Abstract
Object
The aim of the present work was to investigate the correlation of plasma platelet‐derived growth factor (PDGF)‐BB level and single nucleotide polymorphism (SNP, rs1800817 and rs2285094) of PDGF‐B gene with the onset and stability condition of coronary heart disease (CHD).
Methods
Totally, 335 subjects were included in and divided into CHD (n = 247) and control group (n = 88) according to coronary angiography. Besides, the patients in the CHD group were divided into acute coronary syndrome (ACS) group (n = 165) and stable angina pectoria (SAP) group (n = 82), based on CHD stability condition. The plasma PDGF‐BB level was measured by ELISA, and the genotype of PDGF‐B was examined through qPCR assay.
Results
The PDGF‐BB level was positively correlated with hsCRP level (r = 0.149, p < 0.05). The genotype frequencies of SNP rs1800817 and rs2285094 match Hardy–Weinberg equilibrium. There was weak linkage disequilibrium between SNP rs1800817 and rs2285094: D′ = 0.419, r 2 = 0.04, which has no correlation with CHD. There was no statistical difference in plasma PDGF‐BB level among different genotypes in rs1800817 and rs2285094. There were no differences in the plasma PDGF‐BB level among patients with any genotype of SNP rs1800817 and rs2285094, no matter how it was grouped. Logistic regression results indicated that the plasma PDGF‐BB level was the independent risk factor of CHD onset (OR = 1.003, 95% CI 1.001–1.006, p = 0.014).
Conclusions
High plasma PDGF‐BB level is the risk factor of CHD and has correlation with instability of CHD. The plasma PDGF‐BB level change may be related to inflammatory response. PDGF‐B gene rs1800817 and rs2285094 polymorphisms are not correlated with CHD.
Keywords: coronary heart disease, platelet‐derived growth factor, single nucleotide polymorphism
Research Process and Results.
1. INTRODUCTION
Coronary atherosclerotic heart disease, abbreviated as coronary heart disease (CHD), is the major cardiovascular disease with high incidence and fatality rate. The disease is caused by various genetic factors and environmental factors. 1 , 2 , 3 Migration and hyperplasia of smooth muscle cells (SMC) are part of the main mechanisms for the plaque formation of atherosclerosis (AS). Platelet‐derived growth factor (PDGF)‐B is a 24‐ku cation glycoprotein, mainly exists in the platelet α granules and injured endothelial cells, macrophages, smooth muscle cells, fibroblasts, etc., in active form of homozygous dimer PDGF‐BB. PDGF‐BB possesses chemotactic effect, promotion of cell division, and vasoconstriction effect. 4 , 5 Malabanan et al. 6 found that PDGF‐BB could promote the proliferation and migration of smooth muscle cells. Therefore, PDGF‐B plays a very important role in the formation of AS and CHD onset. Human PDGF‐B gene is a coding product of proto‐oncogene c‐sis, located in chromosome 22q12.3‐q13.1, composed of seven exons. Several polymorphic sites of PDGF‐B gene have been reported, but the correlation between PDGF‐B gene polymorphism and disease was few reported. 7 There is still no study on the correlation of either plasma PDGF‐BB level or PDFG‐B gene polymorphism with CHD onset or plaque stability. Therefore, by detecting plasma PDGF‐BB level and PDGF‐B single nucleotide polymorphism (SNP) in CHD patients as well as control group, and exploring the correlation of PDGF‐BB level and PDGF‐B gene polymorphism with CHD onset and disease stability, we hope to provide a new target for early‐stage diagnosis of CHD and recognition on high‐risk patients. In this study, we focused on two SNPs: rs2285094 and rs1800817. rs2285094 (chr22:39226555) is located in the intron region of PDGFB transcript variant 1,2 mRNA, and rs1800817 (chr22:39243848) is located in the intron region of PDGFB transcript variant 1 mRNA and upstream of transcript variant 2 mRNA (https://www.ncbi.nlm.nih.gov/snp/rs2285094#variant_details).
2. MATERIALS AND METHODS
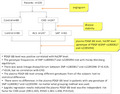
2.1. Subjects inclusion
Three hundred and thirty‐five patients were recruited from the Department of Cardiology, Tianjin Chest Hospital between May 2014 and October 2015. There were 214 male and 121 female (total 335), of 40–78 years old, average age at 60.54 ± 7.64 years. After completing relevant examinations, all the enrolled patients received coronary angiography by Judkins method. 8 Coronary angiography diameter method was used to measure the coronary artery (left main coronary artery, anterior descending branch, circumflex artery, right coronary artery), and its main branches (diagonal branch, obtuse marginal branch, posterior descending coronary artery, posterior branch of left ventricle). Those who have at least one of the branches showing diameter stenosis ≥50% were diagnosed as coronary heart disease (CHD group). The patients without stenosis were considered control group. The patients in CHD group were further divided into stable angina pectoris (SAP) group and acute coronary syndrome (ACS) group according to the stability condition of CHD. 9 The patients with percutaneous coronary intervention history, other cardiovascular diseases, cardiac insufficiency, hepatic and renal insufficiency, coagulation disorders, other systemic diseases, acute infection, administration of inflammation inhibition drugs (such as nonsteroids, anti‐inflammatory analgesic), administration of statin or aspirin within 1 month were excluded.
