Abstract
How do separate sexes originate and evolve? Plants provide many opportunities to address this question as they have diverse mating systems and separate sexes (dioecy) that evolved many times independently. The classic “two-factor” model for evolution of separate sexes proposes that males and females can evolve from hermaphrodites via the spread of male and female sterility mutations that turn hermaphrodites into females and males, respectively. This widely accepted model was inspired by early genetic work in dioecious white campion (Silene latifolia) that revealed the presence of two sex-determining factors on the Y-chromosome, though the actual genes remained unknown. Here, we report identification and functional analysis of the putative sex-determining gene in S. latifolia, corresponding to the gynoecium suppression factor (GSF). We demonstrate that GSF likely corresponds to a Y-linked CLV3-like gene that is specifically expressed in early male flower buds and encodes the protein that suppresses gynoecium development in S. latifolia. Interestingly, GSFY has a dysfunctional X-linked homolog (GSFX) and their synonymous divergence (dS = 17.9%) is consistent with the age of sex chromosomes in this species. We propose that female development in S. latifolia is controlled via the WUSCHEL-CLAVATA feedback loop, with the X-linked WUSCHEL-like and Y-linked CLV3-like genes, respectively. Evolution of dioecy in the S. latifolia ancestor likely involved inclusion of ancestral GSFY into the nonrecombining region on the nascent Y-chromosome and GSFX loss of function, which resulted in disbalance of the WUSCHEL-CLAVATA feedback loop between the sexes and ensured gynoecium suppression in males.
Keywords: sex determination, dioecy evolution, gynoecium suppression, white campion, CLV3
Introduction
Most animal species are comprised of separate male and female individuals, which raise the question: how do separate sexes originate and evolve? Plants provide an excellent opportunity to address this question as separate sexes (dioecy) evolved many times independently and quite recently in different genera (Renner 2014; Charlesworth 2019). Evolution of separate sexes and the diversity of plant mating systems stimulated significant research efforts to understand the drivers of mating system evolution in plants (Charlesworth and Charlesworth 1978; Lloyd 1979; Renner and Ricklefs 1995; Barrett 2010; Yakimowski and Barrett 2016). The evolution of dioecy from hermaphroditism provides self-pollination avoidance mechanisms and helps to balance the investments into male and female functions (Charlesworth and Charlesworth 1981; Aonuma et al. 2021). It is thought that there are two main pathways to evolve dioecy from the ancestral hermaphroditic state: (1) via gynodioecy, when a population consists of females and hermaphrodites and (2) via monoecy, when a plant has separate male and female flowers on the same individual (Lloyd 1980; Dorken and Barrett 2004). The monoecy scenario is thought to be limited to monoecious plants (Lloyd 1980; Renner and Ricklefs 1995; Renner 2016), where flowers are already unisexual, though the relative importance of these two pathways remains unclear (Bawa 1980; Renner and Ricklefs 1995; Weiblen et al. 2000; Spigler and Ashman 2012; Renner 2016). This paper focuses on the gynodioecy pathway that is thought to have been followed by many of the dioecious plants (Dufaÿ et al. 2014).
Many of the species where sex determination and/or sex chromosomes were studied belong to primarily dioecious clades where initial transition to dioecy likely occurred a very long time ago. For example, persimmon (Akagi et al. 2014), papaya (Aryal and Ming 2014), hop (Skof et al. 2012), Cannabis (Van Bakel et al. 2011), Coccinia (Sousa et al. 2016), Mercurialis (Obbard et al. 2006), Populus (Geraldes et al. 2015) all belong to ancestrally dioecious clades (Renner 2014). Although some species in such ancestrally dioecious plant groups may reverse to a cosexual state and reevolve dioecy secondarily, such secondary transitions to dioecy may mis-inform us about the processes accompanying initial evolution of separate sexes. For example, the genomes of nondioecious species in ancestrally dioecious plant genera may be “preadapted” to sexual dimorphism, with regulatory pathways for sexually dimorphic gene expression already in place, minimizing sexual conflict upon secondary transition to dioecy. Silene (Bergero et al. 2015; Papadopulos et al. 2015; Kazama et al. 2016; Qiu et al. 2016) and Rumex (Hough et al. 2014) represent good examples of plant genera with relatively recently evolved derived dioecy, but the molecular bases of sex determination have not been dissected in these genera.
The evolution of dioecy via gynodioecy is thought to involve the spread of at least two separate mutations—a male and a female sterility mutations (Charlesworth and Charlesworth 1978; Charlesworth 2019). The spread (but not fixation) of a male sterility mutation turns a cosexual population into a gynodioecious one (hermaphrodites + male-sterile females), whereas the spread of a female sterility mutation turns hermaphrodites into males and gynodioecy into dioecy. Then, suppression of recombination between male and female sterility mutations to prevent their presence in the same individual (which would be a neuter) is thought to give rise to sex chromosomes (Charlesworth 2015). Although most studied dioecious plants contain only small (∼1 Mb) sex-determining (SD) regions (Obbard et al. 2006; Akagi et al. 2014; Aryal and Ming 2014; Geraldes et al. 2015; Akagi et al. 2019; Veltsos et al. 2019), some species (Westergaard 1958; Charlesworth 2016), such as white campion (Silene latifolia), have evolved large cytologically distinguishable (heteromorphic) X- and Y-chromosomes similar to that in animals such as mammals or Drosophila. The lack of recombination on the sex-specific chromosome (Y- or W-chromosomes) leads to the loss of genes and the accumulation of transposable elements, resulting in genetic degeneration of Y- or W-chromosomes (Charlesworth and Charlesworth 2000, 2005; Bachtrog 2013). In accordance with this scenario, in S. latifolia, which has been estimated to have evolved dioecy and sex chromosomes ∼11 Ma (Krasovec et al. 2018), 45% of Y-linked genes are now not expressed, and 23% have premature stop codons (Papadopulos et al. 2015).
The classic “two-factors” (responsible for male and female sterility) scenario received a lot of attention in the literature (Weiblen et al. 2000; Charlesworth and David 2004; Dorken and Barrett 2004; Renner 2016) and was supported by studies in dioecious white campion (Westergaard 1958), asparagus (Harkess et al. 2020), and kiwi fruit (Akagi et al. 2018; Akagi et al. 2019; Müller et al. 2020; Xue et al. 2020), though the dissection of the molecular basis of sex determination in persimmon (Akagi et al. 2014), poplar (Müller et al. 2020; Xue et al. 2020), and melon (Boualem et al. 2015) appear to contradict the “two-factor” model (Ma and Pannell 2016; Renner 2016). The work on genetics of sex determination and sex chromosomes in S. latifolia has played central role in the development of ideas in this research field (Kejnovsky and Vyskot 2010; Charlesworth 2018). Classic cytogenetic and genetic work in S. latifolia has identified two regions on the Y-chromosome containing sex-determining genes: the gynoecium suppressing factor and the stamen promoting factor (Westergaard 1946; Farbos et al. 1999; Lardon et al. 1999). The third Y-linked region responsible for anther maturation was also identified (Westergaard 1946). The actual genes corresponding to these sex-determining factors remain unknown and the search for the sex-determining genes has been difficult due to the large size of the genome (3 Gb) and the Y-chromosome, which is estimated to be 570 Mb (Široký et al. 2001), with a large nonrecombining region (Bergero et al. 2013; Krasovec et al. 2020). This paper is the first to identify and describe a likely sex-determining gene in S. latifolia. We demonstrate that this gene corresponds to the Y-linked gynoecium suppression factor (GSF).
Results
A Sex-determining Gene GSF Encodes a CLAVATA3 Homolog
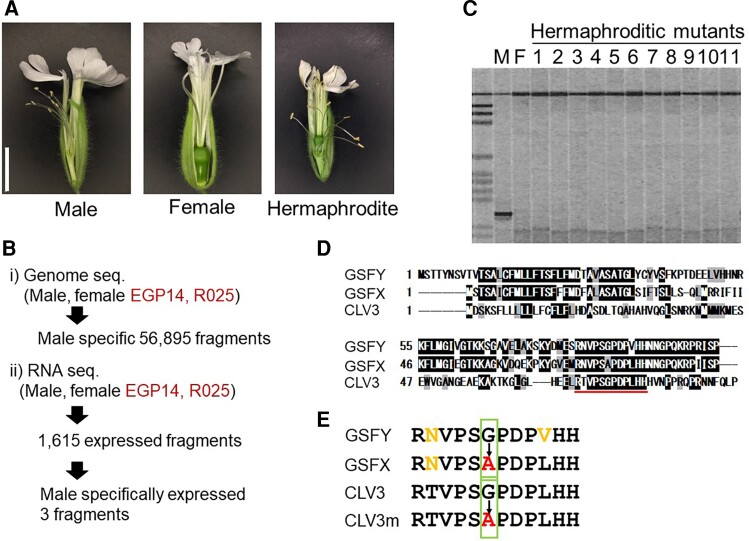
To find the GSF region on the S. latifolia Y-chromosome, we used previously generated (Papadopulos et al. 2015) Illumina genomic sequence data of a male and a female from the highly inbred S. latifolia K-line (Kazama et al. 2003), as well as hermaphroditic Y-deletion mutants (fig. 1A) EGP14 and R025 from that inbred line. These two mutants were previously characterized as having the smallest deletion around the GSF region on the Y-chromosome (Kazama et al. 2016). EGP14 genome has been sequenced previously (Krasovec et al. 2019), whereas R025 was sequenced as part of this study (accession nos. DRR357638 and DRR357639). These sequence data were analyzed by using the k-mer subtraction approach described by Akagi et al. (2014) to identify the sequence reads and assemble genomic contigs corresponding to the Y-specific region deleted in the EGP14 and R025 mutants (see Materials and Methods). Henceforth we will refer to this region as “Ydel.”
