Abstract
Limited proteolysis of the Escherichia coli cell division inhibitor MinC reveals that its dimerization function resides in a structurally autonomous C-terminal domain. We show that cytoplasmic MinC is poised near the monomer-dimer equilibrium and propose that it only becomes entirely dimeric once recruited to the membrane by MinD.
Correct positioning of the division septum in Escherichia coli depends on the coordinated action of the Min proteins (3, 18). MinC and MinD interact to form a division inhibitor that periodically blocks the polar division sites by oscillating from pole to pole in an apparently synchronous manner (8, 15, 17). MinE protects the midcell division site from the effects of MinCD by an unknown mechanism. It was originally thought that MinE formed a static ring-like structure (the “E-ring”) near the center of the cell (16) and that it protected the midcell division site by dissociating the MinCD complex in the vicinity of this ring (10, 12). However, it was recently demonstrated that the E-ring is a dynamic structure that may shield the central division site by piloting MinCD away from midcell (5, 6).
The initiating event in cytokinesis is the formation at midcell of a circumferential ring of polymerized FtsZ (13, 18). MinC interacts directly with FtsZ (9), and it has been proposed that it blocks cell division either by destabilizing FtsZ polymers (9) or inhibiting recruitment of FtsA to the Z ring (11). Genetic dissection of MinC revealed that its MinD- and FtsZ-binding functions are physically separable: N-terminal (residues 1 to 115) and C-terminal (residues 116 to 231) MinC fragments were shown to be sufficient for interaction with FtsZ and MinD, respectively (7). We used limited trypsinolysis to better define the MinD-binding region and to determine whether it resides within a structurally autonomous domain.
It was previously shown that MinC is highly susceptible to degradation by Lon protease (19). However, we were able to stably overexpress E. coli minC as a fusion to the C terminus of Schistosoma japonicum glutathione S-transferase (GST), with a thrombin recognition site inserted between the two protein-coding sequences, in E. coli RC3 (lon+ ΔminCDE). The GST-MinC fusion protein was partially purified by passing the soluble cell fraction over a glutathione-Sepharose affinity column (Amersham Pharmacia Biotech [APB]), and then the fusion protein was cleaved on-column with thrombin to liberate unfused MinC. The semipure MinC was then purified to >95% homogeneity (as judged by sodium dodecyl sulfate-polyacrylamide gel electrophoresis [SDS-PAGE]) by using gel filtration chromatography on a Superdex 75 column (APB). The recombinant MinC is equivalent to wild-type except for an additional Gly-Ser (a vestige of thrombin cleavage) at the N terminus.
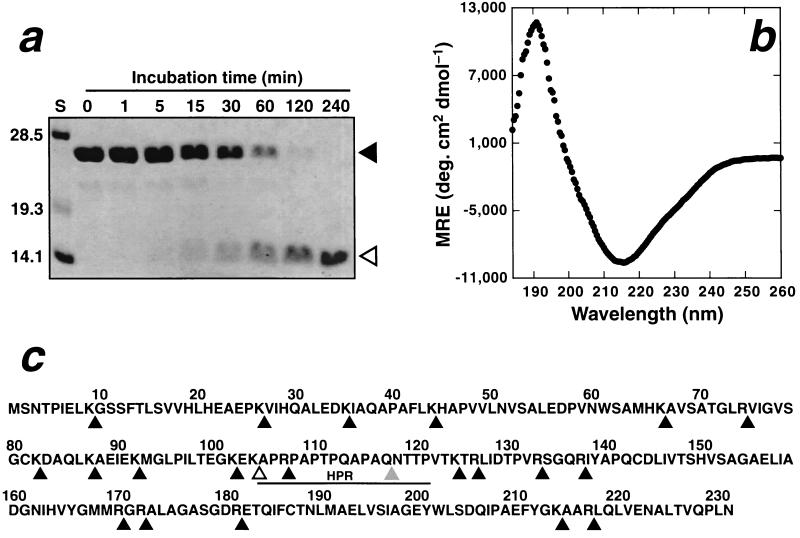
Tryptic digestion of recombinant MinC (25 kDa) led to rapid production of a 14-kDa fragment which remained resistant to further proteolysis for several hours (Fig. 1a). Sequencing and mass spectrometry revealed that this fragment comprised residues 105 to 231, corresponding to cleavage after Lys104 (marked with an unshaded arrowhead in Fig. 1c). This C-terminal fragment contains 10 potential trypsin cleavage sites as illustrated in Fig. 1c. The finding that these sites are protected from trypsinolysis suggests that MinC105–231 forms a tightly folded, structurally autonomous domain. Prolonged storage of MinC without addition of exogenous proteases led to a degradation product of ∼13 kDa (data not shown), which mass spectrometry and sequencing revealed to be residues 118 to 231, thus further delimiting the putative structural domain. Remarkably, this fragment corresponds almost exactly to the MinD-binding fragment (MinC116–231) generated by genetic dissection of MinC (7).
