Abstract
Nipah virus (NiV) and Hendra virus (HeV) are highly pathogenic zoonotic viruses of the genus Henipavirus, family Paramyxoviridae that cause severe disease outbreaks in humans and also can infect and cause lethal disease across a broad range of mammalian species. Another related Henipavirus has been very recently identified in China in febrile patients with pneumonia, the Langya virus (LayV) of probable animal origin in shrews. NiV and HeV were first identified as the causative agents of severe respiratory and encephalitic disease in the 1990s across Australia and Southern Asia with mortality rates reaching up to 90%. They are responsible for rare and sporadic outbreaks with no approved treatment modalities. NiV and HeV have wide cellular tropism that contributes to their high pathogenicity. From their natural hosts bats, different scenarios propitiate their spillover to pigs, horses, and humans. Henipavirus-associated respiratory disease arises from vasculitis and respiratory epithelial cell infection while the neuropathogenesis of Henipavirus infection is still not completely understood but appears to arise from dual mechanisms of vascular disease and direct parenchymal brain infection. This brief review offers an overview of direct and indirect mechanisms of HeV and NiV pathogenicity and their interaction with the human immune system, as well as the main viral strategies to subvert such responses.
Keywords: Henipaviruses, Nipah, Hendra, Langya, Outbreak, Epidemiology, Pathogenesis, Antiviral therapy
General aspects and taxonomy
Nipah henipavirus (NiV) and Hendra henipavirus (HeV) are negative single-stranded RNA viruses that belong to the Paramyxoviridae family and define the genus Henipavirus together with Cedar henipavirus (CedV), Ghanaian bat henipavirus, Mojiang henipavirus, and the very recently identified Langya henipavirus (LayV) in patients from China. NiV and HeV can infect a wide range of wild and domestic animals, as well as humans, in whom they exhibit a high level of pathogenicity mainly causing a pulmonary or encephalitic henipavirus-mediated disease with observed fatality rates of up to 60% and 90% for HeV and NiV, respectively, thus being classified as biosafety level 4 (BSL-4) agents [1]. The major natural reservoir hosts for Henipaviruses appear to be the Pteropus bat species [2–5], as natural hosts do not exhibit any evident disease neither in wild bats nor experimentally infected bats [6].
HeV was isolated and identified in 1994 in an outbreak of fatal cases of respiratory disease in horses and humans in the Brisbane suburb of Hendra, Australia, and was shown to be distantly related to the measles virus and other morbilliviruses [7]. A few years later NiV emerged in a large outbreak of encephalitis among pig farmers in Peninsular Malaysia that began in 1998 and continued into the next year [8]. Very recently, Langya henipavirus (LayV) was identified in China as a new member of the genus most closely phylogenetically related to Mojiang henipavirus. LayV has a probable animal origin (mainly from shrews) but human-to-human transmission has not been documented [9]. LayV infection is associated with febrile illness and pneumonia [10]. Although not associated with a zoonotic event, CedV is another recognized henipavirus species that was identified in a flying-fox colony in Cedar Grove in Queensland, Australia [11]. Finally, the Mojiang henipavirus (MojV) was identified in rats (Rattus flavipectus) in China [12].
Viral particles, genomic organization, and viral tropism
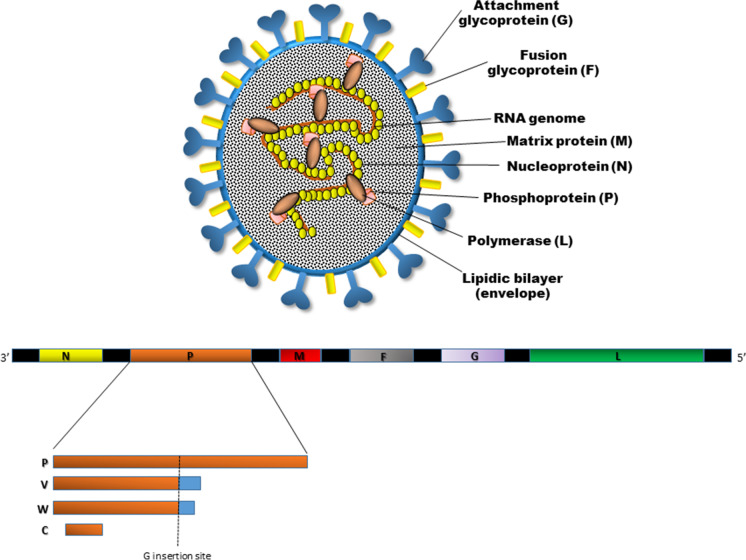
NiV and HeV particles are enveloped and pleomorphic, with a size ranging from 40 to 1900 nm and can vary from spherical to filamentous forms when imaged by electron microscopy. The viral envelope is a lipid bilayer derived from the infected host cell during virus assembly and budding that carries surface projections composed of the viral transmembrane-anchored fusion (F) and attachment (G) glycoproteins. The ribonucleic (RNP) complex shows a characteristic herringbone shape and is responsible for the replication of viral RNA. RNP is formed by N, P, and L proteins and viral RNA [5]. The genetic organization of NiV and HeV resembles that found in viruses in the Respirovirus and Morbillivirus genera in the Orthoparamixovirinae subfamily (Fig. 1). The genome is a non-segmented single-stranded negative-sense RNA of around 18.2 kb, thus around 2700 bases longer than other paramyxovirus genomes. It encodes 6 structural proteins including the nucleocapsid (N), the phosphoprotein (P), the matrix protein (M), the surface glycoprotein (G) and fusion (F) protein, and the viral polymerase (L). In addition, the P gene encodes three non-structural proteins (C, V, W) expressed in infected cells that derive from overlapping open reading frames (ORFs). The V and W proteins are produced through a transcriptional editing mechanism involving the addition of non-templated G nucleotides, while the C protein is encoded by an alternative start site within the P gene [13, 14] (Table 1).
Fig. 1.
Schematic representation of the structure of a Henipavirus particle (A) and the viral RNA genome organization (B). The N, P, and L proteins interact with the viral RNA to form the ribonucleoprotein (RNP) complex, which is bounded by a lipid bilayer envelope containing the viral glycoproteins F and G. The M protein is associated with the inner side of the envelope. The viral proteins and arrangement of genes in the viral genome from 3′–5′. The viral P gene products (V, W, and C proteins) as a result of mRNA editing are explained. The V protein contains a single G insertion, and translation shifts it to a + 1 reading frame. The W protein encompasses two G insertions, shifting the translation to the + 2 reading frame. The C protein is translated from an internal open reading frame of the P gene
Table 1.
