Abstract
The recently discovered bacterial twin-arginine translocation (Tat) pathway was investigated in Streptomyces lividans, a gram-positive organism with a high secretion capacity. The presence of one tatC and two hcf106 homologs in the S. lividans genome together with the several precursor proteins with a twin-arginine motif in their signal peptide suggested the presence of the twin-arginine translocation pathway in the S. lividans secretome. To demonstrate its functionality, a tatC deletion mutant was constructed. This mutation impaired the translocation of the Streptomyces antibioticus tyrosinase, a protein that forms a complex with its transactivator protein before export. Also the chimeric construct pre-TorA-23K, known to be exclusively secreted via the Tat pathway in Escherichia coli, could be translocated in wild-type S. lividans but not in the tatC mutant. In contrast, the secretion of the Sec-dependent S. lividans subtilisin inhibitor was not affected. This study therefore demonstrates that also in general in Streptomyces spp. the Tat pathway is functional. Moreover, this Tat pathway can translocate folded proteins, and the E. coli TorA signal peptide can direct Tat-dependent transport in S. lividans.
In bacteria, three different types of signal peptide-dependent translocation have been described: the Sec-dependent pathway, the signal recognition particle-dependent pathway, and the recently discovered twin-arginine-dependent pathway or Tat pathway, which is structurally and mechanistically related to the ΔpH-dependent pathway in chloroplast thylakoid membranes (11, 32).
The bacterial Tat pathway has been extensively studied in Escherichia coli. At least four proteins—TatA, TatB, TatC, and TatE (7, 36, 43)—are involved in Tat-dependent translocation. The genes coding for the first three components are part of a polycistronic operon, while tatE is located separately on the chromosome. TatA, -B, and -E are sequence-related proteins encoded by homologs of hcf106, a structural gene of the thylakoid ΔpH-dependent pathway (38). They possess a single N-terminal transmembrane helix, followed by a cytoplasmically located amphipatic helix. The TatC protein contains six transmembrane helices. In E. coli, TatB and TatC are essential for Tat-dependent translocation, while TatA and TatE are complements since protein transport is only fully blocked in a tatAE double mutant (7, 36, 37, 43). Very recently, it was shown that the Tat complex contains only TatA, TatB, and TatC and that TatB and TatC act as a unit in both structural and functional terms (8). The precise function of the different components in the translocation of precursor proteins is still to be explained.
Bacterial precursor proteins translocated via the Tat pathway have an unusually long signal peptide with a conserved twin-arginine sequence S/T-R-R-x-φ-φ that is essential for transport (1, 29). The main energy source that drives the translocation process is the proton motive force instead of nucleoside triphosphates, as in the case of Sec-dependent translocation (35). A striking feature of this newly discovered translocation system is its ability to transport folded proteins eventually bound to a cofactor before export (33, 35). In addition to E. coli, the functionality of the Tat pathway has been demonstrated for Zymomonas mobilis (16), Ralstonia eutropha (3), and Bacillus subtilis (17).
We describe here the analysis of the Tat pathway in Streptomyces lividans. This species has a naturally high secretion capacity and is used as an efficient host for heterologous protein production (5, 23, 40). First, the tatA, -B, and -C genes of S. lividans were identified. To demonstrate the functionality of the Tat pathway, we constructed a tatC deletion mutant and tested the translocation of two putative Tat-dependent precursor proteins, the Streptomyces antibioticus tyrosinase and the chimeric pre-TorA-23K, shown to be translocated via the Tat pathway in E. coli (7).
MATERIALS AND METHODS
Bacterial strains and growth conditions.
S. lividans TK24 and derivatives were grown at 27°C with continuous shaking at 300 rpm in phage medium (21) or NM medium (41). For solid medium, R2 medium was used (29). Where necessary, thiostrepton (at 50 μg/ml in solid medium or at 10 μg/ml in liquid medium), apramycin (50 μg/ml), or kanamycin (50 μg/ml) was added. Protoplast formation and transformation of S. lividans was carried out as described by Kieser et al. (19).
E. coli TG1 was used as host for cloning purposes. Cultures were grown in Luria-Bertani medium at 37°C (300 rpm) supplemented with ampicillin (50 μg/ml), tetracycline (15 μg/ml), apramycin (50 μg/ml), kanamycin (50 μg/ml), or chloramphenicol (25 μg/ml), if necessary. Plasmids used in this work are listed in Table 1.
TABLE 1.
Plasmids used in this work
| Plasmid | Relevant propertiesa | Source or reference |
|---|---|---|
| E. coli plasmids | ||
| pBluescript KS(+) | Multiple cloning site; Apr | Stratagene |
| pGEM-T-Easy | Used for cloning of PCR products and for the construction of a tatC deletion mutant; Apr | Promega |
| pBSKAN | pBluescript KS(+) derivative with an inserted neo gene; Kanr Apr | This work |
| pMW18 | pBluescript KS(+) derivative containing pre-TorA-23K; Apr | 7 |
| pBS-CBSS | pBluescript KS(+) derivative containing the S. venezuelae vsi promoter | 23 |
| Streptomyces plasmids | ||
| pIJ486 | Multiple cloning site; Tsrr | 42 |
| pVsiTor23K | pIJ486 derivative; contains pre-TorA-23K after the S. venezuelae vsi promoter; Tsrr | This work |
| pIJ702 | contains the S. antibioticus melC operon; Tsrr | 18 |
| E. coli/Streptomyces shuttle vectors | ||
| pFD666 | Kanr | 13 |
| pGM160 | temperature sensitive replicon; Apr Tsrr | 28 |
| pGMΔtatC | pGM160 derivative used to knock out S. lividans tatC; Kanr Tsrr | This work |
| Integrative vectors containing φC31 att-locus | ||
| pSET152 | Integrative plasmid; Aprr | 4 |
| pSETtatC | pSET152 derivative containing S. lividans tatC | This work |
Apr, ampicillin resistance; Aprr, apramycin resistance; Kanr, kanamycin resistance; Tsrr, thiostrepton resistance.
