Abstract
Three new Anabaena sp. strain PCC 7120 genes encoding group 2 alternative sigma factors have been cloned and characterized. Insertional inactivation of sigD, sigE, and sigF genes did not affect growth on nitrate under standard laboratory conditions but did transiently impair the abilities of sigD and sigE mutant strains to establish diazotrophic growth. A sigD sigE double mutant, though proficient in growth on nitrate and still able to differentiate into distinct proheterocysts, was unable to grow diazotrophically due to extensive fragmentation of filaments upon nitrogen deprivation. This double mutant could be complemented by wild-type copies of sigD or sigE, indicating some degree of functional redundancy that can partially mask phenotypes of single gene mutants. However, the sigE gene was required for lysogenic development of the temperate cyanophage A-4L. Several other combinations of double mutations, especially sigE sigF, caused a transient defect in establishing diazotrophic growth, manifested as a strong and prolonged bleaching response to nitrogen deprivation. We found no evidence for developmental regulation of the sigma factor genes. luxAB reporter fusions with sigD, sigE, and sigF all showed slightly reduced expression after induction of heterocyst development by nitrogen stepdown. Phylogenetic analysis of cyanobacterial group 2 sigma factor sequences revealed that they fall into several subgroups. Three morphologically and physiologically distant strains, Anabaena sp. strain PCC 7120, Synechococcus sp. strain PCC 7002, and Synechocystis sp. strain PCC 6803 each contain representatives of four subgroups. Unlike unicellular strains, Anabaena sp. strain PCC 7120 has three additional group 2 sigma factors that cluster in subgroup 2.5b, which is perhaps specific for filamentous or heterocystous cyanobacteria.
In eubacteria, sigma factors confer promoter-specific transcription initiation on RNA polymerase. Switching of sigma factors permits a precise temporal and spatial activation of particular sets of genes and is a common strategy for regulation of development in such diverse bacterial genera as Bacillus (40), Streptomyces (5, 34, 47), Myxococcus (2, 55), and Caulobacter (9, 42, 60). In many filamentous cyanobacteria, e.g., Anabaena and Nostoc spp., nitrogen starvation triggers the development of highly specialized cells called heterocysts (59). Heterocyst differentiation is thought to involve multiple signaling pathways through which environmental and intercellular cues are integrated to produce a linear developmental pattern of terminally differentiated nitrogen-fixing cells spaced semiregularly along filaments of photosynthetic vegetative cells. There is experimental evidence that regulation of gene expression during heterocyst differentiation occurs primarily, but not exclusively, at the level of transcription and that many genes are expressed in an ordered sequence such that expression at one stage depends upon gene products synthesized at a previous stage (59). A cascade-like activation of several developmental Anabaena genes was detected using the luxAB transcriptional reporter (14). It also has been shown that the activity of certain Anabaena promoters is confined either to vegetative cells or heterocysts, while others are active in both cell types (4, 6, 19, 54, 58). It would be surprising if heterocystous cyanobacteria did not use sigma factor switching as one element of transcriptional control over differentiation.
There are two basic families of eubacterial sigma factors. The ς70 family includes three groups. Group 1, or primary, sigma factors, are highly conserved in sequence, control transcription of housekeeping genes, and are essential for survival. Group 2 sigma factors (41) are similar in sequence to primary sigma factors, especially in their DNA-binding regions and thus probably in promoter specificity, but are dispensable for cell growth. Group 3, or alternative, sigma factors (41) show less similarity in sequence, recognize distinct promoters, and control specific processes such as heat shock and general stress responses (27, 28), motility (29), extracytoplasmic functions (44), and different stages of sporulation (26). Proteins of the ς54 family show no sequence similarity with primary sigma factors, depend on activator proteins that bind to enhancer sequences, recognize highly conserved promoter sequences, and regulate transcription of a variety of specialized groups of genes in different bacteria, e.g., genes involved in nitrogen fixation, synthesis of fimbriae or flagella, chemotaxis, and development (10, 43). No ς54 homologs have been found in cyanobacteria.
An unusual feature of cyanobacteria, shared only with gram-positive Streptomyces spp. (12, 38, 52) and the green sulfur bacterium Chloroflexus aurantiacus (25), is the presence of multiple alternative group 2 sigma factors. Although their exact functions in cyanobacterial cells remain obscure, some data implicate them in the response to nutrient starvation (8, 17, 45), in the response to plant factors in symbiotically competent Nostoc punctiforme (15), in post-exponential-phase growth (24), and in circadian expression of a subset of genes (53). The unicellular cyanobacterium Synechocystis sp. strain PCC 6803, whose entire genome has been sequenced (32), has four genes encoding putative group 2 sigma factors in addition to three genes for group 3 alternative sigma factors.
To date, no heterocyst-specific sigma factor has been identified and it is not known what sigma factor is used to transcribe the nitrogen fixation genes in cyanobacteria. sigA, the gene for the Anabaena sp. strain PCC 7120 principal sigma factor, was cloned and shown to have multiple promoters, several of them functioning exclusively under conditions of nitrogen limitation (7). Two additional Anabaena sp. strain PCC 7120 sigma factor genes have been cloned; sigB and sigC encode putative group 2 members of the ς70 family (8). sigB is expressed only under nitrogen-limiting conditions, while expression of sigC is induced by nitrogen or sulfur limitation. Insertional inactivation of these genes showed that neither of them was required for heterocyst differentiation or nitrogen fixation (8).
In this communication, we describe the identification and analysis of three additional group 2 sigma factor genes, sigD, sigE, and sigF, of Anabaena sp. strain PCC 7120.
MATERIALS AND METHODS
Bacterial strains, plasmids, phages, and growth conditions.
A list of all strains of cyanobacteria, phages, and plasmids used in this study is presented in Table 1. Anabaena sp. strain PCC 7120 and its derivatives were grown as described previously (21). Mutant strains were grown in the presence of appropriate antibiotics at the following final concentrations: neomycin, 25 μg/ml for solid medium and 15 μg/ml for liquid medium; erythromycin, 5 μg/ml; spectinomycin, 5 μg/ml; streptomycin, 2.5 μg/ml. To test for phage sensitivity, fresh, pale green streaks of wild-type and mutant strains grown on BG-11 agar plates were spotted with 3-μl portions of suspensions of cyanophage A-1(L), A-4(L), or A-4C10 (106 to 107 PFU ml−1). After 5 days of incubation at 30°C in the light, clear zones of lysis were observed; in the case of A-4(L), secondary growth of lysogenized cells was also observed. To induce heterocyst formation and diazotrophic growth, portions of exponentially growing cultures were washed twice with BG-110 medium (BG-11 lacking sodium nitrate), diluted 1:2 in BG-110 medium with and without antibiotics, and examined visually and by microscopy during 3 to 6 days following nitrogen stepdown. Micrographs were taken on a Zeiss Axioplan II microscope with differential interference contrast (DIC) optics. Images were captured with a Hamamatsu C5810 camera and processed with Adobe Photoshop version 4.0.
TABLE 1.