Fig. 1.
Isolation of S. latifolia GSF gene and its homology to A. thaliana CLV3 gene. (A) Photographs of flowers of the male, female, and hermaphroditic mutant S. latifolia plants. Bar 1 cm. (B) Scheme of the GSFY gene isolation. (C) Genomic PCR analysis of the GSF candidate in male, female, and 11 hermaphroditic mutants (EGP4, EGP5, EGP6, EGP8, EGP9, EGP10, EGP11, EGP12, EGP13, EGP14, and EGP15). All mutants showed no amplification by the candidate specific primers. (D) Deduced amino acid sequence homology between Y-linked GSF (GSFY), X-linked GSF (GSFX), and A. thaliana CLV3 genes. The conserved CLE domain is underlined. (E) Homology of deduced CLE domain sequences between GSFY, GSFX, CLV3, and its null allele (clv3-1 or clv3-5); CLV3m.
We used the contigs from the Ydel region to blast-search for homologous expressed genes in S. latifolia male transcriptomes published previously (Papadopulos et al. 2015) and newly assembled in this study with Trinity and Drap (Grabherr et al. 2011; Cabau et al. 2017) (accession nos. ICSN01000001–ICSN01003571 and ICSO01000001–ICSO01092457). This identified 1,615 Ydel region sequences that had high identity (e-value < 0.0000001) to the expressed fragments. Mapping RNA-seq reads to these 1,615 expressed Ydel region sequences using RSEM (Li and Dewey 2011) revealed that only three genic fragments (supplementary table S1, Supplementary Material online) were expressed in male flower buds, but not in the female or the deletion mutants (fig. 1B). Genomic polymerase chain reaction (PCR) in male, female, and 11 hermaphroditic mutants showed that only one of these fragments was male-specific (fig. 1C), indicating that this gene is a strong candidate for being the sex-determining gene. Blast-X search of this fragment showed homology to an Arabidopsis thaliana protein, CLAVATA3 (CLV3). The CLV3 gene is well known as a gene that controls the size of the shoot apical meristem and the flower meristem. In the A. thaliana clv3 mutant, not only the size of SAM but also the size of flower meristem was reduced (Fletcher et al. 1999). Therefore, we assumed that the CLV3 homolog was a strong candidate for the GSF gene in S. latifolia and we named it GSFY to denote its location on the Y-chromosome.
To find the homologs of the GSFY gene outside of the Y-chromosome we used blastn to search the female S. latifolia genome (Papadopulos et al. 2015), which revealed a homolog with 87% identity to the GSFY. This blast hit was located only 11 kb away from the gene “contig2443” genetically mapped to the X-chromosome in the previous study (Papadopulos et al. 2015). X-linkage of this genomic region was also confirmed using segregation analysis with genomic sequence data from S. latifolia parents and F1 progeny (Krasovec et al. 2018). Further blastn search against a female S. latifolia transcriptome (Krasovec et al. 2019) identified corresponding cDNA 288 nucleotides long. Henceforth, we will refer to this gene as GSFX. An alignment of the deduced amino acid sequence showed that both GSFY and GSFX were homologous to A. thaliana CLV3, especially in the conserved CLE domain (fig. 1D). The GSFX gene is likely dysfunctional because it contains the same mutation as the A. thaliana clv3-1 and clv3-5 mutants, which causes a single amino acid change (from Gly to Ala) in the CLE domain (fig. 1E). This amino acid change is sufficient to render CLV3 dysfunctional (Fletcher et al. 1999). On the other hand, this Gly residue was conserved in the CLE domains of the GSFY and CLV3 of A. thaliana. These results suggest that GSFY has a function in controlling the size of the flower meristem, but GSFX does not.
GSFY is Expressed in the L3 Layers of Small Flower Buds
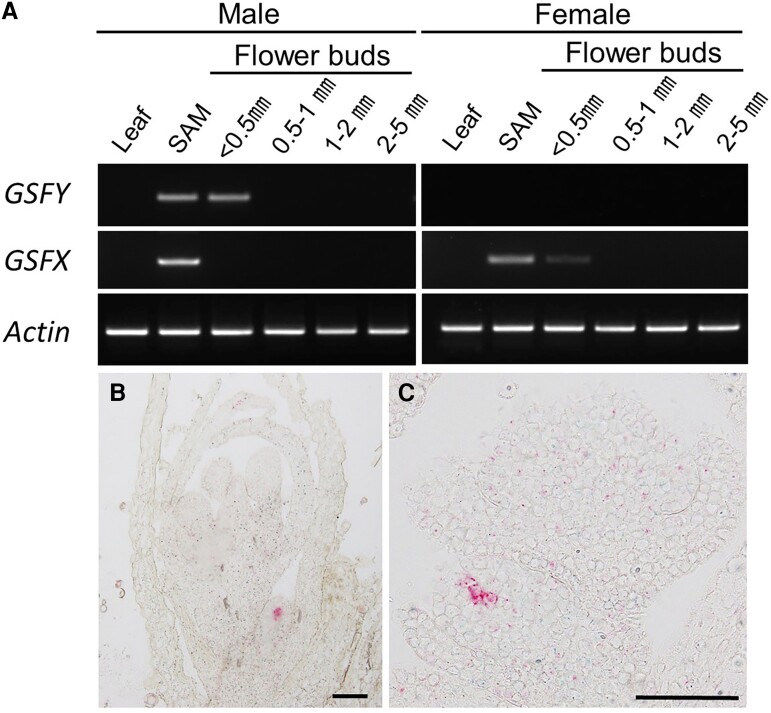
The expression of both GSFY and GSFX was examined by RT-PCR, which revealed that both GSFY and GSFX were expressed in shoot apical meristem, but only GSFY was expressed in small (<0.5 mm) flower buds at the early stages of their development (fig. 2A). To investigate the expression pattern of both genes, RNA in situ hybridization to tissues using BaseScope kit was performed on developmental flower buds (See Materials and Methods). No hybridization was detected for the GSFX probe, whereas for the GSFY probe hybridization signals were detected in the L3 layers of the small flower buds at stages 1–2 (fig. 2B and C). This pattern of GSFY hybridization is consistent with the function of this gene to suppress gynoecium development at early stages of development in male flowers. In A. thaliana, gynoecium primordia originate from L3 layers (Jenik and Irish 2000). Thus, GSFY product may work in L3 layers to repress gynoecium development in S. latifolia.
Fig. 2.
Expression of GSFY and GSFX genes. (A) RT-PCR for GSFY and GSFX on leaf, SAM, and flower buds at different developmental stages (lengths of flower buds are indicated). (B and C) In situ hybridization images for GSFY in developing flower buds. Red signals derived from GSFY probe were detected at stages 1–2 of the developing flower buds, but not at progressed stages. Bars, 100 μm.
