Abstract
Background
Obese type 2 diabetes mellitus (obese T2DM) is one of the prime diseases that endangers human health. Clinical studies have confirmed the ability of the Huanglian Huazhuo capsule to treat obese T2DM; however, its mechanism of action is still unclear. In this study, effects and mechanisms of the Huanglian Huazhuo capsule in obese T2DM were systematically investigated using network pharmacology and molecular docking techniques.
Methods
The active ingredients and targets of the Huanglian Huazhuo capsule were extracted from Traditional Chinese Medicine Systems Pharmacology Database and Analysis Platform (TCMSP). Obese T2DM diabetes-related targets were retrieved from a geographic dataset combined with a gene card database. A protein-protein interaction (PPI) network was constructed to screen core targets. The Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analyses were conducted using Database for Annotation Visualization and Integrated Discovery (DAVID). Interactions between potential targets and active compounds were assessed using molecular docking. Molecular docking was performed on the best core protein complexes obtained using molecular docking.
Results
A total of 89 and 108 active ingredients and targets, respectively, were identified. Seven core targets were obtained using a topological analysis of the PPI network. The GO and KEGG pathway enrichment analyses showed that the effects of the Huanglian Huazhuo capsules were mediated by inflammation, lipid response, oxidative stress-related genes, and HIF-1 and IL-17 signaling pathways. Good binding ability was observed between the active compounds and screened targets using molecular docking.
Conclusions
The active ingredients, potential targets, and pathways of the Huanglian Huazhuo capsule for the treatment of obese T2DM were successfully predicted, providing a new strategy for further investigation of its molecular mechanisms. In addition, the potential active ingredients provide a reliable source for drug screening in obese T2DM.
1. Introduction
Diabetes mellitus (DM) is a metabolic endocrine disease characterized by glucose and fat metabolism disorders and increased plasma glucose levels, and in the symptomatic stage, it is characterized by excessive drinking, polyphagia, polyuria, weakness, wasting, or sweet-tasting urine [1]. Relevant studies in recent years showed that the number of cases of obese type 2 diabetes mellitus (obese T2DM) with insulin resistance is gradually increasing. Obesity in such patients is primarily due to unreasonable diet structure and lack of exercise as well as the cause of insulin resistance [2], which are closely related. In addition, weight gain is an independent risk factor, particularly in central obesity, which predisposes to insulin resistance, leading to a high workload of pancreatic B cells and impaired islet function, resulting in lipolysis insulin inhibition [3].
In Chinese medicine, DM is not named but classifies as a “thirst disorder.” According to Traditional Chinese Medicine (TCM), the etiology of obese T2DM is complex, with congenital deficiency of endowment, emotional disorders, poor diet, and lack of exercise contributing to the disease. However, the congenital deficiency of endowment shows the most impact than others [4]. In the Huanglian Huazhuo capsule, Huanglian can clear away heat and dampness, stop diarrhea, and detoxify; Huangbai can clear away heat and dampness, purge fire, and detoxify; Danshen can activate blood circulation, remove blood stasis, and relieve pain; and Shanzha and Zhiqiao aurantii Immaturus can regulate qi, and relieve stagnation and swelling. Therefore, the Huanglian Huazhuo capsule has the functions of promoting qi and resolving phlegm, invigorating the spleen and eliminating accumulation, and removing dampness and blood stasis [5]. However, systematic studies on the mechanisms underlying the beneficial effects of the Huanglian Huazhuo capsule in T2DM are scarce, including the analyses of potential targets, biological processes, and metabolic pathways.
In Chinese medicine, traditional pharmacological approaches are limited to mechanistic studies [6]. In 2013, Shao Li proposed a new concept called “network pharmacology,” which provides a new strategy to determine the mechanism of action of herbal formulations [7]. Cyberpharmacology of TCM includes virtual computing, high-throughput data analysis, and web-based database search, involving bioinformatics network construction and network topology analysis [8]. This approach emphasizes multicomponent, multichannel, and multiobjective synergies and is well-suited for TCM analysis [8, 9]. Due to recent bioinformatics convergence, computational prediction-based network pharmacology is powerful to systematically reveal the biological mechanisms of complex diseases and drug effects at the molecular level [10, 11]. The potential mechanisms of TCM for the treatment of various diseases are increasingly studied using network pharmacology [12, 13]. Hence, major bioactive compounds, potential targets, and signaling pathways of the Huang Lian Huazhuo capsules were predicted using network pharmacology and molecular docking techniques in obese T2DM. The results provide the basis for investigating the mechanism of action of the Huanglian Huazhuo capsules in obese T2DM.
2. Materials and Methods
2.1. Data Collection
2.1.1. Active Ingredients and Corresponding Targets of the Huanglian Huazhuo Capsule
The active components in the Huanglian Huazhuo capsule were retrieved using Traditional Chinese Medicine Systems Pharmacology Database and Analysis Platform (TCMSP, https://old.tcmsp-e.com/tcmsp.php). “Huanglian, Huangbai, Danshen, Shanzha, and Zhiqiao” were the keywords of the compound. Furthermore, the full name of the target gene was converted into an abbreviation. TCMSP is a systematic pharmacology platform for Chinese herbal medicine, providing interactive data associated with the relationship among drugs, targets, and diseases [14]. In addition, the platform provides data on chemical, target, and drug-target networks and pharmacokinetic effects, including drug similarity (DL), oral bioavailability (OB), intestinal epithelial permeability, water solubility, and blood-brain barrier permeability, of natural compounds. A comprehensive Traditional Chinese Medicine Integrated Database (TCMID, http://www.megabionet.org/tcmid/) provides data for TCM [15]. The OB and DL were ≥30% and ≥0.18, respectively [16], and the active compounds were further analyzed [17]. OB describes the delivery capacity of oral drugs to the systemic circulation [18], and DL is based on the similarities of several known drugs in functional groups and physical properties [19].
2.1.2. Anti-Obese T2DM Targets of the Huanglian Huazhuo Capsule
The mRNA expression profile of an obese T2DM sample (GSE166467) was searched in the gene expression synthesis dataset (GEO: https://www.ncbi.nlm.nih.gov/GEO) [20]. The threshold for identifying differentially expressed genes (DEGs) using SVA and Limma packages in RStudio 4.2.1 (https://www.rstudio.com/products/rstudio/) was |log FC| > 1, P < 0.05 [21]. The DEGs were visualized to generate volcanic and thermal maps. Data on disease targets associated with obese T2DM are available in the GeneCards database (http://www.genecards.org/) [22] using the keyword “Obese T2DM” to screen disease genes and eliminate duplicate targets. The target intersection corresponding to the active ingredient and disease target was selected using Venny 2.1.0 (https://bioinfogp.cnb.csic.es/tools/venny/index.html), and the intersecting target was the action target of the Huanglian Huazhuo capsule in intervening obesity and diabetes.
2.1.3. Constructing a “Drug-Component-Target-Disease” Network
The drugs, active ingredients, and intersection targets of drugs and diseases were input in the Cytoscape 3.8.0 software (https://cytoscape.org/). The network diagram of “drugs-component-targets-diseases” was constructed, which was analyzed using the cytoNCA plug-in.
2.1.4. Construction and Analysis of Protein-Protein Interaction Network
The data of drug-disease intersection genes were imported to the STRING website (https://cn.string-db.org/). The species was restricted to “Homo sapiens,” and data with confidence levels higher than 0.90 were selected. The obtained data were imported into the Cytoscape 3.8.0 software for analysis, and graphs were generated by identifying the core genes of the network.
2.1.5. GO and KEGG Enrichment Analyses
Drug-disease intersection targets were subjected to GO and KEGG analyses using the RStudio 4.2.1 package and bioconductor package (https://mirrors.tuna.tsinghua.edu.cn/bioconductor), respectively. Enrichment analysis statistical filter values were set (P=0.05, Q = 0.05), and the screening results were visualized.
2.1.6. Construction of Compound-Target-Pathway Networks
Compound-target-pathways were constructed and analyzed using Cytoscape 3.8.0 to visualize and elucidate the complex associations among compounds, pathways, and targets.
