Abstract
The colicin A pore-forming domain (pfColA) was fused to a bacterial signal peptide (sp-pfColA). This was inserted into the Escherichia coli inner membrane in functional form and could be coimmunoprecipitated with epitope-tagged immunity protein (EpCai). We constructed a series of fusion proteins in which various numbers of sp-pfColA α-helices were fused to alkaline phosphatase (AP). We showed that a fusion protein made up of the hydrophobic α-helices 8 and 9 of sp-pfColA fused to AP was specifically coimmunoprecipitated with EpCai produced in the same cells. This is the first biochemical evidence that Cai recognizes and interacts with the colicin A hydrophobic helical hairpin.
Pore-forming colicins are plasmid-encoded bacteriocins, synthesized by Enterobacteriaceae, that are lethal to other related strains. Like many toxins, colicins are organized into structural domains that perform different functions: the N-terminal domain is implicated in translocation across the membrane of the target cell, and the central domain recognizes target cells by binding to a specific extracellular surface receptor. The C-terminal domain houses the pore-forming activity of the protein (1, 2).
The soluble form of the colicin A pore-forming domain (pfColA) has been crystallized, and its three-dimensional structure has been determined to a resolution of 2.4 Å (11). The molecule consists of a bundle of eight amphipathic α-helices surrounding two hydrophobic α-helices (H8 and H9) that are completely buried within the protein. Channel formation involves the insertion of significant segments of the protein into the bilayer; however, the structure of the membrane-embedded ion channel is not yet known (10, 15, 17). It is proposed that the hydrophobic helical hairpin is the primary attachment region for channel formation (4, 12, 19).
Colicin-producing cells produce a specific immunity protein that protects them from the action of their own toxin (2, 16). Proteins with immunity to pore-forming colicins are integral inner membrane proteins. Sequence homology studies have separated pore-forming colicins into two groups, type A (colicins A, B, N, and U) and type E1 (colicins E1, 5, K, 10, Ia, and Ib); the corresponding immunity proteins have also been classified into the same two groups. The colicin A immunity protein (Cai) has four transmembrane segments, and both its N and C termini are located in the cytoplasm. The colicin E1 immunity protein (Cei) crosses the cytoplasmic membrane three times, with the N terminus being in the cytoplasm and the C terminus being in the periplasm (7, 18).
Genetic studies of the A-type colicins have indicated that the main determinant recognized by the immunity protein is the hydrophobic helical hairpin (8, 14). Recent studies of E1-type colicins have suggested that the region recognized by the immunity protein is located in the voltage-responsive segment (9, 13). The first biochemical evidence that a colicin physically interacts with its immunity protein was described by Espesset et al. (6), who used a coimmunoprecipitation procedure: pfColA was fused to a prokaryotic signal peptide (sp-pfColA) and coimmunoprecipitated with its cognate epitope-tagged immunity protein (EpCai). This study found that sp-pfColA was functionally inserted into the Escherichia coli inner membrane and inhibited by Cai (6).
In this paper, we used both PhoA fusions and coimmunoprecipitations to determine the minimum size of sp-pfColA that could interact with the immunity protein. We constructed a series of fusion proteins in which varied numbers of pfColA α-helices were fused between a prokaryotic signal peptide and alkaline phosphatase (AP). We demonstrated that a fusion protein consisting of the hydrophobic helical hairpin and AP was specifically coimmunoprecipitated with EpCai.
