staining and microscopic imaging to determine the possible effects of SERINC5 on the nuclear translocation of IRF3. In the absence of SeV infection and SERINC5 (SERINC5-mCherry) expression, IRF3 was predominantly localized in the cytoplasm of transfected HeLa cells, as shown by staining with an anti-IRF3 monoclonal antibody that recognizes both phosphorylated and nonphosphorylated IRF3 species (fig. S4D, top). Upon SeV infection for 8 hours, IRF3 was detected in both the cytosol and nucleus (fig. S4D, middle). Note that in cells that were transfected to express SERINC5-mCherry and then infected with SeV for 24 hours, we observed increased intensity of IRF3 nuclear staining in cells expressing SERINC5 as compared to those not expressing SERINC5 (fig. S4D, bottom). Although it was not possible to quantify the intensity of the IRF3 signal in the nucleus, these results suggest that the expression of SERINC5 may promote the nuclear translocation of IRF3, which is consistent with the potentiating effect of SERINC5 on type I IFN expression and NF-κB signaling.
SERINC5 associates with MAVS upon signaling activation
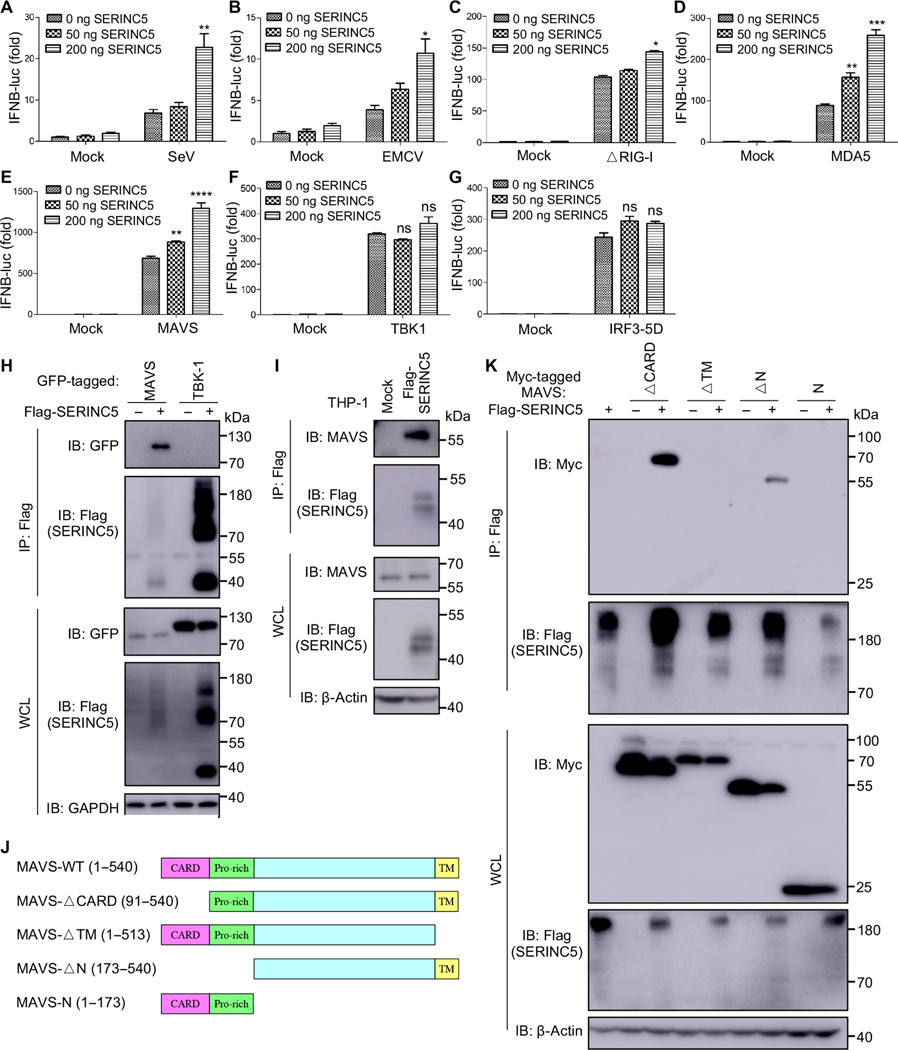
We next investigated at which step SERINC5 enhanced type I IFN production. This was achieved by performing an IFN-β–luc reporter assay in the presence of key molecules that activate the pathway at different levels. We observed that SERINC5 enhanced the IFN-β–luc activities in cells infected with SeV (which stimulates the RIG-I pathway) or encephalomyocarditis virus (EMCV; which stimulates the MDA5 pathway) and in cells overexpressing MDA5, ΔRIG-I (a constitutively active form of RIG-I), and MAVS (Fig. 2, A to E). However, overexpression of TBK1 or IRF3–5D (an active form of IRF3) had no apparent effect on IFN-β–luc activity (Fig. 2, F and G). These results suggest that SERINC5 functions at a step(s) between MAVS and TBK1 to potentiate type I IFN production.
Fig. 2. SERINC5 targets and associates with MAVS.
(A to G) HEK293T cells were transfected with indicated amounts of a SERINC5-encoding and an IFN-β–luc reporter plasmid together with the indicated plasmids of interest (encoding ΔRIG-I, an active form of RIG-I; MDA5; MAVS; TBK1; and IRF3). The cells were then infected with SeV or EMCV, and luc activities were measured. Data are from three to five independent experiments. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001. (H) SERINC5 is associated with MAVS but not TBK1. HEK293T cells were transfected with plasmid encoding Flag-tagged SERINC5, together with plasmids expressing GFP-tagged MAVS or TBK1. Cell lysates were then immunoprecipitated (IP) with anti-Flag beads, and immunoblotting (IB) analysis was performed with an anti-GFP antibody to detect MAVS and TBK1. (I) SERINC5 is associated with endogenous MAVS. Lysates of THP-1 cells stably expressing empty retroviral pQCXIP vector or Flag-tagged SERINC5 were immunoprecipitated with anti-Flag beads, which was followed by immunoblotting analysis with anti-MAVS antibody. (J) Schematic representation of MAVS constructs. (K) HEK293T cells were transfected with plasmid encoding Flag-tagged SERINC5 together with plasmids encoding the indicated Myc-tagged MAVS mutants. Cell lysates were immunoprecipitated with anti-Flag beads, and immunoblotting analysis was performed with anti-Myc and anti-Flag antibodies. Note that SERINC5 displayed distinct molecular weight species, although lysates were treated similarly under 55°C denaturing conditions. Representative results from at least three independent experiments are shown.
We next examined possible interactions between SERINC5 and MAVS or TBK-1 by performing coimmunoprecipitation (co-IP) assays. These were performed by cotransfecting 293T cells with plasmids expressing SERINC5 and green fluorescent protein (GFP)–tagged MAVS or TBK-1. We found that SERINC5 interacted with MAVS, but it did not associate with TBK-1 (Fig. 2H, top). This was despite the relatively low abundance of MAVS compared to that of TBK-1 in the whole-cell lysates (WCLs) (Fig. 2H, bottom). Furthermore, we found that the expression of GFP-tagged TBK1 led to an increase in SERINC5 abundance (Fig. 2H, bottom). We also detected an interaction between Flag-tagged SERINC5 and endogenous MAVS in THP-1 cells (Fig. 2I).
To determine which regions of MAVS are responsible for associating with SERINC5, we examined a panel of Myc-tagged MAVS mutants in which different regions of MAVS were deleted (Fig. 2J). We found that the MAVS-ΔCARD and MAVS-ΔN constructs, which lack the N-terminal CARD and the N-terminal Pro-rich domains of MAVS, respectively, but retain an intact transmembrane (TM) domain, still interacted with SERINC5 (Fig. 2K). Consistent with this pattern, the MAVS-ΔTM and MAVS-N constructs, in which the C-terminal TM domain of MAVS or the TM domain and the middle region of MAVS are deleted, respectively, lost the ability to interact with SERINC5. Results from these mapping experiments suggest that the TM region of MAVS is essential for the association with SERINC5.