FIG. 1.
Trypsin digestion of MinC yields a stable C-terminal domain. (a) Trypsin was added to MinC at a MinC/trypsin mass ratio of 100:1, and the sample was incubated at room temperature for 240 min. Aliquots were removed from the digest mixture at the times indicated, and the reaction was terminated by addition of 10 mM 4-(2-aminoethyl)benzenesulfonyl fluoride, followed by boiling in SDS-PAGE loading buffer. The samples were then electrophoresed on an SDS-PAGE gel and visualized by staining with Coomassie blue. A MinC fragment of ∼14 kDa appeared within 15 min of trypsin addition and remained resistant to further digestion for the next 225 min. The sizes of the standards (lane S) are given in kilodaltons on the left of the gel. The running position of MinC and the trypsin-resistant fragment are indicated with closed and open arrowheads, respectively. (b) Far-UV CD spectrum of MinC118–231 (20 μM in 1 mM NaPi–50 mM NaF [pH 7.6]). The spectrum is the average of 16 transients acquired at 4°C on a Jasco-710 spectropolarimeter. Note the minimum in the mean residue ellipticity (MRE) at 215 nm. (c) Primary structure of E. coli MinC. Potential trypsin cleavage sites are indicated by the black arrowheads below the sequence. The unshaded arrowhead demarcates the N-terminal boundary of the trypsin-generated C-terminal fragment (MinC105–231), while the gray triangle indicates the site of spontaneous MinC cleavage. The HPR is underlined.
In order to test the hypothesis that this fragment corresponds to a structurally autonomous domain, we overproduced and purified recombinant MinC118–231 in exactly the same manner as described above for full-length MinC, except that the strain used for protein overexpression was HMS174 (lon+ minCDE). The far-UV circular dichroic (CD) spectrum of MinC118–231 (Fig. 1b) exhibited pronounced minima and maxima at 215 and 191 nm, respectively, which is characteristic of a predominantly β-sheet protein (20). In contrast, a largely α-helical protein would give rise to a CD spectrum with minima at 208 and 222 nm and a maximum near 190 nm, while an unstructured fragment would be expected to yield a deep minimum near 200 nm (20). A 15N-edited heteronuclear single quantum correlation nuclear magnetic resonance spectrum acquired from a sample of 15N-labeled MinC118–231 was also consistent with this fragment being highly structured (data not shown). Thus, we conclude that MinC118–231 is an autonomous structural domain that contains predominantly β-sheet secondary structure.
