Abstract
Myxococcus xanthus is a gram-negative soil bacterium that undergoes development under starvation conditions. Our previous study identified a new genetic locus, mrp, which is required for both fruiting body formation and sporulation. The locus encodes two transcripts: mrpAB, which consists of a histidine kinase and an NtrC-like response regulator, and mrpC, a cyclic AMP receptor protein family transcription activator. In this study, we used genetic and biochemical analyses to investigate the possible interactions between the mrp genes and other known developmental genes and events. These studies show that the mrp genes possibly function after A-signaling and (p)ppGpp but before C-signaling and that they regulate various early and late developmental genes and events.
Starved Myxococcus xanthus cells initiate a developmental program that involves cellular aggregation and formation of multicellular fruiting bodies. When starvation continues, the rod-shaped cells inside the fruiting bodies differentiate into spherical spores that are resistant to desiccation, high temperature, and radiation. When conditions are favorable, the spores germinate and the vegetative life cycle resumes.
M. xanthus cells regulate their gene expression to bring about physiological changes. By means of cell-cell signaling and transcriptional regulators, the cells coordinate and direct the progression of the developmental program. At the onset of development, nutrient limitation elicits a transient increase in the intracellular guanosine-5′-(tri)di-3′-diphosphate [(p)ppGpp] level (24, 25). Guanosine penta- and tetraphosphate is believed to be a general signal for starvation in M. xanthus as it is in enteric bacteria (11). In enteric bacteria, (p)ppGpp accumulates when tRNAs are uncharged because of amino acid deprivation and serves as a powerful regulator of macromolecular synthesis (for a review of the stringent response, see reference 2). In M. xanthus, ectopic production of (p)ppGpp-activated expression of some early developmental genes and blocking of (p)ppGpp prevented the formation of fruiting bodies (36).
During development, various signaling genes (such as asg, bsg, csg, dsg, and esg) play important roles in regulating developmental gene expression at different time points (for a review, see reference 6). These genes were identified using nonautonomous developmental mutants that could be rescued extracellularly by wild-type or other complementation groups (4, 5, 16). Among them, asg (A-signal) and csg (C-signal) are the best studied. The A-signal is a group of amino acids and small peptides that is generated by proteolysis in the early stages of development (16). The A-signal serves as a cell density signal (16) to ensure that a critical density has been reached for fruiting body formation. The functional C-signal is believed to be a surface-associated protein. It is the 17.7-kDa product of the csgA gene, and its synthesis increases during development (10, 20). csgA positively autoregulates and affects expression of some late developmental genes (37). The addition of purified C-signal to the csgA mutant restores cellular aggregation, and increased levels of C-signal restore sporulation (15). In addition, overproduction of CsgA results in early sporulation or delayed development, and reduced production of CsgA leads to delayed development (20).
Developmental gene expression can be assayed genetically by a set of Tn5lac fusions. A survey using the promoter-probe Tn5lac has identified 36 genetic loci that specifically increase β-galactosidase expression at a particular time during development (19). The expression times range from minutes after starvation to 24 h, when sporulation begins. The dependence of the expression of these Tn5lac fusions on cell-cell interactions has also been investigated (17). Some key developmental genes have been further characterized, and antibodies are available for biochemical analyses for their products, including CsgA, the C-signal; protein S, the spore coat protein S; and FrzCD, a methyl-accepting chemotaxis protein (MCP) that is involved in directional movement of M. xanthus (27, 35, 39).
The directional movement controlled by the frz signal transduction system is required for cellular aggregation during M. xanthus development (for reviews, see references 31 and 41). FrzCD, the MCP, undergoes reversible methylation at specific glutamate residues that are conserved among MCP proteins (29). FrzCD methylation correlates with movement towards attractants (26, 32, 33). During the course of development, FrzCD becomes more methylated, suggesting that an attracting signal(s) might be produced and sensed by the starving M. xanthus cells (28, 32). Developmental FrzCD methylation is abolished in csg mutants, and addition of purified C-signal restores FrzCD methylation (37). Many mutants that are blocked in cellular aggregation (except S-motility mutants) show poor FrzCD methylation, whereas those that are only blocked in sporulation show normal FrzCD methylation, indicating that FrzCD methylation defines a discrete step in the developmental program of M. xanthus (8).
