Abstract
The upper operon of the TOL plasmid pWW0 of Pseudomonas putida encodes a set of enzymes involved in the conversion of toluene and xylenes to their carboxylic acid derivatives. The last gene of the upper operon, xylN, encodes a 465-amino-acid polypeptide which exhibits significant sequence similarity to FadL, an outer membrane protein involved in fatty acid transport in Escherichia coli. To analyze the role of the xylN gene product, xylN on TOL plasmid pWW0 was disrupted by inserting a kanamycin resistance gene, and the phenotypes of P. putida harboring the wild-type and xylN mutant TOL plasmids were characterized. The growth of P. putida harboring the wild-type TOL plasmid was inhibited by a high concentration of m-xylene, while that of P. putida harboring the xylN mutant TOL plasmid was not. The apparent Ks value for the oxidation of m-xylene in intact cells of the xylN mutant was fourfold higher than that of the wild-type strain, although the TOL catabolic enzyme activities in cell extracts from the two strains were almost identical. We therefore presume that the xylN gene product is a porin involved in the transport of m-xylene and its analogues across the outer membrane. Western blot analysis confirmed the localization of XylN in the outer membrane.
TOL plasmids in Pseudomonas putida encode the metabolic pathways for the degradation of toluene, xylenes, and their alcohol and carboxylate derivatives (1). The TOL degradation pathways generally consist of two parts: an upper pathway that converts toluene and xylenes to their carboxylic acid derivatives (11) and a lower (or meta-cleavage) pathway that transforms the carboxylic acids to the precursors of Krebs cycle intermediates (12). Genetic studies have shown that the genes for these catabolic enzymes are organized into two operons, one encoding enzymes for the upper pathway (the upper operon) and the other encoding enzymes for the meta-cleavage pathway (the meta operon) (6, 9, 10, 23).
The upper operon of TOL plasmid pWW0 contains seven genes in the order xylU-xylW-xylC-xylM-xylA-xylB-xylN in a region of about 8 kb (11, 40). The xylU and xylW genes are not required for toluene and xylene catabolism (40). The xylC gene encodes benzaldehyde dehydrogenase (14, 21), and the xylM and xylA genes encode subunits of xylene monooxygenase (31, 33, 34), while the xylB gene encodes benzyl alcohol dehydrogenase (32). The xylN gene, the last gene of the upper operon, synthesizes a 52-kDa protein which is processed to a 47-kDa polypeptide (11); the physiological role of the xylN gene product has remained unknown. In this study, we investigate the function of the xylN gene product.
MATERIALS AND METHODS
Bacterial strains, plasmids, and culture conditions.
The bacterial strains and plasmids used in this study are listed in Table 1. pWW0-161 is a Tn401 insertion derivative of TOL plasmid pWW0 and confers resistance to ampicillin on Escherichia coli hosts (6). The Tn401 insertion occurs outside the catabolic genes in pWW0-161. In this study, an xylN::Kmr mutant of pWW0-161 was isolated and named pWW0-161-xylN. E. coli JM110 was used for routine genetic manipulation. E. coli cells containing recombinant plasmids were maintained on Luria-Bertani plates (29) supplemented with appropriate antibiotics. The concentrations of the antibiotics used were 50 μg/ml for ampicillin and streptomycin and 20 μg/ml for chloramphenicol, while the kanamycin concentration was either 50 μg/ml for E. coli harboring a high-copy-number kanamycin resistance plasmid or 20 μg/ml for E. coli harboring pWW0-161-xylN. P. putida PaW94 (42) harboring pWW0-161 was maintained on M9 minimal medium (29) containing 5 mM m-toluate, while P. putida PaW94 harboring pWW0-161-xylN was maintained on M9 medium containing 5 mM m-toluate and 50 μg of kanamycin/ml. To induce the upper and meta operons on pWW0-161, cells of PaW94 harboring pWW0-116 (hereafter referred to as the wild type) or those harboring pWW0-116-xylN (hereafter referred to as the xylN mutant) were grown for 16 h at 30°C with shaking in 100 ml of M9 minimal medium containing 5 mM benzyl alcohol. m-Xylene vapor was supplemented for the cultures when required. The simultaneous addition of benzyl alcohol and m-xylene vapor to the culture assured the reproducible induction of TOL catabolic enzymes.
TABLE 1.