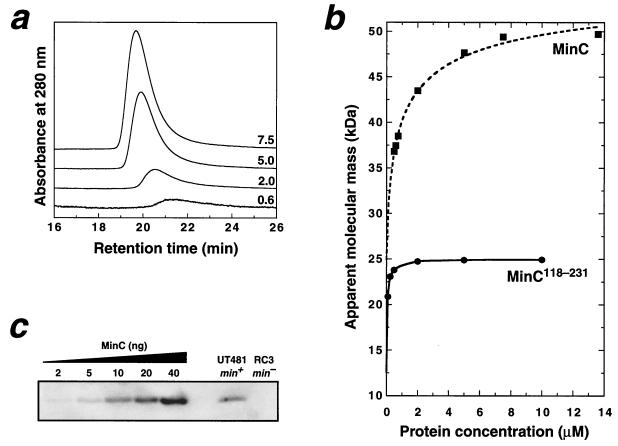
Both MalE-MinC and MalE-MinC116–231 fusion proteins were previously shown to self-associate, but it was uncertain whether the dominant species was dimer or trimer (7). We examined the self-association properties of unfused MinC and MinC118–231 by using gel filtration chromatography (Fig. 2a and b). At concentrations of ≥10 μM, both proteins eluted with retention times that corresponded almost exactly with the predicted dimer molecular weights (49.6 kDa for MinC and 25.0 kDa for MinC118–231). This indicates that the unfused proteins form dimers and not trimers.
FIG. 2.
Dimerization of MinC is mediated by the C-terminal domain. (a) A series of chromatograms resulting from application of different loading concentrations of MinC onto a Superdex 75 gel filtration column (APB). Each chromatogram is labeled with the MinC micromolar loading concentration; the vertical scale of the 0.6 μM data has been multiplied by a factor of 4 for clarity. The elution buffer was 10 mM Tris–50 mM NaCl–1 mM Tris(2-carboxyethyl)phosphine–0.02% NaN3 (pH 8.0), and the flow rate was 0.5 ml min−1. Note the increase in retention time as the protein concentration is reduced. (b) Apparent molecular mass of MinC and MinC118–231 on a Superdex 75 column as a function of protein concentration. The MinC data was fitted (dashed line) by using KaleidaGraph (version 3.04) as described previously (14) to yield a Kd value of 0.5 ± 0.2 μM and apparent monomer and dimer masses of 24.9 ± 0.5 and 57.1 ± 0.8 kDa, respectively. The solid curve is a simple interpolated fit to the MinC118–231 data. (c) Immunoblot of whole-cell extracts of wild-type E. coli UT481 and the isogenic ΔminCDE strain RC3. Note the absence of immunoreactive MinC in the RC3 extract. Recombinant MinC standards run on the same gel (lanes 1 to 5) gave a linear standard curve over the range of 2 to 40 ng of MinC/lane. In order to avoid potential underestimation of the MinC concentration due to limited protein binding capacity of the membranes, the standards were diluted in RC3 extract. Immunoblots were developed by using a chemifluorescent substrate (4) and were quantitated on a Storm 860 Fluorimager (Molecular Dynamics) using ImageQuant software (version 5.0).
As the MinC and MinC118–231 concentrations were lowered, the apparent molecular mass of both proteins decreased (i.e., the gel filtration retention time increased; see Fig. 2a). The change in apparent mass is due to rapid equilibrium between the monomer and dimer states with respect to the separation time (14); thus, as the protein concentration is decreased, the apparent mass shifts toward that expected for the monomer. Even though we could not obtain the entire dissociation profile for either protein because of the sensitivity limits of the detector, we were able to fit the MinC data (dashed line in Fig. 2b) to obtain a Kd estimate of 0.5 ± 0.2 μM, which is similar to the Kd previously determined for MinE dimerization (21). Surprisingly, while we could not obtain a curve fit to the MinC118–231 data, it is apparent from the dissociation profile (Fig. 2b) that the Kd is less than 0.1 μM. This indicates not only that the C-terminal domain is sufficient for MinC dimerization but also implies that the N-terminal domain may inhibit self-association. This contrasts with the observation that the N-terminal domain promotes self-association of a MalE-MinC fusion protein (7).
In order to calculate the oligomerization state of MinC in vivo, we used quantitative immunoblot analysis to determine the cellular concentration of MinC (Fig. 2c). Cells were counted as described for quantitation of MinD (2), and blots were developed, scanned, and quantitated as reported previously (4). We determined that the wild-type E. coli strain UT481 contains 400 ± 80 MinC molecules/cell (mean ± the standard deviation of seven immunoblots), which corresponds to a cellular concentration of ∼0.65 μM (based on a cytoplasmic volume of ∼1 fl) if all MinC molecules are assumed to be cytoplasmic. Interestingly, as found previously for MinE (21), this implies that cytoplasmic MinC is poised near the monomer-dimer equilibrium and only becomes fully dimeric once MinD recruits it to the cell membrane, thereby increasing its concentration 20- to 50-fold (9).
