Abstract
As viral genomic imprints in host genomes, endogenous viral elements (EVEs) shed light on the deep evolutionary history of viruses, ancestral host ranges, and ancient viral–host interactions. In addition, they may provide crucial information for calibrating viral evolutionary timescales. In this study, we conducted a comprehensive in silico screening of a large data set of available mammalian genomes for EVEs deriving from members of the viral family Flaviviridae, an important group of viruses including well-known human pathogens, such as Zika, dengue, or hepatitis C viruses. We identified two novel pestivirus-like EVEs in the reference genome of the Indochinese shrew (Crocidura indochinensis). Homologs of these novel EVEs were subsequently detected in vivo by molecular detection and sequencing in 27 shrew species, including 26 species representing a wide distribution within the Crocidurinae subfamily and one in the Soricinae subfamily on different continents. Based on this wide distribution, we estimate that the integration event occurred before the last common ancestor of the subfamily, about 10.8 million years ago, attesting to an ancient origin of pestiviruses and Flaviviridae in general. Moreover, we provide the first description of Flaviviridae-derived EVEs in mammals even though the family encompasses numerous mammal-infecting members. This also suggests that shrews were past and perhaps also current natural reservoirs of pestiviruses. Taken together, our results expand the current known Pestivirus host range and provide novel insight into the ancient evolutionary history of pestiviruses and the Flaviviridae family in general.
Keywords: endogenous viral element, pestivirus, Flaviviridae, Crocidura, host range, paleovirology
Introduction
Endogenous viral elements (EVEs) are integrations of partial or full-length viral genomic material into the host genome (Katzourakis and Gifford 2010). In addition to retroviruses, which incorporate their genomic sequences into their host genome as an essential part of their replication cycle, many eukaryotic viruses have been found endogenized in various hosts (Feschotte and Gilbert 2012). These non-retroviruses can derive from dsDNA (Aswad and Katzourakis 2014; Li and Li 2015; Liu et al. 2020), ssDNA (Belyi et al. 2010a; Katzourakis and Gifford 2010; Kobayashi et al. 2019), dsRNA (Horie et al. 2010; Katzourakis and Gifford 2010; Liu et al. 2012), and even ssRNA viruses (Crochu et al. 2004; Horie et al. 2010; Lequime and Lambrechts 2017; Flynn and Moreau 2019). Non-retroviral RNA virus-derived EVEs arise from a conjunction of relatively rare events: (i) production of DNA genomic material, using retrotransposon encoded reverse transcriptase (Horie et al. 2010; Horie 2019); (ii) integration in the host chromosome of germ line cells; and (iii) overcoming genetic drift and/or natural selection at the population until fixation (Holmes 2011; Aiewsakun and Katzourakis 2015). EVEs thus reflect long-term and intimate interactions of viruses with their hosts, and their identification can reveal insights into past and present host distributions of viral genera and families. The detection of endogenous bornavirus-like elements in invertebrate genomes (Horie et al. 2013), for example, suggested a broader host range for bornaviruses than previously thought. The study of filovirus-like EVEs in some small mammals offer predictive value for further identifying filovirus reservoirs (Taylor et al. 2010). Similarly, the discovery of flavivirus-derived EVEs in Anopheles mosquito genomes supported the idea that Anopheles mosquitoes could also be natural hosts of flaviviruses (Lequime and Lambrechts 2017), as confirmed by other studies (Colmant et al. 2017; Öncü et al. 2018).
Aside from qualitative insights into host ranges of viruses, EVEs can also shed light on deep evolutionary histories of viruses. EVEs are significant traces of past virus–host interactions; unlike animals or plants, viruses do not leave physical fossil records, limiting our ability to study their deep evolutionary histories. EVEs could thus be considered as ‘genomic fossil records’ that can help to unravel long-term evolutionary dynamics between hosts and viral families. The presence of EVE homologs in different host species hints at an integration event before speciation, here the time to the most recent common ancestor. It thus provides a minimum age estimate for the integration event, and therefore a minimum age for the existence of a specific viral taxonomic group (Aiewsakun and Katzourakis 2015). For example, EVEs derived from adeno-associated viruses appear to be orthologous in African and Asian elephants, indicating an integration event more than 6 million years ago (Kobayashi et al. 2019). Similarly, the discovery of abundant bornavirus-like EVEs across vertebrates reveals that ancient bornaviral infections occurred over a timeframe of about 100 million years before present (Kawasaki et al. 2021). These studies can also provide genetic fossil calibration points for further gauging ancient viral timescales using phylogenetics (Feschotte and Gilbert 2012).
Members of the Flaviviridae family are linear, positive-sense, single-stranded RNA viruses currently classified in four recognized genera (Flavivirus, Hepacivirus, Pestivirus, and Pegivirus). They encompass many significant pathogens, such as dengue, Zika, hepatitis C, or bovine viral diarrhea virus. In addition to human and livestock infections, Flaviviridae viruses have been detected in a broad range of mammalian wildlife hosts, e.g., nonhuman primates (Mares-Guia et al. 2020), rodents (Bletsa et al. 2021), bats (Wu et al. 2018), but also birds (Strand et al. 2018), fish (Hartlage 2016; Soto et al. 2020), and arthropods (Shi et al. 2016). Despite this important host range, current published studies have only identified Flaviviridae-derived EVEs in arthropods, including mosquitos (Crochu et al. 2004; Roiz et al. 2009; Lequime and Lambrechts 2017), ticks (Maruyama et al. 2014), and crustaceans (Parry and Asgari 2019), and these are all related to the Flavivirus genus. Currently, however, convincing evidence for Flaviviridae-derived EVEs in vertebrates remains lacking. Interestingly, potential integration has been suggested in medaka fish (Belyi et al. 2010b) as well as in rabbit and hare genomes (Silva et al. 2012, 2015). While this raises the hypothesis that Flaviviridae viruses have integrated in vertebrate hosts, the origin of these specific genomic sequences remains inconclusive due to their short size and low sequence similarity to the Flaviviridae (Flavivirus and Hepacivirus, respectively). A recent unpublished study also identified Flaviviridae-derived EVEs in a wide variety of hosts, including various invertebrates and fish (Bamford et al. 2021).
In this study, we explored in silico the presence of Flaviviridae-derived EVEs in a comprehensive set of mammalian genomes, and we discovered two novel pesti-like EVEs in the genome of the Indochinese shrew Crocidura indochinensis. We subsequently identified and characterized homologs of these EVEs in vivo in 26 species of the Crocidurinae subfamily and one member of the Soricinae subfamily, establishing the integration event at least 10.8 million years ago. Our results provide the first evidence for an ancient origin of pestiviruses and also contribute to a better understanding of the evolutionary history of the Flaviviridae family in general.
Results
False Positive Flaviviridae-Like Hits from Mammalian Genome Screening
Our initial screening of 689 available mammalian genomes using Flaviviridae and Flaviviridae-like polyproteins yielded 66 positive hits from 49 species, including rodents, nonhuman primates, marsupials, insectivores, carnivores, and bats. Detailed information about our in silico screening results can be found in Supplementary Table S1, Supplementary Material online. All virus-related sequences aligned with three pestiviruses, namely border disease virus, bovine viral diarrhea virus 1, and bovine viral diarrhea virus 2. With the exception of one shrew species (see section below), the position of all hits in the corresponding viral genomic sequence was in the ubiquitin-homolog domain between the nonstructural proteins NS2 and NS3, while some of the hits slightly expanded the alignment to the NS3 region (shown in Supplementary Fig. S1, Supplementary Material online). The ubiquitin domain in some bovine viral diarrhea virus strains is however predicted to originate from cellular derived insertions in cytopathogenic pestivirus (Agapov et al. 2004; Becher and Tautz 2011). The similarity between this viral genomic region and ubiquitin poses a considerable risk for false positives when searching for pestivirus-derived EVEs, and these hits were therefore not further considered.