DNA techniques and vector constructions.
All DNA manipulations were performed by using standard techniques (34), and the restriction enzymes were from Life Technologies (Rockville, Md.). DNA sequence analysis was carried out according to the dideoxy chain termination method with the Thermo Sequenase Primer Cycle Sequencing Kit 7-Deaza-dGTP on an ALFexpress apparatus (Amersham Pharmacia Biotech, Rainham, United Kingdom).
PCR for isolation of tatA, tatB, and tatC was performed with Pfu polymerase (Stratagene, La Jolla, Calif.) in the presence of 10% dimethyl sulfoxide with the primers tat1 (5′-GAACGGCTGAAACCCGCCAC) and tat2 (5′-CCACGTTCCCATCTCGTGCG) for the amplification of tatA, tat3 (5′-GGGGCGGATGCCGCTCGC) and tat4 (5′-CCTCAGGTCACGTCGTCG) for the amplification of tatC, and tat5 (5′-GTGTTCAATGACATAGGCGC) and tat6 (5′-CCGCGCGGGCGTCGCCTTCA) for the amplification of tatB. A DNA fragment containing both tatA and tatC was isolated by using the primers tat1 and tat4. The PCR products were cloned in pGEM-T Easy (Promega, Madison, Wis.), and the DNA sequence of the inserts was determined.
For the construction of a tatC deletion mutant of S. lividans, we used the E. coli-Streptomyces shuttle vector pGM160 (28). First, we amplified the kanamycin resistance gene (neo) from pFD666 (13) with primers KAN1 (5′-CAGGGGGGCGGAGCCTATGG) and KAN2 (5′-TACTGCGGCCGCGATCCAAGC). After cloning of the PCR fragment in pGEM®-T Easy, it was digested with EcoRI and cloned into pBluescript KS(+) (Stratagene), resulting in pBSKAN. Then, via a PCR reaction with primers tat7 (5′-CGCAGGGCCGGTGATCTCC) and tat8 (5′-CGGAGTTCACGCTGTTGT), a DNA fragment containing tatC with neighboring regions was isolated. After cloning of this PCR fragment in pGEM-T Easy, two resulting pGEMtatC vectors with different orientations were digested with SacI and XhoI-SalI. The 0.3-kb SacI fragment consisting of the upstream region of tatC was cloned into the SacI site of pBSKAN, while the 0.6-kb blunted XhoI/SalI fragment containing the downstream region of tatC was cloned into the blunted XhoI site of pBSKAN. The resulting plasmid contains a tatC derivative in which a 730-bp internal fragment was replaced by neo. The complete fragment was amplified by PCR by using the M13 primers, and it was cloned into pGEM-T Easy. Finally, from this vector the 2.2-kb ApaI fragment was cloned into the blunted HindIII site of pGM160, resulting in plasmid pGMΔtatC. After polyethylene glycol-mediated transformation of S. lividans protoplasts with pGMΔtatC, a temperature shift to 39°C promoted the integration of the temperature-sensitive replicon into the chromosomal DNA. The tatC deletion resulting from double homologous recombination was confirmed by PCR analysis.
In order to complement the tatC deletion, a DNA fragment containing the tatC gene was amplified with primers tat9 (5′-CGCAGGGCCGGTGATCTCC) and tat8 (5′-CGGAGTTCACGCTGTTGT) and cloned in pGEM-T Easy. After treatment of the vector with EcoRI, this fragment was cloned into the EcoRI site of the integrative vector pSET152 (4), resulting in pSETtatC.
For the expression of pre-TorA-23K in S. lividans, the gene was placed under control of the Streptomyces venezuelae subtilisin inhibitor (vsi) promoter. Therefore, a PstI/XhoI fragment containing pre-TorA-23K was isolated from pMW18 (7) and cloned into pBS-CBSS (23). From this construct, a BamHI/XhoI fragment containing the vsi promoter and the downstream pre-TorA-23K coding region was cloned into the streptomycete multicopy plasmid pIJ486 (42) to give pVsiTor23K.
Immunoblot analysis.
Western blot analysis was performed to check the translocation of pre-TorA-23K in S. lividans. Extracellular fractions of 30-h S. lividans cultures were obtained by centrifugation (10 min, 4,200 × g, 0°C). Proteins in the growth medium were precipitated with trichloroacetic acid (final concentration of 20%) and separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) (22). Transfer of proteins onto a nitrocellulose Porablot membrane (Macherey-Nagel, Düren, Germany) was performed by using a Bio-Rad Transblot semidry transfer cell (Bio-Rad, Hercules, Calif.) according to the manufacturer's recommendations. The 23K derivatives were visualized with rabbit anti-23K antibodies and alkaline phosphatase-conjugated goat anti-rabbit antibodies (Sigma, St. Louis, Mo.).