Our preceding paper presented experiments showing that two transcripts were essential for M. xanthus development: mrpAB, consisting of a histidine kinase and an NtrC-like response regulator, and mrpC, a cyclic AMP receptor protein family transcriptional activator (38). Deletion of any mrp gene renders M. xanthus cells unable to form fruiting bodies or sporulate. Expression of mrpAB and mrpC is induced upon starvation and is positively autoregulated (38). For mrpAB transcription, MrpB plays a major role and MrpA modulates MrpB activity.
In this study, we further investigated the role of the mrp genes in development. In particular, we examined the expression of mrpAB-lacZ and mrpC-lacZ under various experimental conditions as well as the expression of many developmental genes in mrp mutants to understand the interaction between the mrp genes and other developmental genes and events.
MATERIALS AND METHODS
Bacterial strains, media, and growth conditions.
The bacterial strains used in this study are listed in Table 1. M. xanthus cells were cultured vegetatively in Casitone-yeast extract (CYE) (1). Antibiotics were added when appropriate (kanamycin at 100 μg/ml or tetracycline at 15 μg/ml). The myxophage Mx4 was used for generalized transduction of M. xanthus strains as described previously (1). Liquid cultures were incubated at 32°C with shaking at 250 rpm. Agar plates were incubated at 32°C. Development of M. xanthus was initiated by placing 108 cells on 1.5% agar plates containing clone-fruiting (CF) (9) or MOPS medium (10 mM MOPS [morpholinepropanesulfonic acid, pH 7.6], 8 mM MgSO4). Development of M. xanthus in submerged culture was carried out as described previously (21).
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Relevant features | Source or reference |
|---|---|---|
| M. xanthus | ||
| DK1622 | Wild type | 13 |
| DK4491 | Tn5lacΩ4491 | 19 |
| DK4521 | Tn5lacΩ4521 | 19 |
| DK4300 | Tn5lacΩ4408 (sdeK) | 19 |
| DK4531 | Tn5lacΩ4531 | 19 |
| DK4414 | Tn5lacΩ4414 | 19 |
| DK4500 | Tn5lacΩ4500 | 19 |
| SW2820 | ΔmrpAB | This study |
| SW2851 | Tn5lacΩ4491 ΔmrpAB | This study |
| SW2852 | Tn5lacΩ4408 ΔmrpAB | This study |
| SW2853 | Tn5lacΩ4521 ΔmrpAB | This study |
| SW2854 | Tn5lacΩ4531 ΔmrpAB | This study |
| SW2855 | Tn5lacΩ4414 ΔmrpAB | This study |
| SW2856 | Tn5lacΩ4500 ΔmrpAB | This study |
| SW2808 | ΔmrpC | 38 |
| SW2871 | Tn5lacΩ4491 ΔmrpC | This study |
| SW2872 | Tn5lacΩ4408 ΔmrpC | This study |
| SW2873 | Tn5lacΩ4521 ΔmrpC | This study |
| SW2874 | Tn5lacΩ4531 ΔmrpC | This study |
| SW2875 | Tn5lacΩ4414 ΔmrpC | This study |
| SW2876 | Tn5lacΩ4500 ΔmrpC | This study |
| DK5057 | asgA | 22 |
| DK4398 | asgB | 22 |
| DK2630 | csgA | 34 |
| DK101 ΔrelA | ΔrelA in DK101 background | M. Singer |
| SW2801 | mrpAB-lacZ 201 Kanr | 38 |
| SW2809 | mrpAB-lacZ 201 Tetr | This study |
| SW2831 | asgA mrpAB-lacZ 201 Tetr | This study |
| SW2832 | csgA mrpAB-lacZ 201 Tetr | This study |
| SW2833 | ΔrelA mrpAB-lacZ 201 Kanr | This study |
| SW2803 | mrpC-lacZ 202 Kanr | 38 |
| SW2816 | mrpC-lacZ 202 Tetr | This study |
| SW2834 | asgA mrpC-lacZ 202 Tetr | This study |
| SW2835 | csgA mrpC-lacZ 202 Tetr | This study |
| SW2836 | ΔrelA mrpC-lacZ 202 Kanr | This study |
| Escherichia coli | ||
| Top10 | Cloning host | Invitrogen |
| Plasmids | ||
| pACYC184 | Cloning vector, Tetr | New England Biolabs |
| pBJ113 | In-frame deletion vector, Kanr Gals | B. Julien and D. Kaiser |
| pKY481 | lacZ fusion vector, Kanr | 3 |
| pSH100 | lacZ fusion vector, Tetr | This paper |
| pSH101 | mrpAB-lacZ201 Tetr | This paper |
| pSH102 | mrpC-lacZ202 Tetr | This paper |
| pSH406 | pBJ113 carrying ΔmrpAB | This paper |
Plasmid construction.