2.1.7. Prediction of the Binding Ability of the Huanglian Huazhuo Capsule Core Components to the Target Using Molecular Docking Technique
The 3D structure of the target protein was downloaded from the Research Collaboratory for Structural Bioinformatics, Protein Data Bank (RSCB PDB) database (http://www.rcsb.org), and the water and protein impurities were removed using PyMOL (https://pymol.org/) software. The 2D structure of the active ingredient was downloaded from the PubChem database (https://pubchem.ncbi.nlm.nih.gov/) and imported into Chem3D software (https://www.chemdraw.com.cn/) to convert it into a 3D structure. The target proteins and small molecules were hydrogenated using AutoDockTools 1.5.6 software (https://autodock.scripps.edu/). The Grid Box was set up with the original ligand as the center, and the data were derived, followed by AutoDock Vina application for molecular docking. The target proteins and small molecules were visualized using the PyMoL software.
3. Results
3.1. Active Ingredients and Corresponding Targets of the Huanglian Huazhuo Capsule
From the TCMSP database, 57 active ingredients of “Danshen,” 22 of “Huangbai,” 10 of “Huanglian,” 6 of “Shanzha,” and 5 of “Zhiqiao” were found in the Huanglian Huazhuo capsule after screening. A total of 88 active ingredients were selected after combining and de-weighting all the data (Table 1).
Table 1.
Active ingredients in the Huanglian Huazhuo capsules.
| Name | Pubchem ID | Compound | OB | DL% |
|---|---|---|---|---|
| Danshen | 124416 | MOL0016011,2,5,6-t | 38.75 | 0.36 |
| Danshen | 5281330 | MOL001659Poriferas | 43.83 | 0.76 |
| Danshen | 457801 | MOL001771Poriferas | 36.91 | 0.75 |
| Danshen | 68081 | MOL001942Isoimpera | 45.46 | 0.23 |
| Danshen | 94162 | MOL002222Sugiol | 36.11 | 0.28 |
| Danshen | 128994 | MOL002651Dehydrota | 43.76 | 0.4 |
| Danshen | 64982 | MOL002776Baicalin | 40.12 | 0.75 |
| Danshen | 54711004 | MOL000569Digallate | 61.85 | 0.26 |
| Danshen | 5280445 | MOL000006luteolin | 36.16 | 0.25 |
| Danshen | 11593487 | MOL0070365,6-dihyd | 33.77 | 0.29 |
| Danshen | 135872 | MOL0070412-isoprop | 40.86 | 0.23 |
| Danshen | 1.01E+08 | MOL0070453α-hydro | 44.93 | 0.44 |
| Danshen | 14609851 | MOL0070494-methyle | 34.35 | 0.23 |
| Danshen | 95223221 | MOL0070502-(4-hydr | 62.78 | 0.4 |
| Danshen | 14609847 | MOL007058formyltan | 73.44 | 0.42 |
| Danshen | 10995850 | MOL0070593-beta-Hy | 32.16 | 0.41 |
| Danshen | 105118 | MOL007061Methylene | 37.07 | 0.36 |
| Danshen | 16090911 | MOL007063Przewalsk | 37.11 | 0.65 |
| Danshen | 16102114 | MOL007064Przewalsk | 110.32 | 0.44 |
| Danshen | 622085 | MOL007068Przewaqui | 62.24 | 0.41 |
| Danshen | 56967683 | MOL007069Przewaqui | 55.74 | 0.4 |
| Danshen | 10470747 | MOL007070(6S,7R)-6 | 41.31 | 0.45 |
| Danshen | 126073 | MOL007071Przewaqui | 40.31 | 0.46 |
| Danshen | 163263 | MOL007077Sclareol | 43.67 | 0.21 |
| Danshen | 124268 | MOL007079Tanshinal | 52.47 | 0.45 |
| Danshen | 3083515 | MOL007081Danshenol | 57.95 | 0.56 |
| Danshen | 3083514 | MOL007082Danshenol | 56.97 | 0.52 |
| Danshen | 389885 | MOL007085Salvileno | 30.38 | 0.38 |
| Danshen | 160254 | MOL007088Cryptotan | 52.34 | 0.4 |
| Danshen | 127172 | MOL007093Dan-shenx | 38.88 | 0.55 |
| Danshen | 1.02E+08 | MOL007094Danshensp | 50.43 | 0.31 |
| Danshen | 15690458 | MOL007098Deoxyneoc | 49.4 | 0.29 |
| Danshen | 34754315 | MOL007100Dihydrota | 38.68 | 0.32 |
| Danshen | 11425923 | MOL007101Dihydrota | 45.04 | 0.36 |
| Danshen | 1.02E+08 | MOL007105Epidanshe | 68.27 | 0.31 |
| Danshen | 442027 | MOL007107C09092 | 36.07 | 0.25 |
| Danshen | 626608 | MOL007108Isocrypto | 54.98 | 0.39 |
| Danshen | 44425166 | MOL007111Isotanshi | 49.92 | 0.4 |
| Danshen | 3034394 | MOL007115Manool | 45.04 | 0.2 |
| Danshen | 5319835 | MOL007119Miltionon | 49.68 | 0.32 |
| Danshen | 5319836 | MOL007120Miltionon | 71.03 | 0.44 |
| Danshen | 10086184 | MOL007121Miltipolo | 36.56 | 0.37 |
| Danshen | 160142 | MOL007122Miltirone | 38.76 | 0.25 |
| Danshen | 15690458 | MOL007124Neocrypto | 39.46 | 0.23 |
| Danshen | 389888 | MOL007125Neocrypto | 52.49 | 0.32 |
| Danshen | 10062187 | MOL0071271-methyl- | 34.72 | 0.37 |
| Danshen | 30428202 | MOL007130prolithos | 64.37 | 0.31 |
| Danshen | 65035 | MOL007132(2R)-3-(3 | 109.38 | 0.35 |
| Danshen | 11530200 | MOL007141Salvianol | 45.56 | 0.61 |
| Danshen | 24177556 | MOL007142Salvianol | 43.38 | 0.72 |
| Danshen | 389885 | MOL007143Salvileno | 32.43 | 0.23 |
| Danshen | 389885 | MOL007145Salviolon | 31.72 | 0.24 |
| Danshen | 5321620 | MOL007151Tanshindi | 42.67 | 0.45 |
| Danshen | 126072 | MOL007152Przewaqui | 42.85 | 0.45 |
| Danshen | 164676 | MOL007154Tanshinon | 49.89 | 0.4 |
| Danshen | 9926694 | MOL007155(6S)-6-(h | 65.26 | 0.45 |
| Danshen | 149138 | MOL007156Tanshinon | 45.64 | 0.3 |
| Huangbai | 2353 | MOL001454Berberine | 36.86 | 0.78 |
| Huangbai | 72322 | MOL001458Coptisine | 30.67 | 0.86 |
| Huangbai | 5320517 | MOL002641Phellavin | 35.86 | 0.44 |
| Huangbai | 98608 | MOL002644Phellopte | 40.19 | 0.28 |
| Huangbai | 128994 | MOL002651Dehydrota | 43.76 | 0.4 |
| Huangbai | 65752 | MOL002662Rutaecarp | 40.3 | 0.6 |
| Huangbai | 6760 | MOL002663Skimmiani | 40.14 | 0.2 |
| Huangbai | 2703 | MOL002666Cheleryth | 34.18 | 0.78 |
| Huangbai | 5280794 | MOL000449Stigmaste | 43.83 | 0.76 |
| Huangbai | 20055073 | MOL002668Worenine | 45.83 | 0.87 |
| Huangbai | 193148 | MOL002670Cavidine | 35.64 | 0.81 |
| Huangbai | 222284 | MOL000358Beta-sito | 36.91 | 0.75 |
| Huangbai | 5319198 | MOL000622Magnogran | 63.71 | 0.19 |
| Huangbai | 19009 | MOL000785Palmatine | 64.6 | 0.65 |
| Huangbai | 4970 | MOL000787Fumarine | 59.26 | 0.83 |
| Huangbai | 440229 | MOL000790Isocorypa | 35.77 | 0.59 |
| Huangbai | 5280343 | MOL000098Quercetin | 46.43 | 0.28 |
| Huangbai | 193876 | MOL001131Phellamur | 56.6 | 0.39 |
| Huangbai | 21171 | MOL001455(S)-canad | 53.83 | 0.77 |
| Huangbai | 457801 | MOL001771Poriferas | 36.91 | 0.75 |
| Huangbai | 72703 | MOL002894Berberrub | 35.74 | 0.73 |
| Huangbai | 3084288 | MOL006422Thalifend | 44.41 | 0.73 |
| Huanglian | 5319198 | MOL000622Magnogran | 63.71 | 0.19 |
| Huanglian | 5280343 | MOL000098Quercetin | 46.43 | 0.28 |
| Huanglian | 19009 | MOL000785Palmatine | 64.6 | 0.65 |
| Huanglian | 72703 | MOL002894Berberrub | 35.74 | 0.73 |
| Huanglian | 443422 | MOL002903(R)-canad | 55.37 | 0.77 |
| Huanglian | 160876 | MOL002897Epiberber | 43.09 | 0.78 |
| Huanglian | 2353 | MOL001454Berberine | 36.86 | 0.78 |
| Huanglian | 11066 | MOL002904Berlambin | 36.68 | 0.82 |
| Huanglian | 72322 | MOL001458Coptisine | 30.67 | 0.86 |
| Huanglian | 20055073 | MOL002668Worenine | 45.83 | 0.87 |
| Shanzha | 5281654 | MOL000354Isorhamne | 49.6 | 0.31 |
| Shanzha | 222284 | MOL000359Sitostero | 36.91 | 0.75 |
| Shanzha | 5280863 | MOL000422Kaempfero | 41.88 | 0.24 |
| Shanzha | 5280794 | MOL000449Stigmaste | 43.83 | 0.76 |
| Shanzha | 182232 | MOL000073Ent-epica | 48.96 | 0.24 |
| Shanzha | 5280343 | MOL000098Quercetin | 46.43 | 0.28 |
| Zhiqiao | 6450230 | MOL013381Marmin | 38.23 | 0.31 |
| Zhiqiao | 72281 | MOL002341Hespereti | 70.31 | 0.27 |
| Zhiqiao | 222284 | MOL000358Beta-sito | 36.91 | 0.75 |
| Zhiqiao | 932 | MOL004328Naringeni | 59.29 | 0.21 |
| Zhiqiao | 72344 | MOL005828Nobiletin | 61.67 | 0.52 |
3.2. Differentially Expressed Genes in Obese T2DM
A total of 28 DEGs were identified from this series of analysis (GSE166467). Of these, 19 were upregulated and 9 were downregulated in the obese T2DM (Figure 1(a)). The heat map of the expression patterns of the 28 DEGs is shown in Figure 1(b).
