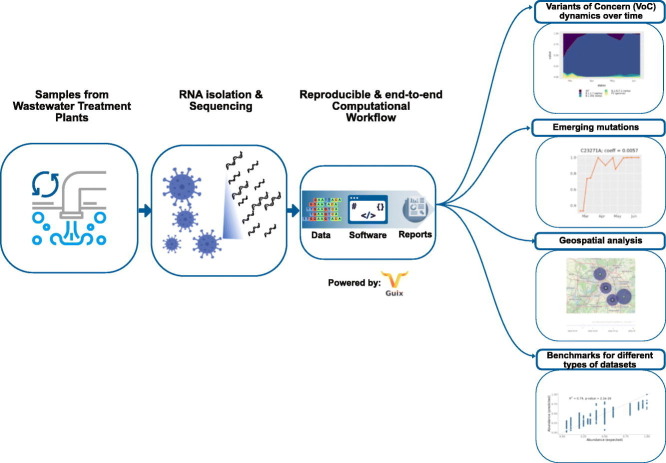
Abstract
The use of RNA sequencing from wastewater samples is a valuable way for estimating infection dynamics and circulating lineages of SARS-CoV-2. This approach is independent from testing individuals and can therefore become the key tool to monitor this and potentially other viruses. However, it is equally important to develop easily accessible and scalable tools which can highlight critical changes in infection rates and dynamics over time across different locations given sequencing data from wastewater. Here, we provide an analysis of lineage dynamics in Berlin and New York City using wastewater sequencing and present PiGx SARS-CoV-2, a highly reproducible computational analysis pipeline with comprehensive reports. This end-to-end pipeline includes all steps from raw data to shareable reports, additional taxonomic analysis, deconvolution and geospatial time series analyses. Using simulated datasets (in silico generated and spiked-in samples) we could demonstrate the accuracy of our pipeline calculating proportions of Variants of Concern (VOC) from environmental as well as pre-mixed samples (spiked-in). By applying our pipeline on a dataset of wastewater samples from Berlin between February 2021 and January 2022, we could reconstruct the emergence of B.1.1.7(alpha) in February/March 2021 and the replacement dynamics from B.1.617.2 (delta) to BA.1 and BA.2 (omicron) during the winter of 2021/2022. Using data from very-short-reads generated in an industrial scale setting, we could see even higher accuracy in our deconvolution. Lastly, using a targeted sequencing dataset from New York City (receptor-binding-domain (RBD) only), we could reproduce the results recovering the proportions of the so-called cryptic lineages shown in the original study. Overall our study provides an in-depth analysis reconstructing virus lineage dynamics from wastewater. While applying our tool on a wide range of different datasets (from different types of wastewater sample locations and sequenced with different methods), we show that PiGx SARS-CoV-2 can be used to identify new mutations and detect any emerging new lineages in a highly automated and scalable way. Our approach can support efforts to establish continuous monitoring and early-warning projects for detecting SARS-CoV-2 or any other pathogen.
Keywords: COVID-19 surveillance, Sewage sampling, Sequencing, Public health risk, Environmental monitoring
Graphical abstract
1. Introduction
The ongoing COVID-19 pandemic highlighted the need for monitoring approaches to track emerging pathogens and pathogenic lineages. Acknowledging the importance and potential impact of wastewater-borne epidemiological analysis, the European Commission has recently recommended to implement continuous monitoring on SARS-CoV-2 through wastewater in all member states (European Comission . Commission Recommendation (EU), 2021). SARS-CoV-2 is a positive strand RNA virus from the family Coronaviridae, genus Betacoronavirus (Fehr and Perlman, 2015; Zhou et al., 2020). As an alternative to individual patient tests that are tedious and expensive, Wastewater Based Epidemiology (WBE) has, before this pandemic, been used for different enteric microorganisms such as vaccine and wildtype polioviruses (Ranta et al., 2001), rotaviruses, hepatitis A, astroviruses, adenoviruses, and noroviruses (Petrinca et al., 2009). In the past two years, wastewater monitoring has been shown to be an effective tool for monitoring incidence rates. Multiple studies showed that it is possible to detect viral RNA even before widespread clinical reports (Wu et al., 2020; Medema et al., 2020; Bar-Or et al., 2022; Xiao et al., 2021), suggesting a potential as an early alert system.
Several WBE initiatives for SARS-CoV-2 monitoring were established worldwide, and currently, the “COVIDpoops19” initiative (Naughton et al., 2021) lists 128 dashboards from 276 universities monitoring 3364 sites. However, many of those studies are based on RT-qPCR analyses, limited to quantifying the viral titer and/or tracking a few known lineages, correlating the results with the reported number of cases in the area. A few studies have been using amplicon sequencing or metagenomics covering the whole viral genome, allowing to track virus lineages through signature mutations (Crits-Christoph et al., 2021; Izquierdo-Lara et al., 2021; Landgraff et al., 2021). However, quantifying Variants of Concern (VOC) by next generation sequencing (NGS) reads remains challenging due to fragmented sequences. Moreover, sequencing and quantifying lineages are just the first steps in understanding the dynamics of the outbreaks. The sequencing results should be easily analyzed and combined with geospatial time series analysis. Tracking of VOCs over time and space can inform policy-making decisions in order to control new outbreaks. In this study, we aimed to develop a reproducible, automatized, open-source pipeline for analyzing continuous sampling of wastewater treatment plants to track signature mutations of SARS-CoV-2 lineages of interest and emerging mutations via wastewater amplicon sequencing. Our main objectives were:
(i) To benchmarked the pipeline using simulated (in silico) data and spiked-in samples (Karthikeyan et al., 2022).
(ii) To sequence and analyze samples from Berlin wastewater using the ARTIC protocol (Pipelines R& et al., 2020) with 2 different sequencing protocols of ~250 bp length (in the following called “dataset-Berlin250”) and under industry conditions of ~35 bp length (in the following called dataset-Berlin35) during the 3ed and 4th pandemic wave in Germany;
(iii) To analyze previously published dataset from New York City, where the sequencing was restricted to the receptor binding domain (RBD) region (Smyth et al., 2022) (in the following called “dataset-NYC(RBD)”), showing the accuracy and usefulness of our methods for SARS-CoV-2 monitoring with data generated from multiple sites and approaches (iSeq and MiSeq).
2. Results
2.1. A reproducible computational pipeline for tracking SARS-CoV-2 in wastewater
We developed a new pipeline - PiGx SARS-CoV-2 - in the framework of our previously published set of pipelines called PiGx (Wurmus et al., 2018). They are designed with a special focus on usability and reproducibility. The new pipeline was added to the PiGx collection of pipelines and it is distributed together, using GNU Guix (See Fig. 1 for a diagram of the workflow). The pipeline comes with all the needed tools and their dependencies and can thus be reproduced on different systems independent of any other installed software. In comparison to other published tools like “Freyja” (Karthikeyan et al., 2022) and some commonly used pipelines for variant analysis like V-pipe (Posada-Céspedes et al., 2021) in combination with “COJAC” (Jahn et al., 2022) or the “ARTIC bioinformatics pipeline” (Pipelines R& et al., 2020), PiGx SARS-CoV-2 additional features (see Table 1 ) improve 1) usability, 2) suitability for environmental samples like wastewater and 3) the deployment for robustness and reproducibility.
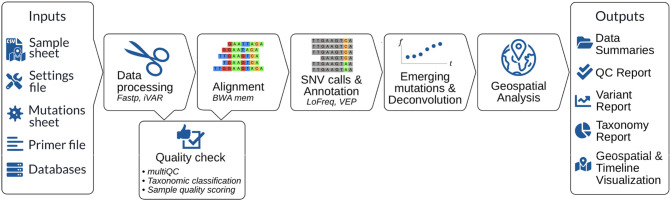
Fig. 1.
Flowchart of PiGx SARS-CoV-2 pipeline describing required input files, the analysis workflow and used tools and output files.
Table 1.
Feature comparison between different available pipelines and analysis tools.
| COJAC (+ V-pipe) | Freyja | ARTIC bioinformatics pipeline | PiGx Sars-Cov-2 | |
|---|---|---|---|---|
| Deployment | Package available through conda, but execution relies on separate jupyter notebooks | Package available through conda | Package available through conda | Package available through GNU Guix, workflow management using snakemake |
| Lineage prediction strategy | Co-occurrence analysis using Maximum-Likelihood-Estimation | Deconvolution using constrained minimization | None, ends at variant calling step | Deconvolution using robust regression |
| Detection/Identification of emerging new/single mutations | + | − | + | + |
| End-to-end |
+ manual execution of multiple notebooks needed |
− | − |
+ fully automatic with options to start at different steps |
| Variable reference ge-mes as Input | + | − | − | + |
| Output summary reports with visualization, stats and data |
− (only separate outputs of each notebook) |
− | − | + |
| Enables geospatial analysis | − | − | − | + |
| Single Mutation trend analysis directly implemented | − | − | − | + |
| Can take Input from different seq strategies and different read length | Limited, performance may vary with read length (Karthikeyan et al., 2022) | Starts only from the BAM files | − | + |
| Bit-by-bit reproducibility | − | − | − | + |
In terms of usability, PiGx SARS-CoV-2 runs all steps end-to-end fully automatic and provides a comprehensive HTML report at the end. It is suitable and tested for a variety of sequencing input formats (variation in read length, paired- and single end format, primer sequence present or absent and different Illumina protocols). Furthermore it allows customizable inputs i.e. the reference genome or tool-specific settings.
