Summary
The epithelial splicing regulatory proteins, ESRP1 and ESRP2, are essential for mammalian development through the regulation of a global program of alternative splicing of genes involved in the maintenance of epithelial cell function. To further inform our understanding of the molecular functions of ESRP1, we performed enhanced crosslinking immunoprecipitation coupled with high-throughput sequencing (eCLIP) in epithelial cells of mouse epidermis. The genome-wide binding sites of ESRP1 were integrated with RNA-Seq analysis of alterations in splicing and total gene expression that result from epidermal ablation of Esrp1 and Esrp2. These studies demonstrated that ESRP1 functions in splicing regulation occur primarily through direct binding in a position-dependent manner to promote either exon inclusion or skipping. In addition, we also identified widespread binding of ESRP1 in 3′ and 5′ untranslated regions (UTRs) of genes involved in epithelial cell function, suggesting that its post-transcriptional functions extend beyond splicing regulation.
Subject areas: Biochemistry, Molecular biology, Cell biology
Graphical abstract
Highlights
-
•
ESRP1 directly binds introns and exons to regulate exon inclusion or exclusion
-
•
ESRP1 binds within introns and broadly within the 3ʹ UTR of protein-coding genes
-
•
Loss of ESRP1 causes a splicing-independent increase in levels of the total CTNND1 protein
-
•
Cytoplasmic ESRP1 appears to regulate CTNND1 protein abundance
Biochemistry; Molecular biology; Cell biology;
Introduction
RNA-binding proteins (RBPs) have diverse post-transcriptional functions that impact cell properties through regulation of alternative splicing, mRNA localization, mRNA translation, and mRNA stability. Whereas some RBPs are ubiquitously expressed and involved in essential cell functions, many are cell type-specific and thereby fine tune functions that contribute to physiologic roles unique to those cells, such as in neurons (Conboy, 2017; Gerstberger et al., 2014; Hakim et al., 2017; Nikonova et al., 2019; Van Nostrand et al., 2020a). We identified ESRP1 and ESRP2 as paralogous epithelial cell type-specific specific splicing regulatory proteins (Warzecha et al., 2009a, 2009b). Studies in epithelial cell lines with ESRP1/2 depletion and in mouse epithelial cells with Esrp1/2 ablation showed that ESRPs regulate a global alternative splicing program that is enriched for genes involved in epithelial cell functions including cytoskeletal organization, cell polarity, and maintenance of adherens junctions, and tight junctions (Bebee et al., 2015; Dittmar et al., 2012; Lee et al., 2018; Warzecha et al., 2009a). Furthermore, we showed that Esrp1 knockout (KO) mice exhibited several developmental defects and perinatal lethality, whereas mice with knockout of both Esrp1 and Esrp2 had substantially greater defects. In contrast, Esrp2 KO mice had no observed abnormalities indicating that whereas there is some functional redundancy of these paralogs, ESRP1 is most essential (Bebee et al., 2015; Lee et al., 2018).
Many RBPs have been shown to regulate multiple steps in RNA processing, and emerging evidence has allocated roles outside of splicing to several splicing factors such as RBFOX1, QK1, MBNL1, PTBP1/2, among others (Hafner et al., 2010; Lee et al., 2016; Masuda et al., 2012; Romanelli et al., 2013). In some instances, the multifunctionality of the RBPs is affected by differential localization wherein alternative splicing leads to the production of RBP protein isoforms with predominantly nuclear or cytoplasmic localization. For example, QKI expresses distinct nuclear and cytoplasmic isoforms that regulate mRNA splicing and translation (Fagg et al., 2017; Hafner et al., 2010; Hall et al., 2013; Wu et al., 1999). RBFOX1, another well-studied splicing regulator, also has both nuclear and cytoplasmic isoforms. It has been shown that nuclear RBFOX1 is largely responsible for regulating splicing in neurons, heart, and muscle, whereas the cytoplasmic isoform regulates mRNA stability and translation (Lee et al., 2016). MBNL1, another RBP with predominantly nuclear and cytoplasmic isoforms, also regulates alternative splicing, mRNA stability, and mRNA localization (Masuda et al., 2012; Wang et al., 2012). Whereas we previously focused on the role of ESRPs in splicing regulation, we also showed that competing 5′ splice sites at the end of Esrp1 exon 12 lead to the production of ESRP1 isoforms with distinct nuclear or cytoplasmic localization (Yang and Carstens, 2017). Furthermore, we also showed that the D. Melanogaster ortholog of ESRP1, Fusilli, also expresses distinct splice variants encoding nuclear and cytoplasmic protein isoforms. These observations suggested that ESRP1 also has a phylogenetically conserved function to regulate post-transcriptional steps in the cytoplasm. However, the targets of ESRP1 in the cytoplasm and its functions in this cell compartment have not been determined.
RBPs, including ESRP1, execute their regulatory functions through sequence-specific binding to RNA. We previously identified a high-affinity binding motif for ESRP1 in vitro (Dittmar et al., 2012). However, the identification of binding motifs for specific RBPs, while providing valuable insight, does not necessarily reflect in vivo binding (Van Nostrand et al., 2020a). Crosslinking immunoprecipitation (CLIP) can be used to interrogate the RBP: RNA relationship in vivo, and when integrated with data from RNA-seq studies on tissues with the modulated expression of RBPs, we can infer functional roles. For example, in the case of MBNL1, separate roles for both nuclear and cytoplasmic were identified using RNA-Seq data from Mbnl1 knockdown and MBNL1 CLIP, which revealed that it not only bound within introns to regulate splicing, but also had significant 3′ untranslated region (UTR) binding through which it regulated mRNA stability (Masuda et al., 2012). Similarly, for RBFOX2, resolution of the functional roles of the cytoplasmic isoform was achieved by interrogation of using crosslinking immunoprecipitations (iCLIP) with transcriptome profiling (Lee et al., 2016). On a global scale, integrative studies combining CLIP and alternative splicing data acquired for a plethora of RBPs from various cell lines have been used to generate RNA maps of how these RBPs bind and regulate alternative splicing (Yee et al., 2018). We previously showed that the binding motif for ESRP1 is enriched in the downstream intron of exons whose splicing is promoted by ESRP1/2, and in the upstream intron and exon body of ESRP-silenced exons (Bebee et al., 2015; Dittmar et al., 2012; Warzecha et al., 2010). However, proof that this position-dependent regulation was owing to the direct binding of ESRPs to most regulated exons required the identification of genome-wide binding sites. In addition, CLIP is needed for the identification of non-splicing related targets of ESRP1. Therefore, to broadly define the post-transcriptional regulatory network of Esrps in vivo we combined RNA-sequencing and enhanced crosslinking immunoprecipitation in the epidermis, where the Esrps are highly expressed. Results from the investigations were used to determine genome-wide in vivo targets of ESRP1, confirm the preferred motif for ESRP1 binding, and generate a transcriptome-wide map of ESRP1 binding and regulation.
Results
Genome-wide profiling of ESRP1-binding sites with enhanced crosslinking immunoprecipitation
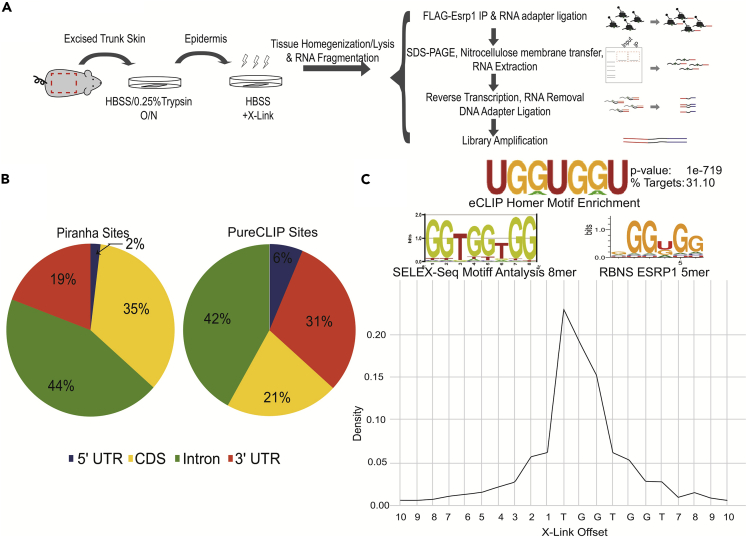
We employed enhanced crosslinking immunoprecipitation (eCLIP) of ESRP1 in mouse epidermis to establish that direct binding of ESRP1 is required for position-dependent splicing regulation and to confirm its binding motif in vivo (Van Nostrand et al., 2016). To facilitate immunoprecipitation of endogenous ESRP1 from mouse tissues we used CRISPR/Cas9 to introduce two tandem copies of the FLAG tag at the endogenous N-terminus of mouse ESRP1 (Esrp1FLAG/FLAG mice)(Lee et al., 2020), an approach previously shown to enable the extraction of high-quality binding data from cells (Van Nostrand et al., 2017). The introduction of an N-terminus FLAG tag to ESRP1 does not affect the maintenance of epithelial splicing by ESRP1 (Figure S1A). The epidermis was separated from the back skin of Esrp1FLAG/FLAG or wild-type ESRP1 (Esrp1WT/WT) mice to obtain a pure population of epithelial cells, which were then crosslinked and ESRP1 bound RNAs were immunoprecipitated using an anti-FLAG antibody for eCLIP (Figure 1A) (see STAR Methods). After determining the optimal RNase concentration to generate libraries of ∼50nt CLIP tags (Figure S1B), we sequenced six replicate CLIP samples from the Esrp1FLAG/FLAG mice with corresponding inputs (see STAR Methods for additional details) to an average depth of 41 million reads per CLIP and 37 million reads per input. As a control, we attempted to generate Flag eCLIP libraries from Esrp1WT/WT mice, but these samples required high cycle numbers for library amplification and predominantly produced primer dimer artifacts, and thus were not sequenced. This result suggested that the libraries made using Esrp1FLAG/FLAG mice were largely specific. Peaks were identified using Piranha (Uren et al., 2012) for each replicate and pooled to identify high-confidence binding sites. This analysis was further supplemented by using PureCLIP (Krakau et al., 2017) on three sets of paired replicates from the Esrp1FLAG/FLAG samples to identify binding sites at single nucleotide resolution. Using PureCLIP and Piranha, we observed that ESRP1 crosslinks were most enriched in introns and abundant in coding exons, consistent with the functions of ESRP1 to regulate splicing through binding in exons and/or flanking intron sequences (Figure 1B). We also noted many binding sites for ESRP1 in both 5′ and 3’ UTRs, consistent with a post-transcriptional role for the cytoplasmic isoform of ESRP1.
Figure 1.
Enhanced crosslinking immunoprecipitation identifies genome wide binding sites for ESRP1
(A) Illustration of workflow for enhanced crosslinking immunoprecipitation using Esrp1FLAG/FLAG neonates.
(B) Pie chart showing of crosslinking sites for ESRP1 identified using PureCLIP and peaks called by Piranha.
(C) (Top) WebLogo of de novo motif analysis performed using Homer identifies UGGUGG as most enriched ESRP1-binding motif bound fragments. WebLogo of position weight matrix of 6- and 8-mers enriched in ESRP1 bound fragments using Selex-seq (Dittmar et al., 2012) WebLogo of aligned enriched 5-mers of ESRP1 (Van Nostrand et al., 2020a). (Bottom) Distribution of crosslinking site frequency relative to the UGGUGG motif determined to be the first nucleotide in the UGGUGG motif (parameters used for calling crosslinks assigns the crosslink to the position upstream of the read start site).
The most enriched motif within the ESRP1 bound RNA fragments consisted of UGG repeats as determined using the HOMER motif discovery algorithm (Heinz et al., 2010). PureCLIP, which facilitates simultaneous peak calling and crosslink site detection at single nucleotide resolution, identified the first position within a UGGUGG motif as the primary site of binding (Figure 1C). This in vivo defined binding motif was highly similar to the one we previously identified in vitro using SELEX-Seq (Figure 1C) (Dittmar et al., 2012). A highly similar motif was also recently identified using RNA bind-n-Seq in the ENCODE project, further validating the accuracy of ESRP1-binding site determination using eCLIP in Esrp1FLAG/FLAG mouse tissues (Van Nostrand et al., 2020a) (Figure 1C).