Figure 1.
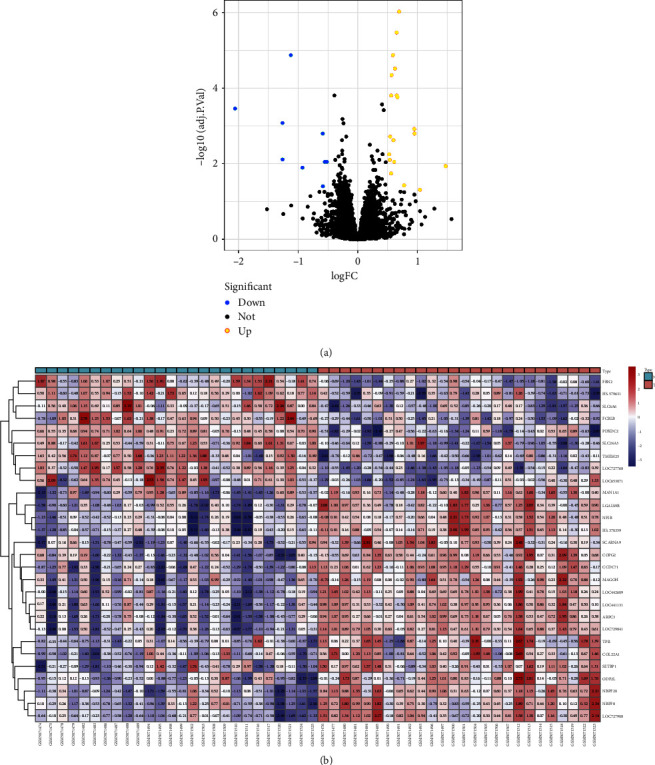
Screening of common targets for the Huanglian Huazhuo capsule-obese T2DM. (a) The differential gene volcano plot shows gene distribution in the disease samples. Red and green represent upregulated and downregulated genes, respectively, and black indicates no significant difference. (b) The heat map shows the expression patterns of 28 DEGs. Columns correspond to samples, and rows correspond to genes.
3.3. Anti-Obese T2DM Action Targets of the Huanglian Huazhuo Capsule
A total of 222 active ingredient targets of the Huanglian Huazhuo capsule were identified using the TCMSP database, and 1056 genes associated with obese T2DM and 108 targets of the drug-disease intersection were obtained using the GeneCards database (Figure 2).
Figure 2.
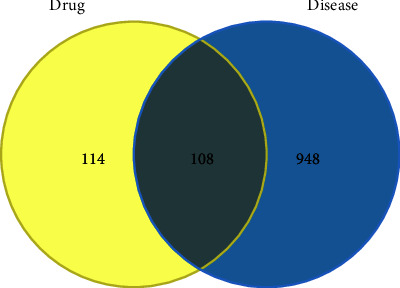
Venn diagram of the Huanglian Huazhuo capsule corresponding to target and disease corresponding to target intersection. Green represents the number of targets of the active components of the drug, and pink represents the number of disease-related genes, which are 108 cross-targeted genes.
3.4. Construction of “Drug-Component-Target-Disease” Network
The “Drug-Component-Target-Disease” network of the Huanglian Huazhuo capsule was mapped using the Cytoscape 3.8.0 software for obesity and DM treatment (Figure 3(a)). The diagram consisted of 298 nodes and 1652 edges, with 88 active ingredients and 108 potential targets. The active components of the Huanglian Huazhuo capsule before the moderate value were quercetin, beta-sitosterol, stigmasterol, kaempferol, luteolin, tanshinone IIA, and naringenin. Furthermore, these seven active components were analyzed to construct a small molecule network map to explore the multitarget properties of the main active components (Figure 3(b)). A total of 124 quercetin targets, 23 beta-sitosterol targets, 23 stigmasterol targets, 46 kaempferol targets, 46 luteolin targets, 32 tanshinone IIA targets, and 27 naringenin targets were found.
Figure 3.
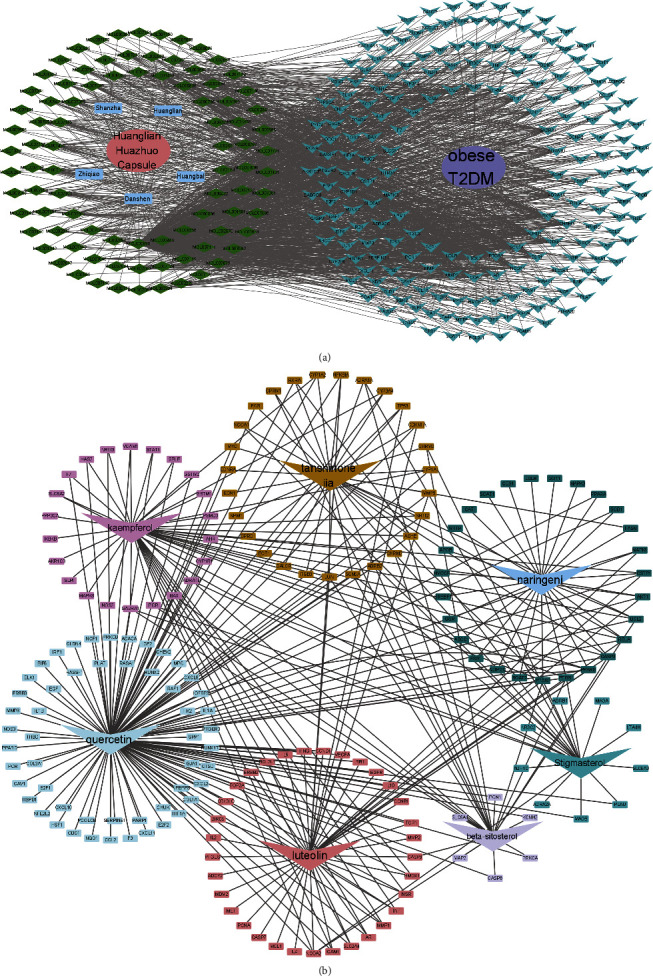
(a) In the “drug-component-target-disease” network, the pink circles represent the Huanglian Huazhuo capsules, the blue rectangles represent drugs, the green diamonds represent drug compounds, the purple circles represent diseases, and the blue triangles represent target proteins. (b) In the small molecule network plot, blue represents quercetin and its corresponding target, purple represents beta-sitosterol and its corresponding target, cyan represents stigmasterol and its corresponding target, red represents kaempferol and its corresponding target, pink represents luteolin and its corresponding target, red represents tanshinone IIA and its corresponding targets, and green represents naringenin and its corresponding targets.