SERINC5 colocalizes with MAVS at mitochondria and enhances MAVS aggregation
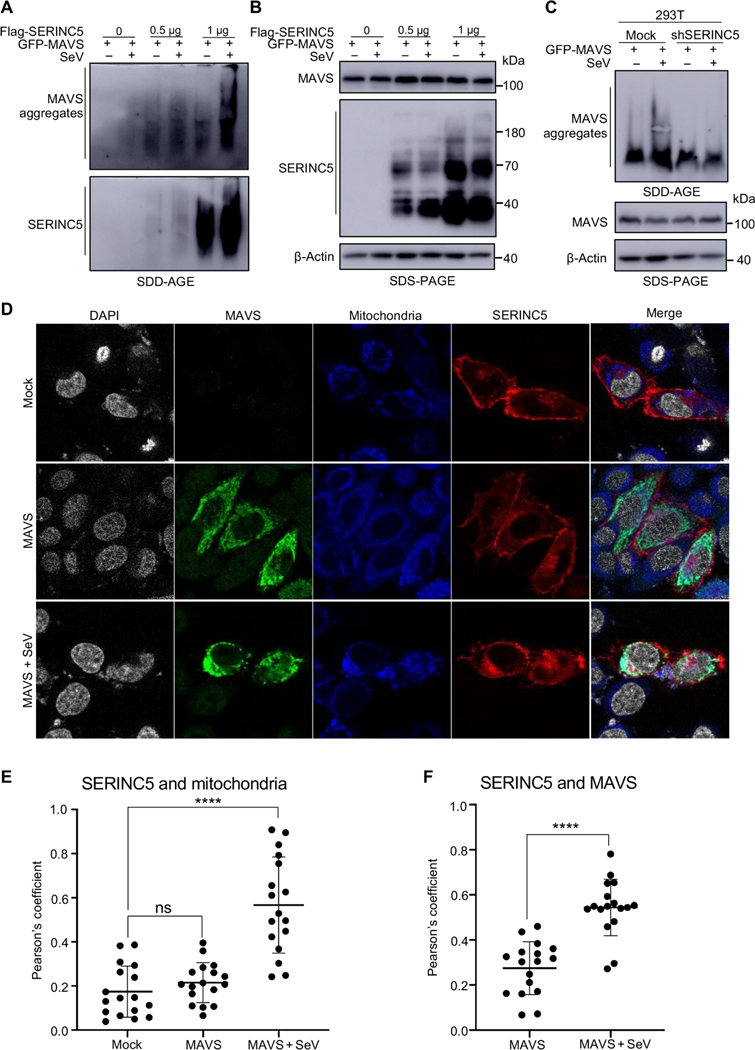
Another important feature of RIG-I–mediated activation of type I IFN production is the formation of prion-like MAVS aggregates at mitochondria (10). We thus explored the effect of SERINC5 on MAVS aggregation by extracting the crude mitochondrial fraction (P5) from HEK293T cells infected with SeV and then subjecting the samples to semi-denaturing detergent agarose gel electrophoresis (SDD-AGE; see Materials and Methods). In the absence of SERINC5, SeV infection resulted in detectable MAVS aggregation in mitochondria (Fig. 3A, top, lane 2). The extent of MAVS aggregation was substantially increased when SERINC5 was expressed (Fig. 3A, top, lanes 3 through 6). We found that SERINC5 itself also formed aggregates in mitochondria (Fig. 3A, bottom, lanes 3 through 6), especially when SERINC5 was more abundant (lanes 5 and 6). The relative amounts of MAVS and SERINC5 proteins in the total lysates of transfected cells were examined by conventional SDS–polyacrylamide gel electrophoresis (PAGE) and Western blotting analysis, which showed that SERINC5 had no apparent effect on MAVS abundance (Fig. 3B, top and middle). These results suggest that both MAVS and SERINC5 can form prion-like aggregates at mitochondria upon signaling activation.
Fig. 3. SERINC5 promotes MAVS aggregation and colocalizes with MAVS at mitochondria.
(A and B) HEK293T cells were transfected with plasmids expressing Flag-SERINC5 and GFP-MAVS and then were left uninfected or were infected with SeV. The mitochondrial P5 fraction was extracted (see Materials and Methods) and analyzed by SDD-AGE (A) and SDS-PAGE (B). β-Actin served as a loading control. Data are representative of three independent experiments. (C) HEK293T cells stably expressing control or SERINC5- specific shRNA were transfected with plasmid encoding GFP-MAVS and then were left uninfected or were infected with SeV. The mitochondrial P5 fraction was analyzed by SDD-AGE and SDS-PAGE as described for (A) and (B). Data are representative of at least three independent experiments. (D to F) HeLa cells were transfected with plasmid expressing mCherry-SERINC5 with or without plasmid expressing Flag-tagged MAVS. Twenty-four hours later, the cells were left uninfected or were infected with SeV for 7 hours and then stained with MitoTracker for 1 hour to label mitochondria (blue). Cells were then fixed and stained with anti-Flag antibody to stain MAVS (green), mounted with Vectashield mounting medium with DAPI to stain nuclei (gray), and examined with a Leica SP8 laser scanning confocal microscope (D). Colocalization between SERINC5 and mitochondria (E) and SERINC5 and MAVS (F) under the indicated conditions was quantified by calculating the Pearson correlation coefficients (R values) with the Fiji ImageJ colocalization analysis module. Data are from at least three independent experiments. ****P < 0.0001.
We next examined MAVS aggregation in SERINC5-KD 293T cells (~60% KD efficiency) (41) and observed that MAVS aggregation was decreased in extent at mitochondria, particularly in cells infected with SeV (Fig. 3C, top, lanes 2 and 4) as compared to that in control uninfected cells. As before, the total amount of MAVS remained unchanged in the SERINC5-KD cells (Fig. 3C, middle). Together, these results indicate that SERINC5 promotes the formation of MAVS aggregates and that SERINC5 itself also forms aggregates at mitochondria when the type I IFN production pathway is activated by MAVS overexpression in the context of SeV infection or ligand stimulation.
To examine the subcellular localizations of SERINC5 and MAVS, we transfected HeLa cells with the pBJ5-SERINC5-mCherry and MAVS-FLAG constructs, separately or in combination, in the presence or absence of SeV, and performed confocal imaging analyses. In uninfected cells coexpressing MAVS (“mock”), SERINC5 was predominantly localized at the plasma membrane (Fig. 3D, top) and did not substantially colocalize with the mitochondrial marker MitoTracker (the Pearson coefficient for SERINC5 and MitoTracker in the absence of SeV, 0.17; Fig. 3E). Upon transfection with the MAVS expression vector, MAVS was localized in the cytoplasm of uninfected cells, as would be expected, and did not colocalize with SERINC5 (Fig. 3D, middle). The Pearson coefficient for SERINC5 and MitoTracker in the presence of MAVS and in the absence of SeV was 0.22 (Fig. 3E). Under these conditions, little colocalization was observed between SERINC5 and MAVS (the Pearson coefficient for SERINC5 and MAVS was 0.27; Fig. 3F). In cells overexpressing MAVS and infected with SeV, a substantial portion of SERINC5 was colocalized with the mitochondrial marker and with MAVS (Fig. 3D, bottom), with Pearson’s coefficients of 0.57 and 0.54, respectively (Fig. 3, E and F). Because of the lack of a reliable anti-SERINC5 antibody, we were unable to assess the translocation of endogenous SERINC5 into the mitochondria. Nonetheless, these results suggest that upon SeV infection, MAVS facilitates the translocation of SERINC5 from the plasma membrane to mitochondria, where SERINC5 and MAVS colocalize.
SERINC5 enhances NF-κB signaling by interacting with TRAF6, which, in turn, stabilizes SERINC5
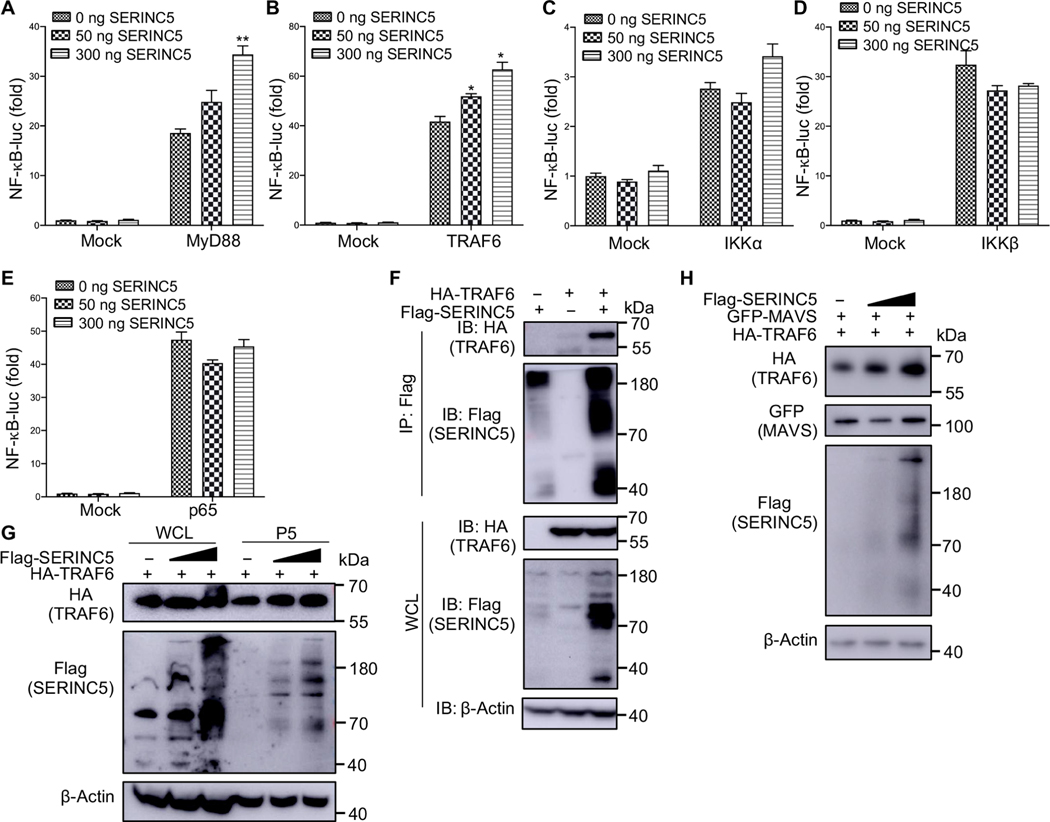
Our NF-κB–luc reporter assays, as well the Western blotting analyses presented earlier, demonstrated that SERINC proteins enhanced the NF-κB signaling pathway. To dissect the steps at which SERINC5 interacted with the NF-κB pathway, we cotransfected 293T cells with a 4×NF-κB-luc reporter vector and a pBJ-HA-SERINC5 construct, together with plasmids encoding key molecules involved in this signaling pathway. We found that SERINC5 enhanced NF-κB signaling activated by TRAF6 and MyD88 but had no effect on signaling activated by the downstream molecules IKKα, IKKβ, or p65 (Fig. 4, A to E). These results suggest that SERINC5 targets a step between TRAF6 and the IKK complex in the NF-κB signaling pathway.
