Abstract
Inorganic pyrophosphate-dependent phosphofructokinase (PPi-PFK) of the amitochondriate eukaryote Mastigamoeba balamuthi was sequenced and showed about 60% identity to PPi-PFKs from two eubacteria, Propionibacterium freudenreichii and Sinorhizobium meliloti. These gene products represent a newly recognized lineage of PFKs. All four lineages of group II PFKs, as defined by phylogenetic analysis, contained both prokaryotic and eukaryotic species, underlining the complex evolutionary history of this enzyme.
We have recently extended our studies on glycolytic enzymes of parasitic amitochondriate eukaryotes (20) to the free-living Mastigamoeba balamuthi (ATCC 30984) (6) with the goal of comparing the metabolic properties of anaerobic and microaerophilic eukaryotes with dramatically different life styles. This species belongs to the pelobionts, a group of amitochondriate amoeboflagellate protists of uncertain evolutionary position (5, 25). We noted that the sequence of its phosphofructokinase (PFK) showed unexpected characteristics, prompting us to revisit the taxonomic distribution and relationships of various PFK types.
Type A PFK, an enzyme of the glycolytic pathway, phosphorylates fructose 6-phosphate to fructose 1,6-bisphosphate. In most organisms, ATP is the phosphoryl donor (ATP-PFK; EC 2.7.1.11) of the irreversible reaction. A number of protists and plants and some eubacteria contain reversible PFKs, which use inorganic pyrophosphate (PPi) instead of ATP (PPi-PFK; EC 2.7.1.90). The assumed significance of PPi as the phosphoryl donor is reflected in an increase of the ATP yield during glycolysis (16, 26). This notion is supported by the predominant occurrence of PPi-PFK in organisms living in hypoxic or anoxic environments, which rely on anaerobic glycolysis (17).
The evolutionary history of PFK does not coincide with accepted notions of organismic relationships and points to past gene duplications and lateral gene transfers. Based primarily on sequence characteristics, type A PFKs are currently assigned to three major groups (groups I, II, and III) (22). Group II can be further subdivided into four subgroups, which appear as robust clades in phylogenetic reconstructions (we are using the tentative nomenclature proposed for clades in group II [18]). Closely related organisms may contain close homologs of PFK which use different phosphoryl donors, indicating that enzyme specificity can change relatively easily (2), a conclusion recently confirmed experimentally (7). Some organisms even contain members of two such subgroups (8, 10–12).
Sequence of M. balamuthi PPi-PFK.
A random clone of an M. balamuthi cDNA library (M. Müller et al., unpublished data), sequenced on both strands, contained a G+C-rich (69.3%) open reading frame putatively encoding a PPi-PFK of 410 amino acids with an Mr of 44,200. The conceptual translation showed that only 36 codons were used and that 98.2% of the nucleotides in the third position of each codon were G or C. This skewed codon usage, characteristic of protein-encoding genes of this organism (Müller et al., unpublished data), and the presence of a typical eukaryotic poly(A) tail support the origin of this message from authentic M. balamuthi DNA. The conceptual translation showed complete colinearity and about 60% amino acid identity to the PPi-PFK of the gram-positive organism Propionibacterium freudenreichii and to the putative PPi-PFK of the α-proteobacterium, Sinorhizobium meliloti (3, 14). The M. balamuthi sequence has been deposited in GenBank under accession number AAF70463.
Phylogenetic analysis.
Sequences of PPi-PFK homologs were retrieved from the National Center for Biotechnology Information protein database. The S. meliloti PFK sequence was retrieved from the website of the corresponding genome project (http://sequence.toulouse.inra.fr/meliloti.html) (3). Sequence sampling for group II encompasses the whole diversity present in the nonredundant National Center for Biotechnology Information database. Sampling of group III was restricted to a few species to provide an outgroup. Group I enzymes were not considered. The alignment was performed using the CLUSTAL X program (23) and adjusted visually. Phylogenetic reconstruction was performed with a maximum-likelihood method (PROTML) (1) on 167 shared amino-terminal amino acid positions. Bootstrap proportions were calculated by a resampling of the estimated log likelihood (RELL) values from the maximum-likelihood method (1). Neighbor joining and maximum-parsimony analyses revealed identical groups and subgroups (data not shown).