Next to monitoring and predicting known lineages, PiGx SARS-CoV-2 does not only allow for reporting a file with new mutations (as most of the other mentioned tools do) but it automatically evaluates trends in all new mutations and reports those with consistently increasing frequency.
In addition, the directly implemented geospatial tracking allows to compare and monitor infection dynamics from different locations (See example reports in Data access section).
2.2. Benchmarking the pipeline using spiked-in and simulated samples
In order to check the accuracy of our pipeline, we analyzed two simulated datasets.
First, we analyzed a spike-in mixture dataset from Karthikeyan et al. (Karthikeyan et al., 2021). We obtained 384 BAM files with reads pre-aligned to a SARS-CoV-2 reference genome (Supplementary Table S2.3, with samples ranging from 1160 to 1,955,791 reads. Four samples did not pass the 90 % reference genome coverage threshold and were discarded. In total, we tracked 99 signature mutations from 5 lineages, which were in almost all samples covered with at least 100 reads per site (Supplementary Table S2.3). Overall, we found 225 ± 54 mutations in this dataset.
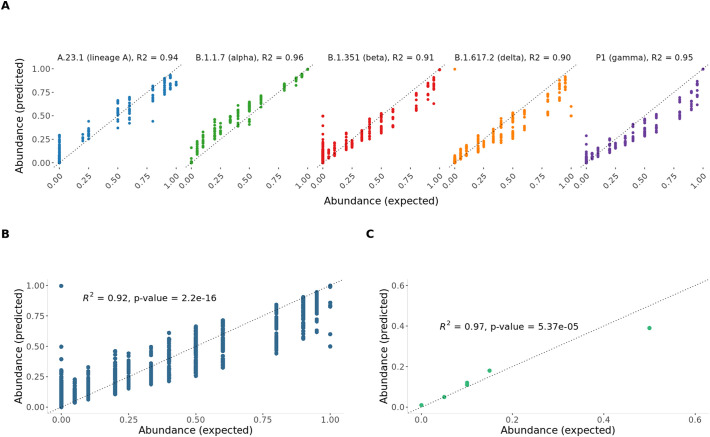
Analyzing the predictions for each lineage with our deconvolution method, we found that we were able to recall the expected proportions of lineages with R2 above 0.9 (Fig. 2B).
Fig. 2.
A) Prediction verification results for the spike-in data simulation per lineage, the dotted line shows the expected trendline; B) Prediction verification results for the spike-in data simulation across all lineages excluding lineage A; C) Prediction verification results in-silico simulation, single-end simulated 40 bp reads from GISAID, 100 k reads.
Additionally, we tested the pipeline on a second simulation data set (see Methods), generated in silico with known proportions of lineages. A total of 100,000 reads were generated. In this comparison, we used a set of 179 signature mutations from 6 lineages, of which 74 were recovered (Supplementary Table 2.4). Overall, our methods were able to get the expected proportions of lineages with R2 of 0.97 (Fig. 2C).
2.3. Wastewater SARS-CoV-2 sequencing and analysis with PiGx SARS-CoV-2
For this study, we sequenced a total of 988,025,456 reads from 171 samples from Berlin, using two different sequencing protocols. Firstly, for dataset-Berlin250 we obtained 74,633,648 reads, from 62 samples collected at four different wastewater treatment plants in Berlin operated by the municipal water authority (“Berliner Wasserbetriebe”) from 09th of February to 10th of June 2021 (Phase I) and from 16th of September 2021 to 19th of January 2022 (Phase II), using paired-end Miseq/Novaseq protocol with 2 × 250 bp reads. Between the two phases, due to low incidence rates, sequencing quality was insufficient. The average number of read-covered signature mutation sites per sample was 105 (SD 33, from a total of 154 tracked signature mutations, see mutation tables in the Supplementary Table S1). Of those 62 samples, 16 samples did not pass the defined quality control threshold (samples for which <90 % of the signature mutation sites were covered).
We were able to align from 11 to 99 % of sequencing reads (1 outlier with only 5 % aligned reads) to the Wuhan reference SARS-CoV-2 genome, and the resulting alignments were used for variant calling. We were able to detect a total of 3210 mutations, of which 133 are signature mutations, across all the samples (See methods for details on alignment and variant calling). The overall frequency of mutations per sample is shown on Supplementary Table S2.1-S2.4. The results of the time-series analysis for mutations and deconvolution of lineages for this dataset is presented in the sections below.
Secondly, industry scale dataset-Berlin35 contains 109 samples from one Berlin wastewater plant and three pumpstations (also operated by “Berliner Wasserbetriebe”). We used a paired-end very-short-read protocol (2 × 35 bp), for fast real time monitoring from 03.08.2021 to 20.01.2022. This data was analyzed in order to test our pipeline in a real time data monitoring system. We obtained a total of 913,391,808 35 bp reads. The average reference genome coverage was 97 % (SD 5.7) with 9 samples not passing the quality control (QC) criteria of >90 % reference genome coverage. The average number of signature mutations found per sample was 27 from the 154 tracked (SD 14.5) mutations and the mean of overall mutations found was 288 per sample (SD 147.5). The results of time-series mutation analysis and deconvolution of lineages for this dataset can be found in Supplementary Table S3.
The third dataset - dataset-NYC(RBD) - originated from published deep sequencing data of the receptor binding domain (RBD) of SARS-CoV-2 on samples from January 31th to June 14th 2021 collected in New York City (NYC) wastewater and published by Smyth et al. (Smyth et al., 2022). In the 94 samples reanalyzed here, we found, on average, 8 of the 12 mutation sites within the RBD (mean number of signature mutations found was 3.5). We did not apply a reference genome coverage cutoff because the sequencing was restricted to a small genomic region.
Smyth et al. described 3 cryptic lineages that were found in New York City wastewater: WWTF #10 (7 mutations), WWTF #11 (8 mutations), WWTF #3 (23 mutations) (see Supplementary Table S4). We tested our pipeline's ability to highlight those “cryptic lineages” early on as well from the purely computational analysis in contrast to the extensive wet lab experiments that it took the authors to discover them. The results of this analysis are shown in the section below.
2.4. Emerging mutations can be teased out from time-series analysis
The time-series nature of the data can not only be used to track SARS-CoV-2 lineages, but also to identify trends for individual mutations. We applied a linear regression model for each mutation using the date of sampling as the independent variable to identify mutations with strong increasing trends over time (see Methods). We considered mutations significant if the t-test p-value is below 0.05 . The full lists can be found in Supplementary Table S5.
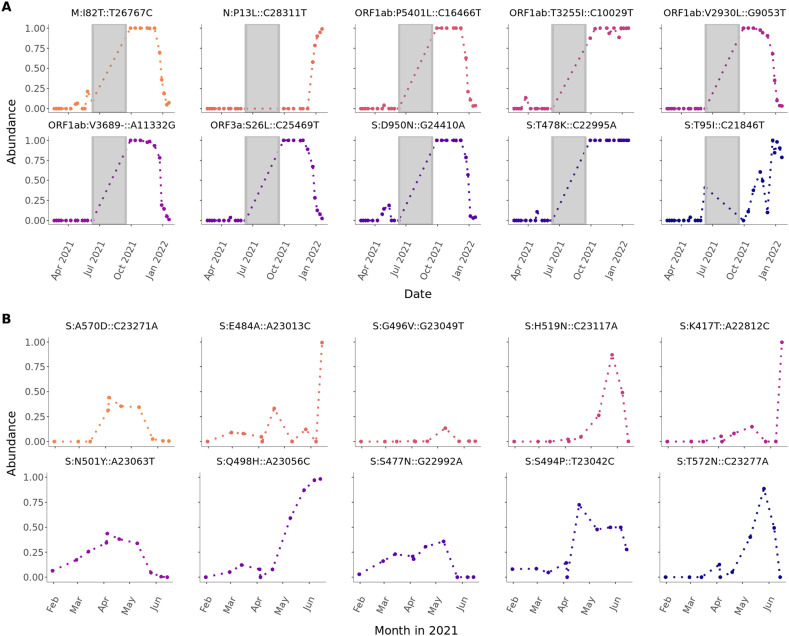
Overall, merging the two sample groups within dataset-Berlin250 from Berlin Phase I and II for a single analysis, 105 mutations were significantly changing over time from February 2021 until January 2022. The top 10 most significantly changed mutations are shown in Fig. 3A.
Fig. 3.
A) Top 10 sequence variants that significantly increase over time in Berlin. The mutations were pooled over locations of four different wastewater treatment plants and daytime and sorted by decreasing coefficients from linear models. Statistical significance was evaluated by a t-test using p ≤ 0.05 as cutoff. Only samples passing the sample quality scoring (>90 % reference genome coverage) were used. There was no sampling between June 11 and September 19, 2021. B) Top 10 sequence variants that significantly increase over time in New York City (NYC) (2021). The mutations were pooled over locations of 14 different wastewater treatment plants in NYC and daytime and sorted by decreasing coefficients from linear models. Statistical significance was evaluated by a t-test using p ≤ 0.05 as cutoff.