3.5. Construction and Analysis of Protein-Protein Interaction Network
Based on the topological analysis, the target data generated from the STRING website was input into the Cytoscape 3.8.0 software. Greater than or equal to the median value was used as a filtering criterion, and the final topological parameter analysis yielded 18 nodes and 192 edges on the way to the PPI network, and STAT3, MAPK1, RELA, IL6, TNF, ESR1, and IL10 were identified as seven core targets (Table 2 and Figure 4).
Table 2.
Seven core targets.
| PDB ID | Gene name | Gene symb | Protein name | Degree |
|---|---|---|---|---|
| 5AX3 | STAT3 | P40763 | Signal transducer and activator of transcription 3 | 18 |
| 7.00E+75 | MAPK1 | P28482 | Transcription factor p65 | 16 |
| 7LEU | RELA | Q04206 | Transcrip | 14 |
| 1IL6 | IL6 | P05231 | Interleuk | 12 |
| 7QLF | TNF | P01375 | Tumor nec | 11 |
| 7RS8 | ESR1 | P03372 | Estrogen | 9 |
| 1ILK | IL10 | P22301 | Interleuk | 9 |
Figure 4.
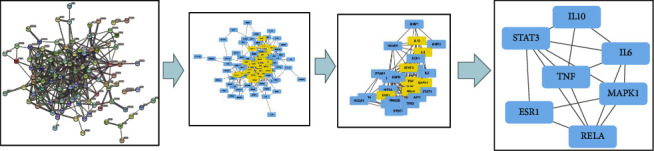
Topological screening process of the PPI network. A total of 85 common targets were screened using degree centrality (DC), betweenness centrality (BC), and closeness centrality (CC), and seven core targets were obtained.
3.6. GO Enrichment Analysis
The common drug-disease targets were input into the R language bioconductor package, and the GO and KEGG analyses were performed. The GO enrichment analysis included molecular function, biological pathway function, and cellular component. A bar chart was created using the top 10 entries (Figure 5), with the lower the bar, the smaller the P. Among them, the target protein molecular functions primarily focused on nuclear receptor activity, ligand activation, cytokine activity, DNA-binding transcription factor binding, and RNA polymerase II-specific DNA-binding transcription factor binding; cell composition, including membrane rafts, membrane microdomains, membrane regions, plasma membrane rafts, and postsynaptic membrane components; and biological processes, including responses to reactive oxygen species, drugs, lipopolysaccharides, and oxidative stress.
Figure 5.
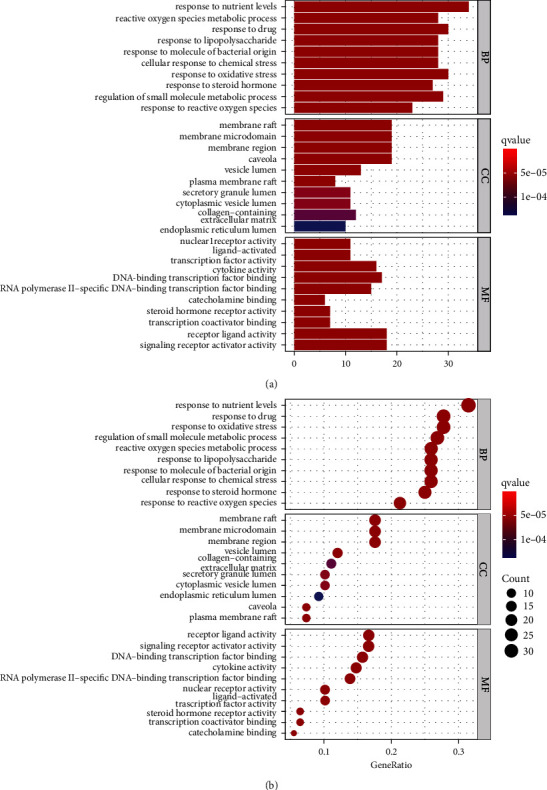
GO enrichment analysis. (a) Histogram of biological process category terms in the GO enrichment analysis. (b) Bubble diagram of biological process category terms in the GO enrichment analysis.
3.7. KEGG Analysis
Based on the gene ratios, the top 20 highly enriched pathways were screened (Table 3, Figures 6(a) and 6(b)). These 20 pathways, P-values from the KEGG enrichment analysis, and the associated targets and compounds were selected to develop a compound-target-pathway network using the Cytoscape 3.8.0 software (Figure 6(c)), containing 161 nodes and 671 edges. Obesity and diabetes treatment using the Huanglian Huazhuo capsules was via multiple pathways associated with various pathways, such as AGE-RAGE signaling pathway, fluid shear stress and atherosclerosis, HIF-1 signaling pathway, IL-17 signaling pathway, and TNF signaling pathway in diabetes complications. We also visualized the distribution of key targets in the most relevant path (Figure 7).
Table 3.
KEGG enrichment analysis of key genes.
| ID | Description | GeneID | Count |
|---|---|---|---|
| hsa04933 | AGE-RAGE signaling pathway in diabetic complications | MMP2, TNF, BCL2, RELA, AKT1, VEGFA, MAPK1, IL6, STAT1, F3, ICAM1, IL1B, CCL2, SELE, VCAM1, CXCL8, PRKCB, NOS3, THBD, SERPINE1, COL1A1, COL3A1, STAT3, EDN1, MAPK8 | 25 |
|
| |||
| hsa05418 | Fluid shear stress and atherosclerosis | MMP2, MMP9, TNF, BCL2, KDR, RELA, AKT1, VEGFA, TP53, HMOX1, CAV1, ICAM1, IL1B, CCL2, SELE, VCAM1, NOS3, PLAT, THBD, IFNG, GSTP1, NFE2L2, NQO1, GSTM1, EDN1, IKBKB, MAPK8 | 27 |
|
| |||
| hsa05417 | Lipid and atherosclerosis | RXRA, PPARG, MMP9, TNF, BCL2, CASP9, MMP3, RELA, AKT1, MAPK1, IL6, TP53, MMP1, CYP1A1, ICAM1, IL1B, CCL2, SELE, VCAM1, CXCL8, NOS3, NFE2L2, CD40LG, GSK3B, STAT3, OLR1, IKBKB, MAPK8, APOB | 29 |
|
| |||
| hsa04066 | HIF-1 Signaling pathway | NOS2, BCL2, RELA, EGFR, AKT1, VEGFA, MAPK1, EGF, IL6, HIF1A, ERBB2, HMOX1, PRKCB, NOS3, SERPINE1, IFNG, INSR, HK2, STAT3, EDN1, TIMP1 | 21 |
|
| |||
| hsa04657 | IL-17 Signaling pathway | PTGS2, MMP9, TNF, IL4, MMP3, RELA, MAPK1, IL6, MMP1, IL1B, CCL2, CXCL8, IFNG, CXCL10, GSK3B, IKBKB, MAPK8 | 17 |
|
| |||
| hsa04668 | TNF Signaling pathway | PTGS2, MMP9, TNF, MMP3, RELA, AKT1, MAPK1, IL6, ICAM1, IL1B, CCL2, SELE, VCAM1, CXCL10, EDN1, IKBKB, MAPK8, CREB1 | 18 |
|
| |||
