Abstract
Several Sphingomonas spp. utilize polyethylene glycols (PEGs) as a sole carbon and energy source, oxidative PEG degradation being initiated by a dye-linked dehydrogenase (PEG-DH) that oxidizes the terminal alcohol groups of the polymer chain. Purification and characterization of PEG-DH from Sphingomonas terrae revealed that the enzyme is membrane bound. The gene encoding this enzyme (pegA) was cloned, sequenced, and expressed in Escherichia coli. The purified recombinant enzyme was vulnerable to aggregation and inactivation, but this could be prevented by addition of detergent. It is as a homodimeric protein with a subunit molecular mass of 58.8 kDa, each subunit containing 1 noncovalently bound flavin adenine dinucleotide but not Fe or Zn. PEG-DH recognizes a broad variety of primary aliphatic and aromatic alcohols as substrates. Comparison with known sequences revealed that PEG-DH belongs to the group of glucose-methanol-choline (GMC) flavoprotein oxidoreductases and that it is a novel type of flavoprotein alcohol dehydrogenase related (percent identical amino acids) to other, so far uncharacterized bacterial, membrane-bound, dye-linked dehydrogenases: alcohol dehydrogenase from Pseudomonas oleovorans (46%); choline dehydrogenase from E. coli (40%); l-sorbose dehydrogenase from Gluconobacter oxydans (38%); and 4-nitrobenzyl alcohol dehydrogenase from a Pseudomonas species (35%).
Polyethylene glycols (PEGs) are industrially produced in large quantities and in a broad spectrum of sizes and derivatives. In view of the application of these xenobiotics, after use they mainly show up in sewage water but are easily biologically degraded by many aerobic bacteria belonging to different genera (3, 19).
PEG metabolization has been reported for several sphingomonads, either in an axenic (e.g., S. macrogoltabidus [8]) or in a mixed culture (e.g., S. terrae with a Rhizobium sp. [4]). In all cases, it has been established that the first step in the degradation pathway involves the oxidation of the terminal alcohol groups with a dye-linked dehydrogenase (5), here called PEG-DH. Originally the enzyme was considered to be a quinoprotein (7), as in Rhodopseudomonas acidophila (20, 21), but this could not be proven because the difficulties encountered in the purification of this membrane-bound enzyme prevented straightforward characterization and cofactor identification of it. However, sufficiently pure enzyme could be prepared for N-terminal amino acid sequence determination. Cloning of the gene (pegA) and overexpression of it in an Escherichia coli recombinant strain provided the quantities of enzyme required for full characterization. It is shown here that PEG-DH is not a quinoprotein but a new type of flavoprotein dehydrogenase with flavin adenine dinucleotide (FAD) as a cofactor.
Purification of PEG-DH from S. terrae and sequencing of its N-terminal amino acids.
PEG-DH from S. terrae was purified from a symbiotic mixed culture (E-1) consisting of S. terrae and a Rhizobium sp. Growth and purification were carried out essentially as described (9). (The organism has been reidentified and deposited as S. terrae IFO 15098 [16].) The final preparation appeared to be pure, as judged from sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) followed by protein staining (not shown). N-terminal amino acid sequencing of the protein was carried out with the Shimadzu protein sequencer PPSQ-10, revealing the sequence MHKFDFVV.
Cofactor analysis.
The final enzyme preparation was incubated at 80°C for 20 min, cooled, and centrifuged to remove the denatured protein. The supernatant was applied to a μBondasphere 5-μm C18 300-Å column (Waters) and chromatographed with an isocratic system of 35% methanol–65% 10 mM sodium phosphate buffer (pH 5.7). The absorbance of the eluate was monitored at 450 nm. The absorption spectrum of the supernatant was measured with a spectrophotometer (Beckman DU-65), and the FAD content was calculated from the absorbance at 450 nm (Ε450 nm = 11,300 M−1 cm−1). The Fe and Zn content in the purified enzyme was measured with a Nippon Jarrell Ash AA-8200 atomic absorption spectrophotometer.
Cloning and sequencing of the gene.
