Abstract
Pruritus is a common symptom of dermatological disorders and has a major negative impact of quality of life. Previously, it was suggested that skin derived β-defensin peptides elicit itch through activation of mast cells. Here we investigated, in more detail, the mechanisms by which β-defensins induce itch by defining the receptors activated by these peptides in humans and mice, by establishing their action in vivo, and examining their expression in dermal diseases. We found in psoriasis and atopic dermatitis, elevated expression of DEFB103 is highly correlated with skin lesions. We showed that the peptide encoded by this and related genes activate Mas-related G protein-coupled receptors with different potencies that are related with their charge density. Furthermore, we establish that although these peptides can activate mast cells, they also activate sensory neurons, with the former cells being dispensable for itch reactions in mice. Together our studies highlight that specific β-defensins are likely endogenous pruritogens that can directly stimulate sensory neurons.
Introduction
Itch is a common symptom in patients with inflammatory skin disorders including atopic dermatitis (AD) and psoriasis (Leader et al., 2015, Lerner, 2018). This unpleasant sensation and the urge to scratch is ultimately triggered by the activation of neural circuits (Carstens and Akiyama, 2016). Even though it is long been known that the degranulation of mast cells and release of histamine evokes itch through stimulation of sensory nerves, antihistamine treatment often fails to prevent chronic itch (Leslie et al., 2015). However, the identity of endogenous pruritic agents in these dermatological diseases is still unknown although it has been speculated that various peptides are involved. These peptides include neuropeptides, somatostatin, VIP and substance P released by dermal nerve fibers, as well as peptides released by immune cells including eosinophil major basic protein (PRG2), and eosinophil major basic proteins (RNASE 2 & 3) and also the antimicrobial peptides, β-defensins and cathelicidin (CAMP)(Takahashi and Yamasaki, 2020). In addition, various other peptides like PAMP-12 (ADM) and proteases, for instance kallikreins and cathepsins that activate protease activated receptors have been suggested to be pruritogenic (Akiyama et al., 2015).
Defensins are short cationic peptides with net charges ranging from +2 to +11 that, based on their structures, have been divided into two groups, α-defensins and β-defensins, (Niyonsaba et al., 2016). These peptides are antimicrobial, their expression is induced during skin inflammation (Dhople et al., 2006), and they are considered to be danger associated molecular patterns (DAMPS) causing inflammation (Niyonsaba et al., 2016).
It was shown that human β-defensin-2 and -3 (HBD-2 and HBD-3), can activate mast cells and cause mast cell degranulation (Subramanian et al., 2013, Zhang and McNeil, 2019) via Mas-related g-protein-coupled receptors (Mrgpr) (Zhang and McNeil, 2019). However, whether β-defensins have roles in human itch has not been fully examined and knowledge for how defensins act mechanistically is fragmentary. Here we assessed, from dermatological samples sequencing data, whether defensin expression is altered in human diseases associated with itch. These analyses showed that the expression of the two β-defensins HBD-2, encoding DEFB4A and DEFB4B, and HBD-3, encoding DEFB103A and DEFB103B, is increased in psoriasis and AD. Although these peptides show elevated expression, we found that they are not equally potent activators of Mrgprs. Further, we found that β-defensins preferentially activate sensory neurons over mast cells. Together our studies provide additional insights into the action of β-defensins in pruritus with DEFB103 being the most potent pruritogen.
Results
Analysis of human transcriptomic data reveals that β-defensins are candidate pruritogens in inflammatory skin diseases.
To look for potential pruritogens in human skin diseases associated with chronic itch, we performed an unbiased analysis of publicly-available RNA-seq data (GSE121212, GSE54456) from skin biopsies of patients with psoriasis and AD comparing them with data from healthy donors (Li et al., 2014, Tsoi et al., 2019) searching for genes that encode secreted peptides with increased expression. This analysis revealed DEFB103 and DEFB4 (encoding human β-defensin 3 and 2, respectively) as the most up-regulated genes in both psoriasis and AD (Figure 1a). Previously it was reported that mast cells can be activated by β-defensins and other basic secretagogues including some peptides (McNeil et al., 2015, Subramanian et al., 2013). Therefore, we used MA-plots to simultaneously examine the levels of up-regulation and expression levels of genes encoding these two β-defensins and other cationic peptides including LL-37 (Cathelicidin, encoded by CAMP), PAMP-12 (ADM), cortistatin-14 (CORT), substance P (TAC1), somatostatin (SST), vasoactive intestinal peptide (VIP), eosinophil major basic protein (PRG2), and eosinophil major basic proteins (RNASE 2 & 3) or other pruritogenic agents including kallikreins (KLK5&7) and cathepsins (CTSS, CTSG) (Figure 1bc). These analyses revealed compared to other basic secretagogues, these two β-defensins are the most differentially and abundantly expressed genes in inflammatory skin samples. The log2 fold changes (log2FC) for DEFB4 were 4.1~8.4 in AD and psoriasis which respectively indicated 17~338 fold increased expression in these samples. Among the genes we examined, we noticed that LL-37 (CAMP) was also modestly upregulated in both AD and psoriasis (log2FC 2.3~3.4). However, the expression levels (counts per million, cpm) of DEFB103 and DEFB4 were 50~1000 folds higher than that of CAMP. Some peptides (kallikreins, cathepsins, PAMP) showed high basal expressions which were not appreciably up regulated in inflamed skin (log2FC ≈ 0). Expression of substance P, on the other hand, was downregulated in both inflammatory skin samples. These analyses suggested that β-defensins may have a more predominant role than LL-37 or other known pruritogenic peptides in itch.
Figure 1. Identification of DEFB4 and DEFB103 as potential pruritogens.