Here, six of the highlighted mutations M:I82T::T26767C, ORF3a:S26L::C25469T, ORF1ab:V3689-::A11332G, ORF1ab:V2930L::G9053T, S:D950N::G24410A, ORF1ab:P5401L::C16466T, are uniquely characteristic for the lineage B.1.617.2 (delta). They show a similar pattern, emerging mostly during the summer of 2021 and decreasing in January 2022. Hereby, S:D950N::G24410A and M:I82T::T26767C already started to appear with increasing frequency in late April 2021, but inconsistently. The mutation S:T478K::C22995A is a shared mutation between the lineages B.1.617.2 (delta), BA.1 and BA.2 (omicron). It showed a consistent increase from July 2021 and reached 100 % of presence until the end of our time-series. However, N:P13L::C28311T and S:T95I::C21846T are unique mutations for the BA.1 lineage where the latter already started to continuously increase in frequency starting in October 2021 which is a month earlier than the B.1.1.529 (omicron) lineage family was started to track by the RKI (Fig. 4C).
Fig. 4.
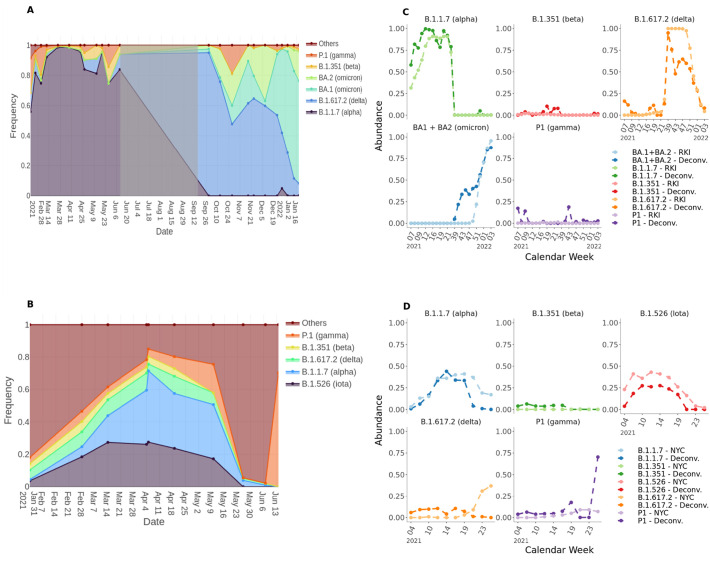
A) Proportion of tracked lineages over time in Berlin wastewater. Only samples passing the sample quality scoring (≥ 90 % reference genome coverage) were considered. Shaded area highlights the non-sampling Phase. B) Proportion of tracked lineages over time in New York City wastewater. The proportions were calculated with a deconvolution model based on the signature mutation frequencies. “Others” denotes a set of reference mutations derived from the deconvolution matrix. Sample results were pooled from four different wastewater treatment plants using weighted mean with read number as weights. In case of undistinguishable lineages the proportion derived for the group was distributed equally for the affected lineages. C,D) Comparison of deconvolution results (dark color) with lineage frequency analysis data from Robert-Koch-Institute (RKI) (C) or NYC Department of Health and Mental Hygiene (NYC) (D) (light color). Deconvolution results were pooled by weeks using weighted mean using sample read numbers as weights. For the data from Berlin only samples passing the sample quality scoring (≥ 90 % reference genome coverage) were used.
Within the dataset-NYC(RBD) (Smyth et al., 2022), we found a total of 69 significantly changing genome variants. The highlighted mutations with the 10 highest correlation values in Fig. 3B point out 8 of the 28 reported mutations of cryptic lineages (see Supplementary Table S4). Additionally S:N501Y::A23063T and S:A570D::C23271A were highlighted which are characteristic mutations for B.1.1.7 (alpha). They show a constant increase already up to 40 % in March which is around 1 month earlier than the reported abundancy for B.1.1.7 (alpha) based on cases (Fig. 4D).
2.5. Deconvolution of mutation frequencies infers SARS-CoV-2 lineage frequencies
In our pipeline, we have implemented methods to deconvolute the frequencies of VOCs from pooled sequencing reads. Briefly, the deconvolution method uses signature mutations for each VOC and tries to discern the proportions of these lineages making up the observed mutation frequencies in the pooled (bulk) sequencing reads obtained from the wastewater. In this study, we tracked 4 lineages which were classified at the time of data collection as VOCs: B.1.1.7 (alpha), B.1.351 (beta), P1 (gamma) and B.1.1617.2 (delta) in both datasets from Berlin and New York City. For the latter we additionally tracked the lineage B.1.526 (Iota). For the samples from Berlin from Phase II we additionally tracked the BA.1 and BA.2 lineages, which taxonomically are classified as sublineages of B.1.1.529 (omicron) and became VOCs in November 2021. We decided to track them separately in order to get a higher resolution on their dynamics. In the following, when comparing to official reported lineage abundances we are adding up our separate abundances for BA.1 and BA.2 to compare to reported values for B.1.1.529 (omicron). We characterized the lineages with a mutation table (Supplementary Table S1) containing signature nucleotide mutations from covidCG (Chen et al., 2021). We took a list of mutations with a sequence consensus threshold of 70 %. We included mutations that are unique for each lineage, as well as mutations that are shared by two or more lineages. Of note, the pipeline is flexible and can track any lineage if the signature mutations are provided in nucleotide format.
We applied this deconvolution method (based on the frequencies of the signature mutations) to infer the proportions of each lineage on each sample (Supplementary Table S3). The lineage frequencies are predicted using a regression model based on the observed frequencies of the signature mutations for each lineage. In the course of the method development, we found that for some datasets - especially those with sparse sampling rate - an additional weighting step improves the prediction results. It is an optional step that was applied to all datasets except those used for benchmarking (see Methods).
Fig. 4A shows VOC proportion changes over time across 4 wastewater treatment plants in Berlin (merged results of Phase I and Phase II). Overall, we predict an increase in B.1.1.7 (alpha) that had 57 % on February 19th (beginning of sampling of Phase I) and increased to 79 % on June 10th (end of sampling of Phase I) with a peak of 99 % on May 25. Also B.1.351 (beta) increased from zero detection in February to 8 % in May with a predicted peak of 10 % on May 25. The B.1.617.2 (delta) lineage was barely detected with 3 % over the sampling time of Phase I increasing to 11 % on May 12. We predicted 16 % of B.1.617.2 (delta) as early as in February 2021 but this result is likely to be inaccurate. For P1 we could predict in Phase I a decrease from 17 % on February 19 to zero in June. However in sampling Phase II, P1 is predicted again with an abundance peak of 18 % on October 28. During winter 2021 the predicted P1 abundance decreases continuously down to 3 % in January. The tracking of the lineages BA.1 and BA.2 started with sampling Phase II in September 2021 where they were initially predicted with a total abundance of 6 %. Their abundance rapidly increased to ~90 % by January 19 with BA.1 at ~70 % and BA.2 with ~20 %. In the timeframe we sampled, the diversity and abundance of lineages that are not VOCs was already very reduced. We only predicted unspecified lineages (labeled as “Others”) with 8 % in February 2021 and it fell below 1 % on March 11 and never increased again.
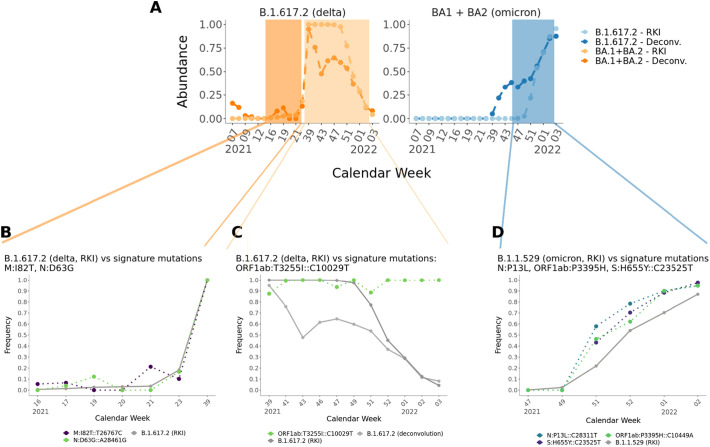
In order to see if the predicted results can reflect the abundances of circulating lineages in Berlin, we compared the deconvolution results with lineage analysis data published by the Robert Koch-Institute (RKI) for Germany (Fig. 4C). Hereby, lineage dynamics for Germany are very comparable to the dynamics within the city of Berlin. We can see that our predicted lineage frequencies are very similar to the reported lineage distribution based patient testing. Only B.1.1.7 (alpha) shows mostly higher predicted values, but with very similar trends. Also the predictions of the lineages BA.1 and BA.2, which are taken together comparable with the reported B.1.1.529 (omicron) values are higher in the beginning (December 2021) than the RKI values, but become very similar in January 2022. This is explainable with the continuous detection of the mutation ORF1ab:T3255I::C10029T, which is listed as unique signature mutation for BA.1 and BA.2, but is also carried by the B.1.617.2 sublineage 21 J (nextstrain.org, n.d.) (but not by the parent clade) that we did not actively track in this analysis.