pSH100, constructed for this study, is a lacZ fusion vector carrying a tetracycline resistance gene. It is a derivative of pKY481 (3) in which the kanamycin resistance cassette was replaced with the tetracycline cassette from the cloning vector pACYC184 (New England Biolabs). pACYC184 was digested with XbaI and ScaI, and the ends were blunt filled by the Klenow extension reaction. pKY481 was digested with SalI and EagI, and the ends were blunt filled by the Klenow extension reaction and then treated with calf intestine alkaline phosphatase (Promega). The two fragments were ligated, and the ligation product was used to transform Top10 cells (Invitrogen), generating vector pSH100. pSH101 is a pSH100 derivative containing an mrpAB-lacZ fusion. The EcoRI-BamHI fragment of pSH201 (mrpAB-lacZ) was ligated into pSH100 partially digested with EcoRI and BamHI. pSH102 is a pSH100 derivative containing an mrpC-lacZ fusion. The EcoRI-BamHI mrpC fragment of pSH202 (mrpC-lacZ) was ligated into pSH100 partially digested with EcoRI and BamHI. pSH101 and pSH102 were introduced into M. xanthus wild-type strain DK1622 through electroporation, and transformants were selected for tetracycline resistance, generating strains SW2809 and SW2816. These two strains were used as recipient strains for transduction of the asg and csg mutations, which have kanamycin resistance markers.
Mutant construction.
The ΔmrpAB mutant SW2820 was constructed as follows. First, pSH406 was constructed, carrying a deleted version of the mrpAB transcript. A 476-bp PCR fragment containing part of the mrpA open reading frame (ORF) was amplified using two oligonucleotides, 5′-TGAATTCCGTGAGCTGGACGCCC-3′ and 5′-ATGGATCCGGTTCCTCGTCCACG-3′. The PCR product was digested with EcoRI and BamHI. Similarly, a 521-bp PCR fragment containing part of the mrpB ORF was amplified using two oligonucleotides, 5′-ATGGATCCTGGACGAGATTCGC-3′ and 5′-TATAAGCTTCGAACATGGCGCTGGC-3′. The PCR product was digested with BamHI and HindIII. The two fragments, part of the mrpA ORF and part of the mrpB ORF, were cloned into pBJ113, generating pSH406. Then, pSH406 was transferred into M. xanthus through electroporation as previously described (14). Chromosomal integration was selected for by plating the cells onto CYE plates containing kanamycin (100 μg/ml) (positive selection). pSH406 could not replicate in M. xanthus, and thus all transformants that were resistant to kanamycin (Kanr) were the result of recombination of the plasmid with the chromosome. Individual Kanr colonies were diluted and plated onto CYE plates containing 1% galactose for negative selection. Southern blot analysis was used to screen the galactose-resistant (Kanr galK) colonies for proper excision of the wild-type copy.
β-Galactosidase and Western blot assays.
For the β-galactosidase assay, M. xanthus cells were harvested at various time points during the course of development by scraping five 20-μl spots from MOPS agar into 100 μl of MOPS buffer or by pipetting the cells off the culture plate wells in the case of submerged culture. All samples were stored at −20°C until completion of the time course. The samples were thawed on ice, and β-galactosidase activity was assayed as described previously (19).
