Abstract
Here, we describe a cystic fibrosis (CF) family with affected siblings, two of whom have a combination of I1234V and 1677delTA variants with classic CF features, the third child with a combination of I1234V and L997F variants with atypical CF, and the apparently healthy mother with a combination of 1677delTA and L997F alleles. Interestingly, the sibling with I1234V and L997F variants had normal sweat test results and had a much milder phenotype than the other two siblings with I1234V and 1677delTA variants, suggesting that this combination is causative for atypical CF. The fact that their mother with the combination of 1677delTA and L997F appears to be healthy suggests that the L997F variant causes different phenotypes in different allele combinations. The current cases show that there is a genotype-phenotype correlation in this disease and underline the importance of genotyping individuals with suspected CF to allow prediction of disease severity and effective treatment.
Keywords: Cystic fibrosis transmembrane conductance regulator (CFTR), Cystic fibrosis (CF), Sweat chloride, CF, L997F, 1677delTA, I1234V
1. Introduction
Cystic fibrosis (CF) is the most common, life-threatening, and autosomal recessive disease in the Caucasian population. It is caused by mutations in the CF transmembrane conductance regulator (CFTR) gene, which encodes a chloride ion channel expressed on the surface of epithelial cells [1]. To date, approximately 2000 variants of the CFTR gene have been reported worldwide, not all of which lead to CF [2].
The clinical picture ranges from multiorgan symptoms such as chronic respiratory infections, bronchiectasis, pancreatic insufficiency, and failure to thrive to single organ manifestations such as male infertility or chronic sinusitis. The first suspicion of CF is often based on an elevated immunoreactive trypsinogen (IRT) in newborn screening (NBS). However, some infants may be negative on screening and only diagnosed with CF later in life due to frequent respiratory infections and/or pancreatic dysfunction. If a newborn tests positive for IRT (higher than 70 μg/L in the second control) or if a child is suspected of having CF due to suggestive clinical findings, a sweat test is classically performed, and if an elevated level (>60 mM/l) is detected, genetic testing is indicated to confirm the diagnosis [3].
Certain CFTR alleles are designated as “CF-causing” alleles based on functional and clinical studies reported in the CFTR2 database [4] and are responsible for progressive pulmonary and gastrointestinal manifestations of the disease. There are also other alleles defined as “risk factor” alleles responsible for CFTR-related disorders (CFTR-RD) and male infertility. For a considerable number of alleles, however, the clinical significance in terms of their pathogenicity has not yet been fully clarified.
The severity of typical CF may be influenced by specific pathogenic variants [5], environmental factors [6], and modifier genes [7,8]. In addition, several authors have shown that atypical and monosymptomatic patients carry at least one non-CF-causing “risk factor” CFTR allele (e.g., L997F, 1525-42G > A, L320V, L967S, R170H) [[9], [10], [11]].
In this study, we describe a familial case of CF in which the broad phenotypic spectrum of the different family members can be strongly attributed to the alleles they carry.
2. Case presentation
The study was approved by the ethics committee of the Tbilisi State Medical University. Informed consent was obtained from all individual participants (or their parents) included in the study.
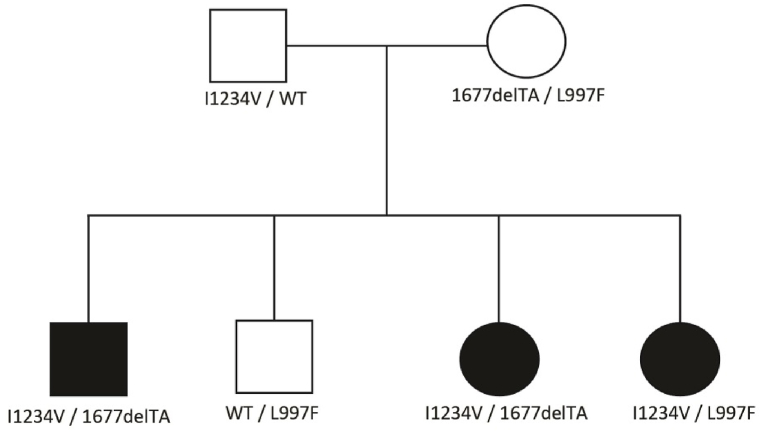
Three affected siblings born to nonblood related, apparently healthy parents were brought to the clinic because of recurrent bronchospasm and upper respiratory tract infections. Subsequent genetic testing revealed that all three siblings carried a common I1234V allele, but two of them had a 1677delTA variant in a different allele, and the third sibling had a different L997F variant. Since it was unusual to find three different alleles in biological siblings, we performed genetic testing on both parents and the fourth asymptomatic brother. Interestingly, parental testing revealed that all three affected children had the same I1234V allele inherited paternally from the heterozygous father. Surprisingly, the mother was found to be compound heterozygous for the 1677delTA and L997F variants. The healthy sibling turned out to be a heterozygous carrier of the L997F allele (Fig. 1).