Main features of Henipavirus (NiV and HeV)
| Virion | Enveloped, pleomorphic (mostly spherical) virions with a diameter of 300–500 nm enclosing a ribonucleoprotein |
|---|---|
| Genome | Non-segmented single-stranded negative-sense RNA |
| Genome size | 18.2 kb (longer than other paramyxoviruses) |
| Genetic organization | Encodes 6 structural proteins including the nucleocapsid (N), the phosphoprotein (P), the matrix protein (M), the surface glycoprotein (G) and fusion (F) protein, and the viral polymerase (L) (similar to Respirovirus, and Morbillivirus genera) |
| Host cell entry | Protein-based receptors (ephrin B2 and B3) |
| Tropism |
Ephrin B2: endothelial vasculature (arteries, arterioles, capillaries), neurons Ephrin B3: central nervous system, the heart, and the prostate |
| Natural host | Bat |
| Host | Pigs, cattle, goats, horses, and humans |
| CNS entry |
Through blood vessels of the brain and/or through olfactory nerves Replication in endothelial cells and disruption of the blood–brain barrier (BBB) |
| Viral mediators involved in the interferon (IFN) response modulation | P, V, W, and C proteins encoded by the P gene (NiV) |
| Neurological sequelae | Severe neuronal necrosis, gliosis, perivascular cuffing, and inflammatory infiltrate are consequences of relapsing |
| Immune damage | Inflammatory damage associated with encephalitis |
NiV, Nipah henipavirus; HeV, Hendra henipavirus
The membrane-anchored envelope glycoproteins (G and F) are the mediators of virus attachment and host cell infection as major determinants of cellular tropism. The G glycoprotein is the attachment glycoprotein and has neither hemagglutinating nor neuraminidase activities. The fusion (F) glycoprotein facilitates the fusion of the viral and host cell membranes and shares several conserved features with other viral fusion glycoproteins.
Different from Respiroviruses and Rubulaviruses, Henipaviruses do not use glycan-based receptors, but instead, like the Morbilliviruses, use protein-based receptors. The NiV and HeV G glycoprotein engage host cell membrane proteins as entry receptors and bind to ephrin-B2 and ephrin-B3, members of a large family of cell surface expressed glycoprotein ligands that bind to Eph receptors, the largest subgroup of receptor tyrosine kinases [15–18] (Table 1).
Ephrin-B2 expression is prominent in arteries, arterioles, and capillaries in multiple organs and tissues while ephrin-B3 is found predominantly in the nervous system and the vasculature. Sequences of ephrin-B2 and -B3 are highly conserved across susceptible hosts including humans, horses, pigs, cats, dogs, mice, and bats with amino acid identities of 95–96% for ephrin-B2 and 95–98% for ephrin-B3 [5]. The membrane fusion process between the virion and plasma membranes is driven by both G and F glycoproteins working together that finally deliver the viral nucleocapsid into the cytoplasm. Likewise, Henipaviruses also feature hallmarks of paramyxovirus infections where virus-infected cells expressing attachment and fusion glycoproteins on their surface can fuse with receptor-bearing cells leading to the formation of multinucleated giant cells or syncytia. Necrosis, vasculitis, and thrombosis are associated with the formation of syncytia. After translation viral surface glycoproteins F and G are inserted into the host cell endoplasmic reticulum for post-translational modifications, particularly glycosylation. The other translated viral proteins—N, P, M, and L—remain in the cytoplasm. When abundant viral mRNA transcripts are produced, full-length anti-genomes are synthesized to generate more copies of the Henipavirus genome. These newly synthesized genome copies assemble with viral proteins near the host cell membrane where F and G proteins are scattered, and the budding of new virions, facilitated by the M protein, will occur [19].
Viral receptors are widely distributed in different tissues thus explaining the systemic nature of NiV and HeV infection; similarly, viral receptors are present in multiple hosts thus supporting a wider host range when compared to most other paramyxoviruses. NiV can infect porcine microvascular endothelial cells and human brain endothelial cells, which express high levels of ephrin-B2, but not other endothelial cells with no detectable ephrin-B2 expression [20]. Both NiV and HeV appear to infect endothelial cells from arteries preferentially, driving the formation of syncytia containing up to 23 nuclei, which rapidly die. The formation of syncytia is associated with necrosis, vasculitis, and thrombosis, as well as with brain parenchyma lesions that then lead to the typical clinical signs observed during virus infection, namely, respiratory disorders, neurological symptoms, and unstable blood pressure [21].
Blood cells—excluding macrophages and dendritic cells in which low NiV replication has been observed—are not permissive to NiV infection. However, they can bind the virus on their surface and transport and deliver the pathogen to new sites of cell recruitment during inflammation processes [22]. Notably, ephrin-B2 is present in arterial (but not venous) endothelial cells, neurons, glial cells, epithelial cells of the upper respiratory tract, alveolar pneumocytes, smooth muscle cells, in macrophages in human spleen and lymph nodes, and in macrophages in pigs [23]. The brain, lungs, placenta, and prostate have high levels of ephrin-B2 [24]. In contrast, ephrin-B3 is mainly expressed in the central nervous system (CNS), the heart, and the prostate [25]. Interestingly, ephrin-B3, but not ephrin-B2, is expressed in the brainstem [26].
Henipavirus pathogenesis
A major challenge is to ascertain the molecular mechanisms of virus replication and immunity associated with protection from infection as well as the development of new antiviral therapeutics and vaccination strategies (Fig. 2). Henipaviruses are designated as biosafety level 4 (BSL-4) agents thus practical constraints of performing functional genomics studies at high levels of containment are evident. Molecular virology methods for genomic characterization (PCR-based amplification techniques, nucleotide sequencing, phylogenetic relationships using bioinformatics approaches) and cell culture isolation have been used for the Henipaviruses study [27].
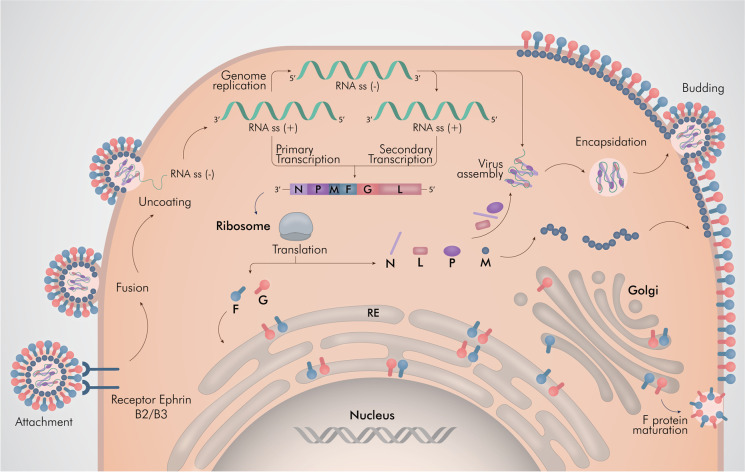
Fig. 2.