Five different adapter-ligated DNA libraries were constructed and subjected to PCR with the primer designed on the basis of the N-terminal amino acid sequence of S. terrae PEG-DH. DNA fragments of about 550 and 300 bp were amplified from the EcoRV- and PvuII-digested DNA libraries, respectively. The nucleotide sequence of the 300-bp fragment was completely included in that of the 550-bp fragment, and the deduced amino acid sequences of both fragments were identical to the determined N-terminal amino acid sequence of PEG-DH (MHKFDFVV). Primers were designed on the basis of the nucleotide sequence of the 550-bp fragment to obtain the complete nucleotide sequence of PEG-DH, and DNA fragments of about 2.2 and 1.5 kbp for downstream and upstream nucleotide sequences from the 550-bp fragment, respectively, were obtained from the EcoRV-digested DNA library.
The nucleotide sequence of the 3,666-bp DNA including the open reading frame of pegA of S. terrae can be found in the DDBJ, EMBL, and GenBank data banks under accession number AB050784. An open reading frame from a start codon ATG at position 1277 to a stop codon TAG at position 2881 encodes a polypeptide of 535 amino acid residues, and a calculated molecular mass of 58,842 Da was deduced.
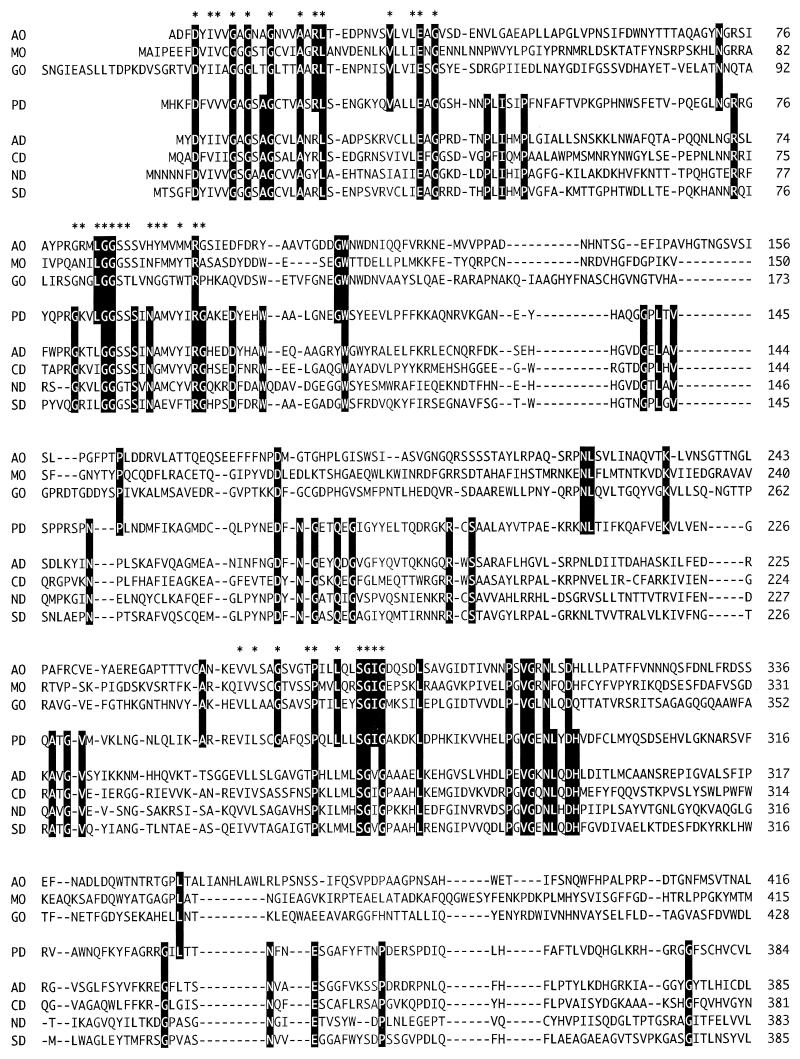
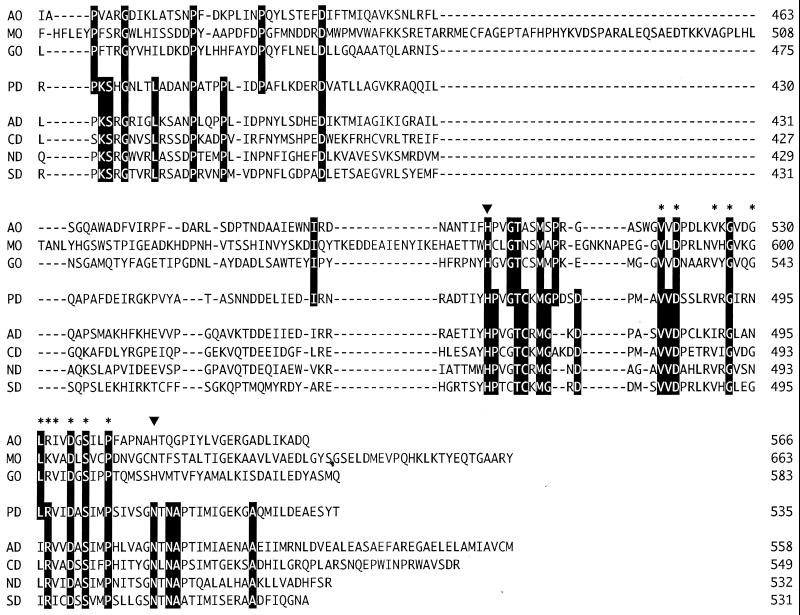
A database search for proteins with significant amino acid sequence similarity to that of S. terrae PEG-DH revealed a number of enzymes (Fig. 1) which are 46 to 35% identical. It appears that PEG-DH has the characteristic signatures of FAD binding and the catalytic residues (H467 and N511) found in glucose-methanol-choline (GMC) oxidoreductases (1), a group of enzymes having noncovalently bound FAD as a cofactor. It also revealed several membrane-bound, dye-linked bacterial alcohol dehydrogenases where this high similarity has remained unnoticed so far. Since the cellular role of oxidases and dehydrogenases is different, it seemed interesting to compare the oxidases and dehydrogenases as a group.
FIG. 1.
Sequence alignment of S. terrae PEG-DH and GMC oxidoreductases. Gaps, indicated by a dash, were introduced in the sequences to maximize homology. Identical amino acid residues are indicated by reversal letters. The amino acid residues of PEG-DH corresponding to the FAD-binding domain and the catalytic residues of GMC oxidoreductases are indicated by an asterisk and an arrowhead, respectively. Abbreviations: AO, aryl-alcohol oxidase from Pleurotus eryngii (18); MO, alcohol (methanol) oxidase from Candida boidinii (15); GO, glucose oxidase from Aspergillus niger (11); PD, PEG-DH from S. terrae; AD, alcohol dehydrogenases from Pseudomonas oleovorans (17); CD, choline dehydrogenase from E. coli (12); ND, 4-nitrobenzyl alcohol dehydrogenase from a Pseudomonas species (2); SD, l-sorbose dehydrogenase from Gluconobacter oxydans (14).