In Silico Identification of Crocidura indochinensis Pesti-Like EVEs
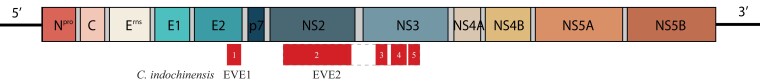
Besides the ubiquitin-related false positive results, our in silico screening identified a series of five Flaviviridae-related EVEs fragments in a single contig of the Crocidura indochinensis reference genome PVKC01 (Table 1). The first EVE (EVE1) is 318 nt long, with its closest BLAST hit being the Linda virus (Pestivirus) envelope glycoprotein E2 region (tBLASTx, 25.5% identity, e-value 3.31E−35), without any stop codon (Fig. 1). The remaining four EVEs fragments are 1,053, 84, 114 and 87 nt long, respectively, with their closest BLAST hits being a classical swine fever virus and a rodent pestivirus (with minimum amino acid identity 24.9%, maximum 62.1%). These four fragments are separated by very short gaps, with lengths of 16, 1, and 4 nucleotides, respectively. The two central fragments of EVE2, fragments 3 (84 nt), and fragment 4 (114 nt), are in a different translation frame than the two others but in the same orientation. Their arrangement in the host contig reflects their relative position in the pestiviral genome, which partially spans the nonstructural NS2 and NS3 genes (Fig. 1). For these reasons, in further analyses, we considered these four fragments as the result of a single pestiviral integration event, and thus a unique EVE (EVE2). Phylogenetic reconstructions of the identified Crocidura indochinensis EVE1, EVE2, and entire concatenated sequence with exogenous Flaviviridae viruses supports the pestivirus-origin of the EVEs (Supplementary Fig. S2, Supplementary Material online).
Table 1.
Newly Detected EVEs in Crocidura Indochinensis Genome.
| Contig accession no. | Contig length (nt) | EVE | GenBank accession no. | EVEs fragments | EVE length (nt) | Position in host contig | Translation frame | Closest BLAST hit | Conserved domain search | |
|---|---|---|---|---|---|---|---|---|---|---|
| Start | End | |||||||||
| PVKC010097735.1 | 6104 | EVE1 | BK014483 | No. 1 | 318 | 2329 | 2646 | 1 | Linda virus | Pestivirus envelope glycoprotein E2 |
| EVE2 | No. 2 | 1053 | 3205 | 4257 | 1 | Classical swine fever virus | Peptidase_C74: Pestivirus NS2 peptidase | |||
| No. 3 | 84 | 4274 | 4357 | 2 | Rodent pestivirus | Peptidase_S31: Pestivirus NS3 polyprotein peptidase S31 | ||||
| No. 4 | 114 | 4359 | 4472 | 2 | Rodent pestivirus | Peptidase_S31: Pestivirus NS3 polyprotein peptidase S31 | ||||
| No. 5 | 87 | 4477 | 4563 | 1 | Rodent pestivirus | Peptidase_S31: Pestivirus NS3 polyprotein peptidase S31 | ||||
Fig. 1.
The positions of the newly detected Crocidura indochinensis EVEs are shown relative to an archetypal Pestivirus genome (classical swine fever virus, NC_002657). Npro, N-terminal protease; C, nucleocapsid core protein; Erns, envelope glycoprotein Erns; E1, envelope glycoprotein E1; E2, envelope glycoprotein E2; p7, nonstructural protein p7; NS2, nonstructural protein NS2; NS3, nonstructural protein NS3; NS4A, nonstructural protein NS4A; NS4B, nonstructural protein NS4B; NS5A, nonstructural protein NS5A; NS5B, nonstructural protein NS5B.
In addition, a fragment prior to EVE1 in the contig shows a strong similarity (tBLASTx, 45.2% identity, e-value 5.51E−19, supplementary Table S1, Supplementary Material online) with the pestivirus ribonuclease T2 gene. However, considering that this enzyme exists in a wide range of organisms (Luhtala and Parker 2010), the virus-derived origin of this sequence in Crocidura indochinensis is not guaranteed.
The position of all pestivirus-like hits, including the ribonuclease T2 gene, in the host contig corresponds to their relative organization in the pestivirus genome. No additional features were detected after a tBLASTx search of the whole contig encompassing the two identified EVEs (Supplementary Table S1, Supplementary Material online). In addition, we did not detect the EVE sequences in reads of publicly available experimental genomic and transcriptomic data from Soricidae species.
Identification and Distribution of Pesti-Like EVEs in Other Soricidae Species
To expand our screening and evaluate the distribution of these newly identified EVEs in additional species that are phylogenetically close to Crocidura indochinensis, we undertook a PCR-based screening of 65 samples from 29 species of the Soricidae family (Supplementary Table S2, Supplementary Material online). These samples belonged to seven different genera (Crocidura, Paracrocidura, Scutisorex, Suncus, Sylvisorex, Neomys, and Sorex), encompassing two subfamilies, Crocidurinae and Soricinae. Cytochrome b (CYTB) genomic sequences were also generated to confirm the species identification (Supplementary Table S3 and Supplementary Fig. S3, Supplementary Material online).
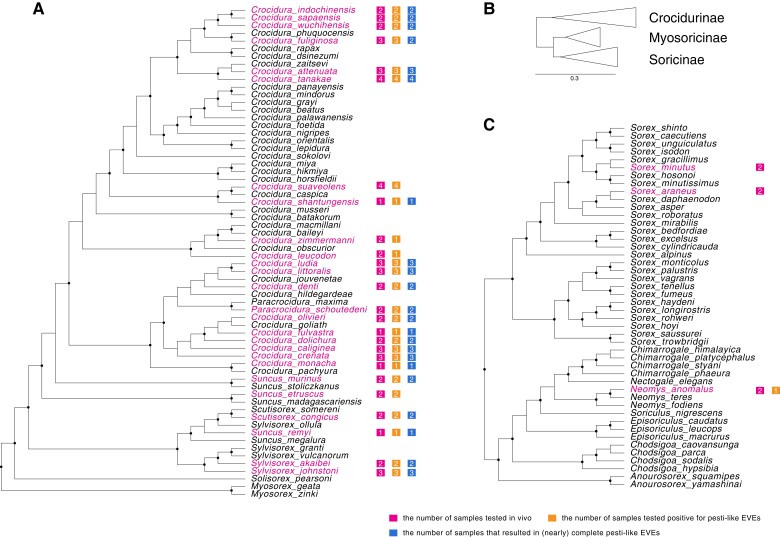
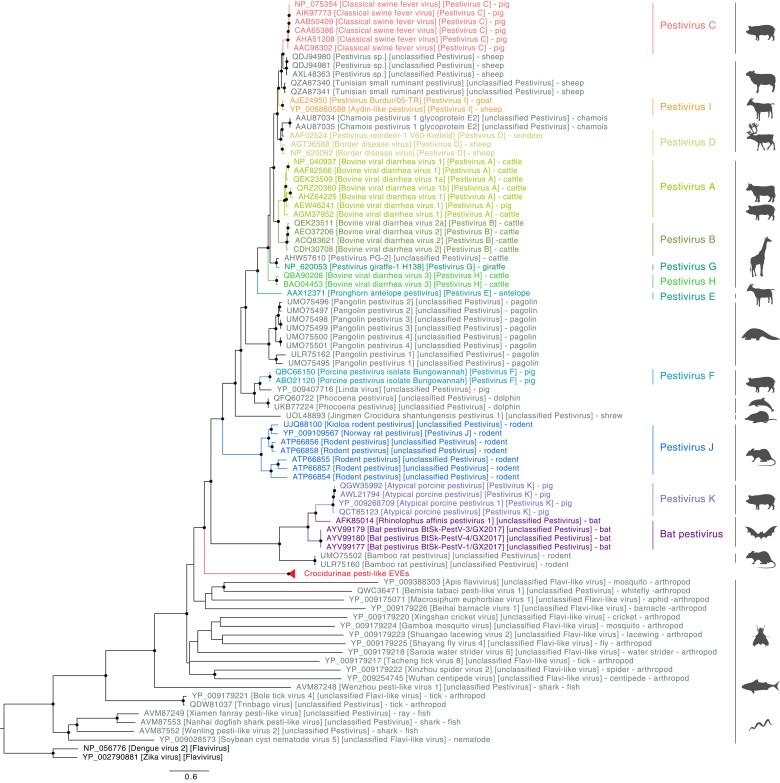
In total, 58 samples contained the newly identified pesti-like EVEs, representing 26 species in the Crocidurinae subfamily and one species (Neomys anomalus) in the Soricinae subfamily. Among them, 48 samples from 22 species in the Crocidurinae subfamily yielded complete or nearly complete EVEs sequences (Fig. 2). All novel EVEs sequences were highly similar to the Crocidura indochinensis EVEs sequences, with a mean identity of 90.38% on amino acid level and 95.12% on nucleotide level, respectively (Supplementary Table S4, Supplementary Material online). Phylogenetic reconstruction indicated that all Crocidurinae pesti-like EVEs clustered together as a sister-lineage of currently recognized Pestivirus species but divergent from the currently sole available Crocidurinae pestivirus (Fig. 3, Supplementary Fig. S4, Supplementary Material online).
Fig. 2.
