Abstract
Systemic vaccination against SARS-CoV-2 elicited high titers of specific antibodies in the blood and in the oral cavity. Preexisting autoimmune diseases, such as rheumatoid arthritis, and biological treatments, like B cell depletion, are known to exhibit higher risk of severe COVID-19 manifestation and increased frequency of breakthrough infections after vaccination. We hypothesized that such increased risk is associated with an aberrant induction of secreted antibodies in the oral cavity. Here we evaluated the levels of secreted antibodies in the oral cavity against the SARS-CoV-2 Spike protein during the course of vaccination in RA patients with or without B cell depletion. We found that total salivary IgG levels were correlated with number of B cells in the blood. Anti-Spike IgG responses 7 days after second vaccination were induced in the oral cavity of all healthy individuals, while only 6 out 23 RA patients exhibited anti-Spike IgG in their saliva regardless of B cell depleting therapy. Importantly, both salivary and serologic anti-Spike IgG and IgA responses towards WT and omicron Spike variants were efficiently induced by third vaccination in RA patients with or without B cell depletion to the levels that were similar to healthy individuals. Altogether, these data advocate for the necessity of three dose vaccination for RA patients to mount anti-Spike antibody responses at the mucosal surfaces and annotate the reduction of secreted salivary IgG by B cell depletion.
Keywords: RA, salivary antibodies, SARS-CoV-2, rituximab
Abbreviations: RA, Rheumatoid arthritis; HC, healthy controls; RTX, Rituximab
1. Introduction
Coronavirus disease 19 (COVID-19) is caused by the infection of epithelial cells of upper respiratory airways and the gut by SARS-CoV-2 virus. SARS-CoV-2 infects cells via interaction of the Spike (S) protein with the ACE2 receptor [[1], [2], [3]]. The Spike protein consists of two subunits, S1 and S2 proteins. In turn, S1 subunit contains a receptor-binding domain (RBD) that mediates its interaction with ACE2 and viral entry into host cells [3,4]. Thus, blocking of Spike-ACE2 interaction by the anti-Spike SARS-CoV-2 antibodies confers protection of the host against infection of target cells [5,6].
Systemic vaccination of healthy individuals with mRNA vaccines encoding the Spike protein of SARS-CoV-2 induce anti-Spike antibodies in nasopharyngeal and oral cavities [7]. Magnitude of vaccine-induced antibody responses is dependent on multiple factors, including age and sex: elderly people exhibit lower antibody responses upon vaccination [8] and females elicit higher anti-Spike antibody titers when compared to males of similar age [9]. However, whether efficient induction occurs in humans with various autoimmune diseases or using biological drugs interfering with immune responses remains unknown. At the same time, rheumatoid arthritis (RA) patients exhibit higher risk of the development of severe COVID-19 disease, most likely due to the pathobiology of the disease, immunocompromising comorbidities and immunosuppressive treatment [10]. Previous studies showed that RA patients develop protective antibody responses against Spike protein of SARS-CoV-2 upon vaccination, albeit at significantly lower titers compared with healthy controls [11]. Vaccinated RA patients have also increased frequency of breakthrough infections after vaccination [12]. Altogether, this suggests that RA may have perturbed antibody responses at their mucosal surfaces.
Multiple biological therapies directed to reduce excessive inflammatory reactions as well as depletion of autoreactive immune cells are actively used for the treatment of various autoimmune diseases. In particular, anti-CD20 antibody (rituximab; RTX) that depletes B cells is used for the treatment of several autoimmune diseases like multiple sclerosis, RA, etc [13]. Patients treated with rituximab demonstrated higher risk of severe COVID-19 upon infection with SARS-CoV-2 [14,15]. Also, the development of protective antibody responses in RTX-treated patients is severely suppressed and correlated with the degree of B cell depletion [11]. However, it remains undetermined how RTX regulates secreted antibody responses in the oral cavity.
Previous studies have demonstrated that mRNA vaccinees exhibit strong salivary IgG, but not IgA production, compared to natural infection [16]. Systemic serum antibodies restrict virus dissemination after infection of the host, while antigen-specific antibodies at the mucosal surfaces are required to prevent initial infection of the host [17,18]. Thus, we evaluated the induction of secreted IgA and IgG antibodies in the oral cavity against the SARS-CoV-2 Spike protein upon vaccination in RA patients with or without B cell depletion.
2. 2 Materials and methods
2.1. Ethics approval
Healthy individual in cohort #1 were consented within the BNT162-01 study (ClinicalTrials.gov Identifier: NCT04380701) and informed consent forms are archived as per 162–01 procedures. Healthy individuals from cohort #2 and all RA patients gave written informed consent according to the approval of the ethics committee at the Charité University Hospital Berlin (EA2/010/21, EA4/188/20). Participants gave informed consent to participate in the study before taking part.