Bacterial strains and plasmids used in this study
| Bacterial strain | Genotype or phenotypea | Reference |
|---|---|---|
| P. putida | ||
| KT2440 | 10 | |
| PaW94 | Benzoate-1,2-dioxygenase-negative mutant | 40 |
| E. coli | ||
| JM110 | rpsL (Strr) thr leu thi-l lacY galK galT ara tonA tsx dam dcm supE44 Δ(lac-proAB) [F′ traD36 proA+B+lacIq ZΔM15] | |
| M15 | F− Nal Str Rif Thi− Lac− Ara+ Gal+ Mtl− RecA+ Uvr+ Lon+ | |
| Plasmids | ||
| pFC121 | xylB+N+; ColE1 replicon | |
| pSHG398 | Cmr; ColE1 replicon | |
| pWW0-161 | Tol+ Apr (Tn401) | 5 |
| pWW0-161-xylN | Tol+ Apr (Tn401) xylN::Kmr | This work |
| pXN960 | Cmr | This work |
| pXN961 | Cmr Kmr | This work |
| pUC4K | Ampr Kmr | 34 |
Tol+ indicates that the cells grow on m-xylene and toluene. Nal, Str, and Rif stand for sensitivity to nalidixic acid, streptomycin, and rifampin, while Apr, Cmr, and Kmr stand for resistance to ampicillin, chloramphenicol, and kanamycin, respectively.
DNA sequence analysis.
DNA sequencing with double-stranded DNA was carried out by using a dye terminator cycle-sequencing kit (Applied Biosystems) according to the manufacturer's instructions, and the products were analyzed with a model 377 DNA sequencer (Applied Biosystems). The computer-assisted sequence analysis was made with the PC/GENE software package (IntelliGenetics).
Fractionation of membranes.
Cells of the wild-type strain were induced as described above, harvested by centrifugation, washed with a 50 mM sodium phosphate buffer (pH 8.6), and resuspended in the same buffer. The cells were disrupted on ice using a sonifier (model 250; Branson) at 100-W output by three 30-s bursts of sonication interspersed with a 30-s cooling. Nondisrupted bacteria and large cell debris were removed by centrifugation at 8,000 × g for 5 min. The supernatant was removed and centrifuged at 138,000 × g for 1 h. After this step, the supernatant was used as the soluble cytoplasmic fraction. The inner and outer membranes were prepared as described by Feilmeier et al. (5) with slight modifications. Cells were suspended in 20% (wt/vol) sucrose in 10 mM Tris (pH 8.0) and disrupted by passing the cell suspension through a French pressure cell (Ohtake) at 137 MPa. After nonlysed bacteria and cell debris were removed, the clarified lysate was layered onto a 60 to 70% (wt/vol) discontinuous sucrose gradient and centrifuged at 247,000 × g for 18 h. The inner membrane fraction was recovered at the top of the 60% (wt/vol) sucrose layer, while the outer membrane fraction was collected at the top of the 70% (wt/vol) sucrose layer. The recovered samples were diluted threefold with distilled water. Each fraction was layered on a second 60 to 70% (wt/vol) discontinuous sucrose gradient and centrifuged as described above. The membrane fractions were again diluted as described above and centrifuged at 100,000 × g for 2 h to pellet the membranes. The pellets were washed once with 1 M KCl followed by an additional centrifugation step at 100,000 × g for 2 h to recover the membranes. The protein concentration was estimated with a DC protein assay kit (Bio-Rad Laboratories).
Western blot analysis.
An xylN gene fragment was amplified by PCR using a set of PCR primers, pXN1 (5′-GCGGATCCATGAAAATAAAAAATTTA) and pXN2 (5′-GCAGATCTGAATGAATAATTATAGGC) (the introduced BamHI and BglII restriction sites are underlined). The 1,411-bp PCR product was separated by electrophoresis on a 0.8% (wt/vol) low-melting-point agarose gel and purified with a QIAquick gel extraction kit (Qiagen). The amplified DNA was subsequently digested with BamHI and BglII and cloned into pQE-60 (Qiagen). The constructed plasmid was transferred to E. coli M15(pREP4). The recombinant XylN with a tail of six histidine residues at the N-terminal end was expressed in the transformant and purified using an Ni-nitrilotriacetic acid spin column (Qiagen) according to the manufacturer's protocol.
For Western blot analysis, proteins in the soluble, inner membrane, and outer membrane fractions were separated by 10% (wt/vol) polyacrylamide gel electrophoresis by the method of Laemmli (20) and transferred to a polyvinylidene difluoride membrane by electroblotting using a Hoefer electroblotter according to the manufacturer's instructions. Prestained protein molecular mass standards (New England BioLabs) were used for molecular mass estimation. Blots were incubated for 1 h with 10,000-fold-diluted rabbit anti-XylN serum and then with horseradish peroxidase-conjugated anti-rabbit donkey immunoglobulin G (Amersham Pharmacia Biotech) for 1 h. Detection of the XylN protein was carried out using ECL Plus Western blotting detection reagents (Amersham Pharmacia Biotech) according to the manufacturer's protocol.