Maximum likelihood phylogeny of the Soricidae (shrews) family based on the dataset of 31 assembled nuclear & mitochondrial genes from (Upham et al. 2019). For the four species (Crocidura denti, Crocidura sapaensis, Scutisorex congicus, Sylvisorex akaibei) tested in this study and not included in the Upham et al. 2019 data set, we used our generated CYTB sequences in the alignment. Nodes labeled in black circles indicate Shimodaira-Hasegawa (SH)-like branch support (%, only values > 80% are shown). The available samples’ species distribution in this study were highlighted in pink: (A) The phylogeny of Crocidurinae subfamily and sample distribution; (B) Subfamilies relationships within the Soricidae family; (C) The phylogeny of Soricinae subfamily and sample distribution.
Fig. 3.
Phylogenetic relationships of pesti-like EVEs with representative Pestivirus species and pesti-like viruses (based on aligned viral E2 and NS2-3 region). Dengue and Zika virus (Flavivirus) are used as outgroup. Clades are colored based on viral species. Nodes labeled in black circles indicate Shimodaira-Hasegawa (SH)-like branch support (%, only values > 80% are shown). Scale bars indicate the number of amino acid substitutions per site.
Although collected in different locations, nearly all species tested in the Crocidurinae subfamily harbored the pesti-like EVEs sequences. For some species, however, such as Crocidura cf. zimmermanni, C. leucodon, C. suaveolens, and Suncus etruscus, we could not always detect or sequence the EVEs in all samples. The failed detection could be explained by the genomic template being of poor quality due to storage conditions associated with the museum specimens. Interestingly, the pesti-like EVEs were also detected in one Neomys anomalus sample, while the remaining Soricinae specimens yielded negative results. The widespread nature of these homolog EVEs in the Crocidurinae species suggests a single endogenization event before their common ancestor about 10.8 million years ago (Dubey et al. 2007). Since we were not able to sequence the complete pesti-like EVEs from the Neomys anomalus specimen, their phylogenetic relationship to the Crocidurinae pesti-like EVEs remain unclear and they may not necessarily derive from the same endogenization event.
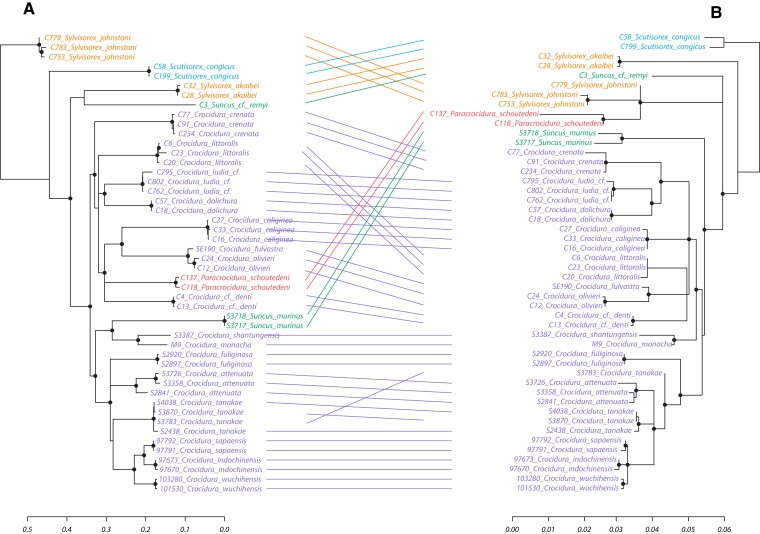
To assess phylogenetic congruence between the EVEs and the shrew host genomes, we performed reconciliation analyses for the evolutionary history of the EVEs with two nuclear and one mitochondrial gene for the available shrew species. All comparisons (EVEs-APOB, EVEs-BRCA1, EVEs-CYTB, CYTB-APOB, CYTB-BRCA1, and APOB-BRCA1) resulted in statistically significant co-phylogenetic patterns with P-values < 0.01. Despite the limited degree of phylogenetic variability observed in some co-phylogenetic plots, overall, the EVEs exhibit a pattern of inheritance as any other host genetic markers and follow the evolutionary history of the shrew host species (Fig. 4, Supplementary Fig. S5, Supplementary Material online).
Fig. 4.
Tanglegram of the cytochrome b phylogeny (A) and the corresponding EVEs phylogeny (B). The cytochrome b tree was inferred for gene sequences from 22 shrew species, and the EVEs tree (right) was inferred using the newly generated EVEs sequences. The clades were colored by shrew genus, orange: Sylvisorex; blue: Scutisorex; green: Suncus; purple: Crocidura; red: Paracrocidura. The clade nodes with SH-like branch support < 50% were collapsed as polytomies. The black circles indicate the SH-like branch support > 80%. Lines connect corresponding tips in the two phylogenies. Scale bars indicate the number of nucleotide substitutions per site.
Selective Pressure on Pesti-Like EVEs
To evaluate potential functional constraints on the EVE, we performed selective pressure analyses. Specifically, we measured selective pressure by estimating the ratio of nonsynonymous substitution rate (dN) and synonymous substitutions rate (dS) in protein-coding sequences using two methods: (i) fixed effects likelihood (FEL) (Kosakovsky Pond and Frost 2005); and (ii) Bayesian renaissance counting (Lemey et al. 2012). It is expected that nonfunctional regions should conform to neutral evolution, whereas functional regions should experience purifying selection. Based on 48 potentially coding sequences for pesti-like EVE1, the FEL method indicates an overall neutral evolution with dN/dS = 0.94. Likewise, the Bayesian renaissance counting model yields a ratio at 1.10 (95% credible interval: 0.66, 1.62), reflecting neutral evolution. The EVE2 coding region was also evaluated using the same methods resulting in a dN/dS ratio of 0.77 using FEL and 0.74 (95% credible interval: 0.58, 0.90) using renaissance counting. While the EVE2 estimates appear to reject strict neutrality for EVE2, the dN/dS estimates remain relatively high and hence do not suggest strong purifying selection. Moreover, the multiple stop codons and frame-shifts in EVE2 sequences strongly support the absence of functional constraints.
Towards a Pestivirus Evolutionary Timescale
Having established that the pesti-like EVE arose through a single endogenization event before the Crocidurinae common ancestor offers an opportunity to gain insights into the timescale of pestiviruses. However, the evolutionary history of pestiviruses and pesti-like EVEs (Fig. 3) represents a complex scenario that combines viral evolution over long time scales, which is known to be subject to time-dependent evolutionary rates (Duchêne et al. 2014; Aiewsakun and Katzourakis 2016, Membrebe et al. 2019), and EVE evolution at the host evolutionary rate. In the absence of molecular clock models that can accommodate such scenarios, we performed a naive divergence time estimation using a standard relaxed molecular clock model and using the estimated Crocidurinae the time to most recent common ancestor (TMRCA) as a calibration on the EVE TMRCA (cfr. Methods). This results in a TMRCA of pestivirus species of 231 (95% HPD: 148–329) MYA, a TMRCA of all pestiviruses and pesti-like viruses of 453 (95% HPD: 296–674) MYA, and a TMRCA of the latter and two flavirus outgroups of 472 (95% HPD: 297–702) MYA.
Discussion
Non-retroviral EVEs are rare traces of the ancient evolutionary history of viruses. These genomic fossils offer valuable insights into host range, ancestral genetic diversity and can provide invaluable information for dating viral evolutionary history (Feschotte and Gilbert 2012; Aiewsakun and Katzourakis 2015). In our study, we screened a comprehensive set of mammalian genomes to discover such Flaviviridae-derived EVEs. We uncovered two Flaviviridae-derived EVEs sequences in the genome of the Indochinese shrew and confirmed their presence in a broad range of shrew species belonging to the Crocidurinae subfamily.