2.2. Methods
The patients received routine blood examination such as fasting blood glucose (Glu), total cholesterol (TC), triglyceride (TG), high‐density lipoprotein (HDLc), low‐density lipoprotein (LDLc), white blood cell count (WBC), platelet count (Plt), high‐sensitivity C‐reactive protein (hsCRP), and ultrasonic cardiography after hospitalization, and their clinical information such as age, gender, body mass index (BMI), hypertension history, diabetes history, CHD family history, and smoking history were all recorded. Venous blood (5 ml) was taken in the morning after fasting for 12 h, anticoagulated with EDTA, centrifugated at 1500 g/min for 15 min to separate plasma and hemocytes, and stored at −20°C. Blood genome DNA extraction system (RelaxGene Blood DNA System (TIANGEN BIOTECH CO. LTD)) was used to extract DNA from leukocytes. The plasma PDGF‐BB levels were measured by ELISA, according to instruction of the kit (BlueGene Biotech).
The gene sequences near rs1800817 and rs2285094 taken from the Genebank, together with the immunofluorescence probe and primer designed by Primer Premier 5.0 and Gene Runner were synthesized by Thermo Fisher Scientific. The primer and probe sequences of rs1800817 loci are shown in Table 1, and the primer and probe of rs2285094 loci refer to Assay ID C_15962706_10. Taqman Quantitative Real‐Time PCR (Prism7500, ABI, USA) was used to detect the genotypes. PCR reaction system (10 μl): 2 × Taqman PCR MIX (5 μl), 20× (Primer‐F, Primer‐R, MGB probe) (0.5 μl), 100 ng/μl DNA (1 μl), deionized water (3.5 μl). PCR reaction condition: [95°C, 10 min] → [95°C, 15 s → 60°C, 60s], 40 cycles.
TABLE 1.
Immunofluorescence probes and primers
| Loci | Sequence (5′–3′) | |
|---|---|---|
| rs1800817 | Upstream primer | GGGCTGGTTCTTCATTCATTACCTT |
| Downstream primer | TGCAAGGGTCCAAAGTTCACT | |
| Probe 1 | CCCCCACCTTCTG | |
| Probe 2 | CCCCCCCCTTCTG | |
Note: rs2285094 probes and primers: Assay ID C_15962706_10, Thermo Fisher Scientific. The mutation sites of rs1800817 are underlined in the sequence of the Prob1 and 2.
Totally 35 samples were randomly selected from different genotypes and verified by Thermo Fisher Scientific. The sequence was compared with the homologous one in the database from http://blast.ncbi.nlm.nih.gov/Blast.cgi. The sequence comparison result proved that the PCR amplification fragment was the target gene one.
2.3. Statistical method
SPSS20.0 software was used to analyze the data. Measurement data were analyzed by normality test. The measurement data meeting with normal distribution were demonstrated with mean ± SD. The measurement data between the two groups were analyzed by t test, and those among groups were analyzed by univariate ANOVA. The pairwise comparison was done by q test. The measurement data that did not match normal distribution were expressed as median and quartile M (P25, P75). The comparison between two groups was analyzed by non‐parametric Mann–Whitney U test, and that among multiple groups was analyzed by nonparametric Kruskal–Wallis univariate ANOVA test. Enumeration data were shown by n (%), and the comparison between groups was analyzed by χ 2 test. The correlation of abnormal distribution was analyzed by Spearman correlation analysis. The correlation of variables with coronary heart disease was analyzed by binomial logistic regression analysis. Bilateral p < 0.05 was considered statistically significant. PDGF‐B gene SNP rs1800817 and rs2285094 allele frequencies and Hardy–Weinberg genetic equilibrium were analyzed by HaploView software and THESIAS 3.1.
3. RESULTS
3.1. General conditions of the included subjects
There were no statistical differences of age, gender, BMI, hypertension history, diabetes history, CHD family history, smoking history, and laboratory examination results (Glu, TG, TC, LDLc, HDLc, WBC, Plt, hsCRP), between CHD group and control group or among control group, SAP group and ACS group (p > 0.05), as shown in Table 2.
TABLE 2.
General condition comparisons between control group and CHD group
| Control group (n = 88) | CHD group (n = 247) | SAP group (n = 82) | ACS group (n = 165) | |
|---|---|---|---|---|
| Age (years) | 60.60 ± 7.95 | 60.52 ± 7.55 | 61.44 ± 7.24 | 60.06 ± 7.67 |
| Male (n (%)) | 54 (61.4) | 160 (64.8) | 56 (68.3) | 104 (63.0) |
| Hypertension (n (%)) | 46 (52.3) | 151 (61.1) | 49 (59.8) | 102 (61.8) |
| Diabetes (n (%)) | 16 (18.2) | 68 (27.5) | 21 (25.6) | 47 (28.8) |
| Smoking history (n (%)) | 30 (34.1) | 109 (44.1) | 32 (39.0) | 77 (46.7) |
| Family history (n (%)) | 22 (25.0) | 77 (31.2) | 23 (28.0) | 54 (32.7) |
| BMI (kg/m2) | 25.51 ± 3.28 | 25.52 ± 3.06 | 25.58 ± 3.38 | 25.49 ± 2.90 |
Note: The measurement data (Age, BMI) were shown by mean ± SD, and the comparison between control group and CHD group was analyzed by t test, and those among control group, SAP group, and ACS group were analyzed by univariate ANOVA. Enumeration data (male, hypertension, diabetes, smoking history, family history) were shown by n (%), and the comparison between groups was analyzed by χ 2 test. BMI = Weight (kg)/height2 (m2).
Abbreviations: ACS, acute coronary syndrome; BMI, body mass index; CHD, coronary heart disease; SAP, stable angina pectoria.
3.2. Laboratory examination results and plasma PDGF‐BB level
The plasma PDGF‐BB level in the CHD group was significantly higher than that in control group (p < 0.05). There were no significant differences in WBC, Plt, Glu, TC, TG, LDLc between the two groups (p > 0.05). The HDLc level in CHD group was significantly lower than that in control group (p < 0.05), and hsCRP level in CHD group was significantly higher than that in control group (p < 0.05), as shown in Table 3.