Henipavirus replication cycle. Henipavirus enters a cell by glycoprotein G attachment and F protein-mediated membrane fusion. The viral nucleocapsid is released in the cytoplasm and the genome is leading to the initiation of transcription and the accumulation of viral mRNA transcripts. In addition, the viral genome is transcribed into a full-length anti-genome, which is used to generate additional copies of the NiV genome. Viral mRNA transcripts are translated into viral proteins, leading to virion assembly, encapsidation, and virus release. Illustrated by @darwid_illustration
Recent loss-of-function (i.e., RNAi) functional genomics screens that shed light on the henipavirus–host interface at a genome-wide level have been carried out [28]. Based on studies of functional genomics, the entry receptor of HeV and NiV, ephrin-B2, was identified by microarray analysis of infection-permissive and infection-resistant cell lines [18]. Transcriptomics and proteomics have been utilized to uncover key differences in cellular responses to HeV infection in HeV disease-susceptible (human) and disease-resistant (bat) cells, and suggest that activation of apoptosis pathways via the innate immune pathway may contribute to the tolerance of Henipaviruses by flying foxes [29] (Fig. 3).
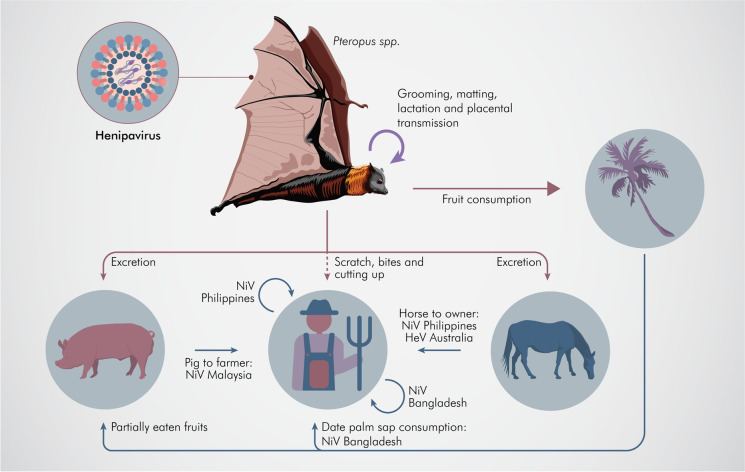
Fig. 3.
Schematic representation of the HeV and NiV transmission from the natural host, fruit bats, to the susceptible species. The arrows represent asymptomatic transmission in the natural reservoir as well as indicate the spillover infection causing a disease. The different contributing factors are indicated adjacent to the arrows. Illustrated by @darwid_illustration
Consistent with widespread ephrin-B2 expression, extensive vasculitis in the lung, kidney, heart, and CNS has been observed during autopsies of NiV deceased patients, as well as necrosis in the highly vascularized spleen [30]. Such extensive distribution of permissive cells explains the wide dissemination of Henipavirus (Fig. 4). Henipavirus transmission may use different pathways such as (i) the bloodstream, after replication in endothelial cells including the disruption of the blood–brain barrier (BBB); (ii) cell-to-cell transmission through syncytia formation; (iii) through trans-infection when the viruses attached to non-permissive blood cells (i.e., lymphocytes, dendritic cells) facilitate its transfer and infection to other susceptible cells (“Trojan horse” model) including by crossing the BBB and delivering NiV into CNS by leukocyte transmigration [22, 31]; and (iv) via CNS spread when replicating in neurons, or using the nervous route through olfactory neurons from the nasal cavity and olfactory epithelial cells [32, 33] (Table 1).
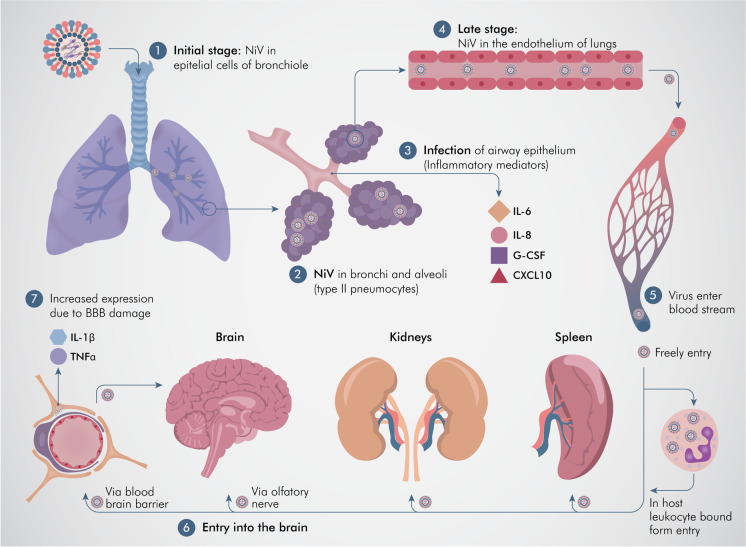
Fig. 4.
Events during organ and cellular pathogenesis of Nipah virus (NiV). 1. NiV can be seen in the epithelial cells of the bronchiole in the initial stage of infection. 2. NiV antigen can be detected in bronchi (epithelial cells) and alveoli, particularly in type II pneumocytes. 3. Inflammatory mediators are induced and released after infection to the airway epithelium. 4. Viral dissemination to the endothelial cells of the lungs in the later stage of the disease. 5, 6. Viruses enter the bloodstream followed by dissemination, either freely or in host leukocyte-bound form (“Trojan horse model”), reaching the brain, spleen, and kidneys. 7. Two pathways are involved in the process of viral entry into the central nervous system (CNS), via the hematogenous route and the anterograde nervous route via the olfactory nerve. 8. The blood–brain barrier (BBB) is disrupted and IL-1β along with tumor necrosis factor (TNF)-α are released due to infection of the CNS by the virus which ultimately leads to the development of neurological signs. Illustrated by @darwid_illustration
In humans and other animal hosts, the main entry route of Henipavirus is oronasal. During NiV infection, epithelial cells and type II pneumocytes from the bronchiole are the primary targets and infection induces the production of inflammatory cytokines such as IL-6, IL-8, IL-1α, and G-CSF, followed by immune cell recruitment. Subsequently, the virus spreads to endothelial cells and eventually gains entry to the bloodstream where it will disseminate to other tissues either freely or attached to the surface of leukocytes, thus leading to multiple-organ-failure syndrome including lungs, spleen, kidneys, and brain [34]. Viral antigens have been localized by immunostaining to alveolar type II pneumocytes, intra-alveolar macrophages, and blood vessels. The virus can also enter the CNS either through blood vessels of the brain and/or through olfactory nerves. Neuropathogenesis appears to arise from dual mechanisms of vascular disease and direct parenchymal brain infection, showing viral inclusion bodies and necrosis in both the gray and the white matter after endothelial infection and vasculitis as a result of platelet activation and thrombi, producing microinfarcts. These direct cytopathic effects of NiV replication in the microvasculature are accompanied by those mediated by inflammatory cytokines (TNF-α and IL1-β) released by microglia or other surrounding cells, promoting BBB disruption [33, 35]. Infiltrating lymphocytes are present during Henipavirus-associated encephalitis but the protective role and/or role as mediators of damage of individual inflammatory cell populations within the brain remains to be determined.