| hsa05215 | Prostate cancer | AR, MMP9, BCL2, CASP9, MMP3, RELA, EGFR, AKT1, MAPK1, EGF, TP53, ERBB2, PLAT, GSTP1, GSK3B, IKBKB, CREB1 | 17 |
|
| |||
| hsa05207 | Chemical carcinogenesis-receptor activation | ESR1, AR, RXRA, ADRB2, CYP3A4, CYP1A2, PGR, ADRB1, BCL2, RELA, EGFR, AKT1, VEGFA, MAPK1, EGF, CYP1A1, PRKCB, BIRC5, PPARA, GSTM1, STAT3, CREB1 | 22 |
|
| |||
| hsa05142 | Chagas disease | NOS2, TNF, RELA, AKT1, MAPK1, IL10, IL6, IL1B, CCL2, CXCL8, IL2, SERPINE1, IFNG, IKBKB, MAPK8 | 15 |
|
| |||
| hsa04931 | Insulin resistance | TNF, RELA, AKT1, IL6, PRKCB, NOS3, SLC2A4, INSR, PPARA, GSK3B, STAT3, IKBKB, MAPK8, SREBF1, CREB1 | 15 |
|
| |||
| hsa05212 | Pancreatic cancer | CASP9, RELA, EGFR, AKT1, VEGFA, MAPK1, EGF, TP53, STAT1, ERBB2, STAT3, IKBKB, MAPK8 | 13 |
|
| |||
| hsa05161 | Hepatitis B | MMP9, TNF, BCL2, CASP9, RELA, AKT1, MAPK1, IL6, TP53, STAT1, CXCL8, PRKCB, BIRC5, STAT3, IKBKB, MAPK8, CREB1 | 17 |
|
| |||
| hsa05144 | Malaria | TNF, IL10, IL6, ICAM1, IL1B, CCL2, SELE, VCAM1, CXCL8, IFNG, CD40LG | 11 |
|
| |||
| hsa04151 | PI3K-Akt signaling pathway | RXRA, IL4, BCL2, CASP9, KDR, RELA, EGFR, AKT1, VEGFA, MAPK1, EGF, IL6, TP53, ERBB2, NOS3, IL2, COL1A1, INSR, SPP1, IGF2, GSK3B, IKBKB, CREB1 | 23 |
|
| |||
| hsa05219 | Bladder cancer | MMP2, MMP9, EGFR, VEGFA, MAPK1, EGF, TP53, MMP1, ERBB2, CXCL8 | 10 |
|
| |||
| hsa04926 | Relaxin signaling pathway | NOS2, MMP2, MMP9, RELA, EGFR, AKT1, VEGFA, MAPK1, MMP1, NOS3, COL1A1, COL3A1, EDN1, MAPK8, CREB1 | 15 |
|
| |||
| hsa05160 | Hepatitis C | RXRA, TNF, CASP9, RELA, EGFR, AKT1, MAPK1, EGF, TP53, STAT1, IFNG, PPARA, CXCL10, GSK3B, STAT3, IKBKB | 16 |
|
| |||
| hsa05145 | Toxoplasmosis | NOS2, TNF, BCL2, CASP9, RELA, AKT1, MAPK1, IL10, STAT1, IFNG, CD40LG, STAT3, IKBKB, MAPK8 | 14 |
|
| |||
| hsa01521 | EGFR tyrosine kinase inhibitor resistance | BCL2, KDR, EGFR, AKT1, VEGFA, MAPK1, EGF, IL6, ERBB2, PRKCB, GSK3B, STAT3 | 12 |
|
| |||
| hsa05167 | Kaposi sarcoma-associated herpesvirus infection | PTGS2, CASP9, RELA, AKT1, VEGFA, MAPK1, IL6, TP53, HIF1A, STAT1, ICAM1, CXCL8, GSK3B, STAT3, IKBKB, MAPK8, CREB1 | 17 |
Figure 6.
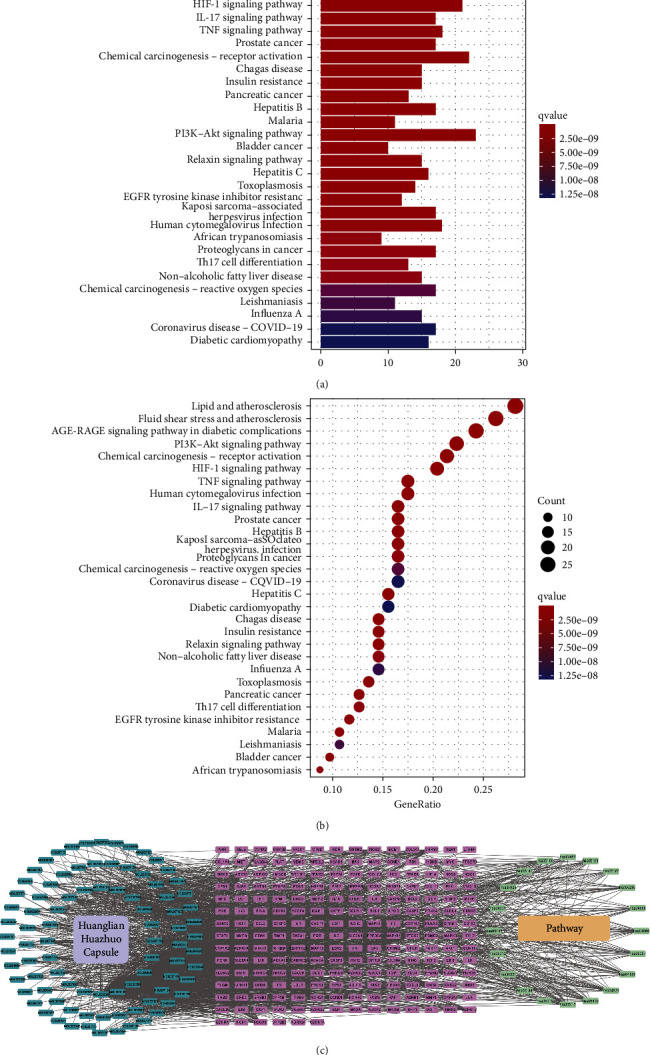
KEGG enrichment analysis and critical path network construction. (a) Histogram of the top 20 pathways based on KEGG enrichment analysis. (b) Bubble diagram of the top 20 pathways based on KEGG enrichment analysis. (c) Compound-target-pathway network associated with the mechanism of the Huanglian Huazhuo capsule for obese T2DM treatment. Purple nodes represent targets, dark green nodes represent compounds, and light green nodes represent pathways.
3.8. Prediction of the Binding Ability of the Huanglian Huazhuo Capsule Core Components to the Target Using Molecular Docking Technique
Molecular docking is a theoretical simulation method to examine the interaction between molecules and predict their binding mode and affinity. The top three target proteins and small molecules were screened for docking. Generally, the lower the energy required for ligand-molecule binding, the easier the docking success. If the binding energy is < 0 kcal/mol−1, the molecules bind by themselves (Table 4, Figure 8). The molecular docking results are shown in Figure 9. Hydrophobic small molecules and target protein active cavity form stable complexes via hydrogen bonding. In this study, tanshinone IIA showed strong binding activity with STAT3, MAPK1, IL6, and IL10, suggesting these as candidate drug molecules. Molecular docking techniques provide a strategy to assess the binding mode between herbal compounds and disease-related targets. However, potential herbal compounds require experimental validation.
Table 4.
Binding energy of active ingredients to target proteins.
| Compound | Chemical formula | Relative molecular weight g/mol | Binding energy / kcal mol–1 | ||||||
|---|---|---|---|---|---|---|---|---|---|
| STAT3 | MAPK1 | RELA | IL6 | TNF | ESR1 | IL10 | |||
| Quercetin | C15H10O7 | 302.23 | −7.4 | −8.9 | −6.8 | −6.9 | −5.2 | −8.1 | −6.7 |
| Beta-sitosterol | C29H50O | 414.7 | −7.0 | −8.2 | −6.3 | −6.6 | −5.3 | −7.3 | −7.8 |
| Stigmasterol | C29H48O | 412.7 | −7.0 | −8.6 | −6.4 | −6.3 | −6.4 | −7.8 | −8.5 |
| Kaempferol | C15H10O6 | 286.24 | −7.4 | −8.5 | −6.7 | −6.8 | −5.0 | −7.9 | −7.0 |
| Luteolin | C15H10O6 | 286.24 | −7.7 | −8.8 | −6.9 | −7.2 | −5.3 | −8.0 | −6.9 |
| Tanshinone IIA | C19H18O3 | 294.3 | −8.2 | −9.0 | −6.5 | −7.8 | −6.0 | −8.0 | −8.6 |
| Naringenin | iC15H12O5 | 272.25 | −7.3 | −8.3 | −6.7 | −7.0 | −5.1 | −7.7 | −6.9 |
Figure 8.