GSFY is Functional But GSFX is Not
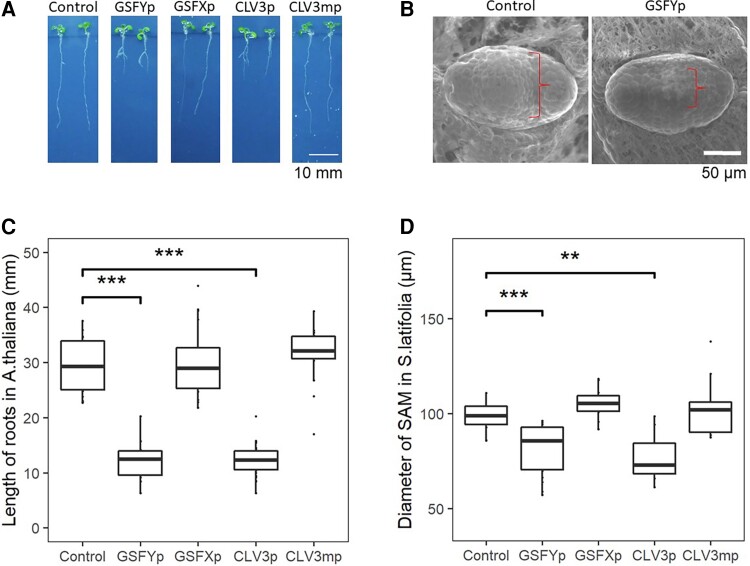
Arabidopsis thaliana CLV3 acts as a small peptide, so the function of the CLV3 variants can be analyzed by a bio assay in which Arabidopsis plants are treated with synthetic peptides (Fiers et al. 2005; Kondo et al. 2006). The peptides encoded by CLV3, the mutated variant of CLV3 found in clv3-1 and clv-3-5 mutants, GSFY, and GSFX were synthesized with replacements of proline residues by hydroxyproline residues, as described previously (Kondo et al. 2006). To investigate the activity of the peptides encoded by GSFY (GSFYp), GSFX (GSFXp), as well as the peptides encoded by CLV3 (CLV3p) and by the mutated CLV3 (CLV3mp), these peptides were synthesized and used to treat A. thaliana. Root elongation was significantly inhibited in the CLV3p and GSFYp treatments (P < 0.01, Wilcoxon rank-sum test), but not at all in the CLV3mp and GSFXp treatments (P > 0.05, Wilcoxon rank-sum test) (fig. 3A and C). The root length after CLV3mp and GSFXp treatments was comparable to that of the nontreated col-0 (fig. 3A). This tendency was also confirmed on the SAM in A. thaliana (supplementary fig. S1, Supplementary Material online). The function of these peptides was also tested in the SAM of S. latifolia. Similar to the bioassay in A. thaliana, the CLV3p and GSFYp treatments significantly decreased the diameter of the S. latifolia SAM in both sexes (P < 0.01, Wilcoxon rank-sum test), whereas the CLV3mp and GSFXp treatments showed almost no effect on the SAM size (P > 0.05, Wilcoxon rank-sum test) (fig. 3B and D). These results indicate that the function of GSFY is similar to CLV3, whereas GSFX gene is likely dysfunctional.
Fig. 3.
The bioassay of CLE peptides. (A and C) CLE peptide treatments on A. thaliana seedlings to investigate their effects on root growth. The roots were measured after 8 days of growth on solid 1/2 MS media with 0.1 μM of different peptides (GSFYp, GSFXp, CLV3p, and CLV3mp). (A) Photographs of two different plants per treatment. (C) Quantitative measurements of the root length (n = 18 for each treatment). GSFYp and CLV3p showed significant defect on the root growth (P < 0.01, Wilcoxon rank-sum test), whereas GSFXp and CLV3mp showed almost no effect. (B and D) CLE peptide treatments on S. latifolia seedlings to investigate their effects on SAM size. The SAMs were observed by SEM after 14 days of growth on liquid 1/2 MS media with 0.1 μM different peptides followed by calculation of SAM diameter by ImageJ. (B) Photographs of representative samples. (D) Quantitative measuring of the SAM diameter (n = 18 for each treatment). Similar to the A. thaliana experiment, GSFYp and CLV3p showed significant defects of the SAM size (P < 0.01, Wilcoxon rank-sum test), whereas GSFXp and CLV3mp showed almost no effect. Box plots, 25th–75th percentile; center line, median; whiskers, full data range in B and D.
GSFY Supresses Growth of Gynoecium
To test the gynoecium suppressing function of GSFY, we performed the GSFYp treatment on the flower buds of female and hermaphroditic mutants of S. latifolia. After twice dropping of 0.5-µM GSFYp on the developing female flower bud, a female flower having two styles with a small ovary opened (fig. 4). The number of carpels also decreased both in females and hermaphrodites (fig. 4 and supplementary fig. S2, Supplementary Material online). These phenotypes were not observed after the treatment of GSFXp.
Fig. 4.
The effect of GSFYp treatment on gynoecium development in S. latifolia female and hermaphroditic mutant plants. Flower buds at stages 1–3 were treated with water containing 5 μM GSFYp. Open flowers were observed under stereo microscopy. GSFYp treatment decreased the size of ovaries and the number of styles in both female and hermaphrodite mutants. Bars, 500 μm.
One of the ways to prove the function of GSFY in S. latifolia is transformation. As transformation of S. latifolia is problematic, we performed transformation of A. thaliana instead. The coding sequences of GSFY and GSFX under control of their native promoters were introduced into A. thaliana. The resulting transgenic line for GSFY showed inhibition of pistil growth and sterility in the T2 generation (fig. 5). Such a phenotype was not observed in the GSFX-transgenic plants (fig. 5). Taken together, we concluded that the Y-linked CLV3 homolog, the GSFY but not the GSFX has a gynoecium suppressing function.
Fig. 5.
Transformation of GSFY and GSFX in A. thaliana. GSFY and GSFX sequences under their native promoters (pGSFY and pGSFX) were introduced into the Col-0 plants. In the T2 generation, the pGSFY plants showed inhibition of the pistil growth and sterility, whereas the phenotype of the pGSFX-transgenic T2 plants was not affected.
GSFY Emerged When the Sex Chromosomes Evolved
Synonymous divergence between the GSFX and GSFY genes is dSx:y = 0.18, which is higher than the average dSx:y = 0.11 for the older stratum of S. latifolia sex chromosomes (Krasovec et al. 2018), but is similar to dSx:y ∼ 0.15–0.20 reported for some of the sex-lined genes in this species, such as SlX4/Y4 (Atanassov et al. 2001) and the genes in the upper range of the dSx:y distribution in the older stratum (Papadopulos et al. 2015). Assuming a mutation rate in S. latifolia is m = 7.31 × 10−9 per nucleotide per generation (Krasovec et al. 2018), dSx:y = 0.18 corresponds to divergence time between the GSFX and GSFY genes Tgenerations = dSx:y/2 m ∼ 12 million generations or Tyears ∼ 18 My, assuming 1.5 years per generation on average (Richards et al. 2003). This estimate of the time since the divergence between the GSFX and GSFY genes is close to the previously estimated upper limit for the age of sex chromosomes in this species (95% CI: 7.83–15.03 My) (Krasovec et al. 2018), which is consistent with the GSFY gene being the sex-determining gene that evolved its sex-determining function prior to or at the same time as the cessation of recombination in the older stratum of the S. latifolia sex chromosomes.
In order to establish the ancestral state of the GSF gene before divergence into the X- and Y-linked gametologs, we identified the homologs of this gene in a nondioecious outgroup—Silene uniflora. For this purpose, we blast-searched a draft assembly of the S. uniflora genome (Papadopulos et al. 2021) with the CDS of the GSFY gene and found two homologs with 93% identity to GSFY, on contigs JAGPOY010206798.1 and JAGPOY010140724.1. The sequences of the two S. uniflora homologs are very similar to each other with only three nucleotide differences (in an alignment 197 nucleotides long) and apparently represent allelic variation in a single gene or a recent duplication in S. uniflora. The predicted protein sequence of the S. uniflora genes in the conserved CLE domain is the same as that in the GSFY and A. thaliana CLV3, indicating that the Gly to Ala mutation in that domain in the GSFX is a derived state. If the GSFX were dysfunctional, we would expect this gene to accumulate amino acid replacements because purifying selection should not eliminate them in a dysfunctional gene. Indeed, GSFX contains 12 amino acid replacements specific to this gene, whereas GSFY contains only three such replacements (P = 0.02 with Tajima’s relative rate test for amino acids). Accordingly, the nonsynonymous to synonymous substitution rate ratio (dN/dS) is much higher for the S. latifolia GSFX (dN/dS = 0.38) compared with the GSFY (dN/dS = 0.08) and the homologs in S. uniflora (dN/dS = 0.09). Thus, multiple lines of evidence indicate that GSFX is dysfunctional and it must have lost its function at least a few million years ago to allow enough time to accumulate multiple amino acid replacements.
Discussion
Here, we report the identification and functional analysis of the gene likely responsible for the sex-determining gynoecium suppressing function (GSF) in S. latifolia—the dioecious plant species that inspired the development of the canonical “two-factor” model of dioecy evolution (Westergaard 1953, 1958; Charlesworth and Charlesworth 1978). This model includes the spread of a male and a female sterility mutations that lead to evolution of gynodioecy and dioecy, respectively, from the ancestral hermaphroditic state (Charlesworth 2015). The putative sex-determining gene, GSFY, identified in this study corresponds to the second step—the loss of female function caused by the GSF identified genetically many decades ago (Westergaard 1946, 1958). We report that the GSF likely corresponds to the Y-linked CLV3-like gene that is specifically expressed in early male flower buds and encodes the protein that suppresses gynoecium development in S. latifolia and A. thaliana.