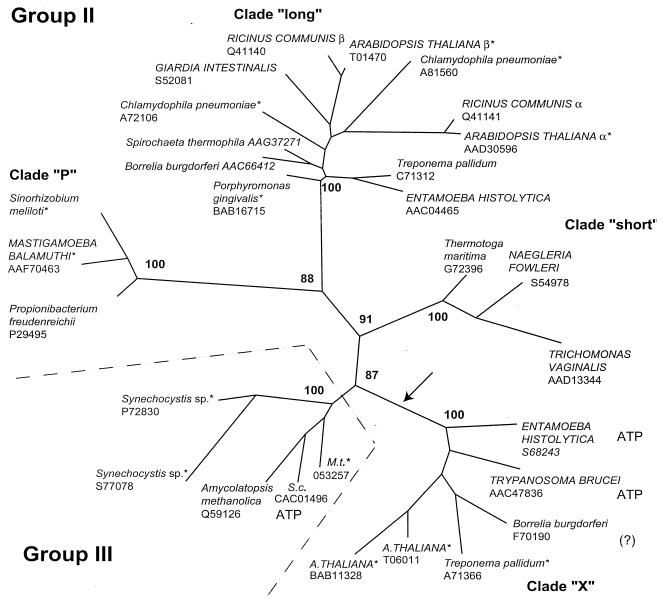
Evolutionary relationships of group II and III PFK sequences.
The phylogenetic tree obtained shows five major clades (Fig. 1). Four clades belong to group II, and one represents group III. The internal branches connecting these clades are relatively long, and the branching pattern is supported by high bootstrap values. The order of separation of lineages within these clades has not been resolved, however. While this subdivision is based only on the easily aligned amino-terminal parts of the sequences, a global comparison of the complete sequences and an analysis of the lengths and positions of insertions and deletions result in an identical subdivision (18).
FIG. 1.
Unrooted phylogenetic tree of group II and III PPi-PFKs based on 167 shared amino-terminal residues (between residues 2 and 204 in the P. freudenreichii sequence). Taxon selection extended over the complete diversity present in available databases. The arrow denotes a proposed gene duplication. Close paralogs among chlamydiae, actinomycetales, and plants were omitted for clarity. Database accession numbers follow the species names. For the S. meliloti sequence, see http://sequence.toulouse.inra.fr/meliloti.html. Eukaryotic species names are in capital letters, and eubacterial species names are in lowercase letters. A., Arabidopsis; M.t., Mycobacterium tuberculosis; S.c., Streptomyces coelicolor. PPi specificity was established biochemically for unmarked species; asterisks mark species not explored biochemically, “ATP” indicates enzymes with ATP specificity, and the question mark indicates a species for which the heterologously expressed enzyme showed no PFK activity with either PPi or ATP (9). For the long clade plant enzymes, α is the regulatory subunit and β is the catalytic one. The maximum-likelihood method with local rearrangements was utilized. Numbers at the nodes give RELL bootstrap values (see text).
The data clearly show that each of the four clades of group II PFKs encompasses both eubacterial and eukaryotic species. Eubacterial species in each clade represent diverse lineages, but the overall taxonomic diversity of group II enzymes is rather limited. Clade X contains sequences from several species that are also present in the long clade. So far, no archaebacterial sequences were noted in PFK group II. While the products of a number of sequences analyzed remain to be studied, available information indicates that the long, short, and P (new designation) clades correspond to PPi-specific enzymes (10, 18) and that clade X contains ATP-specific PFKs (8, 19).
Since only the amino-terminal half of the molecule could be confidently aligned across all groups, the information available for phylogenetic analysis was limited. The robust separation of major groups and clades, however, seems to be sufficient to suggest two major events in the evolution of the enzymes of group II. The first event was probably a gene duplication that separated clade X and the lineage leading to the three other clades (Fig. 1). This duplication was probably accompanied by a change of enzyme specificity in one of the branches. The substrate specificity of the common ancestor remains to be established, however. The second set of events led to differentiation between clades long, short, and P by marked changes in the overall sequence structure but without further change in phosphoryl donor specificity.
The presence of two PFK genes for the chlamydial and plant enzymes within the long clade reveals further gene duplications within this clade. In plants, gene duplication led to the emergence of catalytic (β) and regulatory (α) subunits (4, 24). A similar duplication and functional change also occurred in group I, which contains the classical ATP-linked enzymes (21). The functional significance of the chlamydial paralogs remains unknown.
The limited and peculiar taxonomic distribution of group II sequences makes a coherent reconstruction of events leading to the observed phenomena a daunting task. The relationships seen in the phylogenetic reconstruction do not coincide with accepted organismic relationships. One must account for the presence of both eubacteria and eukaryotes in each of the four clades of group II PFK genes as well as for the existence of sequences from the same organisms that fall into separate clades. While both early gene duplications and subsequent differential losses (15) and lateral gene transfers (13) have probably contributed to the current picture, only a significantly larger taxonomic sampling and functional characterization of the proteins encoded will permit a convincing reconstruction of the peculiar history of PPi-PFK homologs.