Fig. 4. SERINC5 interacts with and stabilizes TRAF6.
(A to E) HEK293T cells were transfected with the indicated amounts of a SERINC5-encoding plasmid together with a NF-κB–luc reporter plasmid and plasmids expressing MyD88, TRAF6, IKKα, IKKβ, or p65, as indicated. The luc activities in the transfected cells were measured, with relative values plotted by setting the value of mock cells not expressing SERINC5 to zero. Data are from three independent experiments. *P < 0.05; **P < 0.01. (F) HEK293T cells were transfected with plasmids encoding Flag-tagged SERINC5 and HA-tagged TRAF6. Cell lysates were then immunoprecipitated (IP) with anti-Flag beads, which was followed by immunoblotting (IB) analysis with anti-HA and anti-Flag antibodies. (G) HEK293T cells were transfected with increasing amounts of plasmid encoding Flag-tagged SERINC5 together with a fixed amount of plasmid encoding HA-tagged TRAF6. Whole-cell lysates (WCLs) and mitochondrial P5 fractions were extracted and subjected to immunoblotting analysis with anti-HA and anti-Flag antibodies. (H) Experiments were performed similarly to those described in (G), except that the HEK293T cells were cotransfected with a plasmid encoding GFP-MAVS to stimulate the type I IFN production. Only samples of the mitochondrial P5 fraction were examined in these experiments. Representative results from three independent experiments are shown.
Because SERINC5 interacted with MAVS (Fig. 2, H and I), which recruits multiple TRAF family members, including TRAF6, through the TRAF-interacting motifs (TIMs) in MAVS that are essential for NF-κB signaling (11, 42), we next investigated whether SERINC5 also interacted with TRAF6. To this end, we performed a co-IP assay and found that SERINC5 interacted with TRAF6 (Fig. 4F, top, lane 3). Furthermore, we found that the relative abundance of SERINC5 was increased in the presence of TRAF6 (Fig. 4F, lane 3).
We next examined whether the relative amount of TRAF6 was increased at mitochondria when SERINC5 was coexpressed through extracting the cellular mitochondrial fraction (P5) as described earlier. With increasing amounts of SERINC5 in this fraction, the relative amount of TRAF6 at mitochondria appeared to increase accordingly (Fig. 4G, top, lanes 5 and 6). To confirm these results, we performed similar cotransfection assays in the presence of GFP-MAVS to enhance the production of type I IFN. As before, the relative amount of mitochondrial TRAF6 appeared to increase in a manner dependent on the amount of overexpressed SERINC5 (Fig. 4H).
SERINC5 potentiates K63-linked, but impairs K48-linked, ubiquitylation of TRAF6
K63-linked polyubiquitination of TRAF6 is required for activation of NF-κB signaling and the expression of genes encoding type I IFNs (43, 44). We examined the polyubiquitylation status of TRAF6 in the presence or absence of SERINC5 in experiments in which 293T cells were transfected with constructs encoding total HA-ubiquitin [HA-Ub-WT (wild type)], HA-K48–specific Ub (HA-Ub-K48), and HA-K63–specific Ub (HA-Ub-K63). Co-IP and Western blotting analyses revealed that SERINC5 enhanced the total polyubiquitylation of TRAF6 in a dose-dependent manner (fig. S5A, top). The expression of SERINC5 decreased the extent of K48-conjugated TRAF6 ubiquitylation (fig. S5B, top) but increased the extent of K63 ubiquitylation of TRAF6 (fig. S5C, top). Consistent with the decreased K48-mediated and increased K63-mediated ubiquitylation of TRAF6 by SERINC5, the relative abundance of TRAF6 was increased in the presence of SERINC5, especially in cells transfected with plasmids encoding HA-Ub-WT and HA-Ub-K63 (fig. S5, A and C, second panel from the bottom). Note that in these experiments, we were unable to measure the abundance of SERINC5 protein because of the lack of a reliable antibody to detect untagged, WT SERINC5. Together, these results support the notion that SERINC5 enhances the K63-linked polyubiquitylation of TRAF6, thus promoting NF-κB signaling and the type I IFN response.
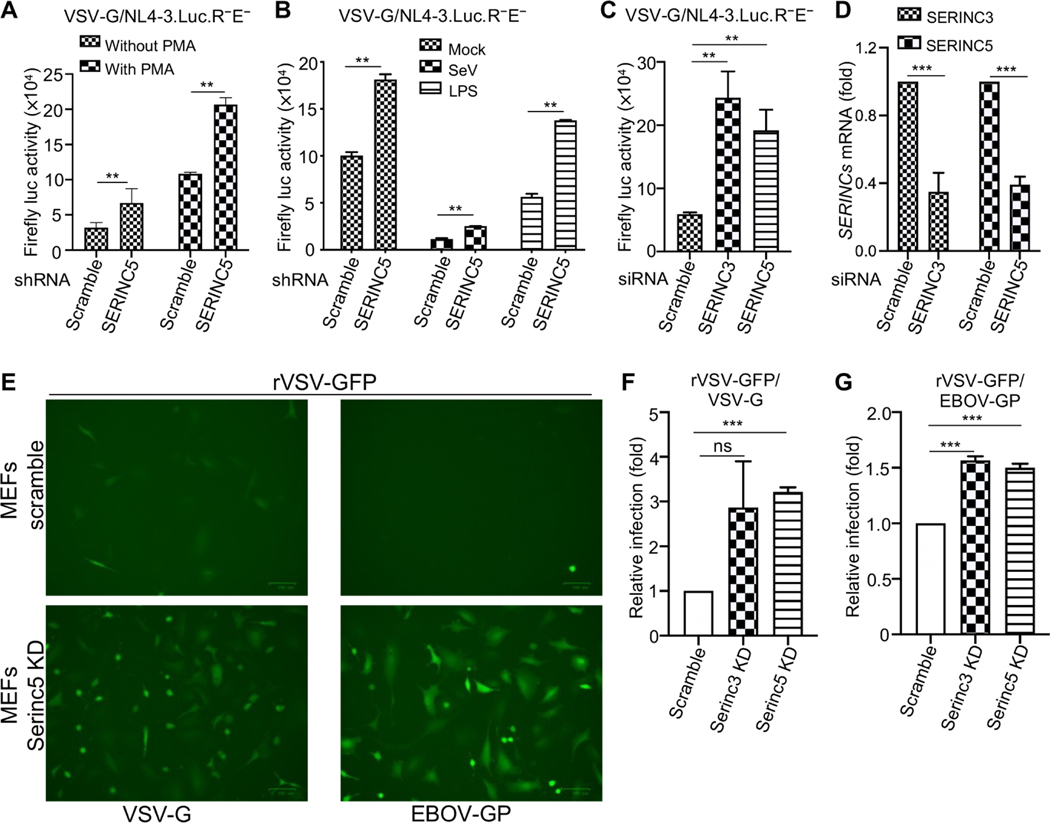
SERINC-induced type I IFN signaling contributes to its antiviral activity
SERINC5 is thought to reduce HIV-1 infectivity through its incorporation into virions, thereby inhibiting viral entry into subsequent target cells (13, 14). We investigated whether the enhanced type I IFN production and NF-κB signaling by SERINCs also contributed to the antiviral effect. We knocked down SERINCs in target cells and then measured the effects on HIV-1 single-round infectivity. We first infected THP-1 cells (which had been left untreated or were treated with PMA) that were stably expressing SERINC5-specific shRNA or scrambled shRNA with NL4–3.Luc.R−E− bearing VSV-G and measured the HIV-1 long terminal repeat (LTR)-driven firefly luc activity. The single-round infection assay was used here to avoid the confounding effect of producer-cell SERINC5 expression on HIV-1 infectivity. We found that KD of SERINC5 in target cells increased HIV-1 single-round infectivity (Fig. 5A), suggesting that endogenous SERINC5 imposed an intrinsic inhibitory effect on HIV-1 infection. We next incubated PMA-treated THP-1 cells either with SeV or LPS to activate the type I IFN response and NF-κB signaling pathways, and as expected, we observed reduced HIV-1 infectivity, especially in cells preinfected with SeV (Fig. 5B). Again, in all cases, shRNA-mediated KD of SERINC5 in THP-1 cells increased HIV-1 single-round infection (Fig. 5B). Last, we used small interfering RNA (siRNA) to knock down SERINC3 or SERINC5 in human MDMs, which was followed by infection of these cells with NL4–3. Luc.R−E− bearing VSV-G. Consistent with earlier results, the single- round infectivity of HIV-1 was enhanced by KD of SERINC3 or SERINC5 as compared to that in control cells treated with scrambled siRNA (Fig. 5, C and D). Together, these results suggest that SERINC proteins in target cells inhibit single-round HIV-1 infection likely by promoting type I IFN production.