TABLE 3.
Comparisons of laboratory examination results and PDGF‐BB levels between control group and CHD group
| Control group (n = 86) | CHD group (n = 247) | t/Z | p | |
|---|---|---|---|---|
| Glu (mmol/L) | 5.50 ± 1.09 | 5.69 ± 1.47 | −1.330 | 0.185 |
| TC (mmol/L) | 4.62 ± 1.13 | 4.59 ± 1.17 | 0.219 | 0.827 |
| TG (mmol/L) | 1.58 ± 0.85 | 1.86 ± 1.39 | −1.761 | 0.079 |
| HDLc (mmol/L) | 1.16 ± 0.33 | 1.05 ± 0.41 | 2.451 | 0.015* |
| LDLc (mmol/L) | 2.88 ± 0.91 | 2.88 ± 1.01 | −0.010 | 0.992 |
| WBC (×109/L) | 6.39 ± 1.50 | 6.67 ± 1.53 | −1.478 | 0.140 |
| Plt (×109/L) | 234.25 ± 54.07 | 228.60 ± 55.82 | 0.823 | 0.411 |
| hsCRP (mg/L) | 0.83 (0.44, 2.32) | 1.41 (0.63, 3.28) | 2.573 | 0.010** |
| PDGF‐BB (pg/ml) | 224.10 ± 111.83 | 258.37 ± 132.40 | −2.168 | 0.031* |
Note: The measurement data (Glu, TC, TG, HDLc, LDLc, WBC, Plt, PDGF‐BB) were shown by mean ± SD, and the comparison between control group and CHD group was analyzed by t test. The data of hsCRP level that did not match normal distribution were expressed as median and quartile M (P25, P75), the comparison between groups was analyzed by nonparametric Mann–Whitney U test. Other measurement data between control group and CHD group were analyzed by t test (*p < 0.05, **p < 0.01).
Abbreviations: CHD, coronary heart disease; Glu, Glucose; HDLc, high‐density lipoprotein cholesterol; hsCRP, high sensitivity C‐reactive protein; LDLc, low‐density lipoprotein cholesterol; PDGF‐BB, platelet‐derived growth factor‐BB; Plt, blood platelet; TC, total cholesterol; TG, triglyceride; WBC, white blood cells.
There were no statistical differences in WBC, Plt, Glu, TC, TG, LDLc levels among control group, SAP group, and ACS group (p > 0.05). There were statistical differences in PDGF‐BB and hsCRP levels among the three groups (p < 0.01). Those in the ACS group were significantly higher than that of the other two groups (p < 0.01). However, the difference between the latter two groups was not statistically different (p > 0.05). There was statistical difference in HDLc level among the three groups (p < 0.05), and SAP group was significantly lower than control group (p < 0.05). Although HDLc level in ACS group was lower than that of control group, there was no statistical significance between the two groups, as shown in Table 4. PDGF‐BB level was positive correlated with hsCRP level (r = 0.149, p = 0.009).
TABLE 4.
Comparison of laboratory examination results and PDGF‐BB levels among control group, SAP group, and ACS group
| Control group (n = 86) | SAP group (n = 79) | ACS group (n = 134) | F/Z | p | |
|---|---|---|---|---|---|
| Glu (mmol/L) | 5.50 ± 1.09 | 5.70 ± 1.53 | 5.69 ± 1.45 | 0.667 | 0.514 |
| TC (mmol/L) | 4.62 ± 1.13 | 4.50 ± 0.89 | 4.63 ± 1.29 | 0.360 | 0.698 |
| TG (mmol/L) | 1.58 ± 0.85 | 1.88 ± 1.34 | 1.85 ± 1.50 | 1.559 | 0.212 |
| HDLc (mmol/L) | 1.16 ± 0.33 | 1.01 ± 0.25Δ | 1.06 ± 0.46 | 3.554 | 0.030* |
| LDLc (mmol/L) | 2.88 ± 0.91 | 2.80 ± 0.81 | 2.92 ± 1.10 | 0.349 | 0.706 |
| WBC (×109/L) | 6.39 ± 1.49 | 6.49 ± 1.57 | 6.76 ± 1.50 | 1.938 | 0.146 |
| Plt (×109/L) | 234.25 ± 54.07 | 222.78 ± 55.04 | 231.48 ± 56.14 | 1.016 | 0.363 |
| hsCRP (mg/L) | 0.83 (0.44, 2.32) | 1.02 (0.49, 2.42) | 1.44 (0.75, 3.91)ab | 10.653 | 0.005** |
| PDGF‐BB (pg/ml) | 224.10 ± 111.83 | 231.00 ± 134.50 | 271.98 ± 129.62ab | 5.262 | 0.006** |
Note: The measurement data (Glu, TC, TG, HDLc, LDLc, WBC, Plt) were shown by mean ± SD, and those among control group, SAP group, and ACS group were analyzed by univariate ANOVA (*p < 0.05, **p < 0.01). The data of hsCRP level that did not match normal distribution were expressed as median and quartile M (P25, P75), that among three groups was analyzed by nonparametric Kruskal–Wallis univariate ANOVA test (*p < 0.05, **p < 0.01). The pairwise comparison was analyzed by q test (Δ compared with control group p < 0.05; Superscript letter a compared with control group, p < 0.01; Superscript letter b compared with SAP group, p < 0.01).
Abbreviations: ACS, acute coronary syndrome; CHD, coronary heart disease; Glu, Glucose; HDLc, high‐density lipoprotein cholesterol; hsCRP, high sensitivity C‐reactive protein; LDLc, low‐density lipoprotein cholesterol; PDGF‐BB, platelet‐derived growth factor‐BB; Plt, blood platelet; SAP, stable angina pectoria; TC, total cholesterol; TG, triglyceride; WBC, white blood cells.