Acknowledgments
We thank Gordona Bothe (GATC-Biotechnology GMBH, Constance, Germany) for providing the S. meliloti sequence before its publication, Frederick Schuster (Brooklyn College, City University of New York, Brooklyn) for the M. balamuthi culture, Rama K. Singh and his team (NRC, Institute for Marine Biosciences, Halifax, Canada) for the DNA sequencing, Robert Kemp (The Chicago Medical School, North Chicago, Ill.) for suggestions and permission to refer to his paper before its publication, and William Martin (Heinrich-Heine Universität, Düsseldorf, Germany) and Lidya B. Sanchez and Dorothy V. Moore of the New York laboratory for comments on the manuscript.
This research was supported by grants to M.M. from the National Science Foundation (MCB9615659) and the National Institutes of Health (AI 11942).
REFERENCES
- 1.Adachi J, Hasegawa M. MOLPHY version 2.3: programs for molecular phylogenetics based on maximum likelihood. Computer Science Monographs, no. 28. Tokyo, Japan: Institute of Statistical Mathematics; 1996. [Google Scholar]
- 2.Alves A M C R, Euverink G J W, Bibb M J, Dijkhuizen L. Identification of ATP-dependent phosphofructokinase as a regulatory step in the glycolytic pathway of the actinomycete Streptomyces coelicolorA3(2) Appl Environ Microbiol. 1997;63:956–961. doi: 10.1128/aem.63.3.956-961.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Capela D, Barloy-Huber F, Gouzy J, Bothe G, Ampe F, Batut J, Boistard P, Becker A, Boutry M, Cadieu E, Dréano S, Gloux S, Godrie T, Goffeau A, Kahn D, Kiss E, Lelaure V, Masuy D, Pohl T, Portetelle D, Purnelle B, Ramsperger U, Renard C, Galibert F, Vandenbol M, Weidner S, Galibert F. Analysis of the chromosome sequence of the legume symbiont Sinorhizobium melilotistrain 1021. Proc Natl Acad Sci USA. 2001;98:9877–9882. doi: 10.1073/pnas.161294398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Carlisle S M, Blakele S D, Hemmingsen S M, Trevanion S J, Hiyoshi T, Kruger N J, Dennis D T. Pyrophosphate-dependent phosphofructokinase. Conservation of protein sequence between the α- and β-subunits and with the ATP-dependent phosphofructokinase. J Biol Chem. 1990;265:18366–18371. [PubMed] [Google Scholar]
- 5.Cavalier-Smith T. Archamoebae: the ancestral eukaryotes? BioSystems. 1991;25:25–38. doi: 10.1016/0303-2647(91)90010-i. [DOI] [PubMed] [Google Scholar]
- 6.Chavez L A, Balamuth W, Gong T. A light and electron microscopical study of a new, polymorphic free-living amoeba, Phreatamoeba balamuthin.g., n. sp. J Protozool. 1986;33:397–404. doi: 10.1111/j.1550-7408.1986.tb05630.x. [DOI] [PubMed] [Google Scholar]
- 7.Chi A, Kemp R G. The primordial high energy compound: ATP or inorganic pyrophosphate. J Biol Chem. 2000;275:35677–35679. doi: 10.1074/jbc.C000581200. [DOI] [PubMed] [Google Scholar]
- 8.Chi A S, Deng Z, Albach R A, Kemp R G. The two phosphofructokinase gene products of Entamoeba histolytica. J Biol Chem. 2001;276:19974–19981. doi: 10.1074/jbc.M011584200. [DOI] [PubMed] [Google Scholar]
- 9.Deng Z, Roberts D, Wang X, Kemp R G. Expression, characterization, and crystallization of the pyrophosphate-dependent phosphofructo-1-kinase of Borrelia burgdorferi. Arch Biochem Biophys. 1999;371:326–331. doi: 10.1006/abbi.1999.1446. [DOI] [PubMed] [Google Scholar]
- 10.Ding Y-H R, Ronimus R S, Morgan H W. Thermotoga maritimaphosphofructokinases: expression and characterization of two unique enzymes. J Bacteriol. 2001;183:791–794. doi: 10.1128/JB.183.2.791-794.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fraser C M, Casjens S, Huang W M, Sutton G G, Clayton R, Lathigra R, White O, Ketchum K A, Dodson R, Hickey E K, Gwinn M, Dougherty B, Tomb J-F, Fleischmann R D, Richardson D, Peterson J, Kerlavage A R, Quackenbush J, Salzberg S, Hanson M, van Vugt R, Palmer N, Adams M D, Gocayne J, Weidman J, Utterback T, Whattey L, McDonald L, Artiach P, Bowman P, Garland S, Fujii C, Cotton M D, Horst K, Roberts K, Hatch B, Smith H O, Venter J C. Genomic sequence of a Lyme disease spirochaete, Borrelia burgdorferi. Nature. 1997;390:580–586. doi: 10.1038/37551. [DOI] [PubMed] [Google Scholar]