For the complementation assay, each strain was cultured in CYE broth to exponential phase, harvested, and resuspended to 5 × 109 cells/ml. An equal volume (10 μl) of the each cell type was mixed and placed on MOPS plates at 32°C for 3 days. Controls done in parallel contained 20 μl of each cell type. Cells were scraped from the plate and sonicated for 20 s at 6 W power output, heated at 50°C for 2 h, and then diluted and plated on CYE plates with kanamycin (100 μg/ml). The colonies were counted to determine the number of spores that germinated from the kanamycin-resistant cells.
Western blot analysis was used to study CsgA and protein S expression. FrzCD methylation pattern was examined following the protocol described previously (29). Cells were allowed to develop in submerged cultures or on MOPS agar for various times, collected, lysed using loading buffer for sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), and boiled for 5 min before loading. Protein concentration was determined for each sample using the Bradford assay (Bio-Rad), and equal amounts of protein were loaded for each sample for CsgA and protein S Western blot analyses. Anti-CsgA antibody was provided courtesy of L. Sogaard-Anderson. Anti-protein S and anti-FrzCD antibodies were provided courtesy of D. Zusman.
RESULTS
(p)ppGpp and A-signaling but not C-signaling affect mrp expression.
relA encodes the enzyme that synthesizes the stringent response signal (p)ppGpp (2). The M. xanthus relA mutant does not accumulate (p)ppGpp after starvation, and it fails to activate developmental gene expression. Consequently, it does not form fruiting bodies or sporulate (11). We examined the effect of relA mutation on the expression of mrp genes. Previously we constructed lacZ reporter strains for the mrp genes (38). SW2801 carries a translational lacZ fusion to mrpB, and SW2803 carries a translational lacZ fusion to mrpC. As mrpA and mrpB are cotranscribed, the mrpB-lacZ expression level would also reflect the expression of the transcript; hence it is referred to as mrpAB-lacZ hereafter.
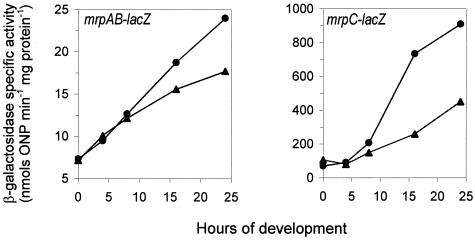
We transduced mrpAB-lacZ and mrpC-lacZ into the ΔrelA mutant and measured β-galactosidase activity in both the wild-type and the ΔrelA mutant backgrounds (Fig. 1). As we found previously, mrpAB-lacZ and mrpC-lacZ expression was induced upon starvation in the wild-type background (38) (Fig. 1). The ΔrelA mutation greatly reduced the expression of mrpAB and mrpC after 8 h of starvation, suggesting that the response of mrpAB and mrpC to starvation may be partially regulated by the (p)ppGpp signaling pathway.
FIG. 1.
Effect of ΔrelA on mrp expression. M. xanthus cells were placed on MOPS starvation plates and incubated at 32°C. The cells were harvested at different time points, and β-galactosidase activity was measured. Each experiment was repeated at least twice, and one set of results is shown. Circles, mrp expression in wild-type background. Triangles, mrp expression in ΔrelA background.
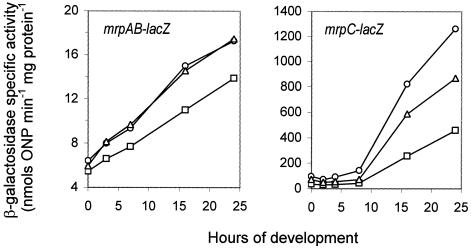
To examine the effect of A-signaling and C-signaling on mrp gene expression, we constructed tetracycline-resistant lacZ reporter strains for mrpAB and mrpC and then transduced asgA and csgA mutations (kanamycin resistance) into these reporter strains. We found that introduction of the asgA mutation reduced the expression of mrpAB and mrpC, whereas introduction of the csgA mutation had no or little effect on mrpAB or mrpC expression (Fig. 2). Therefore, the expression of the mrp genes is affected significantly by A-signaling but little, if at all, by C-signaling.
FIG. 2.
Effect of asgA and csgA on mrp expression. M. xanthus cells were placed on MOPS starvation plates and incubated at 32°C. The cells were harvested at different time points, and β-galactosidase activity was measured. Each experiment was repeated at least twice, and one set of results is shown. Circles, expression in wild-type background. Squares, expression in the asgA mutant. Triangles, expression in the csgA mutant.
mrp mutants still produce A-signal but not C-signal.