Bacterial strains, phages, and plasmids
| Strain, phage, or plasmid | Derivation and/or relevant characteristics | Source or reference |
|---|---|---|
| Anabaena sp. strains | ||
| PCC 7120 | Wild type | R. Haselkorn |
| DR1 | PCC 7120 sigB::Ω Spr Smr cassette | 8 |
| AMC645 | PCC 7120 sigC::pSC1 in their region of homology; Emr | This study |
| AMC646 | PCC 7120 sigD::C.CE3 Cmr Emr cassette at PflMI site of sigD | This study |
| AMC647 | PCC 7120 sigD::luxAB-Spr Smr cassette at PflMI site of sigD | This study |
| AMC648 | PCC 7120 sigE::luxAB-Spr Smr cassette at XbaI site of sigE | This study |
| AMC649 | PCC 7120 sigF::luxAB-Spr Smr cassette at ClaI site of sigF | This study |
| AMC650 | DR1 sigC::pSC1 in their region of homology; Spr Smr Emr | This study |
| AMC651 | AMC646 sigB::pSB1 in their region of homology; Spr Smr Emr | This study |
| AMC652 | AMC646 sigC::pSC1 in their region of homology; Spr Smr Emr | This study |
| AMC653 | AMC646 sigE::pSE2 in their region of homology; Spr Smr Emr | This study |
| AMC654 | AMC646 sigF::pSF26 in their region of homology; Spr Smr Emr | This study |
| AMC655 | AMC648 sigB::pSB2 in their region of homology; Spr Smr Emr | This study |
| AMC656 | AMC648 sigC::pSC1 in their region of homology; Spr Smr Emr | This study |
| AMC657 | AMC648 sigF::pSF25 in their region of homology; Spr Smr Emr | This study |
| AMC658 | AMC649 sigB::pSB2 in their region of homology; Spr Smr Emr | This study |
| AMC659 | AMC649 sigC::pSC1 in their region of homology; Spr Smr Emr | This study |
| Phages | ||
| A-1(L) | Virulent on strains of Anabaena spp. and Nostoc spp. | 30, 39 |
| A-4(L) | Temperate on strains of Anabaena spp. and Nostoc spp. | 30, 36 |
| A-4C10 | Clear-plaque mutant of A-4(L) | 35 |
| Plasmids | ||
| cos3H3 | Cosmid clone from pDUCA7M library containing sigF | 16 |
| cos9E7 | Cosmid clone from pDUCA7M library containing sigD and 3′ portion of sigE | 16 |
| pAM1011 | Shuttle vector | 49 |
| pAM1279 | Derivative of pAM1011 containing ca 0.5-kb deletion in pDU1 portion and AflII-NdeI (blunted) lacZ′-MCS from pUC18 inserted in the EcoRI (blunted) cloning sitea, Kmr Nmr | This study |
| pAM2178 | pARO180 with Ω Spr Smr cassette inserted into unique SspI site; Apr Spr Smr | This study |
| pAM2179 | pARO180 with C.CE3 Cmr Emr cassette inserted into unique SspI site; Apr Cmr Emr | This study |
| pARO180 | Mobilizable pUC18-based vector containing bom site of pSUP2021 and oriT of RP4; Apr | 46 |
| pBH500 | 4.7-kb EcoRI fragment with Anabaena sp. strain PCC 7120 sigB in pUC19; Apr | 8 |
| pBH700 | 3.4-kb HincII fragment with Anabaena sp. strain PCC 7120 sigC in pUC19; Apr | 8 |
| pK18 | pBR322-derived cloning vector; Kmr | 48 |
| pRL58 | Source of luxAB-Spr Smr cassette; Apr Spr Smr | 6 |
| pRL277 | Anabaena suicide vector containing sacB-Spr Smr cassette | 6 |
| pRL1075 | Source of sacB-oriT -Cmr Emr cassette | 6 |
| pRL1567 | Source of C.CE3 Cmr Emr cassette; Apr Cmr Emr | 14, 35 |
| pSB1 | 0.47-kb NheI-SspI fragment from within sigB cloned from pBH500 into pAM2178; Apr Spr Smr | This study |
| pSB2 | 0.54-kb NheI-AlwN fragment from within sigB cloned from pBH500 into pAM2179; Apr Cmr Emr | This study |
| pSC1 | 0.6-kb EcoRV fragment from within sigC cloned from pBH700 into pAM2179; Apr Cmr Emr | This study |
| pSD1 | 0.35-kb PCR fragment containing central part of sigD in pAM2178; Apr Spr Smr | This study |
| pSD7 | 3.1-kb NheI fragment containing sigD from cos9E7 in pUC19; Apr | This study |
| pSD21 | pSD7::luxAB-Spr Smr; SmaI-excised cassette from pRL58 in PflMI site (blunted) of sigD; luxAB oriented parallel to sigD; Apr Spr Smr | This study |
| pSD22 | pSD21 with cassette from pRL1075 inserted; Apr Spr Smr Cmr Emr | This study |
| pSD24 | pSD7::C.CE3; SmaI-excised cassette from pRL1567 in PflMI site (blunted) of sigD; transcription of Cmr and Emr genes parallel to sigD; Apr Cmr Emr | This study |
| pSD25 | XbaI-NheI fragment bearing sigD::C.CE3 from pSD24 ligated into XbaI site of pRL277 polylinker; Spr Smr Cmr Emr | This study |
| pSD27 | 2-kb XbaI-NheI fragment with sigD from cos9E7 in pAM1279; sigD oriented parallel to lacZ; Kmr Nmr | This study |
| pSE1 | 0.95-kb SmaI-NheI fragment from cos9E7 with 3′ part of sigE in pUC19; Apr | This study |
| pSE2 | 0.7-kb EcoRI-SmaI fragment from pSE1 with central part (0.57 kb) of sigE in pAM2178; Apr Spr Smr | This study |
| pSE14 | pSE1::luxAB-Spr Smr; cassette from pRL58 in XbaI site (blunted) of sigE; luxAB oriented parallel to sigE; Apr Spr Smr | This study |
| pSE15 | pSE14 with cassette from pRL1075 inserted; Apr Spr Smr Cmr Emr | This study |
| pSE23 | 0.82-kb XbaI fragment with 3′ end and downstream region of sigE in pAM2178; Apr Spr Smr | This study |
| pSE27 | EcoRV recovery of pAM2178 with flanking DNA from SRpSE23; Apr Spr Smr | This study |
| pSE30 | 2.2-kb HindIII-XbaI fragment containing wild-type sigE from pSE27 cloned into pAM1279; Kmr Nmr | This study |
| pSF5 | 6.5-kb SalI-NheI fragment containing sigF gene region from cos3H3 cloned into pUC19; Apr | This study |
| pSF18 | pSF5::luxAB-Spr Smr; SmaI-excised cassette from pRL58 in ClaI site (blunted) of sigF; luxAB oriented parallel to sigF; Apr Spr Smr | This study |
| pSF24 | pSF18 with cassette from pRL1075 inserted; Apr Spr Smr Cmr Emr | This study |
| pSF25 | 0.49-kb ClaI-BspE fragment from within sigF in pAM2179 | This study |
| pSF26 | Insert from pSF25 cloned into pAM2178 | This study |
| pUC19 | pBR322-derived cloning vector; Apr | 56 |
A detailed description of a series of pAM1011-based shuttle vectors containing pUC18 or pUC19 or pBluescript KS or SK polylinkers is available on request.
DNA manipulations.
Total DNA from cyanobacterial strains was extracted by vortexing cells with glass beads in the presence of phenol (13). Recombinant DNA procedures were performed by standard techniques (50). Enzymes were purchased from New England Biolabs or Promega and used according to the recommendations of the supplier. DNA sequencing from both strands by BigDye terminator cycle sequencing reaction (ABI Prism; Perkin-Elmer Applied Biosystems, Foster City, Calif.) was performed by using a series of fragments subcloned from original cosmid clones with synthetic oligonucleotide primers. Sequencing data were analyzed with Sequencher sequence analysis software (Gene Codes Corp.) and the National Center for Biotechnology Information GenBank BLAST e-mail server (1).