Inactivation of xylN.
Plasmid pFC121 is a pUC18 derivative containing a 4-kb SacI-PstI fragment that incorporates the intact xylN gene. This plasmid was digested with PvuII and ClaI, and the resulting 2.8-kb PvuII-ClaI DNA fragment was extracted after electrophoretic separation in a 0.8% (wt/vol) agarose gel to be subcloned into the SmaI site of pHSG398 (35) after its sticky end was converted to a blunt end with a DNA-blunting kit (Takara Shuzo). The constructed plasmid was called pXN960. The xylN gene was inactivated by excising the 1.3-kb fragment containing a kanamycin resistance gene from plasmid pUC4K (36) with BamHI and inserting it into the DraIII site of xylN on pXN960 to yield pXN961.
The pWW0-161 plasmid in P. putida KT2440 (11) was transferred to E. coli JM110 by conjugation as previously described (18), and ampicillin-resistant derivatives of JM110 were selected on Luria-Bertani plates containing ampicillin and streptomycin. JM110(pWW0-161) thus constructed was subsequently transformed with pXN961, and those transformants resistant to kanamycin and chloramphenicol were selected. One of the transformants, JM110(pWW0-161, pXN961), was conjugated with P. putida PaW94, and kanamycin-resistant transconjugants capable of growing on m-toluate were selected on M9 plates containing 5 mM m-toluate and 50 μg of kanamycin/ml.
PCR for the detection of xylN::Kmr.
PCR for amplifying the DNA fragment containing xylN or xylN::Kmr was carried out in a volume of 50 μl with a GeneAmp kit (Perkin-Elmer). The oligonucleotides used as PCR primers were XYLU7191 (5′-GTTCACTTGATGCCAAGTGGAC-3′) and XYLU3 (5′-CCGCTGTAACAGTCCCCTTC-3′).
Growth inhibition by m-xylene.
TOL catabolic enzymes were induced in cells of the wild-type strain and the xylN mutant as already described, harvested by centrifugation, washed twice with a 50 mM potassium phosphate buffer (pH 7.4), and resuspended in the same buffer. Four-hundred microliters of the suspension containing 6 × 106 cells was then used to inoculate 100-ml bottles containing 20 ml of the M9 medium supplemented with 5 mM benzyl alcohol with or without 2.5 mM m-xylene. The bottles were tightly sealed with a Teflon-lined rubber septum and a crimped aluminum seal to prevent the evaporation of m-xylene. The cultures were grown at 30°C with shaking, and the turbidities of the cultures at 600 nm were periodically measured.
Enzyme assays.
Cells grown under inducing conditions were harvested at the late exponential growth phase by centrifugation, resuspended in 1/10 volume of a 10 mM ethylene diamine buffer (pH 7.4) containing 10% (vol/vol) isopropanol, and disrupted by sonication. After centrifugation at 20,000 × g for 30 min, the supernatants were used as cell-free crude extracts. The protein concentration of each crude extract was estimated with a protein assay kit (Bio-Rad Laboratories). Assays for benzyl alcohol dehydrogenase were carried out at 25°C in a 100 mM glycine-NaOH buffer (pH 9.4) containing 1 mM NAD+, 0.1 mg of bovine serum albumin/ml, and 200 μM benzyl alcohol as a substrate. NADH formation was measured spectrophotometrically at 340 nm (15). Catechol 2,3-dioxygenase was assayed at 25°C in a 100 mM potassium phosphate buffer (pH 7.5) with 33 μM catechol as the substrate; the amount of the reaction product (2-hydroxymunoic semialdehyde) was determined spectrophotometrically at 375 nm (26).
m-Xylene-dependent oxygen consumption by P. putida cells.
Cells of the wild-type strain and the xylN mutant were cultivated in the presence of benzyl alcohol and m-xylene vapor as described previously, harvested, washed twice with a 50 mM potassium phosphate buffer (pH 7.4), and resuspended in the same buffer. The cells were adjusted to a turbidity of 1.0 at 600 nm and incubated for 5 h at 30°C with shaking. For oxygen consumption assays, cells were placed in a Clark-type oxygen electrode (model 5/6 Oxygraph; Gilson), and the oxygen consumption rates in response to the addition of various concentrations of m-xylene were determined. The apparent kinetic constants, Ks and Vmax, were determined by reciprocal plots.
Chemicals and reagents.