The analysis of the dataset-Berlin35 showed similar results as for the dataset-Berlin250 as shown in Supplementary Fig. 1. Of note, the prediction results for the abundances of B.1.617.2 (delta) and B.1.1.529/BA.1 + BA.2 (omicron) are showing less divergence from the RKI values than for the dataset-Berlin250.
The data from New York City (Fig. 4B) shows a more diverse mixture throughout the sampling phase according to the predicted high proportion of “Others”. This proportion was as high as 82 % in January 2021 decreasing to 15 % on April the 5th but then had a predicted increase again to 97 % in June. The most dominant lineages were B.1.1.7 (alpha) which increased from 0 % in January up to 44 % in April and B.1.526 (Iota) which increased from 4 % in January up to predicted 28 % in April. However, the comparison with the data reported from NYC Department of Health and Mental Hygiene (NYC health) (Fig. 4D) suggests that both lineages circulated with similar abundances within the given timeframe and that the differences in the predicted values are due to the expected inaccuracy of the pipeline. The abundance of B.1.351 (beta) increased slightly from 4 % in January to 5 % in April but was not present anymore after. For B.1.617.2 (delta), we predicted a continued increase up to 10 % in April which is also in contrast to the NYC health data where the abundance of delta only starts to increase at the end of May. For P1 (gamma) we predicted an increase from 3 % in January up to 17 % in May. This trend is also shown from the NYC health data. However, we also predicted 70 % P1 in June. For this prediction only 4 signature mutations across all lineages were found and 1 of them is S:K417T::A22812C with a frequency of 1 which is a unique signature mutation of P1 (gamma). Besides both above-mentioned differences, our prediction results are consistent with the NYC health data as shown in Fig. 4D. Unpooled results for single locations for both datasets are attached as Supplemental material (Supplementary Table S3).
In Fig. 5 , we combined the visualization of key mutation frequencies, cases of COVID-19 in Berlin (from RKI), and deconvolution results for B.1.617.2 (delta) and BA.1/BA.2 (omicron) lineages.
Fig. 5.
A) Combination of lineage prediction results (deconvolution) for B.1.617.2 and BA.1/BA.2 (dataset-Berlin250), B,C,D) single key signature mutations M:I82T::T26767C, N:D63G::A28461G, ORF1ab:T3255I::C10029T, ORF1ab:P3395H::C10449A, N:P13L::C28311T, S:H655Y::C23525T and case numbers in Berlin (from RKI).
We can see that the mutations M:I82T and M:D63G showed a strong increase together with RKI case numbers and with B.1.617.2 (delta) proportions from our deconvolution results. The same pattern is shown for N:P13L, ORF1ab:P3395H and S:H655Y, raising together with omicron lineages. However, the mutation ORF1ab:T3255I (tracked as BA.1 and BA.2 signature mutations in our deconvolution) was detected with frequency of 100 % already in September 2021, while Omicron was not present yet. This mutation was in high frequency when B.1.617.2 (delta) was predominant and stayed high while omicron raised. This could have hinted that this mutation was already present in a sub-clade of B.1.617.2 (delta) (nextstrain.org, n.d.) and in fact this mutation is present in delta sub-clade 21J (covariants.org, n.d.).
2.6. RT-qPCR on wastewater samples reflect SARS-CoV-2 incidences
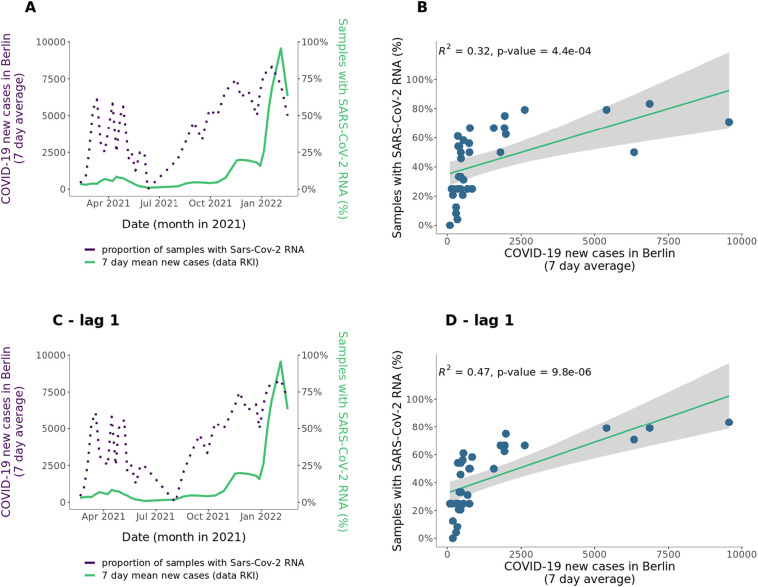
SARS-CoV-2 levels in wastewater have been repeatedly used to monitor and also predict incidence rates in the populations (Isaksson et al., 2022; Lastra et al., 2022). In order to recapitulate this in our samples, we checked if RT-qPCR results correlated with case numbers in the region. For RT-qPCR, we used 4 pairs of primers for SARS-CoV-2 detection (RT-qPCR) on the wastewater samples. Due to the very low amount of viral particles present overall, we decided for a semi-quantitative approach, instead of using the cycle threshold (Ct) values, calculating the number of positive detections divided by the number of total reactions carried, grouping all the samples for each day (See Methods for details). The daily percentage of positive qPCR reactions ranges from 0 to 7 out of 8 (Supplementary Table S6). We also found positive, significant correlation with RT-qPCR results and incidence rates (adjusted R2 = 0.32, t-test p-value = 0.0004, see Fig. 6A-B). In addition, we have also repeated the cross-correlation analysis between incidence rate and RT-qPCR results with different time lags. In this case, lag = −1 week also had positive correlation with the incidence rate (adjusted R2 = 0.47, coefficient = 0.5, t-test p-value = 9.8e-06) (Fig. 6C-D). Overall, this is in agreement with the predictive value of wastewater monitoring detailed in previous studies (Wolfe et al., 2021).
Fig. 6.
A) 7 days average of COVID-19 cases in Berlin, data from Robert Koch-Institute (RKI) (light green, left axis) and proportion of samples positively determined SARS-CoV-2 RNA by RT-qPCR (dark violet, right axis) over Feb - Jan 2022. B) Correlation of 7 days average of COVID-19 cases in Berlin and proportion of samples with positively determined SARS-CoV-2 RNA by RT-qPCR. C) 7 days average of COVID-19 cases in Berlin, data from Robert Koch-Institute (RKI) (light green, left axis) and proportion of samples positively determined SARS-CoV-2 RNA by RT-qPCR (dark violet, right axis) over Feb - Jan 2022 with one time point lag. D) Correlation of 7 days average of COVID-19 cases in Berlin and proportion of samples with positively determined SARS-CoV-2 RNA by RT-qPCR with one time point lag.
3. Discussion
In many countries, epidemiological monitoring of SARS-CoV-2 is largely dependent on PCR-based or antigen detection methods without sequencing which is applied on patient samples. These techniques can be used for variant detection only after a concerning new lineage is detected and an appropriate assay was developed. In order to discover new lineages, we need to be able to call mutations of the SARS-CoV-2 genome which can be done using sequencing methods. However, sequencing-based techniques are deployed on only a fraction of the patient population. Wastewater monitoring emerged as a viable option to track the prevalence of COVID-19 and also for the emergence of different lineages (Lin et al., 2021) at the population level not only because it is faster and cheaper than sequencing of samples derived from patients, but it can also be more representative due to less bias through the choice of which samples are going to be sequenced. Furthermore it can also be used to track early emerging mutations or lineages of SARS-CoV-2. However, sequencing of SARS-CoV-2 material obtained from wastewater presents data analysis challenges as the samples are potentially from numerous patients, and have lower quality than material obtained directly from patients. In addition, the analytical workflows should be able to deal with samples from multiple locations and time points and combine the information in an easily accessible manner.
In order to address these challenges, we have built a reproducible analytics pipeline that takes in raw sequencing reads and provides sharable interactive reports. It contains essential data, analysis results (summarized and per-sample) in commonly used formats (for sharing and postprocessing) and visualizations all in one. Furthermore it directly includes geospatial information, and mutation and lineage tracking features over time. This provides a more straightforward way for discovering lineage divergence and cryptic lineages (as shown on the analysis of the dataset-NYC(RBD)). For monitoring projects, it is important to provide a workflow that can handle both continuous sampling and the need to make slight adjustments to the expected data. Above that, maintaining proper documentation and reproducibility of the results at all times is also an essential feature. All of the tools mentioned in the comparison come as packages deployed e.g. through conda, which is a first step for ensuring this. However, having to manually execute single notebooks for single samples or single sample batches, or executing single commands is only marginally scalable and prone to human error and insufficient documentation behavior which can lead to a lack of reproducibility. PiGx SARS-CoV-2 offers state-of-the-art software bit-by-bit reproducibility thanks to GNU Guix (Wurmus et al., 2018) and workflow management using snakemake. By providing the flexibility to start the workflow at different steps but also keeping as many steps as possible in an automatic way, many samples can be processed over time in a timely and error efficient way.