2.2. Study participants
Outpatient rheumatoid arthritis (RA) patients, who received primary 2x SARS-CoV-2 vaccination between February and May 2021, were asked to participate into this study. We included 23 RA patients (according 2010 ACR Rheumatoid Arthritis Classification Criteria (20)). Serologic and cellular data of RA group after 2nd vaccination have partially been published (11). The majority of RA patients got vaccinated with 2x BNT162b2 (1st and 2nd vaccination 3 weeks apart), one patient got 2x mRNA-1273 (1st and 2nd vaccination 3 weeks apart) and other two RA patients got 2x ChAdOx1 (1st and 2nd vaccination 3 months apart). The patients were scheduled for a 3rd vaccination with BNT162b2, 6 months after 2nd vaccination according to federal state recommendations. As healthy control (HC) groups, we recruited 19 volunteers after 2x vaccination with BNT162b2 (cohort #1) and another 28 volunteers after 3rd SARS-CoV-2 vaccination (cohort #2, the majority received mRNA 1273 6 months after primary 2x vaccination). In general, HC (cohort #1 and #2) were younger than RA patients. Thus, we have also analyzed correlation of age and immune responses. Since we focused initially upon patients treated with rituximab, 11/23 patients included in this study received anti-CD20 therapy with rituximab. Detailed donor information is summarized in Table 1 and Table 2 .
Table 1.
Vaccination study participants -Cohort #1.
| Healthy controls, n = 19 | Rheumatoid arthritis patients (RA), n = 23 | |
|---|---|---|
| Age | ||
| median | 52 | 68 |
| 25% Percentile | 33 | 59 |
| 75% Percentile | 58 | 79 |
| under 50 | 8 | 2 |
| between 50 and 69 | 9 | 11 |
| >70 | 2 | 10 |
| Gender | ||
| female | 9 | 18 |
| male | 10 | 5 |
| Vaccines | ||
| 2x BNT162b2 | 19 | 20 |
| 2x mRNA-1273 | 0 | 1 |
| 2x ChAdOx1 | 0 | 2 |
| Immunosuppression | ||
| RTX | 11 | |
| MTX | 10 | |
| Leflunomid | 1 | |
| JAKI | 1 | |
| TNFI | 1 | |
| Abatacept | 3 | |
| Prednisolone | 5 | |
| SASP | 1 | |
Table 2.
Vaccination study participants -Cohort #2.
| Healthy controls (cohort 2, after 3rd vaccination), n = 28 | Rheumatoid arthritis patients (RA), n = 23 | |
|---|---|---|
| Age | ||
| median (yrs) | 44,5 | 68 |
| 25% Percentile | 35,5 | 59 |
| 75% Percentile | 61 | 79 |
| under 60 | 21 | 5 |
| >60 | 7 | 17 |
| Gender | ||
| female | 21 | 18 |
| male | 7 | 5 |
| Vaccines | ||
| 2x BNT162b2 | 18 | 20 |
| 2x mRNA-1273 | 0 | 1 |
| 1x ChAdOx1 + 1x BNT162b2 | 10 | 0 |
| 2x ChAdOx1 |
0 |
2 |
|
3rd vaccination |
||
| BNT162b2 | 8 | 23 |
| mRNA-1273 | 20 | 0 |
| Time after vaccination (months) | 2,2 ± 2,0 | 1,9 ± 0,5a |
| Immunosuppression | ||
| RTX | 11 | |
| MTX | 10 | |
| Leflunomid | 1 | |
| JAKI | 1 | |
| TNFI | 1 | |
| Abatacept | 3 | |
| Prednisolone | 5 | |
| SASP | 1 | |
serum analysis was performed 21 days after 3rd vaccination.
2.3. Sample processing
We collected whole saliva and peripheral blood samples 7 days after 2nd and 1–2 months after the 3rd vaccination. Whole venous blood was collected in VACUETTE® CAT Serum Separator Clot Activator tubes and centrifuged down at 2000g for 10 min at +4 °C. Serum was stored at −20 °C for further analysis. Whole saliva was collected in collection tube and centrifuged down at 2000g for 15 min at +4 °C. Supernatant was stored at −20 °C for further analysis.
2.4. Analysis of anti-Spike protein responses in saliva
To determine the SARS-CoV-2 specific antibody titers, 96-well plates were coated overnight with either 0,5 μg/ml recombinant SARS-CoV-2 Spike S1 Subunit His-tag Protein (R&D 10522-CV) or 0,5 μg/ml recombinant SARS-CoV-2 Nucleocapsid His Protein, CF (RnD Systems; Cat. No. 10474-CV) or 0.5 μg/ml recombinant SARS-CoV-2 Spike RBD protein (flag-his) (Omicron/B.1.1.529) (SanyouBio, Cat. #PNA055). Plates were washed, blocked and the administration of sera and saliva were done as previously described [19]. To detect S1-specific IgA, a biotinylated anti-human IgA antibody (Southern Biotech, Cat. No. 2050–08) was applied, followed by an incubation for 1 h at room temperature. After washing 5 times with PBST, streptavidin-HRP (Invitrogen, Cat. No. 88-7324-88) was added and after 30 min of incubation at RT and 5 times washing with PBST, Tetramethylbenzidine (TMB) Substrate (Invitrogen, Cat. No. 88-7324-88) was added. The reaction was stopped by addition of 2 N H2SO4 (Sigma-Aldrich: Cat. No. 84736). For anti-S1 IgG detection in saliva anti-human IgG-AP (ICN/Cappel, Cat No. 59289) was applied and plates were incubated for 1 h at room temperature. Subsequently, the plates were washed 5 times with 200 μL of 1x PBST and 100 μL of pNPP (Sigma, Cat. No. N2770) was added to each well. Reactions were stopped by addition of 3 M NaOH (Roth: Cat. No. 6771.1). Optical densities were measured on Spectramax plus 384(Molecular devices). OD values were further plotted against respective saliva dilutions and areas under the curve (AUC) were quantified using Graphpad Prism 9.3.1.