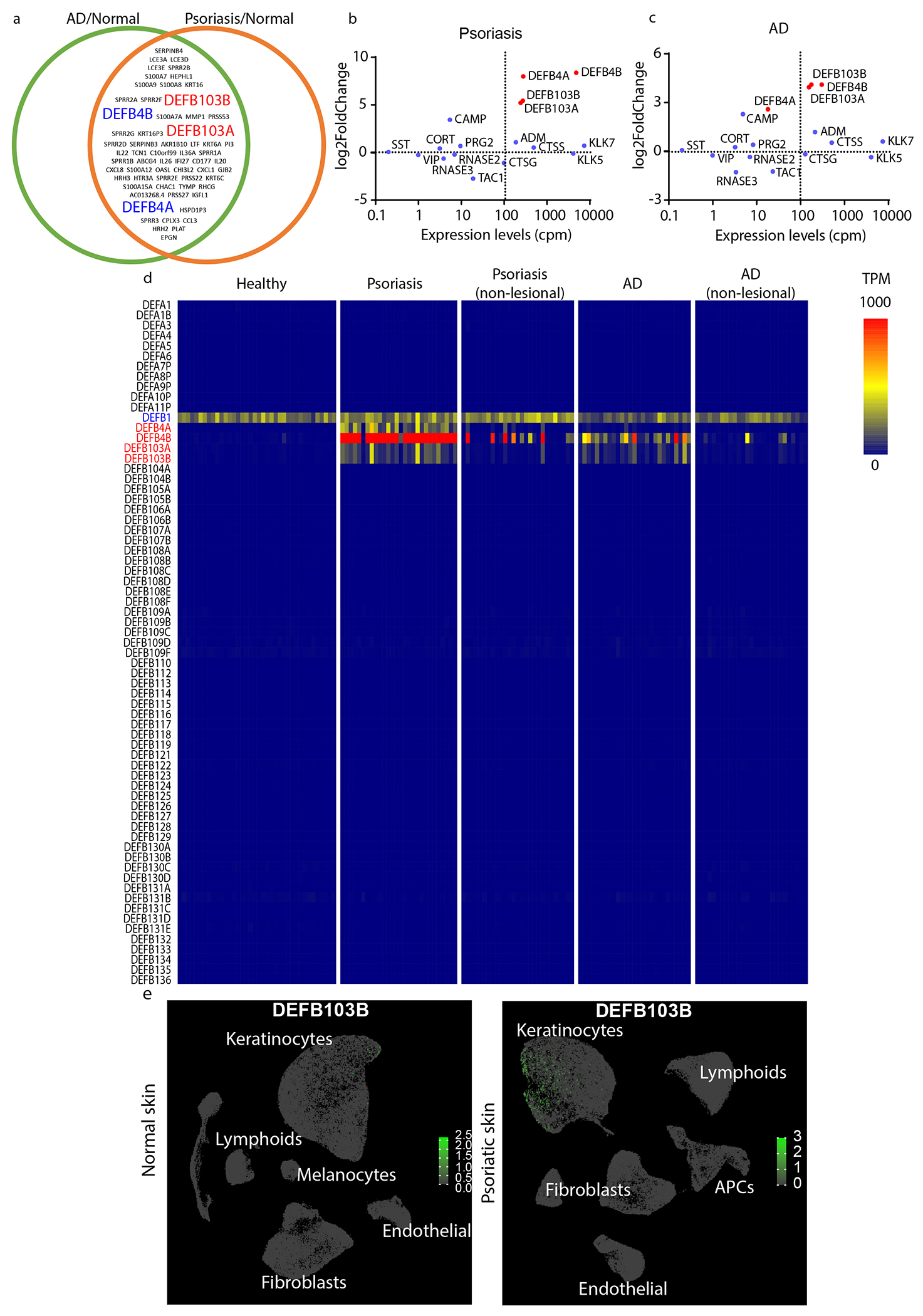
(a) Schematic of the top 100 differentially upregulated genes in skin biopsies from patients with AD and psoriasis. MA-Plot of defensins in skin samples from (b) psoriasis and (c) AD-patients (GSE121212). (d) Heatmap of gene expression levels (as transcript per million, TPM) of all defensin genes in healthy, lesion and non-lesion psoriasis, lesion and non-lesion AD skin samples (GSE121212, n=38, 28, 27, 27, and 27 respectively). (e) Single-cell RNA-sequencing analysis (UMAP) of normal (66,243 cells from 5 donors) and psoriatic skin samples (71,077 cells from 3 donors). Color bar indicates expression levels (TPM) at log 10 scale.
Since the human genome encodes 67 defensin genes, we also examined whether other defensins might be involved in itch by assessing their abundance in skin samples from healthy donors, as well as the samples from lesion sites and non-lesion sites of psoriasis and AD (Figure 1d). This analysis showed DEFB1 is constitutively expressed in healthy, lesion, and non-lesion skin, DEFB4 is highly expressed in psoriatic and AD skin, however, in some patients DEFB4 is also highly expressed in non-lesion skin. By contrast, DEFB103 is selectively expressed in lesion skin, but not in non-lesioned skin. Interestingly, analysis of a third dataset which was recently reported for a very large cohort of patients with psoriasis showed that DEFB103 is again prominently expressed (Reynolds et al., 2021). In addition, since this report provided single cell sequencing of dermal cells, we examined the cells that express DEFB103. This revealed that keratinocytes and not immune cells express DEF103B in psoriasis (Figure 1e)(Reynolds et al., 2021). Together, our analyses showed that DEFB4 and DEFB103 are candidate basic secretagogues in psoriasis and AD, and with regard to inflammation, DEFB103 is the most relevant.
Mouse β-defensins exhibit distinct pro-inflammatory features in vivo.
To examine the activation potencies of DEFB103 and DEFB4 on mast cells, we performed experiments with LAD2-cells, an immortalized human mast-cell cell line (Kirshenbaum et al., 2003). Interestingly, calcium imaging revealed activation by DEFB103 is very different from DEFB4 (Figure 2ab), with DEFB103 being about 10-fold more potent (Figure 2c; EC50s of 0.74±0.23 μM and 7.0±6.3 μM for DEFB103 and DEFB4 respectively) and these recorded potencies likely reflect the extent of degranulation (Subramanian et al., 2013). To investigate whether the different in vitro potencies of these peptides produce similar effects in vivo, we next tested them in mice. Differences in the naming and the low sequence identity of defensins across species makes it challenging to easily define the mouse orthologs of human defensins. Mouse Defb14 has been suggested to be the mouse ortholog of human DEFB103 (Hinrichsen et al., 2008, Rohrl et al., 2008) with 75.6% sequence identity and Defb4 has been suggested to be the mouse ortholog of human DEFB4 (Jia et al., 2000), but its structure is less well conserved with 48.7% identity (Figure 2de). Based on this, we decided to test the effects of mouse Defb14, and Defb4 in vivo on mast cells using Evans Blue to measure the extent of mast cell induced extravasation upon injection of peptide into the footpad. In agreement with our results from LAD2 cells, recombinant Defb14, the ortholog of human DEFB103, caused extensive extravasation (Figure 2f). By contrast, Defb4 caused only minimal extravasation, a level that was not statistically different from saline injection (Figure 2g). Together, these results show that human DEFB103 and mouse Defb14 are effective mast cell activators.
Figure 2. β-defensins exhibit mixed capability for activation of mast cells.
