Abstract
目的
旨在寻找与浸润性乳腺癌(BRCA)发生相关的肿瘤微环境(TME)相关基因,以预测其预后并为临床提供治疗靶点。
方法
从癌症基因组图谱(TCGA)数据库中检索到RNA转录组数据和临床相关数据。利用ESTIMATE算法计算基质评分和免疫评分。然后通过取交集筛选出差异表达基因(DEGs)。利用蛋白质-蛋白质相互作用(PPI)网络和单变量COX回归分析来确定DEGs中的核心基因。选取一个核心基因进行GSEA集富集分析和CIBERSORT分析,以分别区分核心基因表达的功能和肿瘤浸润免疫细胞(TICs)的比例。最终用Western blot和qRT-PCR对CD40LG的表达水平进行临床验证。
结果
从TCGA中提取了1222个样本(124个正常样本和1098个肿瘤样本)进行分析,共获得了487个DEG。这些基因主要富集在与免疫相关的途径中。进一步的交叉分析揭示了11个关键基因,包括CD40LG、ITK、CD5、CD3E、SPN、IL7R、CD48、CCL19、CD2、CD52和CD2711,这些基因被证明与乳腺癌TME状态相关。挑选了CD40LG进行进一步研究,结果表明,CD40LG高表达BRCA患者的总生存期(OS)比低表达BRCA患者更长(P=0.002),并且CD40LG表达在TNM分期、肿瘤大小方面的差异也具有统计学意义(P<0.05)。GSEA和CIBERSORT分析表明CD40LG的表达与TME中的免疫活性有关。Western blot和qRT-PCR结果显示CD40LG在乳腺癌细胞及癌组织中的蛋白及mRNA表达量均低于乳腺正常细胞及癌旁组织(P<0.05)。
结论
CD40LG在TME中高表达与BRCA患者的生存呈正相关,所以CD40LG可能是一种新的用于预后预测的生物标志物。其生物学行为可能促进我们对肿瘤进展的分子机制和靶向治疗的理解。
Keywords: 乳腺癌, 肿瘤微环境, 癌症基因组图谱, 肿瘤浸润免疫细胞, CD40LG
Abstract
Objective
To identify tumor microenvironment (TME)- related genes associated with the occurrence of invasive breast cancer as potential prognostic biomarkers and therapeutic targets.
Methods
RNA transcriptome data and clinically relevant data were retrieved from TCGA database, and the StromalScore and ImmuneScore were calculated using the ESTIMATE algorithm. The differentially expressed genes (DEGs) were screened by taking the intersection. A protein- protein interaction network was established, and univariate COX regression analysis was used to identify the core genes among the DEGs. A core gene was selected for GSEA and CIBERSORT analysis to determine the function of the core gene and the proportion of tumor-infiltrating immune cells, respectively. Western blotting and qRT-PCR were performed to verify the expression level of CD40LG in breast cancer cell lines and clinical specimens.
Results
A total of 1222 samples (124 normal and 1098 tumor samples) were extracted from TCGA for analysis, from which 487 DEGs were identified. These genes were mainly enriched in immune-related pathways, and crossover analysis identified 11 key genes (CD40LG, ITK, CD5, CD3E, SPN, IL7R, CD48, CCL19, CD2, CD52, and CD2711) associated with breast cancer TME status. CD40LG was selected as the core gene, whose high expression was found to be associated with a longer overall survival of breast cancer patients (P=0.002), and its expression level differed significantly with TNM stage and tumor size (P < 0.05). GSEA and CIBERSORT analyses indicated that CD40LG expression level was associated with immune activity in the TME. Western blotting and qRT-PCR showed that the protein and mRNA expression of CD40LG were significantly lower in breast cancer cells and cancer tissues than in normal breast cells and adjacent tissues.
Conclusions
The high expression of CD40LG in TME is positively correlated with the survival of patients with invasive breast cancer, suggesting its value as a potential new biomarker for predicting prognosis of the patients.