The EVEs we identified are related to extant viruses within the Pestivirus genus. Viruses belonging to this genus were initially detected in a variety of artiodactylous hosts, such as ruminants and swine, in which they cause subclinical or clinical infections including hemorrhagic syndrome, abortion, acute fatal mucosal disease. Recent metagenomic studies extended the host range towards rodents (Wu et al. 2018, 2020), bats (Wu et al. 2018), fish (Shi et al. 2018), and ticks (Sameroff et al. 2019), but to some extent, the restricted sampling beyond agriculturally important animals limits our understanding of the real host range. Shrews, for example, have been recently identified as hosts of hepaciviruses, another genus in the Flaviviridae family (Guo et al. 2019; Wu et al. 2020), but limited evidence shows its relationship with pestivirus. The broad detection of pestivirus-derived EVEs reported in our study strongly supports that ancestors of the Crocidurinae shrew subfamily have been hosts of pestiviruses and suggests that their descendants might still be. Indeed, considering the extremely low probability of a non-retroviral endogenization event to occur in the germline, EVEs are strong indicators of frequent interactions between the original exogenous viruses and their hosts (Feschotte and Gilbert 2012; Aiewsakun and Katzourakis 2015). This hypothesis is supported by a recent directly submitted sequence in National Center for Biotechnology Information (NCBI) originating from a Chinese Crocidura shantungensis shrew, strongly suggesting the presence of exogenous pestivirus in Crocidurinae shrew. Interestingly, the newly described EVE sequences are not directly related to this Crocidura pestivirus. The difference might be due to the genetic drift since the historical endogenization event, or the endogenization of another, currently undetected shrew pestivirus clade. Our study thus provides indirect support for a wider and more diverse host range of pestiviruses. Additional efforts for direct detection and characterization of pestiviruses from shrews would still be required to formally demonstrate the natural hosts range of pestiviruses and to characterize the relationship between exogenous pestivirus and pesti-like EVEs.
Given the low probability of endogenization events of non-retroviral RNA viruses and the contiguous nature of the two EVEs on the host and viral genome, our results suggest a single endogenization event followed by genetic drift. One or several insertion events separated the original EVE in two fragments, EVE1 and EVE2. EVE1 shows a short but intact open reading frame to be evolving under neutral evolution while EVE2 exhibits multiple stop codons and frame-shifts due to additional insertions. Many studies have identified the important roles that EVEs can play in host antiviral immunity, both in vertebrates and invertebrates (Ophinni et al. 2019; Blair et al. 2020; Skirmuntt et al. 2020). Flavivirus-like EVEs in Aedes mosquitoes, for example, can produce P-element-induced wimpy testis (PIWI)-interacting RNAs (piRNAs) which limit the cognate virus replication (Suzuki et al. 2020). It is highly unlikely that the pesti-like sequences we discovered currently have a function in shrews because of the absence of negative selection and the disruption of the original viral coding region. It does not, however, exclude that the pesti-like sequences may have served a function in the past, following their integration, but we currently have no evidence to support this hypothesis.
The evolutionary history of the EVEs sequences after integration largely follows the host genetic patterns of inheritance. Despite showing significant phylogenetic consistency with the host mitochondrial and nuclear gene sequences (P-value < 0.01), there is a limited degree of discordance, particularly in the co-phylogenetic plot of CYTB and the EVEs phylogeny (Fig. 4). These discrepancies might be explained by differences in the genetic inheritance of the CYTB gene and EVEs: the CYTB gene is a mitochondrial gene whereas EVEs are integrated in the nuclear genome. Moreover, discordances are highlighted, although to a lesser extent, in the reconciliations of the APOB and BRCA1 nuclear genes with the EVEs phylogeny, as well as in the co-phylogenetic plot of the CYTB with the nuclear genes (Supplementary Fig. S5, Supplementary Material online). These results are in line with the different observed evolutionary patterns between host genetic markers, especially in the case of weak reproductive isolation within species or species complex, allowing hybridization, as has been suggested for some Crocidura species (Vogel et al. 2004; Dubey et al. 2006, 2008).
Dating the ancient evolutionary history of ssRNA viruses such as pestiviruses and Flaviviridae in general is challenging. The most commonly used method for inferring viral divergence time is based on the estimation of evolutionary rates derived from sequence data and their collection dates. However, the applicability of this method is often limited by different rate estimates on different timescales (Aiewsakun and Katzourakis 2016) and rate variation among viral lineages (Duffy et al. 2008; Sanjuán 2012). Not accounting for the former leads to recent estimates for the origins of ssRNA viruses that are often in conflict with other phylogenetic evidence (Holmes 2003). Using suitable molecular clock models, the powerful combination of both tip and node calibrations may help to recover more accurate evolutionary timescales (O’Reilly et al. 2015). Node calibration is however challenging for viruses as no fossil evidence can be found. It thus often relies on known phylogeographic events and other indirect calibrations point, such as ecological events or assumptions of co-divergence as alternative (Bamford et al. 2021; Moureau et al. 2015; Pettersson and Fiz-Palacios 2014). The discovery of ssRNA virus-related EVEs, thus, offers opportunities for estimating deep timescales of virus evolution history by co-phylogenetic analysis of EVE’s orthologs in different hosts (Gilbert and Feschotte 2010). However, when combining EVEs and extant viruses, we need to consider both EVE evolution according to the host evolutionary rate and time-dependent viral evolutionary rates.
The pesti-like EVE sequences characterized in our study are widespread in Crocidurinae species, are monophyletic and exhibit high sequence similarity. Considering the low probability of endogenization events of non-retroviral RNA viruses, this suggests that the pesti-like EVE got integrated before the most recent common ancestor of the subfamily, which is estimated to be over 10.8 million years ago (Dubey et al. 2007). There are only a handful of molecular dating estimates for pestiviruses, and they mostly focus on viral species or clades that are associated with economic losses. Diversification of bovine viral diarrhea virus 1 (Pestivirus A) subtypes was estimated to have started about 363 years ago (Weber et al. 2021), and the divergence of HoBi-like pestivirus (Pestivirus H) was dated back to the 16th century (Silveira et al. 2020). Applying the Crocidurinae TMRCA as a calibration, we estimate that the pestiviruses date back several hundreds of MY ago. However, in the absence of molecular clock models that can tackle complex virus and EVE evolutionary histories, we present our estimates using a standard relaxed molecular clock as naive estimates. Considering that a recent mechanistic model of time-dependent rates assumes that the long-term viral rate aligns to the host evolutionary rate (Ghafari et al. 2021), and that our EVE calibration relies on the host evolutionary rate, it may be that the deep divergence estimates still provide a reasonable indication of the age of pestiviruses (and flaviviruses). Another study estimates, based on a strong co-divergence hypothesis, emergence of pestiviruses at 465 MY ago (Bamford et al. 2021). While exact dating between our naive estimates and another study somewhat diverges, both strongly support an ancient evolutionary history of these viruses going back at least several hundred MY ago.
In conclusion, we discovered and characterized the first Flaviviridae-related EVEs records from mammalian reference genomes, which derived from pestiviruses. The wide EVEs distribution in shrew Crocidurinae subfamily indicates they are a historical host group of pestiviruses and further suggests an ancient origin time of the Pestivirus genus. Our results show the key role of EVEs not only in expanding our knowledge about ancient viral–host interactions, but also their importance in reconstructing the viral evolutionary history, which contributes to our understanding of viral evolutionary dynamics from ancient times to the present.
Materials and Methods
In Silico Survey
Data Collection
To screen for Flaviviridae-like EVEs, 689 mammalian genomes (57 bats, 9 insectivores, 177 rodents, 101 nonhuman primates, 207 even-toed ungulates, 15 odd-toed ungulates, 108 carnivores, and 15 marsupials), were retrieved from the NCBI Whole Genome Shotgun (WGS) database (last accessed in November 2020). A detailed list of all the surveyed mammalian genomes is provided in Supplementary Table S5, Supplementary Material online. A representative group of 306 Flaviviridae or Flaviviridae-like polyprotein sequences was compiled from the NCBI non-redundant protein database (accessed in February 2019). We provide a list of the nucleotide/protein accession numbers in Supplementary Table S6, Supplementary Material online.
Genome Screening
Flaviviridae polyprotein sequences were used as queries in tBLASTn (BLAST+ v2.6.0) (Camacho et al. 2009) searches with mammalian genomes as targets. We only considered hits with an E-value < 10−4. BLAST hits from the same contig and same orientation with gap size < 100 nt were combined into a single fragment, due to the great possibility of sharing the same integration event and to increase the computational efficiency. To avoid potential artifacts, only the hits with length ≥ 250 nt were extracted from mammalian genomes based on the reported position by BLAST in the host contig. These putative EVEs were then used as query in a reciprocal tBLASTx (BLAST+ v2.6.0) (Camacho et al. 2009) against a local NCBI nucleotide (nt) database (accessed in October 2018) and BLASTx (BLAST+ v2.6.0) (Camacho et al. 2009) against a non-redundant protein (nr) database (accessed in October 2018). EVEs were confirmed if the best hits contained Flaviviridae family members with an E-value < 10−4. The presence of conserved viral genetic features within the hits was assessed using the NCBI Conserved Domain Database (Marchler-Bauer et al. 2015).