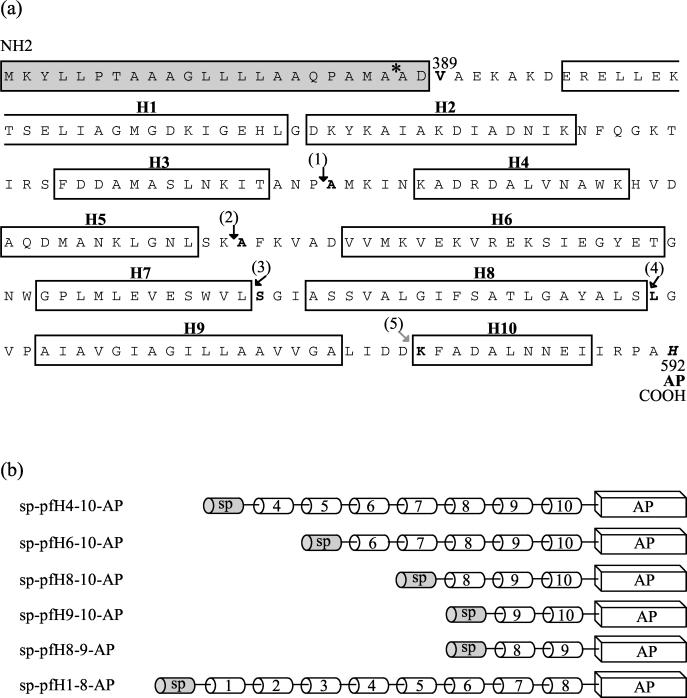
DNA fragments containing codons 459 to 592, 492 to 592, 531 to 592, and 553 to 592 of the colA gene were amplified by PCR and inserted into pPelBPhoA. The resulting plasmids encoded domains of pfColA extending from helix 4 to helix 10 (pfH4–10), helix 6 to helix 10 (pfH6–10), helix 8 to helix 10 (pfH8–10), and helix 9 to helix 10 (pfH9–10). These domains were fused between the PelB signal sequence and AP (Fig. 1). Expression of the sp-pfColA-AP gene was controlled by the tac-inducible promoter. We used immunoblot analysis with a polyclonal antibody directed against AP to show that sp-pfColA-AP hybrid proteins are produced at the same levels in cells producing (or not producing) an epitope-tagged immunity protein (EpCai; the epitope consisting of the 30 N-terminal amino acid residues of colicin A) (data not shown). As expected, hybrid proteins of various sizes were obtained (50 to 68 kDa) according to the number of pfColA α-helices fused to AP (see Fig. 3a; data not shown). Production of EpCai was controlled by the tightly regulated caa promoter.
FIG. 1.
Location of fusion sites of hybrid proteins. (a) Numbers above the amino acid residues indicate the corresponding positions in the entire colicin A (ColA) protein. The positions of the 10 α-helices of pfColA (H1 to H10) refer to the structure determined by X-ray crystallography. The residues of the Erwinia carotovora pectate lyase B (PelB) signal peptide are boxed in grey. The cleavage site of the PelB signal peptide is indicated by a star. Numbers in parentheses with arrows indicate residues at the beginning of the pfColA part of the fusion proteins: (1), sp-pfH4-10-AP; (2), sp-pfH6-10-AP; (3), sp-pfH8-10-AP; (4), sp-pfH9-10-AP. The residue in bold and italics (His 592) indicates the junction between pfColA and AP. The pfColA part of sp-pfH8-9-AP begins at Ser 530 [indicated by “(3)”] and ends at Lys 578 [indicated by “(5)”], where it joins to AP. (b) Representation of the sp-pfColA-AP fusion proteins. The α-helices of pfColA are represented by cylinders, and AP is represented by a rectangle.
FIG. 3.
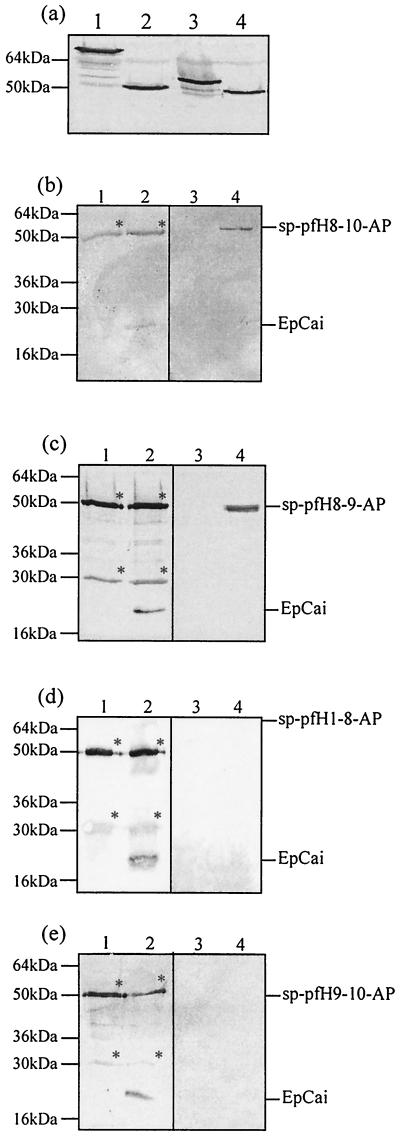