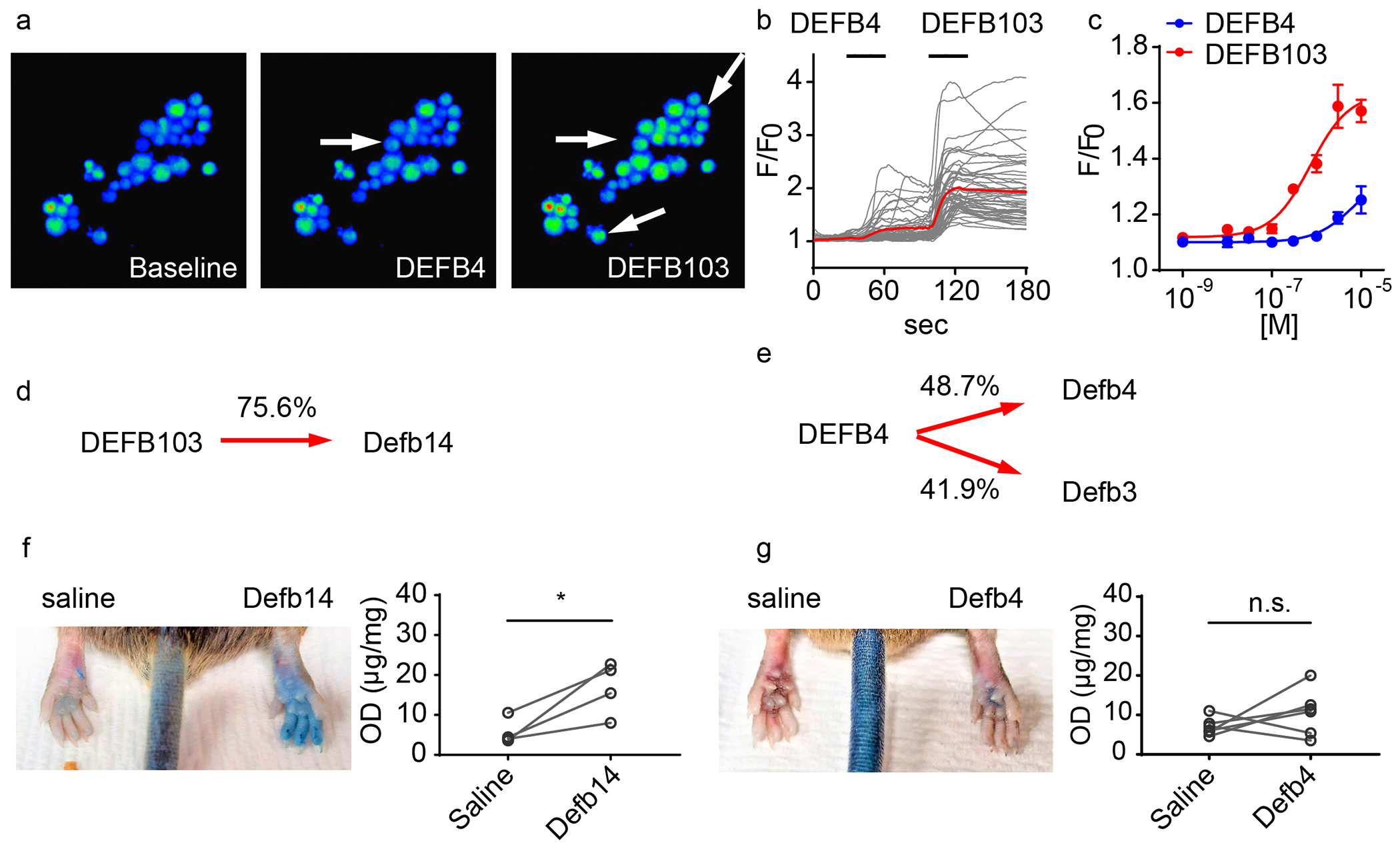
(a) Calcium imaging of fields of LAD2 mast cells treated with recombinant DEFB4 and DEFB103 peptides (1μM). DEFB103 activated more LAD2 cells and evoked stronger calcium influx than the DEFB4 (indicated by arrows). (b) Individual cellular (black lines) and averaged calcium responses (red traces), evoked by β-defensins, in LAD cells (c) Dose-dependent responses of calcium influx evoked by β-defensin peptides in LAD2 cells. (d-e) Primary sequence alignments of human and mouse β-defensins. (f-g) Evans-blue extravasation assay for evaluating mast cell activation in vivo. (e) Plantar subcutaneous injection of recombinant Defb14 (1mM, 10μL) triggered significant extravasation (p=0.036, paired T-test, n=4). (f) In contrast, extravasation evoked Defe4 was not statistically different from saline injection (p=0.29, paired T-test, n=6).
β-defensins evokes itch by stimulating Mrgpra3-sensory neurons.
Given the difference in the ability of β-defensin peptides to activate mast cells, we wondered whether these peptides also exhibit different pruritogenic potencies. We reanalyzed RNA-Seq data of mouse skin samples from the MC903-induced mouse model of atopic-like dermatitis (GSE90883) (Oetjen et al., 2017) and data from imiquimod-induced model of psoriasiform dermatitis (GSE161084) to determine whether β-defensins contribute to inflammatory itch in these mouse models and which ones might be involved. In both models, Defb1 and Defb6 are constitutively highly expressed, while the expression of Defb14, Defb4, and Defb3 is significantly upregulated (Figure 3a–b). These results confirm our analysis of human transcriptomic data by showing in mouse models of chronic itch, like in human dermal skin diseases, the expression of select defensins is upregulated. Since Defb3 is also up-regulated in inflamed skin and shares moderate sequence identity (41.9%) with human DEFB4 (Figure 2e), we examined itch-responses evoked by, Defb14, Defb4, Defb1, as well as Defb3. These peptides induced scratching, but there were major differences in evoked itch-responses. Defb14 (1mM, 10μL) evoked the most intense itch phenotype, while Defb4 and Defb1 peptides induced effects that were not significantly different from saline injection. Intriguingly, Defb3 peptide also evoked a strong itch phenotype despite the fact that it shares a similar sequence to Defb4 which had negligible effects (Figure 3c). The latter result suggests that there are likely factors that contribute to pruritogenic potency which are not solely determined by degree of primary sequence homology (see next Section).
Figure 3. β-defensins directly activate sensory neurons.
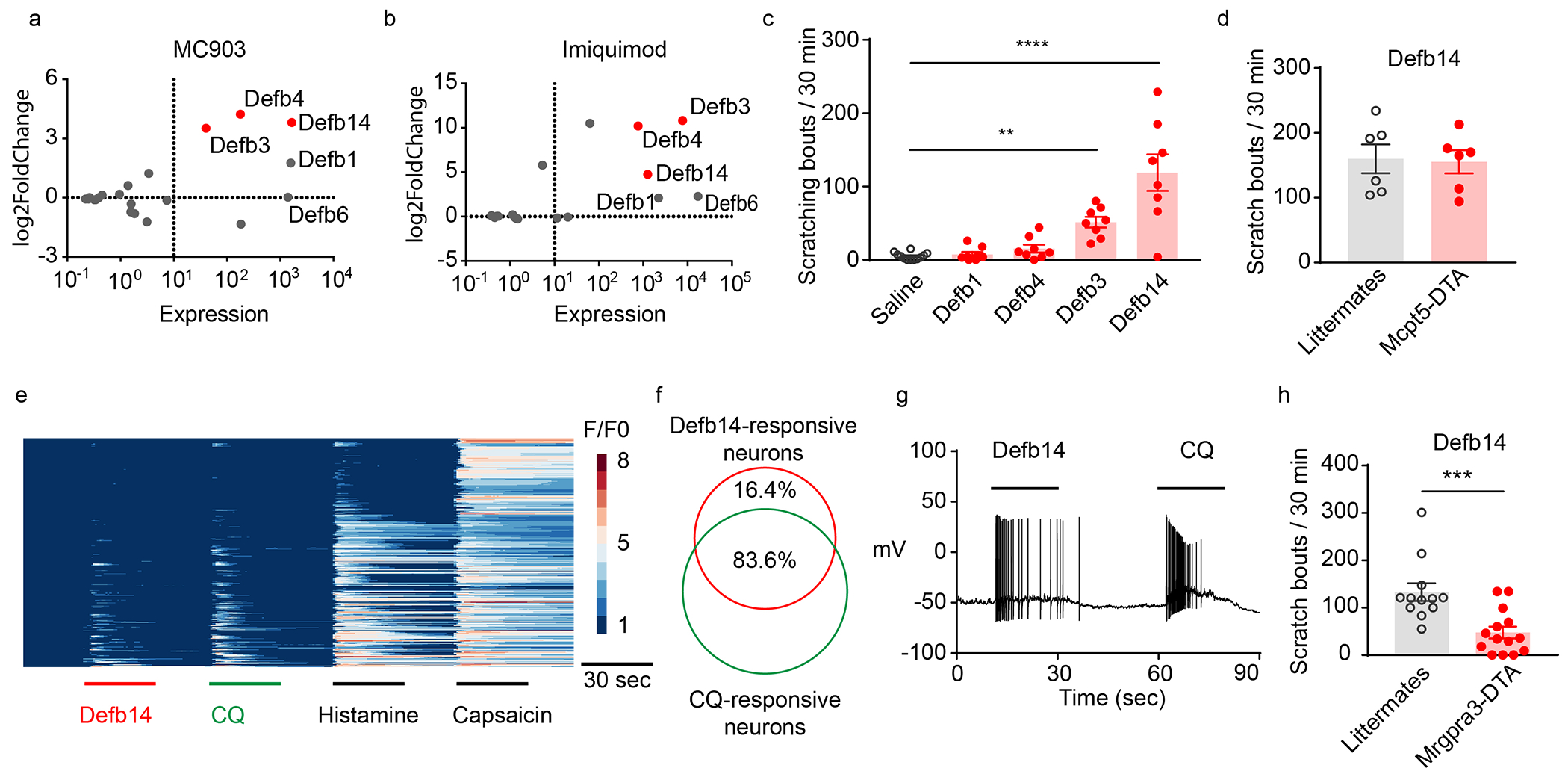
MA-Plot of defensins in skin samples from (a) MC903- (GSE90883) and (b) imiquimod-treated mice (GSE161084). (c) Scratching bouts elicited by intradermal injection of 1mM of the indicated recombinant β-defensins and saline (n=8-12). Defb3 and Defb14 induced scratching was significantly greater than saline, p = 0.003 and p < 0.0001, respectively (ANOVA Fisher’s LSD post-hoc analysis). (d) Mast-cells deficient mice (Mcpt5-DTA) and wild-type controls exhibited similar responses to Defb14 (1mM, p=0.87, two-tailed T-test). (e) Calcium imaging of 352 cultured DRG-neurons transfected with AAV9-GCaMP6f. (f) 46/55 Defb14-responsive neurons responded to CQ. (g) Whole-cell patch-clamp recording of Mrgpra3-Cre-GFP-neurons revealed that Defb14 (1μM) and CQ (100μM) evoked action potential discharges (n =8/8). (h) Scratching bouts of Mrgpra3-DTA mice (n=14) were significantly reduced compared to controls (n=12, p=0.007, two-tailed T-test).