One effective way to examine extracellular signal production is the complementation assay. Many developmental mutants, including the asg and csg mutants, although unable to form fruiting bodies on their own, can be rescued by mixing with wild-type cells (9). It is speculated that these mutants fail to produce extracellular signal molecules but are able to receive them (9). We tested whether the mrp mutants could produce the A- and C-signals by mixing them with asg and csg mutants to see if their developmental defects could be rescued. As shown in Table 2, the mrpAB and mrpC mutants partially rescued the asg mutant, although fivefold less than the wild type did, but they failed to rescue the csg mutant, suggesting that the mrp mutants still produced the A-signal but to a lesser degree and that the mrp mutants produced little C-signal. This result is consistent with the effect of asg and csg mutations on mrp expression (Fig. 2) in that mrp probably functions after A-signaling but before C-signaling. Furthermore, the interaction between mrp and A-signaling may not be one linear pathway, as mrp mutations also reduced A-signal production to 20%.
TABLE 2.
Extracellular complementation of sporulation of asg and csg mutants by mrp mutants
| Strain (relevant genotype) | No. of Kanr sporesa |
|---|---|
| DK4398 (asgB) | <10 |
| DK4398 + DK1622 (asgB + wild type) | 1.0 × 105 |
| DK4398 + SW2820 (asgB + ΔmrpAB) | 1.8 × 104 |
| DK4398 + SW2808 (asgB + ΔmrpC) | 1.9 × 104 |
| DK2630 (csgA) | <10 |
| DK2630 + DK1622 (csgA + wild type) | 6.9 × 104 |
| DK2630 + SW2820 (csgA + ΔmrpAB) | 20 |
| DK2630 + SW2808 (csgA + ΔmrpC) | 30 |
The number of kanamycin-resistant spores represents the number of germinated spores from DK4398 or DK2630, which is kanamycin resistant.
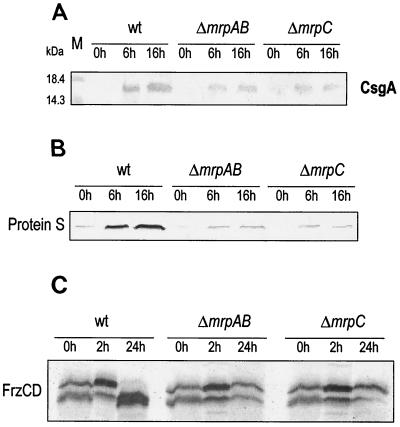
To more directly evaluate C-signal production in the mrp mutants, Western blot analysis was performed using antibody against the CsgA protein. Equal amounts of developing cell extracts were fractionated by gel electrophoresis, and the two forms of CsgA protein were both visualized via a multiclonal antibody (20). Shown in Fig. 3 is the 17-kDa CsgA product, which was believed to be the active C-signal (the 25-kDa form displayed a pattern parallel to that of the 17-kDa form; data not shown) (20). Consistent with previous findings in the wild-type cells, C-signal production was induced during development (10) (Fig. 3). In both mrpAB and mrpC deletion mutants, in contrast, the intensity of the C-signal band is significantly less than that in the wild type at each time point and remained at its 6-h level at 16 h of development. This indicates that csgA expression is directly or indirectly dependent on mrpAB and mrpC.
FIG. 3.
C-signal production (A), protein S production (B), and FrzCD methylation (C) during development in the wild type (wt) and the mrp mutants. (A and B) Cells were allowed to develop in submerged culture or on MOPS agar for various times, collected, and prepared for SDS-PAGE and Western blot analysis. Protein concentration was determined for each sample using the Bradford assay, and equal amounts of protein were loaded into each lane. The blot was probed with anti-CsgA antibody (A) or anti-protein S antibody (B). Lane M, size markers. (C) Cells were collected at different time during development in submerged cultures, lysed, and prepared for SDS-PAGE and Western blot analysis. The blot was probed with anti-FrzCD antibody. The upper band is unmethylated FrzCD, and the lower band is methylated FrzCD.
Effect of mrp on developmental gene expression.