All the chemicals used in this study were of the highest purity commercially available. m-Xylene was purchased from Tokyo Kasei Kogyo; the other chemicals were purchased from Wako Pure Chemical Industries, Difco, Aldrich Chemical, and Gibco BRL. The enzymes and reagents used for nucleic acid manipulation were purchased from Takara Shuzo.
Nucleotide sequence accession number.
The nucleotide sequence data reported in this paper have been submitted to the DDBJ, EMBL, and GenBank libraries under accession number D63341.
RESULTS AND DISCUSSION
Nucleotide sequence analysis of xylN.
Harayama et al. (11) have reported that xylN was the last gene of the upper operon on the TOL plasmid pWW0 and that its product was a 52-kDa protein which was processed to a 47-kDa polypeptide. In this study, the nucleotide sequence of xylN was determined. The gene encoded a 465-amino-acid-long polypeptide with an estimated molecular mass of 49 kDa. A comparison of the deduced amino acid sequence of XylN with translated sequences from the GenBank database suggested that XylN had significant homology with a group of proteins in hydrocarbon-degrading bacteria, including PorA from Pseudomonas sp. strain Y2 (37), CumH from Pseudomonas fluorescens IP01 (8), IpbH from P. putida RE204 (GenBank accession no. AF006691), CymD from P. putida F1 (4), TodX from P. putida F1 (38), TbuX from Ralstonia picketti PKO1 (17), and PhlX from Ralstonia eutropha JMP134 (GenBank accession no. AF06589) (Table 2). These proteins are homologous with FadL from E. coli (2), which is involved in the transport of long-chain fatty acids across the outer membrane (19).
TABLE 2.
Membrane-associated proteins sharing homology with XylN
| Gene product | Bacterial strain | Identity (%) | Reference |
|---|---|---|---|
| CumH | P. fluorescens IP01 | 49.7 | 7 |
| CymD | P. putida F1 | 38.7 | 4 |
| TodX | P. putida F1 | 37.8 | 36 |
| IpbH | P. putida RE204 | 49 | GenBank accession no. AF006691 |
| PorA | Pseudomonas sp. strain Y2 | 50.9 | 35 |
| PhlX | R. eutropha JMP134 | 42.2 | GenBank accession no. AF065891 |
| TbuX | R. eutropha PKO1 | 41.8 | 16 |
| FadL | E. coli | 21.0 | 2 |
Location of the xylN product in the outer membrane of P. putida cells.
The signal sequence was predicted at the N-terminal region of XylN by using both SignalP (version 1.1; Center for Biological Sequence Analysis [http://www.cbs.dtu.dk]) (24) and PSORT (http://psort.nibb.ac.jp) (22). These programs suggested the predicted cleavage site to be at position 24, 25, or 26, and the estimated molecular mass of the mature polypeptide was 47 kDa. This result agrees with that of the previous study: a 52-kDa XylN product was processed to a 47-kDa polypeptide (11). PSORT (22) predicted XylN to be an outer membrane protein. The computer program for predicting bacterial outer membrane β-strand proteins (Faculty of Biology, University of Konstanz [http://loop8.biologie.uni-konstanz.de]) (3) also produced this result. The existence of the signal sequence together with the sequence similarity to the outer membrane protein FadL suggested XylN is an outer membrane protein.
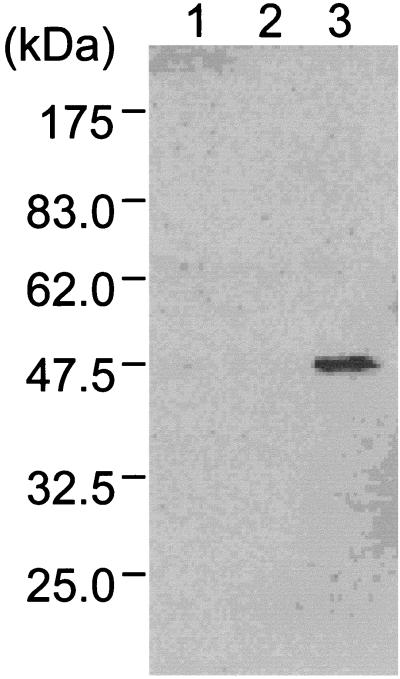
To identify the localization of XylN, cells of the wild-type strain were cultivated under conditions to induce the upper operon and fractionated into cytoplasm, inner membrane, and outer membrane. Western blot analysis detected XylN in the outer membrane (Fig. 1). No XylN was detected among cytoplasmic proteins and inner membrane proteins.
FIG. 1.