Keywords: breast cancer, tumor microenvironment, TCGA, tumor-infiltrating immune cells, CD40LG
乳腺癌是世界上女性最常见的癌症类型之一,也是癌症相关死亡的主要原因[1]。尽管随着技术的发展,乳腺癌的治疗取得了很大的进步,但它仍然是女性恶性肿瘤的常见死因[2]。研究表明仅靠临床病理特征不能为乳腺癌患者提供准确的预后信息,从而导致过度或不充分的治疗[3]。新的证据表明,使用肿瘤微环境(TME)相关免疫评分和TME中肿瘤浸润性免疫细胞(TIC)的比例提高了TNM分期系统的预测效率[4-5]。TME由肿瘤细胞、间质细胞、免疫细胞、细胞外基质(ECM)、信号分子和细胞因子组成[6],其中免疫细胞可以影响肿瘤的生长[7]。同时,基因表达异常与表观遗传学改变显著相关,表达特征可预测临床结果[8, 9]。因此,寻找一种可以预测癌症预后的生物标志物成为研究的热点。
研究显示目前已有CD2、CD52及ITK等基因被证实是乳腺癌免疫相关的预后因子[10-12]。有研究表明CD40LG修饰的肺癌细胞外泌体能有效激活树突状细胞,抑制肺癌的进展,延长小鼠的生存时间[13]。通过整合的生物信息学分析确定了CD40LG是乳腺癌的另一个免疫和TME调节密切相关的预后基因[14]。但已有的研究缺乏相关的临床实验验证。
本研究从肿瘤基因组图谱(TCGA)数据库下载乳腺癌转录组RNA-seq数据和临床相关数据,并在临床收集乳腺癌组织及癌旁组织。通过一系列生物信息学分析,11个基因被认为是乳腺癌的预后基因,其中CD40LG对乳腺癌患者的总生存期和临床预后具有很大的预测价值,进一步运用Western blot和qRT-PCR法进行临床验证,证实CD40LG在癌旁组织中高表达。CD40LG在TME中高表达与生存呈正相关,可能成为乳腺癌患者新的预后生物标志物和治疗靶点。
1. 资料和方法
1.1. 数据收集与处理
1.1.1. 临床资料
从宁夏医科大学总医院肿瘤医院收集2021年1月~2022年1月行乳腺癌改良根治术患者的癌组织24例和相对应的癌旁组织24例为研究对象,患者均为女性,平均年龄51岁。纳入标准:所有患者均经组织病理学确诊为浸润性乳腺癌;癌组织和癌旁组织(距肿瘤组织边缘大于5 cm)均取自同一确诊乳腺癌患者。排除标准:术前行新辅助治疗,如化疗、放疗等;本研究已通过宁夏医科大学总医院医学伦理委员会审查。
1.1.2. 数据库资料
本研究通过从癌症基因组图谱(TCGA)(https://portal.gdc.cancer.gov/repository) 下载乳腺癌数据集,获得了1 222个转录组RNA测序数据和临床数据(124个正常样本和1098个肿瘤样本)。在排除没有随访信息及随访时间较短的患者后,选择了1046名患者进行进一步分析。应用Estimate R软件包中的Expression Data(Estimate)算法,以免疫评分(Immune Score)、基质评分(Stromal Score)和总评分(Estimate Score)3个分数的形式对各样本的免疫成分和基质成分在TME中所占的比例进行估计,得分越高,相应成分在TME中所占的比例越大。
1.2. TME相关性分析
根据Stromal Score和Immune Score的中位数,将患者分为高、低基质评分组和高、低免疫评分组,以总生存期(OS)为主要终点,用Kaplan-Meier(KM)生存分析判断高、低基质和免疫评分组之间是否存在差异,亚组间比较采用对数秩检验,P<0.05认为差异有统计学意义。采用Kruskal- Wallis秩和检验评价Stromal Score、Immune Score和Estimate Score评分在年龄、性别、病理分期、T分期、N分期和M分期上是否存在差异,P<0.05时认为差异有统计学意义。
1.3. 鉴定差异表达基因(DEGs)
按高分和低分分组,采用“LIMMA”软件包进行筛查。筛选条件设置:|log2(FC)|>1和调整后的错误发现率(FDR)P<0.05。用R语言的pheatmap软件包绘制DEGs的热图。用clusterProfile、richlot和ggplot2R软件包对DEGs进行基因本体论(GO)和京都基因与基因组百科全书(KEGG)的富集分析并作图,P<0.05和Q<0.05的富集项目具有统计学意义,结果用气泡图和圆圈图可视化。蛋白质-蛋白质相互作用(PPI)使用交互置信度大于0.95的节点来构建网络。红色代表上调基因,绿色代表下调基因,利用Cytoscape中的CytosHubba插件进行模块分析,根据多网络聚类(MNC)的方法筛选出前30个有意义的基因。
1.4. 单因素COX回归分析
为了确定哪些DEGs与OS相关,我们使用“survival”R包进行单变量COX回归分析,确定与总生存期相关的基因(P<0.05)。
1.5. CD40LG差异表达及生存分析
通过“bee colony”和“ggpubr”R软件包,采用Wilcoxon秩和检验比较肿瘤组织和正常组织中CD40LG mRNA的表达。根据表达中位数将样本分为高表达组和低表达组,两组均采用“survival”R包进行生存分析。通过“ggpubr”R软件包采用Kruskal-Wallis秩和检验进行临床相关性分析,评价CD40LG表达与病理分期、T分期、M分期、N分期的关系。
1.6. GSEA(Gene Set Enrichment Analysis)分析
采用GSEAv4.1.0软件进行基因集富集分析(GSEA),确定CD40LG高、低表达的相关生物学通路。当P<0.05时,认为该通路显著富集。
1.7. 免疫细胞浸润的计算
使用CIBERSORT计算方法来估计肿瘤免疫细胞(即TICs)在所有肿瘤样本中丰度分布的比例,并使用柱状图可视化。采用Wilcoxon秩和检验比较CD40LG高、低表达肿瘤组织中CD40LG表达的比例,并评价CD40LG表达与肿瘤组织比例之间的Pearson相关性。使用“violot”“、ggplot22”和“ggpubr”R软件包制作散点图和小提琴图。