Once Henipavirus primary infection has occurred and virus replication leads to systemic infection, the immune response contributes to generalized symptomatology. Hence, after 3–14 days of infection, the respiratory tract and specifically the bronchi and alveoli are affected as evidenced by clinical symptoms (fever, cough, hypoxia) and even neurological signs that appear with evidence of interstitial pneumonia and ARDS-like disease, vasculitis, and severe meningitis and encephalitis. It is also a concern that infection by both viruses, but more pronouncedly NiV, can also manifest as relapsing encephalitis. This neurological disease is preceded by a chronic and quiescent course (> 10 weeks) that follows a non-encephalitic or asymptomatic infection, or after recovering from an acute infection. The neurological disease follows a recrudescence of virus replication in the CNS involving unknown mechanisms that allow NiV and HeV to escape immunological clearance for an extended period to later result in disease. The pathology and clinical presentation of the acute infection of the brain differ when the disease relapses. Notably, vasculopathy and demyelination are absent, with fewer instances of fever, while seizures and focal signs are more frequent in relapsing encephalitis. Large confluent lesions with extensive viral inclusions both in neurons and the neuropil, but also in glial and ependymal cells are hallmarks of the relapsing form of NiV and HeV infection that trigger severe neuronal necrosis, gliosis, perivascular cuffing, and inflammatory infiltrate [14, 30] (Table 1).
Immune response and evasion strategies
Current uncertainties regarding the immune response to Henipavirus infection in humans include whether this response is either inefficient at dealing with the virus or if the immune response is itself detrimental to the host, thus exacerbating the pathogenic process. Neutrophils are one of the first immune cells to be recruited to the site of infection. These cells produce reactive oxygen species, antimicrobial peptides, and neutrophil extracellular traps (NET) to contain the virus. Following Henipavirus entry via ephrin-B2 or -B3 receptors on the host cellular membrane, virions are engulfed and broken down into viral peptides by antigen-presenting cells such as macrophages and dendritic cells. The presentation of the viral peptides on the MHC molecules activates the T cells through their T-cell receptor (TCR). The activation of the helper T cells subsequently drives B cells to activate, proliferate, and develop a mature antibody response. As a result, plasma cells and memory B cells are formed, producing virus-specific antibodies for protection against infection. On the other hand, the activation of cytotoxic T cells allows them to target and kill the virus-infected cells [36].
The humoral adaptive immune response during NiV infection includes both specific IgM and IgG in high levels on day 1 and day 25 post-admission, respectively [37]. Moreover, marked elevated activation of CD8 T cells (HLADR + /CD38 +) has been detected accompanied by absolute numbers of T lymphocytes within normal levels in NiV-infected patients [38]. It has been shown that reactive IgM and IgG are maintained for 18 months in patients after HeV infection [39].
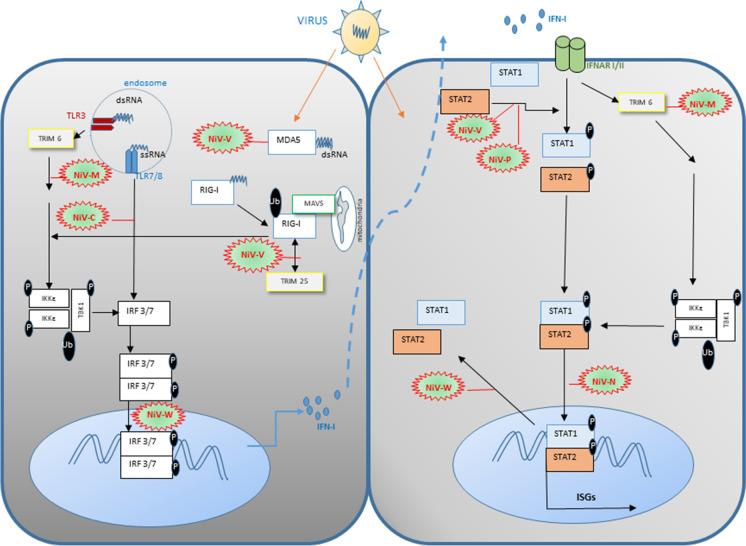
As mentioned above, the NiV P gene encodes 4 viral products: P, V, W, and C. Products of the P gene can antagonize both double-stranded (ds) RNA signaling and interferon (IFN) signaling [40, 41] (Table 1). Similar to that of other paramyxoviruses, the V protein functions as an inhibitor of IFN induction or dsRNA signaling by targeting the helicase encoded by the melanoma differentiation-associated gene 5 (MDA5). Whereas the NiV W protein can also inhibit dsRNA signaling, it does so by nuclear translocation, targeting interferon regulatory factor 3 (IRF-3) and effectively blocking both dsRNA signaling via MDA5 and through the cell surface expressed toll-like receptor 3 (TLR-3) signaling pathway. Henipaviruses also target the paracrine signal transduction pathway that is initiated by the binding of type I IFN to the two cell surface interferon alpha and beta receptors, IFNAR1 and IFNAR2 which assemble into a functional receptor complex leading to the activation of signal transducers and activators of transcription (STAT) factors, that later direct the expression of genes possessing an interferon-stimulated response element (ISRE) within the nuclease [42]. The Henipavirus V, W, and P proteins block the type I IFN signaling pathway, with the NiV V and P proteins forming high-molecular-weight complexes in the cytoplasm with STAT1, and the NiV W protein targeting STAT1 within the nuclease [40, 41]. Altogether, these mechanisms prevent the expression of IFN-I and ISG genes (Fig. 5).
Fig. 5.