EVE Characterization
Upon identification of the EVEs, they were translated and aligned with corresponding polyprotein sequences from several representative Flaviviridae species using MAFFT v7.453 (Katoh et al. 2002). All alignments were trimmed in BMGE v1.12 (Criscuolo and Gribaldo 2010) in order to select for phylogenetic informative regions. The best substitution models were PMB + G4 for the EVE1 alignment, LG + F + G4 for the EVE2 alignment, and LG + F + I + G4 for the concatenated EVEs alignment according to the BIC criterion and were used to construct phylogenetic maximum likelihood (ML) trees with IQ-TREE v1.6.12 (Nguyen et al. 2015).
Flanking Region Analysis
To characterize the EVEs loci and identify potential transposable elements or other genetic features, flanking regions of the identified EVEs were extracted from the host contigs and used as BLAST queries to screen against the NCBI nucleotide (nt) and non-redundant protein (nr) databases (both accessed in October 2018).
Metagenomic Screening
According to the WGS screening results above, some Flaviviridae-related hits were detected in a shrew (Crocidura indochinensis) genome. However, apart from the Crocidura indochinensis and Sorex araneus complete genomes, only a limited number of shrew genomes are currently available in the NCBI WGS database. Therefore, 73 DNA experimental genomic data sets and 6 RNA-Seq transcriptome data sets (Supplementary Table S7, Supplementary Material online) from the Soricidae family were retrieved from NCBI Sequence Read Archive (SRA) database using SRA Toolkit v2.10.8 (Leinonen et al. 2011). Reads were mapped to the identified EVEs nucleotide references using Bowtie2 v2.3.5.1 (Langmead and Salzberg 2012). Alignment files were processed with SAMtools v1.10 (Li et al. 2009) and coverage was determined using bedtools v2.27.1 (Quinlan and Hall 2010) and visualized in RStudio v1.1.463.
In Vivo Validation
Sample Collection
Based on the screening results, to further verify the presence of Flaviviridae-related EVEs in vivo, a total of 65 tissue and DNA samples from species belonging to the Crocidura genus and 6 other related genera of the Soricidae family, namely Paracrocidura, Scutisorex, Suncus, Sylvisorex (subfamily Crocidurinae), Neomys, and Sorex (subfamily Soricinae), were screened for the presence of the identified EVEs. These samples were previously collected in China, Vietnam, Africa, and the Eastern Mediterranean (Supplementary Table S2, Supplementary Material online) as part of other studies (Bannikova et al. 2011; Jenkins et al. 2013; Catalano et al. 2018; Van de Perre et al. 2018, 2019).
Target EVEs and Cytochrome b Amplification
DNA was extracted from tissue samples using the DNeasy Blood & Tissue Kit (Qiagen) following the manufacturer’s instructions.
To screen for the presence of EVEs in vivo, we designed 18 PCR primers (Supplementary Table S8, Supplementary Material online) spanning the 2 EVEs region and a section of the intermediate flanking region from the host genome. Amplicons were generated with DreamTaq DNA Polymerase (ThermoFisher Scientific) using the following cycling conditions: (i) 3 min of denaturation at 95°C; (ii) 35 cycles of 95°C for 30 s, 56°C for 30 s, 72°C extension for 1 min/kb; and (iii) 10 min final extension at 72°C.
To confirm the host species, and to complement available specimen information, the mitochondrial CYTB gene was amplified using general primers (Supplementary Table S8, Supplementary Material online) of the Crocidurinae subfamily. PCR reactions were conducted using the DreamTaq DNA Polymerase (ThermoFisher Scientific) with the following thermal cycling conditions: (i) 3 min of denaturation at 95°C; (ii) 35 cycles of 95°C for 30 s, 56°C for 30 s, 72°C extension for 1 min/kb; and (iii) 10 min final extension at 72°C.
All PCR products were purified using the ExoSAP-IT PCR Product Cleanup (ThermoFisher Scientific) or Zymoclean Gel DNA Recovery Kits (ZYMO Research) to remove primer dimers and unspecific products, following the manufacturer’s instructions.
Sanger Sequencing
The generated PCR products were sequenced by Macrogen Europe. The amplicons were mapped to the whole EVEs region (2,235 nt) from the WGS Crocidura indochinensis contig, and concatenated based on consensus sequence to get the complete EVEs in Geneious Prime® v2020.2.4. CYTB amplicons (∼1,140 nt) were forward and reverse sequenced and a consensus sequence was generated using Geneious Prime® v2020.2.4.
MinION Sequencing
For 12 samples with relatively low-quality Sanger sequencing chromatograms (additional information provided in Supplementary Table S3, Supplementary Material online), MinION sequencing was performed to obtain the complete EVEs region (∼2,235 nt) together with the CYTB gene (∼1,140 nt). The Oxford Nanopore Technologies (ONT) 1D Native barcoding genomic DNA protocol was used without the DNA fragmentation step and the barcoded amplicons were loaded onto the MinION device. We used the MinKNOW software v19.13.5 on the MinIT companion for data acquisition and basecalling. Qcat v1.1.0 (ONT, https://github.com/nanoporetech/qcat) was used to demultiplex reads under the epi2me algorithm and to trim bad quality reads and adapters with min score of 90. The EVE regions extracted from the WGS Crocidura indochinensis contig were used as references to map the reads with Minimap2 v.2.22 (Li 2018) using -ax map-ont parameters. Alignments were converted and indexed using SAMtools v1.10 (Li et al. 2009) and consensus sequences were generated using a custom Python script (Kafetzopoulou et al. 2019).
Phylogenetic Analysis and Visualization
All generated EVEs sequences were translated and aligned with homologous polyproteins from available Pestivirus species using MAFFT v7.453 (Katoh et al. 2002). Sequences of dengue-2 and Zika virus (Flavivirus genus) were used as an outgroup. The alignment was trimmed using BMGE v1.12 (Criscuolo and Gribaldo 2010) and the filtered regions were used to construct ML phylogenetic trees using IQ-TREE v1.6.12 (Nguyen et al. 2015) under the best-fitting models (according to the Bayesian information criterion): LG + G4. Phylogenies were visualized and annotated using FigTree v1.4.4 (Rambaut A; http://tree.bio.ed.ac.uk/software/figtree/). Percent identity matrices were generated using Clustal Omega (Sievers et al. 2011) via EMBL-EBI web services (Madeira et al. 2019).
For species classficiation, CYTB sequences of EVEs-positive specimens (n = 48) were aligned using MAFFT v7.453 (Katoh et al. 2002) together with a data set of n = 393 Soricidae nucleotide sequences downloaded from NCBI. The generated alignment was trimmed in BMGE v1.12 (Criscuolo and Gribaldo 2010) and an ML phylogeny was reconstructed using IQ-TREE v1.6.12 (Nguyen et al. 2015) with the best-fitting model (TIM2 + F + I + G4). The phylogenies were annotated in ggtree v1.14.6 (Yu et al. 2017) and treeio v1.6.2 (Wang et al. 2020) R packages.
To investigate the co-phylogenetic relationships of our newly discovered EVEs and their hosts, we compared the topological structure between the host and EVEs phylogenies using both nuclear and mitochondrial gene information for a variable number of shrew species. For the mitochondrial gene data set, we used our generated CYTB sequences (n = 48), while for the nuclear gene data sets we revisited the most complete mammalian phylogeny up-to-date (Upham et al. 2019) and extracted genomic information for a subset of n = 11 shrew species on the apolipoprotein b gene (APOB) and for n = 10 shrew species on the breast cancer 1 gene (BRCA1). The event-based eMPRess tool (Santichaivekin et al. 2021) was used to assess phylogenetic congruence between the host and EVEs trees. Upon determining the most parsimonious costs for duplications, transfers and losses, we selected the representative reconciliations for the various comparisons and performed a permutation test of 100 randomizations to compute support values. Graphical representation of the congruence level between the EVEs phylogenies and the host trees was performed using the ape v5.0 (Paradis and Schliep 2019) R package.
Characterization of Selective Pressure
We characterized the selective pressure acting on the EVE 1 and EVE 2 region. We respectively aligned the open reading frame of the complete EVE1 (318 nt) and the coding region of EVE 2 fragments (1335 nt) using MEGA v11.0.9 (Tamura et al. 2021). An ML tree was built based on this alignment in IQ-TREE v1.6.12 (Nguyen et al. 2015) under the best-fitting model HKY + F (for EVE1) and HKY + F + G4 (for EVE2). We then conducted two site-specific selection analyses to characterize the selective pressure on each site using estimates of the ratio of non-synonymous/synonymous substitution rate (ω = dN/dS): (i) FEL analyses (Kosakovsky Pond and Frost 2005) using MG94xREV model available in HyPhy software v2.5.3 (Kosakovsky Pond et al. 2005), and (ii) the Bayesian renaissance counting method (Lemey et al. 2012) implemented in BEAST v1.10.5 (Suchard et al. 2018). The value of ω quantifies the selective pressure, with ω > 1 suggesting positive selection, ω = 1 neutral evolution and ω < 1 negative or purifying selection.