3.3. SNP analysis of PDGF‐B
There was linkage disequilibrium between PDGF‐B gene SNP rs1800817 and rs2285094: D′ = 0.419, r 2 = 0.04. Only one case with CC homozygote was detected in the rs1800817 loci, and others were AA and AC genotypes. The allele frequency A 0.940, C 0.060, genotype frequency AA 0.884, AC 0.113, CC 0.003, χ 2 = 0.036, p = 0.850 match the Hardy–Weinberg equilibrium. There was no statistical difference in rs1800817 loci genotype or allele frequency between CHD group and control group, or among control group, SAP group, and ACS group (p > 0.05). rs2285095 loci allele frequency T 0.782, C 0.218, genotype frequency TT 0.603, TC 0.358, CC 0.039, χ 2 = 0.869, p = 0.351 match the Hardy–Weinberg equilibrium. There was no statistical differences in the genotype and allele frequency between CHD group and control group, or among control group, SAP group, and ACS group (p > 0.05) (Tables 5 and 6).
TABLE 5.
Genotype and allele frequency comparison between control group and CHD group
| SNPs | Genotype/allele | Control group (n = 88) | CHD group (n = 247) | χ 2 | p |
|---|---|---|---|---|---|
| rs1800817 | AA | 81 (92.0) | 215 (87.0) | 0.249 | 0.143 |
| AC + CC | 7 (8.0) | 32 (13.0) | |||
| A | 169 (0.960) | 461 (0.933) | 1.702 | 0.266 | |
| C | 7 (0.040) | 33 (0.067) | |||
| rs2285094 | TT | 54 (61.4) | 148 (59.9) | 1.294 | 0.524 |
| TC | 29 (33.0) | 91 (36.8) | |||
| CC | 5 (5.7) | 8 (3.2) | |||
| T | 137 (0.778) | 387 (0.783) | 0.016 | 0.916 | |
| C | 39 (0.222) | 107 (0.217) |
Note: Genotype/allele data were shown by n (%), and the comparison between groups was analyzed by χ 2 test.
Abbreviations: CHD, coronary heart disease; SNP, single nucleotide polymorphism.
TABLE 6.
Genotype and allele frequency comparison among control group, SAP group, and ACS group
| SNPs | Genotype/allele | Control group (n = 86) | SAP group (n = 82) | ACS group (n = 165) | χ 2 | p |
|---|---|---|---|---|---|---|
| rs1800817 | AA | 81 (92.0) | 71 (86.6) | 144 (87.3) | 1.603 | 0.449 |
| AC + CC | 7 (8.0) | 11 (13.4) | 21 (12.7) | |||
| A | 169 (0.960) | 153 (0.933) | 308 (0.933) | 1.689 | 0.430 | |
| C | 7 (0.040) | 11 (0.067) | 22 (0.067) | |||
| rs2285094 | TT | 54 (61.4) | 53 (64.6) | 95 (57.6) | 2.474 | 0.649 |
| TC | 29 (33.0) | 27 (32.9) | 64 (38.8) | |||
| CC | 5 (5.7) | 2 (2.4) | 6 (3.6) | |||
| T | 137 (0.778) | 133 (0.811) | 254 (0.770) | 1.114 | 0.573 | |
| C | 39 (0.222) | 31 (0.189) | 59 (0.230) |
Note: Genotype/allele data were shown by n (%), and the comparison between groups was analyzed by χ 2 test.
Abbreviations: ACS, acute coronary syndrome; CHD, coronary heart disease; SAP, stable angina pectoria; SNP, single nucleotide polymorphism.
3.4. Correlation between PDGF‐B genotype and plasma PDGF‐BB level
There was no statistical difference in the plasma PDGF‐BB level in three different genotypes on both SNP rs1800817 and rs2285094 among all the subjects (p > 0.05), as shown in Table 7.
TABLE 7.
Comparison of PDGF‐BB levels of different genotypes
| SNPs | Genotype | n | PDGF‐BB (pg/ml) | t/F | p |
|---|---|---|---|---|---|
| rs1800817 | AA | 296 | 248.98 ± 126.89 | −0.151 | 0.880 |
| AC + CC | 39 | 252.28 ± 138.24 | |||
| rs2285094 | TT | 202 | 249.10 ± 125.35 | 0.008 | 0.992 |
| TC | 120 | 250.19 ± 136.58 | |||
| CC | 13 | 245.81 ± 90.55 |
Note: PDGF‐BB level was shown by mean ± SD, and the comparison between genotype AA group and AC + CC genotype was analyzed by t test, and that among genotype TT, TC, CC groups was analyzed by univariate ANOVA.
Abbreviations: PDGF‐BB, platelet‐derived growth factor‐BB; SNP, single nucleotide polymorphism.
The plasma PDGF‐BB level of the patients with AA genotype at rs1800817 loci in CHD group was significantly higher than that of control group (p < 0.05). There was no statistical difference in AC + CC genotype between CHD group and control group (p > 0.05). The plasma PDGF‐BB level in CHD group had no statistical difference between AA and AC + CC genotypes (p > 0.05). There is also no such statistical difference in control group (p > 0.05). Because the sample size of AC and CC genotypes in rs1800817 loci was small, the PDGF‐BB levels in control group, SAP group, and ACS group were not compared, as shown in Table 8.
TABLE 8.