Activity assays.
The tyrosinase activity was measured using the dopachrome assay procedure with l-3,4-dihydroxyphenylalanine (l-DOPA) as a substrate (18). We used 48-h precultures in phage medium (21) to inoculate 5-ml cultures of tyrosinase production medium (NM medium supplemented with 0.0375% l-tyrosine, 0.75% tiger milk [19], and 2 μM CuSO4) that were subsequently cultivated for 6 h. Supernatants were diluted in the assay buffer (0.1 M sodium phosphate buffer; pH 6.2), and the tyrosinase activity was measured immediately. The intracellular amount of tyrosinase was determined on cell lysates obtained by sonication (2 min, 20,000 Hz, 0°C) of the mycelia suspended in assay buffer. One unit of tyrosinase was defined as the amount of enzyme that converts 1 μmol of l-DOPA into dopachrome/min.
The inhibitory activity of subtilisin inhibitor was determined in the presence of the substrate N-succinyl-l-Ala-l-Ala-l-Pro-l-Phe-p-nitroanilide as described by Kojima et al. (20). Cultures were grown in tyrosinase production medium, and supernatants and intracellular fractions were obtained in the same way as described for the sampling of the tyrosinase test. One unit of subtilisin inhibitor activity was defined as the percentage of inhibition of 5 μg of subtilisin/ml during 10 min of incubation at 25°C.
RESULTS
Cloning and sequencing of the S. lividans tatA, -B, and -C genes.
In the S. coelicolor genome bank (http://www.sanger.ac.uk), we found a tatA, tatB, and tatC homolog. The tatA and tatC gene are clustered (cosmid SCI41), while tatB is located elsewhere on the chromosome (cosmid SCP8). With primers based on the S. coelicolor sequence, we were able to amplify the corresponding genes from S. lividans TK24. tatA and tatC were isolated on a 1,617-bp fragment and are separated by 92 bp. tatB, on the other hand, was isolated as a 580-bp fragment. While the S. lividans and S. coelicolor tatA genes are identical, the tatC and tatB genes of both strains show 99.8 and 99.6% identities at the nucleotide level and 100 and 98.1% identities at the amino acid level, respectively. CLUSTALW similarity scores with analogous proteins of E. coli and B. subtilis are rather low (19 to 24% for E. coli and 22 to 27% for B. subtilis). All DNA sequences were submitted to the EMBL bank (accession numbers AJ251128, AJ292256, and AJ251149 for tatA, -B, and -C, respectively).
Construction of a tatC deletion mutant.
TatC is an essential component of the E. coli Tat pathway (7). To prove the functionality of the twin-arginine translocation pathway in S. lividans, we therefore decided to knock out tatC. To do this, a 730-bp internal fragment of tatC comprising the six transmembrane domains was replaced with the neo gene in the S. lividans chromosome by using the integrative E. coli-Streptomyces shuttle vector pGMΔtatC. ΔtatC mutants differed phenotypically from the wild type. Colonies grown on R2 medium (30) lacked red pigmentation, and the mycelium grew very dispersed in liquid medium in contrast to the mycelial aggregates of the wild-type strain.
Translocation of pre-TorA-23K is tatC dependent.
Pre-TorA-23K is a chimeric construct that consists of the signal peptide and the first six amino acids of the mature E. coli trimethylamine N-oxide reductase (TMAO) fused to the mature part of the 23-kDa subunit of the plant photosystem II oxygen-evolving complex (7). It has clearly been proven that the pre-TorA-23K protein is translocated via the Tat pathway in E. coli (7) and via the ΔpH pathway in thylakoid membranes (44).
To investigate whether pre-TorA-23K is also Tat dependent in S. lividans, the translocation efficiency was compared in pVsiTor23K transformants of S. lividans TK24 and its ΔtatC mutant. Western blot analysis of extracellular protein fractions from the wild-type strain revealed an immunoreactive band of 26 kDa (Fig. 1), somewhat larger than expected for the fusion protein. On the other hand, no 23K-specific band was detected in the supernatant of the ΔtatC mutant containing pVsiTor23K (Fig. 1). These results showed that pre-TorA-23K could not be translocated in the ΔtatC mutant. Intracellular accumulation of pre-TorA-23K in the ΔtatC mutant could not be detected. To prove that S. lividans ΔtatC(pVsiTor23K) could express but not secrete pre-TorA-23K, a plasmid was constructed to complement the disrupted tatC gene. To do this, wild-type tatC transcribed from its own promoter was inserted into the integrative plasmid pSET152, resulting in pSETtatC. tatC is transcribed from its own promoter and does not constitute an operon with tatA, as revealed by transcription analysis experiments (data to be published). Western blot analysis of supernatants of 30-h cultures of S. lividans ΔtatC(pVsiTor23K) transformed with pSETtatC showed that the 26-kDa band appeared again (Fig. 1). The result indicated that the integrated copy of tatC restored the translocation of pre-TorA-23K in ΔtatC, and tatC was thus essential for translocation of pre-TorA-23K. The 23K-specific immunoreactive band observed in the supernatant of the tatC complemented strain was less intense than in the wild-type strain. Therefore, it is obvious that the integrated tatC copy complemented the deleted tatC, but secretion was not at the same level as in the original wild-type strain.