Comprehensive identification of transcriptomic changes in the epidermis following inducible ESRP1/2 ablation
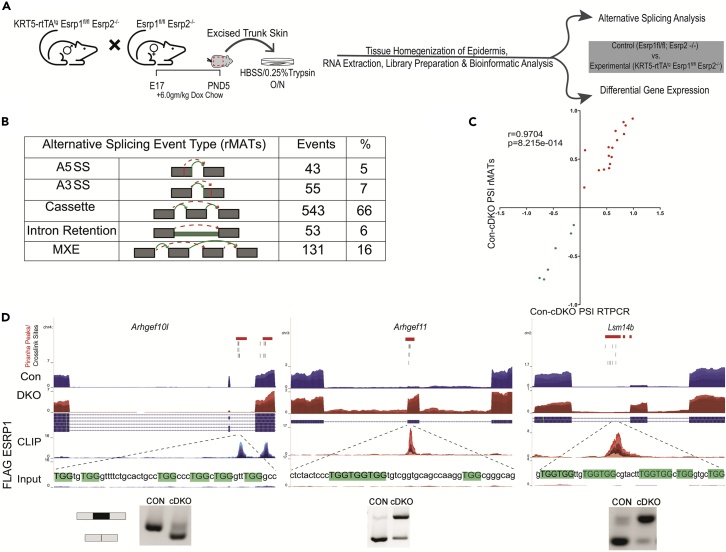
To comprehensively identify changes in splicing and total mRNA expression that result from the acute loss of Esrp1/2 expression we used an inducible knockout strategy to conditionally ablate ESRP1. We previously identified splicing changes in the epidermis of mice with germline deletion of Esrp1 alone or combined deletion of Esrp1 and Esrp2 (Bebee et al., 2015). However, these studies used fewer replicates and a lower read depth. It was also possible that some transcriptomic changes in mice with germline Esrp1/2 ablation were indirect consequences of functional and developmental defects in Esrp ablated epidermis (see below). We therefore used mice with conditional Esrp1 knockout (Esrp1flox/flox) alleles crossed with transgenic K5rTAand tetO-Cre mice to enable doxycycline-inducible ablation of Esrps in neonates by feeding pregnant female mice with a high concentration doxycycline chow and collecting epidermis from P5 offspring for transcriptomic analysis (Figure 2A). Given some functional redundancy of ESRP2, we used mice with homozygous deletion of Esrp2. Littermates were treated as experimental (K5rTA tetOCre Esrp1flox/flox; Esrp2−/−; conditional double knockout (cDKO)) or control (Esrp1flox/flox; Esrp2−/−). We sequenced four control and four experimental replicates to an average depth of 150 million reads and assessed changes in alternative splicing and gene expression.
Figure 2.
ESRP1 is a direct regulator of alternative splicing
(A) Illustration of workflow to generate RNA from control (Esrp1flox/flox; Esrp2−/−) and experimental (K5rTA; tetOCre; Esrp1flox/flox; Esrp2−/−) neonates.
(B) Number and type of alternatively spliced events identified by rMATs between control and experimental epidermis.
(C) Correlation plot of ΔPSI estimated by rMATs skipped exon events validated by RT-PCR, with Pearson Correlation r and p-values. Green dots represent splicing events suppressed by ESRP1 and red dots represent splicing events enhanced by ESRP1.
(D) Genome browser views showing ESRP1-binding peak distribution of pooled replicates for Arhgef10L, Arhgef11, and Lsm14b. Shown are densities in CLIP and size-matched Input. Also shown are read densities for transcript levels from RNA sequencing in epidermis ablated of Esrps and control epidermis. UGG motifs are in uppercase and highlighted in green. Positions of crosslink sites from PureCLIP for three replicate pairs and Piranha peaks are shown at the top.
Alternatively spliced events identified were congruent with a shift from epithelial to mesenchymal splicing patterns, consistent with a loss of Esrps. We used both rMATs (Shen et al., 2012, 2014) and MAJIQ (Vaquero-Garcia et al., 2016) to assess alternative splicing. Using rMATs, we identified 845 splicing changes between control and cDKO epidermis with a |ΔPSI| greater than 10% (FDR ≤ 0.05) (Figure 2B and Table S1). Using a similar stringency MAJIQ identified a total of 512 local splicing variations (LSVs) that included complex and binary changes in splicing with an absolute change in percentage spliced in (|ΔPSI|) greater than 10% (p ≥ 0.95) (Figure S2A and Table S2). We used semiquantitative RT-PCR to validate over 20 exon skipping events identified by either MAJIQ or rMATS and observe a strong correlation between ΔPSI calculated by using RT-PCR and ΔPSI (r = 0.9707, p < 0.005; r = 0.9704, p < 0.005) for MAJIQ and rMATS, respectively (Figures 2C and S2B). Furthermore, we observe a strong correlation of the reported ΔPSI between rMATs and MAJIQ for shared exon skipping events (r = 0.9872, p < 0.005) (Figure S2C).
Consistent with a position-dependent role of ESRP1 in control of splicing through direct binding, we identified ESRP1 eCLIP peaks in several ESRP1-regulated targets. For example, Arhgef10L, Arhgef11, and Lsm14b exons were previously shown to have large splicing changes in response to ESRP1/2 depletion or ablation and we observed robust peaks for ESRP1 in or near these regulated exons. In the case of Arhgef11 and Lsm14b, we identified ESRP1-binding sites either within the exon or in the upstream intron that were associated with increased splicing upon Esrp1/2 deletion. In contrast, we identified ESRP1 binding in the intron downstream of an Arhgef10l exon that is skipped after the loss of ESRP1/2 (Figure 2D). These observations supported our previously proposed model of an “RNA map” wherein Esrp1 binding upstream of or within an exon induces exon skipping, whereas binding in the downstream intron promotes exon splicing (Dittmar et al., 2012; Warzecha et al., 2010). Using MAJIQ, we were also able to detect additional AS events beyond the five major types: Skipped Exon (SE), Mutually Exclusive Exon (MXE), Alternative 5’ Splice Site (A5SS), Alternative 3’ Splice Site (A3SS), and Retained Intron (RI), such as Alternative Last Exon (ALE) events (Figure S2A). For example, we identified a splicing change in Gpatch2 that altered the ratio of short and long isoforms owing to an ALE event (Figure S2D). Overall, the splicing changes we identified with inducible Esrp ablation included many that were previously identified with germline knockout of Esrp1/2 (DKO) (Bebee et al., 2015), but consisted of a significantly larger dataset of ESRP-regulated events as well as detection of more complex splicing alterations.
The positions of ESRP1-binding sites globally establish an RNA map that determines whether it functions to promote exon splicing or skipping
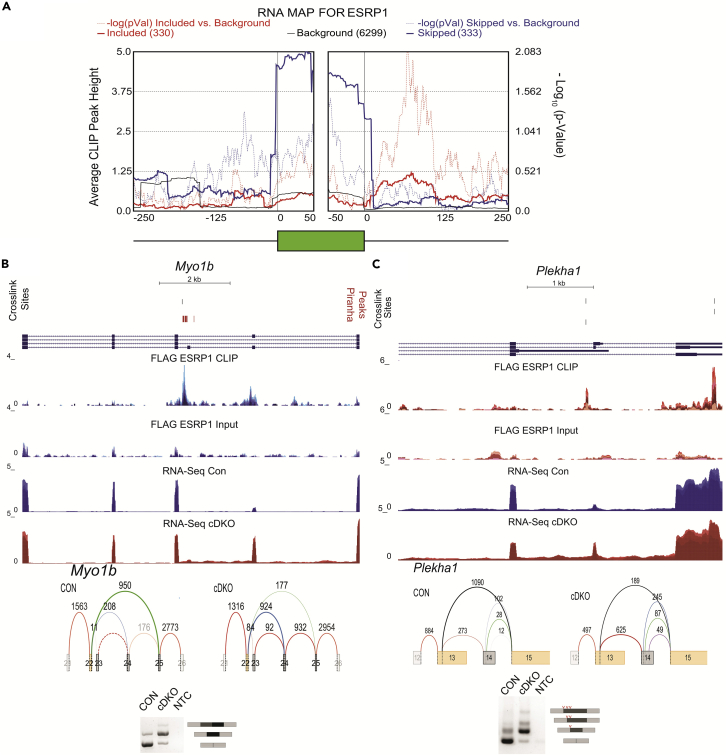
To further characterize the positional effects of ESRP1 binding on splicing at genome-scale, we used rMAPS2, a web server that combines differentially regulated AS events data obtained from RNA-seq as determined by rMATs with peaks called from eCLIP-seq data using Piranha (Hwang et al., 2020; Uren et al., 2012). The output from rMAPS2 visualizes the spatial distributions of Esrp1-binding sites near (within 250 nucleotides) upregulated, downregulated, and background control exons for five major types of AS events: SE, MXE, A5SS, A3SS, and RI. This analysis confirmed that ESRP1 binding in the intron downstream of a regulated exon was generally associated with ESRP1 mediated promotion of exon inclusion, whereas ESRP1-binding sites in the upstream intron or within the regulated exon were associated with exon skipping (Figure 3A). The CLIP data also demonstrated how the position-dependent binding pattern of ESRP1 underlies the regulation of more complex changes in alternative splicing. For example, in the case of MYO1B, we noted ESRP1-binding sites upstream of tandem exons (exons 23 and 24) that illustrate mechanistically how ESRP1 binding coordinates the skipping of both exons (Figure 3B). A similar observation of tandem exon regulation is CD44, where ESRP1-binding sites are present downstream of numerous tandem variable exons cases to coordinate the splicing of these exons in epithelial cells (Figure S3A). We also noted that three changes in cassette exon splicing in PLEKHA1 corresponded to three splice variants of the same penultimate exon that collectively indicated a greater total splicing change in exon skipping than was calculated using each of the three variants separately (Figure 3C).
Figure 3.
A functional splicing RNA Map for ESRP1
(A) Splicing RNA Map of the distribution of ESRP1 CLIP peaks on alternatively spliced exons. The solid blue line shows the ESRP1 CLIP densities over ESRP1 downregulated exons, and the dotted blue line shows the significance of the peak at each position. The solid red line shows the ESRP1 CLIP densities of ESRP1 upregulated exons, with dotted red line indicating significance (p-value) of the binding at the position shown. (B) Voila plots, genome browser views showing ESRP1-binding (CLIP-Seq). and transcript levels (RNA-seq) of (B) Myo1b and (C) Plekha and ethidium bromide-stained agarose gel of RT-PCR products of ESRP1-regulated splicing event (red hatches indicate A5SS in Plekha1). Voila plots show differential exon inclusion between Control (CON) and Experimental (cDKO) conditions. CLIP-seq densities show CLIP and size-matched input, and transcript levels are from RNA sequence in epidermis ablated of Esrps and control epidermis. Positions of crosslink sites from PureCLIP for three replicate pairs and Piranha peaks are shown at the top.
To confirm that ESRP1-regulated events are direct targets, we initially examined a total of 543 cassette exons (SE exons) identified by rMATS with |ΔPSI| ≥ 10% and FDR ≤0.05% (Table S1). For 278 ESRP1-enhanced exons, we identified 47 (17%) that had significant peaks within 250 nucleotides of the downstream intron as determined by Piranha. For 265 exons for which ESRP1 promotes skipping we identified 24 (9%) that had significant peaks in the 250 nucleotides of the upstream exon. We also identified 13 (5%) Esrp1-skipped exons that had peaks within the regulated exon itself (Table S3). Whereas these data supported the RNA map of regulation determined by rMAPS2, we noted that the ESRP1-regulated exons in this total list (Table S3) likely consisted of some false negatives. To assess a broader pattern of regulation among the most robust ESRP1-regulated exons, we examined a more stringent subset of exons that were identified using both rMATS and MAJIQ with an estimated change in |ΔPSI| ≥ 20% (Table 1). For rMATS, we further filtered for only events with the lowest FDR possible (0) and identified a set of 54 non-redundant simple cassette exons (Table S4). With MAJIQ, we identified a set of 59 manually curated non-redundant cassette exons (Table S4). From the refinement of these two data sets we derived a list of 39 events that were identified with both MAJIQ and rMATs with highly stringent criteria (|ΔPSI| ≥ 20%). We then extended our analysis to identify any peaks in the entire intron downstream of ESRP1- enhanced exons and the entire upstream intron or exon of ESRP1-silenced exons that were significant peaks by called by Piranha and/or that contained crosslinked site regions identified with PureCLIP. For the 39 high-confidence events that were identified using both rMATS and MAJIQ, we identified 29 (74%) with ESRP1-binding sites in a position consistent with the RNA map (Table S4). This analysis suggests that most significant ESRP1-regulated exons are directly regulated. This direct regulation of alternative splicing by ESRP1 was not restricted to skipped exon events but was also observed for mutually exclusive exons such as is the case for FGFR2 and FAR1 (Figure S3C). We previously showed that ESRP1 regulates mutually exclusive exons IIIb and IIIc in Fgfr1, Fgfr2, and Fgfr3 (Bebee et al., 2015). Whereas there was complex ESRP1 binding near these exons, a strong binding peak was identified in the intron between exons IIIb and IIIc. This ESRP1-binding site was previously shown in transfected minigene studies to mediate activation of the upstream IIIb exon and repression of the downstream IIIc exon in Fgfr2 (Hovhannisyan and Carstens, 2005; Hovhannisyan et al., 2006; Warzecha et al., 2009a). In this example, it is apparent that the position-dependent function of ESRP1 ensures coordination of mutually exclusive exon regulation. In contrast, in the case of Far1, there is the inclusion of either alternate exon 10A or exon 10B, with binding of ESRP1 appearing to enhance the inclusion of exon 10b. In Fgfr2 and Far1, both the mutually exclusive exons as well as the ESPR1-binding sites show high conservation.