Insertional inactivation of group 2 sigma factor genes.
Details of the construction of conjugative suicide plasmids used to disrupt sigma factor genes are provided in Table 1 and Fig. 1. To facilitate gene disruption by single recombination with an internal gene fragment, we constructed conjugal vectors derived from the mobilizable plasmid pARO180 (46) by inserting an Ω Spr Smr cassette (yielding pAM2178) or a C.CE3 Cmr Emr cassette (yielding pAM2179) into a unique SspI site of pARO180. These vectors contain drug resistance markers readily selectable in cyanobacteria, contain unique cloning sites of the pUC18 polylinker (except AccI and SphI for pAM2178 and AccI and EcoRI for pAM2179), permit blue-white screening for inserts on 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) plates, are convenient for sequencing cloned fragments, and can be used as conjugative suicide vectors for obtaining single recombinants in Anabaena spp. Internal fragments of various sigma factor genes were cloned into pAM2178 or pAM2179, and resulting plasmids (Table 1) were transferred to wild-type Anabaena sp. strain PCC 7120 by conjugation as described by Elhai and Wolk (18a). Insertion mutations of sigD, sigE, and sigF genes were also constructed by double recombination with cloned genes interrupted by C.CE3 or luxAB-Spr Smr cassettes. The conjugative suicide plasmid pSD25 bearing sigD::C.CE3 was produced by digestion of pSD24 with XbaI and NheI and ligation with sacB-containing pRL277. Conjugative suicide plasmids pSD22 (with sigD::luxAB-Spr Smr), pSE15 (with sigE::luxAB-Spr Smr), and pSF24 (with sigF::luxAB-Spr Smr) were produced by ligation of the SphI-excised sacB-containing cassette from pRL1075 into the SphI site of pSD21, pSE14, or pSF18, respectively. Suicide plasmids were conjugated into Anabaena sp. strain PCC 7120, and the next day conjugation plates were underlaid with appropriate antibiotics to select for single recombinants. Subsequent selection for double recombinants using the sacB gene present on the vector was performed as described by Cai and Wolk (13). The genotypes of all constructed mutant strains were confirmed by Southern blot analysis (data not shown).
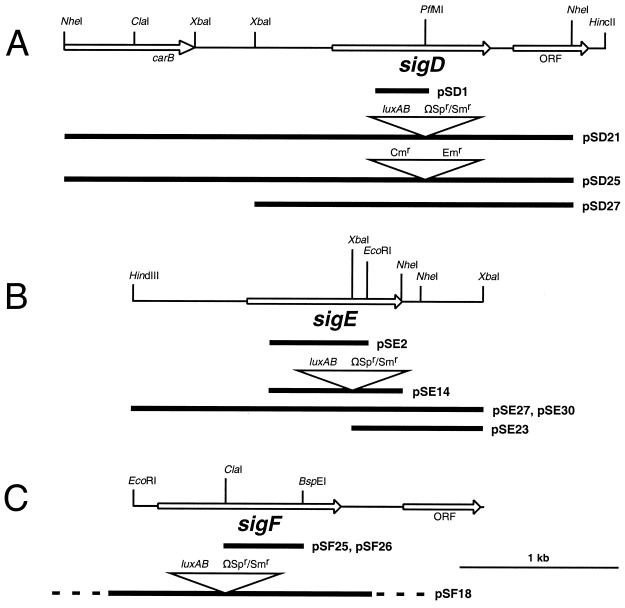
FIG. 1.
Maps for the Anabaena sp. strain PCC 7120 group 2 sigma factor genes sigD (A), sigE (B), and sigF (C), and related plasmids. Suicide plasmids pSD1, pSE2, pSF25, and pSF26 contain internal fragments of sigD, sigE, and sigF, respectively, and were used for gene inactivation by single recombination. Suicide plasmids pSD21, pSD25, pSE14, and pSF18, which contain sigma factor genes interrupted by antibiotic-resistance cassettes and sacB cassettes to allow positive selection for loss of the vector (13), were used for gene inactivation by double recombination. Replicative plasmids pSD27 and pSE30 were used for complementation of sigD sigE double-mutant strains.
Measurements of luciferase expression.
Bioluminescent reporter strains in 200-μl samples of an appropriate dilution of culture medium plus 4 μl of 2% decanal in mineral oil were transferred to scintillation vials and incubated for 3 min in darkness to allow chlorophyll fluorescence to decay. Luciferase expression was measured as light production (counts per second) in a scintillation counter with coincidence counting disabled. To induce heterocyst formation, portions of exponentially growing BG-11 cultures were washed twice with BG-110 medium and resuspended in the original volume of BG-110.
Nucleotide sequence accession numbers.
The nucleotide sequences of the sigD, sigE, and sigF gene regions reported in this paper have been submitted to GenBank under accession no. AF262216, AF262217, and AF262218, respectively.
RESULTS
Cloning and sequence analyses of the sigD, sigE, and sigF genes. During an unsuccessful attempt to clone an Anabaena sp. strain PCC 7120 group 3 sigma factor gene (unpublished data), a PCR fragment was cloned whose sequence showed high similarity to a variety of bacterial principal and group 2 secondary sigma factor genes. This fragment was used to screen an Anabaena sp. strain PCC 7120 pDUCA7M cosmid library (16). Two cosmid clones that hybridized with the probe, cos9E7 and cos3H3, were used for subcloning and sequence analyses. The sigD gene and the 3′ part of the sigE gene were cloned from cos9E7, and sigF was cloned from cos3H3. To obtain the entire sigE gene, the DNA region flanking the cos9E7 insert was cloned by sequential insertion and recovery of a suicide plasmid. pSE23, bearing a 0.82-kb XbaI fragment containing the 3′ end and downstream region of sigE (Fig. 1), was integrated by homologous recombination into the chromosome of wild-type Anabaena sp. strain PCC 7120. DNA from a resulting single recombinant strain (SRpSE23) was digested with EcoRV and self-ligated, and pSE27, which contained the vector plasmid with flanking Anabaena DNA, was recovered. A 2.2-kb HindIII-XbaI fragment from pSE27 containing the whole sigE gene was subcloned into the shuttle vector pAM1279, producing pSE30, and used for sequencing and complementation experiments (see below).
Physical maps of the sigD, sigE, and sigF gene regions are shown in Fig. 1. The sigD sequence has two possible methionine start codons, both preceded by identical potential ribosome-binding sites (RBS) (5′-AAAGAG-3′). The spacing of this RBS is −20 to −15 relative to the first ATG codon and −13 to −8 relative to the second; the lengths of resulting translational products are 332 and 318 amino acids, respectively. A perfect 20-bp palindrome, 5′-AGACGCGATTAATCGCGTCT-3′, is located 4 nucleotides downstream of a TAG stop codon.
The sigE sequence, starting with a methionine codon, encodes a 327-amino-acid protein and is preceded by a putative RBS (5′-AGAGG-3′) at −16 to −12. A second potential start site, TTG, is located farther upstream and, if functional, would produce a 343-amino-acid SigE polypeptide.
For the sigF gene, multiple in-frame methionine codons and a leucine TTG codon that could serve as translational start sites in the nonconserved 5′ region could produce potential SigF polypeptides of 390, 378, 370, and/or 328 amino acids. The best putative RBS, 5′-AAGGA-3′, is located at −9 to −5 before the second methionine codon; weaker potential RBS precede other start codons.