Strategies of the Nipah virus (NiV) to modulate and evade the type I interferon (IFN-I) synthesis and signaling. (a) Once the NiV infection occurs, viral RNA is released, which activates PRRs (TLR and RLR) pathways in the cell, leading to the activation of IFN-I and IFN-stimulated genes (ISGs). Different NiV proteins are able to interfere these pathways using several strategies. NiV-V disrupts MDA5 stimulation and subsequent RIG-I activation. NiV-C protein frustrates the activation of IRF3 and IRF7/8 by affecting the IKK dimerization. NiV-W protein prevents nuclear transport of phosphorylated IRF3/7 dimers. In addition, NiV-V protein inhibits RIG-I activation and its signaling pathway. NiV-M gene expression highlighted the effect of NiV-M could also degrade TRIM6 and IKK-dependent signaling. (b) NiV-induced production of IFN-I leads to the stimulation of IFN-I receptor (IFNAR) and subsequent anti-viral signaling, which could be interrupted by several NiV proteins. NiV-N could impede nuclear import of STAT1/2 dimer, while NiV-M triggers degradation of TRIM6 and disrupts subsequent IKK, TBK1, and STAT1/2 phosphorylation. NiV-P and V could interfere with STAT1 and STAT2 phosphorylation while NiV-W prevents their nuclear exportation
Host range and transmission
The exceptionally broad species tropism of Henipaviruses as represented by NiV and HeV distinguishes them from all other known paramyxoviruses. Among paramyxoviruses, Henipavirus is the only genus that causes a highly pathogenic disease of zoonotic origin. As mentioned above, to date, bats appear to be the predominant natural reservoir hosts for Henipaviruses. Among them, Pteropus bat species are distributed as far west as Madagascar, through the Indian subcontinent to Southeastern Asia and Australia, and eastwards through Oceania [43]. However, there is evidence of Henipaviruses in a wide variety of other bat species [44–46]. Furthermore, these viruses have a very broad range of natural hosts including other multiple types of mammalian such as shrews, pigs, goats, cattle, cats, dogs, and horses, which significantly increases the risk of spillover to humans considering its frequent close proximity [14]. Using serological and/or nucleic acid laboratory approaches to different species, it was possible to reveal the globally expanded distribution of Henipaviruses, including in some of the most populated areas in the world (Australia, Bangladesh, Cambodia, China, Ghana, India, Indonesia, Madagascar, Malaysia, Papua New Guinea, Singapore, Thailand, and throughout South America). These studies also have evidenced sporadic Henipavirus spillover events in domestic livestock (pigs, cattle, goats, horses) and in human populations, also reflecting the presence of less pathogenic-related Henipaviruses [47–49] (Table 1). A disease outbreak in the Philippines offered evidence of horse-to-human and human-to-human transmission with NiV as the likely cause [50].
Henipaviruses can be transmitted both among bats and in spillover events to other species. Between bats, the viruses are shed orally, urogenital, in feces, and in birthing fluids during grooming, mating, and fighting [6]. Such biological fluids and ingestion of contaminated food are mechanisms of transmission [51]. Ingestion of raw contaminated date sap is one of the most common transmission routes for NiV but not for HeV [52], and increasing cases are registered when the palm sap is harvested and infected bats may lick the sweet sap [53]. Henipaviruses’ foodborne zoonotic transmission is rare, but it should not be ruled out since eventual cross-contamination or consumption of the virus-infected comestible product is possible [54]. When fruit trees, pigs, bats, and humans are in the same surroundings, the emergence of Henipaviruses could be facilitated. For instance, close proximity contact with domestic animals may promote transmission of NiV when these animals feed from palm sap or partially eaten fruit contaminated with NiV-containing feces, urine, or saliva. If infected, these domestic animals can then shed the virus and transmit it to humans. In the case of HeV, horses are the only bridge and amplifying hosts that can be directly infected by bats. When infected, horses can excrete HeV up to 3 days before presenting clinical signs [55]. However, HeV infectivity is low and humans are generally infected only when exposure to HeV-infected horse secretions or organs is high. Despite the fact that bat handlers are frequently exposed to potential pathogens from sick bats by scratches, bites, saliva, feces, etc., no direct transmission of HeV from bats to humans has been reported [14].
Multiple NiV outbreaks have been documented where human-to-human transmission took place. Close physical contact and/or contact with respiratory secretions are important for person-to-person NiV transmission. It has been shown that patients coughing are more likely to transmit NiV and that care providers sharing rooms, food, or contact with NiV-infected patients are at very high risk, especially when there is an exposure of care personnel to the saliva of sick patients, thus highlighting close contact as a necessary condition for human-to-human transmission. It has also been shown that the corpses of NiV deceased patients can transmit the virus to people that were in close contact with the body [56]. In the case of HeV, no human-to-human transmission has been detected to date but the potential risk of transmission to humans should not be neglected [57, 58].
Spillover events that cause disease in humans or other animal species may depend on multiple factors that propitiate “jumps” of viruses between species. Changes in the virus, the host, and/or the environment may be involved. When exposure of humans to the pathogen is augmented, the interspecies transmission may occur, enhancing infection in the human host. Likewise, intraspecies transmission is possible when expanded transmission among individuals of the newly targeted host species ensues. Based on studies from different “bridge” animal hosts and from humans, similarities in Henipaviruses RNA sequences would suggest that the main changes influencing spillover events occur in the environment and involve increased interaction between bats and livestock/humans. There are many circumstances by which humans can be exposed to bats including activities in caves, hunting, or exposure to bats roosting in houses. The natural bat roosting sites have been deeply affected by changing landscapes and deforestation, thus forcing colonies to change their ecology and conduct and to look for niche expansion, often closer to human locations [59].
Nipah viruses have long been thought to be a potential cause of future pandemics. Human activity affects a number of elements that favor spillover occurrences, including changes in land use, interactions with other animal species, and the sale and consumption of wild animal meat. Immunological and genetic variables also influence how susceptible a person is to infections, in addition to these sociocultural and biological ones. Current methods for controlling infectious illnesses have focused on prevention but they are used after a disease has spread across the human population. Strategies to stop the spillover of new diseases before they infect humans and the quick reaction mechanisms to control the spread of infection after it has started are lacking. During the last two decades, we have experienced pandemic warnings and the growing pace of new disease development, including influenza viruses, coronaviruses, paramyxoviruses, and retroviruses. Strict public health regulations might be enforced, using lessons from the SARS-CoV-2 pandemic.
Animal and cell culture models for studying Henipaviruses
The development of animal models has been essential for understanding henipavirus virus replication, antibody development, viral shedding, transmission, and pathogenesis as well as for the evaluation of prospective vaccines and anti-viral therapies. Under laboratory conditions, the host range of Henipaviruses can be extended to rodents such as golden hamsters [60], guinea pigs [61–63], ferrets [64], and non-human primates including Old (African green monkeys) and New World monkeys (Saimiri/squirrel monkeys) [65–67]. Notably, while hamsters and ferrets are permissive to infection, and repeat many symptoms reflective of human infections [68], mice are resistant to NiV infection [60] despite the fact that murine ephrin-B2 shares 97% sequence similarity with human ephrin-B2. This murine resistance to henipavirus infection likely arises at a post-entry step, as the henipavirus attachment glycoprotein binds to murine ephrinB2 just as well as human ephrinB2 [69]. At least under laboratory conditions, other animals that are susceptible to henipavirus infection include cats, dogs [70–72], and chicken embryos [73].