Divergence Time Estimation
In order to obtain an estimate of the time-scale of pestivirus evolution, we performed a molecular clock analysis of the pestivirus-EVE amino acid alignment used for phylogenetic analysis (cfr. 4.3). We used a Bayesian approach implemented in BEAST v1.10.5 (Suchard et al. 2018), incorporating an LG + G4 substitution model, a Bayesian skygrid coalescent prior, and an uncorrelated relaxed clock model. Following a divergence time estimate for Crocidurinae, we specify a normal prior with mean 10.8 MYA and standard deviation of 1.6 MY on the EVE most recent common ancestor. We constrained the dengue and Zika virus sequences to form an outgroup in the analyses. Markov chain Monte Carlo (MCMC) analyses were performed for 100 M generations sampling every 10,000th generation. The MCMC runs were diagnosed using Tracer ensuring that all effective sampling sizes were larger than 200.
Supplementary Material
Acknowledgments
The samples from Vietnam were collected during biodiversity surveys carried by the Joint Vietnam-Russian Tropical Research and Technological Centre.
Contributor Information
Yiqiao Li, Department of Microbiology, Immunology and Transplantation, Rega Institute, KU Leuven, 3000 Leuven, Belgium.
Magda Bletsa, Department of Microbiology, Immunology and Transplantation, Rega Institute, KU Leuven, 3000 Leuven, Belgium.
Zafeiro Zisi, Department of Microbiology, Immunology and Transplantation, Rega Institute, KU Leuven, 3000 Leuven, Belgium.
Ine Boonen, Department of Microbiology, Immunology and Transplantation, Rega Institute, KU Leuven, 3000 Leuven, Belgium.
Sophie Gryseels, Department of Microbiology, Immunology and Transplantation, Rega Institute, KU Leuven, 3000 Leuven, Belgium; Belgium Evolutionary Ecology Group, University of Antwerp, 2610 Wilrijk, Belgium.
Liana Kafetzopoulou, Department of Microbiology, Immunology and Transplantation, Rega Institute, KU Leuven, 3000 Leuven, Belgium; Virology Department, Belgium Bernhard Nocht Institute for Tropical Medicine, 20359 Hamburg, Germany.
Joanne P Webster, Department of Pathobiology and Population Science, Royal Veterinary College, University of London, Herts, AL9 7TA, UK.
Stefano Catalano, Department of Pathobiology and Population Science, Royal Veterinary College, University of London, Herts, AL9 7TA, UK.
Oliver G Pybus, Department of Pathobiology and Population Science, Royal Veterinary College, University of London, Herts, AL9 7TA, UK.
Frederik Van de Perre, Belgium Evolutionary Ecology Group, University of Antwerp, 2610 Wilrijk, Belgium.
Haotian Li, Marine College, Shandong University (Weihai), 264209 Weihai, China.
Yaoyao Li, Marine College, Shandong University (Weihai), 264209 Weihai, China.
Yuchun Li, Marine College, Shandong University (Weihai), 264209 Weihai, China.
Alexei Abramov, Laboratory of Theriology, Zoological Institute of the Russian Academy of Sciences, 190121 Saint Petersburg, Russia.
Petros Lymberakis, Natural History Museum of Crete, Iraklio 712 02, Greece.
Philippe Lemey, Department of Microbiology, Immunology and Transplantation, Rega Institute, KU Leuven, 3000 Leuven, Belgium.
Sébastian Lequime, Department of Microbiology, Immunology and Transplantation, Rega Institute, KU Leuven, 3000 Leuven, Belgium; Cluster of Microbial Ecology, Groningen Institute for Evolutionary Life Sciences, University of Groningen, 9747 AG Groningen, the Netherlands.
Supplementary Material
Supplementary data are available at Molecular Biology and Evolution online.
Data Availability
The data underlying this article are available in the article and in its online supplementary material. The EVE and CYTB sequences for all Crocidura species are available in the GenBank Nucleotide Database and can be accessed with the accession numbers available in Supplementary Table S3, Supplementary Material online.
Funding
This work was supported by the European Research Council under the European Union’s Horizon 2020 Framework Programme for research and innovation (grant agreement no. 725422-ReservoirDOCS). P.L. acknowledges support by the Fonds Wetenschappelijk Onderzoek (‘Research Foundation – Flanders’, G066215N, G0D5117N and G0B9317N). Y.Q.L. acknowledges the support of China Scholarship Council. A.A. acknowledges the support by Ministry of Science and Higher Education of the Russian Federation (grant no. 075-15-2021-1069). Y.C.L. acknowledges the support of National Natural Science Foundation of China (grant ID. 31672254). F.V.d.P. acknowledges the support of the Ph.D. fellowship from the Fonds Wetenschappelijk Onderzoek. J.P.W. acknowledges the support of the UK Research and Innovation (grant BB/L018985/1 Zoonoses in Emerging Livestock Systems).
References
- Agapov EV, Murray CL, Frolov I, Qu L, Myers TM, Rice CM. 2004. Uncleaved NS2-3 is required for production of infectious bovine viral diarrhea virus. J Virol. 78:2414–2425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aiewsakun P, Katzourakis A. 2015. Endogenous viruses: connecting recent and ancient viral evolution. Virology. 479–480:26–37. [DOI] [PubMed] [Google Scholar]
- Aiewsakun P, Katzourakis A. 2016. Time-Dependent rate phenomenon in viruses. J Virol. 90:7184–7195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aswad A, Katzourakis A. 2014. The first endogenous herpesvirus, identified in the tarsier genome, and novel sequences from primate rhadinoviruses and lymphocryptoviruses. PLoS Genet. 10(6):e1004332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bamford CGG, de Souza WM, Parry R, Gifford RJ. 2022. Comparative analysis of genome-encoded viral sequences reveals the evolutionary history of flavivirids (family Flaviviridae). Virus Evolution:veac085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bannikova AA, Abramov AV, Borisenko AV, Lebedev VS, Rozhnov VV. 2011. Mitochondrial diversity of the white-toothed shrews (mammalia, eulipotyphla, crocidura) in Vietnam. Zootaxa. 2812:1–20. [Google Scholar]
- Becher P, Tautz N. 2011. RNA Recombination in pestiviruses: cellular RNA sequences in viral genomes highlight the role of host factors for viral persistence and lethal disease. RNA Biol. 8:216–224. [DOI] [PubMed] [Google Scholar]
- Belyi VA, Levine AJ, Skalka AM. 2010a. Sequences from ancestral single-stranded DNA viruses in vertebrate genomes: the parvoviridae and circoviridae are more than 40 to 50 million years old. J Virol. 84:12458–12462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belyi VA, Levine AJ, Skalka AM. 2010b. Unexpected inheritance: multiple integrations of ancient bornavirus and ebolavirus/marburgvirus sequences in vertebrate genomes. PLoS Pathog. 6:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blair CD, Olson KE, Bonizzoni M. 2020. The widespread occurrence and potential biological roles of endogenous viral elements in insect genomes. Curr Issues Mol Biol. 34:13–29. [DOI] [PubMed] [Google Scholar]
- Bletsa M, Vrancken B, Gryseels S, Boonen I, Fikatas A, Li Y, Laudisoit A, Lequime S, Bryja J, Makundi R, et al. 2021. Molecular detection and genomic characterization of diverse hepaciviruses in african rodents. Virus Evol. 7:1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, Madden TL. 2009. BLAST+: architecture and applications. BMC Bioinf. 10:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catalano S, Sène M, Diouf ND, Fall CB, Borlase A, Léger E, Bâ K, Webster JP. 2018. Rodents as natural hosts of zoonotic schistosoma species and hybrids: an epidemiological and evolutionary perspective from West Africa. Journal of Infectious Diseases. 218:429–433. [DOI] [PubMed] [Google Scholar]
- Colmant A, Hobson-Peters J, Bielefeldt-Ohmann H, van den Hurk AF, Hall-Mendelin S, Chow WK, Johansen CA, Fros J, Simmonds P, Watterson D, et al. 2017. A new clade of insect-specific flaviviruses from Australian anopheles mosquitoes displays Species-specific host restriction. mSphere. 2:1–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Criscuolo A, Gribaldo S. 2010. BMGE (Block mapping and gathering with entropy): a new software for selection of phylogenetic informative regions from multiple sequence alignments. BMC Evol Biol. 10:210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crochu S, Cook S, Attoui H, Charrel RN, de Chesse R, Belhouchet M, Lemasson JJ, de Micco P, de Lamballerie X. 2004. Sequences of flavivirus-related RNA viruses persist in DNA form integrated in the genome of aedes spp. Mosquitoes. Journal of General Virology. 85:1971–1980. [DOI] [PubMed] [Google Scholar]