Localization of XylN in the outer membrane. The cytoplasmic, inner membrane, and outer membrane fractions were prepared from cells of the wild-type strain grown under conditions to induce the upper operon. XylN was immunodetected with antibodies raised against XylN. Lanes: 1, cytoplasmic fraction; 2, inner membrane fraction; 3, outer membrane fraction. The positions of the molecular mass markers are shown.
Construction of an xylN mutant of pWW0-161.
To analyze the possible role of the xylN product, PaW94 harboring pWW0-161-xylN was constructed by inserting the DNA fragment containing a kanamycin resistance gene in xylN. To confirm the mutation in this plasmid, total DNAs were extracted from the wild-type strain and the xylN mutant, and partial xylN fragments were amplified from the total DNAs by PCR. As expected, a 1.5-kb-long DNA fragment was amplified from the DNA that had been isolated from the xylN mutant, while a 0.3-kb-long fragment was amplified from the wild-type strain.
Growth inhibition of P. putida by m-xylene.
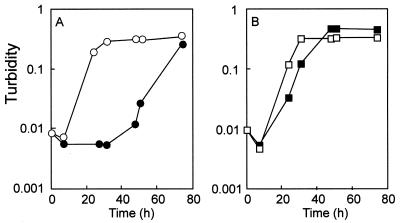
Growth inhibition of the wild-type strain but not the xylN mutant was observed with 2.5 mM m-xylene as described below. Cells grown in the presence of benzyl alcohol and m-xylene vapor were used to inoculate M9 medium containing 5 mM benzyl alcohol with or without 2.5 mM m-xylene in tightly stoppered bottles. They were cultivated at 30°C with shaking, and their growth was monitored by determining the turbidity of the cultures at 600 nm (Fig. 2). When the wild-type strain and the xylN mutant were grown on M9 medium containing 5 mM benzyl alcohol without m-xylene, the exponential growth was observed without an apparent lag phase, and the specific growth rate constants (μ) for these two strains were almost the same, namely, 0.20 and 0.19 h−1, respectively. However, when 2.5 mM m-xylene was added to the culture of the wild-type strain, a lag phase of about 40 h followed by the exponential growth phase (μ = 0.12 h−1) was observed. On the other hand, the growth of the xylN mutant occurred without significant lag, and the growth rate was similar to that in the absence of m-xylene (μ = 0.19 h−1). The same results were obtained when o-xylene was added to the cultures (data not shown). Our results with the wild-type TOL plasmid agree with a previous observation: it has been reported that the direct addition of aromatic hydrocarbons to a culture medium prevented the growth of P. putida harboring the TOL plasmid while growth of this strain was observed when these compounds were supplied in the vapor phase (7, 41).
FIG. 2.
Effect of m-xylene on the growth of the wild-type strain (circles) and the xylN mutant (squares). Cells were grown in M9 medium containing 5 mM benzyl-alcohol at 30°C with (solid) and without (open) 2.5 mM m-xylene. Growth was determined by monitoring the turbidity at 600 nm. The values are averages of three independent experiments. The standard deviation was less than 35% of the averages.
Xylenes, like toluene, are toxic to microorganisms, even at a subsaturated concentration in water (30). It has been suggested that the accumulation of such organic solvents as toluene and xylenes in the cytoplasmic membrane disturbs its structural and functional properties (30). Accumulated aromatic compounds may reduce the integrity of the cytoplasmic membrane and increase its permeability to proteins and ions. Some P. putida strains are tolerant of such organic solvents due to active efflux of the solvents from the cytoplasm and the cytoplasmic membrane (16, 25, 28) or due to solvent-impermeable outer membranes (13, 27, 28, 39). Considering the location of XylN in the outer membrane, we propose that the resistance to xylenes of the xylN mutant was due to a reduction in the permeability of xylenes across the outer membrane, which was caused by the inactivation of the XylN outer membrane porin.
m-Xylene-dependent oxygen consumption by whole cells.
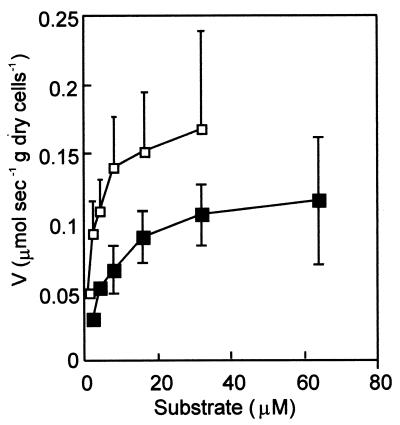
Cells of the wild-type strain and the xylN mutant were grown under inducing conditions and resuspended in a 50 mM potassium phosphate buffer (pH 7.4) with the turbidity of each suspension adjusted to 1.0 at 600 nm. The suspensions were incubated for 5 h at 30°C to reduce the endogenous energy sources. The stimulation of oxygen consumption by whole cells upon the addition of various concentrations of m-xylene was then determined (Fig. 3). The apparent Ks value for m-xylene oxidation in the xylN mutant (6.7 ± 1.9 μM) was four-fold higher than that of the wild-type strain (1.7 ± 1.2 μM), while the apparent Vmax value for the xylN mutant (0.12 ± 0.02 μmol s−1 g of dry cells−1) was 70% of that for the wild-type strain (0.17 ± 0.05 μmol s−1 g of dry cells−1).