The expression of ephrin-B2/B3 has a pivotal role in a cell line’s susceptibility to henipavirus infection in vitro [74, 75]. For instance, the CHO hamster cell line, which lacks native ephrin-B expression, is particularly susceptible to NiV infection when exogenous ephrin-B2 and -B3 (but not -B1) expression is present [16]. The majority of commonly used laboratory cell lines, such as HEK293T [76], Vero [20, 69], and HeLa-CCL2 [18], are capable of supporting henipavirus infection. However, although being known to be endothelium-tropic, NiV does not have the ability to infect all endothelial cell types. PBMECs (porcine microvascular endothelial cells) and HBMECs (human brain endothelial cells), endothelial cells from capillaries and the brain that express ephrin-B2 at high levels, are susceptible to NiV infection, whereas MyEnd (murine myocardium) and PAECs (porcine aorta endothelial cells), which do not express quantifiable amounts of the receptors, are resistant to infection [20, 74]. With the probable exception of P815 (mouse mast cells) and 208f, most cell lines with detectable ephrinB2 expression have so far been proven to be tolerant to NiV infection (rat embryonic fibroblasts). It is not known why these cell lines are unable to sustain NiV replication; however, in the instance of 208f, it is intriguing to note that despite the high quantities of mRNA present, the quantity of ephrinB2 at the cell surface as identified by flow cytometry is minimal [74, 76].
Antiviral therapy and vaccines
At present, there are no approved antiviral therapeutics to treat Henipavirus infections in humans. Ribavirin and chloroquine have been used separately or combined, but with poor clinical benefits. Peptide fusion inhibitors constitute an alternative approach through the utilization of the heptad repeat domains of several paramyxovirus F glycoproteins, including the Henipaviruses, to inhibit membrane fusion by blocking the formation of the trimer-of-hairpins structure [77]. Finally, protective passive immunotherapy based on polyclonal antiserums, or mouse monoclonal antibodies specific for the henipavirus G or F glycoproteins has been tested [78]. All these therapeutic approaches merit further investigation.
A variety of active immunization strategies for Henipavirus have been explored using recombinant virus platforms, protein subunits virus-like particles and DNA vaccines. Several of these strategies have only been examined in terms of their ability to generate a henipavirus-specific neutralizing response [5], whereas other studies characterized immune responses and efficacy in animal challenge models. The horse vaccine against HeV (Equivac® HeV) was the first commercially deployed vaccine developed against a BSL-4 agent and is the only licensed treatment for Henipavirus infection. This is an HeV-sG subunit vaccine for horses which is also expected to provide a substantial health benefit to humans [79].
Since viruses recognize no national borders, international cooperation and quick action are required to close knowledge gaps and halt further outbreaks. Because of the recent henipavirus outbreaks, it may be necessary to coordinate a worldwide effort to develop efficient vaccinations. In the absence of easily accessible prophylaxis or medication, quick case identification is crucial to containment. Henipavirus is no different from other illnesses in that it can manifest itself in a variety of ways, as is customary in clinical practice.
Author contribution
JQ, VG, and MVD have contributed equally to conceptualization, methodology, writing, review, and editing.
Funding
This research was partially funded by the Universidad of Buenos Aires (UBACYT N°20020190100119BA to JQ).
Declarations
Conflict of interest
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Eaton BT, et al. Hendra and Nipah viruses: different and dangerous. Nat Rev Microbiol. 2006;4(1):23–35. doi: 10.1038/nrmicro1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sendow I, et al. Nipah virus in the fruit bat Pteropus vampyrus in Sumatera, Indonesia. PLoS ONE. 2013;8(7):e69544. doi: 10.1371/journal.pone.0069544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yadav PD, et al. Detection of Nipah virus RNA in fruit bat (Pteropus giganteus) from India. Am J Trop Med Hyg. 2012;87(3):576–578. doi: 10.4269/ajtmh.2012.11-0416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wacharapluesadee S, et al. A longitudinal study of the prevalence of Nipah virus in Pteropus lylei bats in Thailand: evidence for seasonal preference in disease transmission. Vector Borne Zoonotic Dis. 2010;10(2):183–190. doi: 10.1089/vbz.2008.0105. [DOI] [PubMed] [Google Scholar]
- 5.Broder, C.W., K.T., Henipaviruses, in Neurotropic viral infections, C.S. Reiss, Editor. 2016, Springer International Publishing: Switzerland. p. 45–83.
- 6.Halpin K, et al. Pteropid bats are confirmed as the reservoir hosts of henipaviruses: a comprehensive experimental study of virus transmission. Am J Trop Med Hyg. 2011;85(5):946–951. doi: 10.4269/ajtmh.2011.10-0567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Murray K, et al. A novel morbillivirus pneumonia of horses and its transmission to humans. Emerg Infect Dis. 1995;1(1):31–33. doi: 10.3201/eid0101.950107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chua KB, et al. Fatal encephalitis due to Nipah virus among pig-farmers in Malaysia. Lancet. 1999;354(9186):1257–1259. doi: 10.1016/S0140-6736(99)04299-3. [DOI] [PubMed] [Google Scholar]
- 9.Choudhary, O.P., et al., Spillover zoonotic ‘Langya virus’: is it a matter of concern? Vet Q, 2022: p. 1–4. [DOI] [PMC free article] [PubMed]
- 10.Zhang XA, et al. A zoonotic Henipavirus in febrile patients in China. N Engl J Med. 2022;387(5):470–472. doi: 10.1056/NEJMc2202705. [DOI] [PubMed] [Google Scholar]
- 11.Marsh GA, et al. Cedar virus: a novel Henipavirus isolated from Australian bats. PLoS Pathog. 2012;8(8):e1002836. doi: 10.1371/journal.ppat.1002836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wu Z, et al. Novel Henipa-like virus, Mojiang Paramyxovirus, in rats, China, 2012. Emerg Infect Dis. 2014;20(6):1064–1066. doi: 10.3201/eid2006.131022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lamb, R.A.P., G.D. , Paramyxoviridae: the viruses and their replication, in Fields virology, D.K. Bernard Fields, Peter Howley, Editor. 2013, Lippincott, Williams, and Wilkins: USA. p. 957–995.
- 14.Lawrence, P. and B. Escudero-Perez, Henipavirus immune evasion and pathogenesis mechanisms: lessons learnt from natural infection and animal models. Viruses, 2022. 14(5). [DOI] [PMC free article] [PubMed]
- 15.Bishop KA, et al. Identification of Hendra virus G glycoprotein residues that are critical for receptor binding. J Virol. 2007;81(11):5893–5901. doi: 10.1128/JVI.02022-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Negrete OA, et al. Two key residues in ephrinB3 are critical for its use as an alternative receptor for Nipah virus. PLoS Pathog. 2006;2(2):e7. doi: 10.1371/journal.ppat.0020007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bowden TA, et al. Structural basis of Nipah and Hendra virus attachment to their cell-surface receptor ephrin-B2. Nat Struct Mol Biol. 2008;15(6):567–572. doi: 10.1038/nsmb.1435. [DOI] [PubMed] [Google Scholar]
- 18.Bonaparte MI, et al. Ephrin-B2 ligand is a functional receptor for Hendra virus and Nipah virus. Proc Natl Acad Sci U S A. 2005;102(30):10652–10657. doi: 10.1073/pnas.0504887102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang, L.-F.M., J.S.; Broder, C.C., Henipaviruses, in Fields virology, H.P. Knipe DM, Editor. 2013, Lippincott Williams & Wilkins: Philadelphia, USA. p. 1070–1085.