FIG. 1.
Western blotting analysis with 23K antibodies to detect TorA-23K in supernatants of 30-h cultures from S. lividans TK24 (lane 1), S. lividans(pVsiTor23K) (lane 2), S. lividans ΔtatC(pVsiTor23K) (lane 3), and S. lividans ΔtatC(pVsiTor23K)(pSETtatC) (lane 4) grown in NB medium. In each lane, the amount of supernatant corresponding to 3 mg of dry weight was loaded. A 23K-specific immunoreactive band is present in wild-type S. lividans carrying pVsiTor23K, absent in S. lividans ΔtatC(pVsiTor23K), but restored in S. lividans ΔtatC(pVsiTor23K)(pSETtatC).
Translocation of S. antibioticus tyrosinase is tatC dependent.
In addition to the heterologous pre-TorA-23K, we also tested the tatC dependence of the S. antibioticus tyrosinase secretion. Tyrosinase (EC 1.14.18.1) is a copper-containing monooxygenase that catalyzes the formation of melanin pigment from tyrosine (25). The S. antibioticus tyrosinase is encoded by melC2, the second open reading frame of the melanin operon (melC). The upstream gene melC1 encodes a transactivator protein with a twin-arginine signal peptide that was demonstrated to be essential for both tyrosinase activation and secretion (10, 26).
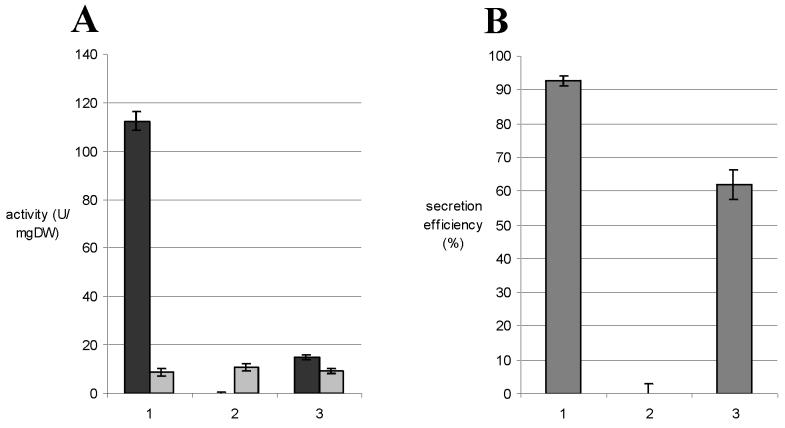
To test the Tat dependence of tyrosinase secretion, S. lividans TK24 and its ΔtatC mutant were transformed with plasmid pIJ702 (18) encoding the S. antibioticus melC operon, and the tyrosinase activity was assayed. Figure 2A shows the tyrosinase activities in cell lysates and supernatants of cultures grown for 6 h in tyrosinase production medium. At this growth stage, the activities reached a maximum value. Thereafter, decreasing values were measured, probably as a consequence of a lack of copper ions in the medium, since after the addition of extra Cu2+ the tyrosinase activity increased again (data not shown). The activity of tyrosinase in the supernatant of the wild-type strain was ca. 112 U/mg (dry weight) (Fig. 2A). In contrast, only trace amounts of tyrosinase activity could be detected in ΔtatC supernatant, indicating that tyrosinase was not translocated in this mutant. The rather low intracellular tyrosinase activity detected in the mutant and as a consequence, a much lower total amount of activity (ca. 11% of the wild type), indicated that there was no intracellular accumulation of tyrosinase in the ΔtatC strain (Fig. 2A), a similar phenomenon as that observed for pre-TorA-23K. In the wild-type strain, 92.7% of the tyrosinase was secreted in contrast to almost 0% in the mutant (Fig. 2B).
FIG. 2.
(A) Tyrosinase activity in cell lysates (shaded bars) and supernatants (solid bars) of S. lividans TK24(pIJ702) (set 1), S. lividans ΔtatC(pIJ702) (set 2), and S. lividans ΔtatC(pIJ702)(pSETtatC) (set 3). The efficient translocation of tyrosinase in the wild-type strain was totally abolished in ΔtatC but could partially be restored by acquisition of a tatC gene. (B) Secretion efficiency of tyrosinase (i.e., ratio extracellular/total amount × 100) in S. lividans TK24(pIJ702) (bar 1), S. lividans ΔtatC(pIJ702) (bar 2), and S. lividans ΔtatC(pIJ702)(pSETtatC) (bar 3).
After complementation of ΔtatC(pIJ702) with pSETtatC, the tyrosinase secretion was restored, although only partially. As mentioned above, this phenomenon was also observed for pre-TorA-23K translocation when ΔtatC was complemented with the intact tatC. The tyrosinase activity in the supernatant was only ca. 15 U/mg (dry weight) (Fig. 2A, set 3), but the efficiency of secretion was restored to 61.9% (Fig. 2B). The lack of tyrosinase secretion in the S. lividans ΔtatC mutant and the complementation experiments confirmed the tatC dependence of the tyrosinase secretion. Therefore, it could be concluded that S. antibioticus tyrosinase is a substrate of the Tat pathway in S. lividans.