Fig. 5. SERINC-induced type I IFN signaling contributes to antiviral activity.
(A) THP-1 cells stably expressing scrambled or SERINC5-specific shRNAs were treated with PMA for 12 hours or were left untreated. The cells were then infected for 48 hours with the HIV-1 reporter virus NL4–3. Luc.R−E− bearing VSV-G before being lysed for measurement of the firefly luc activity. Results are from three independent experiments. (B) Experiments were performed as described for (A) except that the PMA-treated THP-1 cells were preincubated with SeV (at an MOI of 2) or LPS (100 ng/ml) to stimulate type I IFN expression and NF-κB activation, respectively, before they were infected with the VSV-G–pseudotyped HIV-1 reporter virus NL4–3.Luc.R−E−. Data are from three independent experiments. (C and D) Human MDMs were transfected with scrambled siRNA or with siRNAs specific for SERINC3 or SERINC5, which was followed by infection with VSV-G–pseudotyped NL4–3.Luc.-R−E−. Firefly luc activity was then measured to determine HIV-1 infectivity (C). The efficiency of SERINC3 and SERINC5 KD was also assessed (D). Data are from three independent experiments. (E to G) MEFs stably expressing lentiviral vector expressing scrambled control gRNA or gRNAs specific for SERINC3 and SERINC5 were infected with rVSV-GFP bearing VSV-G (rVSV-G) and EBOV GP (EBOV GP), and thes of infection were analyzed by fluorescence microscopy (E) and flow cytometry (F and G). The results in (F) and (G) are pooled from three independent experiments. **P < 0.01; ***P < 0.001.
We also confirmed the direct antiviral effect of SERINCs on other viruses. Previous studies showed that SERINC5 has no inhibitory effect on the infectivity of retroviral pseudotypes bearing VSV-G and even enhances infectivity mediated by Ebola virus (EBOV) glycoprotein (GP) (45). This feature provided an opportunity for us to explore the possible effect of SERINC5-mediated signaling on VSV infection. We observed increased spreading infection of replication-competent, recombinant VSV (rVSV) bearing VSV-G or EBOV GP in SERINC5 KD mouse embryonic fibroblasts (MEFs) compared to that in MEFs treated with scrambled siRNA, suggesting that endogenous mouse SERINC5 suppresses rVSV infection mediated by VSV-G or EBOV GP (Fig. 5, E to G).
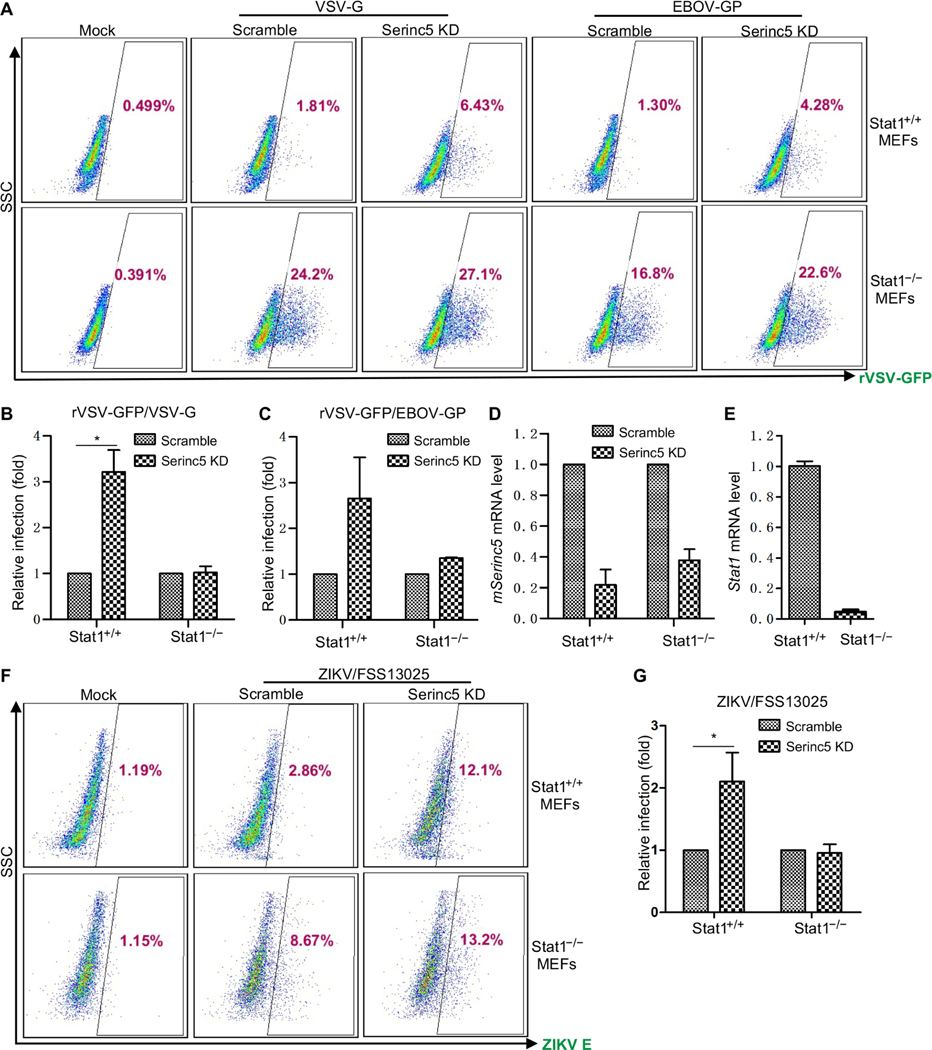
To confirm these results, we compared rVSV infection in SERINC5 KD MEFs, Stat1 knockout (KO) MEFs, and SERINC5 KD/Stat1 KO MEFs. We observed that whereas rVSV infection was increased in both SERINC5 KD MEFs and Stat1 KO MEFs, as expected, the extent of rVSV infection in SERINC5 KD/Stat1 KO MEFs was not as great as that in the Stat1 KO MEFs (Fig. 6, A to C). These results suggest that the enhanced rVSV infection observed in SERINC5 KD MEFs was likely due to impaired production of type I IFNs. We also determined the efficiency of SERINC5 KD and Stat1 KO in these cells (Fig. 6, D and E). Similar results were also obtained by infecting cells with the endemic strain (FSS13025) of ZIKV (Fig. 6, F and G), where the promoting effect of SERINC5 KD on ZIKV infection observed in Stat1+/+ MEFs, especially for FSS13025 ZIKV strain, was reduced in Stat1−/− MEFs. Because the enhancing effect of SERINC5 KD on viral infection was reduced in type I IFN–deficient cells, we conclude that SERINC5 functions to potentiate type I IFN production and NF-κB signaling, thereby inhibiting viral infections.
Fig. 6. SERINC5-mediated inhibition of viral infection is due, in part, to type I IFN signaling.
(A to E) Wild-type (Stat1+/+) and Stat1 knockout (Stat1−/−) MEFs stably expressing lentivirus encoding scrambled control or mouse Serinc5-specific gRNA were left uninfected (mock) or were infected with rVSV-GFP bearing VSV-G or EBOV GP. The GFP-positive cell populations were determined by flow cytometry. (A) Representative flow cytometric profiles of rVSV-GFP–infected MEFs, with the percentages of the GFP-positive cell populations indicated. (B and C) Summary of the relative viral infection efficiencies relative to those of cells expressing scrambled control gRNA. Data are from three or four independent experiments. *P < 0.05. (D and E) The KD efficiency of mouse Serinc5 and Stat1 in the indicated MEFs was determined by qPCR analysis. (F and G) The indicated MEFs were infected with endemic ZIKV strain FSS13025 for 48 hours. ZIKV-positive cells were then determined by flow cytometry with an antibody (4G2) against the ZIKV E protein. (F) Representative flow cytometric profiles of ZIKV infection. (G) Summary of the relative ZIKV infection efficiencies relative to that of cells expressing the scrambled control gRNA. Data are from three independent experiments. *P < 0.05.