FIG. 3.
m-Xylene-dependent oxygen consumption by the wild-type strain (open squares) and the xylN mutant (solid squares). The values were averages of three independent experiments. Each vertical bar represents one standard deviation. The bars appear only above the average values of the data for the wild-type strain to make the graph clearer.
To confirm that the upper and meta operons of TOL plasmids pWW0-161 and pWW0-161-xylN had been induced at the same level, the activities of benzyl-alcohol dehydrogenase and catechol 2,3-dioxygenase in PaW94 harboring these two plasmids were determined. The activities of benzyl-alcohol dehydrogenase in these two strains were 31 ± 11 and 32 ± 10 nmol min−1 mg−1, respectively, while the activities of catechol 2,3-dioxygenase were 1.4 ± 0.5 and 1.4 ± 1.0 μmol min−1 mg−1, respectively.
Although the activities of TOL catabolic enzymes in the wild-type strain and the xylN mutant were not very different, the oxygen consumption rate of the xylN mutant was lower than that of the wild-type strain. These oxygraph data support the interpretation that XylN is an outer membrane porin involved in m-xylene uptake.
Since XylN renders cells sensitive to their growth substrates, this protein places host cells at a disadvantage when the substrate is available at a high concentration. However, in most natural environments, the concentrations of available substrates may be very low, and the XylN function may be indispensable for the survival of the host in such environments. In fact, the growth rate of the xylN mutant was only one-third of that of the wild-type strain when m-xylene was supplied in the vapor phase (data not shown).
ACKNOWLEDGMENTS
We thank Hiroshi Nikaido and Teruko Nakazawa for discussions.
This work was supported by the New Energy and Industrial Technology Development Organization (NEDO).
REFERENCES
- 1.Assinder S J, Williams P A. The TOL plasmids: determinants of the catabolism of toluene and the xylenes. Adv Microb Physiol. 1990;31:1–69. doi: 10.1016/s0065-2911(08)60119-8. [DOI] [PubMed] [Google Scholar]
- 2.Black P N. Primary sequence of the Escherichia coli fadL gene encoding an outer membrane protein required for long-chain fatty acid transport. J Bacteriol. 1991;173:435–442. doi: 10.1128/jb.173.2.435-442.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Diederichs K, Freigang J, Umhau S, Zeth K, Breed J. Prediction by a neural network of outer membrane beta-strand protein topology. Protein Sci. 1998;7:2413–2420. doi: 10.1002/pro.5560071119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Eaton R W. p-Cymene catabolic pathway in Pseudomonas putida F1: cloning and characterization of DNA encoding conversion of p-cymene to p-cumate. J Bacteriol. 1997;179:3171–3180. doi: 10.1128/jb.179.10.3171-3180.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Feilmeier B J, Iseminger G, Schroeder D, Webber H, Phillips G J. Green fluorescent protein functions as a reporter for protein localization in Escherichia coli. J Bacteriol. 2000;182:4058–4076. doi: 10.1128/jb.182.14.4068-4076.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Franklin F C H, Bagdasarian M M, Bagdasarian M, Timmis K N. Molecular and functional analysis of the TOL plasmid pWW0 from Pseudomonas putida and cloning of genes for the entire regulated aromatic ring meta cleavage pathway. Proc Natl Acad Sci USA. 1981;78:7458–7462. doi: 10.1073/pnas.78.12.7458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gibson D T, Cardini G E, Maseles F C, Kallio R E. Incorporation of oxygen-18 into benzene by Pseudomonas putida. Biochemistry. 1970;9:1631–1635. doi: 10.1021/bi00809a024. [DOI] [PubMed] [Google Scholar]