Because it has already been shown that the two components of tyrosinase, MelC1 and MelC2, form a complex before export (10, 26), our results show that, similar to E. coli and some other species (3, 15, 16, 35, 40), the Tat pathway of Streptomyces can translocate folded proteins.
tatC is not involved in Sec-dependent secretion.
As a supplementary proof that S. lividans ΔtatC is a specific mutant that only affects Tat-dependent translocation and not the Sec-dependent secretion process, we investigated the secretion of the S. lividans subtilisin inhibitor, a Sec pathway-dependent substrate (unpublished data). Therefore, S. lividans TK24 and its ΔtatC strain were cultured in tyrosinase production medium. After 15 and 24 h of growth, subtilisin inhibition activity in the culture medium was measured. Values of ca. 26 and 45 U/mg (dry weight) were obtained after 15 and 24 h, respectively, both for wild-type S. lividans and the ΔtatC mutant (Fig. 3). These results showed that a disrupted TatC does not affect the Sec-dependent secretion of subtilisin inhibitor.
FIG. 3.
Extracellular subtilisin inhibitor activity expressed as units per milligram of dry weight (DW) of culture of S. lividans TK24 (shaded bars) and ΔtatC (solid bars) at 15 and 24 h of incubation.
DISCUSSION
The experiments described in this report demonstrate that the translocation of two tested precursor proteins containing a twin-arginine signal peptide, i.e., the heterologous chimeric pre-TorA-23K and the S. antibioticus tyrosinase, is blocked in a tatC deletion mutant of S. lividans TK24. As a consequence, these results prove the existence of a functional twin-arginine-dependent translocation pathway in S. lividans.
The chimeric pre-TorA-23K, known to be translocated via the Tat pathway in E. coli (7), is also translocated through this pathway in S. lividans. Moreover, although the homologous components of the Tat complexes of E. coli and S. lividans have rather low similarity scores (ranging from 19 to 24%), the signal peptide of the E. coli TMAO-reductase can direct Tat-dependent transport in S. lividans. This is an indication of the conserved nature of the Tat mechanism. Also, the signal peptides of the Desulfovibrio vulgaris [NiFe]-hydrogenase (29) and of the high potential iron-sulfur protein of Chromatium vinosum (9) can direct Tat-dependent transport in E. coli. Moreover, signal peptides are functionally interchangeable between the bacterial Tat pathway and the ΔpH pathway in thylakoid membranes of chloroplasts (15, 27). An example of species-specific recognition, however, is observed in Zymomonas mobilis (6). It was shown for this species that the Tat-dependent glucose-fructose oxidoreductase (GFOR) could only be exported to the periplasm in E. coli when its own signal peptide was replaced with an E. coli Tat signal peptide. It was suggested that specific recognition events should exist between Tat signal peptides and one or more components of the Tat translocase. As a consequence, signal peptides of Tat-dependent precursors would be optimally adapted specifically to their cognate export apparatus.
On the other hand, Cristóbal et al. (12) showed that not only the signal peptide but also the mature part of the precursor can be a determinant for Tat-dependent transport. The fusion between the TorA signal peptide and the Sec-dependent leader peptidase, for instance, did not reroute the translocation to the Tat pathway, indicating that Sec-targeting signals in Lep could override the Tat-targeting information in the TorA signal peptide.
The secretion of S. antibioticus tyrosinase has previously been investigated but was not fully understood (26). The melanin operon (melC) consists of two genes coding for MelC1, a transactivator protein, and the apotyrosinase MelC2. Only MelC1 has a signal peptide and can mediate MelC2 activation by a binary complex formation followed by copper insertion (10). A functional MelC1 is also essential for tyrosinase secretion (26). How the MelC1/MelC2 complex could cross the membrane was not evident, because Sec and SRP pathways cannot translocate folded proteins. As experimentally proven in this study and already suggested by Berks et al. (2), tyrosinase is translocated via the Tat pathway. No secretion was detectable in the tatC mutant, while in the wild-type S. lividans secretion of tyrosinase was very efficient and probably occurred cotranslationally or immediately after translation, since only very low amounts of tyrosinase could be detected intracellularly.
The translocation of folded proteins in Streptomyces can now be explained by the functionality of the Tat pathway. E. coli hydrogenase 2 is also encoded by an operon with two genes, of which only one is coding for a protein containing a signal peptide. This type of “hitchhiker” mechanism is also used by S. lividans to cotranslocate the two subunits of tyrosinase.
The complementation of ΔtatC with a wild-type copy was not complete. The reason for this effect is not yet understood. Gene dose effect cannot be the reason because the disrupted tatC gene was replaced by one intact copy. However, positional effects of integration or a lack of regulatory sequences present in the neighborhood of the wild-type tatC gene might result in a different transcription efficiency.