DISCUSSION
SERINCs are HIV restriction factors with antiviral mechanisms that are incompletely defined. In this work, we showed that SERINC3 and SERINC5 enhanced type I IFN production and NF-κB signaling, thus contributing to their antiviral activities. Specifically, we found that SERINC5 increased the phosphorylation of IRF3 and IκBα, as well as promoted the translocation of IRF3 from the cytoplasm to the nucleus. Furthermore, upon infection of cells with SeV or their treatment with poly(I:C), SERINC5 was recruited to mitochondria, where it colocalized and interacted with MAVS, enhancing its polymerization. SERINC5 also interacted with and stabilized TRAF6, suggesting a model in which SERINC5, MAVS, and TRAF6 form a signaling complex at mitochondria, thus cooperatively enhancing the type I IFN production and NF-κB signaling pathways. This previously uncharacterized mode of SERINC activity in inducing antiviral immunity is distinct from the previously reported mechanism in which SERINC3 and SERINC5 are incorporated into HIV-1 virions in virus-producing cells, thereby impeding subsequent entry into target cells (13, 14). Because KD of SERINC3 or SERINC5 alone led to reduced signaling through the type I IFN and NF-κB pathways, we reason that the loss of one SERINC protein does not appear to be compensated by the presence of the other. Because we have examined only SERINC3 and SERINC5 in this study, it is possible that other SERINC family proteins, including those shown not to diminish viral infectivity when expressed in the virus-producing cells, share a similar function to modulate host innate immunity.
How does SERINC5 positively regulate the production of type I IFNs and NF-κB signaling? We found that SERINC5 interacted with MAVS and TRAF6, two adaptor molecules that are essential for transducing type I IFN expression and NF-κB signaling. MAVS is a key adaptor protein that drives the host innate immune response to infection by RNA viruses (46). With a TM anchor located at its C terminus, MAVS is localized in the membranes of peroxisomes and mitochondria and in a subdomain of the endoplasmic reticulum referred to as the mitochondrion-associated membrane (47). TRAF6 is a key E3 Ub ligase that interacts with MAVS through TIMs, thus facilitating the activation of both type I IFN production and NF-κB signaling pathways (43, 44). We showed in this work that SERINC5 enhanced the formation of MAVS polymers at mitochondria (Fig. 3A), suggesting that SERINC5 is a positive regulator of type I IFN expression. We also provided evidence that, in the presence of MAVS, SERINC5 itself was oligomerized and stabilized at mitochondria (Fig. 3A). Thus, the SERINC5-MAVS interaction may generate a positive feedback loop to enhance signaling. However, we must emphasize that the effect of SERINC5 on MAVS oligomerization and the activation of downstream signaling appeared to be dependent on SeV infection or ligand stimulation, because MAVS oligomerization itself did not always lead to the formation of a functional complex required for type I IFN signaling (10). In addition, the role of endogenous SERINC5 and MAVS in this process will need to be examined. It is also currently unclear how SERINC5 is recruited to mitochondria, either from the plasma membrane, intracellular organelles, or both, which will be investigated in future studies. Nevertheless, given that SERINC5 interacts with both MAVS and TRAF6 and that MAVS also intrinsically interacts with TRAF6 (11, 42, 48), we propose a working model in which SERINC5, MAVS, and TRAF6 form a signaling complex and cooperatively promote type I IFN and NF-κB signaling (fig. S6), thus executing antiviral activities.
Supporting the working model described earlier, we demonstrated that the expression of SERINC5 appeared to stabilize TRAF6 at mitochondria (Fig. 4, G and H), likely by promoting its K63-linked polyubiquitylation and also reducing its K48-linked polyubiquitylation (fig. S5). Ubiquitylation of TRAF proteins, especially TRAF2, TRAF5, and TRAF6, is critical for the induction of IFN-I signaling and NF-κB activation (43, 44). We suspect that, in addition to TRAF6, other TRAF proteins are also likely to be involved, including in the SERINC5-mediated enhancement of the noncanonical NF-κB pathway. In this respect, note that TRAF6 together with TRAF2 mediates Tetherin-induced NF-κB activation (33, 34), although another study showed that Tetherin also associates with MARCH8 and mediates the K27-linked ubiquitylation of MAVS, resulting in the suppression of type I IFN signaling (49). Because TRAF6 is an E3 Ub ligase that is itself ubiquitylated, it would be interesting to determine whether posttranslational modifications of SERINC5, such as phosphorylation or ubiquitylation (22, 23, 50), are required for SERINC5 to associate with MAVS and TRAF6 and modulate type I IFN signaling and NF-κB activation.
The type I IFN response plays a crucial role in limiting viral infection, especially at the early stage of infection. This is achieved largely not only by the direct antiviral effects of hundreds of ISGs but also, in many instances, by the regulatory functions of ISGs that modulate IFN sensing and proinflammatory signaling, such as Tetherin and Trim5α (33, 34). In this work, we provided evidence that the SERINC-mediated enhancing of type I IFN production and NF-κB inflammatory signaling protects cells from infection by HIV, VSV (bearing VSV-G or EBOV GP), and ZIKV (Fig. 6). For HIV-1, we applied a single-round infection assay, which bypassed the antiviral effect of SERINC incorporation into viral particles in the virus-producing cell. For other viruses, we knocked down SERINC5 in MEFs and compared the viral infection rate with that in Stat1 KO/SERINC5 KD MEFs. Results from these experiments suggest a role for SERINC-mediated induction of the type I IFN response in combating viral infection. Note that the role of NF-κB in HIV-1 replication is complex. On one hand, NF-κB is required for efficient proviral gene transcription; however, on the other hand, NF-κB inhibits HIV-1 at a late stage of the viral life cycle (51, 52). Furthermore, the HIV-1 accessory protein Nef stimulates the NF-κB signaling pathway, whereas the HIV-1 Vpu protein inhibits it (53). Hence, it is unlikely that the HIV-1 Nef protein could counteract the SERINC-mediated enhancement of the type I IFN response similarly to the way it counteracts SERINC-mediated inhibition of viral infectivity. Ultimately, the role of SERINCs in viral infection, including that of HIV-1, will need to be determined in animals and humans. Given that activation of type I IFN and NF-κB signaling is involved in many physiological and pathological settings, including autophagy, apoptosis, and cancer, the results reported here may also have implications for understanding the normal physiological functions of SERINC proteins.
MATERIALS AND METHODS
Plasmids and constructs
The pQCXIP vector that expresses human SERINC5 with a C-terminal FLAG tag was a gift from C. Liang, McGill University, Canada (54). The pBJ5 vectors expressing SERINC5 with a C-terminal HA or mCherry tag were gifts from H. Gottlinger, University of Massachusetts Medical School (14). The GFP-tagged ΔRIG-I, MDA5, MAVS, TBK1, IRF3–5D, and Flag-tagged TRAF6 constructs (55) were obtained from R. Lin, McGill University, Canada. Truncated MAVS mutants with a Myc tag (ΔCARD, ΔTM, ΔN, and N) (56); the Flag-tagged MAVS, IKKα, IKKβ, and p65; and the HA-tagged MyD88 and TRAF6 constructs (57) were from H.W., The Ohio State University. Lentiviral vectors encoding SERINC3- and SERINC5-specific shRNA or control shRNA were purchased from Sigma-Aldrich. Lentiviral vectors expressing guide RNAs (gRNAs) for mouse SERINC3 or SERINC5, together with Cas9, were purchased from GenScript.
Cells, reagents, and antibodies
HEK293 [American Type Culture Collection (ATCC) CRL-1573, RRID: CVCL_0045], HEK293T (ATCC CRL-11268, RRID: CVCL_1926), HeLa (ATCC CCL-2, RRID: CVCL_0030), and Vero cells (ATCC CCL-81, RRID: CVCL_0059) and MEFs (a gift from H.W.) were grown in Dulbecco’s modified Eagle’s medium (DMEM), supplemented with 1% penicillin/streptomycin and 10% (v/v) fetal bovine serum (FBS). THP-1 cells (ATCC TIB-202, RRID: CVCL_0006) and human primary MDMs (purchased from the National Institutes of Health) were grown in RPMI 1640 medium with 1% penicillin/ streptomycin and 10% (v/v) FBS. THP-1 cells were treated with PMA (12.5 ng/ml) for 24 hours to induce their differentiation into macrophage-like cells. HEK293 cells stably expressing SERINC5 were generated by transduction with pQCXIP retroviral vectors expressing SERINC5, followed by selection in puromycin (1 μg/ml) for 6 days. THP-1 cells stably expressing shRNA against SERINC3 or SERINC5 or a scrambled control shRNA were established by transduction with lentiviral vectors expressing shRNAs targeting SERINC3 or SERINC5 or expressing a scrambled shRNA, respectively, followed by selection in puromycin (2 μg/ml) for 6 days. All cell lines were maintained at 37°C and 5% CO2. LPS was purchased from Sigma-Aldrich (L6529), poly(I:C) was from Sigma-Aldrich (P1530), TNF-α was from Sino Biological (10602-HNAE), 4′,6-diamidino-2-phenylindole (DAPI) was from Vector Laboratories (H-1200), MitoTracker Green FM was from Thermo Fisher Scientific (M-7514), PMA was from Sigma-Aldrich (P1585), and Protease Inhibitor Cocktail and PhosSTOP were from Sigma-Aldrich (P8340 and 0496837001). ZIKV FSS13025 was obtained from the University of Texas Medical Branch (UTMB) and amplified in Vero cells, whereas rVSV-G-GFP and rVSV-GP-GFP (EBOV) were obtained from K. Chandra and amplified in 293T cells, which were all maintained under a humidified atmosphere of 5% CO2 at 37°C in DMEM supplemented with 10% FBS (Thermo Fisher Scientific). SeV (SeV-Cantell, ATCC VR-907) was provided by J. Yount, The Ohio State University; EMCV was purchased from ATCC (VR-1762). Antibodies used for Western blotting included the following: anti-HA (BioLegend, 901503), anti-Flag M2 (Sigma-Aldrich, F1804), anti-GFP (Santa Cruz Biotechnology, sc9996), anti– glyceraldehyde-3-phosphate dehydrogenase (GAPDH; Santa Cruz Biotechnology, sc-47724), anti–β-actin (Sigma-Aldrich, A1978), anti-Myc (Abcam, Mab9106), anti-IRF3 (Santa Cruz Biotechnology, sc-9082), anti–p-IRF3 (Cell Signaling, 4947S), anti–p-IκBα (Cell Signaling, 9246S), anti-IκBα (Abnova, MAB0057), anti–NF-κB p52 (Merck Millipore, 05–361), anti-MAVS (Santa Cruz Biotechnology, sc-166583), anti-mouse immunoglobulin G (IgG)–peroxidase (Sigma-Aldrich, A5278), and anti-rabbit IgG-peroxidase (Sigma-Aldrich, A9169). Secondary antibodies used for immunofluorescence included anti-mouse IgG–fluorescein isothiocyanate (FITC; Sigma-Aldrich, F0257), anti-rabbit IgG-FITC (Sigma-Aldrich, F9887), and anti-mouse IgG–Alexa Fluor 647 (Thermo Fisher Scientific, A-21235).