1.8. Western blot实验
主要试剂:乳腺细胞(MCF10A、MDA-MB-231、MDA-MB-468、MCF-7、SK-BR-3、BT-474)(赛库生物),液氮保存。兔抗人CD40LG单克隆抗体及山羊抗兔二抗(Proteintech);蛋白总浓度测定(BCA法)蛋白定量检测试剂盒、全蛋白提取试剂盒和SDS-PAGE凝胶试剂盒均(南京凯基生物)。
从液氮罐中取出BRCA患者的癌组织及癌旁组织标本,手工充分磨碎后离心取上清液,通过蛋白总浓度测定(BCA法)测蛋白浓度。8% SDS-PAGE凝胶电泳,将凝胶中的蛋白转移到PVDF膜上,5%脱脂奶粉封闭60 min;放入一抗工作液,4℃孵育过夜;TBST洗膜,5 min×5次;放入相应二抗工作液中,室温60 min;TBST洗膜,5 min×5次;通过ECL显影,用Image J软件对条带进行灰度值检测,结果用计算公式:目的蛋白的相对表达量=目的蛋白吸光度值/β-Actin吸光度值。从液氮中取出细胞,解冻后进行离心、培养,将收取的细胞用蛋白裂解液充分裂解提取细胞总蛋白,严格按照全蛋白提取试剂盒操作,后续操作同上。
1.9. qRT-PCR法
首先进行试剂配置及器皿处理,细胞处理同上;分别提取乳腺正常细胞及乳腺癌细胞和癌组织及癌旁组织中总RNA,并对提取的RNA进行质量检测;根据RNA反转录试剂盒的步骤逆转录合成cDNA,以cDNA为模板,按照公司说明书操作配制反应体系,将U6作为内参,实验结果采用2-ΔΔCt法进行表达量相对定量分析。qRT-PCR引物序列:CD40LG上游引物:5'-AATACCCACAGTTCCGCCAAACC-3';下游引物:5'-CATTGACAAACACCGAAGCACCTG-3';U6上游引物:5'-GATTCCTATGTGGGCGACGAG-3',下游引物: 5'-CCATCTCTTGCTCGAAGTCC-3',由上海生工生物工程股份有限公司合成。
2. 结果
图 1是本研究的流程图。我们从TCGA数据库中提取了转录组数据和临床数据(表 1),包括124个正常样本和1098个肿瘤样本。临床数据包括年龄、性别、TNM分期、T分期、N分期、M分期及总生存期和生存状态。利用ESTIMATE和CIBERSORT算法对转录组数据进行分析。通过在高、低Immune Score组和Stromal Score组之间取交集获得了DEGs。建立PPI网络,进行单因素COX回归分析,并对网络核心基因和存活相关基因取交集,共获得了CD40LG、ITK、CD5、CD3E、SPN、IL7R、CD48、CCL19、CD2、CD52、CD27等11个预后基因。我们选择CD40LG进行了进一步的分析,包括差异表达分析、生存分析、临床相关性分析和GSEA富集分析。最后运用Western blot实验和qRT-PCR法分别验证CD40LG在乳腺正常细胞及不同乳腺癌细胞和BRCA组织及癌旁组织中的表达水平。
图 1.

本研究的流程图
Flow chart of this study.
表 1.
TCGA临床基线资料表
TCGA clinical baseline data sheet
| ID | Futime | Fustat | Age | Gender | Stage | T | M | N |
| TCGA-A8-A097 | 30 | 0 | 65 | Female | ⅡB | T2 | M0 | N1 |
| TCGA-BH-A1ET | 30 | 1 | 55 | Female | Ⅰ | T1 | M0 | N0 |
| TCGA-AR-A2LQ | 30 | 0 | 59 | Female | ⅡB | T3 | M0 | N0 |
| TCGA-E9-A5FK | 30 | 0 | 60 | Female | ⅢC | T2 | M0 | N3 |
| TCGA-E9-A22G | 30 | 0 | 47 | Female | ⅡA | T2 | M0 | N0 |
| TCGA-GI-A2C8 | 31 | 0 | 63 | Female | ⅢB | T4b | MX | N0 |
| TCGA-AC-A8OS | 31 | 0 | 71 | Female | ⅡA | T2 | MX | N0 (i-) |
| TCGA-AC-A7VB | 31 | 0 | 51 | Female | ⅡA | T1c | MX | N1a |
| TCGA-A2-A0T1 | 31 | 0 | 55 | Female | ⅢC | T3 | M0 | N3 |
| TCGA-OL-A5RW | 34 | 0 | 40 | Female | ⅡA | T1c | MX | N1 |
| TCGA-AR-A1AJ | 34 | 0 | 83 | Female | Ⅰ | T1 | M0 | N0 |
| TCGA-E9-A243 | 40 | 0 | 52 | Female | ⅡA | T2 | M0 | N0 (i-) |
| TCGA-A2-A0EP | 49 | 0 | 56 | Female | Ⅰ | T1 | M0 | N0 (i+) |
| TCGA-AC-A2FF | 51 | 0 | 40 | Female | ⅡB | T2 | MX | N1 |
| TCGA-AR-A1AL | 52 | 0 | 60 | Female | ⅢA | T3 | M0 | N1 |
| TCGA-A8-A0A4 | 54 | 0 | 73 | Female | ⅡA | T2 | M0 | N0 |
| TCGA-A8-A07G | 64 | 0 | 65 | Female | ⅡA | T1c | M0 | N1a |
| TCGA-E2-A10B | 70 | 0 | 67 | Female | ⅡB | T2 | M0 | N1a |