To assess whether the pruritogenic effects of Defb14 are due to mast cell activation as previously suggested (Subramanian et al., 2013, Zhang and McNeil, 2019), we tested animals in which mast-cells are depleted by expression of diphtheria toxin fragment A under the control of a mast cell specific protease 5 gene, Mcpt5-Cre::Rosa-stop-DTA mice (Peschke et al., 2015). Surprisingly, for a peptide thought to elicit itch-behavior through activation of mast cells, the ablation of mast cells did not attenuate Defb14 induced scratching (Figure 3d). This suggests that Defb14 might stimulate itch sensory neurons. To test this hypothesis, we performed calcium imaging on dissociated DRG neurons expressing calcium indicator GCaMP6f (Mishra et al., 2011, Solinski et al., 2019b). As postulated, Defb14 induced calcium increases in DRG neurons. Interestingly, the subset of DRG-neurons that were Defb14 responsive also responded to chloroquine, (and histamine and capsaicin) (Figure 3ef). The specific receptor for chloroquine is Mrgpra3 (Liu et al., 2009) and the neurons expressing this receptor co-express receptors for histamine and capsaicin, showing that Defb14 specifically stimulates Mrgpra3-neurons (Xing et al., 2020). To test this further, we performed patch-clamp recordings on dissociated genetically marked Mrgpra3-Cre-GFP neurons. As expected, applying Defb14 peptide (1μM) (and chloroquine) triggered action potentials discharges in these cells (Figure 3g). Additionally, corroborating that Mrpgra3-neurons are required for Defb14-induced itch, the ablation of Mrgpra3-neurons (Mrgpra3-Cre::Rosa-stop-DTA mice) significantly attenuated scratching responses evoked to Defb14 (1mM) (Figure 3h).
Mrgpr receptors are activated by specific β-defensins.
Previously, it was reported that human DEFB103 and DEFB4 as well as mouse Defb14 and Defb3 can activate human MRGPRX1 and MRGPRX2 and mouse Mrgpra3 and Mrgprc11 respectively (Zhang and McNeil, 2019). However, whether all these β-defensins share similar potencies across different MRG receptors is unclear. In order to determine the activation thresholds for receptors, we performed fluorescence Imaging Plate Reader (FLIPR) calcium imaging on the eight human MRGPRs. We tested recombinant DEFB103 and DEFB4 on HEK293 cells transiently expressing MRGPRs together with Gα15 subunit and the calcium indicator GCaMP6s. Our results showed that DEFB103 exhibits higher potency on MRGPRX1 than MRGPRX2 with EC50s of 0.2 μM and 0.7 μM, respectively (Figure 4a and Table 1), and with other receptors responding at higher concentrations (>10μM). In contrast, DEFB4 poorly activated MRGPRX2 with an EC50 of 17.5 μM and negligibly stimulated other MRGPRs (Figure 4b and Table 1). These results establish that human β-defensins relatively selectively activate MRGPRX1 and MRGPRX2 and that DEFB103 is considerably more potent at activating these receptors than DEFB4. Next, we performed a similar pharmacological assessment of mouse Mrgprs. We found that Defb14 activated Mrgprb2 as well as Mrgpra1 and Mrgpra3 with EC50s of 1.7μM, 0.6μM and 1.0μM respectively (Figure 4c). Defb14, with lower affinity, also activated other receptors including Mrgpra2b and Mrgprc11 (Table 2). These results show that β-defensins are capable of activating Mrgprb2 expressed in mast cells and can potently stimulate Mrgprs expressed in sensory neurons including several previously unappreciated receptors shown previously to be responsible for chronic itch (Zhu et al., 2017).
Figure 4. Characterization of Mrgprs activated by β-defensins.