According to the canonical two-factor model, the evolution of dioecy and XY sex chromosomes involves a recessive loss-of-function male sterility mutation on the proto-X and a dominant gain-of-function female sterility mutation on the proto-Y-chromosome (Charlesworth 2002). Thus, our finding of an X-linked loss-of-function mutation in GSFX may seem surprising and would be expected for the other sex-determining gene (SPF that remains to be identified in S. latifolia), rather than for the GSF studied in this paper. However, as argued below, the loss-of-function mutation in the GSFX gene may be an important step in dioecy evolution, contributing to the control of gynoecium suppression via the WUSCHEL-CLAVATA feedback loop (Schoof et al. 2000). Sex determination in plants is often unstable, condition-dependent and can be affected by hormone treatments (Khryanin 2002; Masuda et al. 2022). Contrary to this, sex determination in S. latifolia is very stable and completely genetically determined (Heslop-Harrison 1963), with plant hormone treatments having no effect on the sex expression (Ruddat et al. 1991). Recently, androhermaphrodite S. latifolia plants were reported to be produced by the treatment of chemicals affecting epigenetic states (Bacovsky et al. 2022). This is consistent with the fact that WUSCHEL-CLAVATA pathway is not affected by major plant hormones except for the stabilization of WUS proteins by cytokinins (Snipes et al. 2018), but controlled by epigenetic modifications (Cao et al. 2015; Shang et al. 2021).
Within the WUSCHEL-CLAVATA feedback loop, the CLV3 mRNA production is activated by WUS protein, whereas the expression of WUS is repressed by CLV3. Expression of CLV3 is also repressed by the WUS protein when its concentration is high (Perales et al. 2016; Plong et al. 2021). WUSCHEL plays a role in pistil growth and the A. thaliana wus mutant has a smaller floral meristem, frequently resulting in the absence of pistil (Laux et al. 1996). CLV3 inhibits expression of WUS and the A. thaliana clv3 mutant shows an enlarged pistil (Fletcher et al. 1999). In A. thaliana, the termination of floral meristem is induced by the cessation of WUS expression followed by shutting down of CLV3 expression, to produce gynoecium primordia (Laux et al. 1996; Mayer et al. 1998; Shang et al. 2021). Thus, shifting the balance between WUS and CLV3 in the termination of floral meristem, it is possible to ensure suppression of gynoecium development in males and active carpel development in females. Importantly, the GSFY-transgene in Arabidopsis plants specifically affected gynoecium suppression (fig. 5). This may be due to the change of expression pattern of GSFY; it was expressed at the L3 layer of the male floral meristems (fig. 2B and C), whereas CLV3 is expressed in the L1 and L2 layers of the Arabidopsis floral meristems (Brand et al. 2000). The loss of function mutation in GSFX also contributes to the masculinization of the male flower, limiting GSF expression to males and ensuring active carpel development in females. Reducing the number of copies of the WUSCHEL gene in males was likely another contribution to this balance shift. In particular, as reported previously (Kazama et al. 2012), the S. latifolia WUS homolog SlWUS1 is linked to the X chromosome but no Y-linked copy was found. This creates a bias in the number of the SlWUS1 gene copies between the sexes, with females having twice as many SlWUS1 compared with males. As SlWUS1 is not dosage compensated (Kazama et al. 2012), this copy number difference between the sexes likely leads to gynoecium enlargement in females. The results of GSFYp treatment on female and hermaphroditic plants support this hypothesis, as hermaphrodites (having one X chromosome) tended to show smaller gynoecium compared with XX-females that had two copies of SlWUS1 gene.
Gene duplication and their sub- or neofunctionalization are expected to play a significant role in evolution of carpel suppression in dioecious plants (Ohno 1967). Consistent with this concept, genes involved in the suppression of female function, ShyGirl in kiwifruits (Akagi et al. 2018; Akagi et al. 2019) and SOFF in Asparagus (Harkess et al. 2017), are thought to be derived from duplications of autosomal genes. Moreover, the sex-determining gene in persimmon, OGI, which is a small RNA repressing autosomal feminizing gene, MeGI, was also derived from a duplication (Akagi et al. 2014). However, our findings do not support the role of gene duplication in evolution of gynoecium suppression during dioecy evolution in S. latifolia. Identification of an X-linked homolog of the Y-linked carpel suppressor GSFY indicates that the gene ancestral to GSFX and GSFY was already present on the proto-sex chromosomes before the cessation of recombination between the X- and Y-linked alleles of that gene. Furthermore, it is likely that SlWUS1 was also present on the proto-sex chromosomes because the homolog of this gene in nondioecious Silene vulgaris is located on the same chromosome as other homologs of genes sex-linked in S. latifolia (Kazama et al. 2012). As no Y-linked copy of the X-linked SlWUS1 gene was found in S. latifolia (Kazama et al. 2012), this gene was likely lost from the Y-chromosome, whereas the X-linked copy was retained. The analysis of gene expression in males and females revealed no dosage compensation for SlWUS1 gene (Kazama et al. 2012). Thus, the loss of the Y-linked copy of SlWUS1 gene further contributed to the disbalance in WUSCHEL-CLAVATA feedback loop between males and females and thus it could have been an important step in dioecy evolution. Furthermore, given the well-documented functional role of CLAVATA3 and WUSCHEL in regulation of the sizes of shoot and floral meristems (Prunet et al. 2009; Somssich et al. 2016), this disbalance between sexes may play a key role in controlling the sexual dimorphism of this dioecious plant species, where male flowers are about three times smaller than female flowers and male inflorescences are bigger than female inflorescences (Gehring and Linhart 1993).
The estimated synonymous divergence between the GSFX and GSFY is consistent with the maximal estimate of the sex chromosomes age in S. latifolia (95% CI: 7.83–15.03 My) (Krasovec et al. 2018) and indicates that cessation of recombination between the X- and the Y-linked GSF copies has occurred at the very early stage of sex chromosome evolution in this species. It is possible that inclusion of the GSFY into the nonrecombining region on the nascent Y-chromosome (NRY) was nearly simultaneous with the mutation in the GSFX gene that prevents its carpel-suppressing function. We hypothesize that inclusion of the GSFY into the NRY and dysfunctionalization of the X-linked copy of that gene were the two key events that led to evolution of dioecy from gynodioecy in S. latifolia via the WUSCHEL-CLAVATA feedback loop, as described above. Although further studies will be necessary to prove the involvement of this mechanism in carpel suppression in males, our results open the door to investigate the involvement of the X chromosome in sex determination in S. latifolia.
Materials and Methods
Plant Material
The inbred S. latifolia line, the K line (Kazama et al. 2003), and its Y-deletion mutants EGP14, EGP15, and R025 (Kazama et al. 2016) were used in this study. Plants were grown in pots in a regulated chamber at 23°C in a 16 h light/8 h dark cycle. The leaves and flower buds were frozen in liquid nitrogen and stored at −80 °C prior to DNA and RNA extraction. The Columbia (Col-0) ecotype and the clv3-101 mutant of A. thaliana were used in peptide treatment assays. The clv3-101 mutant was originally obtained by the heavy-ion irradiation (Maeda et al. 2014) and kindly provided from Dr. Ali Ferjani of Tokyo Gakugei University. Arabidopsis thaliana seeds were grown in either soil or 0.7% (w/v) agar-containing Murashige-Skoog (MS) medium (Wako-junyaku) supplemented with 3% (w/v) sucrose and Gamborg’s B5 vitamins at 23°C on a 16 h light/8 h dark cycle.
Genomic Sequencing
To extract the candidate gene fragments responsible for the GSF function, we sequenced the genome of the deletion mutant R025 and a female plant. In addition, we used genome sequence data from nonirradiated plants of the inbred K-line and another deletion mutant EGP14 (Krasovec et al. 2019) as well as from parents and F1 progeny published previously (Krasovec et al. 2018).
DNA for genomic sequencing was extracted from fresh leaves using the DNA Plant Easy kit (Qiagen). For high-throughput sequencing, PCR-free Illumina libraries were prepared at Beijing Genomics Institute (BGI, Shenzhen, China). These libraries were sequenced on HiSeq4000 instrument at BGI. An additional PCR-free Illumina library of genomic DNA from R025 was independently prepared at RIKEN, and sequenced on HiSeq2500 instrument at RIKEN. All sequence data generated are listed in supplementary table S2, Supplementary Material online.
Male-specific K-mer Extraction and Assembly of Putative Ydel Contigs
After the trimming of raw reads by Trimmomatic (version 0.36) using the parameters LEADING: 20, TRAILING:20, SLIDINGWINDOW:4:28, MINLEN:40 (Bolger et al. 2014), genomic reads were processed to produce 35-bp k-mers as described previously (Akagi et al. 2014). The python script for the k-mer extraction was kindly provided by Dr. Tsuyoshi Akagi. Briefly, we generated 35-bp k-mer starting with “A” and then kept a set of k-mer with a minimum total count threshold of 10 and a maximum total count threshold of 1000 for male + female, male + R025, or male + EGP14 respectively. Then, male-specific k-mers that were absent in female, R025, and EGP14, were identified. All pair-ended reads containing at least one male-specific k-mer were retained and assembled with SOAPdenovo2 (Luo et al. 2012).