For both pre-TorA-23K and tyrosinase, an intracellular accumulation of the preprotein was not observed when the secretion process was blocked. As a consequence, the total amount of pre-TorA-23K or tyrosinase in the ΔtatC cultures was much lower than in those of the wild type. This effect was also noticed by Jongbloed et al. (17), who investigated the secretion of PhoD in a tatCd mutant of B. subtilis. These authors postulated that the intracellular amount of preprotein is not folded and therefore not stable. In the case of tyrosinase, it has been described that the two subunits of tyrosinase form an intracellular complex (10). The proteolytic breakdown of this complex is unlikely. However, if the total amount of the respective components should be unequal in the S. lividans ΔtatC, as already observed with some signal peptide mutants of MelC1 (26), it might be that the component in excess becomes sensitive to proteases. When Leu et al. (26) demonstrated low intracellular accumulation of tyrosinase, they agreed with the hypothesis suggested by Oliver (31) and Gennity et al. (14) that a coupling between secretion and translation exists. A blocked secretion apparatus would have a negative feedback on translation of the protein. The results obtained in this study and in our previous work with Sec-dependent translocated proteins (23, 24) are in agreement with a negative feedback on the translation, or even transcription, of the precursor as a consequence of inhibiting secretion. The mechanism of this negative feedback has to be further investigated.
The TatC component of the Tat complex in S. lividans is essential for the translocation of the two precursor proteins under investigation, i.e., S. antibioticus tyrosinase and chimeric pre-TorA-23K. In E. coli, the unique tatC copy is essential for the translocation of all Tat pathway precursors studied this far (7). However, in B. subtilis, two tatC copies were detected but only one of these is essential for the Tat-dependent translocation of PhoD, indicating that tatC is a specificity determinant for protein secretion via the Tat pathway (17). In the fully sequenced genomes of Mycobacterium tuberculosis and Mycobacterium leprae, two species classified to be in the same order of the Actinomycetales as Streptomyces, only one tatC copy was identified. The S. coelicolor genome project is now finished, but no second tatC homolog was detected; neither was one observed by hybridization experiments (data not shown). Further studies are required to characterize the components of the S. lividans Tat system at the molecular level and to investigate the interactions between the Tat translocon components and between these proteins and the Tat-dependent precursors. In addition, the underlying reason for the altered phenotype observed for the ΔtatC mutant, indicating that the mutation has a pleiotropic effect, as also noticed for E. coli, where it resulted in an impaired cell separation and a defective cell envelope (39), is not understood.
ACKNOWLEDGMENTS
We thank Tracy Palmer for kindly providing the pMW18 plasmid and Colin Robinson for the 23K antibodies.
Kristien Schaerlaekens and Nick Geukens are research fellows of IWT (Vlaams Instituut voor de Bevordering van het Wetenschappelijk-Technologisch Onderzoek in de Industrie). Michaela Schierová is a Marie Curie Fellow of the European Commission (MCF1-2000-01142). This study was further supported by grants G.0271.98 and 1.5.1.7.01 from FWO (Fonds Wetenschappelijk Onderzoek–Vlaanderen), grant OT/00/37 from K. U. Leuven, and grant QLK3-2000-00122 from the EC.
K.S. and M.S. contributed equally to this study.
REFERENCES
- 1.Berks B C. A common export pathway for proteins binding complex redox cofactors? Mol Microbiol. 1996;22:393–404. doi: 10.1046/j.1365-2958.1996.00114.x. [DOI] [PubMed] [Google Scholar]
- 2.Berks B C, Sargent F, Palmer T. The Tat protein export pathway. Mol Microbiol. 2000;35:260–274. doi: 10.1046/j.1365-2958.2000.01719.x. [DOI] [PubMed] [Google Scholar]
- 3.Bernhard M, Friedrich B, Siddiqui R A. Ralstonia eutropha TF93 is blocked in Tat-mediated protein export. J Bacteriol. 2000;182:581–588. doi: 10.1128/jb.182.3.581-588.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bierman M, Logan R, O'Brien K, Seno E T, Rao R N, Schoner B E. Plasmid cloning vectors for the conjugal transfer of DNA from Escherichia coli to Streptomyces spp. Gene. 1992;116:43–49. doi: 10.1016/0378-1119(92)90627-2. [DOI] [PubMed] [Google Scholar]
- 5.Binnie C, Cossar J D, Stewart D I. Heterologous biopharmaceutical protein expression in Streptomyces. Trends Biotechnol. 1997;15:315–320. doi: 10.1016/s0167-7799(97)01062-7. [DOI] [PubMed] [Google Scholar]