| TCGA-AR-A24U | 72 | 0 | 47 | Female | ⅡA | T1 | M0 | N1 |
| TCGA-B6-A40B | 76 | 0 | 76 | Female | Ⅰ | T1b | M0 | N0 (i-) |
| TCGA-E2-A15K | 78 | 0 | 58 | Female | ⅡB | T2 | M0 | N1a |
| TCGA-AC-A5XS | 78 | 0 | 74 | Female | ⅡA | T2 | MX | N0 |
| TCGA-A7-A4SE | 78 | 0 | 54 | Female | ⅡA | T2 | M0 | N0 |
| TCGA-GM-A5PV | 79 | 0 | 63 | Female | ⅡB | T2 | M0 | N1a |
| TCGA-E9-A295 | 80 | 0 | 71 | Female | ⅡA | T2 | M0 | N0 (i-) |
| TCGA-BH-A0DD | 84 | 0 | 58 | Male | ⅡB | T2 | M0 | N1a |
| TCGA-AR-A2LO | 90 | 0 | 46 | Female | ⅡB | T2 | M0 | N1 |
| TCGA-E9-A1N8 | 92 | 0 | 48 | Female | ⅡA | T2 | M0 | N0 |
| TCGA-A2-A0CW | 98 | 0 | 67 | Female | ⅡB | T2 | M0 | N1a |
| TCGA-A7-A0DC | 116 | 0 | 63 | Female | ⅠA | T1c | M0 | N0 (i-) |
| TCGA-AO-A0J9 | 118 | 0 | 61 | Female | ⅢC | T2 | M0 | N3 |
| TCGA-A8-A09Q | 134 | 0 | 83 | Female | ⅢB | T4b | M0 | N2a |
| TCGA-C8-A26V | 149 | 0 | 47 | Female | ⅢA | T3 | M0 | N2 |
| TCGA-EW-A6S9 | 158 | 0 | 34 | Female | ⅡA | T1c | M0 | N1 |
| TCGA-BH-A1F2 | 160 | 1 | 53 | Female | ⅢB | T4b | M0 | N1b |
| TCGA-E2-A1B5 | 160 | 0 | 46 | Female | ⅡA | T2 | M0 | N0 |
| TCGA-AO-A0JD | 162 | 0 | 59 | Female | ⅢA | T3 | M0 | N1a |
| TCGA-E9-A1RI | 163 | 0 | 43 | Female | ⅢA | T1c | M0 | N2 |
| ... | ... | ... | ... | ... | ... | ... | ... | ... |
| TCGA-E2-A1LK | 8605 | 1 | 84 | Female | ⅢC | T4b | M0 | N3a |
2.1. ImmuneScore与乳腺癌患者生存率相关
根据各评分中位数(ImmuneScore、StromalScore、EstimatScore)将1046份BRCA样本分为高、低评分组。采用Kaplan-Meier生存分析,探讨各评分与总生存率的相关性。结果显示,免疫评分与OS显著相关,评分高的患者生存率高于评分低的患者(P=0.011,图 2A)。然而,患者的StromalScore、EstimatScore和OS之间无显著差异(图 2B、C)。
图 2.

ImmuneScore, StromalScore, and ESTIMATEScore与BRCA患者生存率的关系
Association of ImmuneScore, StromalScore, and ESTIMATEScorewith survival rate of BRCA patients. Kaplan-Meier survival analysis was performed in BRCA patients with high and low scores. A: ImmuneScores (P=0.011). B: StromalScores (P=0.784). C: ESTIMATEScores (P=0.142).
2.2. ImmuneScore、StromalScore和ESTIMATEScore与临床病理特征的关系
为了确定免疫成分和基质成分的比例与临床病理特征之间的关系,我们从TCGA数据库中分析了BRCA样本的临床信息,包括年龄、性别、原发肿瘤大小(T分期)、区域淋巴结状态(N分期)、远处转移(M分期)和病理分期。ImmuneScore与年龄、性别显著相关(P= 0.048;P=0.014),但在T分期、N分期、M分期及病理分期中差异无统计学意义(P>0.05,图 3A)。StromalScore与年龄(P=0.033)、T分期、N分期、Ⅱ期与Ⅰ期(P= 0.0082)、Ⅱ期与Ⅲ期(P=0.0077)相关,但与性别和M期无显著相关性(P>0.05,图 3B)。EstimateScore与N分期、M分期、病理分期差异均无统计学意义(P>0.05),与年龄、性别、T分期相关性显著(图 3C)。
图 3.

BRCA患者ImmuneScore(A), StromalScore(B)and ESTIMATEScore(C)与年龄、性别、T分期、N分期、M分期和病理分期的关系
Correlation of ImmuneScore (A), StromalScore (B), ESTIMATEScore (C) with age, sex, T stage, N stage, M stage and pathological stage of the patients with BRCA.