This comparison showed that although the two groups have many residues in common (mainly the FAD binding regions), probably related to their functional differences, they also have their own characteristic residues as a group (Fig. 1). Although, with the exception of PEG-DH, the dehydrogenases have scarcely been characterized, based on the conserved signatures and active-site residues, it is clear that they contain FAD and belong to the group for which the name flavoprotein alcohol dehydrogenases has been proposed. The conserved amino acids for this group revealed now by the comparison could be used as targets for protein engineering, leading to better understanding of this new type of alcohol dehydrogenase.
Expression and purification of recombinant PEG-DH.
The pegA gene was inserted into the upstream region of a histidine tag gene and into the downstream region of a T7 promoter in the pET23d vector, using appropriate primers which are available on request. After IPTG (isopropylthiogalactoside) addition, E. coli cells harboring pPEGDH-EX produced an extra protein of 58 kDa whose expression level attained about 8% of the total protein. The recombinant PEG-DH was expressed in soluble form by the recombinant E. coli strain, but purification of the enzyme led to large losses in activity. This could be prevented by adding detergent to the buffers, dodecyl-β-maltoside (0.2%) giving the best results. Previous studies on the localization of PEG-DH showed that it is bound to membrane particles (6, 8, 9). This is in line with the present findings, i.e., the hydrophobic character requiring the presence of detergent for solubilization of the enzyme from S. terrae and stabilization of the recombinant enzyme in chromatography. Since no leader sequence is present, PEG-DH could be attached to the membrane, but in the absence of hydrophobic stretches in the sequence, the underlying binding mechanism for this is unclear. Purification of the recombinant PEG-DH with Ni-nitrilotriacetic acid affinity chromatography in the indicated way yielded a preparation that showed only one band (at 58 kDa) with SDS-PAGE. An 11-fold purification with an 89% yield was obtained with this purification step.