- 6.Blaudeck N, Sprenger G A, Freudl R, Wiegert T. Specificity of signal peptide recognition in Tat-dependent bacterial protein translocation. J Bacteriol. 2001;183:604–610. doi: 10.1128/JB.183.2.604-610.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bogsch E G, Sargent F, Stanley N R, Berks B C, Robinson C, Palmer T. An essential component of a novel bacterial protein export system with homologues in plastids and mitochondria. J Biol Chem. 1998;273:18003–18006. doi: 10.1074/jbc.273.29.18003. [DOI] [PubMed] [Google Scholar]
- 8.Bolhuis A, Mathers J E, Thomas J D, Barrett C L, Robinson C. TatB and TatC form a functional and structural unit of the twin-arginine translocase from Escherichia coli. J Biol Chem. 2001;276:20213–20219. doi: 10.1074/jbc.M100682200. [DOI] [PubMed] [Google Scholar]
- 9.Brüser T, Deutzmann R, Dahl C. Evidence against the double-arginine motif as the only determinant for protein translocation by a novel Sec-independent pathway in Escherichia coli. FEMS Microbiol Lett. 1998;164:329–336. doi: 10.1111/j.1574-6968.1998.tb13106.x. [DOI] [PubMed] [Google Scholar]
- 10.Chen L Y, Leu W M, Wang K T, Lee Y H. Copper transfer and activation of the Streptomyces apotyrosinase are mediated through a complex formation between apotyrosinase and its trans-activator MelC1. J Biol Chem. 1992;267:20100–20107. [PubMed] [Google Scholar]
- 11.Cline K, Henry R, Li C, Yuan J. Multiple pathways for protein transport into or across the thylakoid membrane. EMBO J. 1993;12:4105–4114. doi: 10.1002/j.1460-2075.1993.tb06094.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cristóbal S, de Gier J-W, Nielsen H, von Heijne G. Competition between Sec- and TAT-dependent protein translocation in Escherichia coli. EMBO J. 1999;18:2982–2990. doi: 10.1093/emboj/18.11.2982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Denis F, Brzezinski R. A versatile shuttle cosmid vector for use in Escherichia coli and actinomycetes. Gene. 1992;111:115–118. doi: 10.1016/0378-1119(92)90611-r. [DOI] [PubMed] [Google Scholar]
- 14.Gennity J, Goldstein J, Inouye M. Signal peptide mutants of Escherichia coli. J Bioenerg Biomembr. 1990;22:233–269. doi: 10.1007/BF00763167. [DOI] [PubMed] [Google Scholar]
- 15.Halbig D, Hou B, Freudl R, Sprenger G A, Klösgen R B. Bacterial proteins carrying twin-R signal peptides are specifically targeted by the delta pH-dependent transport machinery of the thylakoid membrane system. FEBS Lett. 1999;447:95–98. doi: 10.1016/s0014-5793(99)00269-0. [DOI] [PubMed] [Google Scholar]
- 16.Halbig D, Wiegert T, Blaudeck N, Freudl R, Sprenger G A. The efficient export of NADP-containing glucose-fructose oxidoreductase to the periplasm of Zymomonas mobilis depends both on an intact twin-arginine motif in the signal peptide and on the generation of a structural export signal induced by cofactor binding. Eur J Biochem. 1999;263:543–551. doi: 10.1046/j.1432-1327.1999.00536.x. [DOI] [PubMed] [Google Scholar]
- 17.Jongbloed J D H, Martin U, Antelmann H, Hecker M, Tjalsma H, Venema G, Bron S, van Dijl J M, Müller J. TatC is a specificity determinant for protein secretion via the twin-arginine translocation pathway. J Biol Chem. 2000;275:41350–41357. doi: 10.1074/jbc.M004887200. [DOI] [PubMed] [Google Scholar]
- 18.Katz E, Thompson C J, Hopwood D A. Cloning and expression of the tyrosinase gene from Streptomyces antibioticus in Streptomyces lividans. J Gen Microbiol. 1983;129:2703–2714. doi: 10.1099/00221287-129-9-2703. [DOI] [PubMed] [Google Scholar]
- 19.Kieser T, Bibb M J, Buttner M J, Chater K F, Hopwood D A. Practical Streptomyces genetics. Norwich, United Kingdom: John Innes Centre; 2000. [Google Scholar]
- 20.Kojima S, Obata S, Kumagai I, Miura K. Alteration of the specificity of the Streptomyces subtilisin inhibitor by gene engineering. Biotechnology. 1990;8:449–452. doi: 10.1038/nbt0590-449. [DOI] [PubMed] [Google Scholar]
- 21.Korn F, Weingärtner B, Kutzner H J. A study of twenty actinophages: morphology, serological relationship and host range. In: Freechsen E, Tarnak I, Thumin J H, editors. Genetics of the Actinomycetales. Stuttgart, Germany: Fisher G; 1978. pp. 251–270. [Google Scholar]
- 22.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 23.Lammertyn E, Van Mellaert L, Schacht S, Dillen C, Sablon E, Van Broekhoven A, Anné J. Evaluation of a novel subtilisin inhibitor gene and mutant derivatives for the expression and secretion of mouse tumor necrosis factor alpha by Streptomyces lividans. Appl Environ Microbiol. 1997;63:1808–1813. doi: 10.1128/aem.63.5.1808-1813.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lammertyn E, Desmyter S, Schacht S, Van Mellaert L, Anné J. Influence of charge variation in the Streptomyces venezuelae alpha-amylase signal peptide on heterologous protein production by Streptomyces lividans. Appl Microbiol Biotechnol. 1998;49:424–430. doi: 10.1007/s002530051193. [DOI] [PubMed] [Google Scholar]