The pipeline comes with built-in flexible quality control metrics since samples from wastewater pipeline can have more frequent quality issues. In our analysis, we applied a strict cutoff for reference genome coverage (≥ 90 %) for whole-genome sequencing data to reduce noise in our predictions. Our pipeline also allows the user to input their own reference genome and their own set of signature mutations and lineages. As an additional step for QC, we implemented a taxonomic classification of reads that did not align to the SARS-CoV-2 reference genome. Since we used a PCR based protocol, we expect some degree of nonspecific amplifications, so it is of great help to have an additional control by means of the taxonomic classification to assess potential biases. Also since Kraken2 is a k-mer classifier, this method can reveal reads that match SARS-CoV-2 but are not aligned by stringent alignment tools. This is important to know because it provides insights about potential loss of new mutations missed on the alignment. This step allows the user to investigate potential issues and, if necessary, to adjust the alignment stringency.
Aiming to benchmark our pipeline, we tested it on two different simulated datasets, allowing us to estimate the error rates from our methods. For the dataset generated in silico, we were able to show that our predicted results are in high agreement with the expected values. This shows that our pipeline has potential to track accurately VOC from sequencing data. However, we are aware that real sequencing data can offer further challenges, such as low quality sequencing, presence of many other microorganisms and untracked lineages of SARS-CoV-2. In order to better benchmark our tool, we also analyzed spiked-in samples generated by (Karthikeyan et al., 2021). Our results overall outperformed those of the original publication.
One of the primary features of our approach is built-in tracking of emerging mutations. This feature allowed, for example, early prediction of lineages such as B.1.617.2 (delta) from a single signature mutation M:I82T::T26767C (Fig. 5) in the dataset-Berlin250. We were able to detect the lineage characterizing mutations before the lineage itself was detected in the population (Fig. 5). This specific mutation was described to be associated with critically increased viral fitness (Shen et al., 2021). The analysis and results are also visualized without the need for any additional steps directly in the summarizing report. We showed that our pipeline and its reports can be a valuable tool for early warning predictions and to guide additional targeted analysis.
Another key feature of our approach is the deconvolution method that helps us identify the proportion of lineages present in environmental samples such as wastewater samples. By making use of a weighted regression method, we were able to provide accurate estimates of lineage proportions for our samples over time. For the VOCs that we tracked with signature mutations, we show in Fig. 4 that our model can accurately predict the composition of lineages when comparing with abundances of circulating lineages reported during the same time frame, even with very low frequencies. This method was able to predict the rapid increase of the lineages BA.1 and BA.2 in the winter (Fig. 4).
It is important to note that the mutations commonly used for tracking B.1.1.7 in other studies, S:N501Y::A23063T and del69/70 (Sandoval Torrientes et al., 2021; Vega-Magaña et al., 2021) were rare or not found in our Berlin dataset, but they were detected in NYC dataset (Fig. 3B), and this might be explained by PCR bias differences between the datasets, because the NYC dataset only sequenced the RBD genomic region, having a higher resolution on the mutations in this genomic region.
Additionally, with the dataset-Berlin35, we showed that our pipeline can be used in an industrial production system for real-time monitoring. The results obtained were comparable with the dataset-Berlin250 for the same time frame (Phase 2). Interestingly, for the dataset-Berlin35, BA.1 + BA.2 (omicron) predictions are followed by RKI incidence cases closer in time than for the dataset-Berlin250, where we detect omicron and delta one week in advance. For B.1.617.2 (delta), the dataset-Berlin35 shows more similar proportions than for the dataset-Berlin250 (See Supplementary Fig. 2). These results can suggest that the inaccuracies found in our dataset-Berlin250 can be explained by differences in the data generation (read length, internal sequencing validations or differences on sampling sites) rather than in data processing with our pipeline.
As reported in previous studies in other cities around the globe (Ahmed et al., 2020), we showed that also for Berlin, the quantification from wastewater can reveal the prevalence of infections on a community scale earlier than it is possible from clinical testing. Although RT-qPCR results are not fully quantitative, observing this expected trend was important and paved the way for more robust lineage and mutation trend analysis using sequencing.
Regardless of the methods used on wastewater, as previously published reports also indicate, wastewater monitoring may provide early warning for future case numbers and emerging mutations even post-pandemic when populations are not tested and monitored that thoroughly as during the pandemic.
In conclusion, we present a reproducible and comprehensive workflow with a strong emphasis on usability and reproducibility that has features for tracking mutations and VOC over time and geographical locations. We stress-tested the tool with simulated data and real world data from different locations and with different methods, showing the usefulness of our tool but also the importance of keeping lineage nomenclature and mutations tracked consistent, for comparable results.
4. Methods
4.1. Experimental methods
4.1.1. Enrichment of viral particles from raw wastewater and RNA extraction
For the dataset-Berlin250, raw wastewater samples were collected from four different wastewater treatment plants across Berlin, serving a population of approximately 3.4 million people in total. They were collected as composite 2 h samples (8–10 pm and 10–12 pm) at the primary influent collector at the indicated wastewater treatment plants. Typical characteristics of Berlin wastewater treatment plant effluents are 500–1500 mg/L chemical oxygen demand, 200–600 mg/L suspended solids, 40–80 mg/L ammonium-N, 2–8 mg/L orthophosphate-P, 1500–2000 μS/cm electrical conductivity.
Samples were stored and transported at four degrees, and processed about 12 h after collection. The samples were enriched for viral RNA as previously described (Jahn et al., 2021). About 100 mL sample was filtered through 2 glass fiber and 0.2 μM PVDF filters (Millipore, cat# AP2007500 and S2GVU02RE). Of this filtrate, 60 mL was transferred to a 10 kDa cutoff centricon unit, that was previously pre-conditioned with 50 mL ultrapure water and centrifuged with 3000 g for 15 min at 4 °C. After centrifugation of the samples for 30 min at 4 °C and again 3000 g, the unit was inverted and about 400 μL concentrate was collected by centrifugation for 1000 g at 4 °C for 3 min. The concentrate was mixed with 3 volumes of Trizol LS (ThermoFisher cat# 10296-010), and the RNA extracted using the Direct-zol RNA miniprep kit (Zymo cat# R2052) including the DNase treatment and elution with 50 μL ultrapure water according to the manufacturer's instruction. Absence of PCR inhibitors was confirmed by mixing the sample 1:1 with total RNA from human cells followed by amplification of a human transcript. Not detectable in waste water alone by RT-qPCR.
4.1.2. Reverse transcription/quantitative polymerase chain reaction (RT-qPCR)
The extracted RNA was amplified using the LunaScript reverse transcription mix (NEB cat# E3010L), with 16 μL RNA and 4 μL reaction master mix according to the manufacturer's instructions, except for a 20 min incubation at 55 °C instead of 10 min. Afterwards, the cDNA was diluted 1:10 with ultrapure water, and 3.75 μL diluted cDNA used per qPCR reaction, using a SYBR green master mix (ThermoFisher cat# 43-643-46), and final concentrations of 250 nM of the primers on Supplementary Table S7. The presence of the proper amplicon was verified using a 2.5 % TAE agarose gel. If the expected amplicon was not detected on the gel, the sample was counted as negative even if a qPCR signal was observed.
4.1.3. ARTIC-seq of the SARS-CoV-2 genome
Amplicon sequencing libraries of the SARS-CoV-2 genome were generated using the ARTIC protocol v3 (phase I) and modified version of ARTIC protocol v4 (Phase II) (Pipelines R& et al., 2020), using 6 μL of the cDNA generated as described above as an input. The primer pools were obtained from IDT. Amplicon libraries were sequenced on an Illumina Miseq or Novaseq device with 2 × 250 paired-end sequencing and 20 % phiX spike-in. The modified ARTIC v4 primer can be found in Supplementary Table S8.
4.2. Berlin wastewater samples processing for very-short-reads
In order to test if the pipeline would perform reliably under industry conditions we also used it with so-called production data from the amedes analytical company. The sequencing was performed as follows:
45 mL of raw wastewater was centrifuged for 10 min with 10,000 x g at 4 °C. Subsequently, the supernatant was prefiltered using Filtropur S 0.45 μm filter units (Sarstedt, Darmstadt, Germany), further transferred to 100 kDa cutoff Amicon Ultra-15 units (PLHK Ultracel-PL Membran, 100 kDa Centrifugation units; Merck Sigma Aldrich Chemie GmbH, Taufkirchen, Germany) and processed according to the manufacturer's manual.
Automated RNA isolation was accomplished using a Qia-Cube HT Extractor using the QIAamp 96 DNA QIAcube HT Kit according to the manufacturer's protocol (Qiagen, Hildesheim, Germany).
Library preparation for NGS sequencing was performed following the complete Illumina SARS-CoV-2 sequencing workflow (Illumina COVIDSeq Test, Illumina, San Diego, USA) including RNA-to-cDNA conversion and SARS-CoV-2 targeted PCR using the ARTIC V3 primer set. The generated libraries were analyzed using NextSeq 550 and 550Dx sequencers with NextSeq 500/550 High Output Kits (v2.5; Illumina #20024906) generating 2 × 37 bp paired-end output.
4.3. Computational methods
4.3.1. General pipeline description
The PiGx SARS-CoV-2 pipeline provides end-to-end data processing and analysis for wastewater RNA sequencing.