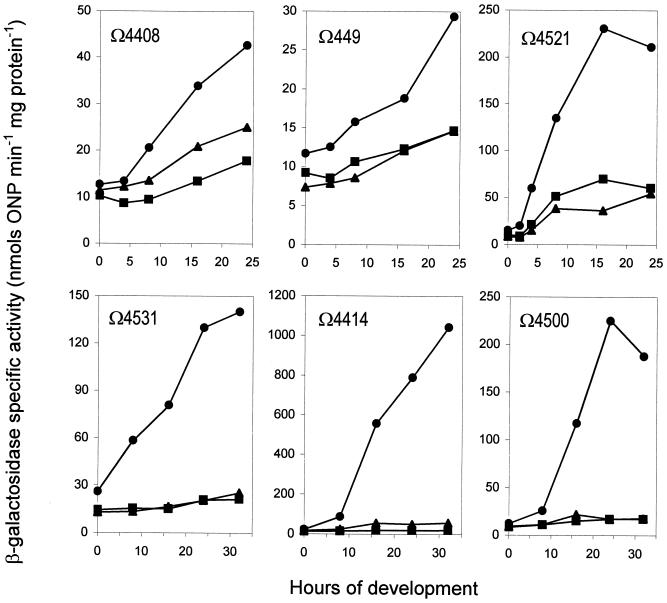
Both mrpAB and mrpC encode putative transcriptional regulators (38). To examine possible genes regulated by mrpAB and mrpC, we looked at the expression of known developmental markers. Kroos et al. used a promoter probe, Tn5lac, and identified 36 strains that specifically increase β-galactosidase expression at particular times during development (19). We chose several such Tn5lac markers based on the time of their expression, their importance to development, and their dependence on intercellular signaling. These markers include Ω4491, Ω4408 (sdeK) (7), Ω4521, Ω4531, Ω4414 (devR) (40), and Ω4500. According to previous studies, the first three markers were expressed early, at about 2 h of development. Tn5lacΩ4531 was expressed at about 7 h, Tn5lacΩ4414 at about 11.5 h, and Tn5lac Ω4500 at around 14 h (19). Strains containing Ω4408 (sdeK), Ω4491, or Ω4414 lose their capacity to complete development, meaning that the gene disrupted by the transposon is required for development (7, 18, 40). Τn5lacΩ4521 is usually used as a marker for A-signaling (23). Tn5lacΩ4414 expression requires C-signaling and is an indicator of the production of the active C-signal (40). Tn5lacΩ4408 expression is independent of A- and C-signaling (17). Tn5lacΩ4531 and Tn5lacΩ4500 have not been tested for A- or C-signaling dependence (7).
We found that the expression of all Tn5lac markers tested was reduced in the mrp mutation backgrounds (Fig. 4). We compared the expression levels of the Tn5lac markers at 24 h of development in the mrp mutants to those in the wild-type background and found that the later the markers are normally expressed, the more they were reduced by the mrp mutations. For the Tn5lac markers in the order Ω4491, Ω4408 (sdeK), Ω4521, Ω4531, Ω4414 (devR), and Ω4500, 50, 42, 29, 16, 3, and 8% of wild-type-level expression was left in the mrpAB mutant and 50, 59, 26, 16, 6, and 8% of wild-type level expression was left in the mrpC mutant, respectively. This shows that mrpAB and mrpC directly or indirectly regulate these Tn5lac markers and indicates that the mrp genes function early during development.
FIG. 4.
Effect of mrp on Tn5lac marker expression: Ω4491, Ω4408 (sdeK), Ω4521, Ω4531, Ω4414 (devR), and Ω4500. M. xanthus cells were placed on MOPS starvation plates and incubated at 32°C. The cells were harvested at different time points, and β-galactosidase activity was measured. Each experiment was repeated at least twice, and one set of results is shown. Circles, expression in wild-type background. Squares, expression in the ΔmrpAB mutant. Triangles, expression in the ΔmrpC mutant.