- 25.Lerch K. Amino acid sequence of tyrosinase from Neurospora crassa. Proc Natl Acad Sci USA. 1978;75:3605–3609. doi: 10.1073/pnas.75.8.3635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Leu W M, Chen L Y, Liaw L L, Lee Y H. Secretion of the Streptomyces tyrosinase is mediated through its trans-activator protein, MelC1. J Biol Chem. 1992;267:20108–20113. [PubMed] [Google Scholar]
- 27.Mori H, Cline K. A signal peptide that directs non-Sec transport in bacteria also directs efficient and exclusive transport on the thylakoid ΔpH pathway. J Biol Chem. 1998;273:11405–11408. doi: 10.1074/jbc.273.19.11405. [DOI] [PubMed] [Google Scholar]
- 28.Muth G, Nussbaumer B, Wohlleben W, Pühler A. A vector system with temperature-sensitive replication for gene disruption and mutational cloning in streptomycetes. Mol Gen Genet. 1989;219:341–348. [Google Scholar]
- 29.Nivière V, Wong S-L, Voordouw G. Site-directed mutagenesis of the hydrogenase signal peptide consensus box prevents export of a β-lactamase fusion protein. J Gen Microbiol. 1992;138:2173–2183. doi: 10.1099/00221287-138-10-2173. [DOI] [PubMed] [Google Scholar]
- 30.Okanishi M, Suzuki K, Umezawa H. Formation and reversion of Streptomycete protoplasts: cultural condition and morphological study. J Gen Microbiol. 1974;80:389–400. doi: 10.1099/00221287-80-2-389. [DOI] [PubMed] [Google Scholar]
- 31.Oliver D. Protein secretion in Escherichia coli. Annu Rev Microbiol. 1985;39:615–648. doi: 10.1146/annurev.mi.39.100185.003151. [DOI] [PubMed] [Google Scholar]
- 32.Robinson C, Cai D, Hulford A, Brock I W, Michl D, Hazell L, Schmidt I, Herrmann R G, Klösgen R B. The presequence of a chimeric construct dictates which of two mechanisms are utilized for translocation across the thylakoid membrane: evidence for the existence of two distinct translocation systems. EMBO J. 1994;13:279–285. doi: 10.1002/j.1460-2075.1994.tb06260.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rodrigue A, Chanal A, Beck K, Müller M, Wu L F. Co-translocation of a periplasmic enzyme complex by a hitchhiker mechanism through the bacterial Tat pathway. J Biol Chem. 1999;274:13223–13228. doi: 10.1074/jbc.274.19.13223. [DOI] [PubMed] [Google Scholar]
- 34.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 35.Santini C L, Ize B, Chanal A, Muller M, Giordano G, Wu L F. A novel sec-independent periplasmic protein translocation pathway in Escherichia coli. EMBO J. 1998;17:101–112. doi: 10.1093/emboj/17.1.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sargent F, Bogsch E G, Stanley N R, Wexler M, Robinson C, Berks B C, Palmer T. Overlapping functions of components of a bacterial Sec-independent protein export pathway. EMBO J. 1998;17:3640–3650. doi: 10.1093/emboj/17.13.3640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sargent F, Stanley N R, Berks B C, Palmer T. Sec-independent protein translocation in Escherichia coli. J Biol Chem. 1999;274:36073–36082. doi: 10.1074/jbc.274.51.36073. [DOI] [PubMed] [Google Scholar]
- 38.Settles A M, Yonetani A, Baron A, Bush D R, Cline K, Martienssen R. Sec-independent protein translocation by the maize Hcf106 protein. Science. 1997;278:1467–1470. doi: 10.1126/science.278.5342.1467. [DOI] [PubMed] [Google Scholar]
- 39.Stanley N R, Findlay K, Berks B C, Palmer T. Escherichia coli strains blocked in Tat-dependent protein export exhibit pleiotropic defects in the cell envelope. J Bacteriol. 2001;183:139–144. doi: 10.1128/JB.183.1.139-144.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Thomas J D, Daniel R A, Errington J, Robinson C. Export of active green fluorescent protein to the periplasm by the twin-arginine translocase (Tat) pathway in Escherichia coli. Mol Microbiol. 2001;39:47–53. doi: 10.1046/j.1365-2958.2001.02253.x. [DOI] [PubMed] [Google Scholar]
- 41.Van Mellaert L, Dillen C, Proost P, Sablon E, DeLeys R, Van Broekhoven A, Heremans H, Van Damme J, Eyssen H, Anné J. Efficient secretion of biologically active mouse tumor necrosis factor alpha by Streptomyces lividans. Gene. 1994;150:153–158. doi: 10.1016/0378-1119(94)90876-1. [DOI] [PubMed] [Google Scholar]
- 42.Ward J M, Janssen G R, Kieser T, Bibb M J, Buttner M J, Bibb J. Construction and characterisation of a series of multi-copy promoter-probe plasmid vectors for Streptomyces using the aminoglycoside phosphotransferase gene from Tn5 as indicator. Mol Gen Genet. 1986;203:468–478. doi: 10.1007/BF00422072. [DOI] [PubMed] [Google Scholar]
- 43.Weiner J H, Bilous P T, Shaw G M, Lubitz S P, Frost L, Thomas G H, Cole J A, Turner R J. A novel and ubiquitous system for membrane targeting and secretion of cofactor-containing proteins. Cell. 1998;93:93–101. doi: 10.1016/s0092-8674(00)81149-6. [DOI] [PubMed] [Google Scholar]
- 44.Wexler M, Bogsch E G, Klösgen R B, Palmer T, Robinson C, Berks B C. Targeting signals for a bacterial Sec-independent export system direct plant thylakoid import by the delta pH pathway. FEBS Lett. 1998;431:339–342. doi: 10.1016/s0014-5793(98)00790-x. [DOI] [PubMed] [Google Scholar]