Comparison of plasma PDGF‐BB levels of different genotypes between control group and CHD group
| SNPs | Genotype | Control group | CHD group | t | p | ||
|---|---|---|---|---|---|---|---|
| n | PDGF‐BB (pg/ml) | n | PDGF‐BB (pg/ml) | ||||
| rs1800817 | AA | 81 | 220.02 ± 105.72 | 215 | 259.89 ± 132.60 | −2.430 | 0.016* |
| AC + CC | 7 | 271.27 ± 171.76 | 32 | 248.13 ± 132.73 | 0.397 | 0.694 | |
| T | −1.166 | 0.468 | |||||
| p | 0.247 | 0.640 | |||||
| rs2285094 | TT | 54 | 231.92 ± 129.79 | 148 | 255.37 ± 123.55 | −1.178 | 0.240 |
| TC | 29 | 215.09 ± 79.30 | 91 | 261.37 ± 148.94 | −1.600 | 0.112 | |
| CC | 5 | 191.81 ± 45.43 | 8 | 279.56 ± 97.42 | −1.868 | 0.089 | |
| F | 0.429 | 0.163 | |||||
| p | 0.653 | 0.850 | |||||
Note: PDGF‐BB level was shown by mean ± SD, and that between control group and CHD group of different genotypes was analyzed by t test (*p < 0.05). In control group or CHD group, the comparison between genotype AA group and genotype AC + CC group was analyzed by t test, and that among genotype TT, TC, CC groups was analyzed by univariate ANOVA (*p < 0.05).
Abbreviations: CHD, coronary heart disease; PDGF‐BB, platelet‐derived growth factor‐BB; SNP, single nucleotide polymorphism.
There is no statistical difference of plasma PDGF‐BB level of different genotypes at rs2285094 between control group and CHD group, or within either CHD group or control group (p > 0.05). Among control group, SAP group, and ACS group, the plasma PDGF‐BB level of TT genotype at rs2285094 showed the trend of ACS group > SAP group > control group, which did not achieve statistical difference (p > 0.05). However, the CC genotype sample size was small, we combined TC genotype with CC genotype. There was statistical difference of plasma PDGF‐BB level of TC + CC genotypes at rs2285094 among control group, SAP group, and ACS group. The plasma PDGF‐BB level of ACS group is higher than SAP group and control group. There was no statistical significance between SAP group and control group (p > 0.05). The plasma level of different genotypes within each group was not statistically different (p > 0.05) as shown in Tables 8 and 9.
TABLE 9.
Comparison of PDGF‐BB levels of different genotypes in rs2285094 among control group, SAP group and ACS group
| Genotype | Control group | SAP group | ACS group | F | p | |||
|---|---|---|---|---|---|---|---|---|
| n | PDGF‐BB (pg/ml) | n | PDGF‐BB (pg/ml) | n | PDGF‐BB (pg/ml) | |||
| TT | 54 | 231.92 ± 129.79 | 53 | 235.99 ± 132.29 | 95 | 266.18 ± 117.73 | 1.690 | 0.187 |
| TC + CC | 34 | 211.67 ± 75.20 | 29 | 221.81 ± 140.33 | 70 | 279.84 ± 144.71Δ$ | 4.030 | 0.020* |
| t | 0.826 | 0.454 | −0.668 | |||||
| p | 0.411 | 0.651 | 0.505 | |||||
Note: PDGF‐BB level was shown by mean ± SD, and that among control group, SAP group, and ACS group of different genotypes were analyzed by univariate ANOVA (*p < 0.05). The pairwise comparison was analyzed by q test (Δcompared with control group p < 0.05; $compared with SAP group, p < 0.05). The comparison of PDGF‐BB level between TT and TC + CC genotypes was analyzed by t test (*p < 0.05).
Abbreviations: ACS, acute coronary syndrome; CHD, coronary heart disease; PDGF‐BB, platelet‐derived growth factor‐BB; SAP, stable angina pectoria.
3.5. Logistic regression analysis
Coronary heart disease onset was taken as a dependent variable, and age, gender, BMI, hypertension history, diabetes history, CHD family history, smoking history, Glu, blood fat levels (TG, LDLc, HDLc), WBC, Plt, hsCRP, PDGF‐BB, genotypes of rs1800817 loci and rs2285094 loci were taken as independent variables. The result of binary logistic regression analysis indicated that plasma PDGF‐BB level is the risk factor of CHD onset (OR = 1.003, 95% CI 1.001 ⁓ 1.006, p = 0.014).
4. DISCUSSION
Platelet‐derived growth factor‐BB plays very important roles in various physiological and pathological processes, such as trauma tissue repair, onset and treatment of malignant tumors, formation of pulmonary arterial hypertension, etc. 10 , 11 , 12 Leitzel 13 reported that the normal plasma PDGF‐BB level was 0.32 ± 0.14 ng/ml (0.10–0.69 ng/ml), and the level of 15% of 131 cancer patients was significantly increased >0.69 ng/ml. The level was not related to age or gender in both cancer patients and normal people. The plasma PDGF‐BB level was proven to have positive correlation with diabetic foot syndromes, pulmonary fibrosis, etc.; it has negative correlation with hepatic fibrosis caused by hepatitis B. 14 , 15 , 16 The study of relationship between the plasma PDGF‐BB level and AS or CHD was few reported.
There was no statistical difference in hypertension, diabetes, blood fat level between CHD group and control group in our study. Our results showed that PDGF‐BB level and hsCRP level in the CHD group were significantly higher than the control group. AS plaque fibrous cap thinning, rupture, and thrombosis are considered as the main pathological basis for instability of coronary artery disease, as well as the major reasons for ACS occurrence. According to the stability of AS plaque, the CHD group was further divided into ACS and SAP groups. The PDGF‐BB level and hsCRP level among the control group, SAP group, and ACS group had statistical differences. The levels in ACS group were significantly higher than those in SAP group, but there was no statistical difference between SAP group and control group. PDGF‐BB level was positive correlated with hsCRP level. The results indicated that high plasma PDGF‐BB level was related to CHD onset and instability condition, and the change of plasma PDGF‐BB level was related to inflammatory response.