Sp-pfH8-9-AP and EpCai are coimmunoprecipitated. (a) Production of sp-pfColA-AP fusion proteins in cells producing EpCai before immunoprecipitation. Total cell extracts from strain C600 containing plasmids encoding hybrid proteins and EpCai (optical density at 600 nm [OD600], 0.2) were analyzed by immunoblot analysis for the presence of sp-pfH1-8-AP (lane 1), sp-pfH8-9-AP (lane 2), sp-pfH8-10-AP (lane 3), and sp-pfH9-10-AP (lane 4). (b) sp-pfH8-10-AP and EpCai were coimmunoprecipitated. C600 cells (OD600, 0.3) containing plasmids encoding EpCai and sp-pfH8-10-AP or (ΔEp)Cai and sp-pfH8-10-AP were incubated for 60 min with 300 ng of mitomycin C/ml; the bacteria were then grown for 15 min at 37°C with 100 μM IPTG. Cells (50 × 109) were solubilized with detergent, and the proteins were immunoprecipitated with MAb 1C11. Half of the immunoprecipitated fraction was immunoblotted with the MAb 1C11 used to detect EpCai (lanes 1 and 2), and the other half was immunoblotted with the anti-AP serum used to detect sp-pfH8-10-AP (lanes 3 and 4). The samples loaded were immunoprecipitated proteins from strain C600 producing (ΔEp)Cai and sp-pfH8-10-AP (lanes 1 and 3) and EpCai and sp-pfH8-10-AP (lanes 2 and 4). EpCai and sp-pfH8-H10-AP are indicated. The sample buffer contained 5% (vol/vol) β-mercaptoethanol, which dissociates the light and heavy chains of the immunoglobulins; both the heavy and light chains were revealed by the secondary anti-mouse antibody (indicated by ∗). The same result was obtained with sp-pfH4-10-AP and sp-pfH6-10-AP. (c) sp-pfH8-9-AP and EpCai were coimmunoprecipitated as described above. The samples loaded were immunoprecipitated proteins from strain C600 producing (ΔEp)Cai and sp-pfH8-9-AP (lanes 1 and 3) and EpCai and sp-pfH8-9-AP (lanes 2 and 4). EpCai and sp-pfH8-9-AP are indicated. (d) sp-pfH1-8-AP and EpCai were not coimmunoprecipitated. The samples loaded were immunoprecipitated proteins from strain C600 producing (ΔEp)Cai and sp-pfH1-8-AP (lanes 1 and 3) and EpCai and sp-pfH1-8-AP (lanes 2 and 4). EpCai and sp-pfH1-8-AP are indicated. (e) sp-pfH9-10-AP and EpCai were not coimmunoprecipitated. The samples loaded were immunoprecipitated proteins from strain C600 producing (ΔEp)Cai and sp-pfH9-10-AP (lanes 1 and 3) and EpCai and sp-pfH9-10-AP (lanes 2 and 4). EpCai and sp-pfH9-10-AP are indicated.