In addition to β-galactosidase assays, we used Western blot analysis to examine the expression level of developmentally regulated genes. Protein S is the major component of the outmost layer of fruiting body spores. Expression of its encoding gene, tps, is induced by starvation and begins at about 5 h into development; accumulation of protein S peaks at about 24 h (12, 30). We examined the effect of mrp mutation on protein S production (Fig. 3B). In agreement with previous findings, we saw a substantial increase in the production of protein S from the onset of development to 6 h in wild-type cells. After 16 h, more protein S had accumulated. In contrast, in the mrpAB and mrpC mutants, there was only a slight increase from 0 to 6 h, much less than the increase in the wild type. From 6 to 16 h, the protein S level did not increase and remained the same as at 6 h. This result suggests that protein S synthesis is regulated, directly or indirectly, by mrpAB and mrpC. Furthermore, as the increase in protein S from 0 to 6 h did not happen in the mrp mutants, very likely mrp functions before 6 h during development.
Effect of mrp on FrzCD methylation.
The mrpAB and mrpC mutants are defective in aggregation and fail to form fruiting bodies (38). Many developmental mutants that are defective in fruiting body formation show abnormal FrzCD methylation (8, 37). When the mrp mutants were tested, we found that they were defective in FrzCD methylation also (Fig. 3C). Consistent with previous findings (28), FrzCD of wild-type cells was largely methylated during vegetative growth in rich CYE medium. After 2 h of starvation in submerged culture, FrzCD became more demethylated. After 16 h, FrzCD was fully methylated. In the ΔmrpAB and ΔmrpC mutants, however, FrzCD methylation did not follow the same pattern. During vegetative growth, the cells were normal in sensing CYE as the attractant. After 2 h of starvation, they appeared normal in FrzCD demethylation, indicating that the cells sensed the removal of the nutrients. After 16 h, however, FrzCD extracted from these mutants did not become as fully methylated as that extracted from the wild-type cells. The methylation pattern of FrzCD remained essentially as it was at 2 h. This result suggests that the mrp genes function early in development and that the mrp mutants are unable to produce the putative attractant to methylate FrzCD. This is consistent with their nonaggregating phenotype (38) and also with the finding that the mrp mutants produce little C-signal during development (Fig. 3A), as FrzCD methylation is also under control of the C-signal (37).
DISCUSSION
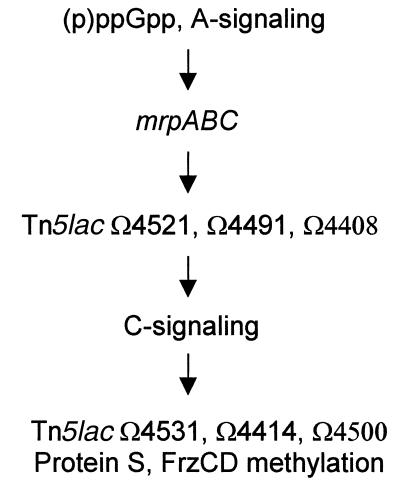
Based on the foregoing data, we tentatively assigned a position to the role of the mrp genes during development (Fig. 5). Since the expression of mrpAB and mrpC was affected by (p)ppGpp and A-signaling but little by C-signaling, and since the mrp mutants produced A-signal but little C-signal, it is likely that the mrpAB and mrpC products function after (p)ppGpp and A-signal but before C-signal. Consistently, we found that the mrpAB and mrpC mutations have a mild inhibitory effect on expression of A-signal-dependent genes but have a strong effect on expression of C-signal-dependent genes. These findings suggested that the mrp genes function early in development and are required for many key developmental gene expression and events. It should be noted, though, that the developmental program may very well not be a linear pathway, and Fig. 5 only represents our current understanding of the order of developmental events in relation to the mrp genes. The direct pathway of mrp regulation, such as the putative signaling events leading to MrpB phosphorylation and activation (38) and the genes directly activated by MrpC, wait to be revealed by further studies. From there, more specifc interactions between mrp regulation and other regulatory events such as (p)ppGpp and A- and C-signaling might be speculated. Our results so far strongly suggest that the mrp genes are an important part of the complex regulatory circuitry controlling the M. xanthus developmental program.
FIG. 5.
Timing of mrpABC action in M. xanthus development.