2.3. 共享DEGs主要富含免疫相关功能
为了确定TME中免疫成分和基质成分之间的差异,我们分析了TCGA数据库中1046例BRCA患者的转录组数据,并比较了ImmuneScore高低组和StromalScore组之间的基因表达,ImmuneScore高组和低组之间有1446个DEGs(1256个上调,190个下调)(图 4A、B),StromalScore高、低组之间有1330个DEGs(1118个上调,212个下调)(图 4C、D)。分别于两组中上调(图 4E)和下调(图 4F)基因取交集,最终获得487个共享DEG(s其中447个上调,40个下调)。
图 4.

DEGs的热图、火山图、维恩图及GO和KEGG富集分析
Heatmap, Volcano plots, Venn diagrams, and GO and KEGG enrichment analyses of the differentially expressed genes (DEGs). A-C: Heatmap and Volcano plots of DEGs were obtained by comparing the high score group ImmuneScore (A, B) and the low score group StromalScore(C, D) (FDR adjustment P < 0.05, | log FC| > 1). E, F: Venn diagrams of upregulated (E) and downregulated (F) DEGs shared by the ImmuneScore and StromalScore analyses. G, H: Bubble diagram of GO (G) and KEGG (H) enrichment analysis (P < 0.05 vs Q < 0.05).
利用GO和KEGG对这些共享基因进行分析,预测其功能。GO富集分析结果表明,DEGs主要富集在免疫应答激活细胞表面受体和淋巴细胞免疫等免疫相关信号通路中(图 4G)。KEGG富集分析表明,DEGs富集于细胞因子-细胞因子受体相互作用、细胞黏附分子、造血细胞系等(图 4H)。
2.4. 通过PPI网络和单变量COX回归分析获得预后基因
利用487个DEGs通过STRING数据库建立PPI网络,最小置信值为0.9,由Cytoscape软件显示(图 5A)。然后按照节点数的顺序得到前30个网络核心基因(5B)。为了确定提取的基因是否为影响乳腺癌预后的关键基因,对487个DEGs进行单因素COX回归分析,截断值P<0.05(图 5C)。最后,通过对PPI网络中前30个网络核心基因和单变量COX回归分析的结果取交集(图 5D),获得了CD40LG、ITK、CD5、CD3E、SPN、IL7R、CD48、CCL19、CD2、CD52、CD27共11个预后基因。
图 5.

PPI网络与COX回归分析
PPI network and COX regression analysis. A, B: The PPI network (A) is established with a minimum confidence value of 0.9, and the visualization was carried out using Cytoscape (B). MNC algorithm was used to identify the core genes in PPI network. C, D: The red node represents the gene with higher MNC score, the yellow node represents the gene with lower MNC score (C), and the first 30 genes with higher score (D). E: Univariate COX regression analysis with P < 0.05 as standard. F: Venn diagram after intersection between the first 30 genes from PPI network and the results of univariate COX regression analysis.
2.5. CD40LG的差异表达、生存分析及临床病理特征分析
我们的研究显示,CD40LG在正常组织和配对组织中的表达明显高于肿瘤样本(P<0.05,图 6A、B)。根据CD40LG中位表达情况,将肿瘤标本分为高表达组和低表达组。与CD40LG低表达组相比,BRCA患者高表达组OS显著延长(图 6C)。在临床病理分析中,CD40LG表达与病理分期、T分期显著相关。CD40LG在TNM Ⅳ期的表达低于其他分期(Ⅳ期vs Ⅰ期、Ⅱ期、Ⅲ期,P值分别为0.021、0.046、0.021)(图 6D)。同样,CD40LG的表达也与T分期呈负相关(T4 vs T1、T2、T3,P值分别为0.0024、0.01、0.0037)(图 6E)。但在N分期和M分期两组间差异无统计学意义(P>0.05,图 6F、G)。
图 6.
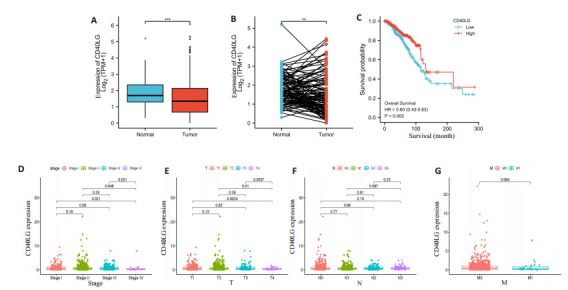
CD40LG的表达与患者生存时间及临床特征的关系
Correlation of CD40LG expression with survival time and clinical characteristics of patients. A, B: There was a significant difference in the expression of CD40LG between tumor tissues and normal tissues. C: Survival analysis of BRCA patients with low and high expression of CD40LG. D-G: Kruskal-Wallis rank sum test for the correlation between CD40LG expression and pathological stage, T stage, N stage and M stage. ***P < 0.001, **P < 0.01.
2.6. CD40LG相关的生物学途径
GSEA用于研究CD40LG表达的生物学途径,CD40LG的高表达主要集中在免疫相关通路(图 7),如B细胞受体信号通路、T细胞受体信号通路、JAK-STAT信号通路、趋化因子信号通路、Toll样受体信号通路等。
图 7.

CD40LG高表达BRCA样本的GSEA分析
GSEA of BRCA samples with high CD40LG expression.