The pipeline needs local databases (downloaded by the user) for some of the annotation and alignment tools, while the tools themselves are automatically installed. Furthermore, the user needs to provide: (i) a sample sheet (CSV format) containing information about sampling date and location; (ii) a settings file (YAML format) for specifying the experimental setup and optional custom parameter adjustments, (iii) a mutation sheet containing the lineages of interest and their signature mutations in nucleotide notation and BED file containing their genomic coordinates; (iv) the reference genome of the target species; (v) BED file containing the PCR primer locations (provided with the pipeline for ARTIC protocol).
In the first step, primer trimming is done with iVAR (Grubaugh et al., 2019), and fastp (Chen et al., 2018) is used for adapter trimming and filtering. To ensure reliable variant calling and robust lineage abundance prediction, the sample has to match stringent quality control measures. For this, information about the sequencing primers, adapters, and also a BED file containing the sites of the signature mutations is necessary. Specifically the latter is important to ensure comparability of the called variants across all processed samples.
In order to make the read quality process comprehensible, fastQC reports are generated after each step and summarized with additional MultiQC reports. The processed reads are aligned to the reference genome by BWA Mem ( Li, 2014 ) and various coverage statistics are taken by SAMtools coverage/bedcov (Li et al., 2009). The alignment is used further for single nucleotide variant (SNV) calling using LoFreq (Wilm et al., 2012). For predicting the lineage abundances, a deconvolution matrix is generated by matching the set of mutations called by LoFreq against the provided mutation table. The SNVs are translated to protein mutations by Ensemble VEP (McLaren et al., 2016). Kraken2 (Wood et al., 2019) is used to get taxonomic classification of the unaligned reads as an additional quality measure and further insight in the samples. The mutations were filtered for a minimum read coverage, then a deconvolution method was used to calculate the proportion of lineages representing Variants of Concern over time (more details in the section Deconvolution analysis) for each sample. For summarizing and visualizing the deconvolution results as a time series, by default, samples with SARS-CoV-2 reference genome coverage below 90 % are discarded. For each mutation, linear regression models are used (more details in the section Regression analysis for mutations) to detect if any mutation is significantly increasing over time. Here discarded samples were also not included.
For each sample a set of four reports (multiQC, general QC report, taxonomic classification report, lineage report) is generated using Rmarkdown and knitr. The R-package of plotly is used for generating interactive visualizations. The relevant results across all provided samples are summarized by an extra report that provides insightful visualizations and accessible navigation linking to all the single reports. The samples from different timepoints are used to produce time-series reports that track trending mutations over time. Furthermore, all per-sample results are summarized as tables and also combined to visualize time-series and geo-location plots, making the pipeline suitable for continuous sampling.
In this way the pipeline output provides an easily accessible overview about lineage and mutation dynamics in a communicable format but also enables extensive data exploration and access to sample-wise tables and summaries without the need for running extra scripts. PiGx SARS-CoV-2 uses snakemake (Mölder et al., 2021) to define and run the workflow.
4.3.2. Deconvolution analysis
4.3.2.1. Model description
With m being a system of linear equations built by using B being a signature matrix constructed from the signature mutations provided as input and f being the proportions for the lineages the deconvolution approach can be represented as m = f x B. Similar to what has been shown before for deconvolution of cell types from gene expression profiles or methylation profiles (Newman et al., 2015), we follow the assumption that the frequency of signature mutations corresponds with the frequency of the actual lineage which is characterized by it. The difference in our approach is that we use sequence mutations and apply weights to the signature matrix in order to get more realistic prediction results.
4.3.2.2. Signature matrix construction
The signature matrix is obtained by matching the set of mutations found in the sample against the set of signature mutations provided as input. In case the mutation table contains mutations that are shared between lineages, it is possible that multiple lineages cannot be distinguished from each other. In this case, the signature matrix will be deduplicated leaving only one column of the duplicated lineages which will be renamed with the grouped names of all lineages showing this duplicated signature mutation “pattern”.
To make the matrix more robust, additional “reference mutations” are added as well as a reference column denoted as “Others”. Bulk frequencies for the “reference mutations” are the difference between 1 and the value of the related signature mutation.
We propose the assumption that the more signature mutations can be found for a specific lineage the higher the probability that this lineage is present with a higher proportion within the sample. We therefore weigh the signature matrix (without the reference mutations) for each lineage with the proportion of signature mutations that has been found for each specific lineage from the total number of signature mutations that was given to characterize it. For “Others” the weight was calculated proportionally to the number of mutations of the mutation table that were found. Applying weights results in less variation and more accurate predictions for the datasets obtained from actual wastewater. For the artificial datasets however, applying the deconvolution without the additional weighting step resulted in more accurate prediction. Such differences can result due to the different composition of artificial data and real data. This weighting step is therefore optional and whether it is used or not should depend on the dataset in question.
4.3.2.3. Regression
To deconvolute the lineage abundance we performed robust regression analysis on the signature matrix and the bulk frequency values of the signature mutations using the “Robust Fitting of Linear Models” - rlm() function from the R library MASS (Venables and Ripley, 2010) (default settings, maxit = 100). Similar to the deconvolution method CIBERSORT (Newman et al., 2015), we set negative coefficients to 0 and normalized all coefficients to add up to 1, which then form the output value providing the predicted lineage frequency values for the provided lineages and an additional “Others” estimation.
PCR bias as well as the number of detected signature mutations influence the robustness of the results. We therefore added the additional constraint to only perform the deconvolution analysis on samples matching a minimum quality score.
4.3.2.4. Dealing with indistinguishable variants
After deconvolution, grouped indistinguishable lineages have to be split again. There are three possible outcomes for those groups:
Firstly, when no signature mutations for a lineage could be found, the group includes the “Others” column and is in fact “Others” only. So the grouped lineages are getting the proportion value 0, “Others” gets the deconvoluted value. Secondly, the grouped lineages are deconvoluted to 0. In this case both lineages are assigned with the value 0. Thirdly, the grouped lineages are not equal to “Others” and are getting a deconvolution value above 0. In this case the assumption for normal distribution of the lineage abundances is applied and the deconvolution value is divided by the number of grouped lineages. Each lineage is assigned this adjusted value.
4.3.3. Regression analysis for mutations time-series
For the regression analysis on mutation frequencies we applied a linear regression model using the “Fitting Linear Models” - lm - function of R base. The test was only performed on mutations if N(x > 0) > 5 being the number of frequency values x that are above 0 across all samples. To get only increasing trends, the coefficient values were filtered for values x > 0 only. P-values were calculated by the lm-function using t-test and were filtered for p < 0.05. We report the mutation trend analysis together with and sorted by the regression coefficient as a comparable value for unstandardized effect size.
4.3.4. Pooling of samples for time series analysis and plots
For summarizing across daytime and location, the lineage frequencies are pooled by calculating the weighted average using the total number of reads of each sample as weights. The mutation frequencies are pooled by using the simple mean (without removing missing values). Figures and deconvolution plots are done with ggplot2 ( Wickham, 2016 ). For the cross-correlation analysis samples were pooled by week and the pooled unique set of non-signature mutations was counted.
4.3.5. Sample scoring for quality check
For reference genome coverage quality control, the pipeline uses the BEDtools coverage (Quinlan and Hall, 2010), using a BED file with the tracked signature mutation sites as input. For the regression analysis and time series plots only samples are taken in account that cover >90 % SARS-CoV-2 genome (except for the NYC dataset).
4.3.6. In silico data simulation
In order to qualify and test the accuracy of the pipeline under industrial sequencing parameters, an artificial dataset containing only short single-end sequencing was simulated. The simulated dataset was generated in-silico using full genomes of 6 SARS-COV-2 lineages obtained from GISAID (Shu and McCauley, 2017). The genomes were used to simulate Illumina sequencing using InSilicoSeq (Gourlé et al., 2019) and Seqtk (github.com, n.d.) was used to trim sequences down to 40 bp of length and subsample reads. A total of 100.000 reads was generated using the following proportions: 10 % P1 (gamma), 10 % B.1.1.7 (alpha), 10 % B.1.621 (mu), 50 % C.37 (lambda), 15 % Delta (B.1.617.2) and 5 % B.1.1.529 (omicron).
The data was processed without primer trimming and without an additional filter for read coverage.
Accessions from the genomes used to simulate sequencing can be found on Table 2 .
Table 2.
SARS-CoV-2 genomes used for in silico simulations.
| WHO Lineage/Pango ID | GISAID accession |
|---|---|
| Gamma/P1 | >hCoV-19/Brazil/AM-FIOCRUZ-21890579EMP/2021|EPI_ISL_4520422|2021-07-14 |
| Alpha/B.1.7.7 | >hCoV-19/Kenya/KEM-CVR-3EL/2021|EPI_ISL_4506017|2021-04-21 |
| Lambda/C37 | >hCoV-19/Denmark/DCGC-151255/2021|EPI_ISL_3450383|2021-08-11 |
| Delta/B.1.617.2 | >hCoV-19/Poland/CovSeq215/2021|EPI_ISL_4551640|2021-09-08 |
| Mu/B.1.621 | >hCoV-19/Colombia/ATL-UNIANDES-G029686/2021|EPI_ISL_4566376|2021-08-20 |
| Omicron/B.1.1.529 | >hCoV-19/Belgium/rega-20,174/2021|EPI_ISL_6794907.2|2021-11-24 |
4.3.7. Spike-in samples data acquisition
Spike-in sequencing bam files were generated by Karthikeyan et al. (Karthikeyan et al., 2021) Data was downloaded from: https://console.cloud.google.com/storage/browser/search-reference_data. The data was processed without primer trimming and but the filter for minimal read coverage was set to 100.