In summary, we demonstrated that the dimerization function of E. coli MinC resides in a structurally autonomous C-terminal domain. Both of the proteolytic cleavages which produced a stable C-terminal domain occurred within a hypervariable proline-rich region (HPR) of MinC marked by the solid line in Fig. 1c. All known MinC sequences from gram-negative bacteria can be aligned with minimal additions or deletions in the regions N- and C-terminal of the HPR. In contrast, the HPR is poorly conserved and varies from as few as 10 residues in Vibrio cholerae to as many as 42 residues in Pseudomonas aeroginosa. Thus, we postulate that all of the gram-negative MinC proteins comprise two structurally autonomous domains, an N-terminal FtsZ-binding domain (residues 1 to 104 in the E. coli sequence) and a C-terminal MinD-binding and/or dimerization domain (residues 118 to 231 in the E. coli sequence), separated by a flexible, hypervariable linker.
Following submission of this study for publication, the crystal structure of MinC from Thermotoga maritima was reported (1), and it is consistent with many of the results presented here. First, as we predicted, the protein comprises N- and C-terminal domains separated by a short flexible linker. Second, despite limited sequence identity (26%), the T. maritima C-terminal domain (residues 103 to 206, which correspond to residues 126 to 231 in the E. coli protein) corresponds very closely to the protease-resistant C-terminal fragment of E. coli MinC (i.e., residues 118 to 231). Third, the T. maritima C-terminal domain consists entirely of β-sheet secondary structure, a finding consistent with the CD spectrum of the E. coli domain. Finally, as we found for E. coli MinC, the T. maritima protein was found to dimerize via its C-terminal domain. Thus, the T. maritima MinC structure appears to be a good model for E. coli MinC despite the limited sequence identity between the two proteins.
Acknowledgments
This work was supported by grant AI48583 from the National Institutes of Health (to G.F.K.).
We thank Lawrence Rothfield for supplying anti-MinC antiserum and Assen Marintchev for help with Kd measurements.
REFERENCES
- 1.Cordell S C, Anderson R E, Löwe J. Crystal structure of the bacterial cell division inhibitor MinC. EMBO J. 2001;20:2454–2461. doi: 10.1093/emboj/20.10.2454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.de Boer P A J, Crossley R E, Hand A R, Rothfield L I. The MinD protein is a membrane ATPase required for the correct placement of the Escherichia coli division site. EMBO J. 1991;10:4371–4380. doi: 10.1002/j.1460-2075.1991.tb05015.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.de Boer P A J, Crossley R E, Rothfield L I. A division inhibitor and a topological specificity factor coded for by the minicell genetic locus determine proper placement of the division septum in E. coli. Cell. 1989;56:641–649. doi: 10.1016/0092-8674(89)90586-2. [DOI] [PubMed] [Google Scholar]
- 4.Feucht A, Lucet I, Yudkin M D, Errington J. Cytological and biochemical characterization of the FtsA cell division protein of Bacillus subtilis. Mol Microbiol. 2001;40:115–125. doi: 10.1046/j.1365-2958.2001.02356.x. [DOI] [PubMed] [Google Scholar]