Table 1.
Highly significant changes in cassette exons detected by rMATS and MAJIQ
| Gene symbol | Exon coordinates | Exon size | ΔPSI rMATS | ΔPSI Majiq |
|---|---|---|---|---|
| Atp6v1c2 | 12:17288995–17289133 | 138 | 0.918 | 0.854 |
| Arhgef10l | 4:140568755–140568770 | 15 | 0.882 | 0.892 |
| Slk | 19:47627647–47627740 | 93 | 0.847 | 0.878 |
| Flnb | 14:7922674–7922746 | 72 | 0.791 | 0.77 |
| Grhl1 | 12:24582884–24582961 | 77 | 0.759 | 0.743 |
| Scrib | 15:76061738–76061801 | 63 | −0.755 | −0.796 |
| Arhgef11 | 3:87734423–87734552 | 129 | −0.739 | −0.724 |
| Lsm14b | 2:180031793–180031871 | 78 | −0.725 | −0.913 |
| Nf2 | 11:4780577–4780622 | 45 | 0.696 | 0.602 |
| Myo9a | 9:59875201–59875414 | 213 | 0.647 | 0.674 |
| Myo1b | 1:51766821–51766908 | 87 | −0.639 | −0.692 |
| Slc37a2 | 9:37233052–37233109 | 57 | 0.623 | 0.51 |
| Mgll | 6:88813879–88813963 | 84 | 0.618 | 0.723 |
| Ralgps2 | 1:156821386–156821464 | 78 | 0.599 | 0.588 |
| Epb41 | 4:131937014–131937464 | 450 | 0.592 | 0.603 |
| Usp4 | 9:108365829–108365970 | 141 | −0.577 | −0.542 |
| Myo6 | 9:80303272–80303299 | 27 | 0.572 | 0.514 |
| Uap1 | 1:170147995–170148046 | 51 | 0.535 | 0.506 |
| Eea1 | 10:95978160–95978286 | 126 | 0.522 | 0.495 |
| Plekha1 | 7:130909587–130909628 | 41 | −0.52 | −0.587 |
| Ctnnd1 | 2:84624269–84624558 | 289 | −0.516 | −0.466 |
| Atp6v1c2 | 12:17307470–17307500 | 30 | 0.459 | 0.52 |
| Tsc2 | 17:24606223–24606352 | 129 | 0.454 | 0.253 |
| Vps39 | 2:120346856–120346889 | 33 | 0.449 | 0.577 |
| Timm17b | X:7900960–7901060 | 100 | −0.418 | −0.471 |
| Osbpl3 | 6:50346010–50346103 | 93 | −0.417 | −0.418 |
| Itga6 | 2:71853533–71853663 | 130 | 0.407 | 0.335 |
| Dock9 | 14:121577346–121577424 | 78 | 0.402 | 0.318 |
| Enah | 1:181911596–181911659 | 63 | 0.393 | 0.413 |
| Arhgap17 | 7:123294471–123294705 | 234 | −0.382 | −0.438 |
| Myl6 | 10:128491719–128491764 | 45 | 0.375 | 0.343 |
| Flnb | 14:7934563–7934701 | 138 | −0.371 | −0.341 |
| Sorbs1 | 19:40321792–40321960 | 168 | 0.354 | 0.229 |
| Atp2c1 | 9:105494414–105494597 | 183 | −0.339 | −0.293 |
| Csnk1d | 11:120964947–120965010 | 63 | 0.29 | 0.428 |
| Arfgap1 | 2:180971573–180971683 | 110 | −0.282 | −0.262 |
| Inf2 | 12:112612547–112612604 | 57 | 0.256 | 0.254 |
| Golgb1 | 16:36893315–36893438 | 123 | 0.235 | 0.291 |
| Lsr | 7:30962097–30962244 | 147 | −0.233 | −0.252 |
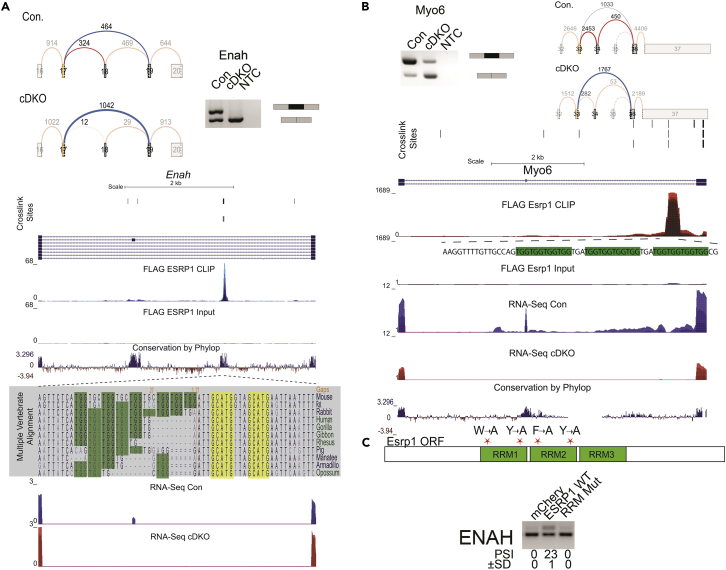
In addition to the expected observation that binding of ESPR1 proximal to exons controls splicing choices, we identified multiple examples of definitive CLIP peaks located outside the 250-nucleotide window in adjacent introns. We identified some cases in which the binding sites were more distal, such as an ESRP-enhanced exon in ENAH (Figure 4A) and MYO6 (Figure 4B). In both ENAH and MYO6, the peak of ESRP1 lies over 1 kb downstream of the regulated exon, but the position of ESRP1 binding is consistent with position-dependent regulation by ESRP1. Furthermore, both the regulated exon as well as the region of ESRP1 binding shows high conservation among placental mammals and vertebrates. This suggested that a larger percentage of ESRP1- regulated events were regulated by direct binding than indicated by analysis of the peak densities within the 250 nucleotides of the regulated exon. Additional examples of ESRP1 enhanced exons with binding sites in the downstream intron, including far distal sites, in Slc37a2, Atp6v1c2, and Uap1 are shown in Figure S4. We also illustrate additional examples of ESRP1 silenced exons in Flnb, Timm17b, Scrib, and Magi1 that have ESRP1-binding sites in the upstream intron and/or exon (Figure S5).
Figure 4.
ESRP1 binding and regulation of distal exons
(A) ESRP1 binding and regulation of Enah. Voila plots and ethidium bromide-stained agarose gel of RT-PCR products of the ESRP1-regulated skipped exon splicing event in Enah. Genome browser views showing ESRP1-binding (CLIP Seq) and transcript levels (RNA-seq) of for Enah. CLIP-seq densities show CLIP and size-matched input, and transcript levels are from RNA sequence in epidermis ablated of Esrps and control epidermis. Crosslink sites identified by PureCLIP are shown for three replicate pairs at the top of the browser views. Conservation scores showing Placental mammal conservation by PhyloP at the base. Multiple alignment of ∼50 nucleotide regions bound by ESRP1 for several vertebrate species shown. Highlighted in green are UGG motifs recognized by ESRP1 and highlighted in yellow is the RBFOX2 motif. (B) ESRP1 binding and regulation of Myo6. Voila plots and ethidium bromide-stained agarose gel of RT-PCR products of ESRP1-regulated skipped exon splicing event in Myo6. Genome browser views showing ESRP1-binding (CLIP Seq) and transcript levels (RNA-seq) of for Myo6. CLIP-seq densities show CLIP and size-matched input, and transcript levels are from the RNA sequence in epidermis ablated of Esrps and control epidermis. Crosslink sites identified by PureCLIP are shown for three replicate pairs at the top of the browser views. Conservation scores showing Placental mammal conservation by PhyloP at the base. Highlighted in green are UGG motifs recognized by ESRP. (C) Schematic of ESRP1 RRM mutant indicating mutated amino acids. Ethidium bromide-stained agarose gel of ENAH splicing in 293T cells transfected with either wild-type ESRP1 or RNA-binding deficient ESRP1 RRM mutant.
ESRP1 contains three RRM motifs, and it has been previously shown that RRM1 and RRM2 are highly similar to hnRNP F and hnRNP H (Warzecha et al., 2009a). Furthermore, RRM1 and 2 in ESRP1 contain conserved residues tryptophan and tyrosine (in RRM1) and phenylalanine and tyrosine (in RRM2), which, in hnRNP F and hnRNP H, have been shown to be essential for RNA binding and/or splicing activity (Dominguez and Allain, 2005; Mauger et al., 2008). Therefore, we mutated the conserved residues (Figure 3C) to generate an ESRP1 RRM mutant, which is not expected to bind RNA. To support our observation that ESRP1 binding to several of these distal motifs was directly regulating these splicing events, we assessed splicing changes in HEK293T cells which were transfected with ESRP1 or the ESRP1 RRM Mutant. (Figures 4C and S4). We first demonstrate that the ESRP1 RRM mutant does not promote splicing of FGFR2 IIIIb exon (Figure S4), which is a well-validated ESRP1/2 target (Bebee et al., 2015; Warzecha et al., 2009a). We similarly observed that splicing of the regulated exon in ENAH (Figure 4), SLC37A2, RALGPS, UAP1, and ATP6V1C2 was responsive to ESRP1, in a manner that was dependent on its ability to bind RNA (Figure S4). Thus, ESRP1 directly regulates splicing in a position-dependent manner, in a manner dependent upon its ability to bind RNA, and this regulation is highly conserved.