Semi-denaturing detergent agarose gel electrophoresis
Preparation of crude mitochondrial (P5) fractions was performed according to a modified Abcam protocol. Briefly, cells were resuspended with buffer A [10 mM tris-HCl (pH 7.5), 10 mM KCl, 1.5 mM MgCl2, and protease inhibitor cocktail] by repeated douncing (15 times), followed by centrifugation at 700g for 10 min at 4°C. The supernatant was then centrifuged at 10,000g for 30 min at 4°C to obtain the intact crude mitochondrial pellet (P5). The crude mitochondria (P5) were resuspended in 1× sample buffer [25 mM tris-Cl (pH 7.5), 1% SDS, 10% glycerol, and 0.01% bromophenol blue] and loaded onto a vertical 1.5% agarose gel (Bio-Rad), which was run in normal SDS running buffer at 50 V for 40 min before the proteins were transferred to the polyvinylidene difluoride membrane for Western blotting analysis.
Reverse transcription polymerase chain reaction
Total cellular RNA was extracted with TRIzol (Thermo Fisher Scientific, 15596018). RNA was digested with deoxyribonuclease to eliminate genomic DNA. Complementary DNA was synthesized with Maxima H Minus Reverse Transcriptase (Thermo Fisher Scientific, EP0751) at 50°C for 30 min. Reverse transcription polymerase chain reaction (RT-PCR) was performed with SYBR Green (Applied Biosystems, A25742) in a QuantStudio 3 detection system (Applied Biosystems). The fold change between the abundances of target and control mRNAs was determined with the ΔΔCt method. GAPDH mRNA was used as an internal control. The primer pair sequences are listed in table S1.
Determination of human IFN-β in cell culture medium by ELISA
HEK293T cells were transfected with different amounts of plasmid encoding SERINC5 and then infected with SeV. Thirty-six hours later, the cell culture medium was harvested, and the amount of IFN-β in the medium was determined with an enzyme-linked immunosorbent assay (ELISA) kit (PBL, 41410–1) according to the manufacturer’s instructions.
Transfections and infections
Transfections of HEK293T cells for IFN-β–luc reporter assays were performed in 12- or 24-well plates using the calcium phosphate method. The plasmids were qQCXIP- or pBJ-SERINC5, together with IFN-β-luc and pRLTK. Where appropriate, the cells were also transfected with plasmids encoding GFP-tagged MDA5, ΔRIG-I, MAVS, TBK1, and IRF3–5D, which was followed by infection for the cells with SeV [at a multiplicity of infection (MOI) of 2] for 16 hours. The cells were then lysed, and luc activity was measured with a dual-luc reporter assay system (Promega, E1960). For NF-κB–luc reporter assays, HEK293T cells were cotransfected with plasmids encoding qQCXIP- or pBJ-S ERINC5 and NF-κB–luc. Where appropriate, the cells were cotransfected with plasmids encoding MyD88, TRAF6, IKKα, IKKβ, or p65. The cells were then treated with TNF-α (10 ng/ml) for 8 hours before being lysed and subjected to luc activity measurement. For co-IP assays, HEK293T cells in six-well plates were cotransfected with the appropriate plasmids. To transfect MDMs with siRNAs, we used Lipofectamine 2000 (Invitrogen, 11668019) with 40 nM scrambled control siRNA or SERINC3- or SERINC5-specific siRNAs (Ambion). The cell culture medium was replaced 24 hours after transfection, and the cells were maintained in culture for another 24 hours before being infected with SeV. For pseudotyped vector production, we transfected HEK293T cells with retroviral or lentiviral vectors encoding the constructs of interest (SERINC5, SERINC5- or SERINC3-specific shRNA, NL4–3.Luc.R−E−, etc.) together with plasmids expressing MLV Gag-Pol or HIV-1 Gag-PolΔ8.2 and VSV-G. The cell culture medium was harvested at 24 and 48 hours after transfection and used for infection of target cells in the presence of polybrene. For the infection of THP-1 cells, we performed spinoculation at 1680g at 4°C for 1 hour. All infections were in the presence of polybrene (5 μg/ml; Sigma-Aldrich).
Co-IPs and Western blotting
Western blotting analysis was performed as previously described (58). Briefly, cells were collected and lysed in radioimmunoprecipitation assay (RIPA) buffer [50 mM tris (pH 7.5), 150 mM NaCl, 1 mM EDTA, 1% NP-40, 0.1% SDS, and protease inhibitor cocktail], which disrupts membrane-associated proteins. Cell lysates were clarified by centrifugation at 12,000g for 10 min at 4°C. For detection of protein phosphorylation, lysates were additionally treated with the phosphatase inhibitor PhosSTOP (Sigma-Aldrich, 0496837001). To detect SERINC5, cell lysates were incubated at 55°C for 5 min, whereas for the detection of other proteins, samples were boiled at 100°C for 10 min. Treated samples were resolved on 10% SDS-PAGE gels, transferred to membranes according to standard protocols, and incubated with the appropriate primary antibodies. To examine possible interactions between SERINC5 and proteins of interest, we cotransfected HEK293T cells with plasmids expressing Flag-tagged SERINC5 and target genes. Thirty-six hours later, the cells were collected and lysed in RIPA buffer, immunoprecipitated with anti-Flag antibody beads (Sigma-Aldrich, F2426), and analyzed by Western blotting with the appropriate antibodies.
Immunofluorescence microscopy and confocal imaging
To visualize IRF3 localization, HeLa cells were transfected with empty plasmid or plasmid encoding pBJ5-SERINC5-mCherry and then were left uninfected or were infected with SeV. Thirty-six hours after transfection, the cells were washed with phosphate-buffered saline (PBS), fixed in 4% paraformaldehyde (PFA) for 10 min, permeabilized with 1× intracellular fixation (IC) buffer, and incubated with an anti-IRF3 antibody (Santa Cruz Biotechnology, sc-9082) in 1× IC buffer (at a 1:100 dilution) for 1 hour. After three washes with PBS, the cells were incubated with FITC-conjugated secondary antibody (Sigma-Aldrich, F9887) and examined with a Leica microscope (DMi8). To analyze the colocalization between SERINC5 and MAVS, HeLa cells cultured in chamber slides were cotransfected with a plasmid encoding mCherry-SERINC5 in the absence or presence of a plasmid expressing Flag-tagged MAVS with the PEI transfection reagent. Seven hours later, the medium was changed, and the cells were cultured overnight. Twenty-four hours after transfection, the cells were mock-infected or infected with SeV at an MOI of 20. Seven hours after SeV infection, the cells were treated with MitoTracker Green FM (Thermo Fisher Scientific, M-7514) at a final concentration of 250 nM to stain mitochondria. After 1 hour of incubation, the cells were rinsed with PBS and fixed with 3.7% PFA in PBS for 1 hour at room temperature, which was followed by incubation at 4°C overnight. The next day, the cells were rinsed with PBS, permeabilized with methanol at −20°C for 4 min, washed in PBS, and incubated with 0.1 M glycine-PBS for 10 min to quench the remaining aldehyde residues. After being blocked with 3% bovine serum albumin (BSA)–PBS for 30 min, the cells were incubated with anti-FLAG antibody diluted in 3% BSA-PBS for 2 hours to stain MAVS. The cells were then washed with PBS three times and incubated for 1 hour with Alexa Fluor 647–conjugated secondary antibody diluted in 3% BSA-PBS. The cells were then mounted with Vectashield mounting medium with DAPI (Vector Laboratories, H-1200) and examined with a Leica SP8 laser scanning confocal microscope.