Construction and phenotypes of single and double mutants.
To obtain disruption mutations of sigD, sigE, and sigF genes, we used either insertion of an antibiotic resistance cassette into the corresponding open reading frame by double recombination or inactivation by single recombination with the suicide vector pAM2178 or pAM2179 carrying an internal fragment of the corresponding gene (Table 1; Fig. 1). Inactivation of any of these three genes by single or double recombination resulted in mutant strains that did not differ from the wild type in their appearance or growth on nitrate-containing media. However, inactivation of the sigD or sigE gene transiently impaired the ability to establish diazotrophic growth (Table 2). Depending on growth conditions, there was a variable fraction of filaments in sigD and sigE mutant strains that fragmented extensively upon nitrogen stepdown, and detached mature heterocysts were abundant after 3 to 4 days of diazotrophic growth.
TABLE 2.
Phenotypic characteristics of single and double mutants after removal of combined nitrogen
| Strain | Genotypea | Diazotrophic growth | Differentiation timeb (h) | Bleaching intensity, duration (days) | Lysogenization with A-4(L) | Other characteristics |
|---|---|---|---|---|---|---|
| PCC 7120 | wt | + | 12–18 | Slight, 1 | + | |
| DR1 | sigB | + | 18–24 | Moderate, 2 | + | |
| AMC645 | sigC | + | 18–24 | Moderate, 2 | + | |
| AMC646 | sigD | + | 12–18 | Moderate, 2 | + | |
| AMC648 | sigE | + | 12–18 | Moderate, 2 | − | |
| AMC649 | sigF | + | 12–18 | Slight, 1 | + | |
| AMC650 | sigB sigC | + | 24–30 | Moderate, 2 | + | |
| AMC651 | sigD sigB | + | 12–18 | Moderate, 2 | + | |
| AMC652 | sigD sigC | + | 12–18 | Strong, 2 | + | |
| AMC653 | sigD sigE | − | 12–18 | Severe | − | Fra, extensive fragmentation |
| AMC654 | sigD sigF | + | 12–18 | Strong, 3 | + | |
| AMC655 | sigE sigB | + | 18–24 | Strong, 3 | − | |
| AMC656 | sigE sigC | + | 24–30 | Strong, 3 | − | |
| AMC657 | sigE sigF | + | 24–30 | Severe, 4–5 | − | Transient Fra; turns light brown on old sealed BG110 plates |
| AMC658 | sigF sigB | + | 24–30 | Moderate, 3 | + | |
| AMC659 | sigF sigC | + | 24–30 | Strong, 3 | + |
wt, wild type. See Table 1 for details.
Time after nitrogen stepdown when morphologically distinct proheterocysts can be seen.
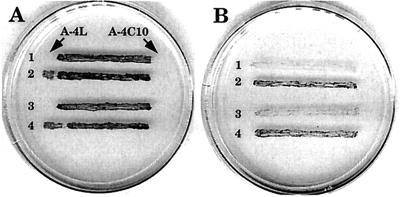
To test the possibility that phage development in Anabaena sp. strain PCC 7120 might be affected by inactivation of a particular group 2 sigma factor, streaks of different single and double mutants (see below) were spotted with drops of phage A-1(L), A-4(L), and A-4C10 lysates. All phages produced zones of lysis on all mutant strains, indicating that lytic development was not significantly impaired in any of the mutants; however, in sigE single and double mutants (see below), the ability to support lysogenic growth of the temperate phage A-4(L) was abolished or severely reduced (Table 2). This ability was restored when sigE mutants were complemented with a cloned sigE gene (Fig. 2).
FIG. 2.
Complementation of Fox Fra and lysogenization deficiency phenotypes of sigD sigE double mutants with the pSE30 plasmid containing a wild-type copy of sigE in the pAM1279 shuttle vector. Control strains of two independent double mutants, strain 1 [sigD sigE-10 (pAM1279)] and strain 3 [sigD sigE-12 (pAM1279)], and complemented strain 2 [sigD sigE-10 (pSE30)] and strain 4 [sigD sigE-12 (pSE30)] were streaked onto BG-11 (nitrate) (A) and BG-110 (nitrogen-free) (B) plates. The ends of streaks on plate A (arrows) were spotted with the temperate phage A-4L (left) and its clear-plaque mutant A-4C10 (right). Note the secondary growth of lysogenic cells at the left ends of streaks 2 and 4 on plate A and restoration of diazotrophic growth in streaks 2 and 4 on plate B.
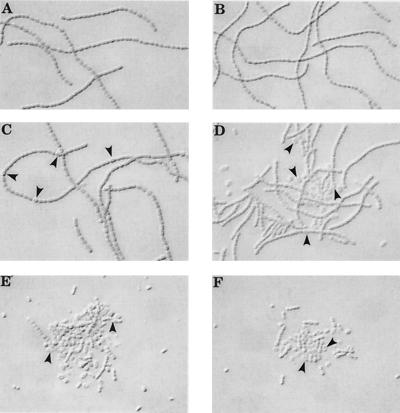
We constructed different combinations of double mutants (Table 2) and found that a sigD sigE mutant strain, though able to differentiate heterocysts, was unable to grow diazotrophically, possibly due to extensive fragmentation of filaments starting about 24 h after nitrogen deprivation (Fig. 3). Three independently isolated sigD sigE mutant clones exhibited the same phenotype, which parallels those of several Fra (fragmentation of filaments upon nitrogen deprivation) mutants described previously (3, 11). During exponential growth in a nitrate-containing liquid medium, cultures of the sigD sigE mutants grew at approximately the same growth rate as the wild type, but upon entering post-exponential phase they started to fragment and lyse; stationary-phase cultures tended to lyse completely. Complementation of sigD sigE mutants with a wild-type copy of sigE restored both the Fox+ phenotype (Fox denotes the inability to fix nitrogen in the presence of oxygen [20]) and the ability to be lysogenized by A-4(L) (Fig. 2). Plasmid pSD27, containing a wild-type copy of sigD, also complemented the Fox Fra phenotype (data not shown). However, SigD cannot substitute for SigE for efficient establishment and/or maintenance of lysogeny by temperate phage A-4(L). Perhaps both SigD and SigE participate in the regulation of fra genes, which are implicated in the synthesis of modified cell junctions in response to nitrogen starvation (3, 11), and exhibit some degree of functional redundancy. An alternative explanation is that different fra genes, each recognized by a different sigma factor, are redundant or have overlapping functions.
FIG. 3.
Fragmenting phenotype of sigD sigE double-mutant strain AMC653. (A) uninduced nitrate-grown wild-type strain; (B) uninduced nitrate-grown AMC653; (C) wild-type strain 26 h after nitrogen stepdown; (D) AMC653 26 h after nitrogen stepdown; (E) AMC653 48 h after nitrogen stepdown; (F) AMC653 72 h after nitrogen stepdown. Arrowheads point to representative heterocysts.
Strains with other combinations of double mutations were able to grow on N2; however, several combinations yielded a transient Fox phenotype revealed by prolonged bleaching and variable lags before resuming normal pigmentation and diazotrophic growth (Table 2). The most impaired (but still Fox+) double mutant, sigE sigF, lacks those sigma factors whose homologs in Synechococcus sp. strain PCC 7002, SigB and SigC, respectively, have been implicated in transcription modification in response to changes in nitrogen and carbon availability (17).