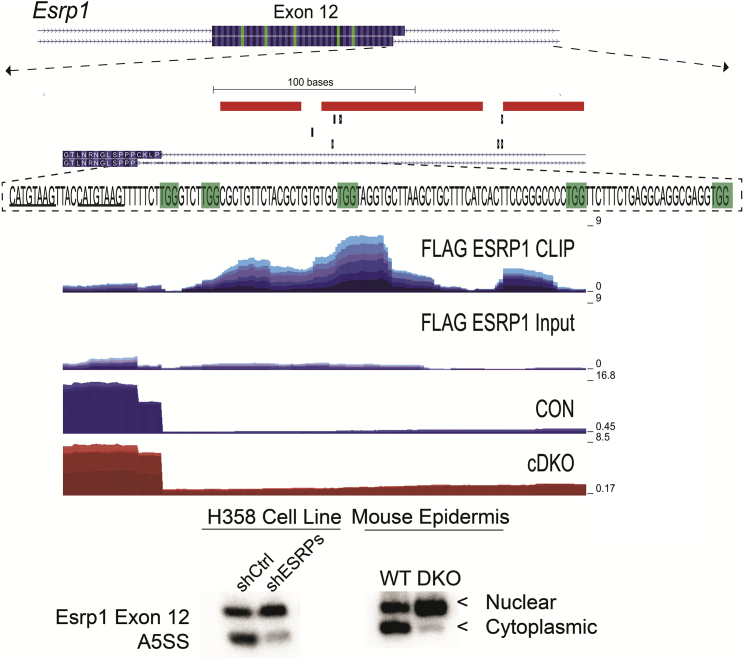
ESRP1 autoregulates the ratio of cytoplasmic and nuclear isoforms
ESRP1 peaks were also found within or near alternative 3′ and 5′ splice sites (A3SS or A5SS). We generated a splicing RNA MAP for the A3SS and A5SS for ESRP1-regulated transcripts (Figure S6). Although we do not observe many examples of A3SS or A5SS events, limiting our ability to conclude strong position-dependent effects, at least one of these has clear functional implications in that we observe a high confidence peak near competing alternative 5′ splice sites of exon 12 of ESRP1 (Figure 5 and Table S1). We previously showed that the use of alternative 5′ splice sites (with identical 5′ splice site consensus sequences) associated with exon 12 of ESRP1 that are 12 nucleotides apart generates distinct isoforms of ESRP1. The Use of the downstream 5′ splice site results in the inclusion of the amino acids CKLP that are a component of a nuclear localization signal (NLS) that directs the nuclear import of the splice isoform that includes these sequences (ESRP1 +CKLP or Nuc-ESRP1). However, the use of the upstream 5′ splice site generates a protein isoform (ESRP1 –CKLP or Cyto-ESRP1) lacking this essential component of the NLS resulting in cytoplasmic localization (Yang and Carstens, 2017). The observed peak of ESRP1 lies downstream of the A5SS, close to the second competing splice site, and congruent with our RNA Map we would expect that ESRP1 binding in this region would promote the use of the upstream 5′ splice site. To assess the functional relevance of the peak near the competing 5′ splice sites of exon 12, we assessed the splicing in both mouse epidermis ablated of Esrp1 and in human cell lines with knockdown of ESRP1. As predicted, we observe that depletion of ESRP1 shifts the cytoplasmic: nuclear ratio toward the production of the nuclear isoform (Figure 5). This finding is consistent with the RNA MAP and indicates an autoregulatory function of ESRP1 in the nucleus to balance the production of both isoforms, in which high levels of ESRP1 lead to inhibition of the downstream A5SS and a shift to the further upstream A5SS leading to the production of the cytoplasmic ESRP1 isoform. However, in limiting conditions of ESRP1, the second splice site is not impaired facilitating greater production of the nuclear ESRP1.
Figure 5.
Autoregulation of splicing by ESRP1 balances the expression of nuclear and cytoplasmic isoforms
(Top) Genome browser view of Esrp1 exon 12 with CLIP seq densities pooled for CLIP and Sm Input showing the peak of ESRP1 downstream of the 5′ splice site of Esrp1 with crosslink sites and Piranha peaks at top. Highlighted in green text are UGG motifs. The underlined text indicates 5′ splice sites, and the use of the more distal 5′ splice site results in the inclusion of the CKLP nuclear localization signal. Transcript levels (RNA-seq) show the increase in the use of the more distal 5′ splice site in cDKO conditions compared with control. (Bottom) Ethidium bromide-stained agarose gel of RT-PCR products of ESRP1-regulated splicing event at exon 12 of Esrp1 in mouse epidermis ablated of Esrps and control epidermis and H358 cells with or without knockdown of ESRP1.
ESRP1-binding sites are located in 3′ and 5′ UTRs of genes relevant to epidermal function
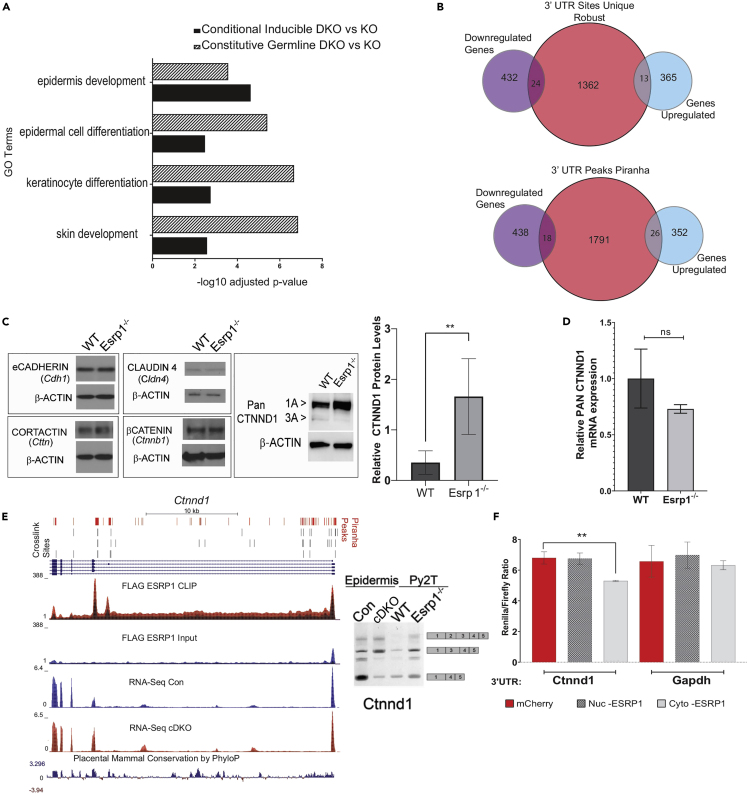
We observed substantial numbers of ESRP1-binding sites in 3′ and 5′ UTRs (Figure 1B), suggestive of a function of the cytoplasmic ESRP1 isoform to regulate gene expression in the cytoplasm or additional roles of nuclear ESRP1 beyond splicing regulation. We first sought to determine whether ESRP1 binding in UTRs might regulate mRNA stability by evaluating changes in total mRNA levels of these binding targets. However, this analysis was complicated by epidermal barrier defects that result from germline or conditional Esrp ablation (Bebee et al., 2015; Lee et al., 2018). For example, many of the gene expression changes overlap substantially with those that have been described in other mutant mice with epidermal barrier defects such as the knockout models of Loricrin, Klf4, and Grhl3 (Koch et al., 2000; Segre et al., 1999; Yu et al., 2006). These gene expression alterations included components of the epidermal differentiation complex (EDC), including Small proline-rich proteins (SPRR proteins) and Late cornified envelope proteins (LCE proteins) like SPRR1B, SPRR2D, SPRR2E, LCE3C, LCE3E, LCE3F (Bebee et al., 2015). Consequently, the large changes in total gene expression we observed in Esrp1/2 KO epidermis may likely be indirect effects of altered barrier function (Kypriotou et al., 2012; Patel et al., 2003). In order to assess a potential direct role for ESRP1 in modulating gene expression changes, we used our inducible conditional knockout model of Esrp1/2 (Figure 2A). We evaluated changes in gene expression using DeSeq2. A total of 1,494 genes were differentially expressed between neonatal epidermis with Esrps ablated and control epidermis, of which there were 378 genes upregulated more than 2-fold and 456 genes had at least 2-fold or more downregulation and an adjusted p-value <0.05. The gene expression changes observed in the cDKO epidermis were similar to that previously reported in the constitutive ablation of Esrps (Figure 6A) (Bebee et al., 2015), which is more consistent with secondary effects of the KO attributable to compensatory changes skin barrier relevant genes as seen in similar mouse models, such as those aforementioned. We explored whether genes with peaks in their 5′ or 3′ UTR were enriched for genes that were altered at the mRNA level. We observed that the vast majority of genes with ESRP1-binding sites in either the 3′ and/or 5′ UTR did not show changes in mRNA levels following inducible Esrp ablation (Figure 6B). Hence, many if not most changes in total mRNA levels may be attributed to indirect effects of ESRP1 ablation.
Figure 6.
ESRP1-binding in the 3′ UTR shows the minimal overlap with changes in transcript levels for after Esrp1 ablation
(A) Gene ontology analysis from EnrichR showing the top 4 processes affected in the constitutive Germline knockout of Esrp1 (hatched box) compared with the conditional inducible of knockout Esrp1 (solid black boxes).
(B) Venn diagram showing overlap of genes up- or downregulated during Esrp1 ablation in the mouse epidermis with genes that have peaks of ESRP1 in the 3′ UTR as identified by CLIP sites in two or more replicate pairs from PureCLIP or from peaks identified from pooled data from Piranha.
(C) Western blot analysis of Py2T cell lines, WT, and Esrp1−/−, assessing protein levels of eCadherin, CLAUDIN4, CTNND1, CTNNB1, and CORTACTIN as well as quantification of the CTNND1 western blot. Data are represented as the mean ± SD.
(D) qRT-PCR analysis of transcript levels for CTNND1 from the same cells. Data are represented as the mean ± SD.
(E) Genome browser views showing ESRP1-binding (CLIP Seq) and transcript levels (RNA-seq). CLIP-seq densities show CLIP and size-matched input, and transcript levels are from the RNA sequence in epidermis ablated of Esrps and control epidermis. Crosslink sites identified by PureCLIP are shown for three replicate pairs at top of the browser views as are peaks identified using the Piranha peak caller. Conservation scores showing Placental mammal conservation by PhyloP at the base. Ethidium bromide-stained agarose gel of RT-PCR products of ESRP1-regulated splicing event for Ctnnd1 in control (CON) compared with conditional double knockout epidermis (cDKO or EXP), and in Py2T epithelial cell lines.
(F) Luciferase reporter assay comparing effects of Nuc-ESRP1 (+CKLP) or Cyto-ESRP1 (-CKLP) on Ctnnd1 UTR and Gapdh UTR. Data are represented as the mean ± SD. ∗∗, p < 0.01.
Esrp1 binds within 3′ UTR of cadherin-binding genes and contributes to altering protein expression
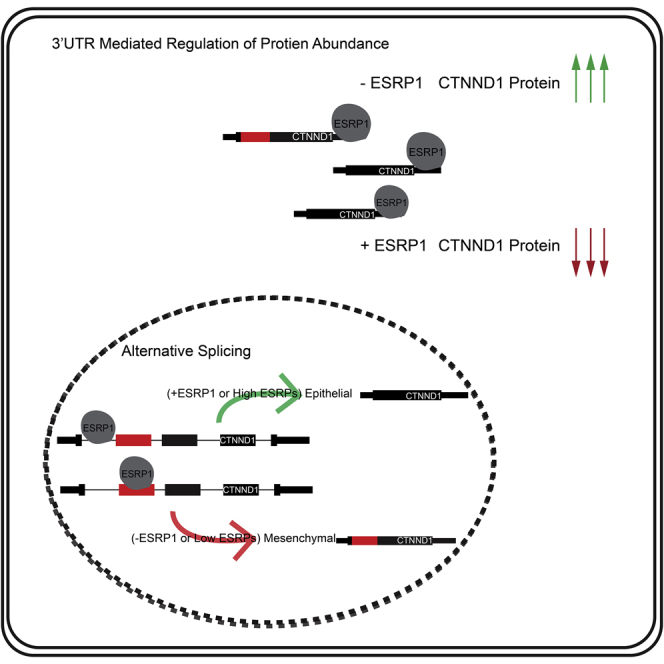
To expand our assessment, we performed gene ontology (GO) analysis of genes with strong binding peaks in the 3′ UTR using the web server EnrichR (Chen et al., 2013; Kuleshov et al., 2016). We observed that these genes are enriched for Cadherin binding, which is a gene ontology molecular classification for genes that interact selectively and non-covalently with membrane protein, cadherin (Table S5). Furthermore, the biological function of several of these genes was associated with ubiquitin-dependent/proteasomal protein catabolic processes. To assess the functional consequences of ESRP1 binding in the 3′ UTR, we used a cellular model, the Py2T mouse epithelial cell line. We previously used CRISPR/Cas9 technology to generate Py2T Esrp1−/− clonal cell lines (Lee et al., 2018). Consistent with the general lack of correlation between ESRP1 binding in 3′ UTRs and mRNA abundance our RNA-seq data revealed no significant difference in total RNA levels for eCadherin (Cdh1), Claudin 4 (Cldn4), β-Catenin (Ctnnb1), Cortactin (Cttn), and p120-Catenin, also known as δ-catenin (Ctnnd1) (Figure S7A), despite the fact that these transcripts exhibit crosslinking peaks for ESRP1 in their 3′ UTRs. Thus, our data provide no evidence for the role of ESRP1 in controlling mRNA stability. However, upon performing western blots in wild-type versus ESRP1 deficient Py2T cells, we do observe a striking increase in total protein abundance of CTNND1 upon ESRP1 depletion despite no increase in mRNA (Figures 6C, 6D, and S7B).