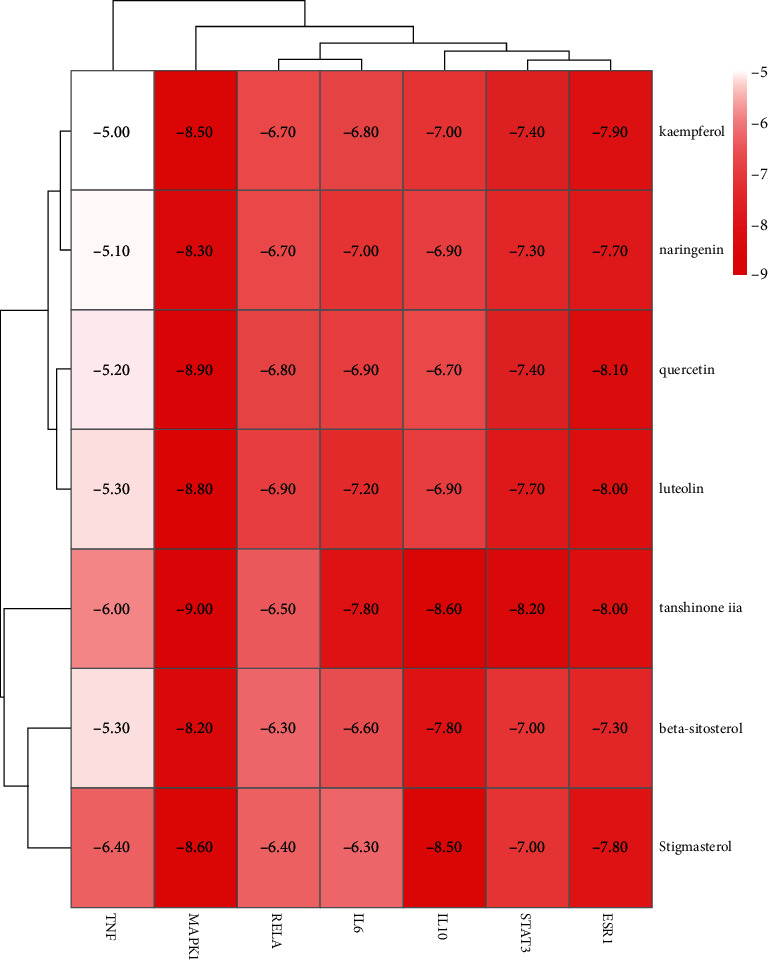
Thermogram of molecular docking fractions. Binding energy of key targets and herbal active compounds (kcal/mol).
Figure 9.
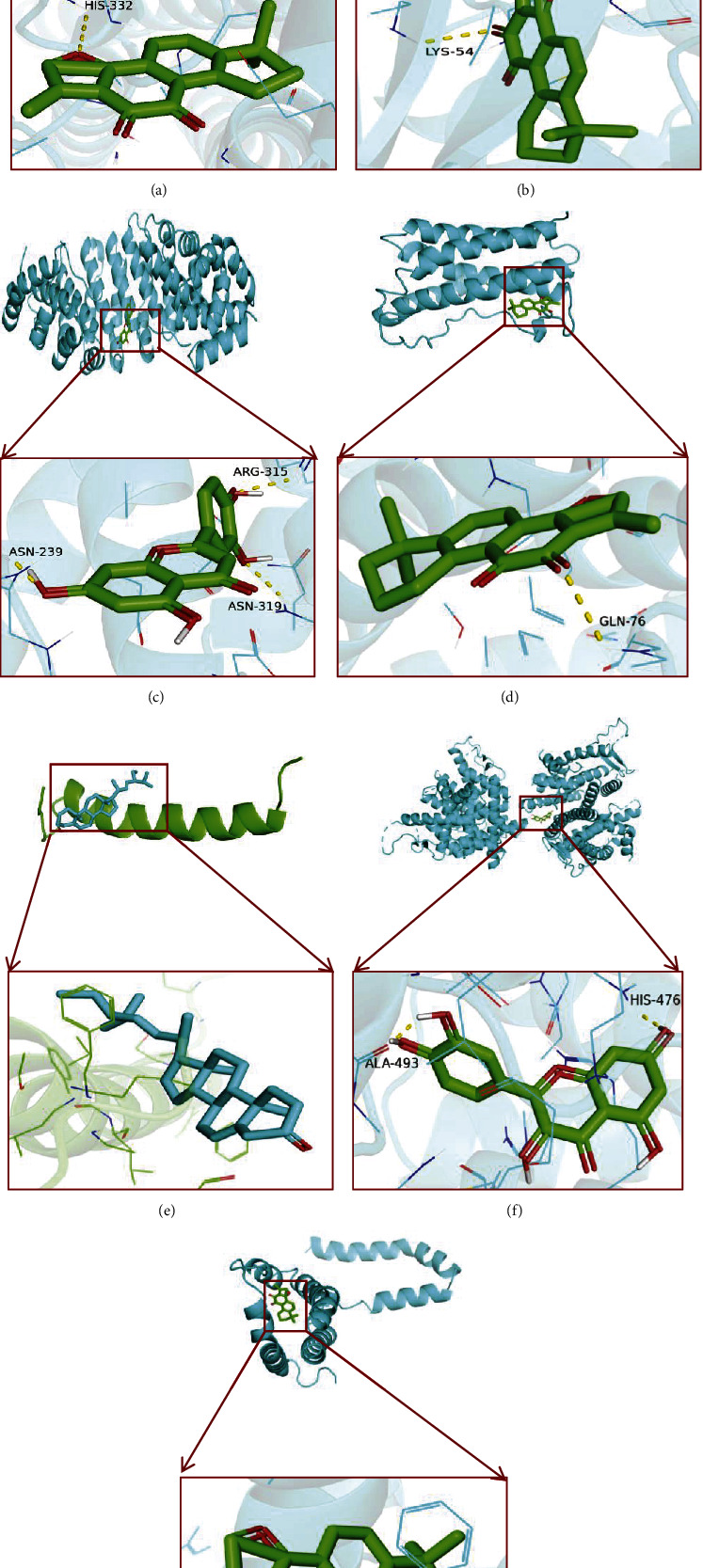
Docking patterns of key targets and specific active compounds. Tanshinone IIA-STAT3 (a), tanshinone IIA-MAPK1 (b), luteolin-RELA (c), tanshinone IIA-IL6 (d), stigmasterol-TNF (e), quercetin-ESR1 (f), and tanshinone IIA-IL10 (g).
4. Discussion
DM is a common chronic disease with increasing annual incidence. The number of patients with T2DM primarily characterized by insulin resistance is increasing [1]. Therefore, active intervention is required for the prognosis of T2DM. Currently, no specific drugs exist for T2DM treatment, bringing the clinical focus to glycemic control. Furthermore, the ideal efficacy of pure Western medical treatment is difficult to obtain, and new treatment methods are needed. Nevertheless, the positive role of Chinese medicine in diabetes prevention and treatment has been affirmed [6].
The Huanglian Huazhuo capsule removes phlegm, accumulation, dampness, and blood stasis and invigorates spleen. Among them, Huanglian and Huangbai removes heat and dampness and releases fire and detoxify; Shanzha digests and removes accumulation, strengthens stomach, and removes stasis by eliminating Qi; Zhiqiao detoxifies and removes turbidity; and Danshen promotes blood circulation and removes blood stasis. However, the bioactive compounds of the Huanglian Huazhuo capsules and their mechanisms of action against obese T2DM remain unclear. Therefore, the potential targets and mechanisms of action of the Huanglian Huazhuo capsules were identified in obese T2DM cases using a network pharmacology strategy and molecular docking.
A total of 88 active compounds from the TCMSP database were screened for the Huanglian Huazhuo capsules. Furthermore, 1056 obese T2DM disease targets were obtained from the geographic database. Hence, 108 putative Huanglian Huazhuo capsule-obese T2DM targets were identified. Based on the degree in the drug-component-target-disease network, the top seven active compounds of the Huanglian Huazhuo capsule were quercetin, beta-sitosterol, stigmasterol, kaempferol, luteolin, tanshinone IIA, and naringenin. Their effectiveness is supported by previous studies. Reportedly, quercetin attenuates lipid peroxidation, platelet aggregation, and capillary permeability and contains anti-inflammatory effects with therapeutic efficacy in obesity and T2DM [23]. Beta-sitosterol ameliorates the IKKβ/NF-κB and c-Jun-N-terminal kinase signaling pathways in the adipose tissue by downregulating inflammatory events, thereby inhibiting obesity-induced insulin resistance [24]. Stigmasterol and phytosterol-rich diets control glucolipid metabolism and insulin resistance [25]. Kaempferol increases lipid metabolism by downregulating PPAR-γ and SREBP-1c, thereby reducing adipose tissue accumulation and improving hyperlipidemia in mice with obesity and diabetes [26]. Luteolin attenuates neuroinflammation, oxidative stress, and neuronal insulin resistance in the mouse brain and normalizes blood adipocytokine levels [27]. Tanshinone IIA improves hepatic steatosis by inhibiting excess endoplasmic reticulum stress, endoplasmic reticulum stress-induced apoptosis, and hepatic steatosis [28]. Naringenin promotes adipose tissue in insulin receptor expression, GLUT4, lipocalin, and antidiabetic effects [29]. The strength of the protein gene role in the entire network is proportional to the degree value, and the protein genes with larger degree values play a significant role [30]. STAT3, MAPK1, RELA, IL6, TNF, ESR1, and IL10 were the seven core targets identified by the degree in the PPI network.