2.5. Flow cytometric assay for analysis of antibody responses against wild-type variant of spike protein
For the detection of “Wuhan” Spike specific immunoglobulins in serum HEK293T cells were transfected with a plasmid expressing wild-type variant SARS-CoV-2 Spike protein. Next day, the proportion of transfected cells was determined by staining with anti-SARS-CoV-2 Spike Glycoprotein S1 antibody (clone: CR3022, Abcam, Cat. No. ab273073) for 30 min, wash cells once with PBS/0.2% BSA and subsequent staining with goat anti-human IgG-Alexa-647 (Southern Biotech, Cat. No. 2014–31). Further transfected cells were collected and incubated with sera for 30 min, washed twice with PBS/BSA and stained with goat anti-human IgG Alexa647 (Southern Biotech, Cat. No. 2014–31), and anti-human IgA FITC (Sothern Biotech, Cat. No. 2052–02) in the presence of Fixable Viability Dye eFluor 450 (Invitrogen, Cat. No. 50-112-8817). Cells were washed with PBS/0.2% BSA and either measured directly or fixed in 2% paraformaldehyde solution for 20 min at 4 °C. Samples were acquired on a FACSCanto II (BD Biosciences) or a MACS Quant 16 (Miltenyi Biotec) and analyzed using FlowJo v10 (Tree Star Inc.) analysis software. In the respective fluorescent channels, geometric mean of fluorescent intensity (MFI) Spike expressing cells and non-expressing cells was quantified and ΔMFI = MFI (S+)-MFI (S−) for IgG and IgA was determined. ΔMFI values were further plotted against respective serum dilutions and AUC were quantified using Graphpad Prism 9.3.1.
2.6. Flow cytometric assay for analysis of antibody responses against omicron variant of spike protein
For the detection of Omicron Spike specific immunoglobulins in serum, sequence coding for omicron variant SARS-CoV-2 Spike protein was cloned into the vector carrying GFP protein under CAG promoter. HEK293T cells were transfected using newly generated plasmid. Next day, the proportion of transfected cells was determined by by GFP detection using flow cytometry. Further transfected cells were collected and incubated with serial sera dilutions for 30 min, washed twice with PBS/BSA and stained with anti-human IgG Alexa647 (Southern Biotech, Cat. No. 2014–31), and biotinylated goat anti-human IgA (Sothern Biotech, Cat. No. 2050–08) in the presence of Fixable Viability Dye eFluor 450 (Invitrogen, Cat. No. 50-112-8817). Cells were further stained using streptavidin conjugated with PE-Cy7 (Thermofisher Scientific: Ct. No.: SA1012), washed with PBS/0.2% BSA and either measured directly or fixed in 2% paraformaldehyde solution for 20 min at 4 °C. Samples were acquired on a FACSCanto II (BD Biosciences) or a MACS Quant 16 (Miltenyi) and analyzed using FlowJo v10 (Tree Star Inc.) analysis software. In the respective fluorescent channels, geometric mean of fluorescent intensity (MFI) Spike expressing cells and non-expressing cells was quantified and ΔMFI = MFI (S+)-MFI (S−) for IgG and IgA was determined. ΔMFI values were further plotted against respective serum dilutions and the area under the curve (AUC) was quantified using Graphpad Prism 9.3.1.
2.7. Statistical analysis
Statistical analysis was performed using Graphpad Prism 9.3.1. All the data were analyzed using Kruskal-Wallis non-parametric one-way ANOVA test or Mann-Whitney test. Correlation analysis in was performed using spearman rank correlation. *, p < 0.05, **, p < 0.01, ***, p < 0.001, not significant differences are not shown.