In addition to the novel regulation of protein abundance via binding to the 3′ UTR, we and others have shown that ESRP1/2 also regulates Ctnnd1 splicing (Cox et al., 2018; Faux et al., 2021; Warzecha et al., 2009a, 2009b). Two predominant isoforms of CTNND1 are 3A (predominantly epithelial) and 1A (predominantly mesenchymal) that differ in the difference in start codon utilization in exon 3 that is included in mesenchymal cells but skipped in epithelial cells (Keirsebilck et al., 1998; Markham et al., 2014; Mo and Reynolds, 1996). As a result, the mesenchymal isoform has a longer N-terminus. We previously showed that ESRP1 induces the skipping of tandem exons 2 and 3 to maintain the expression of the epithelial 3A isoform (Warzecha et al., 2009a). Consistent with direct regulation by ESRP1, we observe ESRP1 binding within the introns upstream of the exons skipped in the 3A isoform and validated that in the epidermis tissue ablated of Esrps and in the Esrp1−/− knockout cell line, that there is increased inclusion of the regulated tandem exons (Figure 6E). Further, in the western blot analysis of CTNND1, we observe there was a reduction in the amount of the predominantly epithelial isoform of CTNND1 3A (Figure 6C). However, the increase we observe in CTNND1 protein expression in the ESRP1-depleted cells cannot be explained simply by a bias in splicing as the loss of the 3A isoform is not sufficient to compensate for the increase in the 1A isoform and the total protein (accounting for both isoforms) increases by over 3-fold upon the loss of ESRP1.
The increase in CTNND1 protein in the absence of any increase in CTNND1 mRNA (Figures 6C and 6D), suggests a role of ESRP1 in translation control. To further explore a functional consequence of ESRP1 on protein expression of CTNND1, we ectopically expressed Cytoplasmic Esrp1 (ESRP1 -CKLP) and Nuclear Esrp1 (ESRP1 +CKLP) in HEK293T cells transiently transfected with a luciferase reporter bearing the 3′ UTR of Ctnnd1, or the 3′ UTR of Gapdh (which lacks binding sites for ESRP1) as a control. We observed that ESRP1-CLKP specifically caused a modest but significant reduction in the total luciferase activity from the reporter with the Ctnnd1 3′ UTR but not the Gapdh 3′ UTR control (Figure 6F), suggesting that ESRP1 functions inhibit protein translation when bound to the 3′ UTR. Importantly, a similar result was obtained with a distinct luciferase reporter background (Figure S7B). Furthermore, the fact that the cytoplasmic form of ESPR1, not the nuclear form, exhibits translation repression activity of a cDNA reporter underscores that the ability of ESRP1 to regulate protein expression of CTNND1 is likely a direct effect in the cytoplasm and independent of its role in splicing. This demonstrates a two-tier regulation of CTNND1 by ESRP1, in which ESRP1-regulated splicing and translation control affect the production and expression level of CTNND1 protein isoforms.
Discussion
Comprehensive analysis of 150 RBPs using eCLIP showed that analysis of RBP binding modalities can infer RBP function (Van Nostrand et al., 2020a, 2020b). It was shown that RBP-binding patterns and cellular localization correlated with function (Van Nostrand et al., 2020b). Many RBPs have multifunctional roles in the regulation of RNA processing, and this multifunctionality can result from differences in the cellular localization of the RBP (Hafner et al., 2010; Lee et al., 2016; Masuda et al., 2012). We have previously shown that Esrp1 regulates the splicing of a large set of epithelial genes and other studies suggest that ESRP1 may also regulate alternative polyadenylation (Bebee et al., 2015; Dittmar et al., 2012).
We generated a high-resolution RNA map of ESRP1binding sites in the epidermis by integrating peaks from crosslinking immunoprecipitation of ESRP1 from mouse epidermis and analysis of differential alternative splicing between control and Esrp1/2-deleted epidermis. We confirmed that many ESRP regulated events are directly regulated by ESRP1. Moreover, we provided global evidence supporting the RNA map model wherein ESRP1 binding downstream of the regulated exon promotes exon inclusion, whereas binding of ESRP1 within or upstream of the regulated exon suppresses exon inclusion. Over 70% of the regulated skipped exon events between control and cDKO epidermis with |ΔPSI| ≥ 20% (quantified by MAJIQ and rMATs using the most stringent criteria) had a peak of ESRP1 near the regulated exon consistent with the position-dependent splicing RNA MAP for ESRP1 (Figure 3A and Table S4). Whereas many of the alternatively spliced transcripts directly regulated by ESRP1 had peaks proximal to the regulated exon, we observe instances of more distal regulation by ESRP1. We observed several instances of genes with intronic ESRP1-binding peaks that are far (>1 kb) from the regulated exon, such as in Enah, Myo9a, and Uap1 (Figures 4A and S4). In these genes, the peaks of ESRP1 are detected with crosslink sites over 1kb downstream of the regulated exon. Nonetheless, congruent with the position-dependent regulation of alternative splicing of ESRP1 the regulated exons in these genes are upregulated by the presence of ESRP1. In the example of Enah exon 11a, the function of a distal splicing enhancer was previously proposed to involve the formation of “RNA bridges” or secondary structures. This distal enhancer contained binding sites for RBFOX2 that, like ESRP1, promotes exon splicing. The formation of stem loop secondary structures was proposed to bring the regulated ENAH exon and the RBFOX2 binding sites in the distal enhancer within closer spatial proximity (Lovci et al., 2013). The identification of ESRP1-binding sites within the same region of this enhancer strongly suggests that the same stem loops similarly facilitate exon splicing regulation by ESRP1. We previously showed that ESRP1 and RBFOX2 both promote splicing of the ENAH exon and that combined knockdown of ESRP1/2 and RBFOX2 in an epithelial cell line causes a greater increase in exon skipping that ESRP1/2 or RBFOX2 knockdown alone (Dittmar et al., 2012). These observations together with our demonstration of ESRP1 and RBFOX2 binding sites in this distal enhancer region suggest that they cooperatively regulate this splicing event (Dittmar et al., 2012). Furthermore, in addition to identifying binding sites for ESRP1 in the distal intron of Enah we show that the ability of the ESRP1 to bind RNA is critical for splicing enhancement of the upstream exon. This observation of ESRP1 regulation from distal sites within the exon was also observed for ATP6V1C2 and SLC37A2 among other genes, further suggesting that the effect is unlikely to be indirect. Other instances of stem loops within introns bridging genomic distances and facilitating regulated alternative splicing has been shown for additional shared ESRP and RBFOX target genes, as we and others showed for FGFR2 (Baraniak et al., 2006; Muh et al., 2002). Our findings add further support to the proposal that there are likely thousands of deep intronic splicing regulatory elements (Conboy, 2021). Moreover, we identified ESRP1-binding sites located near tandem alternatively spliced exons, such as those of Cd44 (Figure S3A), Myo1b (Figure 3B), and Ctnnd1 (Figure 6), supporting a mechanism to account for coordinated regulation of these exons.
We observe many large changes in total gene expression upon Esrp ablation, but there is little overlap between these genes and those bound by ESRP1 in the 3′ UTR. Deletion of Esrp1/2 causes an epidermal barrier defect, and many large changes in total gene expression, including those of the epidermal differentiation complex, are highly similar to those observed in other mouse models associated with epidermal barrier defects (Bebee et al., 2015; Koch et al., 2000; Segre et al., 1999). As such we posit that these gene expression changes may be secondary to Esrp ablation, as the majority of these gene transcripts were not directly bound by ESRP1 in the 3′ UTR. Alternatively, we observed that some of these genes (late cornified genes like LCE3C, LCE3E, LCE3F) showed binding within the CDS, providing another avenue for regulation by ESRP1. Using CLIP, we identity robust binding of ESRP1 within the UTRs of genes involved in epithelial cell function. We observe peaks for ESRP1 in the 3′ UTR of Ctnnb1 and, Cttn, in the CDS and 3′ UTR of Cdh1, and in the 5′ UTR, 3′ UTR, and CDS of Ctnnd1. We previously showed that Esrp1 ablation is associated with Cleft Lip with or without Cleft Palate (CL/P), skin loss, and hair follicle abnormalities (Bebee et al., 2015; Lee et al., 2018). Further, mutations in CTNND1 and CDH1 have been implicated in the CL/P (Cox et al., 2018). In the case of CTNND1, we observe that ESRP1 ablation leads to loss of the epithelial isoform of CTNND1, but also an increase in the protein expression of the predominantly mesenchymal isoform of CTNND1 (p120-1) (Figure 6D). It has been shown that whereas p120-3 is almost exclusively epithelial, the p120-1 isoform, although predominantly mesenchymal, is also expressed in the epithelial cells with lower differentiation or rapid turnover (Venhuizen et al., 2019). The increased protein expression of p120-1 seen with Esrp1 deletion may be attributable to a lack of repression by Cytoplasmic ESRP1. One possibility for the regulation of CTNND1 by ESRP1, is not only to generate the CTNND1 splice isoform that stabilizes membrane association of CDH1, but ESRP1 may also maintain a balance of these isoforms by repressing the expression of mesenchymal isoforms that associate with other cadherins or that have different functional properties. In epithelial cells, CTNND1 is co-localized with CDH1 at adherences junctions and has been proposed to promote cell adhesion and maintain epithelial barriers (Smalley-Freed et al., 2010). This association appears to occur in a tissue- and isoform-specific manner (Venhuizen et al., 2019). In contrast, the mesenchymal CTNND1 isoform has been shown to have opposing functions to promote cell motility and tissue invasion (Yanagisawa et al., 2008). Some of these differences in the function of epithelial and mesenchymal CTNND1 isoforms are likely owing to differences in protein–proteins interactions as shown for CTNND1 interacting proteins (Markham et al., 2014; Yu et al., 2016) CDH1 is a transmembrane protein and its membrane association is stabilized by CTNND1; however, we have previously shown that the loss of ESRPs (which results in a loss of the epithelial CTNND1 (p120-3) isoform) results in diffuse localization of CDH1 in the cytoplasm (Davis et al., 2003; Warzecha et al., 2010). We observe the change in protein abundance of CTNND1, without a robust change in the other tested genes. However, ESRP1 ablation alone does not cause a loss in expression of the prototypical epithelial cell marker E-cadherin, although cells acquire some mesenchymal cell properties (Warzecha et al., 2010) Our work supports the proposition that the loss of Esrps may prime epithelial cells for further cellular transitions and that ESRPs work in concert with other RBPs and transcription factors to maintain the epithelial state (Bebee et al., 2014; Warzecha and Carstens, 2012; Warzecha et al., 2010; Yang et al., 2016).
Overall, we have demonstrated that ESRP1 directly binds and regulates an epithelial splicing network, further that ESRP1 has apparent roles outside of splicing as indicated by its binding within the UTR. Whereas we currently have some evidence in support of the role of ESRP1 in modulating protein abundance, further studies are needed to characterize global changes in protein levels that result from loss of ESRP1 and gain mechanistic insight into whether ESRP1 directly or indirectly affects translation or other protein catabolic processes. Such studies would include proteomic studies as well as analysis of changes in ribosome association. Another intriguing possibility is that ESRP1 controls mRNA localization of mRNAs with 5′ or 3′ binding sites and localized translation. Previous studies have suggested that mRNA localization in epithelial cells, and localized translation are important for epithelial cell homeostasis (Moor et al., 2017). Given the well-established roles of RBPS binding in 3′ UTRs to regulate RNA stability, and burgeoning evidence of RBPs wearing multiple hats to regulate mRNA metabolism it would stand to reason that ESRP1, which is more specifically expressed in epithelial cells than any other RBP, would be uniquely poised to regulate mRNA stability, localization or translation to better support epithelial cell functions. Future studies will explore this hypothesis.