Flow cytometry
MEFs infected with rVSV-GFP-G or rVSV-GFP-EBOV GP were detached with 0.05% trypsin-EDTA, washed with cold PBS, and fixed with 3.7% formaldehyde for 10 min. After being washed with PBS, the cells were resuspended in PBS and analyzed by flow cytometry. WT (Stat1+/+) and Stat1 KO (Stat1−/−) MEFs were infected with ZIKV FSS13025 for 48 hours, digested, and fixed in 4% PFA for 10 min. The samples were permeabilized with 1× IC buffer and incubated with an anti–flavivirus group antigen (4G2) antibody (Millipore, MAB10216) in 1× IC buffer (1:200) for 1 hour. The cells were then washed with PBS and incubated with FITC-conjugated secondary antibody (Sigma-Aldrich, F0257) for 45 min, which was followed by analysis by flow cytometry.
Statistical analysis
Data were analyzed and presented as means with standard deviation (SD). All experiments were performed at least three times independently, and the number of biological replicates for each dataset is given by “n” and is provided in the respective figure legend. Statistical analyses were performed with GraphPad Prism 5.0 software as follows: One-way analysis of variance (ANOVA) with Bonferroni’s posttests was used to compute statistical significance between multiple groups for multiple comparisons, or the t test was used for two groups for a single comparison. A P value of <0.05 was considered to be significant and is indicated by an asterisk (*P < 0.05).
Supplementary Material
Acknowledgments:
We thank H. Gottlinger, R. Lin, and C. Liang for the provision of plasmids, cells, constructs, and reagents. We also thank the NIH AIDS Reagent Program for supplying important reagents that made this work possible.
Funding:
This work was supported by NIH grants to S.-L.L. (R01 AI112381 and R01 AI150473) and to H.W. (R01GM120496 and R01GM135234). Research in the Freed laboratory is supported by the Intramural Research Program of the Center for Cancer Research, National Cancer Institute, NIH.
Footnotes
Competing interests: The authors declare that they have no competing interests.
Data and materials availability:
All data needed to evaluate the conclusions in the paper are present in the paper or the Supplementary Materials.
REFERENCES AND NOTES
- 1.Chan YK, Gack MU, Viral evasion of intracellular DNA and RNA sensing. Nat. Rev. Microbiol 14, 360–373 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Akira S, Uematsu S, Takeuchi O, Pathogen recognition and innate immunity. Cell 124, 783–801 (2006). [DOI] [PubMed] [Google Scholar]
- 3.Schlee M, Hartmann G, Discriminating self from non-self in nucleic acid sensing. Nat. Rev. Immunol 16, 566–580 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tan X, Sun L, Chen J, Chen ZJ, Detection of microbial infections through innate immune sensing of nucleic acids. Annu. Rev. Microbiol 72, 447–478 (2018). [DOI] [PubMed] [Google Scholar]
- 5.Yoneyama M, Kikuchi M, Matsumoto K, Imaizumi T, Miyagishi M, Taira K, Foy E, Loo YM, Gale M Jr., S. Akira, S. Yonehara, A. Kato, T. Fujita, Shared and unique functions of the DExD/H-box helicases RIG-I, MDA5, and LGP2 in antiviral innate immunity. J. Immunol 175, 2851–2858 (2005). [DOI] [PubMed] [Google Scholar]
- 6.Seth RB, Sun L, Ea CK, Chen ZJ, Identification and characterization of MAVS, a mitochondrial antiviral signaling protein that activates NF-κB and IRF 3. Cell 122, 669–682 (2005). [DOI] [PubMed] [Google Scholar]
- 7.Xu LG, Wang YY, Han KJ, Li LY, Zhai Z, Shu HB, VISA is an adapter protein required for virus-triggered IFN-β signaling. Mol. Cell 19, 727–740 (2005). [DOI] [PubMed] [Google Scholar]
- 8.Meylan E, Curran J, Hofmann K, Moradpour D, Binder M, Bartenschlager R, Tschopp J, Cardif is an adaptor protein in the RIG-I antiviral pathway and is targeted by hepatitis C virus. Nature 437, 1167–1172 (2005). [DOI] [PubMed] [Google Scholar]
- 9.Kawai T, Takahashi K, Sato S, Coban C, Kumar H, Kato H, Ishii KJ, Takeuchi O, Akira S, IPS-1, an adaptor triggering RIG-I- and Mda5-mediated type I interferon induction. Nat. Immunol 6, 981–988 (2005). [DOI] [PubMed] [Google Scholar]
- 10.Hou F, Sun L, Zheng H, Skaug B, Jiang QX, Chen ZJ, MAVS forms functional prion-like aggregates to activate and propagate antiviral innate immune response. Cell 146, 448–461 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu S, Chen J, Cai X, Wu J, Chen X, Wu YT, Sun L, Chen ZJ, MAVS recruits multiple ubiquitin E3 ligases to activate antiviral signaling cascades. eLife 2, e00785 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brubaker SW, Bonham KS, Zanoni I, Kagan JC, Innate immune pattern recognition: A cell biological perspective. Annu. Rev. Immunol 33, 257–290 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rosa A, Chande A, Ziglio S, De Sanctis V, Bertorelli R, Goh SL, McCauley SM, Nowosielska A, Antonarakis SE, Luban J, Santoni FA, Pizzato M, HIV-1 Nef promotes infection by excluding SERINC5 from virion incorporation. Nature 526, 212–217 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Usami Y, Wu Y, Gottlinger HG, SERINC3 and SERINC5 restrict HIV-1 infectivity and are counteracted by Nef. Nature 526, 218–223 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chande A, Cuccurullo EC, Rosa A, Ziglio S, Carpenter S, Pizzato M, S2 from equine infectious anemia virus is an infectivity factor which counteracts the retroviral inhibitors SERINC5 and SERINC3. Proc. Natl. Acad. Sci. U.S.A 113, 13197–13202 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Aiken C, HIV: Antiviral action countered by Nef. Nature 526, 202–203 (2015). [DOI] [PubMed] [Google Scholar]
- 17.Fackler OT, Spotlight on HIV-1 Nef: SERINC3 and SERINC5 identified as restriction factors antagonized by the pathogenesis factor. Viruses 7, 6730–6738 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Firrito C, Bertelli C, Vanzo T, Chande A, Pizzato M, SERINC5 as a new restriction factor for human immunodeficiency virus and murine leukemia virus. Annu. Rev. Virol 5, 323–340 (2018). [DOI] [PubMed] [Google Scholar]
- 19.Heigele A, Kmiec D, Regensburger K, Langer S, Peiffer L, Sturzel CM, Sauter D, Peeters M, Pizzato M, Learn GH, Hahn BH, Kirchhoff F, The potency of Nef-mediated SERINC5 antagonism correlates with the prevalence of primate lentiviruses in the wild. Cell Host Microbe 20, 381–391 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Trautz B, Pierini V, Wombacher R, Stolp B, Chase AJ, Pizzato M, Fackler OT, The antagonism of HIV-1 Nef to SERINC5 particle infectivity restriction involves the counteraction of virion-associated pools of the restriction factor. J. Virol 90, 10915–10927 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sood C, Marin M, Chande A, Pizzato M, Melikyan GB, SERINC5 protein inhibits HIV-1 fusion pore formation by promoting functional inactivation of envelope glycoproteins. J. Biol. Chem 292, 6014–6026 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shi J, Xiong R, Zhou T, Su P, Zhang X, Qiu X, Li H, Li S, Yu C, Wang B, Ding C, Smithgall TE, Zheng YH, HIV-1 Nef antagonizes SERINC5 restriction by downregulation of SERINC5 via the endosome/lysosome system. J. Virol 92, e00196-18 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ahmad I, Li S, Li R, Chai Q, Zhang L, Wang B, Yu C, Zheng YH, The retroviral accessory proteins S2, Nef, and glycoMA use similar mechanisms for antagonizing the host restriction factor SERINC5. J. Biol. Chem 294, 7013–7024 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stoneham CA,Ramirez PW, Singh R, Suarez M, Debray A, Lim C, Jia X, Xiong Y, Guatelli J, A conserved acidic-cluster motif in SERINC5 confers partial resistance to antagonism by HIV-1 Nef. J. Virol 94, e01554–19 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pye VE, Rosa A, Bertelli C, Struwe WB, Maslen SL, Corey R, Liko I, Hassall M, Mattiuzzo G, Ballandras-Colas A, Nans A, Takeuchi Y, Stansfeld PJ, Skehel JM, Robinson CV, Pizzato M, Cherepanov P, A bipartite structural organization defines the SERINC family of HIV-1 restriction factors. Nat. Struct. Mol. Biol 27, 78–83 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lim ES, Malik HS, Emerman M, Ancient adaptive evolution of tetherin shaped the functions of Vpu and Nef in human immunodeficiency virus and primate lentiviruses. J. Virol 84, 7124–7134 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McNatt MW, Zang T, Hatziioannou T, Bartlett M, Fofana IB, Johnson WE, Neil SJ, Bieniasz PD, Species-specific activity of HIV-1 Vpu and positive selection of tetherin transmembrane domain variants. PLOS Pathog. 