4.3.8. Processing of wastewater data from New York City
The data was downloaded from the Sequence Read Archive SBI with the accession number # PRJNA715712. The MiSeq data was processed with primer trimming, using the primer sequences published by the authors. The iSeq data was processed without primer trimming. For both datasets a minimal read coverage filter of 100 was applied. No genomic coverage percent cutoff was used for those datasets.
4.3.9. Data/code availability
The pipeline can be installed with GNU Guix and is executed with the command [pigx-sars-cov2-ww -s {sample_sheet} {settings_file}]. A cloud version is also being developed. Information about other alternatives like building from source or potentially a Docker image can be found on the repository. We recommend installing the pipeline with GNU Guix for its reproducibility guarantees (Courtès and Wurmus, 2015). For installation advice, documentation and code please visit the pipeline's repository: https://github.com/BIMSBbioinfo/pigx_sars-cov-2.
4.3.9.1. Reproducible environment
The presented results were produced using PiGx SARS-CoV-2 version 0.0.5.
-
●
dataset-Berlin250, dataset-NYC(RBD) (MiSeq data and all samples merged) - commit 524ed4832a6972fd695c0eeec25264188710a143
-
●
dataset-Berlin35, dataset-NYC(RBD) (iSeq data), insilico-simulation - commit 0a150c4bec58a5a8296c870586e225e49ee2b6f8
-
●
UCSD-spike in - commit bd87e7f2d83317e9d83f6fd81abb631af95476f6
The repository also contains the Guix manifest for this analysis (commit 4ded8c5bdc755391360e5695003d6d4085110d08). The detailed and up-to-date information about reproducing the analysis in this manuscript is available at: https://github.com/BIMSBbioinfo/pigx_sars-cov-2/blob/main/README.md#reproducing-the-analysis.
4.3.9.2. Data access
The raw sequencing read data from Berlin wastewater samples is deposited to the Sequence Read Archive (SRA) available using the accession number #PRJNA827160.
The interactive reports that were used and produced for this pipeline can be found here:
-
●
dataset-Berlin250 - https://bimsbstatic.mdc-berlin.de/akalin/AAkalin_pathogenomics/sarscov2_ww_reports/220225_dataset_Berlin250/index.html
-
●
dataset-Berlin35 - https://bimsbstatic.mdc-berlin.de/akalin/AAkalin_pathogenomics/sarscov2_ww_reports/220310_dataset_Berlin35/index.html
-
●
dataset-NYC(RBD) - https://bimsbstatic.mdc-berlin.de/akalin/AAkalin_pathogenomics/sarscov2_ww_reports/220225_dataset_NYC_RBD/index.html
-
●
UCSD-spike in - https://bimsbstatic.mdc-berlin.de/akalin/AAkalin_pathogenomics/sarscov2_ww_reports/220309_ucsd_spikeIn/index.html
-
●
Insilico-simulation- https://bimsbstatic.mdc-berlin.de/akalin/AAkalin_pathogenomics/sarscov2_ww_reports/220310_insilico_simulation/insilico_simulation.html
The following are the supplementary data related to this article.
Supplementary tables S1, S2.1, S2.4, S3, S4, S5, S6, S7, S8
mutations found Berlin35.csv.
mutations found ucsd.csv.
Supplementary Figures S1–S4
CRediT authorship contribution statement
AA designed and supervised the whole course of the project including the computational pipeline and provided the initial deconvolution algorithm. NR initialised the study and collaboration. ML supervised the wet-lab sample processing and sequencing further “performed” by EW and planned the study with AA. Sampling was supervised and performed by FZ, RG, UB. NB contributed to the design and refinement of the ARTIC v4 primer.
EW contributed to the data analysis and designed and supervised the sample preparation and sequencing experiments for the dataset-Berlin250 which were performed by him, AD, CQ. TB, JA supervised the sequencing experiments. MM, KL and TH conceptualized, developed and carried out the production 35 bp sequencing strategy for Berlin wastewater. JG and KL curated very short read data and helped with the calibration, analysis and interpretation of the results, as well as producing in-silico sequencing simulations. MF contributed to the pipeline by developing the taxonomic analysis part. JD contributed to the pipeline by building the whole automation backbone with snakemake and the initial test dataset. JD and VS did the initial exploration of the data and the available tools and pipelines which lead to the set of tools used for this study. BU, AB, VF contributed partially to the pipeline with the code backbone on which parts of the pipeline are built and also supported through discussions about the choice of tools. BU additionally provided a critical review of methods, continuous support in the development process and a critical review of the manuscript. JF provided support on code development, method improvements and generated data used after review rounds. RW did the major work on the pipeline's backbone, its implementation and packaging. He also led the development process bringing the parts written by JD, MF, VS, RC together. VS did most of the downstream analysis and additional pipeline development with continuous support and consultation from RC and AA. VS, RC and AA wrote the paper with input from EW and all other authors.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
We thank Mrs. Burzyk, Cytner, Darre, Flatau, Göldner, Grunow, Heinig, Horn, Kapczynski, Klawonn, Koch, Krug, Meyer, Neideck, Schmidt, Schwarzenberg, Stroede, Zühlsdorff and Messrs. Armbrecht, Dombrowski, Frankenstein, Halatta, Hambarsomian, Linnek, Muss, of the Berliner Wasserbetriebe for sampling and logistic support; as well as Dr. Selinka of the Umweltbundesamt and also Mrs. Schumacher for helpful discussions. Also, we would like to thank Friederike Dündar for consultation on best practices for visualization techniques.
EW and ML are supported by the Project “Virological and immunological determinants of COVID-19 pathogenesis – lessons to get prepared for future pandemics (KA1-Co-02 ‘COVIPA’)”, a grant from the Helmholtz Association's Initiative and Networking Fund.
Editor: Damià Barceló
Data availability
Data and code is shared, links on the manuscript
References
- Ahmed W., Bertsch P.M., Bivins A., Bibby K., Farkas K., Gathercole A., et al. Comparison of virus concentration methods for the RT-qPCR-based recovery of murine hepatitis virus, a surrogate for SARS-CoV-2 from untreated wastewater. Sci. Total Environ. 2020;739 doi: 10.1016/j.scitotenv.2020.139960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bar-Or I., Yaniv K., Shagan M., Ozer E., Erster O., Mendelson E., et al. Regressing SARS-CoV-2 sewage measurements onto COVID-19 burden in the population: a proof-of-concept for quantitative environmental surveillance. Front. Public Health. 2022;9(56171) doi: 10.3389/fpubh.2021.561710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen A.T., Altschuler K., Zhan S.H., Chan Y.A., Deverman B.E. COVID-19 CG enables SARS-CoV-2 mutation and lineage tracking by locations and dates of interest. elife. 2021;10 doi: 10.7554/eLife.63409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S., Zhou Y., Chen Y., Gu J. fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics. 2018;34:i884–i890. doi: 10.1093/bioinformatics/bty560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courtès L., Wurmus R. 2015. Reproducible and User-Controlled Software Environments in HPC with Guix. arXiv:150602822 [cs] [Google Scholar]
- covariants.org. n.d. https://covariants.org/variants/21J.Delta
- Crits-Christoph A., Kantor R.S., Olm M.R., Whitney O.N., Al-Shayeb B., Lou Y.C., et al. Genome sequencing of sewage detects regionally prevalent SARS-CoV-2 variants. mBio. 2021;12 doi: 10.1128/mBio.02703-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- European Commission. Commission Recommendation (EU) 2021/472 of 17 March 2021 on a common approach to establish a systematic surveillance of SARS-CoV-2 and its variants in wastewaters in the EU.