Fig. 1.
Pedigree of the family shows the distribution of the variants in different family members. Abbreviations: WT - wile type.
3. Sibling 1. 12-year-old boy: I1234V/1677delTA
G1P1, born at term, BW 4400 gr. The perinatal period was uneventful. NBS was not performed. At 3 months of age, he was found to have persistent productive bronchial mucus and constipation. At 6 months of age, there was severe vomiting, dehydration, and lethargy requiring immediate hospitalization. Since 8 months, the child had recurrent respiratory infections and episodes of pneumonia (5 in total) with productive cough. Some of them were treated in the intensive care unit. The patient was treated with various antibiotics. A mucus culture test performed at 24 months was positive for Staphylococcus aureus infection. At 6 years of age, a sweat test was performed for the first time, which showed an elevated chloride level of 105 mmol/L, and the second sweat test was also abnormal, with a value of 114 mmol/L. In addition, stool pancreatic elastase was measured, which was low (<200 mcg/g), and pancreatic enzyme replacement therapy (PERT) was initiated immediately. Despite the PERT, the child continued to have breathing problems. It is noteworthy that the patient always had adequate weight gain even before starting PERT. At 7 years of age, the sputum smear was positive for Pseudomonas aeruginosa. At the age of 10, genetic testing confirmed a combination of the I1234V and 1677delTA variants. In the same year, a C. difficile infection (Glutamate Dehydrogenase (GDH) and toxin A-positive) was treated with oral vancomycin. Ultrasonography of the abdomen showed thickening of the gallbladder walls and sludge. Currently, the child continues to suffer from periodic productive cough and periumbilical abdominal pain. His current treatment includes PERT, ursodeoxycholic acid, regular administration of DEKAs vitamins, and inhalation with 3% NaCl.
4. Sibling 2. 9-year-old-girl: I1234V/1677delTA
G2P2, term birth with a birth weight of 3500 g. The perinatal period was uneventful. The NBS was negative. At 2 months of age, she started having frequent bronchospasms due to productive mucus in the airways. Some of the episodes were treated in the intensive care unit. Since 2 years of age, she had a protruding abdomen with severe flatulence. At the age of 3 years and 9 months, she had intussusception, where the intestines were forced back into position, and no bowel resection was required. Three weeks after surgery, she developed severe pneumonia and was hospitalized again. At the age of 4, CF was suspected, and the sweat test revealed a markedly elevated chloride level (120 mmol/L). Despite complicated health problems, her weight gain was always reasonable. Stool pancreatic elastase was low, and PERT was initiated. A sputum smear was positive for Pseudomonas aeruginosa since she was 6 years old, which was treated regularly with antibiotics. At the age of 7, a genetic test was performed which confirmed that she had a combination of the I1234V and 1677delTA variants. At the same age, she experienced severe abdominal pain, an abdominal ultrasound showed colonopathy (thickened wall of ileum and cecum), and the stool test was positive for C. difficile, which was treated with colomycin. The patient is currently suffering from periodic cough, abdominal pain, episodes of constipation, and enuresis. Her treatment includes PERT, ursodeoxycholic acid, regular administration of DEKAs vitamins, and inhalation with 3% NaCl.