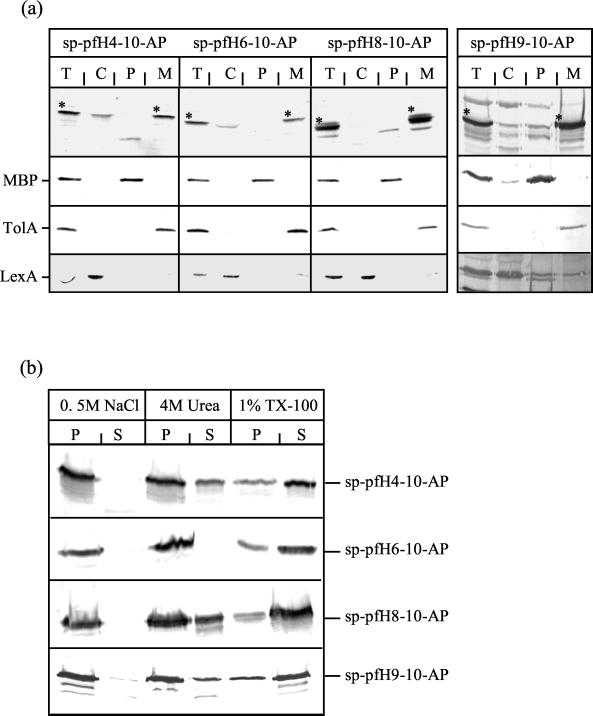
Bacteria coproducing the various fusion proteins were fractionated into cytoplasmic, periplasmic, and membrane fractions as previously described (3). The hybrid proteins were detected in membrane fractions by immunoblot analysis with antisera directed against AP (Fig. 2a). Each fraction was tested for the presence of the periplasmic maltose binding protein, the cytoplasmic LexA protein, or the membrane TolA protein as a control (Fig. 2a). The membrane association of the hybrid proteins was analyzed by a series of membrane extraction treatments. Membrane fractions were incubated in 0.5 M NaCl, 4 M urea, or 1% Triton X-100 for 4 h at 4°C with stirring. The content of each fraction was analyzed by immunoblot analysis with antisera directed against AP. The solubilization of each hybrid protein required 1% Triton X-100, indicating that the sp-pfColA-AP fusion proteins were tightly associated with the membrane (Fig. 2b).
FIG. 2.
Intracellular location of sp-pfColA-AP hybrid proteins. (a) The sp-pfColA-AP hybrid proteins are located in the membrane. Cells (lanes T) were induced with 100 μM IPTG for 15 min at 37°C and separated into cytoplasmic (lanes C), periplasmic (lanes P), and membrane (lanes M) fractions as previously described (3). Fractions (optical density at 600 nm, 0.3) were tested for the presence of the sp-pfColA-AP proteins (indicated by ∗) by immunoblot analysis with the anti-AP antibody. The same fractions were tested for the presence of maltose binding protein, TolA, and LexA markers of the periplasmic, membrane, and cytoplasmic fractions, respectively. (b) The sp-pfColA-AP hybrid proteins are tightly associated with the membrane. The membrane fractions were subjected to various treatments (4 h at 4°C with 0.5 M NaCl, 4 M urea, or 1% Triton X-100) and were centrifuged for 30 min at 18,000 × g. sp-pfColA-AP fusion proteins were detected in the resulting pellets (lanes P) or supernatants (lanes S) by immunoblot analysis with the anti-AP antibody.
To determine which pfColA α-helices are recognized by the immunity protein, we first attempted to coimmunoprecipitate EpCai and sp-pfH4-10-AP, sp-pfH6-10-AP, and sp-pfH8-10-AP produced in the same cells, as previously described (Fig. 3, lane 3). EpCai production was induced with 300 ng of mitomycin C/ml for 1 h, and then sp-pfColA-AP hybrid proteins were induced with 100 μM IPTG (isopropyl-β-d-thiogalactopyranoside) for 15 min. Total cell proteins were immunoprecipitated with the monoclonal antibody (MAb) 1C11 (directed against the epitope tag of EpCai) as previously described (6). The immunoprecipitates were tested by immunoblotting for the presence of sp-pfColA-AP hybrid proteins and EpCai. We found that each sp-pfColA-AP hybrid protein was specifically coimmunoprecipitated with EpCai (Fig. 3b and data not shown). It was estimated by immunoblot quantification that 500 to 800 ng of 10 μg of the membrane-associated sp-pfColA-AP fusion proteins was recovered in the immunoprecipitates (data not shown). To confirm that the 1C11 MAb did not cross-react with the sp-pfColA-AP fusion proteins, we coproduced sp-pfColA-AP proteins with EpCai from which its epitope tag had been deleted [(ΔEp)Cai] and repeated the immunoprecipitation assay. Both proteins, EpCai and (ΔEp)Cai, had the same immunity activities (data not shown), and the amount of sp-pfColA-AP fusion proteins coproduced with (ΔEp)Cai was identical to that coproduced with EpCai (data not shown). The sp-pfColA-AP fusion proteins were not detected in the immunoprecipitates from cells producing both (ΔEp)Cai and sp-pfColA-AP fusion proteins (Fig. 3b and data not shown). Taken together, these results indicate that helices 8, 9, and 10 of pfColA interact with the immunity protein inside the membrane.