Characterization of recombinant PEG-DH.
PEG-DH activity was assayed by measuring the initial rate of 2,6-dichlorophenolindophenol (DCIP) reduction at 600 nm at 30°C. The reaction mixture consisted of 0.1 M Tris-HCl, pH 9.0, containing 0.2 M KCl, 0.1 mM DCIP, 0.4 mM benzyl alcohol, and an appropriate amount of enzyme. One unit is defined as the amount of enzyme that reduces 1 μmol of DCIP per min (ε600 = 22,000 M−1 cm−1).
Using the indicated assay system, a pH optimum of 9.0 was found. The optimum is caused by the pH-dependent variation of the Km value for DCIP, showing a minimum value at 9.0. Although, even at this pH value, the standard concentration of DCIP (0.1 mM) is far from saturating (a threefold higher activity was found with 0.4 mM DCIP), for practical reasons no higher concentration was used. The addition of salts increased the activity, so that 0.2 M KCl was always included in the assay buffer. Since benzyl alcohol is one of the best substrates (Table 1), it was used as standard substrate (0.4 mM). Using the indicated assay system, the purified recombinant PEG-DH showed a specific activity of 18.2 U/mg of protein (with 5 mM PEG 6000, a value of 11.1).
TABLE 1.
Apparent kinetic parameter valuesa
| Substrate | Vmax (U/mg of protein) | Km (mM) |
|---|---|---|
| Benzyl alcohol | 17.8 | 0.6 |
| 1-Heptanol | 23.5 | 0.4 |
| 1-Hexanol | 17.0 | 0.5 |
| 1-Propanol | 3.3 | 6.6 |
| TEG | 14.7 | 14 |
| TEG-monoethyl ether | 6.6 | 4.2 |
| PEG 300 | 10.8 | 0.8 |
| PEG 400 | 10.2 | 0.7 |
| PEG 1000 | 12.1 | 1.6 |
| PEG 6000 | 10.5 | 2.4 |
Values were determined with the standard assay in which various concentrations of substrate replaced benzyl alcohol.
Insignificant activities (below 0.5% of that obtained with benzyl alcohol) were observed for ethylene glycol, glycerol, 2,3-butanediol, 2,4-pentanediol, sorbitol, l-sorbose, choline, or secondary alcohols. A low activity (approximately 3%) was observed for diglycolic acid (but only using the extremely high concentration of 0.15 M), octanal, and benzaldehyde.
Aliphatic alcohols with a size of about C6 and benzyl alcohol appeared to be the best substrates (Table 1). However, substituting the phenyl ring or extending the aliphatic chain strongly decreased the activity (p-methoxybenzyl alcohol, vanillyl alcohol, and 2-phenyl ethanol showing 3, 3, and 25% of the activity for benzyl alcohol, respectively). The smaller PEGs, PEG 300 and PEG 400, were better substrates than the larger ones, PEG 1000 and PEG 6000, mainly because the latter have higher apparent Km values (Table 1), the observations suggesting a size restriction to the substrate. High apparent Km values were also observed for tetraethylene glycol (TEG = PEG 200) and its monoethyl ether. Since the value was somewhat better for the latter (Table 1), this suggests that in addition to a maximum, a minimum size is also required for adequate substrate binding. The SH group reagent p-chloromercuribenzoic acid (0.1 mM) and Cu2+ (1 mM) and Pb2+ (1 mM) strongly inhibited the enzyme. No inhibition was found with pyrazole or DACA (trans-4-[N,N-dimethylamino]-cinnamaldehyde).
Since PEG-DH is the sole PEG-oxidizing enzyme that could be found in cell extracts of PEG-utilizing sphingomonads (6, 8, 10), it is clear that this enzyme catalyzes the first step in oxidative PEG degradation by these bacteria. As shown here, besides alcohols, the enzyme also oxidizes aldehydes. Oxidation of aldehydes has been reported for several different types of alcohol dehydrogenase, but just as for PEG-DH, the rates for aldehydes are always low compared to those for the corresponding alcohols.