5. Sibling 3. 7-year-old girl: I1234V/L997F
G4P4, term birth with a birth weight of 3300 g. The perinatal period was uneventful. NBS was negative. Up to 5 years of age, 5 episodes of pneumonia occurred, all of which were treated with antibiotics but never required hospitalization. However, the sputum smear was never positive for Pseudomonas aeruginosa infection. Her weight gain was always reasonable. Based on her family history and frequent pneumonia with productive cough, CF was also suspected in her. The sweat test performed was in the normal range (25 mmol/L). Subsequent genetic testing revealed a heterozygous linkage for the variants I1234V and L997F. Pancreatic elastase in stool was normal. Immediately after diagnosis, PERT was initiated, and since then she has had no respiratory infections, productive cough, or pneumonia for three years. Her current treatment includes PERT and regular doses of DEKAs vitamins.
6. Mother. 43-Year-old: 1677delTA/L997F
G2P2, born of nonsanguineous parents. She didn't undergo NBS. She had pneumonia before the age of 12 months. Since then, she had no lung infections until she had mild left-sided pneumonia 40 years ago, which was treated with oral amoxicillin-clavulanic acid and recovered quickly. There is no history of bronchospasm or persistent cough. She weighs 60 kg and is 165 cm tall. At the age of 43, a sweat chloride test and stool elastase measurement were performed, both of which gave normal results: SCL 19 mmol/L and stool pancreatic elastase >200 mcg/g. She is not taking any medication currently.
The details of the different family members are summarized in Table 1. The characteristics of the variants according to the American College of Medical Genetics and genomics (ACMG) classification and the CFTR2 database are summarized in Table 2.
Table 1.
Demographics and clinical data of family members.
| Family member | Current age/Age at diagnosis (years) | Clinical diagnosis | Cause of diagnosis | Allele 1 | Allele 2 | NBS | SCL (value) | Pancreatic status | Weight | Excessive mucus | Cough | Pseudomonas colonization |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| S 1 | 12/10 | CF | Respiratory | c.3700A > G p.(Ile1234Val) | c.1545_1546del p.(Tyr515fs*) | N/A | 105 | PI | N | Yes | No | Yes |
| S 2 | 9/7 | CF | Respiratory | c.3700A > G p.(Ile1234Val) | c.1545_1546del p.(Tyr515fs*) | Negative | 120 | PI | N | Yes | No | Yes |
| S 3 | 7/5 | ACF | Familiarity | c.3700A > G p.(Ile1234Val) | c.2991G > C p.(Leu997Phe) | Negative | 15 | PI | N | Yes | Yes | No |
| M | 43/40 | Healthy | Familiarity | c.1545_1546del p.(Tyr515fs*) | c.2991G > C p.(Leu997Phe) | N/A | 19 | PS | N | No | No | No |
| F | 43/40 | Healthy | Familiarity | c.3700A > G p.(Ile1234Val) | WT | N/A | N/A | PS | N | No | No | No |
Abbreviations: ACF - atypical cystic fibrosis; CF - cystic fibrosis; F - Father; M - Mother; N - normal; N/A - not applicable; NBS - newborn screening; PI - pancreatic insufficient; PS - pancreatic sufficient; S - Sibling; SCL - sweat chloride level; WT - wild type.
Table 2.
Current classification of CFTR alleles according to the ACMG classification and CFTR2 database.
| Allele | ACMG Classification | CFTR2 Database |
|---|---|---|
| c.3700A > G p.(Ile1234Val) | P | CF-causing |
| c.1545_1546del p.(Tyr515fs*) | P | N/A |
| c.2991G > C p.(Leu997Phe) | P | Non CF-causing |
Abbreviations: P - pathogenic, N/A - not available.
7. Discussion
So far, more than 2000 CFTR variants have been identified. However, the clinical consequences of all variants have not yet been clarified. A small number of disease-causing mutations account for about 85% of all known variants worldwide. F508del is the most common CF-causing mutation, with a prevalence of 30–80% depending on the ethnic group [12]. In addition, there are many other mutations that are rare elsewhere but common in certain ethnic groups, due to geographic isolation or founder effects [13,14]. In addition, there are an increasing number of newly identified variants that complicate the interpretation of consequences as some changes in the gene may cause little or no clinical complications. These alleles may not cause any variation in sweat chloride levels and clearly do not cause respiratory infections, but they have been identified as responsible for CFTR-RD. However, the influence of environmental factors, modifier genes, and the heterogeneity of the large number of variant combinations in the compound heterozygous state make it difficult to predict an accurate genotype-phenotype correlation [7,8]. It is clear that there is a complex relationship between CFTR genotype and clinical phenotype.