Ho et al. 17 found that SMC treated by CRP was more sensitive to PDGF‐BB, suggesting inflammatory response could improve the effects of PDGF‐BB. After inflammation and mechanical injury caused arterial intimal injury, the coagulation mechanism will be activated. The thrombin stimulates large amount of PDGF‐BB release from platelet at the same time of activation of clotting cascade. 18 , 19 PDGF‐BB changes the SMC phenotype, turns contraction type into synthetic type that possesses synthesis, secretion, migration, and proliferation functions. It migrates from media to intima, and secretes lots of extracellular matrix, protease, various cytokines, which is beneficial to vessel remodeling and AS plaque formation and stability. 20 , 21 , 22 Cui et al. 23 carried out exogenous PDGF‐BB transfected with plasmid to rat acute myocardial infarction (AMI) model. After infarction, at Day 28, neonatal vessels with mature tissue structure were formed in the exogenous PDGF‐BB group, and the infraction area shrunk. Compared with the control group, LVDd and LVDs significantly reduced, and LVEF significantly increased, suggesting PDGF‐BB played a very important role in post‐infraction myocardium repairing and cardiac function improvement. Therefore, increase in plasma PDGF‐BB level in the ACS patients might be the manifestation of body repair after AS plaque instability and fracture.
Results of many clinical investigations supported above opinions. Koizumi et al. 24 reported that the plasma PDGF‐BB level in coronary circulation increased in the AMI patients receiving emergent PCI, suggesting PDGF‐BB participated in the instability process of AS plaque, and the increase of the level was the manifestation during AMI acute phase. Nakagawa et al. 25 found that in the carcass after ACS stenting, during Day 24 and 55, the PFGF‐B was highly expressed in both the macrophages accumulated under neonatal intima and highly differentiated SMC. At Month 6, the expression significantly decreased and could not be detected in the neonatal intima after 1–5 years, suggesting that PDGF‐B was related to the intima repair after PCI surgery within time limit. However, Rattik et al. 26 found that the high PDGF‐BB level was related to stable plaque during the investigation on 202 patients receiving carotid endarterectomy, 384 patients with coronary artery events, and 409 control patients. Yeboah et al. 27 believed that low plasma PDGF‐BB level was the risk factor of cardiovascular diseases. The investigation on patients with Type II diabetes and CHD indicated that the plasma PDGF‐BB level is higher in diabetes patients without cardiovascular disease history than those with cardiovascular disease.
Up to now, the reported polymorphic sites of PDGF‐B gene SNP mainly included rs1800817, rs1800818, rs2285094, rs2040399, rs2267406, rs2285097, rs130650. SNP rs1800817 (+1135C/A) is located in the first intron, and rs2285094 is located in the intron near the mRNA splice site. 7 Ben‐Ari et al. 28 found in post‐orthotopic liver transplantation patients with PDGF‐B gene SNP rs1800817 AA genotype, the hepatic C recurrence rate was high; liver fibrosis tends to be more serious. While all these were not related to SNP rs1800818. Bicanski et al. 29 investigated IgA nephropathy patients in Germany and Italy and found that four SNP polymorphism sites of PDGF‐B gene (rs2285094, rs2040399, rs2267406, rs2285097), allele distributions matched the Hardy–Weingerg linkage equilibrium, and each SNP genotype distribution was not related to disease onset and severity. Anat et al. 30 studied 70 patients with heart transplantation and found that patients with GG genotype at PDGF‐B gene SNP rs1800818 had lower cardiac allograft vasculopathy (CAV) rate, longer survival time, and lower fatality rate. The occurrence risk of CAV in AA patients was higher, suggesting GG genotype might be the protective factor. The occurrence risk of CAV in patients with rs1800817 CC genotype was relatively low, and CC genotype was the protective factor. Osadnik et al. 27 followed up 265 patients with stable CHD implanted with bare metal stent and found that late lumen loss of patients with PDGF‐B gene rs2285094 CC genotype was larger than those with CT and TT genotypes. The results of Gensini score indicated that the genotype of rs2285094 was not related to coronary artery disease severity.
The PDGF‐B gene SNP rs1800817 and rs2285094 genotypes and allele frequency distribution match the Hardy–Weinberg linkage equilibrium. The detection rate of CC homozygote in rs1800817 was low, and only one patient of CC genotype was detected in CHD group (CC and AC genotypes were combined to perform the statistical analysis). This was similar with our results of investigation on the patients with restenosis after stenting. No CC homozygous genotype was detected, and there was no statistical difference in AA and AC genotype distribution between the instent restenosis group and non‐instent restenosis group. 31 The genotypes of rs1800817 and rs2285094 had no statistical significance among the control group, SAP group, and ACS group. The haplotype analysis indicated that SNP rs1800818 and rs2285094 presented weak linkage disequilibrium, but it was not related to coronary heart disease onset.
Up to now, there is no relevant report found on the correlation between PDGF‐B gene polymorphism and plasma PDGF‐BB level, as well as the correlation between CHD onset and instability condition. To compare different genotype plasma levels in PDGF‐B gene SNP rs1800817 and rs2285094, we found that there was no difference in PDGF‐BB level among different genotypes in rs2285094. The level of TC + CC of CHD group was significantly higher than control group (because the number of cases with CC genotype was small, the cases with TC and CC types were combined for analysis). There was no statistical significance in the plasma PDGF‐BB level of TT genotype between the two groups. The PDGF‐BB level of TC + CC genotype had statistical difference among the control group, SAP group, and ACS group. The level in the ACS group was significantly higher than those in the SAP group and control group, but there was no statistical difference between the latter two groups. For the TT genotype, the level in the ACS group was higher than the other two groups but without statistical difference. Moreover, there was no statistical difference in the level between the different genotypes in each group. The above results suggested that plasma PDGF‐BB level had no correlation with rs2285094 genotype, but correlated with different types of coronary heart disease. High‐plasma PDGF‐BB level might be related to the instability of coronary lesions.