Limitation of the study
Our novel finding that cytoplasmic ESRP1 alters protein expression of CTNND1 demonstrates that it has a regulatory role beyond the regulation of splicing in the nucleus. However, to date, we have not yet been able to identify other ESRP1 targets that are regulated at the translational level. This may in part owing to multifaceted regulation by ESRP1, wherein ESRP1 binding in the 3′ UTR functions has other regulatory functions, such as RNA stability or localization, which were not identified in this study. A goal for future investigations will be to comprehensively identify the global translational targets of ESRP1 as well as other potential roles in the cytoplasm. Studies using methods such as polysome sequencing and translating ribosome affinity purification (TRAP) should be able to extend the list of mRNAs that are translationally regulated by ESRP1. This study also does not uncover the mechanism by which ESRP1 binding within the UTR regulates translation. Further studies are currently underway to address these shortcomings and provide a high-level understanding of ESRP1 functions that are essential for the maintenance of epithelial cell functions.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| ESRP1 Antibody, mouse monoclonal | Generated by the Carstens Lab (Warzecha et al., 2010) (Also available commercially from Rockland) | 27H12 Rockland Cat# #210-301-B89 |
| Anti-β-Actin antibody, Mouse monoclonal | Sigma Aldrich | Cat# A2228, RRID:AB_476697 |
| p120-Catenin Antibody, Rabbit polyclonal | Gift from Albert Reynolds (Vanderbilt University, Tennessee) | F1SH |
| Anti-Cldn4 | Thermo Fisher | Cat# 36–4800, RRID: AB_2533262 |
| Anti-Cdh1 | Cell Signalling | Cat # 610182 |
| Anti-Ctnnb1 | Abcam | Cat # ab22656 |
| Anti-Cttn | Abcam | Cat # ab81208 |
| ECL Anti-mouse IgG | GE Healthcare | Cat# NA931, RRID:AB_772210 |
| ECL Anti-rabbit IgG | GE Healthcare | Cat# NA934, RRID:AB_772206 |
| Chemicals, peptides, and recombinant proteins | ||
| RPMI 1640 | Thermo Fisher | Cat # 72400047 |
| DMEM | Gibco | Cat # 11885 |
| TransIT-293 | Mirus | Cat#MIR2700 |
| Lipofectamine 3000 | Thermo Fisher | Cat#11668019 |
| Dynabeads Protein G | Thermo Fisher | Cat # 10004D |
| Dynabeads MyOne Silane | Thermo Fisher | Cat # 37002D |
| AMPure XP | Beckman Coulter | Cat # A63880 |
| Pierce ECL Western | Thermo Fisher | Cat #32106 |
| Critical commercial assays | ||
| Dual Luciferase Reporter Assay System | Promega | Cat#E1910 |
| RNeasy Mini Kit | Qiagen | Cat#74104 |
| Deposited data | ||
| CLIP Seq and RNA Seq Data Sets | This study | GEO Accession: GSE212307 |
| Experimental models: Cell lines | ||
| Human: ESRP1/2 KD H358 cells | (Yang et al., 2016) | N/A |
| Human: HEK293T cells | ATCC | CRL-3216 |
| Mouse: Py2T cells | Gerhard Christofori (Waldmeier et al., 2012) | N/A |
| Mouse: ESRP1 KO Py2T cells | (Lee et al., 2018) | Py2T Clone H6 |
| Human: H358 cells | ATCC | CRL-5807 |
| Experimental models: Organisms/strains | ||
| Esrp1flox/flox; Esrp2−/− | (Bebee et al., 2015) | N/A |
| K5rTA | Sarah Millar (Ichan School of Medicine) | Mucenski et al. (2003) |
| Esrp1flox/flox; Esrp2−/−; K5rTA tetO-Cre | (Lee et al., 2018) | N/A |
| Esrp1FLAG/FLAG | (Lee et al., 2020); Also available from the Mutant Mouse Resource and Research Centers (MMRRC) | RRID:MMRRC_068071-UCD |
| Esrp1−/−; Esrp2−/− | (Bebee et al., 2015) | N/A |
| Tg(tetO-cre)1Jaw/J (tetO-Cre) | The Jackson Laboratory | JAX stock #006224 |
| Oligonucleotides | ||
| Primers for qPCR | See Table S6 | See Table S6 |
| Primers for RT-PCR | See Table S6 | See Table S6 |
| Primers for UTR Cloning | See Table S6 | See Table S6 |
| Recombinant DNA | ||
| Plasmid: psiCheck2 | Promega | C8021 |
| Plasmid: pIS0 | Addgene | 121718 |
| Plasmid: pIBX-ESRP1 2A + CKLP | (Yang and Carstens, 2017) | N/A |
| Plasmid: pIBX-ESRP1 2A- CKLP | (Yang and Carstens, 2017) | |
| Plasmid: pIBX-ESRP1 2A + CKLP RRM Mutant | This publication | N/A |
| Plasmid: pIBX-mCherry | (Yang and Carstens, 2017) | N/A |
| Plasmid: pdp19 | Ambion | N/A |
| Software and algorithms | ||
| MAJIQ algorithm | (Vaquero-Garcia et al., 2016) | https://majiq.biociphers.org |
| rMATs Tool | (Shen et al., 2012, 2014) | http://rnaseq-mats.sourceforge.net/ |
| EnrichR | (Chen et al., 2013; Kuleshov et al., 2016) | https://maayanlab.cloud/Enrichr/ |
| rMAPS2 | (Hwang et al., 2020) | http://rmaps.cecsresearch.org/ |
| Piranha Peak Caller | (Uren et al., 2012) | http://smithlabresearch.org/software/piranha/ |
| PureCLIP | (Krakau et al., 2017) | https://github.com/skrakau/PureCLIP |
| Prism, version 8 | Graphpad | N/A |
| ImageLab, version 6.1 | Bio-Rad | N/A |
| ImageJ, version ImageJ 1.153f51. | NIH | N/A |
| ImageQuantTL, version 7 | GE Healthcare | N/A |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Natoya Peart (npeart@upenn.edu).
Materials availability
This study did not generate new unique reagents.
Experimental model and subject details
Mouse strains
Generation of Esrp1 KO (Esrp1−/−) and conditional floxed Esrp1 (Esrp1flox/flox) were described previously (Bebee et al., 2015) as was the Esrp1 FLAG/FLAG (Lee et al., 2020), and Esrp1flox/flox; Esrp2−/−; K5rTA tetO-Cre mice (Lee et al., 2018). Transgenic Keratin-5 rtTA (K5-rtTA) (Mucenski et al., 2003) mice were obtained from Sarah Millar (Icahn School of Medicine) and tetO-Cre strains (Diamond et al., 2000) were obtained from JAX. Esrp1 deletion in epidermis was induced by Doxycycline (600mg/kg) (Bio-Serv) feeding of nursing mothers at E17.5 until PND5. Genomic DNA for genotyping was derived from tail biopsies and genotyping was performed using standard procedures. Both male and female mice and were used in this study. Experiments with Esrp1 FLAG/FLAG mice were performed with pups age P1-P0. Experiments with Esrp1flox/flox; Esrp2−/−; K5rTA tetO-Cre mice were performed with pups at age P5. All animal procedures and experiments were approved by the Institutional Animal Care and Use Committee (IACUC) at the University of Pennsylvania.
Cell Culture
Py2T (Waldmeier et al., 2012) and HEK293T cells were grown in DMEM with 10% FBS, and Human non-small cell lung cancer cell line H358 (obtained from the American Type Culture Collection) were maintained in RPMI1640 with 10% FBS at 37°C and 5% CO2. Esrp1 KO Py2T cells were previously described (Lee et al., 2018). H358 knockdown cells are from our previous work (Yang et al., 2016).
Method details
Transfections
For luciferase reporter assays, HEK293T cells were transfected (using Mirus Transit 293 reagent) in a 96 well plate with indicated plasmids (total of 20ng DNA) in biological triplicates. Cells were harvested 48hrs after transfections with 100uL Passive Lysis Buffer. Dual Luciferase assay were performed using the Dual-Luciferase Reporter Assay (Promega) according to manufactures instructions. Reporter renilla luciferase activity was normalized to firefly luciferase activity. For splicing analysis, HEK293T cells were transfected (using Mirus Transit 293 reagent) in a 24-well plate in triplicate with plasmids containing either WT ESRP1 or ESRP1 RRM Mutant cells were harvested 48hrs after transfections with TriZol.
Real-time RT-PCR and RT-PCR
Real-time RT-PCR and RT-PCR were performed as described (Bebee et al., 2015; Lee et al., 2018, 2020). RT-PCR was quantified using ImageLab, Version 6.1 Standard Edition (Biorad). Splicing ratios are represented as PSI for cassette exons and were normalized to RT-PCR product size. Real-time RT-PCR and RT-PCR primer sequences are available.
Western Blot
Total cell lysates were harvested in RIPA buffer and immunoblotting was performed as described previously (Bebee et al., 2015).Briefly, Total proteins were separated by 4–12% SDS-PAGE and then transferred to nitrocellulose membranes. Membranes were blocked in 5% Non-fat dry milk powder in phosphate buffered saline-tween 20 (PBST), then incubated overnight at 4°C with the primary antibodies. Subsequently, membranes were washed in PBST and incubated for 1 h with the appropriate secondary antibodies. After washing three times for 10 min with PBST, proteins were visualized by chemiluminescent detection (Thermo Scientific). Western blots were quantified using ImageJ 1.153f51 or ImageLab, Version 6.1 Standard Edition.
RNA extraction
RNA was extracted from the epidermis as described previously (see STAR Methods in (Bebee et al., 2015)) with the following modifications. The pups aged P5 were cryoeuthanized and decapitated. Trunk skin was removed and floated dermis side down on 0.25% trypsin/HBSS at 4°C for 16–18 h. Epidermis and dermis were manually separated using forceps, rinsed HBSS and snap frozen on liquid nitrogen. Epidermis was lysed for RNA isolation in TriZol (Invitrogen, Carlsbad, CA) and RNA was isolated with RNeasy mini kit (Qiagen).
RNA sequencing and data analysis
Total RNA from epidermis was used for RNA-Seq and samples were processed at Genewiz.
Sequencing averaged 147 × 106 reads (range: 104–179 × 106 reads) per sample. To assess the RNA sequencing data using MAJIQ, adapters were trimmed from RNA-Seq samples using BBDuk, aligned to the mouse GRCm38 genome assembly using STAR v.2.5.1B (Dobin et al., 2013)and sorted and indexed using samtools v.1.9 (Li et al., 2009). Samples averaged 88.93% uniquely mapped reads (range: 87.28–90.06%) for the epidermis. For gene expression quantification, salmon v.0.14.0 (Patro et al., 2017) was used in mapping-based mode with selective alignment on trimmed fastq files using GENCODE vM23 annotation to create the index. Differential gene expression analysis was performed with DESeq2 v.1.22.2 (Love et al., 2014). We identified differentially expressed genes by retaining genes that had an adjusted p-value lower than 0.05 and 2-fold difference between genotypes. Differential splicing analysis was performed with MAJIQ v.2.1 using GENCODE vM23 reference transcriptome annotation (Vaquero-Garcia et al., 2016). We identified differentially spliced junctions by retaining junctions that had a delta PSI of at least 10% with a probability of at least 95%. Gene enrichment analysis was performed with EnrichR v.1.0 using a 2018 release of the GO Consortium annotations.