We reported a familial case of CF with rare variant combinations. In our cases, siblings 2 and 3 had negative NBS results, while sibling 1 did not undergo NBS because the national NBS programme in Georgia was initiated after his birth. Compound heterozygous patients (siblings 1 and 2) with variants I1234V and 1677delTA had recurrent bronchopulmonary infections, an insufficient pancreas, normal growth and development and were diagnosed as classical CF. Sibling 3 with variants I1234V and L997F had episodes of pneumonia, but sweat test and stool pancreatic elastase were normal. It is noteworthy that in our report, we detected three different alleles in a single family where asymptomatic carrier parents have three affected children, and to the best of our knowledge, none of the allele combinations (1677delTA/I1234V, I1234V/L997F, and 1677delTA/L997F) present in these family members have been reported in the CFTR2 database.
Both 1677delTA and I1234V have been reported as pathogenic alleles causing PI CF [1]. The 1677delTA variant is found with high frequency in countries bordering or historically associated with the Black Sea region [[14], [15], [16]]. A recent study found that the heterozygous state of the F508del/1677delTA linkage was associated with a less severe disease course than that of F508del homozygotes, providing strong evidence that the 1677delTA mutation is much milder than the F508del mutation [14]. The I1234V allele is the second most common in the Middle East and is thought to originate from Bedouin tribes. In a recent retrospective study of 9 homozygous or compound heterozygous patients, the I1234V variant was clearly associated with the CF phenotype, and interestingly, these patients had a more favorable outcome than F508del homozygotes, despite both mutations being misfolding defects with formation of the dysfunctional CFTR protein [17].
For the L997F variant, it is even more complicated to consider it as causative of atypical CF, CFTR-RD, or even benign [9]. In another study, this allele in combination with a severe F508del variant was associated with recurrent pancreatitis in normal sweat chloride testing [18]. Other cases of L997F alleles in combination with other variants were also found with high frequency in patients with recurrent idiopathic pancreatitis and neonates with hypertrypsinaemia [10,19], but not in the classical CF phenotype. Therefore, some authors even advised against inclusion in population-based screening panels, considering that these patients might have a functional CFTR allele (10Strom et al., 2011) and even considered it benign [20].
In a recent study, Atag and colleagues [21] genotyped 150 CF patients in Turkey who were diagnosed based on two positive sweat chloride tests and the presence of typical pulmonary and gastrointestinal symptoms. None of these patients had the L997F variant, while other Turkish authors [22] have genotyped 1595 NBS-positive infants since 2017. Interestingly, the second most common allele in this cohort was L997F (after the V470M allele, which is classified as a polymorphism according to the ACMG). Such a dramatic difference in the frequency of the L997F allele in two cohorts of Turkish patients, one of whom are clinically and biochemically diagnosed CF patients with a mean age of 11.3 ± 7.5 years and the other NBS-positive neonates, suggests that the L997F allele is not associated with the CF phenotype and that these individuals are indeed revealed because of a tendency to recurrent idiopathic pancreatitis rather than CF.
In the present study, both the mother and her daughter (sibling 3) are compound heterozygotes who share an L997F allele, but in different combinations with 1677delTA and I1234V. Of note, the mother with the 1677delTA/L997F combination is asymptomatic, while sibling 3 with the I1234V/L997F combination has nonclassical CF with recurrent pneumonia that improved after starting treatment. We performed genetic testing only in sibling 3 based on family history. Delayed or missed diagnosis of patients with nonclassical or atypical CF may result in failure to provide appropriate genetic counseling and treatment. Therefore, in our study, we focus on the importance of identifying individuals who carry a CF-causing mutation and a risk factor allele, who may be asymptomatic or develop a mild CF phenotype later in life.
8. Conclusion
These cases illustrate the existence of genotype-phenotype correlation for CFTR variants and highlight the importance of genotyping individuals with suspected CF or idiopathic chronic pancreatitis. Understanding the clinical consequences of specific CFTR variants is critical for predicting the severity of the disease and for effective treatment. As more individuals are diagnosed with the above allele combinations in the future, we will be able to better understand the true significance and effect of these alleles on health and disease.