5, e1000300 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sawyer SL, Wu LI, Emerman M, Malik HS, Positive selection of primate TRIM5α identifies a critical species-specific retroviral restriction domain. Proc. Natl. Acad. Sci. U.S.A 102, 2832–2837 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sawyer SL, Emerman M, Malik HS, Ancient adaptive evolution of the primate antiviral DNA-editing enzyme APOBEC3G. PLoS Biol. 2, E275 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lahouassa H, Daddacha W, Hofmann H, Ayinde D, Logue EC, Dragin L, Bloch N, Maudet C, Bertrand M, Gramberg T, Pancino G, Priet S, Canard B, Laguette N, Benkirane M, Transy C, Landau NR, Kim B, Margottin-Goguet F, SAMHD1 restricts the replication of human immunodeficiency virus type 1 by depleting the intracellular pool of deoxynucleoside triphosphates. Nat. Immunol 13, 223–228 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Murrell B, Vollbrecht T, Guatelli J, Wertheim JO, The evolutionary histories of antiretroviral proteins SERINC3 and SERINC5 do not support an evolutionary arms race in primates. J. Virol 90, 8085–8089 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hotter D, Sauter D, Kirchhoff F, Emerging role of the host restriction factor tetherin in viral immune sensing. J. Mol. Biol 425, 4956–4964 (2013). [DOI] [PubMed] [Google Scholar]
- 33.Galão RP, Le Tortorec A, Pickering S, Kueck T, Neil SJ, Innate sensing of HIV-1 assembly by Tetherin induces NFκB-dependent proinflammatory responses. Cell Host Microbe 12, 633–644 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cocka LJ, Bates P, Identification of alternatively translated Tetherin isoforms with differing antiviral and signaling activities. PLOS Pathog. 8, e1002931 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pertel T, Hausmann S, Morger D, Zuger S, Guerra J, Lascano J, Reinhard C, Santoni FA, Uchil PD, Chatel L, Bisiaux A, Albert ML, Strambio-De-Castillia C, Mothes W, Pizzato M, Grutter MG, Luban J, TRIM5 is an innate immune sensor for the retrovirus capsid lattice. Nature 472, 361–365 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Aiken C, Joyce S, Immunology: TRIM5 does double duty. Nature 472, 305–306 (2011). [DOI] [PubMed] [Google Scholar]
- 37.Yamasoba D, Sato K, Ichinose T, Imamura T, Koepke L, Joas S, Reith E, Hotter D, Misawa N, Akaki K, Uehata T, Mino T, Miyamoto S, Noda T, Yamashita A, Standley DM, Kirchhoff F, Sauter D, Koyanagi Y, Takeuchi O, N4BP1 restricts HIV-1 and its inactivation by MALT1 promotes viral reactivation. Nat. Microbiol 4, 1532–1544 (2019). [DOI] [PubMed] [Google Scholar]
- 38.Li S, Wang L, Berman M, Kong YY, Dorf ME, Mapping a dynamic innate immunity protein interaction network regulating type I interferon production. Immunity 35, 426–440 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu S, Cai X, Wu J, Cong Q, Chen X, Li T, Du F, Ren J, Wu Y-T, Grishin NV, Chen ZJ, Phosphorylation of innate immune adaptor proteins MAVS, STING, and TRIF induces IRF3 activation. Science 347, aaa2630 (2015). [DOI] [PubMed] [Google Scholar]
- 40.Sun SC, Non-canonical NF-κB signaling pathway. Cell Res. 21, 71–85 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li M, Waheed AA, Yu J, Zeng C, Chen HY, Zheng YM, Feizpour A, Reinhard BM, Gummuluru S, Lin S, Freed EO, Liu SL, TIM-mediated inhibition of HIV-1 release is antagonized by Nef but potentiated by SERINC proteins. Proc. Natl. Acad. Sci. U.S.A 116, 5705–5714 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Saha SK, Pietras EM, He JQ, Kang JR, Liu SY, Oganesyan G, Shahangian A, Zarnegar B, Shiba TL, Wang Y, Cheng G, Regulation of antiviral responses by a direct and specific interaction between TRAF3 and Cardif. EMBO J. 25, 3257–3263 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hacker H, Tseng PH, Karin M, Expanding TRAF function: TRAF3 as a tri-faced immune regulator. Nat. Rev. Immunol 11, 457–468 (2011). [DOI] [PubMed] [Google Scholar]
- 44.Shi JH, Sun SC, Tumor necrosis factor receptor-associated factor regulation of nuclear factor κB and mitogen-activated protein kinase pathways. Front. Immunol 9, 1849 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ahi YS, Zhang S, Thappeta Y, Denman A, Feizpour A, Gummuluru S, Reinhard B, Muriaux D, Fivash MJ, Rein A, Functional interplay between murine leukemia virus Glycogag, Serinc5, and surface glycoprotein governs virus entry, with opposite effects on gammaretroviral and ebolavirus glycoproteins. MBio 7, e01985–16 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wu B, Hur S, How RIG-I like receptors activate MAVS. Curr. Opin. Virol 12, 91–98 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Qian J, Le Duff Y, Wang Y, Pan Q, Ding S, Zheng YM, Liu SL, Liang C, Primate lentiviruses are differentially inhibited by interferon-induced transmembrane proteins. Virology 474, 10–18 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shi Z, Zhang Z, Zhang Z, Wang Y, Li C, Wang X, He F, Sun L, Jiao S, Shi W, Zhou Z, Structural insights into mitochondrial antiviral signaling protein (MAVS)-tumor necrosis factor receptor-associated factor 6 (TRAF6) signaling. J. Biol. Chem 290, 26811–26820 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jin S, Tian S, Luo M, Xie W, Liu T, Duan T, Wu Y, Cui J, Tetherin suppresses type i interferon signaling by targeting MAVS for NDP52-mediated selective autophagic degradation in human cells. Mol. Cell 68, 308–322.e4 (2017). [DOI] [PubMed] [Google Scholar]
- 50.Sharma S, Lewinski MK, Guatelli J, An N-glycosylated form of SERINC5 is specifically incorporated into HIV-1 virions. J. Virol 92, e00753–18 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sauter D, Hotter D, Van Driessche B, Sturzel CM, Kluge SF, Wildum S, Yu H, Baumann B, Wirth T, Plantier JC, Leoz M, Hahn BH, Van Lint C, Kirchhoff F, Differential regulation of NF-κB-mediated proviral and antiviral host gene expression by primate lentiviral Nef and Vpu proteins. Cell Rep. 10, 586–599 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sauter D, Kirchhoff F, Multilayered and versatile inhibition of cellular antiviral factors by HIV and SIV accessory proteins. Cytokine Growth Factor Rev. 40, 3–12 (2018). [DOI] [PubMed] [Google Scholar]
- 53.Langer S, Hammer C, Hopfensperger K, Klein L, Hotter D, De Jesus PD, Herbert KM, Pache L, Smith N, van der Merwe JA, Chanda SK, Fellay J, Kirchhoff F, Sauter D, HIV-1 Vpu is a potent transcriptional suppressor of NF-κB-elicited antiviral immune responses. eLife 8, e41930 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Beitari S, Ding S, Pan Q, Finzi A, Liang C, The effect of HIV-1 Env on SERINC5 antagonism. J. Virol 91, e02214–16 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Belgnaoui SM, Paz S, Samuel S, Goulet ML, Sun Q, Kikkert M, Iwai K, Dikic I, Hiscott J, Lin R, Linear ubiquitination of NEMO negatively regulates the interferon antiviral response through disruption of the MAVS-TRAF3 complex. Cell Host Microbe 12, 211–222 (2012). [DOI] [PubMed] [Google Scholar]
- 56.Liu B, Zhang M, Chu H, Zhang H, Wu H, Song G, Wang P, Zhao K, Hou J, Wang X, Zhang L, Gao C, The ubiquitin E3 ligase TRIM31 promotes aggregation and activation of the signaling adaptor MAVS through Lys63-linked polyubiquitination. Nat. Immunol 18, 214–224 (2017). [DOI] [PubMed] [Google Scholar]
- 57.Li T, Li X, Attri KS, Liu C, Li L, Herring LE, Asara JM, Lei YL, Singh PK, Gao C, Wen H, O-GlcNAc transferase links glucose metabolism to MAVS-mediated antiviral innate immunity. Cell Host Microbe 24, 791–803.e6 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Côté M, Zheng YM, Li K, Xiang S-H, Albritton LM, Liu S-L, Critical role of leucinevaline change in distinct low pH requirements for membrane fusion between two related retrovirus envelopes. J. Biol. Chem 287, 7640–7651 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data needed to evaluate the conclusions in the paper are present in the paper or the Supplementary Materials.