- 20.Erbar S, Diederich S, Maisner A. Selective receptor expression restricts Nipah virus infection of endothelial cells. Virol J. 2008;5:142. doi: 10.1186/1743-422X-5-142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ang, L.T., et al., Generating human artery and vein cells from pluripotent stem cells highlights the arterial tropism of Nipah and Hendra viruses. Cell, 2022. 185(14): p. 2523–2541 e30. [DOI] [PMC free article] [PubMed]
- 22.Mathieu C, et al. Nipah virus uses leukocytes for efficient dissemination within a host. J Virol. 2011;85(15):7863–7871. doi: 10.1128/JVI.00549-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Maisner A, Neufeld J, Weingartl H. Organ- and endotheliotropism of Nipah virus infections in vivo and in vitro. Thromb Haemost. 2009;102(6):1014–1023. doi: 10.1160/TH09-05-0310. [DOI] [PubMed] [Google Scholar]
- 24.Liebl DJ, et al. mRNA expression of ephrins and Eph receptor tyrosine kinases in the neonatal and adult mouse central nervous system. J Neurosci Res. 2003;71(1):7–22. doi: 10.1002/jnr.10457. [DOI] [PubMed] [Google Scholar]
- 25.Benson MD, et al. Ephrin-B3 is a myelin-based inhibitor of neurite outgrowth. Proc Natl Acad Sci U S A. 2005;102(30):10694–10699. doi: 10.1073/pnas.0504021102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Negrete OA, et al. Single amino acid changes in the Nipah and Hendra virus attachment glycoproteins distinguish ephrinB2 from ephrinB3 usage. J Virol. 2007;81(19):10804–10814. doi: 10.1128/JVI.00999-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zemanova, S., et al., Opportunities and limitations of molecular methods for studying bat-associated pathogens. Microorganisms, 2022. 10(9). [DOI] [PMC free article] [PubMed]
- 28.Stewart CR, et al. A functional genomics approach to Henipavirus research: the role of nuclear proteins, microRNAs and immune regulators in infection and disease. Curr Top Microbiol Immunol. 2018;419:191–213. doi: 10.1007/82_2017_28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wynne JW, et al. Proteomics informed by transcriptomics reveals Hendra virus sensitizes bat cells to TRAIL-mediated apoptosis. Genome Biol. 2014;15(11):532. doi: 10.1186/s13059-014-0532-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wong KT, et al. Nipah virus infection: pathology and pathogenesis of an emerging paramyxoviral zoonosis. Am J Pathol. 2002;161(6):2153–2167. doi: 10.1016/S0002-9440(10)64493-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tiong V, et al. Nipah virus infection of immature dendritic cells increases its transendothelial migration across human brain microvascular endothelial cells. Front Microbiol. 2018;9:2747. doi: 10.3389/fmicb.2018.02747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.de Wit E, Munster VJ. Animal models of disease shed light on Nipah virus pathogenesis and transmission. J Pathol. 2015;235(2):196–205. doi: 10.1002/path.4444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dawes, B.E. and A.N. Freiberg, Henipavirus infection of the central nervous system. Pathog Dis, 2019. 77(2). [DOI] [PMC free article] [PubMed]
- 34.Escaffre O, Borisevich V, Rockx B. Pathogenesis of Hendra and Nipah virus infection in humans. J Infect Dev Ctries. 2013;7(4):308–311. doi: 10.3855/jidc.3648. [DOI] [PubMed] [Google Scholar]
- 35.Rockx B, Winegar R, Freiberg AN. Recent progress in henipavirus research: molecular biology, genetic diversity, animal models. Antiviral Res. 2012;95(2):135–149. doi: 10.1016/j.antiviral.2012.05.008. [DOI] [PubMed] [Google Scholar]
- 36.Liew, Y.J.M., et al., The immunobiology of Nipah virus. Microorganisms, 2022. 10(6). [DOI] [PMC free article] [PubMed]
- 37.RAMASUNDPUM, V., et al., Kinetics of IgM and IgCî seroconversion in Nipah virus infection. Malay, 2000. 6: p. 0.
- 38.Arunkumar G, et al. Adaptive immune responses in humans during Nipah virus acute and convalescent phases of infection. Clin Infect Dis. 2019;69(10):1752–1756. doi: 10.1093/cid/ciz010. [DOI] [PubMed] [Google Scholar]
- 39.Taylor C, et al. No evidence of prolonged Hendra virus shedding by 2 patients, Australia. Emerg Infect Dis. 2012;18(12):2025. doi: 10.3201/eid1812.120722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shaw ML. Henipaviruses employ a multifaceted approach to evade the antiviral interferon response. Viruses. 2009;1(3):1190–1203. doi: 10.3390/v1031190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Basler CF. Nipah and Hendra virus interactions with the innate immune system. Curr Top Microbiol Immunol. 2012;359:123–152. doi: 10.1007/82_2012_209. [DOI] [PubMed] [Google Scholar]
- 42.de Weerd NA, Samarajiwa SA, Hertzog PJ. Type I interferon receptors: biochemistry and biological functions. J Biol Chem. 2007;282(28):20053–20057. doi: 10.1074/jbc.R700006200. [DOI] [PubMed] [Google Scholar]
- 43.Clayton BA, Wang LF, Marsh GA. Henipaviruses: an updated review focusing on the pteropid reservoir and features of transmission. Zoonoses Public Health. 2013;60(1):69–83. doi: 10.1111/j.1863-2378.2012.01501.x. [DOI] [PubMed] [Google Scholar]
- 44.Peel AJ, et al. Continent-wide panmixia of an African fruit bat facilitates transmission of potentially zoonotic viruses. Nat Commun. 2013;4:2770. doi: 10.1038/ncomms3770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hasebe F, et al. Serologic evidence of Nipah virus infection in bats. Vietnam Emerg Infect Dis. 2012;18(3):536–537. doi: 10.3201/eid1803.111121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Drexler JF, et al. Bats host major mammalian paramyxoviruses. Nat Commun. 2012;3:796. doi: 10.1038/ncomms1796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chowdhury S, et al. Serological evidence of henipavirus exposure in cattle, goats and pigs in Bangladesh. PLoS Negl Trop Dis. 2014;8(11):e3302. doi: 10.1371/journal.pntd.0003302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pernet O, et al. Evidence for henipavirus spillover into human populations in Africa. Nat Commun. 2014;5(1):1–10. doi: 10.1038/ncomms6342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hayman DT, et al. Antibodies to henipavirus or henipa-like viruses in domestic pigs in Ghana, West Africa. PLoS ONE. 2011;6(9):e25256. doi: 10.1371/journal.pone.0025256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ching PK, et al. Outbreak of henipavirus infection, Philippines, 2014. Emerg Infect Dis. 2015;21(2):328–331. doi: 10.3201/eid2102.141433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Epstein JH, et al. Nipah virus dynamics in bats and implications for spillover to humans. Proc Natl Acad Sci U S A. 2020;117(46):29190–29201. doi: 10.1073/pnas.2000429117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nahar N, et al. Raw sap consumption habits and its association with knowledge of Nipah virus in two endemic districts in Bangladesh. PLoS ONE. 2015;10(11):e0142292. doi: 10.1371/journal.pone.0142292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Salah Uddin Khan, M., et al., Use of infrared camera to understand bats’ access to date palm sap: implications for preventing Nipah virus transmission. Ecohealth, 2010. 7(4): p. 517–525. [DOI] [PubMed]