Therefore, to determine both the minimum number of sp-pfColA α-helices which interact with EpCai and the role of helix 8 and helix 9 in this interaction, a new series of immunoprecipitation assays was performed. It was previously demonstrated that the pfColA domain extending from helix 1 to helix 8 and the pfColA hydrophobic helical hairpin fused to the PelB signal sequence and AP (sp-pfH1-8-AP and sp-pfH8-9-AP, respectively) were inserted into the inner membrane of E. coli (3). Here we show that sp-pfH9-10-AP was also inserted into the membrane, presumably by its helix 9, which has sufficient hydrophobic residues to act as a stop transfer sequence (Fig. 2b). So we tried to coimmunoprecipitate sp-pfH1-8-AP and EpCai, sp-pfH8-9-AP and EpCai, or sp-pfH9-10-AP and EpCai produced in the same cell. sp-pfH8-9-AP was detected in the immunoprecipitates from cells producing both EpCai and sp-pfH8-9-AP and was not detected in the immunoprecipitates from cells producing both (ΔEp)Cai and sp-pfH8-9-AP (Fig. 3c). In contrast, sp-pfH1-8-AP and sp-pfH9-10-AP were not coimmunoprecipitated with EpCai (Fig. 3d and e), although all proteins were produced at the same levels (Fig. 3a). In conclusion, Cai can interact with a membrane peptide that is as short as the pfColA hydrophobic helical hairpin. This indicates that the α-helices 8 and 9 of pfColA carry the information required for the recognition but that helix 8 or helix 9 alone does not. This recognition does not require any other region of pfColA and must occur in the inner membrane, because the hydrophobic helical hairpin and Cai are inserted into the membrane and AP is located in the periplasm.
We previously showed that the membrane insertion of sp-pfColA was independent of the Tol proteins that are normally required for the transport of the colicin A from its receptor on the outer membrane surface to its target, the inner membrane (5). Moreover, Cai inhibited the channel formed by sp-pfColA, confirming that immunity proteins function independently of colicin translocation systems (8, 20). In fact, lateral diffusion of the immunity proteins in the membrane would ensure rapid recognition of colicin pore-forming domains (20). Based on previous results from the last two decades, it seems that the A-type immunity proteins diffuse laterally in the membrane and then recognize and interact with their cognate hydrophobic helical hairpins just prior to colicin channel opening (6, 13, 14, 20). The mode of action of the E1-type immunity proteins is quite similar, except that they interact with the voltage-responsive segment of the E1-type colicin instead of the hydrophobic helical hairpin (9, 13). However, our results do not indicate that this first interaction between Cai and the pfColA hydrophobic helical hairpin is sufficient to prevent channel formation.
Acknowledgments
We gratefully acknowledge S. Slatin and V. Géli for advice and discussions. The excellent technical help of M. Chartier was very much appreciated.
REFERENCES
- 1.Baty D, Frenette M, Lloubès R, Géli V, Howard S P, Pattus F, Lazdunski C. Functional domains of colicin A. Mol Microbiol. 1988;2:807–811. doi: 10.1111/j.1365-2958.1988.tb00092.x. [DOI] [PubMed] [Google Scholar]
- 2.Bénédétti H, Frenette M, Baty D, Knibiehler M, Pattus F, Lazdunski C. Individual domains of colicins confer specificity in colicin uptake in pore-properties and in immunity requirement. J Mol Biol. 1991;217:429–439. doi: 10.1016/0022-2836(91)90747-t. [DOI] [PubMed] [Google Scholar]
- 3.Duché D, Corda Y, Géli V, Baty D. Integration of the colicin A pore-forming domain into the cytoplasmic membrane of Escherichia coli. J Mol Biol. 1999;285:1965–1975. doi: 10.1006/jmbi.1998.2423. [DOI] [PubMed] [Google Scholar]
- 4.Elkins P, Bunker A, Cramer W A, Stauffacher C V. A mechanism for toxin insertion into membranes is suggested by the crystal structure of the channel-forming domain of colicin E1. Structure. 1997;5:443–458. doi: 10.1016/s0969-2126(97)00200-1. [DOI] [PubMed] [Google Scholar]