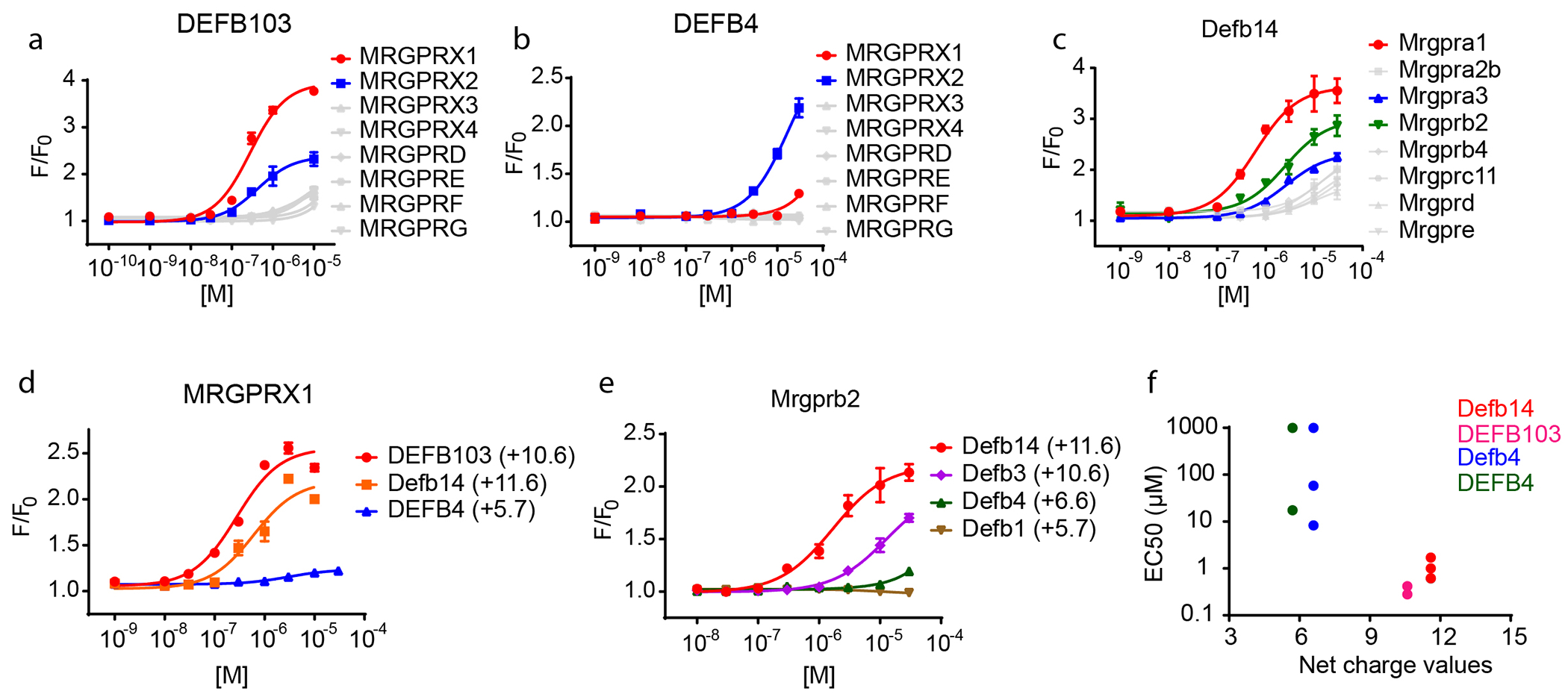
(a) FLIPR screening of HEK-293 cells heterologously expressing human MRG receptors revealed that DEFB103 potently activated human MRGPRX1 and MRGPRX2, (b) DEFB4 weakly activated MRGPRX2, and (c) Defb14 potently stimulated mouse Mrgpra1, Mrgpra3 and Mrgprb2. (d-f) The potency for activation of Mrgprs by defensins is related to the net charge values of peptides. (d) Human MRGPRX1 displayed higher affinity for DEFB103 (+10.6) and Defb14 (+11.6) than DEFB4 (+5.7). (e) Mouse Mrgpra1 exhibited higher affinity for Defb14, Defb3 (+10.6), than Defb4 (+6.6) and Defb1 (+5.7). All results are averages from 3–4 replicates ±SEM. (f) DEFB103 and Defb14 exhibited higher potency (≤ 1μM) in activating MGPRX1, MRGPRX2, Mrgpra1, Mrgpra3 and Mrgprb2. By contrast, DEFB4 and Defb4 exhibited low potency (≥ 10μM).
Table 1.
Summary of EC50s of human MRGPCRs (shown in μM) on β-defensin; n.d. indicates an EC50 that could not be estimated from the fitted data.
| Mrgpr/peptide | X1 | X2 | X3 | X4 | D | E | F | G |
|---|---|---|---|---|---|---|---|---|
| DEFB103 | 0.28±0.05 | 0.42±0.11 | 7.4±1.9 | n.d. | n.d. | n.d. | n.d. | n.d. |
| DEFB4 | 58.3±18 | 17.5±3.8 | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. |
| Defb14 | 0.61±0.17 |
Table 2.
Summary of EC50s of mouse Mrgpcrs (shown in μM) of mouse β-defensin; n.d. indicates an EC50 that could not be estimated from the fitted data.
| Mrgpr/peptide | A1 | A3 | B2 | A2b | C11 | B4 | D | E |
|---|---|---|---|---|---|---|---|---|
| Defb14 | 0.62±0.16 | 1.0±0.18 | 1.7±0.45 | 19.4±8.6 | 21.6±11.7 | n.d. | n.d. | n.d. |
| Defb3 | 2.7±0.6 | 17.5±3.8 | 11.5±3.5 | |||||
| Defb4 | n.d. | 58±12 | >999 | |||||
| Defb1 | n.d. | n.d. | n.d. |
It has been suggested that the bactericidal activity of β-defensins is related to their charge density (Bai et al., 2009, Kluver et al., 2005). To see if the charge density of β-defensins is also related with their potencies in activating MRG receptors, we calculated the net charge values (Moore, 1985) of the β-defensins we studied and found that the peptides with greatest net positive charge had the greatest potency for activation of MRG receptors. The β-defensins with net charge values higher than +10 (+10.6 ~ +11.6) exhibited higher potency than those with net charge values around +6 (+5.7 ~ +6.6) (Figure 4d–f). These results provide a rationale for the relative higher potency of Defb3 over Defb4 to elicit scratching in mice (Figure 3a), and to activate mast cells (Figure 2) and suggest that the net charge of β-defensins is a major determinant for inducing pruritus.
Co-expression of MRGPRX1 and histamine receptor in human sensory neurons.
Since, we found that DEFB103 is a potent ligand for MRGPRX1 and it is found at high levels in the skin of patients with AD and psoriasis this suggests that it is a strong candidate pruritogen in humans. It has also been reported that MRGPRX1 is expressed in human DRG (Flegel et al., 2015, Ray et al., 2018, Solinski et al., 2019a) and in neurons that express the peripheral itch neurotransmitter Nppb. To more fully characterize the expression of MRGPRX1 in sensory neurons, we examined its co-expression with HRH1 (histamine receptor) using in situ hybridization (ISH). As shown in Figure 5, all MRGPRX1 co-express HRH1 (122/122 neurons, 3 donors). Not all HRH1-positive neurons co-express MRGPRX1 (122/168 neurons).
Figure 5: MRGPRX1 is co-expressed with histamine receptor in DRG-neurons.