Expression of sigD, sigE, and sigF genes.
Expression of Anabaena sp. strain PCC 7120 genes can be monitored with a luxAB transcriptional reporter during heterocyst development synchronously induced by the removal of fixed nitrogen (14). We examined the expression of sigma factor genes sigD, sigE, and sigF with luxAB transcriptional fusions in strains AMC647, AMC648, and AMC649, respectively. For all three strains during a 24-h period after nitrogen stepdown, luminescence gradually decreased to 1/2 to 1/3 that of nitrogen-replete control cultures. The absolute levels of luxAB reporter expression (measured as luminescence) varied from experiment to experiment but always decreased after heterocyst induction. In a representative experiment, relative luxAB expression levels (expressed as the ratio of luminescence in an induced culture to that in uninduced control cultures) at 13 and 25 h after nitrogen stepdown were 1.05 and 0.65 for sigD::luxAB, 0.92 and 0.33 for sigE::luxAB, and 0.59 and 0.45 for sigF::luxAB fusions, respectively.
Phylogenetic analysis.
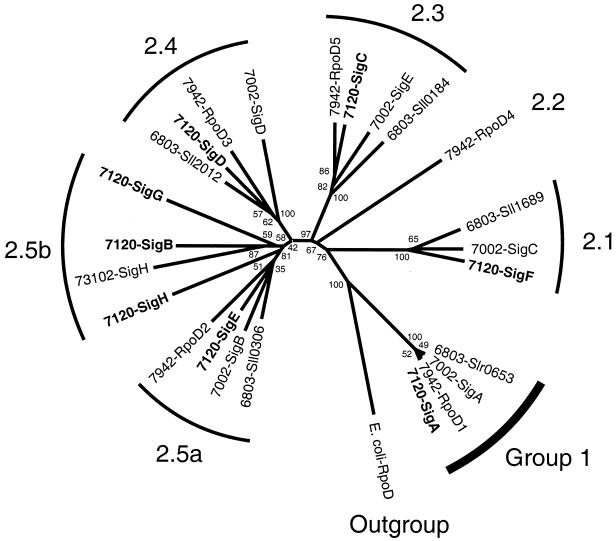
To determine the evolutionary relationships among various cyanobacterial group 2 sigma factors, we aligned predicted amino acid sequences from Anabaena sp. strain PCC 7120, Synechococcus sp. strain PCC 7942, Synechococcus sp. strain PCC 7002, and Synechocystis sp. strain PCC 6803 and constructed a phylogenetic tree (Fig. 4). We have included two additional Anabaena sigma factors, SigG and SigH, which we identified in the Anabaena sp. strain PCC 7120 preliminary genome sequence database (http://www.kazusa.or.jp/cyano/anabaena/). It is evident that the majority of cyanobacterial group 2 sigma factors fall into four distinct clusters, which we have designated here subgroups 2.1, 2.3, 2.4, and 2.5a. Each of the cyanobacterial strains contains one representative from each of these four subgroups, with the exception of Synechococcus sp. strain PCC 7942. RpoD4 from PCC 7942 is placed in a separate subgroup, 2.2, in the analysis shown here because three additional group 2 sigma factors from the marine organism Prochlorococcus marinus also fall into this subgroup (I. Khudyakov, unpublished data). The high conservation, not only of amino acid sequence, but also of overall length and characteristic gaps in the alignment, among proteins in each of these subgroups (data not shown) strongly suggests that these four subgroups constitute an evolutionarily conserved cyanobacterial complement of group 2 sigma factors shared by strains from diverse genera.
FIG. 4.
Unrooted phylogenetic tree of group 1 and group 2 sigma factors from Anabaena sp. strain PCC 7120, Synechocystis sp. strain PCC 6803, Synechococcus sp. strain PCC 7002, Synechococcus sp. strain PCC 7942, and N. punctiforme PCC 73102. The Escherichia coli principal sigma factor RpoD sequence was included as an outgroup. Arches indicate cyanobacterial sigma factors belonging to group 1 and to group 2 subgroups 2.1, 2.2, 2.3, 2.4, 2.5a, and 2.5b. Amino acid positions corresponding to sigma factor structural region 1.2 and from region 2.1 to the carboxy terminus were used for alignments. Multiple-alignment analysis was performed with the PHYLIP software package, and the unrooted phylogenetic tree was drawn with DRAWTREE. Numbers at the nodes are bootstrap values obtained with 1,000 replications. Designations and GenBank accession numbers for sequences of sigma factors are as follows: E. coli-RpoD for Escherichia coli RpoD principal sigma factor (J01687); 7120-SigA, 7120-SigB, 7120-SigC, 7120-SigD, 7120-SigE, and 7120-SigF for Anabaena sp. strain PCC 7120 sigma factors (M60046, M95760, M95759, AF262216, AF262217, and AF262218, respectively); 7120-SigG and 7120-SigH for predicted products of Anabaena sp. strain PCC 7120 genes located on contigs C369 (bp 66478 to 67428) and C380 (bp 91346 to 90375 bp), respectively, in CyanoBase (http://www.kazusa.or.jp/cyano/anabaena/); 73102-SigH for N. punctiforme PCC 73102 sigma factor (AF022822); 7002-SigA, 7002-SigB, 7002-SigC, 7002-SigD, and 7002-SigE for Synechococcus sp. strain PCC 7002 sigma factors (U15574, U82435, U82436, U82484, and U82485, respectively); PCC 7942-RpoD1, 7942-RpoD2, 7942-RpoD3, 7942-RpoD4, and 7942-RpoD5 for Synechococcus sp. strain PCC 7942 sigma factors (D10973, D78583, AB024709, AB024710, and AF288784, respectively); and 6803-Slr0653, 6803-Sll1689, 6803-Sll0184, 6803-Sll2012, and 6803-Sll0306 for Synechocystis sp. strain PCC 6803 sigma factors in CyanoBase (http://www.kazusa.or.jp/cyanobase/index.html).
Unlike the unicellular strains, Anabaena sp. strain PCC 7120 has three additional sigma factors, SigB, SigG, and SigH, that cluster in subgroup 2.5b. SigH, a group 2 sigma factor from the heterocystous strain N. punctiforme (15), also falls into this subgroup.
DISCUSSION
The aim of this work was to examine the possibility of involvement of group 2 sigma factors in the regulation of heterocyst development and diazotrophic growth of Anabaena sp. strain PCC 7120. We found that, as with the mutants with inactivated sigB and sigC genes isolated by Brahamsha and Haselkorn (8), single mutations in sigD, sigE, and sigF had little or no effect on the abilities of the mutant strains to establish efficient diazotrophic growth under standard laboratory conditions. However, phenotypic analyses of double mutants revealed that certain combinations of inactivated sigma factor genes produced distinct Fox Fra or transient Fox phenotypes (Table 2).