2.7. CD40LG表达与TICs的关系
CD40LG高表达组和低表达组共获得22种免疫细胞(图 8A)。通过差异分析和相关分析,发现14种免疫细胞与CD40LG的表达有关,包括B细胞、T细胞CD8、T细胞CD4记忆静息、T细胞CD4记忆激活、T细胞滤泡辅助细胞、T细胞调节因子(Tregs)、T细胞γδ、NK细胞静息、巨噬细胞M0、M1、M2、树突状细胞静息、肥大细胞静息、中性粒细胞。
图 8.

CD40LG高表达组与低表达组TIC比例的差异及其与CD40LG表达的相关性
Differences in proportions of TICs between high and low CD40LG expression groups and correlations of TICs with CD40LG expression. A: The proportions of 22 immune cell types in tumor tissues with high CD40LG expression (red) and low expression (green) are compared. B: Scatter plot showing the Pearson correlation between the proportion of the 12 most significant TICs and CD40LG expression. The blue line represents the most suitable linear model (B).
2.8. 实验测定CD40LG在细胞及临床标本中的表达
qRT-PCR显示,CD40LG在BRCA组织中mRNA表达水平显著低于癌旁组织(P<0.05,图 9A);同时,Western blot结果显示,CD40LG在BRCA组织中蛋白表达水平也显著低于癌旁组织(P<0.05,图 9B、C)。
图 9.

CD40LG的差异表达验证
Validation of differential expression of CD40LG. A, B: CD40LG mRNA expression level in cancer tissue and adjacent tissue (A) and the corresponding proliferation curve and dissolution curve (B). C, D: CD40LG mRNA expression levels in normal breast cells and different breast cancer cells (C) and the corresponding proliferation curve and dissolution curve (D). E, F: Protein expression level of CD40LG in normal breast cells and different breast cancer cells determined by Western blotting. G, H: Protein expression level of CD40LG in cancer tissues and adjacent tissues determined by Western blotting. *P < 0.05, ***P < 0.001.
3. 讨论
越来越多的证据表明,分子靶向治疗是肿瘤治疗的一个很有前途的研究方向[15],因此迫切需要探索乳腺癌发生发展的详细分子机制[16, 17]。目前,TME的研究因其在治疗和预后方面的重要意义而成为研究热点[18, 19],乳腺癌的TME也引起了许多专家的关注。
本研究的目的是基于CIBERSORT和ESTIMATE两种算法,试图确定与BRCA患者肿瘤发生相关的TME核心基因,并可能成为临床治疗的靶点。通过综合生物信息学分析,我们得到了以下结果:ImmuneScore与BRCA患者的OS显著相关,评分高的患者生存率高于评分低的患者;StromalScore与T分期、N分期及病理分期有关,ImmuneScore及ESTIMATEScore与年龄、性别相关;DEGs主要富集在免疫相关的信号通路中;CD40LG在正常及配对组织中的表达高于肿瘤组织;高表达组患者的OS优于低表达组,且CD40LG在TNM Ⅳ期和T分期T4期的表达均低于其他各分期;通过差异分析及相关性分析,我们发现14种免疫细胞与CD40LG表达相关。
尽管已有的研究[14]表明CD40LG是BRCA患者有价值的预后分子标志物,其低表达与预后不良有关,但缺乏相关的临床实验验证,我们的研究从细胞和组织两个层面验证了CD40LG在BRCA中的表达水平;其次,Yuan等[14]只关注了免疫成分在乳腺癌中的作用,而忽视了基质成分,本研究发现StromalScore与BRCA患者的临床病理特征也具有相关性。
ESTIMATE算法可以预测肿瘤纯度并使用ImmuneScore和StromalScore描述肿瘤组织中浸润的免疫细胞、基质细胞的比例[20, 21]。基于TCGA转录组数据,我们观察到高ImmuneScore组患者的OS显著延长,这表明免疫成分是预测预后的潜在因素。这与以往的研究[22, 23]一致。已发表的研究表明[24, 25],在结肠癌中ImmuneScore是预测结肠癌患者复发风险的可靠、有效的临床方法,其对预后的评估优于包括TNM、MSI在内的其他预后因素。在临床相关性方面,本研究显示StromalScore与T分期、N分期及TNM分期有关,这提示基质细胞含量在一定程度上影响着乳腺癌的侵袭、迁移等方面的病程发展。在乳腺癌的小鼠模型中,基质细胞的生长促进作用因糖酵解抑制而受损,表明基质通过分泌乳酸盐为恶性细胞提供营养支持[26]。但值得注意的是ImmuneScore与患者的各分期并不相关,考虑可能是由不同肿瘤阶段免疫细胞浸润类型的差异导致的。
通过GO及KEGG富集分析发现DEGs主要富集在免疫相关的途径中。为了进一步研究预后基因,我们建立了PPI网络,并用DEGs进行了单变量COX回归分析,在此基础上确定了CD40LG、ITK、CD5、CD3E、SPN、IL7R、CD48、CCL19、CD2、CD52和CD2711个预后基因。因CD40LG的HR最小,提示可能是更好的保护因素,所以选择该基因作进一步分析。
CD40LG是与CD40结合的细胞因子,一种Ⅱ型跨膜蛋白,属于肿瘤坏死因子(TNF)基因超家族[27]。CD40-CD40LG相互作用是B细胞增殖、活化标志物表达、免疫球蛋白产生和同型转换的重要信号,可直接抑制CD40阳性癌细胞的生长,并可能通过协调免疫反应间接抑制肿瘤的生长[28, 29]。本研究中CD40LG在正常及配对组织中的表达高于肿瘤组织,且其高表达BRCA患者的OS显著延长;通过qRT-PCR和Western blot实验也证实了癌组织中的CD40LG在mRNA和蛋白水平的表达低于癌旁组织。有研究报道了一种利用神经干细胞的肿瘤归巢特性将CD40LG分子运送到肿瘤组织中的治疗方法,即用编码CD40LG的杆状病毒载体体外转导,静脉注射到免疫活性良好的原位乳腺癌和转移性乳腺癌小鼠体内,发现治疗阻碍了肿瘤的生长,使荷瘤小鼠的存活时间延长[30]。在临床病理特征方面,CD40LG在TNM Ⅳ期和T分期T4期的表达均低于其他各期,表明CD40LG的低表达可能与BRCA的侵袭和转移密切相关,导致预后不良。
为了探讨CD40LG表达与BRCA患者TME的关系,我们进行了GSEA富集分析。分析发现CD40LG高表达主要涉及免疫相关信号通路,如B细胞受体信号通路、T细胞受体信号通路、JAK-STAT信号通路、趋化因子信号通路、Toll样受体信号通路等。这些结果证实了CD40LG可以通过不同的途径调控BRCA的免疫细胞。此后,通过CIBERSORT计算方法从BRCA样本中获得了22种免疫细胞,其中14种与CD40LG的表达相关。这表明CD40LG的生物学行为取决于表达水平和TIC表型,这可能为靶向治疗和分子生物学进展提供新的见解。但具体的作用机制目前并不清楚,有待进一步的研究。
综上所述,本研究采用多种算法证实TME中免疫成分高的BRCA患者具有更好的总生存期。此外,我们还筛选出了一个免疫相关基因CD40LG,该基因在TME中高表达与BRCA患者的生存呈正相关,且临床实验证实与癌组织相比,CD40LG在正常组织中高表达。因此,CD40LG可能是一种新的用于预后预测的生物标志物。另外,CD40LG主要富集在免疫相关的信号通路中,有望成为免疫治疗的靶点。
Biography
郭丽,博士,E-mail: 18309588393@163.com
Funding Statement
国家自然科学基金(82060479)
Supported by National Natural Science Foundation of China (82060479)