Funding statement
Genetic testing of the patients was performed at Centogene GmbH within BioCyFi study (ClinicalTrials.gov Identifier: NCT02710383, March 16, 2016). All authors had full access to all the data in the present study and had final responsibility for the decision to submit for publication.
Data availability
The datasets used and analyzed during the current report are available from the corresponding author (EK) on request.
Author contributions
All authors contributed to the study conception and design. TT and MG supported clinical monitoring of patients. Material preparation, data collection, and analysis were performed by TT, EK, and VS. The first draft of the manuscript was written by TT and EK. ML, AR, and EA were involved in interpreting cases and revising drafts. All authors read and approved the final manuscript.
Declaration of competing interest
The authors declare no conflicts of interest in association with the present study.
References
- 1.O'Sullivan B.P., Freedman S.D. Cystic fibrosis. Lancet. 2009;373:1891–1904. doi: 10.1016/S0140-6736(09)60327-5. [DOI] [PubMed] [Google Scholar]
- 2.Cystic fibrosis mutation database (CFTR1) http://www.genet.sickkids.on.ca
- 3.Rosenfeld M., Sontag M.K., Ren C.L. Cystic fibrosis diagnosis and newborn screening. Pediatr. Clin. 2016;63(4):599–615. doi: 10.1016/j.pcl.2016.04.004. [DOI] [PubMed] [Google Scholar]
- 4.CFTR2; clinical and functional translation of CFTR. http://www.cftr2.org
- 5.Rowntree R.K., Harris A. The phenotypic consequences of CFTR mutations. Ann. Hum. Genet. 2003;67(Pt 5):471–485. doi: 10.1046/j.1469-1809.2003.00028.x. [DOI] [PubMed] [Google Scholar]
- 6.Szczesniak R., Rice J.L., Brokamp C., Ryan P., Pestian T., Ni Y., Andrinopoulou E.R., Keogh R.H., Gecili E., Huang R., Clancy J.P., Collaco J.M. Influences of environmental exposures on individuals living with cystic fibrosis. Expet Rev. Respir. Med. 2020;14(7):737–748. doi: 10.1080/17476348.2020.1753507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sepahzad A., Morris-Rosendahl D.J., Davies J.C. Cystic fibrosis lung disease modifiers and their relevance in the new era of precision medicine. Genes. 2021:13. doi: 10.3390/genes12040562. 12(4):562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kvaratskhelia E., Ghughunishvili M., Gagua M., Tkemaladze T., Surmava S., Kvintradze M., Abzianidze E. Hypomethylation of TNF-alpha gene promoter in patients with cystic fibrosis. Abstracts from the 52nd European society of human genetics (ESHG) conference: posters. Eur. J. Hum. Genet. 2019;27:1910–1911. doi: 10.1038/s41431-019-0493-3. [DOI] [Google Scholar]
- 9.Derichs N., Schuster A., Grund I., Ernsting A., Stolpe C., Körtge-Jung S., Gallati S., Stuhrmann M., Kozlowski P., Ballmann M. Homozygosity for L997F in a child with normal clinical and chloride secretory phenotype provides evidence that this cystic fibrosis transmembrane conductance regulator mutation does not cause cystic fibrosis. Clin. Genet. 2005;67(6):529–531. doi: 10.1111/j.1399-0004.2005.00437.x. [DOI] [PubMed] [Google Scholar]
- 10.Strom C.M., Redman J.B., Peng M. The dangers of including nonclassical cystic fibrosis variants in population-based screening panels: p.L997F, further genotype/phenotype correlation data. Genet. Med. 2011;13(12):1042–1044. doi: 10.1097/GIM.0b013e318228efb2. [DOI] [PubMed] [Google Scholar]