Genomic reads of male, female, R025, and EGP14 were mapped to the resulting male-specific assembly by BWA-MEM (Li 2013). After normalizing the total counts between genomic reads, mapped count ratio of female to male and EGP14 to male were calculated. To narrow down the male-specific contigs we selected the contigs with female/male ratio of <20% or an EGP14/male ratio of <40% These were used as the Ydel assembly in the following analysis.
Identification of the Candidate GSF Gene
GSF gene is expected to be expressed at the early stages of flower development. Thus, we analyzed transcriptome sequence data from flower buds <0.5 mm in diameter of male and female plants from the K-line, as well as from the deletion mutants R025, EGP14, and EGP15. Transcripts of R025 and male were sequenced as part of this study (accession nos. DRR359968, DRR359969, and DRR360400), whereas other transcriptome data were published previously (NCBI bioproject PRJNA474609) (Krasovec et al. 2019). For transcriptome sequencing, total RNA extraction was done with the RNA Easy kit (Qiagen) and sequenced at WTCHG on Illumina HiSeq4000 with 75 bp paired-ends reads. The four sets of reads from male flower buds (accession nos. DRR359968 and DRR360400; NCBI bioproject PRJNA474609) were assembled with Trinity and Drap, respectively (Grabherr et al. 2011; Cabau et al. 2017). The resulting contigs were BLAST-searched against the male-specific assembly to identify 1,615 expressed Ydel fragments (fig. 1B). All RNA-seq reads were aligned to the expressed Ydel fragments using RSEM (Li and Dewey 2011) to obtain expression value of each fragment in the male, female, R025, EGP14, and EGP15. This analysis identified three fragments that were not homologous to TE and expressed in male but not in female, R025, EGP14, and EGP15. Three primer sets were designed for the three fragments (supplementary table S3, Supplementary Material online), and PCR was performed on the genomic DNAs of male, female, and 11 hermaphroditic plants (EGP4, EGP5, EGP6, EGP8, EGP9, EGP10, EGP11, EGP12, EGP13, EGP14, and EGP15) to test the male specificity of their fragments. Only one fragment was found to be amplified from male but not from the female or the deletion mutants.
Segregation Analysis to Confirm X-linkage of GSFX gene
To test whether GSFX is X-linked we reused the sequence data from a S. latifolia genetic cross published previously (Krasovec et al. 2018). That data included Illumina paired end sequences of two parents and 10 F1 progenies (4 males and 6 females). Read mapping to female genome reference and SNP calling were done as described previously (Krasovec et al. 2018). X-linkage was tested by SNP segregation, checking whether the sons always inherit the maternal allele, as expected for the X-linked genetic variants.
Expression Analyses of the GSFX and GSFY
Total RNA was extracted from SAMs, leaves, flower buds, and open flower of male and female plants, respectively, by using the RNeasy Plant Mini kit (Qiagen). cDNA synthesis from 1 µg of total RNA was performed by using Superscript IV reverse transcriptase (Invitrogen). The cDNAs were then subjected to touch down PCR (98°C for 10 sec., Annealing temperature for 15 sec., and 68°C for 2 sec.), in which the annealing temperature was reduced from 68°C by 1°C every cycle until the calculated Tm range was reached, followed by 30 cycles (98°C for 10 sec., Calculated Tm for 15 sec., and 68°C for 2 sec.) by using KOD One polymerase (TOYOBO), with primers of GSFY_F7 and C279090_F2 for GSFY, and GSFX_F3 and GSFX_R2 for GSFX, and Slactin01 and Slactin02 for the Actin gene, respectively (supplementary table S3, Supplementary Material online). For in situ expression analysis, BaseScope Duplex Detection Reagent Kit (ACD, Hayward, CA, USA) was used according to the manufacturer's instructions with some modifications. Probes for GSFY and GSFX were produced by ACD, respectively. Fixation and embedding of the flower buds were performed as described previously (Kazama et al. 2009). Semithin 8-μm sections of male flower buds were made by a Microm HM 340E microtome, and transferred to Fisherbrand Superfrost Plus slides (Thermo Fisher Scientific, Pittsburg, PA, U.S.A.). The sections were dried out overnight at 37°C. After baking for 60 min at 60°C, the slides were treated with xylene, and dehydrated in ethanol. The slides were then pretreated with hydrogen peroxide (provided by the BaseScope Duplex Detection Reagent Kit) for 10 min at room temperature, incubated in a target retrieval buffer (provided by the BaseScope Duplex Detection Reagent Kit) for 15 min maintained at boiling temperature (98°C to 102°C), washed by sterilized water, dehydrated in ethanol, then dried out overnight at room temperature. The slides were treated with protease III (provided by the BaseScope Duplex Detection Reagent Kit) for 30 min at 40°C in a HybEZ hybridization oven (ACD). Hybridization and detection of the signals were performed as described in the manufacturer’s protocol. Slides were imaged on an Olympus BX53 microscope (Olympus, Tokyo, Japan).
Peptide Treatment of the GSFX and GSFY
Following the usual CLV3 production protocol, four dodecapeptides (CLV3p, CLV3mp, GSFYp, and GSFXp) were synthesized at the RIKEN Centre for Brain Science with the modification that two proline residues were replaced with hydroxyproline residues (Shinohara and Matsubayashi 2013). In the root elongation assay with peptide treatments, the surface-sterilized Col-0 seeds were sawn on 0.7% agar-containing 1/2 MS medium (Murashige and Skoog 1962) supplemented with 3% (w/v) sucrose and 0.1 µM synthetic peptides. After 8 days growing at 23 °C on a 16 h light/8 h dark cycle, the root length was measured from the base of the hypocotyl to the tip of the primary root by using the ImageJ software. The peptide treatment assay on the SAM in the A. thaliana clv3-101 mutant was performed as described previously (Shinohara and Matsubayashi 2013, 2015). In the peptide treatment assay on the SAM in S. latifolia, the surface-sterilized S. latifolia seeds were placed in the liquid 1/2 MS medium supplemented with 1% (w/v) sucrose and 0.1 µM synthetic peptides, and incubated at 4 °C for 3 days followed by the incubation at 23 °C on a 16 h light/8 h dark cycle for 10 days. These incubations were performed in 50 ml Falcon tubes (100 seeds/tube) with 20 mL of the liquid media on the RT-50 rotator (TAITEC, Saitama, Japan). After these incubations, the SAM of the peptide-treated seedling was observed and photographed by scanning electron microscopy (SEM) with a cool stage at −20 °C (S-3000N, Hitachi, Tokyo, Japan). The SEM was operated at 4 kV. The diameter of SAM was measured by the ImageJ software.
Peptide Treatment on S. Latifolia Flower Buds
By using a micro-pipet, 0.5 µM synthetic GSFYp was dropped onto female and hermaphroditic mutants (EGP15 and EGP14) at stages 1–3 determined as described previously (Grant et al. 1994; Farbos et al. 1997). After 1 day, additional 0.5 µm synthetic GSFY was dropped onto the same flower buds. These peptide-treated plants were grown in a growth chamber at 23 °C on a 16 h light/8 h dark cycle. After 10–14 days, the developing flower buds at stage 8 were observed and photographed by SEM with a cool stage at −20 °C (S-3000N, Hitachi).
Transformation of A. thaliana
The GSFY fragment was amplified by RT-PCR with the primers (GSFY_cF2 and GSFY_cR2, supplementary table S3, Supplementary Material online) by using cDNA from male flower buds as a template. The resulting fragment was subcloned in the EcoRV site of the pJET2.1 vector (Thermo Fisher Scientific, Waltham, MA, USA). The upstream region was amplified by PCR with primers GSFY_gF1b and GSFY_gcR1b (supplementary table S3, Supplementary Material online), by using the genomic DNA of K-line female as a template and inserted in the EcoRV site of the subclone by using the In-Fusion HD Cloning Kit (TaKaRa Bio. Inc., Shiga, Japan). The resulting subclone was used as a template for PCR amplification with primers (GSFY_gF1b and GSFY_gcR1b, supplementary table S3, Supplementary Material online) to obtain the fragment including the native promoter followed by the cDNA sequence. The amplified fragment was inserted between HindIII and SacI sites of the pSMAH621 binary vector, which was kindly provided by Dr. H. Ichikawa (National Institute of Agrobiological Sciences, Tsukuba, Japan), by using the In-Fusion HD Cloning Kit, to generate pgGSFY. For the cloning of the GSFX gene, its genomic sequence was amplified by PCR with primers (GSFX_gF1 and GSFX_gcR1, supplementary table S3, Supplementary Material online) by using the genomic DNA of K-line female as a template. The amplified fragment was inserted between HindIII and SacI sites of the pSMAH621 binary vector by using the In-Fusion HD Cloning Kit, to generate pgGSFX.
These constructs were introduced into Agrobacterium tumefaciens strain C58, respectively, and the resulting bacteria strains were used to transform wild-type A. thaliana (Col-0) by the floral-dip method (Clough and Bent 1998). Hygromycin-resistant T1 plants were self-pollinated and then the phenotype of T2 plants were observed under stereomicroscopy.