- Dubey S, Diker E, Kurtonur C, Vogel P. 2008. Secondary contact zones and hybridizations: the case of the lesser white-toothed shrew (crocidura suaveolens group, soricidae). Biological Journal of the Linnean Society. 95:557–565. [Google Scholar]
- Dubey S, Salamin N, Ohdachi SD, Barrière P, Vogel P. 2007. Molecular phylogenetics of shrews (mammalia: soricidae) reveal timing of transcontinental colonizations. Mol Phylogenet Evol. 44:126–137. [DOI] [PubMed] [Google Scholar]
- Dubey S, Zaitsev M, Cosson JF, Abdukadier A, Vogel P. 2006. Pliocene and pleistocene diversification and multiple refugia in a eurasian shrew (crocidura suaveolens group). Mol Phylogenet Evol. 38:635–647. [DOI] [PubMed] [Google Scholar]
- Duchêne S, Holmes EC, Ho SY. 2014. Analyses of evolutionary dynamics in viruses are hindered by a time-dependent bias in rate estimates. Proceedings Biological sciences. 281:20140732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duffy S, Shackelton LA, Holmes EC. 2008. Rates of evolutionary change in viruses: patterns and determinants. Nature Reviews Genetics. 9:267–276. [DOI] [PubMed] [Google Scholar]
- Feschotte C, Gilbert C. 2012. Endogenous viruses: insights into viral evolution and impact on host biology. Nature Reviews Genetics. 13:283–296. [DOI] [PubMed] [Google Scholar]
- Flynn PJ, Moreau CS. 2019. Assessing the diversity of endogenous viruses throughout ant genomes. Front Microbiol. 10:1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghafari M, Simmonds P, Pybus OG, Katzourakis A. 2021. A mechanistic evolutionary model explains the time-dependent pattern of substitution rates in viruses. Curr Biol. 31:4689–4696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert C, Feschotte C. 2010. Genomic fossils calibrate the long-term evolution of hepadnaviruses. PLoS Biol. 8:e1000495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo H, Cai C, Wang B, Zhuo F, Jiang R, Wang N, Li B, Zhang W, Zhu Y, Fan Y, et al. 2019. Novel hepacivirus in Asian house shrew, China. Science China Life Sciences. 62:701–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartlage AS. 2016. The strange, expanding world of animal hepaciviruses. Annu Rev Virol. 176:53–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes EC. 2003. Molecular clocks and the puzzle of RNA virus origins. J Virol. 77:3893–3897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes EC. 2011. The evolution of endogenous viral elements. Cell Host and Microbe. 10:368–377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horie M. 2019. Interactions among eukaryotes, retrotransposons and riboviruses: endogenous riboviral elements in eukaryotic genomes. Genes and Genetic Systems. 94:253–267. [DOI] [PubMed] [Google Scholar]
- Horie M, Honda T, Suzuki Y, Kobayashi Y, Daito T, Oshida T, Ikuta K, Jern P, Gojobori T, Coffin JM, et al. 2010. Endogenous non-retroviral RNA virus elements in mammalian genomes. Nature. 463:84–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horie M, Kobayashi Y, Suzuki Y, Tomonaga K. 2013. Comprehensive analysis of endogenous bornavirus-like elements in eukaryote genomes. Phil Trans R Soc B. 368:20120499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkins PD, Abramov AV, Bannikova AA, Rozhnov V. 2013. Bones and genes: resolution problems in three Vietnamese species of crocidura (mammalia, soricomorpha, soricidae) and the description of an additional new species. ZooKeys. 313:61–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kafetzopoulou LE, Pullan ST, Lemey P, Suchard MA, Ehichioya DU, Pahlmann M, Thielebein A, Hinzmann J, Oestereich L, Wozniak DM, et al. 2019. Metagenomic sequencing at the epicenter of the Nigeria 2018 Lassa fever outbreak. Science. 363:74–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Misawa K, Kuma KI, Miyata T. 2002. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 30:3059–3066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katzourakis A, Gifford RJ. 2010. Endogenous viral elements in animal genomes. PLoS Genet. 6:e1001191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawasaki J, Kojima S, Mukai Y, Tomonaga K, Horie M. 2021. 100-My History of bornavirus infections hidden in vertebrate genomes. Proceedings of the National Academy of Sciences. 118:e2026235118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi Y, Shimazu T, Murata K, Itou T, Suzuki Y. 2019. An endogenous adeno-associated virus element in elephants. Virus Res. 262:10–14. [DOI] [PubMed] [Google Scholar]
- Kosakovsky Pond SL, Frost SD. 2005. Not so different after all: a comparison of methods for detecting amino acid sites under selection. Mol Biol Evol. 22:1208–1222. [DOI] [PubMed] [Google Scholar]
- Kosakovsky Pond SL, Frost SD, Muse SV. 2005. Hyphy: hypothesis testing using phylogenies. Bioinformatics. 21:676–679. [DOI] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with bowtie 2. Nat Methods. 9:357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leinonen R, Sugawara H, Shumway M. 2011. The sequence read archive. Nucleic Acids Res. 39:2010–2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemey P, Minin VN, Bielejec F, Pond SL, Suchard MA. 2012. A counting renaissance: combining stochastic mapping and empirical Bayes to quickly detect amino acid sites under positive selection. Bioinformatics. 28:3248–3256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lequime S, Lambrechts L. 2017. Discovery of flavivirus-derived endogenous viral elements in anopheles mosquito genomes supports the existence of anopheles-associated insect-specific flaviviruses. Virus Evol. 3:vew035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H. 2018. Minimap2: pairwise alignment for nucleotide sequences. Bioinformatics. 34:3094–3100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R. 2009. The sequence alignment/map format and SAMtools. Bioinformatics. 25:2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li N, Li Q. 2015. Identification and characterization of endogenous viral elements for the three key schistosomes of humans. Pak J Pharm Sci. 28:375–382. [PubMed] [Google Scholar]
- Liu S, Coates BS, Bonning BC. 2020. Endogenous viral elements integrated into the genome of the soybean aphid, aphis glycines. Insect Biochem Mol Biol. 123:103405. [DOI] [PubMed] [Google Scholar]
- Liu H, Fu Y, Xie J, Cheng J, Ghabrial SA, Li G, Yi X, Jiang D. 2012. Discovery of novel dsRNA viral sequences by in silico cloning and implications for viral diversity, host range and evolution. PLoS ONE. 7:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luhtala N, Parker R. 2010. T2 family ribonucleases: ancient enzymes with diverse roles. Trends Biochem Sci. 35:253–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madeira F, Park YM, Lee J, Buso N, Gur T, Madhusoodanan N, Basutkar P, Tivey ARN, Potter SC, Finn RD, et al. 2019. The EMBL-EBI search and sequence analysis tools APIs in 2019. Nucleic Acids Res. 47:W636–W641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchler-Bauer A, Derbyshire MK, Gonzales NR, Lu S, Chitsaz F, Geer LY, Geer RC, He J, Gwadz M, Hurwitz DI, et al. 2015. CDD: nCBI’s conserved domain database. Nucleic Acids Res. 43:D222–D226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mares-Guia M, Horta MA, Romano A, Rodrigues C, Mendonça M, Dos Santos CC, Torres MC, Araujo E, Fabri A, de Souza ER, et al. 2020. Yellow fever epizootics in non-human primates, southeast and northeast Brazil (2017 and 2018). Parasites and Vectors. 13:1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruyama SR, Castro-Jorge LA, Ribeiro JM, Gardinassi LG, Garcia GR, Brandão LG, Rodrigues AR, Okada MI, Abrão EP, Ferreira BR, et al. 2014. Characterisation of divergent flavivirus NS3 and NS5 protein sequences detected in rhipicephalus microplus ticks from Brazil. Mem Inst Oswaldo Cruz. 109:38–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Membrebe JV, Suchard MA, Rambaut A, Baele G, Lemey P. 2019. Bayesian Inference of evolutionary histories under time-dependent substitution rates. Mol Biol Evol. 36:1793–1803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moureau G, Cook S, Lemey P, Nougairede A, Forrester NL, Khasnatinov M, Charrel RN, Firth AE, Gould EA, de Lamballerie X. 2015. New insights into flavivirus evolution, taxonomy and biogeographic history, extended by analysis of canonical and alternative coding sequences e0117849. PLoS ONE. 10:e0117849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32:268–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Öncü C, Brinkmann A, Günay F, Kar S, Öter K, Sarıkaya Y, Nitsche A, Linton YM, Alten B, Ergünay K. 2018. West Nile virus, anopheles flavivirus, a novel flavivirus as well as merida-like rhabdovirus Turkey in field-collected mosquitoes from thrace and anatolia. Infect Genet Evol. 57:36–45. [DOI] [PubMed] [Google Scholar]