The KEGG and GO functional analyses showed that the effect of the Huanglian Huazhuo capsule in obese T2DM is associated with many biological processes, including inflammatory, lipopolysaccharide, and oxidative stress responses. In this study, the targets were enriched for lipid and inflammatory response pathways, such as HIF-1, IL-17, and TNF signaling pathways. HIF-1 reportedly promotes changes in the adipose tissues of patients with obesity, leading to the inhibition of adipocyte differentiation, adipocyte dysfunction, inflammation, insulin resistance, and T2D [31]. An adipose tissue from patients with obesity and T2D produced specific enrichment of CD4+ T cells for IL-17 and IL-22, which is pathologically relevant to obesity-induced T2D [32]. Furthermore, TNF is associated with obesity and T2D and correlates with glycated hemoglobin [33].
Subsequently, seven key target proteins, such as STAT3, MAPK1, RELA, IL6, TNF, ESR1, IL10, and active compounds, including quercetin, beta-sitosterol, stigmasterol, kaempferol, luteolin tanshinone IIA, naringenin, were evaluated using molecular docking techniques. The binding affinities ranged from −5.1 to −9.0 kcal/mol, indicating that all targets possibly had good docking ability with the active compounds. The result suggests that these compounds may contribute to the effectiveness of the Huanglian Huazhuo capsules in obese T2DM treatment. Chronic low-grade inflammation with elevated levels of nonspecific inflammatory factors, T2DM is an important factor in T2DM development and its complications [34]. The major signaling pathways involved in the inflammatory response include the NF-ΚB, JAK/STAT, MAPK, and PI3K/AKT signaling pathways. Obesity caused by T2MD was treated using the Huanglian Huazhuo capsule mainly through the MAPK, PI3K/AKT, and Wnt signaling pathways (Figure 7). MAPK1, IL6, IL10, STAT3, and ESR1, which bind to small molecules, were stable (Figure 8). MAPK1 is one of the crucial molecules in the MAPK signaling pathway, and its activation regulates the downstream inflammatory response and glucose and lipid metabolisms, as cytokines involved in inflammation and immunosuppression [35, 36]. IL6 and IL10 are regulated by the PI3K/AKT, tgf-β, and Wnt signaling pathways, and they closely associate with the inflammatory signaling pathway [37–39]. These results are consistent with our finding that the Huanglian Huazhuo capsules treat obese T2DM via the inflammatory response pathway. However, in vitro experiments are required to validate these results.
Figure 7.
Distribution of key targets in the most relevant paths.
However, the present study has some limitations. First, bioactive compounds and target data were retrieved from the literature and databases; hence, the reliability and accuracy of the predictions depend on the data quality. The active compounds in the Huanglian Huazhuo capsules can be analyzed using the lC/MS technique. In addition, metabolomics and pharmacokinetic studies may be advantageous. Second, data mining methods that require clinical trials and animal studies to confirm these findings were used.
5. Conclusions
It is the first time that the pharmacological and molecular mechanisms of action of the Huanglian Huazhuo capsule have been systematically explored to treat obese T2DM using network pharmacology and molecular docking techniques. These bioinformatics and computational analyses suggest that quercetin, beta-sitosterol, stigmasterol, and kaempferol are possibly the main active compounds of the Huanglian Huazhuo capsule in obese T2DM treatment. In addition, the Huanglian Huazhuo capsule could treat obese T2DM by reducing pathological damage, inflammatory response, and oxidative stress via various pathways, such as HIF1, IL-17, and TNF. Overall, the present study focused on the multicomponent and multipathway nature and mechanism of action of the Huanglian Huazhuo capsule. These findings can guide the application and further develop the Huanglian Huazhuo capsules in obese T2DM treatment.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (82160884).
Data Availability
The data in the study are obtained from TCMSP, CNKI, PubMed, GEO, RSCB PDB, and PubChem.
Conflicts of Interest
The authors of this work have no conflicts of interest to disclose.
Authors' Contributions
Na Wang and Xin Feng contributed equally to this study.
References
- 1.Zhou Z. Internal Medicine in Chinese Medicine . 2. Beijing, China: China Chinese Medicine Press; 2007. p. p. 407. [Google Scholar]
- 2.Saimaiti M., yisrieeli M., Aili A. Research progress on the relationship between obesity and type 2 diabetes. Chinese Electronic Journal of obesity and metabolic diseases . 2020;6(2):130–134. [Google Scholar]
- 3.Zheng K., Geng J., Liu Q. Effect of insulin treatment of primary obese T2DM patients combined with liraglutide on microinflammation and B-cell function. Chinese Journal of Gerontology . 2019;39(20):4917–4920. [Google Scholar]
- 4.Zhong Q. Clinical Study on the Treatment of Obesity-type Diabetes Mellitus with Liver and Stomach Stagnation and Heat with Stasis . Jiangsu, China: Nanjing University of Traditional Chinese Medicine; 2012. [Google Scholar]
- 5.An S., Cui Q. R., Kang X. D., Wang D. K. Clinical study on the combination of Huanglian Huahuo formula and metformin in the treatment of obese type 2 diabetes mellitus combined with dyslipidemia. Western Chinese Medicine . 2021;34(09):99–102. [Google Scholar]
- 6.Ma Y., Zhou K., Fan J., Sun S. Traditional Chinese medicine: potential approaches from modern dynamical complexity theories. Frontiers of Medicine . 2016;10(1):28–32. doi: 10.1007/s11684-016-0434-2. [DOI] [PubMed] [Google Scholar]
- 7. Kibble M., Saarinen N., Tang J., Wennerberg K., Makela S., Aittokallio T. Network pharmacology applications to map the unexplored target space and therapeutic potential of natural product. Natural Product Reports . 2021;32:1249–1266. doi: 10.1039/c5np00005j. [DOI] [PubMed] [Google Scholar]
- 8.Hopkins A. L. Network pharmacology: the next paradigm in drug discovery. Nature Chemical Biology . 2008;4(11):682–690. doi: 10.1038/nchembio.118. [DOI] [PubMed] [Google Scholar]
- 9.Zhang H., Zhang S., Hu M., et al. An integrative metabolomics and network pharmacology method for exploring the effect and mechanism of Radix Bupleuri and Radix Paeoniae Alba on anti- depression. Journal of Pharmaceutical and Biomedical Analysis . 2020;189 doi: 10.1016/j.jpba.2020.113435.113435 [DOI] [PubMed] [Google Scholar]
- 10.Niu B., Xie X., Xiong X., Jiang J. Network pharmacology-based analysis of the anti-hyperglycemic active ingredients of roselle and experimental validation. Computers in Biology and Medicine . 2022;141 doi: 10.1016/j.compbiomed.2021.104636.104636 [DOI] [PubMed] [Google Scholar]
- 11.Wu Y., Liu X., Li G. Integrated bioinformatics and network pharmacology to identify the therapeutic target and molecular mechanisms of Huangqin decoction on ulcerative Colitis. Scientific Reports . 2022;12(1):p. 159. doi: 10.1038/s41598-021-03980-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jin D., Zhang J., Zhang Y., et al. Network pharmacology-based and molecular docking prediction of the active ingredients and mechanism of ZaoRenDiHuang capsules for application in insomnia treatment. Computers in Biology and Medicine . 2021;135 doi: 10.1016/j.compbiomed.2021.104562.104562 [DOI] [PubMed] [Google Scholar]