Sequence Divergence Analyses
The alignments of GSFX, GSFY, and its homologs from S. uniflora were done with muscle (Edgar 2004) and checked visually in ProSeq3 (Filatov 2009). The CDS alignments from these genes were analyzed in MEGA (Kumar et al. 2018) to measure the pairwise synonymous and nonsynonymous divergence (Nei and Gojobori 1986) and conduct the Tajima’s relative rates test (Tajima 1993) with S. uniflora used as an outgroup.
Statistical Analysis
Statistical calculations were conducted using R (v4.03). Statistical analyses were performed using two-sided Wilcoxon rank-sum test. The exact sample sizes (n) are given as discrete numbers in fig. 3 and supplementary fig. S1, Supplementary Material online.
Supplementary Material
Acknowledgements
We thank Dr. Hidefumi Shinohara (Fukui Prefectural University) for his help in performing A. thaliana SAM observation, Dr. Takashi Akagi (Okayama University) for providing the k-mer extraction script, Dr. Ali Ferjani (Tokyo Gakugei University) for providing clv3-101 seeds, and Dr. Hiroaki Ichikawa (National Institute of Agrobiological Sciences, Japan) for providing pSMAH621 vector. We also acknowledge the staff at the Wellcome Trust Centre (Oxford) and the RIKEN CBS (Center for Brain Science) Research Resources Center for high-throughput sequencing and initial data processing. This work was supported by JSPS KAKENHI Grant Numbers JP20H03297, JP20K21449, JP21KK0128, and 22H05071 to Y.K. and BBSRC grant BB/P009808/1 to D.A.F. The data that support the findings of this study are available from the corresponding authors on request. All sequence data generated in the context of this manuscript have been deposited in NCBI/EMBL/DDBJ (accession nos. DRR357638, DRR357639, DRR359968, DRR359969, DRR360300, DRR360400, ICSN01000001–ICSN01003571, and ICSO01000001–ICSO01092457).
Contributor Information
Yusuke Kazama, Graduate School of Bioscience and Biotechnology, Fukui Prefectural University, 4-1-1 Kenjojima, Matsuoka, Eiheiji-cho, Japan; RIKEN Nishina Center, 2-1 Hirosawa, Wako, Saitama 351-0198, Japan.
Moe Kitoh, Graduate School of Bioscience and Biotechnology, Fukui Prefectural University, 4-1-1 Kenjojima, Matsuoka, Eiheiji-cho, Japan.
Taiki Kobayashi, Graduate School of Bioscience and Biotechnology, Fukui Prefectural University, 4-1-1 Kenjojima, Matsuoka, Eiheiji-cho, Japan.
Kotaro Ishii, RIKEN Nishina Center, 2-1 Hirosawa, Wako, Saitama 351-0198, Japan; National Institutes for Quantum and Radiological Science and Technology, 4-9-1 Anagawa, Inage-ku, Chiba 263-8555, Japan.
Marc Krasovec, Department of Plant Sciences, University of Oxford, Oxford OX1 3RB, UK; Sorbonne Université, CNRS, UMR 7232 Biologie Intégrative des Organismes Marins (BIOM), Observatoire Océanologique, 66650 Banyuls-sur-Mer, France.
Yasuo Yasui, Graduate School of Agriculture, Kyoto University, Kitashirakawa Oiwake-cho, Sakyo-ku, Kyoto 606-8502, Japan.
Tomoko Abe, RIKEN Nishina Center, 2-1 Hirosawa, Wako, Saitama 351-0198, Japan.
Shigeyuki Kawano, Department of Integrated Biosciences, Graduate School of Frontier Sciences, The University of Tokyo, Kashiwa, FSB-601, 5-1-5 Kashiwanoha, Kashiwa, Chiba 277-8562, Japan; Future Center Initiative, The University of Tokyo, 178-4-4 Wakashiba, Kashiwa, Chiba 277-0871, Japan.
Dmitry A Filatov, Department of Plant Sciences, University of Oxford, Oxford OX1 3RB, UK.
Author contributions
Y.K. and D.A.F. conceived the project, designed the work, and drafted the text of the manuscript. Y.K., K.I., M.Kr., and D.A.F. analyzed the data. Y.K., M.Ki., T.K., K.I., and Y.Y. conducted experiments. All the authors contributed to writing and editing the manuscript.
Supplementary material
Supplementary data are available at Molecular Biology and Evolution online. The datasets generated during and/or analysed during this study are available from the corresponding authors on reasonable request.
Data Availability
The datasets generated during and/or analysed during this study are available from the corresponding authors on reasonable request.
References
- Akagi T, Henry IM, Ohtani H, Morimoto T, Beppu K, Kataoka I, Tao R. 2018. A Y-encoded suppressor of feminization arose via lineage-specific duplication of a cytokinin response regulator in kiwifruit. Plant Cell. 30:780–795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akagi T, Henry IM, Tao R, Comai L. 2014. Plant genetics. A Y-chromosome-encoded small RNA acts as a sex determinant in persimmons. Science. 346:646–650. [DOI] [PubMed] [Google Scholar]
- Akagi T, Pilkington SM, Varkonyi-Gasic E, Henry IM, Sugano SS, Sonoda M, Firl A, McNeilage MA, Douglas MJ, Wang T, et al. 2019. Two Y-chromosome-encoded genes determine sex in kiwifruit. Nat Plants. 5:801–809. [DOI] [PubMed] [Google Scholar]
- Aonuma W, Kawamoto H, Kazama Y, Ishii K, Abe T, Kawano S. 2021. Male/female trade-off in hermaphroditic Y-chromosome deletion mutants of the dioecious plant Silene latifolia. Cytologia (Tokyo). 86:329–338. [Google Scholar]
- Aryal R, Ming R. 2014. Sex determination in flowering plants: papaya as a model system. Plant Sci. 217–218. 56–62. [DOI] [PubMed] [Google Scholar]
- Atanassov I, Delichere C, Filatov DA, Charlesworth D, Negrutiu I, Moneger F. 2001. Analysis and evolution of two functional Y-linked loci in a plant sex chromosome system. Mol Biol Evol. 18:2162–2168. [DOI] [PubMed] [Google Scholar]
- Bachtrog D. 2013. Y-chromosome evolution: emerging insights into processes of Y-chromosome degeneration. Nat Rev Genet. 14:113–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bacovsky V, Cegan R, Tihlarikova E, Nedela V, Hudzieczek V, Smrza L, Janicek T, Benes V, Hobza R. 2022. Chemical genetics in Silene latifolia elucidate regulatory pathways involved in gynoecium development. J Exp Bot. 73:2354–2368. [DOI] [PubMed] [Google Scholar]
- Barrett SC. 2010. Understanding plant reproductive diversity. Philos Trans R Soc Lond B Biol Sci. 365:99–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bawa KS. 1980. Evolution of dioecy in flowering plants. Annu Rev Ecol Evol Syst. 11:15–39. [Google Scholar]
- Bergero R, Qiu S, Charlesworth D. 2015. Gene loss from a plant sex chromosome system. Curr Biol. 25:1234–1240. [DOI] [PubMed] [Google Scholar]
- Bergero R, Qiu S, Forrest A, Borthwick H, Charlesworth D. 2013. Expansion of the pseudo-autosomal region and ongoing recombination suppression in the Silene latifolia sex chromosomes. Genetics. 194:673–686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for illumina sequence data. Bioinformatics. 30:2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boualem A, Troadec C, Camps C, Lemhemdi A, Morin H, Sari MA, Fraenkel-Zagouri R, Kovalski I, Dogimont C, Perl-Treves R, et al. 2015. A cucurbit androecy gene reveals how unisexual flowers develop and dioecy emerges. Science. 350:688–691. [DOI] [PubMed] [Google Scholar]
- Brand U, Fletcher JC, Hobe M, Meyerowitz EM, Simon R. 2000. Dependence of stem cell fate in Arabidopsis on a feedback loop regulated by CLV3 activity. Science. 289:617–619. [DOI] [PubMed] [Google Scholar]
- Cabau C, Escudié F, Djari A, Guiguen Y, Bobe J, Klopp C. 2017. Compacting and correcting TRinity and oases RNA-seq de novo assemblies. PeerJ. 5:e2988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao X, He Z, Guo L, Liu X. 2015. Epigenetic mechanisms are critical for the regulation of WUSCHEL expression in floral meristems. Plant Physiol. 168:1189–1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth D. 2002. Plant sex determination and sex chromosomes. Heredity (Edinb). 88:94–101. [DOI] [PubMed] [Google Scholar]
- Charlesworth D. 2015. Plant contributions to our understanding of sex chromosome evolution. New Phytol. 208:52–65. [DOI] [PubMed] [Google Scholar]
- Charlesworth D. 2016. Plant sex chromosomes. Annu Rev Plant Biol. 67:397–420. [DOI] [PubMed] [Google Scholar]
- Charlesworth D. 2018. Mogens Westergaard's contributions to understanding sex chromosomes. Genetics. 210:1143–1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth D. 2019. Young sex chromosomes in plants and animals. New Phytol. 224:1095–1107. [DOI] [PubMed] [Google Scholar]