Contributor Information
郭 丽 (Li GUO), Email: 18309588393@163.com.
李 金平 (Jinping LI), Email: 2634497264@qq.com.
References
- 1.Shachar SS, Deal AM, Weinberg M, et al. Body composition as a predictor of toxicity in patients receiving anthracycline and taxane-based chemotherapy for early-stage breast cancer. Clin Cancer Res. 2017;23(14):3537–43. doi: 10.1158/1078-0432.CCR-16-2266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ferreira MA, Gamazon ER, Al-Ejeh F, et al. Genome-wide association and transcriptome studies identify target genes and risk loci for breast cancer. Nat Commun. 2019;10(1):1741. doi: 10.1038/s41467-018-08053-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pashayan N, Antoniou AC, Ivanus U, et al. Personalized early detection and prevention of breast cancer: ENVISION consensus statement. Nat Rev Clin Oncol. 2020;17(11):687–705. doi: 10.1038/s41571-020-0388-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ma CX, Kang WY, Yu L, et al. AUNIP expression is correlated with immune infiltration and is a candidate diagnostic and prognostic biomarker for hepatocellular carcinoma and lung adenocarcinoma. Front Oncol. 2020;10(2):590006. doi: 10.3389/fonc.2020.590006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang J, Bo XB, Wang CC, et al. Low immune index correlates with favorable prognosis but with reduced benefit from chemotherapy in gallbladder cancer. Cancer Sci. 2020;111(1):219–28. doi: 10.1111/cas.14239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen R, Lu XY, Li Z, et al. Dihydroartemisinin prevents progression and metastasis of head and neck squamous cell carcinoma by inhibiting polarization of macrophages in tumor microenvironment. Onco Targets Ther. 2020;13(4):3375–87. doi: 10.2147/OTT.S249046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hegde PS, Chen DS. Top 10 challenges in cancer immunotherapy. Immunity. 2020;52(1):17–35. doi: 10.1016/j.immuni.2019.12.011. [DOI] [PubMed] [Google Scholar]
- 8.Deng JL, Zhang HB, Zeng Y, et al. Effects of CORO2A on cell migration and proliferation and its potential regulatory network in breast cancer. Front Oncol. 2020;10(7):916. doi: 10.3389/fonc.2020.00916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ruan HL, Li S, Tong JW, et al. The screening of pivotal gene expression signatures and biomarkers in renal carcinoma. J Cancer. 2019;10(25):6384–94. doi: 10.7150/jca.30656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen YZ, Meng ZS, Zhang L, et al. CD2 is a novel immune-related prognostic biomarker of invasive breast carcinoma that modulates the tumor microenvironment. Front Immunol. 2021;12(8):664845. doi: 10.3389/fimmu.2021.664845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ma YF, Chen YC, Fang DL, et al. The immune-related gene CD52 is a favorable biomarker for breast cancer prognosis. Gland Surg. 2021;10(2):780–98. doi: 10.21037/gs-20-922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huang YZ, Chen LZ, Tang ZY, et al. A novel immune and stroma related prognostic marker for invasive breast cancer in tumor microenvironment: a TCGA based study. Front Endocrinol (Lausanne) 2021;12:774244. doi: 10.3389/fendo.2021.774244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang C, Huang X, Wu YJ, et al. Tumor cell-associated exosomes robustly elicit anti-tumor immune responses through modulating dendritic cell vaccines in lung tumor. Int J Biol Sci. 2020;16(4):633–43. doi: 10.7150/ijbs.38414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yuan MQ, Pei JY, Li RH, et al. CD40LG as a prognostic molecular marker regulates tumor microenvironment through immune process in breast cancer. Int J Gen Med. 2021;14:8833–46. doi: 10.2147/IJGM.S336813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li C, Liu SW, Yan RP, et al. CD54- NOTCH1 axis controls tumor initiation and cancer stem cell functions in human prostate cancer. Theranostics. 