A pronounced characteristic of the sigD sigE double mutant is rapid and extensive fragmentation in response to fixed-nitrogen deprivation. Fragmentation mutants constitute a significant proportion of induced (11) and spontaneous (I. Khudyakov, unpublished results) Fox mutants in Anabaena sp. strain PCC 7120, and several distinct Fra phenotypes have been described. Upon nitrogen stepdown, some mutants, typified by genetically uncharacterized strain N5 (20), fragment to very short filaments without any sign of heterocyst differentiation. Strain 129 (11), which contains a frameshift in the fraC gene (3), shows rapid fragmentation and partial differentiation. Strain 415 (11) fragments extensively and forms many proheterocysts but few heterocysts. Fragmentation of filaments in nitrate-containing medium and detachment of differentiating heterocysts were also observed when an adenylate cyclase gene of Anabaena cylindrica was introduced on a replicating plasmid into Anabaena sp. strain PCC 7120 (33). In all these cases the fragmenting phenotype was attributed to weakness of vegetative cell junctions and/or an even more pronounced defect in heterocyst junctions, which are more prone to breakage in wild-type filaments. A unicellular mutant of the Het− Anabaena sp. strain PCC 7118 is an extreme example of fragmentation under nitrate-replete conditions (37).
The variety of different Fra phenotypes and genetic lesions causing such phenotypes probably reflects the complexity of the process of septum formation during vegetative growth and its modification during heterocyst differentiation. In the sigD sigE double mutant, the regulation of these processes is obviously impaired. Altered expression of fraC, fraH, and/or other genes whose products are involved in septum formation may contribute to its phenotype. Cell lysis of this double mutant during post-exponential phase and after nitrogen stepdown points also to some imbalances in production or regulation of autolysin(s). This divergent group of enzymes has been shown (or inferred) to be involved in a number of cellular processes including bacterial cell growth, cell division and separation, cell wall turnover, protein secretion, and differentiation (51).
Although our data indicate that at least some group 2 sigma factors participate in the establishment of diazotrophic growth, at present we have no evidence as to whether they contribute to regulation of heterocyst-specific transcription or to a vegetative cell-specific response to nitrogen deprivation. We examined the expression of sigma factor genes using luxAB transcriptional fusions in strains AMC647, AMC648, and AMC649 and found no evidence for an increase of sigD, sigE, or sigF transcription during a 24-h period following nitrogen stepdown.
In non-N2-fixing cyanobacteria, nitrogen starvation causes a dramatic decrease in phycocyanin content (23). In heterocystous cyanobacteria, heterocyst differentiation after deprivation of fixed nitrogen is accompanied by a transient bleaching response which includes a decrease in the level of mRNA coding for phycocyanin and allophycocyanin (31, 57). After heterocysts mature and start fixing nitrogen, normal levels of cpcBA and apcAB mRNAs are reestablished in vegetative cells but not in heterocysts (31). The delay in reestablishment of normal pigmentation observed in the different sigma factor mutant strains (Table 2) may result from a delay in heterocyst maturation and assembly of a functional nitrogenase complex, or vegetative cells may initially fail to sustain adequate reductant flow to heterocysts. Alternatively, overexpression or inability to rapidly shut down a homolog of nblA, a gene that triggers phycobilisome degradation in Synechococcus sp. strain PCC 7942 (18), could cause the same effect.
Our results show that none of the three Anabaena sigma factor genes sigD, sigE, and sigF is absolutely required for growth or development, and they suggest that these genes encode sigma subunits with overlapping promoter specificities. However, it appears that the complete complement of several group 2 sigma factors is required for rapid establishment of diazotrophic growth. The eventual recovery of all single and most double mutants to nearly normal growth suggests that the transition period is the most sensitive to imbalances in sigma factors and that the cells can eventually adapt, possibly by modifying the expression of remaining sigma factors with partially overlapping functions. Such compensatory adjustment has been suggested to explain the increase in SigB protein in a sigC mutant of Synechococcus sp. strain PCC 7002 (17).
Phylogenetic analysis of deduced amino acid sequences of cyanobacterial group 2 sigma factor proteins has shown that most of them fall into several distinct subgroups (Fig. 4). Representatives of four of these subgroups, 2.1, 2.3, 2.4, and 2.5a, are present in all strains thoroughly examined so far and probably constitute a set of group 2 sigma factors that has been conserved throughout the evolution of cyanobacterial species. Such conservation implies that this complement was highly beneficial for survival and that individual group 2 sigma factors from each subgroup are functionally specialized, each presumably having its own adaptive value. At the same time, such specialization must not be absolute, because our reverse genetics experiments indicate that there is functional redundancy. A clear example of such redundancy is the ability of sigD and sigE to substitute for one another in the regulation of a function essential for filament integrity during the establishment of diazotrophy. Recently, Goto-Seki et al. compared promoter recognition in vitro and found specificity cross talk among group 1 and group 2 sigma factors in Synechococcus sp. strain PCC 7942 (22). Their phylogenetic analysis, as well as a previous phylogenetic analysis conducted by Gruber and Bryant (25), also showed that cyanobacterial group 2 sigma factors form a clade that appears to be further divided into four clusters. The inclusion in our analysis of five new sigma factors from Anabaena sp. strain PCC 7120 revealed that this strain harbors three additional sigma factors which form a loose cluster close to the typical cyanobacterial subgroup 2.5a and constitute subgroup 2.5b, which is perhaps specific for filamentous or heterocystous cyanobacteria.
ACKNOWLEDGMENTS
We thank Bianca Brahamsha for supplying the DR1 mutant strain and pBH500 and pBH700 plasmids. We thank members of our laboratory for critical reading of the manuscript.
This work was supported by National Institutes of Health grant GM36890 and Department of Energy grant DE-FG03-98ER020309.
REFERENCES