- 54.Simons RR, et al. Potential for introduction of bat-borne zoonotic viruses into the EU: a review. Viruses. 2014;6(5):2084–2121. doi: 10.3390/v6052084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Marsh GA, et al. Experimental infection of horses with Hendra virus/Australia/horse/2008/Redlands. Emerg Infect Dis. 2011;17(12):2232–2238. doi: 10.3201/eid1712.111162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chakraborty A, et al. Evolving epidemiology of Nipah virus infection in Bangladesh: evidence from outbreaks during 2010–2011. Epidemiol Infect. 2016;144(2):371–380. doi: 10.1017/S0950268815001314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tian J, et al. Emerging viruses: Cross-species transmission of coronaviruses, filoviruses, henipaviruses, and rotaviruses from bats. Cell Rep. 2022;39(11):110969. doi: 10.1016/j.celrep.2022.110969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Weatherman S, Feldmann H, de Wit E. Transmission of henipaviruses Curr Opin Virol. 2018;28:7–11. doi: 10.1016/j.coviro.2017.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gortazar C, et al. Crossing the interspecies barrier: opening the door to zoonotic pathogens. PLoS Pathog. 2014;10(6):e1004129. doi: 10.1371/journal.ppat.1004129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wong KT, et al. A golden hamster model for human acute Nipah virus infection. Am J Pathol. 2003;163(5):2127–2137. doi: 10.1016/S0002-9440(10)63569-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Williamson MM, et al. Experimental Hendra virus infection in pregnant guinea-pigs and fruit bats (Pteropus poliocephalus) J Comp Pathol. 2000;122(2–3):201–207. doi: 10.1053/jcpa.1999.0364. [DOI] [PubMed] [Google Scholar]
- 62.Williamson MM, et al. A guinea-pig model of Hendra virus encephalitis. J Comp Pathol. 2001;124(4):273–279. doi: 10.1053/jcpa.2001.0464. [DOI] [PubMed] [Google Scholar]
- 63.Williamson MM, Torres-Velez FJ. Henipavirus: a review of laboratory animal pathology. Vet Pathol. 2010;47(5):871–880. doi: 10.1177/0300985810378648. [DOI] [PubMed] [Google Scholar]
- 64.Bossart KN, et al. A neutralizing human monoclonal antibody protects against lethal disease in a new ferret model of acute Nipah virus infection. PLoS Pathog. 2009;5(10):e1000642. doi: 10.1371/journal.ppat.1000642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Geisbert TW, et al. Development of an acute and highly pathogenic nonhuman primate model of Nipah virus infection. PLoS ONE. 2010;5(5):e10690. doi: 10.1371/journal.pone.0010690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Rockx B, et al. A novel model of lethal Hendra virus infection in African green monkeys and the effectiveness of ribavirin treatment. J Virol. 2010;84(19):9831–9839. doi: 10.1128/JVI.01163-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Marianneau P, et al. Experimental infection of squirrel monkeys with Nipah virus. Emerg Infect Dis. 2010;16(3):507–510. doi: 10.3201/eid1603.091346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Weingartl HM, Berhane Y, Czub M. Animal models of henipavirus infection: a review. Vet J. 2009;181(3):211–220. doi: 10.1016/j.tvjl.2008.10.016. [DOI] [PubMed] [Google Scholar]
- 69.Bossart KN, et al. Functional studies of host-specific ephrin-B ligands as Henipavirus receptors. Virology. 2008;372(2):357–371. doi: 10.1016/j.virol.2007.11.011. [DOI] [PubMed] [Google Scholar]
- 70.Hooper P, et al. Comparative pathology of the diseases caused by Hendra and Nipah viruses. Microbes Infect. 2001;3(4):315–322. doi: 10.1016/S1286-4579(01)01385-5. [DOI] [PubMed] [Google Scholar]
- 71.Mills JN, et al. Nipah virus infection in dogs, Malaysia, 1999. Emerg Infect Dis. 2009;15(6):950–952. doi: 10.3201/eid1506.080453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Mungall BA, et al. Feline model of acute Nipah virus infection and protection with a soluble glycoprotein-based subunit vaccine. J Virol. 2006;80(24):12293–12302. doi: 10.1128/JVI.01619-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Tanimura N, et al. Distribution of viral antigens and development of lesions in chicken embryos inoculated with Nipah virus. J Comp Pathol. 2006;135(2–3):74–82. doi: 10.1016/j.jcpa.2006.05.001. [DOI] [PubMed] [Google Scholar]
- 74.Pernet O, Wang YE, Lee B. Henipavirus receptor usage and tropism. Curr Top Microbiol Immunol. 2012;359:59–78. doi: 10.1007/82_2012_222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Negrete OA, et al. EphrinB2 is the entry receptor for Nipah virus, an emergent deadly paramyxovirus. Nature. 2005;436(7049):401–405. doi: 10.1038/nature03838. [DOI] [PubMed] [Google Scholar]
- 76.Yoneda M, et al. Establishment of a Nipah virus rescue system. Proc Natl Acad Sci U S A. 2006;103(44):16508–16513. doi: 10.1073/pnas.0606972103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Bossart KN, Fusco DL, Broder CC. Paramyxovirus entry. Adv Exp Med Biol. 2013;790:95–127. doi: 10.1007/978-1-4614-7651-1_6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Broder CC. Henipavirus outbreaks to antivirals: the current status of potential therapeutics. Curr Opin Virol. 2012;2(2):176–187. doi: 10.1016/j.coviro.2012.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Middleton D, et al. Hendra virus vaccine, a one health approach to protecting horse, human, and environmental health. Emerg Infect Dis. 2014;20(3):372–379. doi: 10.3201/eid2003.131159. [DOI] [PMC free article] [PubMed] [Google Scholar]