- 12.Fraser C M, Norris S J, Weinstock G M, White O, Sutton G G, Dodson R, Gwinn M, Hickey E K, Clayton R, Ketchum K A, Sodergren E, Hardham J M, McLeod M P, Salzberg S, Peterson J, Khalak H, Richardson D, Howell J K, Chidambaram M, Utterback T, McDonald L, Artiach P, Bowman C, Cotton M D, Fujii C, Garland S, Hatch B, Horst K, Roberts K, Sandusky M, Weidman J, Smith H O, Venter J C. Complete genome sequence of Treponema pallidum, the syphilis spirochete. Science. 1998;281:375–388. doi: 10.1126/science.281.5375.375. [DOI] [PubMed] [Google Scholar]
- 13.Groisman F A, Ochman H. Pathogenicity islands: bacterial evolution in quantum leaps. Cell. 1996;87:791–794. doi: 10.1016/s0092-8674(00)81985-6. [DOI] [PubMed] [Google Scholar]
- 14.Ladror U, Gollapudi L, Tripathi R L, Latshaw S L, Kemp R G. Cloning, sequencing and expression of pyrophosphate-dependent phosphofructokinase from Propionibacterium freudenreichii. J Biol Chem. 1991;266:16550–16555. [PubMed] [Google Scholar]
- 15.Martin W, Schnarrenberger C. The evolution of the Calvin cycle from prokaryotic to eukaryotic chromosomes: a case study of functional redundancy in ancient pathways through endosymbiosis. Curr Genet. 1997;32:1–18. doi: 10.1007/s002940050241. [DOI] [PubMed] [Google Scholar]
- 16.Mertens E. Pyrophosphate-dependent phosphofructokinase, an anaerobic glycolytic enzyme? FEBS Lett. 1991;285:1–5. doi: 10.1016/0014-5793(91)80711-b. [DOI] [PubMed] [Google Scholar]
- 17.Mertens E. ATP versus pyrophosphate: glycolysis revisited in parasitic protists. Parasitol Today. 1993;9:122–126. doi: 10.1016/0169-4758(93)90169-g. [DOI] [PubMed] [Google Scholar]
- 18.Mertens E, Ladror U S, Lee J A, Miretsky A, Morris A, Rozario C, Kemp R G, Müller M. The pyrophosphate-dependent phosphofructokinase of the protist, Trichomonas vaginalis, and the evolutionary relationships of protist phosphofructokinases. J Mol Evol. 1998;47:739–750. doi: 10.1007/pl00006433. [DOI] [PubMed] [Google Scholar]
- 19.Michels P A M, Chevalier N, Opperdoes F R, Rider M H, Rigden D. The glycosomal ATP-dependent phosphofructokinase of Trypanosoma bruceimust have evolved from an ancestral pyrophosphate-dependent enzyme. Eur J Biochem. 1997;250:698–704. doi: 10.1111/j.1432-1033.1997.00698.x. [DOI] [PubMed] [Google Scholar]
- 20.Müller M. Enzymes and compartmentation of core energy metabolism of anaerobic protists—a special case in eukaryotic evolution? In: Coombs G H, Vickerman K, Sleigh M A, Warren A, editors. Evolutionary relationships among protozoa. Dordrecht, The Netherlands: Kluwer; 1998. pp. 109–131. [Google Scholar]
- 21.Poorman R A, Randolph A, Kemp R G, Heinrickson R L. Evolution of phosphofructokinase—gene duplication and creation of new effector sites. Nature. 1984;309:467–469. doi: 10.1038/309467a0. [DOI] [PubMed] [Google Scholar]
- 22.Siebers B, Klenk H-P, Hensel R. PPi-dependent phosphofructokinase from Thermoproteus tenax, an archeal descendant of an ancient line in phosphofructokinase evolution. J Bacteriol. 1998;180:2137–2143. doi: 10.1128/jb.180.8.2137-2143.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Thompson J D, Gibson T J, Plewniak F, Jeanmougin F, Higgins D G. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 2000;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Todd J F, Blakeley S D, Dennis D T. Structure of the genes encoding the α- and β-subunits of castor pyrophosphate-dependent phosphofructokinase. Gene. 1995;152:181–186. doi: 10.1016/0378-1119(94)00646-a. [DOI] [PubMed] [Google Scholar]
- 25.Walker G, Simpson A G B, Edgcomb V, Sogin M L, Patterson D J. Ultrastructural identities of Mastigamoeba punctachora, Mastigamoeba simplex and Mastigella commutansand assessment of hypotheses of relatedness of the pelobionts (Protista) Eur J Protistol. 2001;37:25–49. [Google Scholar]
- 26.Wood H G. Some reactions in which inorganic pyrophosphate replaces ATP and serves as a source of energy. Fed Proc. 1977;36:2197–2205. [PubMed] [Google Scholar]