Determination of the molecular mass of the enzyme by standard gel filtration gave a value of 120 kDa. Since a value of 58 kDa was found with SDS-PAGE, and the enzyme is encoded by a single gene, recombinant PEG-DH is a homodimeric enzyme. On the other hand, it has been reported that PEG-DH directly isolated from S. terrae is a tetramer (9). However, since it is clear now that rapid aggregation and inactivation of the enzyme occur (which is not prevented by the presence of the previously used detergent lauryl betaine), the molecular mass was reassessed with fresh S. terrae enzyme.
Cofactor identification.
The absorption spectrum of the enzyme showed maxima at 375 and 450 nm (not shown), indicative of a flavoprotein. After heat denaturation, the compound in the supernatant showed an absorption spectrum and a retention time on high-pressure liquid chromatography identical to that of authentic FAD. From the absorbance at 450 nm, it was calculated that the enzyme contains 1.7 molecules of FAD per enzyme molecule. No Zn or Fe was detected in the enzyme.
An interesting feature is the presence of eight cysteine residues in PEG-DH, one of them (C472) being conserved in all the other alcohol dehydrogenases (Fig. 1). Zn plays a crucial role in certain types of NAD+-dependent alcohol dehydrogenases (a cysteinyl residue being one of its ligands) and Fe in another group (13). Since both are absent in PEG-DH, and addition of these metals to the assay mixture did not stimulate activity and addition of metal chelators did not inhibit it, apparently this type of alcohol dehydrogenase does not need the assistance of these metal ions in catalysis, in line with the observation that pyrazole and DACA were not inhibitors.
Acknowledgments
This work was supported by a grant-in-aid from the Ministry of Education, Science and Culture, the Ohara Foundation in Kurashiki, the Nagase Science Foundation, and the Rhobi-Teien Foundation.
We are grateful to H. Nishizaki for technical assistance in atomic absorption spectrophotometry.
M. Sugimoto and J. A. Duine contributed to an equal extent.
REFERENCES
- 1.Cavener D R. GMC oxidoreductases: a newly defined family of homologous proteins with diverse catalytic activities. J Mol Biol. 1992;223:811–814. doi: 10.1016/0022-2836(92)90992-s. [DOI] [PubMed] [Google Scholar]
- 2.James K D, Hughes M A, Williams P A. Cloning and expression of ntnD, encoding a novel NAD(P)+-independent 4-nitrobenzyl alcohol dehydrogenase from Pseudomonas sp. strain TW3. J Bacteriol. 2000;182:3136–3141. doi: 10.1128/jb.182.11.3136-3141.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kawai F. Breakdown of plastics and polymers by microorganisms. Adv Biochem Eng. 1995;52:151–194. doi: 10.1007/BFb0102319. [DOI] [PubMed] [Google Scholar]
- 4.Kawai F, Yamanaka H. Biodegradation of polyethylene glycol by symbiotic mixed culture (obligate mutualism) Arch Microbiol. 1986;146:125–129. doi: 10.1007/BF00402338. [DOI] [PubMed] [Google Scholar]
- 5.Kawai F, Kimura T, Tani Y, Yamada H, Ueno T, Fukami H. Identification of reaction products of polyethylene glycol dehydrogenase. Agric Biol Chem. 1983;47:1669–1671. [Google Scholar]
- 6.Kawai F, Yamanaka H. Inducible or constitutive polyethylene glycol dehydrogenase involved in the aerobic metabolism of polyethylene glycol. J Ferment Bioeng. 1989;67:300–302. [Google Scholar]
- 7.Kawai F, Yamanaka H, Ameyama M, Shinagawa E, Matsushita K, Adachi O. Identification of the prosthetic group and further characterization of a novel enzyme, polyethylene glycol dehydrogenase. Agric Biol Chem. 1985;49:1071–1076. [Google Scholar]