To assess the RNA sequencing data with rMATs, RNA-seq reads were also mapped to the GRCm38.p6 mouse reference genome and the GRCm38.93 transcriptome (Ensembl GTF release 91) using STAR aligner (v2.6.0c) (Shen et al., 2012, 2014). Differential gene expression between the two cell types were calculated using cuffdiff (v2.2.1) (Trapnell et al., 2010), based on the average FPKM at FDR <5%, > 2-fold when maximum average FPKM from at least one of cell types is larger than 1. Then, to identify differential AS events between the control and conditional knock-out cells, we used rMATS (v3.2.5) that examines all five basic types of AS events (SE, MXE, A5SS, A3SS, and RI). rMATS uses the reads mapped to the splice junctions and the reads mapped to the exon body to estimate the exon usage (PSI: percent-spliced-in (ψ)). The conditional knock-out was compared to the control to identify differentially spliced events with an associated change in PSI (ΔPSI or Δψ) of these events. To compute p-values and FDRs of splicing events with |Δψ| > 0.01% cutoff, we ran rMATS using -c 0.0001 parameter.
eCLIP and data analysis
Library production
Esrp1FLAG/FLAG pups aged P0-P1 were cryoeuthanized and decapitated. Epidermis was isolated as described earlier and after rinsing in HBSS was placed flat on petri dish with HBSS and crosslinked thrice at 400mJ/cm2. The epidermis was minced with a scalpel, and then flash frozen in liquid Nitrogen and stored at −80C until use. Each sample for eCLIP was comprised of the epidermis from two pups (Esrp1FLAG/FLAG or Esrp1WT/WT) which were mixed and dounced with a loose pestle in eCLIP lysis buffer. All other processes after lysis were performed as previously described (see Supplementary Protocols in (Van Nostrand et al., 2016)).
eCLIP sequencing analysis and ESRP1 binding site peak calling using piranha
eCLIP sequencing for ESRP1 protein by enhanced crosslinking and immunoprecipitation was performed for 6 replicates each for both CLIP and Input. eCLIP sequencing reads from 6 replicates each for both sample group were pooled together, respectively, then trimmed to remove adaptor sequences using Cutadapt (v1.16), the 6 individual Input controls were not pooled. The trimmed reads were mapped to the grcm38_snp_tran mouse reference genome using Hisat2 aligner (v2.1.0). After pooling and alignment, CLIP samples had 67.9% uniquely mapped tags and Input controls had an average of 35.5% unique mapped tags. For Esrp1 peak calling, we used Piranha (v.1.2.1), a peak-caller for CLIP-seq data. We input the mapped reads from the pooled CLIP sample in BED format to Piranha for identifying regions of statistically significant read enrichment having the mapped reads from Input sample as covariate for running Piranha. Using Piranha we detected a total 5,730,968, after enrichment for statistically significant peaks (FDR <0.05) 12,982 peaks remained.
eCLIP and motif enrichment analysis of ESRP1 binding using PureCLIP and HOMER
Processed read counts from 6 replicate for both CLIP and Input were used to call peaks and crosslink sites using PureCLIP. As PureCLIP does not support analysis on more than 2 replicates, the 6 biological replicates were pooled into 3 replicate pairs and PureCLIP analysis was performed as previously described using default parameters (see STAR Methods in (Krakau et al., 2017)). CLIP samples had an overall average alignment to the mouse genome of 71% of which there were 43% uniquely mapped reads. Input controls had an average alignment of 38% of which there were 78% uniquely mapped reads. The sites identified in the three replicate pairs were then combined and the redundancies removed. In sum, we identified a total of 81115 unique CLIP sites mapping to the coding transcripts. The HOMER software package was used to identify the motifs four to seven nucleotides in length, enriched on the transcript strand within 10 nucleotides of the cross-link sites and calculate the cross-link densities relative to the identified consensus motif (Heinz et al., 2010).
Quantification and statistical analysis
Statistical analyses of the data were performed in GraphPad Prism. Error bars where present represent ±SD (standard deviation) of experiments performed on biological replicates; n ≥ 3 biological replicates. Statistical significance for the Pearson r correlation was calculated using two tailed t-test. Statistical significance where indicated was calculated by Student’s T test and denoted as follows: ∗, = p < 0.05, ∗∗, p < 0.01; ∗∗∗, p < 0.001; ∗∗∗∗, p < 0.0001.
Acknowledgments
We thank Fange Liu and Kathryn Hamilton for constructive feedback during manuscript revision. We thank Albert Reynolds for providing us with the p120-Catenin antibodies. We are grateful to the Transgenic and Chimeric Mouse Core of the University of Pennsylvania for generation of Esrp1FLAG/FLAG mice (supported by NIH center grants P30DK050306, P30DK019525, and P30CA016520). We are grateful to the Center for Molecular Studies in Digestive and Liver Diseases (NIH-P30-DK050306) and its core facilities (Molecular Pathology and Imaging Core, Host-Microbial Analytic and Repository Core, Genetically-Modified Mouse Core, and the Cell Culture and iPS Core) for use of luminometer for luciferase experiments. Funding for this work was supported by NIH/NIAMS, 1R56AR066741 (R.P.C), NIH/NIAMS, R01 AR066741 (R.P.C), NIGMS/NIH, P20GM103436 (J.W.P), and NIEHS/NIH, P30ES030283 (J.W.P), NIH/NIGMS R35118048 (K.W.L.), and a subproject of a Penn Skin Biology and Diseases Core grant, NIAMS/NIH, 5-P30-AR-057217.
Author contributions
Conceptualization, N.P and R.P.C.; software, J.Y.H., M.Q.V., P.S., Y.B., and J.W.P.; formal analysis, N.P., R.P.C., P.S., J.Y.H., and J.W.P.; investigation, N.P., Y.Y., and M.J.S.; resources, Y.B., J.W.P. K.W.L., and R.P.C.; writing – original draft, N.P. and R.P.C.; supervision, K.W.L., R.P.C.; funding acquisition, R.P.C., K.W.L.; writing – review and editing, N.P., J.Y.H., M.Q.V., M.J.S., Y.Y., P.S., K.W.L. Y.B., J.W.P., and R.P.C.
Declaration of interests
R.P.C is currently employed at Merck. The authors otherwise declare no competing interests.
Published: October 21, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2022.105205.
Contributor Information
Natoya J. Peart, Email: npeart@upenn.edu.
Russ P. Carstens, Email: russcars@upenn.edu.
Supplemental information
Data and code availability
-
•
RNA-seq and CLIP-seq data have been deposited at GEO and are publicly available as of the date of publication. Accession numbers are listed in the key resources table.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- Baraniak A.P., Chen J.R., Garcia-Blanco M.A. Fox-2 mediates epithelial cell-specific fibroblast growth factor receptor 2 exon choice. Mol. Cell Biol. 2006;26:1209–1222. doi: 10.1128/MCB.26.4.1209-1222.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bebee T.W., Cieply B.W., Carstens R.P. Genome-wide activities of RNA binding proteins that regulate cellular changes in the epithelial to mesenchymal transition (EMT) Adv. Exp. Med. Biol. 2014;825:267–302. doi: 10.1007/978-1-4939-1221-6_8. [DOI] [PubMed] [Google Scholar]
- Bebee T.W., Park J.W., Sheridan K.I., Warzecha C.C., Cieply B.W., Rohacek A.M., Xing Y., Carstens R.P. The splicing regulators Esrp1 and Esrp2 direct an epithelial splicing program essential for mammalian development. Elife. 2015;4 doi: 10.7554/eLife.08954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen E.Y., Tan C.M., Kou Y., Duan Q., Wang Z., Meirelles G.V., Clark N.R., Ma'ayan A. Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinf. 2013;14:128. doi: 10.1186/1471-2105-14-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conboy J.G. Developmental regulation of RNA processing by Rbfox proteins. WIREs. RNA. 2017;8:e1398. doi: 10.1002/wrna.1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conboy J.G. Wiley interdisciplinary reviews RNA; 2021. Unannotated Splicing Regulatory Elements in Deep Intron Space; p. e1656. [DOI] [PubMed] [Google Scholar]
- Cox L.L., Cox T.C., Moreno Uribe L.M., Zhu Y., Richter C.T., Nidey N., Standley J.M., Deng M., Blue E., Chong J.X., et al. Mutations in the epithelial cadherin-p120-catenin complex cause mendelian non-syndromic cleft lip with or without cleft palate. Am. J. Hum. Genet. 2018;102:1143–1157. doi: 10.1016/j.ajhg.2018.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis M.A., Ireton R.C., Reynolds A.B. A core function for p120-catenin in cadherin turnover. J. Cell Biol. 2003;163:525–534. doi: 10.1083/jcb.200307111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diamond I., Owolabi T., Marco M., Lam C., Glick A. Conditional gene expression in the epidermis of transgenic mice using the tetracycline-regulated transactivators tTA and rTA linked to the keratin 5 promoter. J. Invest. Dermatol. 2000;115:788–794. doi: 10.1046/j.1523-1747.2000.00144.x. [DOI] [PubMed] [Google Scholar]
- Dittmar K.A., Jiang P., Park J.W., Amirikian K., Wan J., Shen S., Xing Y., Carstens R.P. Genome-wide determination of a broad ESRP-regulated posttranscriptional network by high-throughput sequencing. Mol. Cell Biol. 2012;32:1468–1482. doi: 10.1128/MCB.06536-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T.R. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominguez C., Allain F.H.T. Resonance assignments of the two N-terminal RNA recognition motifs (RRM) of the human heterogeneous nuclear ribonucleoprotein F (HnRNP F) J. Biomol. NMR. 2005;33:282. doi: 10.1007/s10858-005-4287-2. [DOI] [PubMed] [Google Scholar]
- Fagg W.S., Liu N., Fair J.H., Shiue L., Katzman S., Donohue J.P., Ares M. Autogenous cross-regulation of Quaking mRNA processing and translation balances Quaking functions in splicing and translation. Genes Dev. 2017;31:1894–1909. doi: 10.1101/gad.302059.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faux M.C., King L.E., Kane S.R., Love C., Sieber O.M., Burgess A.W. APC regulation of ESRP1 and p120-catenin isoforms in colorectal cancer cells. Mol. Biol. Cell. 2021;32:120–130. doi: 10.1091/mbc.E20-05-0321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerstberger S., Hafner M., Tuschl T. A census of human RNA-binding proteins. Nat. Rev. Genet. 2014;15:829–845. doi: 10.1038/nrg3813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hafner M., Landthaler M., Burger L., Khorshid M., Hausser J., Berninger P., Rothballer A., Ascano M., Jungkamp A.-C., Munschauer M., et al. Transcriptome-wide identification of RNA-binding protein and microRNA target sites by PAR-CLIP. Cell. 2010;141:129–141. doi: 10.1016/j.cell.2010.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hakim N.H.A., Majlis B.Y., Suzuki H., Tsukahara T. Neuron-specific splicing. Biosci. Trends. 2017;11:16–22. doi: 10.5582/bst.2016.01169. [DOI] [PubMed] [Google Scholar]
- Hall M.P., Nagel R.J., Fagg W.S., Shiue L., Cline M.S., Perriman R.J., Donohue J.P., Ares M., Jr. Quaking and PTB control overlapping splicing regulatory networks during muscle cell differentiation. RNA. 2013;19:627–638. doi: 10.1261/rna.038422.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinz S., Benner C., Spann N., Bertolino E., Lin Y.C., Laslo P., Cheng J.X., Murre C., Singh H., Glass C.K. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell. 2010;38:576–589. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hovhannisyan R.H., Carstens R.P. A novel intronic cis element, ISE/ISS-3, regulates rat fibroblast growth factor receptor 2 splicing through activation of an upstream exon and repression of a downstream exon containing a noncanonical branch point sequence. Mol. Cell Biol. 2005;25:250–263. doi: 10.1128/MCB.25.1.250-263.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hovhannisyan R.H., Warzecha C.C., Carstens R.P. Characterization of sequences and mechanisms through which ISE/ISS-3 regulates FGFR2 splicing. Nucleic Acids Res. 2006;34:373–385. doi: 10.1093/nar/gkj407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang J.Y., Jung S., Kook T.L., Rouchka E.C., Bok J., Park J.W. rMAPS2: an update of the RNA map analysis and plotting server for alternative splicing regulation. Nucleic Acids Res. 2020;48:W300–W306. doi: 10.1093/nar/gkaa237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keirsebilck A., Bonné S., Staes K., van Hengel J., Nollet F., Reynolds A., van Roy F. Molecular cloning of the human p120ctn catenin gene (CTNND1): expression of multiple alternatively spliced isoforms. Genomics. 1998;50:129–146. doi: 10.1006/geno.1998.5325. [DOI] [PubMed] [Google Scholar]