In rs1800817 loci, the plasma PDGF‐BB level between AA and AC + CC genotype had no statistical difference. The plasma PDGF‐BB level in the AA genotype of CHD group was significantly higher than that of the control group. The level of AC + CC genotype between CHD and control group had no statistical difference. The level of different genotypes in both the CHD group and control group had no statistical difference. Because there were only seven patients with AC + CC genotype in the control group, and 32 in the CHD group, we did not compared the plasma levels of different genotypes among the three groups. The above results indicated that plasma PDGF‐BB level was not related to rs1800817 genotype.
The CHD onset was taken as a dependent variable, and gender, age, BMI, hypertension history, diabetes history, CHD family history, smoking history, Glu, blood fat levels (TG, LDLc, HDLc), WBC, Plt, hsCRP, PDGF‐BB levels, PDGF‐B genes rs1800817 and rs2285094 were taken as the independent variables. The result of binary logistic regression analysis indicated that plasma PDGF‐BB level was the risk factor of CHD onset.
5. CONCLUSION
The study indicated that high plasma PDGF‐BB level was the risk factor for CHD onset, and the manifestation of coronary artery lesion instability. PDGF‐B gene rs1800817 and rs2285094 polymorphisms were not correlated with the plasma PDGF‐BB level, CHD onset, and coronary artery lesion instability. Up to now, the correlation between plasma PDGF‐BB level and CHD is still controversial. The causality between the PDGF‐BB and occurrence of CHD and ACS is still unclear, nor is the mechanism, which need further exploration.
FUNDING INFORMATION
This article was funded by Tianjin Key Medical Discipline (Specialty) Construction Project.
CONFLICT OF INTEREST
Not reported.
Lu Y, Liu H, Dong B, Yang J, Kou L, Qin Q. Correlation between platelet‐derived growth factor‐B gene polymorphism and coronary heart disease. J Clin Lab Anal. 2022;36:e24683. doi: 10.1002/jcla.24683
Yaru Lu and Hui Liu contributed equally to this article.
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are openly available in https://pan.baidu.com/s/1_v9NlNi5NLKQ3C_wLwiLNg, password: en1h.
REFERENCES
- 1. Xiao‐Ying L, Lin W, Pu‐Lin Y, et al. Present situation on therapy and secondary prevention of coronary heart disease in the elderly. Chin J Geriatr. 2012;31(10):909‐914. [Google Scholar]
- 2. Li J, Li X, Wang Q, et al. ST‐segment elevation myocardial infarction in China from 2001 to 2011 (the China PEACE‐Retrospective Acute Myocardial Infarction Study): a retrospective analysis of hospital data. Lancet. 2015;385:441‐451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Erbilgin A, Civelek M, Romanoski CE, et al. Identification of CAD candidate genes in GWAS loci and their expression in vascular cells. J Lipid Res. 2013;54:1894‐1905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Martins M, Warren S, Kimberley C, et al. Activity of PLCε contributes to chemotaxis of fibroblasts towards PDGF. J Cell Sci. 2012;125(Pt 23):5758‐5769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Zhang ZW, Yanamoto H, Nagata I, et al. Platelet‐derived growth factor‐induced severe and chronic vasoconstriction of cerebral arteries: proposed growth factor explanation of cerebral vasospasm. Neurosurgery. 2010;66(4):728‐735. [DOI] [PubMed] [Google Scholar]
- 6. Malabanan KP, Sheahan AV, Khachigian LM. Platelet‐derived growth factor‐BB mediates cell migration through induction of activating transcription factor 4 and tenascin‐C. Am J Pathol. 2012;180(6):2590‐2597. [DOI] [PubMed] [Google Scholar]
- 7. Osadnik T, Strzelczyk JK, Lekston A, et al. The association of functional polymorphisms in genes encoding growth factors for endothelial cells and smooth muscle cells with the severity of coronary artery disease. BMC Cardiovasc Disord. 2016;16(1):218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Judkins MP. Selective coronary arteriography. A percutaneous transfemoral technic. Radiology. 1967;89(5):815‐824. [DOI] [PubMed] [Google Scholar]
- 9. Makki N, Brennan TM, Girotra S. Acute coronary syndrome. J Intensive Care Med. 2015;30(4):186‐200. [DOI] [PubMed] [Google Scholar]
- 10. Friedlaender GE, Lin S, Solchaga LA, Snel LB, Lynch SE. The role of recombinant human platelet‐derived growth factor‐BB (rhPDGF‐BB) in orthopaedic bone repair and regeneration. Curr Pharm Des. 2013;19(19):3384‐3390. [DOI] [PubMed] [Google Scholar]
- 11. Hosaka K, Yang Y, Seki T, et al. Tumour PDGF‐BB expression levels determine dual effects of anti‐PDGF drugs on vascular remodelling and metastasis. Nat Commun. 2013;4:2129. [DOI] [PubMed] [Google Scholar]
- 12. Takahashi J, Orcholski M, Yuan K, de Jesus Perez V. PDGF‐dependent β‐catenin activation is associated with abnormal pulmonary artery smooth muscle cell proliferation in pulmonary arterial hypertension. FEBS Lett. 2016;590(1):101‐109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Leitzel K, Bryce W, Tomita J, et al. Elevated plasma platelet‐derived growth factor B‐chain levels in cancer patients. Cancer Res. 1991;51(16):4149‐4154. [PubMed] [Google Scholar]