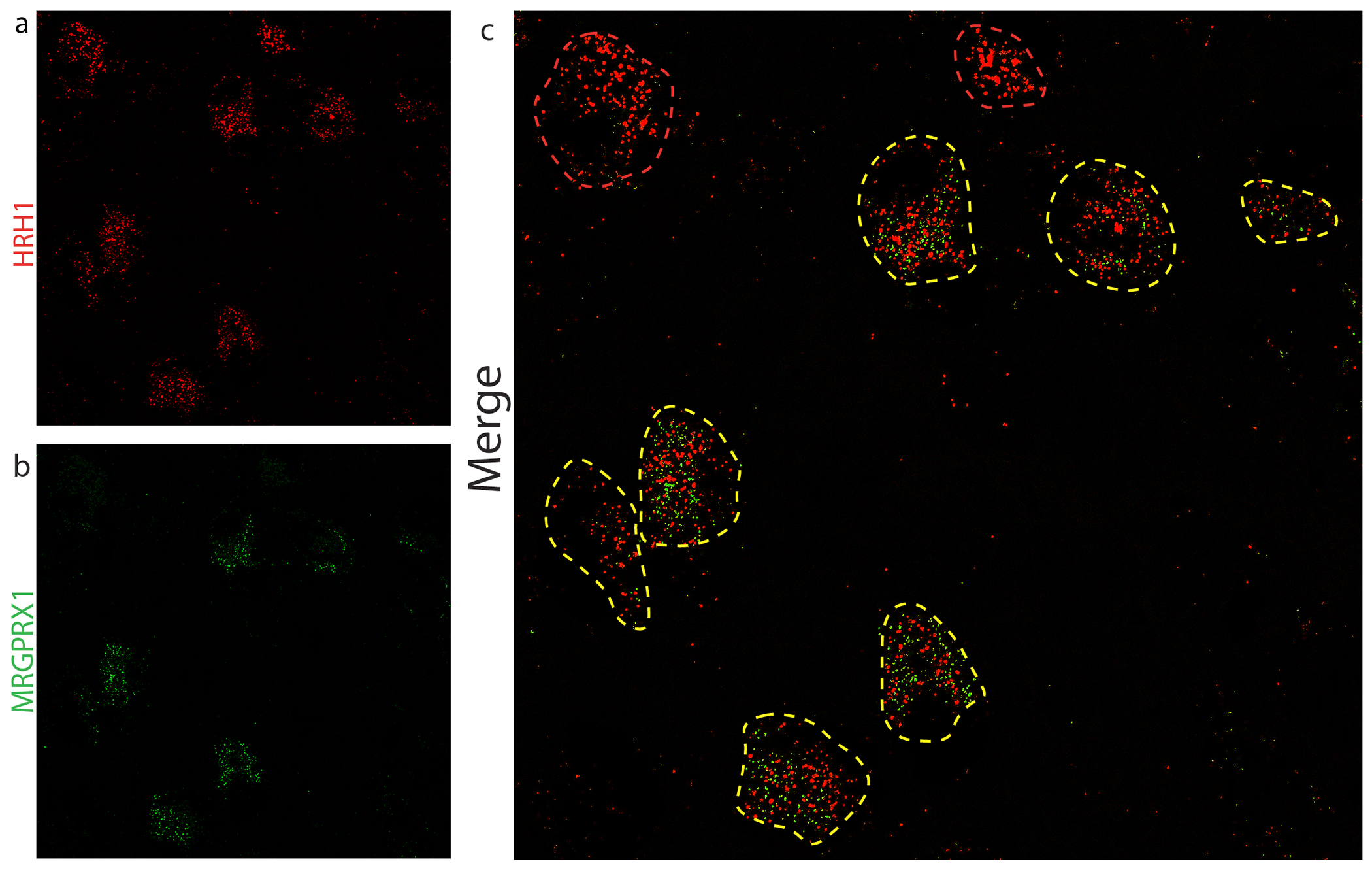
ISH of human DRG revealed that MRGPRX1-positive neurons (a) co-express HRH1 (b) as shown in merged image (c). Yellow outlines indicate double positive neurons and red profiles HRH1-alone-neurons.
Discussion
The skin is the first barrier protecting our bodies from environmental pathogens and the production of antimicrobial peptides (AMPs) is a primary method of defense. In addition, some of the AMP peptides are DAMPs (Seong and Matzinger, 2004). Here we investigated the potential mechanisms of action of the DAMP, β-defensins and uncovered several unappreciated and critical features of the way these molecules work. Unexpectedly, we exposed that, at least in mouse models of chronic itch, they act predominantly on sensory neurons. We further showed that β-defensins are not equally pruritogenic by examining the properties of several defensins which were previously not fully studied. Specifically, we show that DEFB103 and the mouse homolog Defb14 are the most pertinent with regard itch inducing effects with other β-defensins, such as Defb1 and Defb4, being much less effective. In addition, we showed that net charge is an important determinant in predicting the itch potency of these peptides. Lastly and importantly, we provide evidence that, in human conditions associated with chronic itch, β-defensins expression is elevated and we show that these peptides can activate both sensory neurons expressing MRGPRX1 (where it is co-expressed with histamine receptor) and immune cells expressing MRGPRX2 providing strong evidence that DEFB103 is likely an endogenous pruritogen in AD and psoriasis.
Previously, it was suggested that β-defensins can activate mast cells, however, whether β-defensins activate rodent mast cells is inconclusive (Chen et al., 2007, Niyonsaba et al., 2001, Subramanian et al., 2013). We speculate that these inconsistent results could be partly due to different β-defensins that were tested.
In addition, we found that mast cells are unnecessary to trigger scratching in mice. This may account for why anti-histaminergic treatment are largely ineffective at alleviating itch in many dermal diseases. However, we note that basophils and eosinophils, cells not eliminated in our studies, are reported to express MRGPRX2 (Wedi et al., 2020). These cell-types might also contribute to itch in the mouse model we used and may be active in certain itch exacerbations (Wang et al., 2021). Additionally, activation of Mrgprb2 in mast cells was shown to elicit itch through non-histaminergic processes (Meixiong et al., 2019). Interestingly, it was recently reported that, tick-secreted β-defensins can activate MRG receptors on sensory neurons to induce itch. The EC50 of the described defensin for human MRGPRX1 is 4.32 μM (Li et al., 2021), a potency about 15 folds lower than DEFB103. In agreement with the idea that charge ratio is a critical determinant in potency, the net charge value for tick defensin is +5.8 versus +10.6 for DEFB103. Therefore overall, our studies and the findings of others show that β-defensins directly activate sensory neurons but may also stimulate various immune cells to indirectly induce itch. Our findings provide clarification of the mechanisms involved in β-defensin induced pruritus and offer further confirmation that they are endogenous pruritogens.
Materials and Methods
Transcriptome analyses
RNA sequence reads were mapped to Ensembl reference transcriptomes GRCh38.v98 (Homo sapiens) and GRCm38.v98 (Mus musculus) and counted by kallisto (Bray et al., 2016). Raw counts were imported into Rx64 and RStudio by tximport (Soneson et al., 2015) for differential gene expression analysis (DGEA) by DESeq2 (Love et al., 2014) and differentially expressed genes were plotted using http://jvenn.toulouse.inra.fr/app/index.html.
Animals
Experiments using mice followed NIH guidelines and were approved by the National Institute of Dental and Craniofacial Research ACUC. 6-12 week old male and female mice were used; C57BL/6N (Envigo), ROSA-stop-DTA (Jax)(Ivanova et al., 2005), Mrgpra3-Cre-eGFP (Han et al., 2013), and Mcpt5-Cre (Peschke et al., 2015).
Extravasation
Hind paws were injected subcutaneously with 10 μL β-defensin peptides (GenScript USA Inc. and Biomatek USA LLC), and an equal volume saline was injected into contralateral paw. Evans blue was used to assay extravasation as described (Zhang and McNeil, 2019).
Itch behavior
Behavioral assessment of scratching was conducted as described (Solinski et al., 2019b). Bouts were defined as scratching events directed toward the site of injection from lifting hind leg from the ground to returning it.
Mast cell culture and imaging
LAD2 cells were maintained in StemPro-34 (GIBCO) supplemented with recombinant human stem cell factor (100 ng/ml), penicillin/streptomycin and as described previously (Kirshenbaum et al., 2003). LAD2 cells were loaded with Fluo-8 AM for 30 minutes at 37°C, washed twice in HBSS and incubated for 30-minute at room temperature.