3. Results
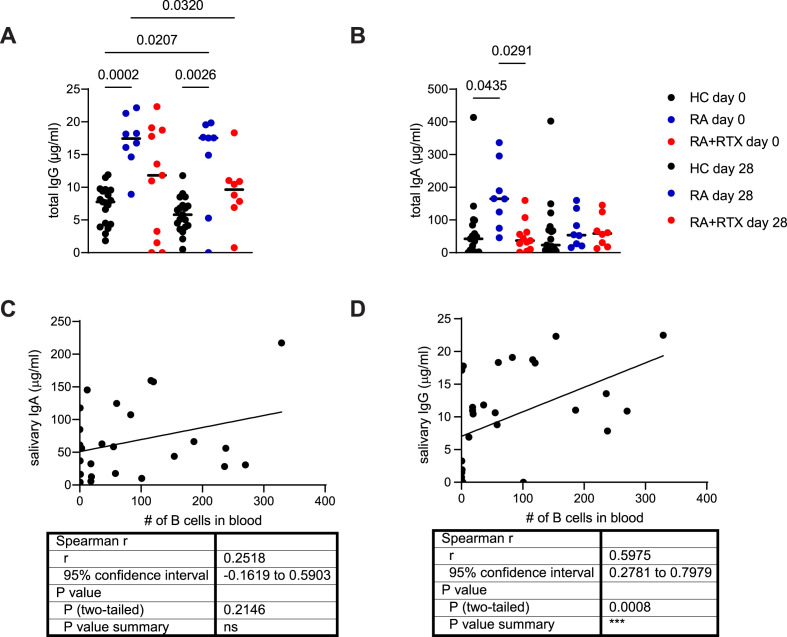
To analyze antibody responses in the oral cavity we collected saliva and serum of healthy individuals (HC) (n = 19) and RA patients (n = 23) before vaccination, 7 days after two injections of vaccine (Cohort #1) and 1–2 months after 3rd vaccination (Cohort #2). All RA patients had a confirmed diagnosis according to the 2010 ACR Rheumatoid Arthritis Classification Criteria guidelines [20], characteristics of all participants are summarized in Table 1, Table 2. Given that B-cell depleting therapy is associated with higher risks for severe infections and vaccination failure, we have initially analyzed the presence of various immunoglobulins in the saliva of RA patients with or without rituximab treatment and corresponding healthy donors (Fig. 1 ). As reported previously, both IgG and IgA can be secreted into the oral cavity via FcRn and pIgR, respectively [21,22] and amount of total IgA and IgG in the saliva is not dependent on the age of healthy volunteers (Fig. S1A, B). All groups exhibited both IgA and IgG in their saliva (Fig. 1A and B). Interestingly, RA patients had increased total IgG levels when compared to controls (Fig. 1A). RTX treatment normalized salivary IgG during RA at the day of vaccination (Fig. 1A). The same pattern was observed 7 days after vaccination (Fig. 1 A). Notably, no consistent differences has been observed for salivary IgA (Fig. 1B). This suggested that salivary IgG, but not IgA, can be affected by B cell depleting therapy. Furthermore, salivary IgA levels did not show any correlation with the efficiency of B cell depletion (Fig. 1C), estimated by quantification of B cell numbers in the peripheral blood. Total IgG concentrations were strongly associated with peripheral B cell numbers (Fig. 1D). Altogether, this indicates that B cell depletion in RA patients reduces total IgG levels in the oral cavity.
Fig. 1.
B cell depletion in rheumatoid arthritis patients diminished total IgG levels in saliva. IgG (A) and IgA (B) concentrations in saliva before vaccination (day 0) and 7 days after (day 28) second vaccination in the first cohort of healthy individuals (HC) and RA patients. Correlation between salivary IgA (C) and IgG (D) and B cell numbers in the blood of RA patients with or without RTX treatment (cohort #1). Data (A, B) were analyzed using Kruskal-Wallis non-parametric one-way ANOVA test. Correlation analysis (C, D) was performed using Spearman rank correlation.
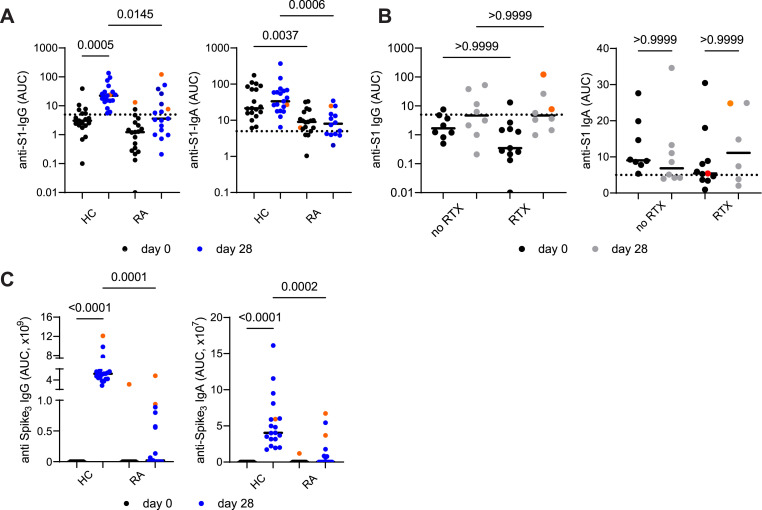
Taking into account that B cell depletion affected total salivary IgG levels, we next quantified their antigen-specific antibody response upon vaccination. All healthy participants developed salivary IgG responses 7 days after second vaccination towards S1 subunit of Spike protein of SARS-CoV-2 (Fig. 2 A), however, only 6 RA patients exhibited anti-S1 IgG in their saliva (Fig. 2A), including two patients who contracted SARS-CoV-2 infections before as determined by IgG antibodies against nucleocapsid protein of SARS-CoV-2 (data not shown). Of note, preexisting salivary anti-S1 IgA were higher in naive HC when compared to RA, but were not further amplified in this time frame in both HC and RA groups (Fig. 2A). Consistently, both subgroups of RA patients with or without B cell depletion therapy did not mount anti-S1 antibody responses at this time point (Fig. 2B). Similar results about salivary Ig responses were obtained against trimeric Spike protein expressed on 293 T cells (data not shown). Further, no correlation between number of B cells and anti-S1 immunoglobulin titers were observed in RA patients at day 7 after vaccination (Fig. S2A, B). Consistent with the lack of salivary responses, RA patients exhibited reduced anti-Spike trimer (Spike3) IgG serum responses when compared to healthy cohort: albeit half of the patients (8 out of 16) seroconverted (Fig. 2C). Magnitude of salivary anti-Spike IgG responses did not correlate with the age of healthy and RA patients (Fig. S2C). Salivary anti-Spike IgA was reduced with age in healthy individuals, but was not age-dependent in RA patients (Fig. S2D). This suggests that other factors may contribute to the antibody secretion to the oral cavity in RA. Also salivary anti-Spike responses in RA patients appeared to be less dependent on other immunosuppressive treatment modalities (MTX, prednisolone) (Fig. S2E). Thus, RA patients have reduced antibody responses elicited by vaccination regardless B cell depleting therapy.