- 8.Habe H, Kasuga K, Nojiri H, Yamane H, Omori T. Analysis of cumene (isopropylbenzene) degradation genes from Pseudomonas fluorescens IP01. Appl Environ Microbiol. 1996;62:4471–4477. doi: 10.1128/aem.62.12.4471-4477.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Harayama S, Lehrbach P R, Timmis K N. Transposon mutagenesis analysis of meta-cleavage pathway operon genes of the TOL plasmid of Pseudomonas putida mt-2. J Bacteriol. 1984;160:251–255. doi: 10.1128/jb.160.1.251-255.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Harayama S, Leppik R A, Rekik M, Mermod N, Lehrbach P R, Reineke W, Timmis K N. Gene order of the TOL catabolic plasmid upper pathway operon and oxidation of both toluene and benzyl alcohol by the xylA product. J Bacteriol. 1986;167:455–461. doi: 10.1128/jb.167.2.455-461.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Harayama S, Rekik M, Wubbolts M, Rose K, Leppik R A, Timmis K N. Characterization of five genes in the upper-pathway operon of TOL plasmid pWW0 from Pseudomonas putida and identification of the gene products. J Bacteriol. 1989;171:5048–5055. doi: 10.1128/jb.171.9.5048-5055.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Harayama S, Rekik M. The meta cleavage operon of TOL degradative plasmid pWW0 comprises 13 genes. Mol Gen Genet. 1990;221:113–120. doi: 10.1007/BF00280375. [DOI] [PubMed] [Google Scholar]
- 13.Heipieper H J, Diefenbach R, Keweloh H. Conversion of cis unsaturated fatty acids to trans, a possible mechanism for the protection of phenol-degrading Pseudomonas putida P8 from substrate toxicity. Appl Environ Microbiol. 1992;58:1847–1852. doi: 10.1128/aem.58.6.1847-1852.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Inoue J, Shaw J P, Rekik M, Harayama S. Overlapping substrate specificities of benzaldehyde dehydrogenase (the xylC gene product) and 2-hydroxymuconic semialdehyde dehydrogenase (the xylG gene product) encoded by TOL plasmid pWW0 of Pseudomonas putida. J Bacteriol. 1995;177:1196–1201. doi: 10.1128/jb.177.5.1196-1201.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Inoue J, Tomioka N, Itai A, Harayama S. Proton transfer in benzyl alcohol dehydrogenase during catalysis: alternate proton-relay routes. Biochemistry. 1998;37:3305–3311. doi: 10.1021/bi970726g. [DOI] [PubMed] [Google Scholar]
- 16.Isken S, de Bont J A. Active efflux of toluene in a solvent-resistant bacterium. J Bacteriol. 1996;178:6056–6058. doi: 10.1128/jb.178.20.6056-6058.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kahng H-Y, Byrne A M, Olsen R H, Kukor J J. Characterization and role of tbuX in utilization of toluene by Ralstonia picketti PKO1. J Bacteriol. 2000;182:1232–1242. doi: 10.1128/jb.182.5.1232-1242.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Köhler T, Harayama S, Ramos J L, Timmis K N. Involvement of Pseudomonas putida RpoN sigma factor in regulation of various metabolic functions. J Bacteriol. 1989;71:4326–4333. doi: 10.1128/jb.171.8.4326-4333.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kumar G B, Black P N. Bacterial long-chain fatty acid transport. J Biol Chem. 1993;268:15469–15476. [PubMed] [Google Scholar]
- 20.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 21.Lebens M R, Williams P A. Complementation of deletion and insertion mutants of TOL plasmid pWW0: regulatory implications and location of xylC gene. J Gen Microbiol. 1985;131:3261–3269. [Google Scholar]
- 22.Nakai K, Kanehisa M. Expert system for predicting protein localization sites in gram-negative bacteria. Proteins. 1991;11:95–110. doi: 10.1002/prot.340110203. [DOI] [PubMed] [Google Scholar]
- 23.Nakazawa T, Inouye S, Nakazawa A. Physical and functional mapping of RP4-TOL plasmid recombination: analysis of insertion and deletion mutants. J Bacteriol. 1980;144:222–231. doi: 10.1128/jb.144.1.222-231.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nielsen H, Engelbrecht J, Brunak S, von Heijne G. Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng. 1997;10:1–6. doi: 10.1093/protein/10.1.1. [DOI] [PubMed] [Google Scholar]
- 25.Nikaido H. Multiple antibiotic resistance and efflux. Curr Opin Microbiol. 1998;1:516–523. doi: 10.1016/s1369-5274(98)80083-0. [DOI] [PubMed] [Google Scholar]
- 26.Nozaki M, Kotani S, Ono K, Seno S. Metapyrocatechase. 3. Substrate specificity and mode of ring fission, Biochim. Biophys Acta. 1970;220:213–223. doi: 10.1016/0005-2744(70)90007-0. [DOI] [PubMed] [Google Scholar]