- 13.Liu Z. W., Luo Z. H., Meng Q. Q., Zhong P. C., Hu Y. J., Shen X. L. Network pharmacology-based investigation on the mechanisms of action of Morinda officinalis How. in the treatment of osteoporosis. Computers in Biology and Medicine . 2020;127 doi: 10.1016/j.compbiomed.2020.104074.104074 [DOI] [PubMed] [Google Scholar]
- 14.Ru J., Li P., Wang J., et al. TCMSP: a database of systems pharmacology for drug discovery from herbal medicines. Journal of Cheminformatics . 2014;6(1):p. 13. doi: 10.1186/1758-2946-6-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen C. Y. C. TCM Database@Taiwan: the world’s largest traditional Chinese medicine database for drug screening in silico. PLoS One . 2011;6(1) doi: 10.1371/journal.pone.0015939.e15939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xu X., Zhang W., Huang C., et al. A novel chemometric method for the prediction of human oral bioavailability. International Journal of Molecular Sciences . 2012;13(6):6964–6982. doi: 10.3390/ijms13066964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ban C., Jo M., Park Y. H., et al. Enhancing the oral bioavailability of curcumin using solid lipid nanoparticles. Food Chemistry . 2020;302 doi: 10.1016/j.foodchem.2019.125328.125328 [DOI] [PubMed] [Google Scholar]
- 18.Zhang Y. F., Huang Y., Ni Y. H., Xu Z. M. Systematic elucidation of the mechanism of geraniol via network pharmacology. Drug Design, Development and Therapy . 2019;13:1069–1075. doi: 10.2147/DDDT.S189088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tao W., Xu X., Wang X., et al. Network pharmacology- based prediction of the active ingredients and potential targets of Chinese herbal Radix Curcumae formula for application to cardiovascular disease. Journal of Ethnopharmacology . 2013;145:1–10. doi: 10.1016/j.jep.2012.09.051. [DOI] [PubMed] [Google Scholar]
- 20.Wang Y., Yuan Y., Wang W., et al. Mechanisms underlying the therapeutic effects of Qingfeiyin in treating acute lung injury based on GEO datasets, network pharmacology and molecular docking. Computers in Biology and Medicine . 2022;145 doi: 10.1016/j.compbiomed.2022.105454.105454 [DOI] [PubMed] [Google Scholar]
- 21.Gu S., Xue Y., Gao Y., et al. Mechanisms of indigo naturalis on treating ulcerative colitis explored by GEO gene chips combined with network pharmacology and molecular docking. Scientific Reports . 2020;10(1) doi: 10.1038/s41598-020-71030-w.15204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rebhan M., Chalifa-Caspi V., Prilusky J., Lancet D. GeneCards: integrating information about genes, proteins and diseases. Trends in Genetics . 1997;13(4):p. 163. doi: 10.1016/s0168-9525(97)01103-7. [DOI] [PubMed] [Google Scholar]
- 23.Chen S., Jiang H., Wu X., Fang J. Therapeutic effects of quercetin on inflammation, obesity, and type 2 diabetes. Mediators of Inflammation . 2016;2016:5. doi: 10.1155/2016/9340637.9340637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jayaraman S., Devarajan N., Rajagopal P., et al. β-Sitosterol circumvents obesity induced inflammation and insulin resistance by down-regulating IKKβ/NF-κB and JNK signaling pathway in adipocytes of type 2 diabetic rats. Molecules . 2021;26(7):p. 2101. doi: 10.3390/molecules26072101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Prasad M., Jayaraman S., Eladl M. A., et al. A comprehensive review on therapeutic perspectives of phytosterols in insulin resistance: a mechanistic approach. Molecules . 2022;27(5):p. 1595. doi: 10.3390/molecules27051595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zang Y., Zhang L., Igarashi K., Yu C. The anti-obesity and anti-diabetic effects of kaempferol glycosides from unripe soybean leaves in high-fat-diet mice. Food & Function . 2015;6(3):834–841. doi: 10.1039/c4fo00844h. [DOI] [PubMed] [Google Scholar]
- 27.Liu Y., Fu X., Lan N., et al. Luteolin protects against high fat diet-induced cognitive deficits in obesity mice. Behavioural Brain Research . 2014;267:178–188. doi: 10.1016/j.bbr.2014.02.040. [DOI] [PubMed] [Google Scholar]
- 28.Wang J., Hu R., Yin C., Xiao Y. Tanshinone IIA reduces palmitate-induced apoptosis via inhibition of endoplasmic reticulum stress in HepG2 liver cells. Fundamental & clinical Pharmacology . 2020;34(2):249–262. doi: 10.1111/fcp.12510. [DOI] [PubMed] [Google Scholar]
- 29.Ahmed O. M., Hassan M. A., Abdel-Twab S. M., Abdel Azeem M. N. Navel orange peel hydroethanolic extract, naringin and naringenin have anti-diabetic potentials in type 2 diabetic rats. Biomedicine & Pharmacotherapy . 2017;94:197–205. doi: 10.1016/j.biopha.2017.07.094. [DOI] [PubMed] [Google Scholar]
- 30.Lv T., Luo Y. H., Luo S. H. A., et al. Exploring the potential molecular mechanism of Gui Zhi Fen Zi Tang for rheumatoid arthritis based on network pharmacology and molecular docking. New Drugs in Chinese Medicine and Clinical Pharmacology . 2022;33(07):915–926. doi: 10.19378/j.issn.1003-9783.2022.07.009. [DOI] [Google Scholar]
- 31.Messineo S., Laria A. E., Arcidiacono B., et al. Cooperation between HMGA1 and HIF-1 contributes to hypoxia-induced VEGF and visfatin gene expression in 3T3-L1 adipocytes. Frontiers in Endocrinology . 2016;7:p. 73. doi: 10.3389/fendo.2016.00073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dalmas E., Venteclef N., Caer C., et al. T cell-derived IL-22 amplifies IL-1β-driven inflammation in human adipose tissue: relevance to obesity and type 2 diabetes. Diabetes . 2014;63(6):1966–1977. doi: 10.2337/db13-1511. [DOI] [PubMed] [Google Scholar]
- 33.Alzamil H. Elevated serum TNF-α is related to obesity in type 2 diabetes mellitus and is associated with glycemic control and insulin resistance. Journal of Obesity . 2020;2020:5. doi: 10.1155/2020/5076858.5076858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Halim M., Halim A. The effects of inflammation, aging and oxidative stress on the pathogenesis of diabetes mellitus (type 2 diabetes) Diabetes & Metabolic Syndrome: Clinical Research Reviews . 2019;13(2):1165–1172. doi: 10.1016/j.dsx.2019.01.040. [DOI] [PubMed] [Google Scholar]
- 35.Lim A., Nikolic-Paterson D. J. Role of MKK3 -p38 MAPK signalling in the development of type 2 diabetes and renal injury in obese db/db mice. Diabetologia . 2009;52(2):347–358. doi: 10.1007/s00125-008-1215-5. [DOI] [PubMed] [Google Scholar]
- 36.Shi Y., Wan X., Shao N., Ye R., Zhang N., Zhang Y. Protective and antiangiopathy effects of ginsenoside Re against diabetes mellitus via the activation of p38 MAPK, ERK1/2 and JNK signaling. Molecular Medicine . 2016;14(5):4849–4856. doi: 10.3892/mmr.2016.5821. [DOI] [PubMed] [Google Scholar]
- 37.Rehman K., Akash M. S. H., Liaqat A., Kamal S., Qadir M. I., Rasul A. Role of interleukin-6 in development of insulin resistance and type 2 diabetes mellitus. Critical Reviews in Eukaryotic Gene Expression . 2017;27(3):229–236. doi: 10.1615/CritRevEukaryotGeneExpr.2017019712. [DOI] [PubMed] [Google Scholar]
- 38.Ribeiro I. S., Pereira ÍS., Santos D. P., et al. Association between body composition and inflammation: a central role of IL-17 and IL-10 in diabetic and hypertensive elderly women. Experimental Gerontology . 2019;127 doi: 10.1016/j.exger.2019.110734.110734 [DOI] [PubMed] [Google Scholar]
- 39.Kibble M., Saarinen N., Tang J., Wennerberg K., Makela S., Aittokallio T. Network pharmacology applications to map the unexplored target space and therapeutic potential of natural products. Natural Product Reports . 2015;32(8):1249–1266. doi: 10.1039/c5np00005j. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data in the study are obtained from TCMSP, CNKI, PubMed, GEO, RSCB PDB, and PubChem.