- 14. Drela E, Kulwas A, Jundziłł W, et al. VEGF‐A and PDGF‐BB—angiogenic factors and the stage of diabetic foot syndrome advancement. Endokrynol Pol. 2014;65(4):306‐312. [DOI] [PubMed] [Google Scholar]
- 15. Ziora D, Jastrzebski D, Adamek M, et al. Circulating concentration of markers of angiogenic activity in patients with sarcoidosis and idiopathic pulmonary fibrosis. BMC Pulm Med. 2015;15:113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Zhou J, Deng Y, Yan L, Zhao H, Wang G, China HepB‐Related Fibrosis Assessment Research Group . Serum platelet‐derived growth factor BB levels: a potential biomarker for the assessment of liver fibrosis in patients with chronic hepatitis B. Int J Infect Dis. 2016;49:94‐99. [DOI] [PubMed] [Google Scholar]
- 17. Ho KJ, Owens CD, Longo T, Sui XX, Ifantides C, Conte MS. C‐reactive protein and vein graft disease: evidence for a direct effect on smooth muscle cell phenotype via modulation of PDGF receptor‐β. Am J Physiol Heart Circ Physiol. 2008;295(3):H1132‐H1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Cha BY, Shi WL, Yonezawa T, Teruya T, Nagai K, Woo JT. An inhibitory effect of chrysoeriol on platelet‐derived growth factor (PDGF)‐induced proliferation and PDGF receptor signaling in human aortic smooth muscle cells. J Pharmacol Sci. 2009;110(1):105‐110. [DOI] [PubMed] [Google Scholar]
- 19. Qi YX, Jiang J, Jiang XH, et al. PDGF‐BB and TGF‐{beta} 1 on cross‐talk between endothelial and smooth muscle cells in vascular remodeling induced by low shearstress. Proc Natl Acad Sci U S A. 2011;108(5):1908‐1913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Chen S, Liu B, Kong D, et al. Atorvastatin calcium inhibits phenotypic modulation of PDGF‐BB‐induced VSMCs via down‐regulation the Akt signaling pathway. PLoS One. 2015;10(4):e0122577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Dong LH, Wen JK, Liu G, et al. Blockade of the Ras‐extracellular signal‐regulated kinase 1/2 pathway is involved in smooth muscle 22 alpha‐mediated suppression of vascular smooth muscle cell proliferation and neointima hyperplasia. Arterioscler Thromb Vasc Biol. 2010;30(4):683‐691. [DOI] [PubMed] [Google Scholar]
- 22. Alexander MR, Moehle CW, Johnson JL, et al. Genetic inactivation of IL‐1 signaling enhances atherosclerotic plaque instability and reduces outward vessel remodeling in advanced atherosclerosis in mice. J Clin Invest. 2012;122(1):70‐79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Cui K, Zhou X, Luo J, et al. Dual gene transfer of bFGF and PDGF in a single plasmid for the treatment of myocardial infarction. Exp Ther Med. 2014;7(3):691‐696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Koizumi T, Komiyama N, Nishimura S. In‐vivo higher plasma levels of platelet‐derived growth factor and matrix metalloproteinase‐9 in coronary artery at the very onset of myocardial infarction with ST‐segment elevation. Ann Vasc Dis. 2015;8(4):297‐301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Nakagawa M, Naruko T, Ikura Y, et al. A decline in platelet activation and inflammatory cell infiltration is associated with the phenotypic redifferentiation of neointimal smooth muscle cells after bare‐metal stent implantation in acute coronary syndrome. J Atheroscler Thromb. 2010;17(7):675‐687. [DOI] [PubMed] [Google Scholar]
- 26. Rattik S, Wigren M, Björkbacka H, et al. High plasma levels of heparin‐binding epidermal growth factor are associated with more stable plaque phenotype and reduced incidence of coronary events. Arterioscler Thromb Vasc Biol. 2015;35(1):222‐228. [DOI] [PubMed] [Google Scholar]
- 27. Yeboah J, Sane DC, Crouse JR, Herrington DM, Bowden DW. Low plasma levels of FGF‐2 and PDGF‐BB are associated with cardiovascular events in type II diabetes mellitus (diabetes heart study). Dis Markers. 2007;23(3):173‐178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ben‐Ari Z, Tambur AR, Pappo O, et al. Platelet‐derived growth factor gene polymorphism in recurrent hepatitis C infection after liver transplantation. Transplantation. 2006;81(3):392‐397. [DOI] [PubMed] [Google Scholar]
- 29. Bicanski B, Wenderdel M, Mertens PR, et al. PDGF‐B gene single‐nucleotide polymorphisms are not predictive for disease onset or progression of IgA nephropathy. Clin Nephrol. 2007;67(2):65‐72. [DOI] [PubMed] [Google Scholar]
- 30. Anat R, Tambur D, Pamboukian S, et al. Genetic polymorphism in platelet‐derived growth factor and vascular endothelial growth factor are significantly associated with cardiac allograft vasculopathy. J Heart Lung Transplant. 2006;25(6):690‐698. [DOI] [PubMed] [Google Scholar]
- 31. Lu Yaru Y, Zhen LG. Association between platelet‐derived growth factor‐B and in‐stent restenosis in elderly patients. Chin J Geriatr. 2014;33(2):138‐142. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data that support the findings of this study are openly available in https://pan.baidu.com/s/1_v9NlNi5NLKQ3C_wLwiLNg, password: en1h.