- 1.Altschul S F, Madden T L, Schaffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Apelian D, Inouye S. A new putative sigma factor of Myxococcus xanthus. J Bacteriol. 1993;175:3335–3342. doi: 10.1128/jb.175.11.3335-3342.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bauer C C, Buikema W J, Black K, Haselkorn R. A short-filament mutant of Anabaena sp. strain PCC 7120 that fragments in nitrogen-deficient medium. J Bacteriol. 1995;177:1520–1526. doi: 10.1128/jb.177.6.1520-1526.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bauer C C, Haselkorn R. Vectors for determining the differential expression of genes in heterocysts and vegetative cells of Anabaena sp. strain PCC 7120. J Bacteriol. 1995;177:3332–3336. doi: 10.1128/jb.177.11.3332-3336.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bibb M J, Molle V, Buttner M J. ςBldN, an extracytoplasmic function RNA polymerase sigma factor required for aerial mycelium formation in Streptomyces coelicolor A3(2) J Bacteriol. 2000;182:4606–4616. doi: 10.1128/jb.182.16.4606-4616.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Black T A, Cai Y, Wolk C P. Spatial expression and autoregulation of hetR, a gene involved in the control of heterocyst development in Anabaena. Mol Microbiol. 1993;9:77–84. doi: 10.1111/j.1365-2958.1993.tb01670.x. . (Erratum, 10:1153.). [DOI] [PubMed] [Google Scholar]
- 7.Brahamsha B, Haselkorn R. Isolation and characterization of the gene encoding the principal sigma factor of the vegetative cell RNA polymerase from the cyanobacterium Anabaena sp. strain PCC 7120. J Bacteriol. 1991;173:2442–2450. doi: 10.1128/jb.173.8.2442-2450.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brahamsha B, Haselkorn R. Identification of multiple RNA polymerase sigma factor homologs in the cyanobacterium Anabaena sp. strain PCC 7120: cloning, expression, and inactivation of the sigB and sigC genes. J Bacteriol. 1992;174:7273–7282. doi: 10.1128/jb.174.22.7273-7282.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brun Y V, Shapiro L. A temporally controlled sigma-factor is required for polar morphogenesis and normal cell division in Caulobacter. Genes Dev. 1992;6:2395–2408. doi: 10.1101/gad.6.12a.2395. [DOI] [PubMed] [Google Scholar]
- 10.Buck M, Gallegos M T, Studholme D J, Guo Y, Gralla J D. The bacterial enhancer-dependent ς54 (ςN) transcription factor. J Bacteriol. 2000;182:4129–4136. doi: 10.1128/jb.182.15.4129-4136.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Buikema W J, Haselkorn R. Isolation and complementation of nitrogen fixation mutants of the cyanobacterium Anabaena sp. strain PCC 7120. J Bacteriol. 1991;173:1879–1885. doi: 10.1128/jb.173.6.1879-1885.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Buttner M J, Chater K F, Bibb M J. Cloning, disruption, and transcriptional analysis of three RNA polymerase sigma factor genes of Streptomyces coelicolor A3(2) J Bacteriol. 1990;172:3367–3378. doi: 10.1128/jb.172.6.3367-3378.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cai Y, Wolk C P. Use of a conditionally lethal gene in Anabaena sp. strain PCC 7120 to select for double recombinants and to entrap insertion sequences. J Bacteriol. 1990;172:3138–3145. doi: 10.1128/jb.172.6.3138-3145.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cai Y, Wolk C P. Anabaena sp. strain PCC 7120 responds to nitrogen deprivation with a cascade-like sequence of transcriptional activations. J Bacteriol. 1997;179:267–271. doi: 10.1128/jb.179.1.267-271.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Campbell E L, Brahamsha B, Meeks J C. Mutation of an alternative sigma factor in the cyanobacterium Nostoc punctiforme results in increased infection of its symbiotic plant partner Anthoceros punctatus. J Bacteriol. 1998;180:4938–4941. doi: 10.1128/jb.180.18.4938-4941.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Carrasco C D, Buettner J A, Golden J W. Programmed DNA rearrangement of a cyanobacterial hupL gene in heterocysts. Proc Natl Acad Sci USA. 1995;92:791–795. doi: 10.1073/pnas.92.3.791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Caslake L F, Gruber T M, Bryant D A. Expression of two alternative sigma factors of Synechococcus sp. strain PCC 7002 is modulated by carbon and nitrogen stress. Microbiology. 1997;143:3807–3818. doi: 10.1099/00221287-143-12-3807. [DOI] [PubMed] [Google Scholar]
- 18.Collier J L, Grossman A R. A small polypeptide triggers complete degradation of light-harvesting phycobiliproteins in nutrient-deprived cyanobacteria. EMBO J. 1994;13:1039–1047. doi: 10.1002/j.1460-2075.1994.tb06352.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18a.Elhai J, Wolk C P. Conjugal transfer of DNA to cyanobacteria. Methods Enzymol. 1988;167:747–754. doi: 10.1016/0076-6879(88)67086-8. [DOI] [PubMed] [Google Scholar]
- 19.Elhai J, Wolk C P. Developmental regulation and spatial pattern of expression of the structural genes for nitrogenase in the cyanobacterium Anabaena. EMBO J. 1990;9:3379–3388. doi: 10.1002/j.1460-2075.1990.tb07539.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ernst A, Black T, Cai Y, Panoff J M, Tiwari D N, Wolk C P. Synthesis of nitrogenase in mutants of the cyanobacterium Anabaena sp. strain PCC 7120 affected in heterocyst development or metabolism. J Bacteriol. 1992;174:6025–6032. doi: 10.1128/jb.174.19.6025-6032.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Golden J W, Whorff L L, Wiest D R. Independent regulation of nifHDK operon transcription and DNA rearrangement during heterocyst differentiation in the cyanobacterium Anabaena sp. strain PCC 7120. J Bacteriol. 1991;173:7098–7105. doi: 10.1128/jb.173.22.7098-7105.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Goto-Seki A, Shirokane M, Masuda S, Tanaka K, Takahashi H. Specificity crosstalk among group 1 and group 2 sigma factors in the cyanobacterium Synechococcus sp. PCC7942: in vitro specificity and a phylogenetic analysis. Mol Microbiol. 1999;34:473–484. doi: 10.1046/j.1365-2958.1999.01608.x. [DOI] [PubMed] [Google Scholar]
- 23.Grossman A R, Schaefer M R, Chiang G G, Collier J L. The responses of cyanobacteria to environmental conditions: light and nutrients. In: Bryant D A, editor. The molecular biology of cyanobacteria. Vol. 1. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1994. pp. 641–675. [Google Scholar]
- 24.Gruber T M, Bryant D A. Characterization of the alternative sigma-factors SigD and SigE in Synechococcus sp. strain PCC 7002. SigE is implicated in transcription of post-exponential-phase-specific genes. Arch Microbiol. 1998;169:211–219. doi: 10.1007/s002030050563. [DOI] [PubMed] [Google Scholar]
- 25.Gruber T M, Bryant D A. Characterization of the group 1 and group 2 sigma factors of the green sulfur bacterium Chlorobium tepidum and the green non-sulfur bacterium Chloroflexus aurantiacus. Arch Microbiol. 1998;170:285–296. doi: 10.1007/s002030050644. [DOI] [PubMed] [Google Scholar]
- 26.Haldenwang W G. The sigma factors of Bacillus subtilis. Microbiol Rev. 1995;59:1–30. doi: 10.1128/mr.59.1.1-30.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hecker M, Schumann W, Volker U. Heat-shock and general stress response in Bacillus subtilis. Mol Microbiol. 1996;19:417–428. doi: 10.1046/j.1365-2958.1996.396932.x. [DOI] [PubMed] [Google Scholar]
- 28.Hecker M, Volker U. Non-specific, general and multiple stress resistance of growth-restricted Bacillus subtilis cells by the expression of the ςB regulon. Mol Microbiol. 1998;29:1129–1136. doi: 10.1046/j.1365-2958.1998.00977.x. [DOI] [PubMed] [Google Scholar]
- 29.Helmann J D. Alternative sigma factors and the regulation of flagellar gene expression. Mol Microbiol. 1991;5:2875–2882. doi: 10.1111/j.1365-2958.1991.tb01847.x. [DOI] [PubMed] [Google Scholar]