- 5.Fu X, Shih Y-L, Zhang Y, Rothfield L I. The MinE ring required for proper placement of the division site is a mobile structure that changes its cellular location during the Escherichia coli division cycle. Proc Natl Acad Sci USA. 2001;98:981–985. doi: 10.1073/pnas.031549298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hale C A, Meinhardt H, de Boer P A. Dynamic localization cycle of the cell division regulator MinE in Escherichia coli. EMBO J. 2001;20:1563–1572. doi: 10.1093/emboj/20.7.1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hu Z, Lutkenhaus J. Analysis of MinC reveals two independent domains involved in interaction with MinD and FtsZ. J Bacteriol. 2000;182:3965–3971. doi: 10.1128/jb.182.14.3965-3971.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hu Z, Lutkenhaus J. Topological regulation of cell division in Escherichia coli involves rapid pole to pole oscillation of the division inhibitor MinC under the control of MinD and MinE. Mol Microbiol. 1999;34:82–90. doi: 10.1046/j.1365-2958.1999.01575.x. [DOI] [PubMed] [Google Scholar]
- 9.Hu Z, Mukherjee A, Pichoff S, Lutkenhaus J. The MinC component of the division site selection system in Escherichia coli interacts with FtsZ to prevent polymerization. Proc Natl Acad Sci USA. 1999;96:14819–14824. doi: 10.1073/pnas.96.26.14819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Huang J, Cao C, Lutkenhaus J. Interaction between FtsZ and inhibitors of cell division. J Bacteriol. 1996;178:5080–5085. doi: 10.1128/jb.178.17.5080-5085.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Justice S S, García-Lara J, Rothfield L I. Cell division inhibitors SulA and MinC/MinD block septum formation at different steps in the assembly of the Escherichia coli division machinery. Mol Microbiol. 2000;37:410–423. doi: 10.1046/j.1365-2958.2000.02007.x. [DOI] [PubMed] [Google Scholar]
- 12.King G F, Shih Y-L, Maciejewski M W, Bains N P S, Pan B, Rowland S L, Mullen G P, Rothfield L I. Structural basis for the topological specificity function of MinE. Nat Struct Biol. 2000;7:1013–1017. doi: 10.1038/80917. [DOI] [PubMed] [Google Scholar]
- 13.Lutkenhaus J. The regulation of bacterial cell division: a time and place for it. Curr Opin Microbiol. 1998;1:210–215. doi: 10.1016/s1369-5274(98)80013-1. [DOI] [PubMed] [Google Scholar]
- 14.Marintchev A, Robertson A, Dimitriadis E K, Prasad R, Wilson S H, Mullen G P. Domain specific interaction in the XRCC1-DNA polymerase β complex. Nucleic Acids Res. 2000;28:2049–2059. doi: 10.1093/nar/28.10.2049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Raskin D M, de Boer P A J. MinDE-dependent pole-to-pole oscillation of division inhibitor MinC in Escherichia coli. J Bacteriol. 1999;181:6419–6424. doi: 10.1128/jb.181.20.6419-6424.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Raskin D M, de Boer P A J. The MinE ring: an FtsZ-independent cell structure required for selection of the correct division site in E. coli. Cell. 1997;91:685–694. doi: 10.1016/s0092-8674(00)80455-9. [DOI] [PubMed] [Google Scholar]
- 17.Raskin D M, de Boer P A J. Rapid pole-to-pole oscillation of a protein required for directing division to the middle of Escherichia coli. Proc Natl Acad Sci USA. 1999;96:4971–4976. doi: 10.1073/pnas.96.9.4971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rothfield L I, Justice S, Garcia-Lara J. Bacterial cell division. Annu Rev Genet. 1999;33:423–448. doi: 10.1146/annurev.genet.33.1.423. [DOI] [PubMed] [Google Scholar]
- 19.Sen M, Rothfield L I. Stability of the Escherichia coli division inhibitor protein MinC requires determinants in the carboxy-terminal region of the protein. J Bacteriol. 1998;180:175–177. doi: 10.1128/jb.180.1.175-177.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Woody R W. Theory of circular dichroism of proteins. In: Fasman G D, editor. Circular dichroism and the conformational analysis of biomolecules. New York, N.Y: Plenum Press; 1996. pp. 25–67. [Google Scholar]
- 21.Zhang Y, Rowland S L, King G F, Braswell E, Rothfield L I. The relationship between hetero-oligomer formation and function of the topological specificity domain of the Escherichia coli MinE protein. Mol Microbiol. 1998;30:265–273. doi: 10.1046/j.1365-2958.1998.01059.x. [DOI] [PubMed] [Google Scholar]