- Koch P.J., de Viragh P.A., Scharer E., Bundman D., Longley M.A., Bickenbach J., Kawachi Y., Suga Y., Zhou Z., Huber M., et al. Lessons from loricrin-deficient mice: compensatory mechanisms maintaining skin barrier function in the absence of a major cornified envelope protein. J. Cell Biol. 2000;151:389–400. doi: 10.1083/jcb.151.2.389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krakau S., Richard H., Marsico A. PureCLIP: capturing target-specific protein–RNA interaction footprints from single-nucleotide CLIP-seq data. Genome Biol. 2017;18:240. doi: 10.1186/s13059-017-1364-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuleshov M.V., Jones M.R., Rouillard A.D., Fernandez N.F., Duan Q., Wang Z., Koplev S., Jenkins S.L., Jagodnik K.M., Lachmann A., et al. Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 2016;44:W90–W97. doi: 10.1093/nar/gkw377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kypriotou M., Huber M., Hohl D. The human epidermal differentiation complex: cornified envelope precursors, S100 proteins and the ‘fused genes’ family. Exp. Dermatol. 2012;21:643–649. doi: 10.1111/j.1600-0625.2012.01472.x. [DOI] [PubMed] [Google Scholar]
- Lee J.A., Damianov A., Lin C.H., Fontes M., Parikshak N.N., Anderson E.S., Geschwind D.H., Black D.L., Martin K.C. Cytoplasmic Rbfox1 regulates the expression of synaptic and autism-related genes. Neuron. 2016;89:113–128. doi: 10.1016/j.neuron.2015.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S., Cieply B., Yang Y., Peart N., Glaser C., Chan P., Carstens R.P. Esrp1-Regulated splicing of Arhgef11 isoforms is required for epithelial tight junction integrity. Cell Rep. 2018;25:2417–2430.e5. doi: 10.1016/j.celrep.2018.10.097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S., Sears M.J., Zhang Z., Li H., Salhab I., Krebs P., Xing Y., Nah H.-D., Williams T., Carstens R.P. Cleft lip and cleft palate in Esrp1 knockout mice is associated with alterations in epithelial-mesenchymal crosstalk. Development. 2020;147 doi: 10.1242/dev.187369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R., 1000 Genome Project Data Processing Subgroup The sequence alignment/map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovci M.T., Ghanem D., Marr H., Arnold J., Gee S., Parra M., Liang T.Y., Stark T.J., Gehman L.T., Hoon S., et al. Rbfox proteins regulate alternative mRNA splicing through evolutionarily conserved RNA bridges. Nat. Struct. Mol. Biol. 2013;20:1434–1442. doi: 10.1038/nsmb.2699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markham N.O., Doll C.A., Dohn M.R., Miller R.K., Yu H., Coffey R.J., McCrea P.D., Gamse J.T., Reynolds A.B. DIPA-family coiled-coils bind conserved isoform-specific head domain of p120-catenin family: potential roles in hydrocephalus and heterotopia. Mol. Biol. Cell. 2014;25:2592–2603. doi: 10.1091/mbc.E13-08-0492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masuda A., Andersen H.S., Doktor T.K., Okamoto T., Ito M., Andresen B.S., Ohno K. CUGBP1 and MBNL1 preferentially bind to 3′ UTRs and facilitate mRNA decay. Sci. Rep. 2012;2:209. doi: 10.1038/srep00209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mauger D.M., Lin C., Garcia-Blanco M.A. hnRNP H and hnRNP F complex with Fox2 to silence fibroblast growth factor receptor 2 exon IIIc. Mol. Cell Biol. 2008;28:5403–5419. doi: 10.1128/MCB.00739-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mo Y.Y., Reynolds A.B. Identification of murine p120 isoforms and heterogeneous expression of p120cas isoforms in human tumor cell lines. Cancer Res. 1996;56:2633–2640. [PubMed] [Google Scholar]
- Moor A.E., Golan M., Massasa E.E., Lemze D., Weizman T., Shenhav R., Baydatch S., Mizrahi O., Winkler R., Golani O., et al. Global mRNA polarization regulates translation efficiency in the intestinal epithelium. Science (New York, NY) 2017;357:1299–1303. doi: 10.1126/science.aan2399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mucenski M.L., Wert S.E., Nation J.M., Loudy D.E., Huelsken J., Birchmeier W., Morrisey E.E., Whitsett J.A. β-Catenin is required for specification of proximal/distal cell fate during lung morphogenesis. J. Biol. Chem. 2003;278:40231–40238. doi: 10.1074/jbc.M305892200. [DOI] [PubMed] [Google Scholar]
- Muh S.J., Hovhannisyan R.H., Carstens R.P. A non-sequence-specific double-stranded RNA structural element regulates splicing of two mutually exclusive exons of fibroblast growth factor receptor 2 (FGFR2) J. Biol. Chem. 2002;277:50143–50154. doi: 10.1074/jbc.M207409200. [DOI] [PubMed] [Google Scholar]
- Nikonova E., Kao S.-Y., Ravichandran K., Wittner A., Spletter M.L. Conserved functions of RNA-binding proteins in muscle. Int. J. Biochem. Cell Biol. 2019;110:29–49. doi: 10.1016/j.biocel.2019.02.008. [DOI] [PubMed] [Google Scholar]
- Patel S., Kartasova T., Segre J.A. Mouse Sprr locus: a tandem array of coordinately regulated genes. Mamm. Genome. 2003;14:140–148. doi: 10.1007/s00335-002-2205-4. [DOI] [PubMed] [Google Scholar]
- Patro R., Duggal G., Love M.I., Irizarry R.A., Kingsford C. Salmon provides fast and bias-aware quantification of transcript expression. Nat. Methods. 2017;14:417–419. doi: 10.1038/nmeth.4197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romanelli M.G., Diani E., Lievens P.M.-J. New insights into functional roles of the polypyrimidine tract-binding protein. Int. J. Mol. Sci. 2013;14:22906–22932. doi: 10.3390/ijms141122906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segre J.A., Bauer C., Fuchs E. Klf4 is a transcription factor required for establishing the barrier function of the skin. Nat. Genet. 1999;22:356–360. doi: 10.1038/11926. [DOI] [PubMed] [Google Scholar]
- Shen S., Park J.W., Huang J., Dittmar K.A., Lu Z.-x., Zhou Q., Carstens R.P., Xing Y. MATS: a Bayesian framework for flexible detection of differential alternative splicing from RNA-Seq data. Nucleic Acids Res. 2012;40:e61. doi: 10.1093/nar/gkr1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen S., Park J.W., Lu Z.X., Lin L., Henry M.D., Wu Y.N., Zhou Q., Xing Y. rMATS: robust and flexible detection of differential alternative splicing from replicate RNA-Seq data. Proc. Natl. Acad. Sci. USA. 2014;111:E5593–E5601. doi: 10.1073/pnas.1419161111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smalley-Freed W.G., Efimov A., Burnett P.E., Short S.P., Davis M.A., Gumucio D.L., Washington M.K., Coffey R.J., Reynolds A.B. p120-catenin is essential for maintenance of barrier function and intestinal homeostasis in mice. J. Clin. Invest. 2010;120:1824–1835. doi: 10.1172/JCI41414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C., Williams B.A., Pertea G., Mortazavi A., Kwan G., van Baren M.J., Salzberg S.L., Wold B.J., Pachter L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 2010;28:511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uren P.J., Bahrami-Samani E., Burns S.C., Qiao M., Karginov F.V., Hodges E., Hannon G.J., Sanford J.R., Penalva L.O.F., Smith A.D. Site identification in high-throughput RNA–protein interaction data. Bioinformatics. 2012;28:3013–3020. doi: 10.1093/bioinformatics/bts569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Nostrand E.L., Freese P., Pratt G.A., Wang X., Wei X., Xiao R., Blue S.M., Chen J.-Y., Cody N.A.L., Dominguez D., et al. A large-scale binding and functional map of human RNA-binding proteins. Nature. 2020;583:711–719. doi: 10.1038/s41586-020-2077-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Nostrand E.L., Gelboin-Burkhart C., Wang R., Pratt G.A., Blue S.M., Yeo G.W. CRISPR/Cas9-mediated integration enables TAG-eCLIP of endogenously tagged RNA binding proteins. Methods. 2017;118–119:50–59. doi: 10.1016/j.ymeth.2016.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Nostrand E.L., Pratt G.A., Shishkin A.A., Gelboin-Burkhart C., Fang M.Y., Sundararaman B., Blue S.M., Nguyen T.B., Surka C., Elkins K., et al. Robust transcriptome-wide discovery of RNA-binding protein binding sites with enhanced CLIP (eCLIP) Nat. Methods. 2016;13:508–514. doi: 10.1038/nmeth.3810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Nostrand E.L., Pratt G.A., Yee B.A., Wheeler E.C., Blue S.M., Mueller J., Park S.S., Garcia K.E., Gelboin-Burkhart C., Nguyen T.B., et al. Principles of RNA processing from analysis of enhanced CLIP maps for 150 RNA binding proteins. Genome Biol. 2020;21:90. doi: 10.1186/s13059-020-01982-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaquero-Garcia J., Barrera A., Gazzara M.R., González-Vallinas J., Lahens N.F., Hogenesch J.B., Lynch K.W., Barash Y. A new view of transcriptome complexity and regulation through the lens of local splicing variations. Elife. 2016;5:e11752. doi: 10.7554/eLife.11752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venhuizen J.-H., Sommer S., Span P.N., Friedl P., Zegers M.M. Differential expression of p120-catenin 1 and 3 isoforms in epithelial tissues. Sci. Rep. 2019;9:90. doi: 10.1038/s41598-018-36889-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waldmeier L., Meyer-Schaller N., Diepenbruck M., Christofori G. Py2T murine breast cancer cells, a versatile model of tgfβ-induced EMT in vitro and in vivo. PLoS One. 2012;7:e48651. doi: 10.1371/journal.pone.0048651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang E.T., Cody N.A.L., Jog S., Biancolella M., Wang T.T., Treacy D.J., Luo S., Schroth G.P., Housman D.E., Reddy S., et al. Transcriptome-wide regulation of pre-mRNA splicing and mRNA localization by muscleblind proteins. Cell. 2012;150:710–724. doi: 10.1016/j.cell.2012.06.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warzecha C.C., Carstens R.P. Complex changes in alternative pre-mRNA splicing play a central role in the epithelial-to-mesenchymal transition (EMT) Semin. Cancer Biol. 2012;22:417–427. doi: 10.1016/j.semcancer.2012.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warzecha C.C., Jiang P., Amirikian K., Dittmar K.A., Lu H., Shen S., Guo W., Xing Y., Carstens R.P. An ESRP-regulated splicing programme is abrogated during the epithelial-mesenchymal transition. The EMBO journal. 2010;29:3286–3300. doi: 10.1038/emboj.2010.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warzecha C.C., Sato T.K., Nabet B., Hogenesch J.B., Carstens R.P. ESRP1 and ESRP2 are epithelial cell-type-specific regulators of FGFR2 splicing. Mol. Cell. 2009;33:591–601. doi: 10.1016/j.molcel.2009.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warzecha C.C., Shen S., Xing Y., Carstens R.P. The epithelial splicing factors ESRP1 and ESRP2 positively and negatively regulate diverse types of alternative splicing events. RNA Biol. 2009;6:546–562. doi: 10.4161/rna.6.5.9606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J., Zhou L., Tonissen K., Tee R., Artzt K. The quaking I-5 protein (QKI-5) has a novel nuclear localization signal and shuttles between the nucleus and the cytoplasm. J. Biol. Chem. 1999;274:29202–29210. doi: 10.1074/jbc.274.41.29202. [DOI] [PubMed] [Google Scholar]
- Yanagisawa M., Huveldt D., Kreinest P., Lohse C.M., Cheville J.C., Parker A.S., Copland J.A., Anastasiadis P.Z. A p120 catenin isoform switch affects rho activity, induces tumor cell invasion, and predicts metastatic disease. J. Biol. Chem. 2008;283:18344–18354. doi: 10.1074/jbc.M801192200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y., Carstens R.P. Alternative splicing regulates distinct subcellular localization of Epithelial splicing regulatory protein 1 (Esrp1) isoforms. Sci. Rep. 2017;7:3848. doi: 10.1038/s41598-017-03180-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y., Park J.W., Bebee T.W., Warzecha C.C., Guo Y., Shang X., Xing Y., Carstens R.P. Determination of a comprehensive alternative splicing regulatory network and combinatorial regulation by key factors during the epithelial-to-mesenchymal transition. Mol. Cell Biol. 2016;36:1704–1719. doi: 10.1128/MCB.00019-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yee B.A., Pratt G.A., Graveley B.R., Van Nostrand E.L., Yeo G.W. RBP-Maps enables robust generation of splicing regulatory maps. RNA. 2018;25:193–204. doi: 10.1261/rna.069237.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H.H., Dohn M.R., Markham N.O., Coffey R.J., Reynolds A.B. p120-catenin controls contractility along the vertical axis of epithelial lateral membranes. J. Cell Sci. 2016;129:80–94. doi: 10.1242/jcs.177550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Z., Lin K.K., Bhandari A., Spencer J.A., Xu X., Wang N., Lu Z., Gill G.N., Roop D.R., Wertz P., Andersen B. The Grainyhead-like epithelial transactivator Get-1/Grhl3 regulates epidermal terminal differentiation and interacts functionally with LMO4. Dev. Biol. 2006;299:122–136. doi: 10.1016/j.ydbio.2006.07.015. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
RNA-seq and CLIP-seq data have been deposited at GEO and are publicly available as of the date of publication. Accession numbers are listed in the key resources table.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.