- 8.Kawai F, Enokibara S. Role of novel dye-linked dehydrogenases in the metabolism of polyethylene glycol by pure cultures of Sphingomonas sp. N6. FEMS Microbiol Lett. 1996;141:45–50. [Google Scholar]
- 9.Kawai F, Kimura T, Yamada H, Kurachi M. Purification and characterization of polyethylene glycol dehydrogenase involved in the bacterial metabolism of polyethylene glycol. Appl Environ Microbiol. 1980;40:701–705. doi: 10.1128/aem.40.4.701-705.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kawai F, Kimura T, Fukaya M, Tani Y, Ogata K, Ueno T, Fukami H. Bacterial oxidation of polyethylene glycol. Appl Environ Microbiol. 1978;35:679–684. doi: 10.1128/aem.35.4.679-684.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kriechbaum M, Heilmann H J, Wientjes F J, Hahn M, Jany K D, Gassen H G, Alaeddinoglu G. Cloning and DNA sequence analysis of the glucose oxidase gene from Aspergillus niger NRRL-3. FEBS Lett. 1989;255:63–66. doi: 10.1016/0014-5793(89)81061-0. [DOI] [PubMed] [Google Scholar]
- 12.Lamark T, Kaasen I, Eshoo M W, Falkenberg P, McDougall J, Strom A R. DNA sequence and analysis of the bet genes encoding the osmoregulatory choline-glycine betaine pathway of Escherichia coli. Mol Microbiol. 1991;5:1049–1064. doi: 10.1111/j.1365-2958.1991.tb01877.x. [DOI] [PubMed] [Google Scholar]
- 13.Reid M F, Fewson C A. Molecular characterization of microbial alcohol dehydrogenases. Crit Rev Microbiol. 1994;20:13–56. doi: 10.3109/10408419409113545. [DOI] [PubMed] [Google Scholar]
- 14.Saito Y, Ishii Y, Hayashi H, Imao Y, Akashi T, Yoshikawa K, Noguchi Y, Soeda S, Yoshida M, Niwa M, Hosoda J, Shimomura K. Cloning of genes coding for l-sorbose and l-sorbosone dehydrogenase from Gluconobacter oxydans and microbial production of 2-keto-l-gulonate, a precursor of l-ascorbic acid, in a recombinant G. oxydans strain. Appl Environ Microbiol. 1997;63:454–460. doi: 10.1128/aem.63.2.454-460.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sakai Y, Tani Y. Cloning and sequencing of the alcohol oxidase-encoding gene (AOD1) from the formaldehyde-producing asporogenous methylotrophic yeast Candida boidinii S2. Gene. 1992;114:67–73. doi: 10.1016/0378-1119(92)90708-w. [DOI] [PubMed] [Google Scholar]
- 16.Takeuchi M, Kawai F, Shimada Y, Yokota A. Taxonomic study of polyethylene glycol-utilizing bacteria: emended description of the genus Sphingomonas and new description of Sphingomonas macrogoltabidus sp. nov., Sphingomonas sanguis sp. nov., and Sphingomonas terrae sp. nov. Syst Appl Microbiol. 1993;16:227–238. [Google Scholar]
- 17.van Beilen J B, Eggink G, Enequist H, Bos R, Witholt B. DNA sequence determination and functional characterization of the OCT-plasmid-encoded alkJKL genes of Pseudomonas oleovorans. Mol Microbiol. 1992;6:3121–3136. doi: 10.1111/j.1365-2958.1992.tb01769.x. [DOI] [PubMed] [Google Scholar]
- 18.Varela E, Martínez M J, Martínez A T. Aryl-alcohol oxidase protein sequence: a comparison with glucose oxidase and other FAD oxidoreductases. Biochim Biophys Acta. 2000;1481:202–208. doi: 10.1016/s0167-4838(00)00127-8. [DOI] [PubMed] [Google Scholar]
- 19.White G F, Russell N J, Tidswell E C. Bacterial scission of ether bonds. Microbiol Rev. 1996;60:216–232. doi: 10.1128/mr.60.1.216-232.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yamanaka K. Polyethylene glycol dehydrogenase activity demonstrated by dye-linked alcohol dehydrogenase of Rhodopseudomonas acidophila M402. Agric Biol Chem. 1991;55:837–844. [Google Scholar]
- 21.Yasuda M, Cherepanov A, Duine J A. Polyethylene glycol dehydrogenase activity of Rhodopseudomonas acidophila derives from a type I quinohaemoprotein alcohol dehydrogenase. FEMS Microbiol Lett. 1996;138:23–28. [Google Scholar]