- Ophinni Y, Palatini U, Hayashi Y, Parrish NF. 2019. piRNA-Guided CRISPR-like immunity in eukaryotes. Trends Immunol. 40:998–1010. [DOI] [PubMed] [Google Scholar]
- O’Reilly JE, Dos Reis M, Donoghue P. 2015. Dating tips for divergence-time estimation. Trends Genet. 31:637–650. [DOI] [PubMed] [Google Scholar]
- Paradis E, Schliep K. 2019. Ape 5.0: an environment for modern phylogenetics and evolutionary analyses in R. Bioinformatics. 35:526–528. [DOI] [PubMed] [Google Scholar]
- Parry R, Asgari S. 2019. Discovery of novel crustacean and cephalopod flaviviruses: insights into the evolution and circulation of flaviviruses between marine invertebrate and vertebrate hosts. J Virol. 93:1–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersson JH, Fiz-Palacios O. 2014. Dating the origin of the genus flavivirus in the light of beringian biogeography. Journal of General Virology. 95:1969–1982. [DOI] [PubMed] [Google Scholar]
- Quinlan AR, Hall IM. 2010. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 26:841–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roiz D, Vázquez A, Seco MP, Tenorio A, Rizzoli A. 2009. Detection of novel insect flavivirus sequences integrated in aedes albopictus (Diptera: culicidae) in northern Italy. Virol J. 6:1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sameroff S, Tokarz R, Charles RA, Jain K, Oleynik A, Che X, Georges K, Carrington CV, Lipkin WI, Oura C. 2019. Viral diversity of tick Species parasitizing cattle and dogs in Trinidad and Tobago. Sci Rep. 9:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanjuán R. 2012. From molecular genetics to phylodynamics: evolutionary relevance of mutation rates across viruses. PLoS Pathog. 8:1–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santichaivekin S, Yang Q, Liu J, Mawhorter R, Jiang J, Wesley T, Wu YC, Libeskind-Hadas R. 2021. eMPRess: a systematic cophylogeny reconciliation tool. Bioinformatics. 37:2481–2482. [DOI] [PubMed] [Google Scholar]
- Shi M, Lin XD, Chen X, Tian JH, Chen LJ, Li K, Wang W, Eden JS, Shen JJ, Liu L, et al. 2018. The evolutionary history of vertebrate RNA viruses. Nature. 561:E6-E6. [DOI] [PubMed] [Google Scholar]
- Shi M, Lin XD, Vasilakis N, Tian JH, Li CX, Chen LJ, Eastwood G, Diao XN, Chen MH, Chen X, et al. 2016. Divergent viruses discovered in arthropods and vertebrates revise the evolutionary history of the flaviviridae and related viruses. J Virol. 90:659–669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, Lopez R, McWilliam H, Remmert M, Söding J, et al. 2011. Fast, scalable generation of high-quality protein multiple sequence alignments using clustal Omega. Mol Syst Biol. 7:539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva E, Marques S, Osório H, Carvalheira J, Thompson G. 2012. Endogenous hepatitis C virus homolog fragments in European rabbit and hare genomes replicate in cell culture. PLoS ONE. 7:e49820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva E, Osório H, Thompson G. 2015. Hepatitis C-like viruses are produced in cells from rabbit and hare DNA. Sci Rep. 5:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silveira S, Cibulski SP, Junqueira DM, Mósena ACS, Weber MN, Mayer FQ, Canal CW. 2020. Phylogenetic and evolutionary analysis of HoBi-like pestivirus: insights into origin and dispersal. Transbound Emerg Dis. 67:1909–1917. [DOI] [PubMed] [Google Scholar]
- Skirmuntt EC, Escalera-Zamudio M, Teeling EC, Smith A, Katzourakis A. 2020. The potential role of endogenous viral elements in the evolution of bats as reservoirs for zoonotic viruses. Annu Rev Virol. 7:103–119. [DOI] [PubMed] [Google Scholar]
- Soto E, Camus A, Yun S, Kurobe T, Leary JH, Rosser TG, Dill-Okubo JA, Nyaoke AC, Adkison M, Renger A, et al. 2020. First isolation of a novel aquatic flavivirus from chinook salmon (oncorhynchus tshawytscha) and its in vivo replication in a piscine animal model. J Virol. 94:e00337–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strand TM, Lundkvist Å, Olsen B, Gustafsson L. 2018. Breeding consequences of flavivirus infection in the collared flycatcher. BMC Evol Biol. 18:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suchard MA, Lemey P, Baele G, Ayres DL, Drummond AJ, Rambaut A. 2018. Bayesian Phylogenetic and phylodynamic data integration using BEAST 1.10. Virus Evol. 4:1–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki Y, Baidaliuk A, Miesen P, Frangeul L, Crist AB, Merkling SH, Fontaine A, Lequime S, Moltini-Conclois I, Blanc H, et al. 2020. Non-retroviral endogenous viral element limits cognate virus replication in aedes aegypti ovaries. Curr Biol. 30:3495–3506.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Stecher G, Kumar S. 2021. MEGA11: molecular evolutionary genetics analysis version 11. Mol Biol Evol. 38:3022–3027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor DJ, Leach RW, Bruenn J. 2010. Filoviruses are ancient and integrated into mammalian genomes. BMC Evol Biol. 10. Article no. 193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Upham NS, Esselstyn JA, Jetz W. 2019. Inferring the mammal tree: species-level sets of phylogenies for questions in ecology, evolution, and conservation. PLoS Biol. 17:e3000494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van de Perre F, Leirs H, Cigar J, Mbalitini SG, Itoka JCM, Verheyen E. 2019. Shrews (soricidae) of the lowland forests around kisangani (DR Congo). Biodivers Data J. 7:e46948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van de Perre F, Willig MR, Presley SJ, Andemwana FB, Beeckman H, Boeckx P, Cooleman S, de Haan M, de Kesel A, Dessein S, et al. 2018. Reconciling biodiversity and carbon stock conservation in an afrotropical forest landscape. Sci Adv. 4:eaar6603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel P, Maddalena T, Sarà M. 2004. Crocidura cossyrensis contoli, 1989 (mammalia, soricidae): karyotype, biochemical genetics and hybridization experiments. Revue Suisse de Zoologie. 111:925–934. [Google Scholar]
- Wang LG, Lam TT, Xu S, Dai Z, Zhou L, Feng T, Guo P, Dunn CW, Jones BR, Bradley T, et al. 2020. Treeio: an R package for phylogenetic tree input and output with richly annotated and associated data. Mol Biol Evol. 37:599–603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber MN, Wolf JM, da Silva MS, Mosena AC, Budaszewski RF, Lunge VR, Canal CW. 2021. Insights into the origin and diversification of bovine viral diarrhea virus 1 subtypes. Arch Virol. 166:607–611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Z, Han Y, Liu B, Li H, Zhu G, Latinne A, Dong J, Sun L, Du J, Zhou S, et al. 2021. Decoding the RNA viromes of rodent lungs provides new visions into the origin and evolution pattern of rodent-borne diseases in mainland Southeast Asia. Microbiome 9:18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Z, Liu B, Du J, Zhang J, Lu L, Zhu G, Han Y, Su H, Yang L, Zhang S, et al. 2018. Discovery of diverse rodent and bat pestiviruses with distinct genomic and phylogenetic characteristics in several Chinese provinces. Front Microbiol. 9:1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu G, Smith DK, Zhu H, Guan Y, Lam TT. 2017. Ggtree: an R package for visualization and annotation of phylogenetic trees with their covariates and other associated data. Methods in Ecology and Evolution. 8:28–36. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data underlying this article are available in the article and in its online supplementary material. The EVE and CYTB sequences for all Crocidura species are available in the GenBank Nucleotide Database and can be accessed with the accession numbers available in Supplementary Table S3, Supplementary Material online.