- 27.Pinkart H C, Wolfram J W, Rogers R, White D C. Cell envelope changes in solvent-tolerant and solvent-sensitive Pseudomonas putida strains following exposure to p-xylene. Appl Environ Microbiol. 1996;62:1129–1132. doi: 10.1128/aem.62.3.1129-1132.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ramos J L, Duque E, Rodríguez-Herva J J, Godoy P, Haïdour A, Reyes F, Fernández-Barrero A. Mechanisms for solvent tolerance in bacteria. J Biol Chem. 1997;272:3887–3890. doi: 10.1074/jbc.272.7.3887. [DOI] [PubMed] [Google Scholar]
- 29.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 30.Sikkema J, de Bont J A M, Poolman B. Mechanisms of membrane toxicity of hydrocarbons. Microbiol Rev. 1995;59:201–222. doi: 10.1128/mr.59.2.201-222.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shaw J P, Harayama S. Purification and characterisation of the NADH:acceptor reductase component of xylene monooxygenase encoded by the TOL plasmid pWW0 of Pseudomonas putida mt-2. Eur J Biochem. 1992;209:51–61. doi: 10.1111/j.1432-1033.1992.tb17260.x. [DOI] [PubMed] [Google Scholar]
- 32.Shaw J P, Rekik M, Schwager F, Harayama S. Kinetic studies on benzyl alcohol dehydrogenase encoded by TOL plasmid pWW0. A member of the zinc-containing long-chain alcohol dehydrogenase family. J Biol Chem. 1993;268:10842–10850. [PubMed] [Google Scholar]
- 33.Shaw J P, Harayama S. Characterization in vitro of the hydroxylase component of xylene monooxygenase, the first enzyme of the TOL-plasmid-encoded pathway for the mineralization of toluene and xylenes. J Ferment Bioeng. 1995;79:195–199. [Google Scholar]
- 34.Suzuki M, Hayakawa T, Shaw J P, Rekik M, Harayama S. Primary structure of xylene monooxygenase: similarities to and differences from the alkane hydroxylation system. J Bacteriol. 1991;173:1690–1695. doi: 10.1128/jb.173.5.1690-1695.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Takeshita S, Sato M, Toba M, Masahashi W, Hashimoto-Gotoh T. High-copy-number and low-copy-number plasmid vectors for lacZ alpha-complementation and chloramphenicol- or kanamycin-resistance selection. Gene. 1987;61:63–74. doi: 10.1016/0378-1119(87)90365-9. [DOI] [PubMed] [Google Scholar]
- 36.Taylor L A, Rose R E. A correction in the nucleotide sequence of the Tn903 kanamycin resistance determinant in pUC4K. Nucleic Acids Res. 1988;16:358. doi: 10.1093/nar/16.1.358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Velasco A, Alonso S, García J L, Perera J, Díaz E. Genetic and functional analysis of the styrene catabolic cluster of Pseudomonas sp. strain Y2. J Bacteriol. 1998;180:1063–1071. doi: 10.1128/jb.180.5.1063-1071.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang Y, Rawlings M, Gibson D T, Labbe D, Bergeron H, Brousseau R, Lau P C K. Identification of a membrane protein and a truncated LysR-type regulator associated with the toluene degradation pathway in Pseudomonas putida F1. Mol Gen Genet. 1995;246:570–579. doi: 10.1007/BF00298963. [DOI] [PubMed] [Google Scholar]
- 39.Weber F J, Isken S, de Bont J A. cis/trans isomerization of fatty acids as a defense mechanism of Pseudomonas putida strains to toxic concentrations of toluene. Microbiology. 1994;140:2013–2017. doi: 10.1099/13500872-140-8-2013. [DOI] [PubMed] [Google Scholar]
- 40.Williams P A, Shaw L M, Pitt C W, Vrecl M. xylUW, two genes at the start of the upper pathway operon of TOL plasmid pWW0, appear to play no essential part in determining its catabolic phenotype. Microbiology. 1997;143:101–107. doi: 10.1099/00221287-143-1-101. [DOI] [PubMed] [Google Scholar]
- 41.Worsey M J, Williams P A. Metabolism of toluene and xylenes by Pseudomonas putida (arvilla) mt-2: evidence for a new function of the TOL plasmid. J Bacteriol. 1975;124:7–13. doi: 10.1128/jb.124.1.7-13.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Worsey M J, Franklin F C, Williams P A. Regulation of the degradative pathway enzymes coded for by the TOL plasmid (pWW0) from Pseudomonas putida mt-2. J Bacteriol. 1978;134:757–764. doi: 10.1128/jb.134.3.757-764.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]