2017;7(1):67–80. doi: 10.7150/thno.16752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chang KC, Diermeier SD, Yu AT, et al. MaTAR25 lncRNA regulates the Tensin1 gene to impact breast cancer progression. Nat Commun. 2020;11(1):6438. doi: 10.1038/s41467-020-20207-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang J, He YT, Yu Y, et al. Upregulation of miR-374a promotes tumor metastasis and progression by downregulating LACTB and predicts unfavorable prognosis in breast cancer. Cancer Med. 2018;7(7):3351–62. doi: 10.1002/cam4.1576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lei X, Lei Y, Li JK, et al. Immune cells within the tumor microenvironment: biological functions and roles in cancer immunotherapy. Cancer Lett. 2020;470(6):126–33. doi: 10.1016/j.canlet.2019.11.009. [DOI] [PubMed] [Google Scholar]
- 19.Jiang YT, Wang CD, Zhou ST. Targeting tumor microenvironment in ovarian cancer: premise and promise. Biochim Biophys Acta Rev Cancer. 2020;1873(2):188361. doi: 10.1016/j.bbcan.2020.188361. [DOI] [PubMed] [Google Scholar]
- 20.Conway JR, Herrmann D, Evans TJ, et al. Combating pancreatic cancer with PI3K pathway inhibitors in the era of personalised medicine. Gut. 2019;68(4):742–58. doi: 10.1136/gutjnl-2018-316822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mao M, Yu QL, Huang RZ, et al. Stromal score as a prognostic factor in primary gastric cancer and close association with tumor immune microenvironment. Cancer Med. 2020;9(14):4980–90. doi: 10.1002/cam4.2801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Azizi E, Carr AJ, Plitas G, et al. Single-cell map of diverse immune phenotypes in the breast tumor microenvironment. Cell. 2018;174(5):1293–308.e36. doi: 10.1016/j.cell.2018.05.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fox NS, Haider S, Harris AL, et al. Landscape of transcriptomic interactions between breast cancer and its microenvironment. Nat Commun. 2019;10(1):3116. doi: 10.1038/s41467-019-10929-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.PagèS F, Mlecnik B, Marliot F, et al. International validation of the consensus Immunoscore for the classification of colon cancer: a prognostic and accuracy study. Lancet. 2018;391(10135):2128–39. doi: 10.1016/S0140-6736(18)30789-X. [DOI] [PubMed] [Google Scholar]
- 25.Wu ZY, Shen W, Yue JQ, et al. Combining immunoscore with clinicopathologic features in cholangiocarcinoma: an influential prognostic nomogram. Onco Targets Ther. 2020;13(2):11359–76. doi: 10.2147/OTT.S274754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Faubert B, Li KY, Cai L, et al. Lactate metabolism in human lung tumors. Cell. 2017;171(2):358–71.e9. doi: 10.1016/j.cell.2017.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kuhn NF, Purdon TJ, van Leeuwen DG, et al. CD40 ligand-modified chimeric antigen receptor T cells enhance antitumor function by eliciting an endogenous antitumor response. Cancer Cell. 2019;35(3):473–88.e6. doi: 10.1016/j.ccell.2019.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Laman JD, Claassen E, Noelle RJ. Functions of CD40 and its ligand, gp39 (CD40L) Crit Rev Immunol. 2017;37(2/3/4/5/6):371–420. doi: 10.1615/CritRevImmunol.v37.i2-6.100. [DOI] [PubMed] [Google Scholar]
- 29.Jansen MF, Hollander MR, van Royen N, et al. CD40 in coronary artery disease: a matter of macrophages? Basic Res Cardiol. 2016;111(4):38. doi: 10.1007/s00395-016-0554-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhu DT, Chen C, Purwanti YI, et al. Induced pluripotent stem cell-derived neural stem cells transduced with baculovirus encoding CD40 ligand for immunogene therapy in mouse models of breast cancer. Hum Gene Ther. 2014;25(8):747–58. doi: 10.1089/hum.2013.160. [DOI] [PubMed] [Google Scholar]