- Fehr A.R., Perlman S. Coronaviruses: an overview of their replication and pathogenesis. Methods Mol. Biol. 2015;1282:1–23. doi: 10.1007/978-1-4939-2438-7_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- github.com. n.d. https://github.com/lh3/seqtk
- Gourlé H., Karlsson-Lindsjö O., Hayer J., Bongcam-Rudloff E. Simulating Illumina metagenomic data with InSilicoSeq. Bioinformatics. 2019;35:521–522. doi: 10.1093/bioinformatics/bty630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grubaugh N.D., Gangavarapu K., Quick J., Matteson N.L., De Jesus J.G., Main B.J., et al. An amplicon-based sequencing framework for accurately measuring intrahost virus diversity using PrimalSeq and iVar. Genome Biol. 2019;20:8. doi: 10.1186/s13059-018-1618-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isaksson F., Lundy L., Hedström A., Székely A.J., Mohamed N. Evaluating the use of alternative normalization approaches on SARS-CoV-2 concentrations in wastewater: experiences from two catchments in Northern Sweden. Environments. 2022;9:39. [Google Scholar]
- Izquierdo-Lara R., Elsinga G., Heijnen L., Munnink B.B.O., Schapendonk C.M.E., Nieuwenhuijse D., et al. Monitoring SARS-CoV-2 circulation and diversity through community wastewater sequencing, the Netherlands and Belgium. Emerg. Infect. Dis. 2021;27:1405–1415. doi: 10.3201/eid2705.204410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jahn K., Dreifuss D., Topolsky I., Kull A., Ganesanandamoorthy P., Fernandez-Cassi X., et al. Detection and surveillance of SARS-CoV-2 genomic variants in wastewater. MedRxiv. 2021 doi: 10.1101/2021.01.08.21249379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jahn K., Dreifuss D., Topolsky I., Kull A., Ganesanandamoorthy P., Fernandez-Cassi X., et al. Early detection and surveillance of SARS-CoV-2 genomic variants in wastewater using COJAC. Nat. Microbiol. 2022;7:1151–1160. doi: 10.1038/s41564-022-01185-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karthikeyan S., Levy J.I., De Hoff P., Humphrey G., Birmingham A., Jepsen K., et al. Wastewater sequencing uncovers early, cryptic SARS-CoV-2 variant transmission. MedRxiv. 2021 doi: 10.1101/2021.12.21.21268143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karthikeyan S., Levy J.I., De Hoff P., Humphrey G., Birmingham A., Jepsen K., et al. Wastewater sequencing reveals early cryptic SARS-CoV-2 variant transmission. Nature. 2022 doi: 10.1038/s41586-022-05049-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landgraff C., Wang L.Y.R., Buchanan C., Wells M., Schonfeld J., Bessonov K., et al. Metagenomic sequencing of municipal wastewater provides a near-complete SARS-CoV-2 genome sequence identified as the B.1.1.7 variant of concern from a Canadian municipality concurrent with an outbreak. MedRxiv. 2021 doi: 10.1101/2021.03.11.21253409. [DOI] [Google Scholar]
- Lastra A., Botello J., Pinilla A., Urrutia J.I., Canora J., Sánchez J., et al. SARS-CoV-2 detection in wastewater as an early warning indicator for COVID-19 pandemic.Madrid region case study. Environ. Res. 2022;203 doi: 10.1016/j.envres.2021.111852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H. figshare; 2014. Aligning Sequence Reads, Clone Sequences and Assembly con*gs With BWA-MEM. [Google Scholar]
- Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., et al. The sequence alignment/map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin X., Glier M., Kuchinski K., Ross-Van Mierlo T., McVea D., Tyson J.R., et al. Assessing multiplex tiling PCR sequencing approaches for detecting genomic variants of SARS-CoV-2 in municipal wastewater. mSystems. 2021;6 doi: 10.1128/mSystems.01068-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLaren W., Gil L., Hunt S.E., Riat H.S., Ritchie G.R.S., Thormann A., et al. The ensembl variant effect predictor. Genome Biol. 2016;17:122. doi: 10.1186/s13059-016-0974-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medema G., Heijnen L., Elsinga G., Italiaander R., Brouwer A. Presence of SARS-Coronavirus-2 RNA in sewage and correlation with reported COVID-19 prevalence in the early stage of the epidemic in the Netherlands. Environ. Sci. Technol. Lett. 2020;7:511–516. doi: 10.1021/acs.estlett.0c00357. [DOI] [PubMed] [Google Scholar]
- Mölder F., Jablonski K.P., Letcher B., Hall M.B., Tomkins-Tinch C.H., Sochat V., et al. Sustainable data analysis with Snakemake. F1000Res. 2021;10:33. doi: 10.12688/f1000research.29032.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naughton C.C., Roman F.A., Alvarado A.G.F., Tariqi A.Q., Deeming M.A., Bibby K., et al. Show us the data: global COVID-19 wastewater monitoring efforts, equity, and gaps. MedRxiv. 2021 doi: 10.1101/2021.03.14.21253564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman A.M., Liu C.L., Green M.R., Gentles A.J., Feng W., Xu Y., et al. Robust enumeration of cell subsets from tissue expression profiles. Nat. Methods. 2015;12:453–457. doi: 10.1038/nmeth.3337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- nextstrain.org. n.d. https://nextstrain.org/ncov/gisaid/global?gt=nuc.10029T
- Petrinca A.R., Donia D., Pierangeli A., Gabrieli R., Degener A.M., Bonanni E., et al. Presence and environmental circulation of enteric viruses in three different wastewater treatment plants. J. Appl. Microbiol. 2009;106:1608–1617. doi: 10.1111/j.1365-2672.2008.04128.x. [DOI] [PubMed] [Google Scholar]
- Pipelines R&. D D. Farr B., Rajan D., Betteridge E., Shirley L., Quail M. 2020. COVID-19 ARTIC v3 Illumina library construction and sequencing protocol v4. preprint. [Google Scholar]
- Posada-Céspedes S., Seifert D., Topolsky I., Jablonski K.P., Metzner K.J., Beerenwinkel N. V-pipe: a computational pipeline for assessing viral genetic diversity from high-throughput data. Bioinformatics. 2021;37(12):1673–1680. doi: 10.1093/bioinformatics/btab015. btab015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan A.R., Hall I.M. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranta J., Hovi T., Arjas E. Poliovirus surveillance by examining sewage water specimens: studies on detection probability using simulation models. Risk Anal. 2001;21:1087–1096. doi: 10.1111/0272-4332.t01-1-216174. [DOI] [PubMed] [Google Scholar]
- Sandoval Torrientes M., Castelló Abietar C., Boga Riveiro J., Álvarez-Argüelles M.E., Rojo-Alba S., Abreu Salinas F., et al. A novel single nucleotide polymorphism assay for the detection of N501Y SARS-CoV-2 variants. J. Virol. Methods. 2021;294 doi: 10.1016/j.jviromet.2021.114143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen L., Bard J.D., Triche T.J., Judkins A.R., Biegel J.A., Gai X. Emerging variants of concern in SARS-CoV-2 membrane protein: a highly conserved target with potential pathological and therapeutic implications. Emerg. Microbes Infect. 2021;10:885–893. doi: 10.1080/22221751.2021.1922097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu Y., McCauley J. GISAID: global initiative on sharing all influenza data – from vision to reality. Eurosurveillance. 2017;22 doi: 10.2807/1560-7917.ES.2017.22.13.30494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth D.S., Trujillo M., Gregory D.A., Cheung K., Gao A., Graham M., et al. Tracking cryptic SARS-CoV-2 lineages detected in NYC wastewater. Nat. Commun. 2022;13:635. doi: 10.1038/s41467-022-28246-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vega-Magaña N., Sánchez-Sánchez R., Hernández-Bello J., Venancio-Landeros A.A., Peña-Rodríguez M., Vega-Zepeda R.A., et al. RT-qPCR assays for rapid detection of the N501Y, 69–70del, K417N, and E484K SARS-CoV-2 mutations: a screening strategy to identify variants with clinical impact. Front. Cell. Infect. Microbiol. 2021;11 doi: 10.3389/fcimb.2021.672562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venables W.N., Ripley B.D. 4. ed. Springer; [Nachdr.]. New York: 2010. Modern Applied Statistics With S. [Google Scholar]
- Wickham H. Springer-Verlag; New York: 2016. ggplot2: Elegant Graphics for Data Analysis. [Google Scholar]
- Wilm A., Aw P.P.K., Bertrand D., Yeo G.H.T., Ong S.H., Wong C.H., et al. LoFreq: a sequence-quality aware, ultra-sensitive variant caller for uncovering cell-population heterogeneity from high-throughput sequencing datasets. Nucleic Acids Res. 2012;40:11189–11201. doi: 10.1093/nar/gks918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfe M.K., Topol A., Knudson A., Simpson A., White B., Vugia D.J., et al. High-frequency, high-throughput quantification of SARS-CoV-2 RNA in wastewater settled solids at eight publicly owned treatment works in Northern California shows strong association with COVID-19 incidence. mSystems. 2021;6 doi: 10.1128/mSystems.00829-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood D.E., Lu J., Langmead B. Improved metagenomic analysis with Kraken 2. Genome Biol. 2019;20:257. doi: 10.1186/s13059-019-1891-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu F., Zhang J., Xiao A., Gu X., Lee W.L., Armas F., et al. SARS-CoV-2 titers in wastewater are higher than expected from clinically confirmed cases. mSystems. 2020;5 doi: 10.1128/mSystems.00614-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wurmus R., Uyar B., Osberg B., Franke V., Gosdschan A., Wreczycka K., et al. PiGx: reproducible genomics analysis pipelines with GNU Guix. Gigascience. 2018;7 doi: 10.1093/gigascience/giy123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao A., Wu F., Bushman M., Zhang J., Imakaev M., Chai P.R., et al. Metrics to relate COVID-19 wastewater data to clinical testing dynamics. medRxiv. 2021 doi: 10.1101/2021.06.10.21258580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P., Yang X.-L., Wang X.-G., Hu B., Zhang L., Zhang W., et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary tables S1, S2.1, S2.4, S3, S4, S5, S6, S7, S8
mutations found Berlin35.csv.
mutations found ucsd.csv.
Supplementary Figures S1–S4
Data Availability Statement
Data and code is shared, links on the manuscript