- Charlesworth B, Charlesworth D. 1978. A model for the evolution of dioecy and gynodioecy. Am Midl Nat. 112:975–997. [Google Scholar]
- Charlesworth D, Charlesworth B. 1981. Allocation of resources to male and female functions in hermaphrodites. Biol J Linn Soc. 15:57–74. [Google Scholar]
- Charlesworth B, Charlesworth D. 2000. The degeneration of Y chromosomes. Philos Trans R Soc Lond B Biol Sci. 355:1563–1572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth D, Charlesworth B. 2005. Sex chromosomes: evolution of the weird and wonderful. Curr Biol. 15:R129–R131. [DOI] [PubMed] [Google Scholar]
- Charlesworth D, David SG. 2004. The evolution of dioecy and plant sex chromosome systems. In. Sex determination in plants: Garland Science. p. 25–50. [Google Scholar]
- Clough SJ, Bent AF. 1998. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16:735–743. [DOI] [PubMed] [Google Scholar]
- Dorken ME, Barrett SC. 2004. Sex determination and the evolution of dioecy from monoecy in Sagittaria latifolia (Alismataceae). Proc Biol Sci. 271:213–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dufaÿ M, Champelovier P, Käfer J, Henry J-P, Mousset S, Marais G. 2014. An angiosperm-wide analysis of the gynodioecy–dioecy pathway. Annal Bot. 114:539–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32:1792–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farbos I, Oliveira M, Negrutiu I, Mouras A. 1997. Sex organ determination and differentiation in the dioecious plant Melandrium album (Silene latifolia): a cytological and histological analysis. Sex Plant Reprod. 10:155–167. [Google Scholar]
- Farbos I, Veuskens J, Vyskot B, Oliveira M, Hinnisdaels S, Aghmir A, Mouras A, Negrutiu I. 1999. Sexual dimorphism in white campion: deletion on the Y chromosome results in a floral asexual phenotype. Genetics. 151:1187–1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiers M, Golemiec E, Xu J, van der Geest L, Heidstra R, Stiekema W, Liu CM. 2005. The 14-amino acid CLV3, CLE19, and CLE40 peptides trigger consumption of the root meristem in Arabidopsis through a CLAVATA2-dependent pathway. Plant Cell. 17:2542–2553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filatov DA. 2009. Processing and population genetic analysis of multigenic datasets with ProSeq3 software. Bioinformatics. 25:3189–3190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fletcher JC, Brand U, Running MP, Simon R, Meyerowitz EM. 1999. Signaling of cell fate decisions by CLAVATA3 in Arabidopsis shoot meristems. Science. 283:1911–1914. [DOI] [PubMed] [Google Scholar]
- Gehring JL, Linhart YB. 1993. Sexual dimorphisms and response to low resources in the dioecious plant Silene latifolia (Caryophyllaceae). Int J of Plant Sci. 154:152–162. [Google Scholar]
- Geraldes A, Hefer CA, Capron A, Kolosova N, Martinez-Nunez F, Soolanayakanahally RY, Stanton B, Guy RD, Mansfield SD, Douglas CJ, et al. 2015. Recent Y chromosome divergence despite ancient origin of dioecy in poplars (Populus). Mol Ecol. 24:3243–3256. [DOI] [PubMed] [Google Scholar]
- Grabherr MG, Haas BJ, Yassour M, Levin JZ, Thompson DA, Amit I, Adiconis X, Fan L, Raychowdhury R, Zeng Q, et al. 2011. Full-length transcriptome assembly from RNA-seq data without a reference genome. Nat Biotechnol. 29:644–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant S, Hunkirchen B, Saedler H. 1994. Developmental differences between male and female flowers in the dioecious plant Silene latifolia. Plant J. 6:471–480. [Google Scholar]
- Harkess A, Huang K, van der Hulst R, Tissen B, Caplan JL, Koppula A, Batish M, Meyers BC, Leebens-Mack J. 2020. Sex determination by two Y-linked genes in garden asparagus. Plant Cell. 32:1790–1796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harkess A, Zhou J, Xu C, Bowers JE, Van der Hulst R, Ayyampalayam S, Mercati F, Riccardi P, McKain MR, Kakrana A. 2017. The asparagus genome sheds light on the origin and evolution of a young Y chromosome. Nat Commun. 8:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heslop-Harrison J, editor. 1963. Symposia of the society for experimental biology. [PubMed] [Google Scholar]
- Hough J, Hollister JD, Wang W, Barrett SC, Wright SI. 2014. Genetic degeneration of old and young Y chromosomes in the flowering plant Rumex hastatulus. Proc Natl Acad Sci U S A. 111:7713–7718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenik PD, Irish VF. 2000. Regulation of cell proliferation patterns by homeotic genes during Arabidopsis floral development. Development. 127:1267–1276. [DOI] [PubMed] [Google Scholar]
- Kazama Y, Fujiwara MT, Koizumi A, Nishihara K, Nishiyama R, Kifune E, Abe T, Kawano S. 2009. A SUPERMAN-like gene is exclusively expressed in female flowers of the dioecious plant Silene latifolia. Plant Cell Physiol. 50:1127–1141. [DOI] [PubMed] [Google Scholar]
- Kazama Y, Ishii K, Aonuma W, Ikeda T, Kawamoto H, Koizumi A, Filatov DA, Chibalina M, Bergero R, Charlesworth D, et al. 2016. A new physical mapping approach refines the sex-determining gene positions on the Silene latifolia Y-chromosome. Sci Rep. 6:18917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazama Y, Nishihara K, Bergero R, Fujiwara MT, Abe T, Charlesworth D, Kawano S. 2012. SlWUS1; an X-linked gene having no homologous Y-linked copy in Silene latifolia. G3. 2:1269–1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazama Y, Sugiyama R, Matsunaga S, Shibata F, Uchida W, Hizume M, Kawano S. 2003. Organization of the KpnI family of chromosomal distal-end satellite DNAs in Silene latifolia. J Plant Res. 116:317–326. [DOI] [PubMed] [Google Scholar]
- Kejnovsky E, Vyskot B. 2010. Silene latifolia: the classical model to study heteromorphic sex chromosomes. Cytogenet Genome Res. 129:250–262. [DOI] [PubMed] [Google Scholar]
- Khryanin V. 2002. Role of phytohormones in sex differentiation in plants. Russ J Plant Physiol. 49:545–551. [Google Scholar]
- Kondo T, Sawa S, Kinoshita A, Mizuno S, Kakimoto T, Fukuda H, Sakagami Y. 2006. A plant peptide encoded by CLV3 identified by in situ MALDI-TOF MS analysis. Science. 313:845–848. [DOI] [PubMed] [Google Scholar]
- Krasovec M, Chester M, Ridout K, Filatov DA. 2018. The mutation rate and the age of the sex chromosomes in Silene latifolia. Curr Biol. 28:1832–1838. e1834. [DOI] [PubMed] [Google Scholar]
- Krasovec M, Kazama Y, Ishii K, Abe T, Filatov DA. 2019. Immediate dosage compensation is triggered by the deletion of Y-linked genes in Silene latifolia. Curr Biol. 29:2214–2221. e2214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krasovec M, Zhang Y, Filatov DA. 2020. The location of the pseudoautosomal boundary in Silene latifolia. Genes (Basel). 11:610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 35:1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lardon A, Georgiev S, Aghmir A, Le Merrer G, Negrutiu I. 1999. Sexual dimorphism in white campion: complex control of carpel number is revealed by Y chromosome deletions. Genetics. 151:1173–1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laux T, Mayer K, Berger J, Jurgens G. 1996. The WUSCHEL gene is required for shoot and floral meristem integrity in Arabidopsis. Development. 122:87–96. [DOI] [PubMed] [Google Scholar]
- Li H. 2013. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. Preprint at arXiv https://arxiv.org/abs/1303.3997. [Google Scholar]
- Li B, Dewey CN. 2011. RSEM: accurate transcript quantification from RNA-seq data with or without a reference genome. BMC Bioinform. 12:1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lloyd DG. 1979. Parental strategies of angiosperms. N Z J Bot. 17:595–606. [Google Scholar]
- Lloyd DG. 1980. The distributions of gender in four angiosperm species illustrating two evolutionary pathways to dioecy. Evolution. 34:123–134. [DOI] [PubMed] [Google Scholar]
- Luo R, Liu B, Xie Y, Li Z, Huang W, Yuan J, He G, Chen Y, Pan Q, Liu Y. 2012. SOAPdenovo2: an empirically improved memory-efficient short-read de novo assembler. Gigascience. 1:18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma W-J, Pannell JR. 2016. Sex determination: separate sexes are a double turnoff in melons. Curr Biol. 26:R171–R174. [DOI] [PubMed] [Google Scholar]