Fig. 2.
Diminished salivary anti-Spike responses in rheumatoid arthritis patients upon 2x vaccination against SARS-CoV-2 irrespective of B cell depletion.
(A) Anti-Spike S1 IgG and IgA in saliva 7 days after (day 28) second vaccination in HCs and RA patients (cohort #1). (B) Anti-Spike S1 IgG and IgA levels in saliva 7 days after (day 28) second vaccination in RA patients with or without B cell depleting therapy (cohort #1). (C) Anti-Spike (trimer) IgG and IgA levels in serum 7 days after (day 28) second vaccination in HCs and RA patients (cohort #1). Previously infected individuals are shown in orange. AUC - area under curve. Data were analyzed using Kruskal-Wallis non-parametric one-way ANOVA test. *- p < 0,05; ** - p < 0,01; *** - p < 0,001; **** - p < 0,0001.
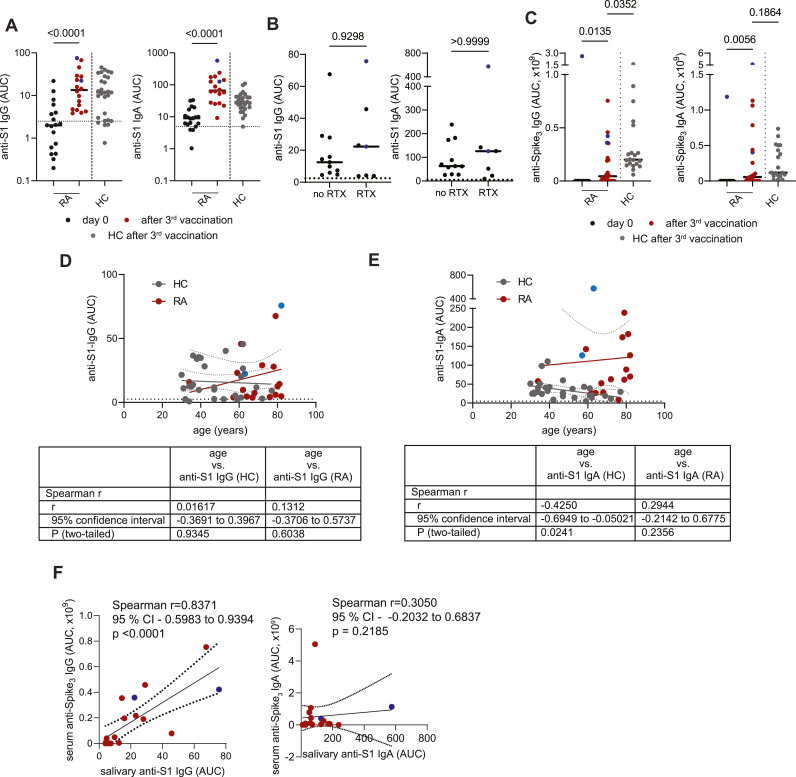
Since two rounds of vaccination induced poor antibody response in the oral cavity of RA patients, we next analyzed whether additional vaccination would induce Spike-specific antibodies secreted to the saliva. Indeed, both salivary and serologic anti-Spike IgG and IgA responses were induced by third vaccination in RA patients in the levels similar to HCs (Fig. 3 A–C), showing that three vaccinations are absolutely needed in RA patients to elicit a salivary response. Similarly to the 2nd vaccination, salivary IgG titers did not correlate with the age of neither healthy or RA patients (Fig. 3D). In case of anti-Spike IgA in the oral cavity, levels were reduced in older HC participants, while no correlation between magnitude of antibody response and age was observed in RA (Fig. 3E). Furthermore, 4 out 7 RA patients using B cell depleting therapy also showed anti-S1 IgG response, while all RA patients on other immunosuppressive drugs elicited anti-S1 IgG in their saliva (Fig. 3B). Anti-S1 IgA was detected in all RA patients with or without B cell depleting therapy (Fig. 3B). Salivary IgG levels after third vaccination also correlated with serum IgG antibody titers (Fig. 3F), suggesting that magnitude of systemic IgG antibody responses influences the appearance of anti-Spike antibodies in the oral cavity. Correlation between IgA levels in saliva and serum was not observed (Fig. 3F).
Fig. 3.