Dorsal root ganglion neuron culture and imaging
Primary cultures of DRG neurons were generated from C57BL/6N mice as described previously (Li et al., 2017). Briefly, DRG were incubated in 5 mg/mL Collagenase/Dispase (10269638001, Millipore-Sigma) for 30 minutes, cells mechanically dissociated, seeded on poly-D lysine-coated coverslips and cultured for 48 hours (DMEM/F12, 10% FBS, penicillin/streptomycin, 100 ng/mL NGF, 50 ng/mL GDNF). For calcium imaging, DRG neurons were transfected with AAV9-Syn-GCaMP6.
Electrophysiology
Whole cell current and voltage clamp were performed on Mrgrpa3-Cre-GFP neurons as described (Tseng, 2019). Extracellular solution contained (in mM) 140 NaCl, 4 KCl, 2 CaCl2,1MgCl2, 10HEPES, and 10 glucose, pH 7.4 and osmolality 310 mOsm/kg. Pipette solution contained 140 KCl, 1MgCl2, 1 EGTA, 10 HEPES, 3 ATP, and 0.5 GTP, pH 7.4 and osmolality of 300 mOsm/kg.
Fluorescence Imaging Plate Reader (FLIPR) Screening
HEK293 cells were cultured in DMEM/F12 supplemented with 10% FBS, 100 U/ml penicillin, and 100 μg/ml streptomycin. For transient expression, 8 x 105 cells were seeded, cultured for 24 hours, and transfected using TransIT-293 (Mirus Bio). Cells were transfected with GCaMP6s, Gα15 subunit, together with individual MRG receptor. Expression vectors for MRGPRX1, MRGPRX2, MRGPRX3, MRGPRX4, MRGPRD, MRGPRE, MRGPRF, MRGPRG, were obtained from Addgene. Mouse Mrgpr expression vectors for Mrgpra1, Mrgpra2b, Mrgpra3, Mrgprb2, Mrgprb4, Mrgprc11, Mrgprd were obtained from Dr. Xinzhong Dong at Johns Hopkins University (Dong et al., 2001) and for Mrgpre (GenScript Inc). After 48 hours, cells were plated at 20,000 cells per well in 96-well plates with HBSS (with calcium and magnesium) and measurements made.
Peptide net charge value calculation
The net charge Z of a peptide at a certain pH can be estimated by calculating
where Ni are the number, and pKi the pKa values, of the N-terminus and the side chains of arginine, lysine, and histidine. The j-index pertain to the C-terminus and the aspartate, glutamate, cysteine, tyrosine (Moore, 1985).
In situ hybridization
Multi-labeling ISH was performed using the RNAscope® technology (ACD, Newark, CA) according to the manufacturer’s instructions on human DRG. Images were collected on an Eclipse Ti (Nikon, Melville, NY) confocal laser-scanning microscope. Multiple sections were imaged and counted from three donors.
Statistical analysis
Prism 7.0 (GraphPad Software, La Jolla, CA) was used for statistical analyses and generation of figures. Differences were considered significant for *p < 0.05. Exact p values, definition and number of replicates as well as definitions of center and dispersion are given in the respective figure legends. The sample sizes used were similar to those generally used in the field. Heatmaps were generated with R package pheatmap.
Acknowledgements
This work was supported by the intramural research program of the National Institute of Dental and Craniofacial Research, National Institutes of Health, project ZIADE000721-20 (MAH). We thank Dr. Dean Metcalfe (NIAID, NIH) for providing LAD2 cells, and Dr Xinzhong Dong for Mrgpr expression vectors and MrgprA3-GFP-cre mice. This work utilized the computational resources of the NIH HPC Biowulf cluster (http://hpc.nih.gov) and human DRGs were obtained from the NIH NeuroBioBank at the University of Maryland, Baltimore, MD.
Footnotes
Conflicts of interest
The author(s) declare no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Data Availability Statement
Dataset related to this article can be found at https://www.ncbi.nlm.nih.gov/geo/ accession numbers GSE121212, GSE54456, GSE90883, GSE161084, and https://developmentcellatlas.ncl.ac.uk/datasets/hca_skin_portal .
References
- Akiyama T, Lerner EA, Carstens E. Protease-activated receptors and itch. Handb Exp Pharmacol 2015;226:219–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai Y, Liu S, Jiang P, Zhou L, Li J, Tang C, et al. Structure-dependent charge density as a determinant of antimicrobial activity of peptide analogues of defensin. Biochemistry 2009;48(30):7229–39. [DOI] [PubMed] [Google Scholar]
- Bray NL, Pimentel H, Melsted P, Pachter L. Near-optimal probabilistic RNA-seq quantification. Nat Biotechnol 2016;34(5):525–7. [DOI] [PubMed] [Google Scholar]
- Carstens E, Akiyama T. Central Mechanisms of Itch. Curr Probl Dermatol 2016;50:11–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Niyonsaba F, Ushio H, Hara M, Yokoi H, Matsumoto K, et al. Antimicrobial peptides human beta-defensin (hBD)-3 and hBD-4 activate mast cells and increase skin vascular permeability. Eur J Immunol 2007;37(2):434–44. [DOI] [PubMed] [Google Scholar]
- Dhople V, Krukemeyer A, Ramamoorthy A. The human beta-defensin-3, an antibacterial peptide with multiple biological functions. Biochim Biophys Acta 2006;1758(9):1499–512. [DOI] [PubMed] [Google Scholar]
- Dong X, Han S, Zylka MJ, Simon MI, Anderson DJ. A diverse family of GPCRs expressed in specific subsets of nociceptive sensory neurons. Cell 2001;106(5):619–32. [DOI] [PubMed] [Google Scholar]
- Flegel C, Schobel N, Altmuller J, Becker C, Tannapfel A, Hatt H, et al. RNA-Seq Analysis of Human Trigeminal and Dorsal Root Ganglia with a Focus on Chemoreceptors. PLoS One 2015;10(6):e0128951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han L, Ma C, Liu Q, Weng HJ, Cui Y, Tang Z, et al. A subpopulation of nociceptors specifically linked to itch. Nat Neurosci 2013;16(2):174–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinrichsen K, Podschun R, Schubert S, Schroder JM, Harder J, Proksch E. Mouse beta-defensin-14, an antimicrobial ortholog of human beta-defensin-3. Antimicrob Agents Chemother 2008;52(5):1876–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanova A, Signore M, Caro N, Greene ND, Copp AJ, Martinez-Barbera JP. In vivo genetic ablation by Cre-mediated expression of diphtheria toxin fragment A. Genesis 2005;43(3):129–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia HP, Wowk SA, Schutte BC, Lee SK, Vivado A, Tack BF, et al. A novel murine beta -defensin expressed in tongue, esophagus, and trachea. J Biol Chem 2000;275(43):33314–20. [DOI] [PubMed] [Google Scholar]
- Kirshenbaum AS, Akin C, Wu Y, Rottem M, Goff JP, Beaven MA, et al. Characterization of novel stem cell factor responsive human mast cell lines LAD 1 and 2 established from a patient with mast cell sarcoma/leukemia; activation following aggregation of FcepsilonRI or FcgammaRI. Leuk Res 2003;27(8):677–82. [DOI] [PubMed] [Google Scholar]