- 30.Hu N T, Thiel T, Giddings T H, Jr, Wolk C P. New Anabaena and Nostoc cyanophages from sewage settling ponds. Virology. 1981;114:236–246. doi: 10.1016/0042-6822(81)90269-5. [DOI] [PubMed] [Google Scholar]
- 31.Johnson T R, Haynes II J I, Wealand J L, Yarbrough L R, Hirschberg R. Structure and regulation of genes encoding phycocyanin and allophycocyanin from Anabaena variabilis ATCC 29413. J Bacteriol. 1988;170:1858–1865. doi: 10.1128/jb.170.4.1858-1865.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kaneko T, Sato S, Kotani H, Tanaka A, Asamizu E, Nakamura Y, Miyajima N, Hirosawa M, Sugiura M, Sasamoto S, Kimura T, Hosouchi T, Matsuno A, Muraki A, Nakazaki N, Naruo K, Okumura S, Shimpo S, Takeuchi C, Wada T, Watanabe A, Yamada M, Yasuda M, Tabata S. Sequence analysis of the genome of the unicellular cyanobacterium Synechocystis sp. strain PCC6803. II. Sequence determination of the entire genome and assignment of potential protein-coding regions. DNA Res. 1996;3:109–136. doi: 10.1093/dnares/3.3.109. [DOI] [PubMed] [Google Scholar]
- 33.Katayama M, Ohmori M. Expression of an adenylate cyclase gene of Anabaena cylindrica in the cyanobacterium Anabaena sp. strain PCC 7120. Plant Cell Physiol. 1998;39:786–789. [Google Scholar]
- 34.Kelemen G H, Brown G L, Kormanec J, Potuckova L, Chater K F, Buttner M J. The positions of the sigma-factor genes, whiG and sigF, in the hierarchy controlling the development of spore chains in the aerial hyphae of Streptomyces coelicolor A3(2) Mol Microbiol. 1996;21:593–603. doi: 10.1111/j.1365-2958.1996.tb02567.x. [DOI] [PubMed] [Google Scholar]
- 35.Khudyakov I, Wolk C P. Evidence that the hanA gene coding for HU protein is essential for heterocyst differentiation in, and cyanophage A-4(L) sensitivity of, Anabaena sp. strain PCC 7120. J Bacteriol. 1996;178:3572–3577. doi: 10.1128/jb.178.12.3572-3577.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Khudyakov I Y, Gromov B V. The temperate cyanophage of the blue-green alga Anabaena variabilis. Mikrobiologiya. 1973;42:904–907. [PubMed] [Google Scholar]
- 37.Khudyakov I Y, Pinevich A V. Unicellular mutant of the filamentous cyanobacterium Anabaena sp. PCC 7118. Mikrobiologiya. 1991;60:704–708. [Google Scholar]
- 38.Kormanec J, Farkasovsky M, Potuckova L. Four genes in Streptomyces aureofaciens containing a domain characteristic of principal sigma factors. Gene. 1992;122:63–70. doi: 10.1016/0378-1119(92)90032-k. [DOI] [PubMed] [Google Scholar]
- 39.Koz'yakov S Y, Gromov B V, Khudyakov I Y. A-1(L) cyanophage of the blue-green alga Anabaena variabilis. Mikrobiologiya. 1972;41:555–559. [PubMed] [Google Scholar]
- 40.Kroos L, Zhang B, Ichikawa H, Yu Y T. Control of sigma factor activity during Bacillus subtilis sporulation. Mol Microbiol. 1999;31:1285–1294. doi: 10.1046/j.1365-2958.1999.01214.x. [DOI] [PubMed] [Google Scholar]
- 41.Lonetto M, Gribskov M, Gross C A. The ς70 family: sequence conservation and evolutionary relationships. J Bacteriol. 1992;174:3843–3849. doi: 10.1128/jb.174.12.3843-3849.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Marczynski G T, Shapiro L. The control of asymmetric gene expression during Caulobacter cell differentiation. Arch Microbiol. 1995;163:313–321. doi: 10.1007/BF00404203. [DOI] [PubMed] [Google Scholar]
- 43.Merrick M J. In a class of its own—the RNA polymerase sigma factor ς54 (ςN) Mol Microbiol. 1993;10:903–909. doi: 10.1111/j.1365-2958.1993.tb00961.x. [DOI] [PubMed] [Google Scholar]
- 44.Missiakas D, Raina S. The extracytoplasmic function sigma factors: role and regulation. Mol Microbiol. 1998;28:1059–1066. doi: 10.1046/j.1365-2958.1998.00865.x. [DOI] [PubMed] [Google Scholar]
- 45.Muro-Pastor A M, Herrero A, Flores E. Nitrogen-regulated group 2 sigma factor from Synechocystis sp. strain PCC 6803 involved in survival under nitrogen stress. J Bacteriol. 2001;183:1090–1095. doi: 10.1128/JB.183.3.1090-1095.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Parke D. Construction of mobilizable vectors derived from plasmids RP4, pUC18 and pUC19. Gene. 1990;93:135–137. doi: 10.1016/0378-1119(90)90147-j. [DOI] [PubMed] [Google Scholar]
- 47.Potuckova L, Kelemen G H, Findlay K C, Lonetto M A, Buttner M J, Kormanec J. A new RNA polymerase sigma factor, ςF, is required for the late stages of morphological differentiation in Streptomyces spp. Mol Microbiol. 1995;17:37–48. doi: 10.1111/j.1365-2958.1995.mmi_17010037.x. [DOI] [PubMed] [Google Scholar]
- 48.Pridmore R D. New and versatile cloning vectors with kanamycin-resistance marker. Gene. 1987;56:309–312. doi: 10.1016/0378-1119(87)90149-1. [DOI] [PubMed] [Google Scholar]
- 49.Ramaswamy K S, Carrasco C D, Fatma T, Golden J W. Cell-type specificity of the Anabaena fdxN-element rearrangement requires xisH and xisI. Mol Microbiol. 1997;23:1241–1249. doi: 10.1046/j.1365-2958.1997.3081671.x. [DOI] [PubMed] [Google Scholar]
- 50.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 51.Smith T J, Blackman S A, Foster S J. Autolysins of Bacillus subtilis: multiple enzymes with multiple functions. Microbiology. 2000;146:249–262. doi: 10.1099/00221287-146-2-249. [DOI] [PubMed] [Google Scholar]
- 52.Tanaka K, Shiina T, Takahashi H. Nucleotide sequence of genes hrdA, hrdC, and hrdD from Streptomyces coelicolor A3(2) having similarity to rpoD genes. Mol Gen Genet. 1991;229:334–340. doi: 10.1007/BF00267453. [DOI] [PubMed] [Google Scholar]
- 53.Tsinoremas N F, Ishiura M, Kondo T, Andersson C R, Tanaka K, Takahashi H, Johnson C H, Golden S S. A sigma factor that modifies the circadian expression of a subset of genes in cyanobacteria. EMBO J. 1996;15:2488–2495. [PMC free article] [PubMed] [Google Scholar]
- 54.Tumer N E, Robinson S J, Haselkorn R. Different promoters for the Anabaena glutamine synthetase gene during growth using molecular or fixed nitrogen. Nature (London) 1983;306:337–342. [Google Scholar]
- 55.Ueki T, Inouye S. A new sigma factor, SigD, essential for stationary phase is also required for multicellular differentiation in Myxococcus xanthus. Genes Cells. 1998;3:371–385. doi: 10.1046/j.1365-2443.1998.00197.x. [DOI] [PubMed] [Google Scholar]
- 56.Vieira J, Messing J. The pUC plasmids, an M13mp7-derived system for insertion mutagenesis and sequencing with synthetic universal primers. Gene. 1982;19:259–268. doi: 10.1016/0378-1119(82)90015-4. [DOI] [PubMed] [Google Scholar]
- 57.Wealand J L, Myers J A, Hirschberg R. Changes in gene expression during nitrogen starvation in Anabaena variabilis ATCC 29413. J Bacteriol. 1989;171:1309–1313. doi: 10.1128/jb.171.3.1309-1313.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wolk C P, Elhai J, Kuritz T, Holland D. Amplified expression of a transcriptional pattern formed during development of Anabaena. Mol Microbiol. 1993;7:441–445. doi: 10.1111/j.1365-2958.1993.tb01135.x. [DOI] [PubMed] [Google Scholar]
- 59.Wolk C P, Ernst A, Elhai J. Heterocyst metabolism and development. In: Bryant D A, editor. The molecular biology of cyanobacteria. Vol. 1. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1994. pp. 769–823. [Google Scholar]
- 60.Wu J, Newton A. Regulation of the Caulobacter flagellar gene hierarchy; not just for motility. Mol Microbiol. 1997;24:233–239. doi: 10.1046/j.1365-2958.1997.3281691.x. [DOI] [PubMed] [Google Scholar]