Induction of salivary anti-Spike responses in RA patients upon third vaccination against SARS-CoV-2.
(A) Anti-Spike S1 IgG and IgA levels in saliva after third vaccination in HCs and RA patients (cohort #2). (B) Anti-Spike S1 IgG and IgA levels in saliva after third vaccination in RA patients with or without B cell depleting therapy (cohort #2). (C) Anti-Spike (trimer) IgG and IgA levels in serum after third vaccination in HCs and RA patients (cohort #2). Correlation between salivary IgG (D), IgA (E) responses and age after third vaccination (cohort #2). (F) Correlation between salivary and serum IgG and IgA responses in RA patients after third vaccination (cohort #2). Previously infected individuals are shown in blue. AUC - area under curve. Data presented in (A, C) were analyzed using Kruskal-Wallis non-parametric one-way ANOVA test. Data presented in (B) were analyzed using Mann-Whitney test. Correlation analysis (D, E, F) was performed using Spearman rank correlation.
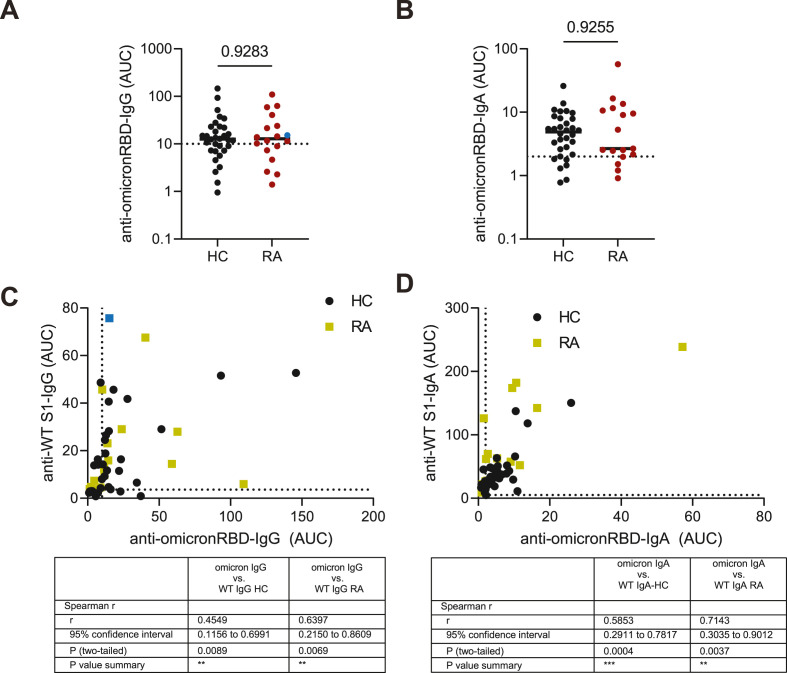
Taking into account that SARS-CoV-2 has already evolved and current dominant variant of SARS-CoV-2 has acquired multiple mutations in RBD domain of Spike protein [23], we next quantified whether salivary antibody response induced after third vaccination is specific against RBD from omicron SARS-CoV-2. RA patients who received third dose and developed serum antibody response mounted IgG and IgA antibodies against wild type and omicron Spike proteins (Fig. S1F). Importantly, saliva from both HC and RA individuals contained similar levels of anti-omicron RBD IgG and IgA levels (Fig. 4 A and B). Finally, strong correlation between salivary IgG and IgA antibody responses towards WT and omicron Spike SARS-CoV-2 has been observed (Fig. 4C and D). Thus, third vaccination mounted IgG and IgA antibodies reactive against WT and omicron SARS-CoV-2 in saliva in RA patients. No correlation was seen between salivary antibodies after 2nd or 3rd vaccination with time since last RTX therapy (Fig. S2G, H).
Fig. 4.
Cross-reactive salivary anti-omicron RBD responses in RA patients upon third vaccination against SARS-CoV-2.
Anti-omicron RBD IgG (A) and IgA (B) in saliva after third vaccination in the second cohort of HCs and RA patients. Correlation between salivary WT and omicron IgG (C) and IgA (D) responses in RA patients after third vaccination (cohort #2). AUC - area under curve. Data presented in (A, B) were analyzed using Mann-Whitney test. Correlation analysis was performed using Spearman rank correlation. *- p < 0,05; ** - p < 0,01; *** - p < 0,001; **** - p < 0,0001.
4. Discussion
Here we describe the oral antibody responses to SARS-CoV-2 vaccines in RA patients using various immunosuppressive medications with a special focus on the B cell depleting therapy. First of all, RTX in RA patients reduced total IgG, but not IgA, levels in the saliva and correlated with number of peripheral B cells in the blood, suggesting that B cell depletion reduce protective salivary IgG and thus, may reduce protection against air-borne pathogens.
Longitudinal antibody analysis further revealed that two shots of vaccine failed to induce salivary anti-Spike antibody response in RA patients with or without RTX in comparison to HCs. Strikingly, while salivary anti-S1 IgG response was quite heterogeneous after the third dose in RA patients with or without RTX therapy, anti-S1 IgA was significantly induced in both subgroups of the RA patients. It is of particular importance since anti-Spike IgA is associated with the protection from infection [24]. Finally, third vaccination induced also cross-reactive antibodies against omicron SARS-CoV-2 variant.