- Maeda S, Gunji S, Hanai K, Hirano T, Kazama Y, Ohbayashi I, Abe T, Sawa S, Tsukaya H, Ferjani A. 2014. The conflict between cell proliferation and expansion primarily affects stem organogenesis in Arabidopsis. Plant Cell Physiol. 55:1994–2007. [DOI] [PubMed] [Google Scholar]
- Masuda K, Ikeda Y, Matsuura T, Kawakatsu T, Tao R, Kubo Y, Ushijima K, Henry IM, Akagi T. 2022. Reinvention of hermaphroditism via activation of a RADIALIS-like gene in hexaploid persimmon. Nat Plants. 8:217–224. [DOI] [PubMed] [Google Scholar]
- Mayer KF, Schoof H, Haecker A, Lenhard M, Jürgens G, Laux T. 1998. Role of WUSCHEL in regulating stem cell fate in the Arabidopsis shoot meristem. Cell. 95:805–815. [DOI] [PubMed] [Google Scholar]
- Müller NA, Kersten B, Leite Montalvão AP, Mähler N, Bernhardsson C, Bräutigam K, Carracedo Lorenzo Z, Hoenicka H, Kumar V, Mader M. 2020. A single gene underlies the dynamic evolution of poplar sex determination. Nat Plants. 6:630–637. [DOI] [PubMed] [Google Scholar]
- Murashige T, Skoog F. 1962. A revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiol Plant. 15:473–497. [Google Scholar]
- Nei M, Gojobori T. 1986. Simple methods for estimating the numbers of synonymous and nonsynonymous nucleotide substitutions. Mol Biol Evol. 3:418–426. [DOI] [PubMed] [Google Scholar]
- Obbard DJ, Harris SA, Buggs RJ, Pannell JR. 2006. Hybridization, polyploidy, and the evolution of sexual systems in Mercurialis (Euphorbiaceae). Evolution. 60:1801–1815. [PubMed] [Google Scholar]
- Ohno S. 1967. Sex chromosomes and sex-linked genes. Berlin, Heidelberg, New York: Springer Science & Business Media. [Google Scholar]
- Papadopulos AS, Chester M, Ridout K, Filatov DA. 2015. Rapid Y degeneration and dosage compensation in plant sex chromosomes. Proc Natl Acad Sci U S A. 112:13021–13026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papadopulos AS, Helmstetter AJ, Osborne OG, Comeault AA, Wood DP, Straw EA, Mason L, Fay MF, Parker J, Dunning LT. 2021. Rapid parallel adaptation to anthropogenic heavy metal pollution. Mol Biol Evol. 38:3724–3736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perales M, Rodriguez K, Snipes S, Yadav RK, Diaz-Mendoza M, Reddy GV. 2016. Threshold-dependent transcriptional discrimination underlies stem cell homeostasis. Proc Natl Acad Sci U S A. 113:E6298–E6306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plong A, Rodriguez K, Alber M, Chen W, Reddy GV. 2021. CLAVATA3 mediated simultaneous control of transcriptional and post-translational processes provides robustness to the WUSCHEL gradient. Nat Commun. 12:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prunet N, Morel P, Negrutiu I, Trehin C. 2009. Time to stop: flower meristem termination. Plant Physiol. 150:1764–1772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu S, Bergero R, Guirao-Rico S, Campos J, Cezard T, Gharbi K, Charlesworth D. 2016. RAD mapping reveals an evolving, polymorphic and fuzzy boundary of a plant pseudoautosomal region. Mol Ecol. 25:414–430. [DOI] [PubMed] [Google Scholar]
- Renner SS. 2014. The relative and absolute frequencies of angiosperm sexual systems: dioecy, monoecy, gynodioecy, and an updated online database. Am J Bot. 101:1588–1596. [DOI] [PubMed] [Google Scholar]
- Renner SS. 2016, Pathways for making unisexual flowers and unisexual plants: moving beyond the “two mutations linked on one chromosome” model. Am J Bot. 103:587-589. [DOI] [PubMed] [Google Scholar]
- Renner SS, Ricklefs RE. 1995. Dioecy and its correlates in the flowering plants. Am J Bot. 82:596–606. [Google Scholar]
- Richards C, Emery S, McCauley D. 2003. Genetic and demographic dynamics of small populations of Silene latifolia. Heredity (Edinb). 90:181–186. [DOI] [PubMed] [Google Scholar]
- Ruddat M, Kokontis J, Birch L, Garber E, Chiang K, Campanella J, Dai H. 1991. Interactions of Microbotryum violaceum (Ustilago violacea) with its host plant Silene alba. Plant Sci. 80:157–165. [Google Scholar]
- Schoof H, Lenhard M, Haecker A, Mayer KF, Jürgens G, Laux T. 2000. The stem cell population of Arabidopsis shoot meristems is maintained by a regulatory loop between the CLAVATA and WUSCHEL genes. Cell. 100:635–644. [DOI] [PubMed] [Google Scholar]
- Shang E, Wang X, Li T, Guo F, Ito T, Sun B. 2021. Robust control of floral meristem determinacy by position-specific multifunctions of KNUCKLES. Proc Natl Acad Sci U S A. 118:e2102826118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinohara H, Matsubayashi Y. 2013. Chemical synthesis of Arabidopsis CLV3 glycopeptide reveals the impact of hydroxyproline arabinosylation on peptide conformation and activity. Plant Cell Physiol. 54:369–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinohara H, Matsubayashi Y. 2015. Reevaluation of the CLV3-receptor interaction in the shoot apical meristem: dissection of the CLV3 signaling pathway from a direct ligand-binding point of view. Plant J. 82:328–336. [DOI] [PubMed] [Google Scholar]
- Široký J, Lysák MA, Doležel J, Kejnovský E, Vyskot B. 2001. Heterogeneity of rDNA distribution and genome size in Silene spp. Chromosome Res 9:387–393. [DOI] [PubMed] [Google Scholar]
- Skof S, Cerenak A, Jakse J, Bohanec B, Javornik B. 2012. Ploidy and sex expression in monoecious hop (Humulus lupulus). Botany. 90:617–626. [Google Scholar]
- Snipes SA, Rodriguez K, DeVries AE, Miyawaki KN, Perales M, Xie M, Reddy GV. 2018. Cytokinin stabilizes WUSCHEL by acting on the protein domains required for nuclear enrichment and transcription. PLoS Genet. 14:e1007351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somssich M, Je BI, Simon R, Jackson D. 2016. CLAVATA-WUSCHEL signaling in the shoot meristem. Development. 143:3238–3248. [DOI] [PubMed] [Google Scholar]
- Sousa A, Bellot S, Fuchs J, Houben A, Renner SS. 2016. Analysis of transposable elements and organellar DNA in male and female genomes of a species with a huge Y chromosome reveals distinct Y centromeres. Plant J. 88:387–396. [DOI] [PubMed] [Google Scholar]
- Spigler RB, Ashman T-L. 2012. Gynodioecy to dioecy: are we there yet? Ann Bot. 109:531–543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tajima F. 1993. Simple methods for testing the molecular evolutionary clock hypothesis. Genetics. 135:599–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Bakel H, Stout JM, Cote AG, Tallon CM, Sharpe AG, Hughes TR, Page JE. 2011. The draft genome and transcriptome of Cannabis sativa. Genome Biol. 12:1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veltsos P, Ridout KE, Toups MA, González-Martínez SC, Muyle A, Emery O, Rastas P, Hudzieczek V, Hobza R, Vyskot B. 2019. Early sex-chromosome evolution in the diploid dioecious plant Mercurialis annua. Genetics. 212:815–835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiblen GD, Oyama RK, Donoghue MJ. 2000. Phylogenetic analysis of dioecy in monocotyledons. Am Nat. 155:46–58. [DOI] [PubMed] [Google Scholar]
- Westergaard M. 1946. Aberrant Y chromosomes and sex expression in Melandrium album. Hereditas. 32:419–443. [DOI] [PubMed] [Google Scholar]
- Westergaard M. 1953. Über den mechanismus der geschlechtsbestimmung bei Melandrium album. Naturwissenschaften. 40:253–260. [Google Scholar]
- Westergaard M. 1958. The mechanism of sex determination in dioecious flowering plants. In. Advances in genetics: Elsevier. p. 217–281. [DOI] [PubMed] [Google Scholar]
- Xue L, Wu H, Chen Y, Li X, Hou J, Lu J, Wei S, Dai X, Olson MS, Liu J. 2020. Evidences for a role of two Y-specific genes in sex determination in Populus deltoides. Nat Commun. 11:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yakimowski SB, Barrett SC. 2016. The role of hybridization in the evolution of sexual system diversity in a clonal, aquatic plant. Evolution. 70:1200–1211. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated during and/or analysed during this study are available from the corresponding authors on reasonable request.