- 5.Espesset D, Corda Y, Cunningham K, Bénédétti H, Lloubès R, Lazdunski C, Géli V. The colicin A pore-forming domain fused to mitochondrial intermembrane space sorting signals can be functionally inserted into the Escherichia coli plasma membrane by a mechanism that bypasses the Tol proteins. Mol Microbiol. 1994;13:1121–1131. doi: 10.1111/j.1365-2958.1994.tb00503.x. [DOI] [PubMed] [Google Scholar]
- 6.Espesset D, Duché D, Baty D, Géli V. The channel domain of colicin A is inhibited by its immunity protein through direct interaction in the Escherichia coli inner membrane. EMBO J. 1996;15:2356–2364. [PMC free article] [PubMed] [Google Scholar]
- 7.Géli V, Baty D, Pattus F, Lazdunski C. Topology and function of the integral membrane protein conferring immunity to colicin A. Mol Microbiol. 1989;3:679–687. doi: 10.1111/j.1365-2958.1989.tb00216.x. [DOI] [PubMed] [Google Scholar]
- 8.Géli V, Lazdunski C. An α-helical hydrophobic hairpin as a specific determinant in protein-protein interaction occurring in Escherichia coli colicin A and B immunity systems. J Bacteriol. 1992;174:6432–6437. doi: 10.1128/jb.174.20.6432-6437.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lindeberg M, Cramer W A. Identification of specific residues in colicin E1 involved in immunity protein recognition. J Bacteriol. 2001;183:2132–2136. doi: 10.1128/JB.183.6.2132-2136.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Merrill A R, Cramer W A. Identification of a voltage-responsive segment of the potential-gated colicin E1 ion channel. Biochemistry. 1990;29:8529–8534. doi: 10.1021/bi00489a004. [DOI] [PubMed] [Google Scholar]
- 11.Parker M W, Pattus F, Tucker A D, Tsernoglou D. Structure of the membrane pore-forming fragment of colicin A. Nature. 1989;337:93–96. doi: 10.1038/337093a0. [DOI] [PubMed] [Google Scholar]
- 12.Parker M W, Postma J P M, Pattus F, Tucker A, Tsernoglou D. Refined structure of the pore-forming domain of colicin A at 2.4 Å resolution. J Mol Biol. 1992;224:639–657. doi: 10.1016/0022-2836(92)90550-4. [DOI] [PubMed] [Google Scholar]
- 13.Pilsl H, Braun V. Evidence that the immunity protein inactivates colicin 5 immediately prior to the formation of the transmembrane channel. J Bacteriol. 1995;177:6966–6972. doi: 10.1128/jb.177.23.6966-6972.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pilsl H, Šmajs D, Braun V. The tip of the hydrophobic hairpin of colicin U is dispensable for colicin U activity but is important for interaction with the immunity protein. J Bacteriol. 1998;180:4111–4115. doi: 10.1128/jb.180.16.4111-4115.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Qui X Q, Jakes K S, Kienker P K, Finkelstein A, Slatin S L. Major transmembrane movement associated with colicin Ia channel gating. J Gen Physiol. 1996;103:313–328. doi: 10.1085/jgp.107.3.313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schramm E, Olschlager T, Troger W, Braun V. Sequence, expression and localization of the immunity protein for colicin B. Mol Gen Genet. 1988;211:176–182. doi: 10.1007/BF00338410. [DOI] [PubMed] [Google Scholar]
- 17.Slatin S L, Qiu X Q, Jakes K S, Finkelstein A. Identification of a translocated protein segment in a voltage-dependent channel. Nature. 1994;371:158–161. doi: 10.1038/371158a0. [DOI] [PubMed] [Google Scholar]
- 18.Song H Y, Cramer W A. Membrane topography of ColE1 gene products: the immunity protein. J Bacteriol. 1991;173:2935–2943. doi: 10.1128/jb.173.9.2935-2943.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Song H Y, Cohen F S, Cramer W A. Membrane topography of ColE1 gene products: the hydrophobic anchor of colicin E1 channel is a helical hairpin. J Bacteriol. 1991;173:2927–2934. doi: 10.1128/jb.173.9.2927-2934.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang Y L, Cramer W A. Intramembrane helix-helix interactions as the basis of inhibition of the colicin E1 ion channel by its immunity protein. J Biol Chem. 1993;268:10176–10184. [PubMed] [Google Scholar]