These data also suggest that vaccination efficacy may not be considered solely via serum titers in immunosuppressed RA patients, but more on the levels of mucosal antibodies, since mucosal antibodies represents the first defense barrier for SARS-CoV-2 infection. This study has several limitations including small sample size and that most RA patients continued immunosuppressive therapy during vaccination, which may confound results and limit interpretation of our findings. However, in our cohort we did not see a correlation between the most immunosupressant medications used and salivary titers, which suggests that the observed phenomenon is due to RA immunopathology. Also RA patients were older than healthy participants. To address the impact of age to the salivary anti-viral Ig responses, we have analyzed age-dependency of salivary anti-Spike responses both after 2nd and 3rd vaccinations. We did not observe correlation between salivary anti-Spike IgG antibody reponses and age of the participants. Only Spike-reactive IgA was found to be decreased in old helathy vaccinees, while RA patients did not show such correlation. This suggests that other factors mediating antibody secretion, like expression of receptors by epithelial cells or microbiota composition, may contribute to antibody secretion. Further studies addressing these mechanisms are needed.
Altogether, these data advocate for the necessity of three dose vaccination for RA patients to mount anti-Spike antibody responses at the mucosal surfaces.
Authors statement
Marina Bondareva: Investigation, Formal analysis, Conceptualization, Methodology, Writing – original draft, review & editing. Philine Letz, Kirsten Karberg, Eva Vanessa Schrezenmeier, Iaroslav Semin, Hector Rincon-Arevalo: Investigation, Resources, review & editing. Thomas Dörner, Mir-Farzin Mashreghi: Conceptualization, Methodology, Writing – original draft, review & editing. Ana-Luisa Stefanski, Andrey Kruglov: Conceptualization, Methodology, Writing – original draft, review & editing, Data curation, Formal analysis, Investigation, Project administration.
Patient consent
Participants gave informed consent to participate in the study before taking part.
Patient and public involvement
Patients and/or the public were not involved in the design, or conduct, or reporting, or dissemination plans of this research.
Funding
ALS was funded by a grant from the German Society of Rheumatology. Work was supported by DFG (TRR241 A04, A.K.), and Russian Science Foundation grant # 21-14-00223 (A.K.). This Work was supported by the state of Berlin and the “European Regional Development Fund” to M.F.M. (ERDF 2014–2020, EFRE 1.8/11, Deutsches Rheuma-Forschungszentrum). AK, IS are members of the Interdisciplinary Scientific and Educational School of Moscow University « Molecular Technologies of the Living Systems and Synthetic Biology».
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
We would like to thank Ugur Sahin and Özlem Türeci (Biontech) for the set-up of clinical study. Also we acknowledge Mona Massoud, Cristopher Skopnik for the help with samples preparation and are very grateful to members of DRFZ for the useful discussions.
Footnotes
Supplementary data related to this article can be found at https://doi.org/10.1016/j.jaut.2022.102918.
Appendix A. Supplementary data
The following is the supplementary data related to this article:
Data availability
Data will be made available on request.
References
- 1.Zhou P., Yang X.L., Wang X.G., Hu B., Zhang L., Zhang W., et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hoffmann M., Kleine-Weber H., Schroeder S., Kruger N., Herrler T., Erichsen S., et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181:271–280 e8. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shang J., Wan Y., Luo C., Ye G., Geng Q., Auerbach A., et al. Cell entry mechanisms of SARS-CoV-2. Proc. Natl. Acad. Sci. U. S. A. 2020;117:11727–11734. doi: 10.1073/pnas.2003138117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Walls A.C., Park Y.J., Tortorici M.A., Wall A., McGuire A.T., Veesler D. Structure, function, and antigenicity of the SARS-CoV-2 Spike Glycoprotein. Cell. 2020;181:281–292 e6. doi: 10.1016/j.cell.2020.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cao Y., Su B., Guo X., Sun W., Deng Y., Bao L., et al. Potent neutralizing antibodies against SARS-CoV-2 identified by high-throughput single-cell sequencing of convalescent patients' B cells. Cell. 2020;182:73–84 e16. doi: 10.1016/j.cell.2020.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ju B., Zhang Q., Ge J., Wang R., Sun J., Ge X., et al. Human neutralizing antibodies elicited by SARS-CoV-2 infection. Nature. 2020;584:115–119. doi: 10.1038/s41586-020-2380-z. [DOI] [PubMed] [Google Scholar]