- 11.Salinas D.B., Sosnay P.R., Azen C., Young S., Raraigh K.S., Keens T.G., Kharrazi M. Benign and deleterious cystic fibrosis transmembrane conductance regulator mutations identified by sequencing in positive cystic fibrosis newborn screen children from California. PLoS One. 2016;11(5) doi: 10.1371/journal.pone.0155624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cutting G.R. Cystic fibrosis genetics: from molecular understanding to clinical application. Nat. Rev. Genet. 2015;16(1):45–56. doi: 10.1038/nrg3849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Leung G.K., Ying D., Mak C.C., Chen X.Y., Xu W., Yeung K.S., Wong W.L., Chu Y.W., Mok G.T., Chau C.S., McLuskey J., Ong W.P., Leong H.Y., Chan K.Y., Yang W., Chen J.H., Li A.M., Sham P.C., Lau Y.L., Chung B.H., Lee S.L. CFTR founder mutation causes protein trafficking defects in Chinese patients with cystic fibrosis. Mol Genet Genomic Med. 2016;5(1):40–49. doi: 10.1002/mgg3.258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Petrova N.V., Kashirskaya N.Y., Saydaeva D.K., Polyakov A.V., Adyan T.A., Simonova O.I., Gorinova Y.V., Kondratyeva E.I., Sherman V.D., Novoselova O.G., Vasilyeva T.A., Marakhonov A.V., Macek M., Jr., Ginter E.K., Zinchenko R.A. Spectrum of CFTR mutations in Chechen cystic fibrosis patients: high frequency of c.1545_1546delTA (p.Tyr515X; 1677delTA) and c.274G>A (p.Glu92Lys, E92K) mutations in North Caucasus. BMC Med. Genet. 2019;20(1):44. doi: 10.1186/s12881-019-0785-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.lvaschenko T., White M.B., Dean M., Baranov V.S. A deletion of two nucleotides in exon 10 of the CFTR gene in a Soviet family with cystic fibrosis causing early infant death. Genomics. 1991;10:298–299. doi: 10.1016/0888-7543(91)90517-I. [DOI] [PubMed] [Google Scholar]
- 16.Angelicheva D., Boteva K., Jordanova A., Savov A., Kufardjieva A., Tolun A., Telatar M., Akarsubaşi A., Köprübaşi F., Aydoğdu S., et al. Cystic fibrosis patients from the Black Sea region: the 1677delTA mutation. Hum. Mutat. 1994;3(4):353–357. doi: 10.1002/humu.1380030405. [DOI] [PubMed] [Google Scholar]
- 17.El Bar Aluma B., Sarouk I., Senderowitz H., Cohen-Cymberknoh M., Khazanov N., Dagan A., Bezalel Y., Ashkenazi M., Keler S., Efrati O. Phenotypic and molecular characteristics of CF patients carrying the I1234V mutation. Respir. Med. 2020;170 doi: 10.1016/j.rmed.2020.106027. [DOI] [PubMed] [Google Scholar]
- 18.Conklin L., Zeitlin P.L., Cuffari C. Cystic fibrosis presenting as recurrent pancreatitis in a young child with a normal sweat test and pancreas divisum: a case report. J. Med. Case Rep. 2008;2:176. doi: 10.1186/1752-1947-2-176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gomez Lira M., Benetazzo M.G., Marzari M.G., Bombieri C., Belpinati F., Castellani C., Cavallini G.C., Mastella G., Pignatti P.F. High frequency of cystic fibrosis transmembrane regulator mutation L997F in patients with recurrent idiopathic pancreatitis and in newborns with hypertrypsinemia. Am. J. Hum. Genet. 2000;66(6):2013–2014. doi: 10.1086/302928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Salinas D.B., Sosnay P.R., Azen C., Young S., Raraigh K.S., Keens T.G., Kharrazi M. Benign outcome among positive cystic fibrosis newborn screen children with non-CF-causing variants. J. Cyst. Fibros. 2015;14(6):714–719. doi: 10.1016/j.jcf.2015.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Atag E., Bas Ikizoglu N., Ergenekon A.P., Gokdemir Y., Eralp E.E., Ata P., Ersu R., Karakoc F., Karadag B. Novel mutations and deletions in cystic fibrosis in a tertiary cystic fibrosis center in Istanbul. Pediatr. Pulmonol. 2019;54(6):743–750. doi: 10.1002/ppul.24299. [DOI] [PubMed] [Google Scholar]
- 22.Bozdogan S.T., Mujde C., Boga I., Sonmezler O., Hanta A., Rencuzogullari C., Ozcan D., Altintas D.U., Bisgin A. Current status of genetic diagnosis laboratories and frequency of genetic variants associated with cystic fibrosis through a newborn-screening program in Turkey. Genes. 2021;12(2):206. doi: 10.3390/genes12020206. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and analyzed during the current report are available from the corresponding author (EK) on request.