- Kluver E, Schulz-Maronde S, Scheid S, Meyer B, Forssmann WG, Adermann K. Structure-activity relation of human beta-defensin 3: influence of disulfide bonds and cysteine substitution on antimicrobial activity and cytotoxicity. Biochemistry 2005;44(28):9804–16. [DOI] [PubMed] [Google Scholar]
- Leader B, Carr CW, Chen SC. Pruritus epidemiology and quality of life. Handb Exp Pharmacol 2015;226:15–38. [DOI] [PubMed] [Google Scholar]
- Lerner EA. Pathophysiology of Itch. Dermatol Clin 2018;36(3):175–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leslie TA, Greaves MW, Yosipovitch G. Current topical and systemic therapies for itch. Handb Exp Pharmacol 2015;226:337–56. [DOI] [PubMed] [Google Scholar]
- Li B, Tsoi LC, Swindell WR, Gudjonsson JE, Tejasvi T, Johnston A, et al. Transcriptome analysis of psoriasis in a large case-control sample: RNA-seq provides insights into disease mechanisms. J Invest Dermatol 2014;134(7):1828–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Yang H, Han Y, Yin S, Shen B, Wu Y, et al. Tick peptides evoke itch by activating MrgprC11/MRGPRX1 to sensitize TRPV1 in pruriceptors. J Allergy Clin Immunol 2021;147(6):2236–48 e16. [DOI] [PubMed] [Google Scholar]
- Li Z, Tseng PY, Tiwari V, Xu Q, He SQ, Wang Y, et al. Targeting human Mas-related G protein-coupled receptor X1 to inhibit persistent pain. Proc Natl Acad Sci U S A 2017;114(10):E1996–E2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q, Tang Z, Surdenikova L, Kim S, Patel KN, Kim A, et al. Sensory neuron-specific GPCR Mrgprs are itch receptors mediating chloroquine-induced pruritus. Cell 2009;139(7):1353–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 2014;15(12):550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNeil BD, Pundir P, Meeker S, Han L, Undem BJ, Kulka M, et al. Identification of a mast-cell-specific receptor crucial for pseudo-allergic drug reactions. Nature 2015;519(7542):237–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meixiong J, Anderson M, Limjunyawong N, Sabbagh MF, Hu E, Mack MR, et al. Activation of Mast-Cell-Expressed Mas-Related G-Protein-Coupled Receptors Drives Non-histaminergic Itch. Immunity 2019;50(5):1163–71 e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra SK, Tisel SM, Orestes P, Bhangoo SK, Hoon MA. TRPV1-lineage neurons are required for thermal sensation. EMBO J 2011;30(3):582–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore DS. Amino acid and peptide net charges: A simple calculational procedure. Bioch Educ 1985;13 (1):10–1. [Google Scholar]
- Niyonsaba F, Kiatsurayanon C, Ogawa H. The role of human beta-defensins in allergic diseases. Clin Exp Allergy 2016;46(12):1522–30. [DOI] [PubMed] [Google Scholar]
- Niyonsaba F, Someya A, Hirata M, Ogawa H, Nagaoka I. Evaluation of the effects of peptide antibiotics human beta-defensins-1/−2 and LL-37 on histamine release and prostaglandin D(2) production from mast cells. Eur J Immunol 2001;31(4):1066–75. [DOI] [PubMed] [Google Scholar]
- Oetjen LK, Mack MR, Feng J, Whelan TM, Niu H, Guo CJ, et al. Sensory Neurons Co-opt Classical Immune Signaling Pathways to Mediate Chronic Itch. Cell 2017;171(1):217–28 e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peschke K, Dudeck A, Rabenhorst A, Hartmann K, Roers A. Cre/loxP-based mouse models of mast cell deficiency and mast cell-specific gene inactivation. Methods Mol Biol 2015;1220:403–21. [DOI] [PubMed] [Google Scholar]
- Ray P, Torck A, Quigley L, Wangzhou A, Neiman M, Rao C, et al. Comparative transcriptome profiling of the human and mouse dorsal root ganglia: an RNA-seq-based resource for pain and sensory neuroscience research. Pain 2018;159(7):1325–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds G, Vegh P, Fletcher J, Poyner EFM, Stephenson E, Goh I, et al. Developmental cell programs are co-opted in inflammatory skin disease. Science 2021;371(6527). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohrl J, Yang D, Oppenheim JJ, Hehlgans T. Identification and Biological Characterization of Mouse beta-defensin 14, the orthologue of human beta-defensin 3. J Biol Chem 2008;283(9):5414–9. [DOI] [PubMed] [Google Scholar]
- Seong SY, Matzinger P. Hydrophobicity: an ancient damage-associated molecular pattern that initiates innate immune responses. Nat Rev Immunol 2004;4(6):469–78. [DOI] [PubMed] [Google Scholar]
- Solinski HJ, Dranchak P, Oliphant E, Gu X, Earnest TW, Braisted J, et al. Inhibition of natriuretic peptide receptor 1 reduces itch in mice. Sci Transl Med 2019a;11(500). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solinski HJ, Kriegbaum MC, Tseng PY, Earnest TW, Gu X, Barik A, et al. Nppb Neurons Are Sensors of Mast Cell-Induced Itch. Cell Rep 2019b;26(13):3561–73 e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian H, Gupta K, Lee D, Bayir AK, Ahn H, Ali H. beta-Defensins activate human mast cells via Mas-related gene X2. J Immunol 2013;191(1):345–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi T, Yamasaki K. Psoriasis and Antimicrobial Peptides. Int J Mol Sci 2020;21(18). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tseng P-YZ, Qin; Li Zhea; Dong Xinzhong. MrgprX1 mediates neuronal excitability and itch through tetrodotoxin-resistant sodium channels. Itch 2019;4(3):1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsoi LC, Rodriguez E, Degenhardt F, Baurecht H, Wehkamp U, Volks N, et al. Atopic Dermatitis Is an IL-13-Dominant Disease with Greater Molecular Heterogeneity Compared to Psoriasis. J Invest Dermatol 2019;139(7):1480–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F, Trier AM, Li F, Kim S, Chen Z, Chai JN, et al. A basophil-neuronal axis promotes itch. Cell 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wedi B, Gehring M, Kapp A. The pseudoallergen receptor MRGPRX2 on peripheral blood basophils and eosinophils: Expression and function. Allergy 2020;75(9):2229–42. [DOI] [PubMed] [Google Scholar]
- Xing Y, Chen J, Hilley H, Steele H, Yang J, Han L. Molecular Signature of Pruriceptive MrgprA3(+) Neurons. J Invest Dermatol 2020;140(10):2041–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, McNeil BD. Beta-defensins are proinflammatory pruritogens that activate Mrgprs. J Allergy Clin Immunol 2019;143(5):1960–2 e5. [DOI] [PubMed] [Google Scholar]
- Zhu Y, Hanson CE, Liu Q, Han L. Mrgprs activation is required for chronic itch conditions in mice. Itch (Phila) 2017;2(3). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Dataset related to this article can be found at https://www.ncbi.nlm.nih.gov/geo/ accession numbers GSE121212, GSE54456, GSE90883, GSE161084, and https://developmentcellatlas.ncl.ac.uk/datasets/hca_skin_portal .