- 7.Chan R.W.Y., Liu S., Cheung J.Y., Tsun J.G.S., Chan K.C., Chan K.Y.Y., et al. The mucosal and serological immune responses to the novel coronavirus (SARS-CoV-2) vaccines. Front. Immunol. 2021;12 doi: 10.3389/fimmu.2021.744887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Muller L., Andree M., Moskorz W., Drexler I., Walotka L., Grothmann R., et al. Age-dependent immune response to the biontech/pfizer BNT162b2 coronavirus disease 2019 vaccination. Clin. Infect. Dis. 2021;73:2065–2072. doi: 10.1093/cid/ciab381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Terpos E., Trougakos I.P., Apostolakou F., Charitaki I., Sklirou A.D., Mavrianou N., et al. Age-dependent and gender-dependent antibody responses against SARS-CoV-2 in health workers and octogenarians after vaccination with the BNT162b2 mRNA vaccine. Am. J. Hematol. 2021;96:E257–E259. doi: 10.1002/ajh.26185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.England B.R., Roul P., Yang Y., Kalil A.C., Michaud K., Thiele G.M., et al. Risk of COVID-19 in rheumatoid arthritis: a national veterans affairs matched cohort study in at-risk individuals. Arthritis Rheumatol. 2021;73:2179–2188. doi: 10.1002/art.41800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stefanski A.L., Rincon-Arevalo H., Schrezenmeier E., Karberg K., Szelinski F., Ritter J., et al. Arthritis Rheumatol; 2022. B Cell Numbers Predict Humoral and Cellular Response upon SARS-CoV-2 Vaccination Among Patients Treated with Rituximab; pp. 934–947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sun J., Zheng Q., Madhira V., Olex A.L., Anzalone A.J., Vinson A., et al. Association between immune dysfunction and COVID-19 breakthrough infection after SARS-CoV-2 vaccination in the US. JAMA Intern. Med. 2022:153–162. doi: 10.1001/jamainternmed.2021.7024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dorner T., Radbruch A., Burmester G.R. B-cell-directed therapies for autoimmune disease. Nat. Rev. Rheumatol. 2009;5:433–441. doi: 10.1038/nrrheum.2009.141. [DOI] [PubMed] [Google Scholar]
- 14.Andersen K.M., Bates B.A., Rashidi E.S., Olex A.L., Mannon R.B., Patel R.C., et al. Long-term use of immunosuppressive medicines and in-hospital COVID-19 outcomes: a retrospective cohort study using data from the National COVID Cohort Collaborative. Lancet Rheumatol. 2022;4:e33–e41. doi: 10.1016/S2665-9913(21)00325-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Simpson-Yap S., De Brouwer E., Kalincik T., Rijke N., Hillert J.A., Walton C., et al. Associations of disease-modifying therapies with COVID-19 severity in multiple sclerosis. Neurology. 2021;97:e1870–e1885. doi: 10.1212/WNL.0000000000012753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Becker M., Dulovic A., Junker D., Ruetalo N., Kaiser P.D., Pinilla Y.T., et al. Immune response to SARS-CoV-2 variants of concern in vaccinated individuals. Nat. Commun. 2021;12:3109. doi: 10.1038/s41467-021-23473-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Renegar K.B., Small P.A., Jr., Boykins L.G., Wright P.F. Role of IgA versus IgG in the control of influenza viral infection in the murine respiratory tract. J. Immunol. 2004;173:1978–1986. doi: 10.4049/jimmunol.173.3.1978. [DOI] [PubMed] [Google Scholar]
- 18.Huang N., Perez P., Kato T., Mikami Y., Okuda K., Gilmore R.C., et al. SARS-CoV-2 infection of the oral cavity and saliva. Nat. Med. 2021;27:892–903. doi: 10.1038/s41591-021-01296-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ferreira-Gomes M., Kruglov A., Durek P., Heinrich F., Tizian C., Heinz G.A., et al. SARS-CoV-2 in severe COVID-19 induces a TGF-beta-dominated chronic immune response that does not target itself. Nat. Commun. 2021;12:1961. doi: 10.1038/s41467-021-22210-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Aletaha D., Neogi T., Silman A.J., Funovits J., Felson D.T., Bingham C.O., 3rd, et al. Rheumatoid arthritis classification criteria: an American College of Rheumatology/European League against Rheumatism collaborative initiative. Arthritis Rheum. 2010;62:2569–2581. doi: 10.1002/art.27584. 2010. [DOI] [PubMed] [Google Scholar]
- 21.Wijburg O.L., Uren T.K., Simpfendorfer K., Johansen F.E., Brandtzaeg P., Strugnell R.A. Innate secretory antibodies protect against natural Salmonella typhimurium infection. J. Exp. Med. 2006;203:21–26. doi: 10.1084/jem.20052093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Spiekermann G.M., Finn P.W., Ward E.S., Dumont J., Dickinson B.L., Blumberg R.S., et al. Receptor-mediated immunoglobulin G transport across mucosal barriers in adult life: functional expression of FcRn in the mammalian lung. J. Exp. Med. 2002;196:303–310. doi: 10.1084/jem.20020400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Planas D., Saunders N., Maes P., Guivel-Benhassine F., Planchais C., Buchrieser J., et al. Considerable escape of SARS-CoV-2 Omicron to antibody neutralization. Nature. 2022;602:671–675. doi: 10.1038/s41586-021-04389-z. [DOI] [PubMed] [Google Scholar]
- 24.Sheikh-Mohamed S., Isho B., Chao G.Y.C., Zuo M., Cohen C., Lustig Y., et al. Mucosal Immunol; 2022. Systemic and Mucosal IgA Responses Are Variably Induced in Response to SARS-CoV-2 mRNA Vaccination and Are Associated with Protection against Subsequent Infection. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data will be made available on request.