ACKNOWLEDGMENTS
We thank D. R. Zusman, Z. Yang, R. Lux, and M. Kempf for helpful discussions. We also thank D. R. Zusman, K. Cho, D. Kaiser, M. Singer, and L. Sogaard-Anderson for kindly providing us with experimental materials. We are grateful to L. Tong and Sue Jeong Choi for providing excellent technical support. We also thank S. Hunt Gerardo for careful editing of the manuscript.
This work is supported by NIH grant GM54666 to W. Shi.
REFERENCES
- 1.Campos J M, Geisselsoder J, Zusman D R. Isolation of bacteriophage Mx4, a generalized transducing phage for Myxococcus xanthus. J Mol Biol. 1978;119:167–178. doi: 10.1016/0022-2836(78)90431-x. [DOI] [PubMed] [Google Scholar]
- 2.Cashel M, Rudd K E. The stringent response. In: Neidhardt F C, Low K, Magasanick B, Schaechter M, Umbarger H, editors. Esherichia coli and Salmonella typhimurium: cellular and molecular biology. Vol. 2. Washington, D.C.: ASM Press; 1987. pp. 1410–1438. [Google Scholar]
- 3.Cho K, Zusman D R. AsgD, a new two-component regulator required for A-signaling and nutrient sensing during early development of Myxococcus xanthus. Mol Microbiol. 1999;34:268–281. doi: 10.1046/j.1365-2958.1999.01594.x. [DOI] [PubMed] [Google Scholar]
- 4.Downard J, Ramaswamy S V, Kil K S. Identification of esg, a genetic locus involved in cell-cell signaling during Myxococcus xanthus development. J Bacteriol. 1993;175:7762–7770. doi: 10.1128/jb.175.24.7762-7770.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Downard J, Toal D. Branched-chain fatty acids: the case for a novel form of cell-cell signalling during Myxococcus xanthus development. Mol Microbiol. 1995;16:171–175. doi: 10.1111/j.1365-2958.1995.tb02290.x. [DOI] [PubMed] [Google Scholar]
- 6.Dworkin M, Kaiser D, editors. Myxobacteria II. Washington, D.C.: ASM Press; 1993. [Google Scholar]
- 7.Garza A G, Pollack J S, Harris B Z, Lee A, Keseler I M, Licking E F, Singer M. SdeK is required for early fruiting body development in Myxococcus xanthus. J Bacteriol. 1998;180:4628–4637. doi: 10.1128/jb.180.17.4628-4637.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Geng Y, Yang Z, Downard J, Zusman D, Shi W. Methylation of FrzCD defines a discrete step in the developmental program of Myxococcus xanthus. J Bacteriol. 1998;180:5765–5768. doi: 10.1128/jb.180.21.5765-5768.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hagen D C, Bretscher A P, Kaiser D. Synergism between morphogenetic mutants of Myxococcus xanthus. Dev Biol. 1978;64:284–296. doi: 10.1016/0012-1606(78)90079-9. [DOI] [PubMed] [Google Scholar]
- 10.Hagen T J, Shimkets L J. Nucleotide sequence and transcriptional products of the csg locus of Myxococcus xanthus. J Bacteriol. 1990;172:15–23. doi: 10.1128/jb.172.1.15-23.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Harris B Z, Kaiser D, Singer M. The guanosine nucleotide (p)ppGpp initiates development and A-factor production in Myxococcus xanthus. Genes Dev. 1998;12:1022–1035. doi: 10.1101/gad.12.7.1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Inouye M, Inouye S, Zusman D R. Biosynthesis and self-assembly of protein S, a development-specific protein of Myxococcus xanthus. Proc Natl Acad Sci USA. 1979;76:209–213. doi: 10.1073/pnas.76.1.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kaiser D. Social gliding is correlated with the presence of pili in Myxococcus xanthus. Proc Natl Acad Sci USA. 1979;76:5952–5956. doi: 10.1073/pnas.76.11.5952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kashefi K, Hartzell P L. Genetic suppression and phenotypic masking of a Myxococcus xanthus frzF− defect. Mol Microbiol. 1995;15:483–494. doi: 10.1111/j.1365-2958.1995.tb02262.x. [DOI] [PubMed] [Google Scholar]
- 15.Kim S K, Kaiser D. C-factor has distinct aggregation and sporulation thresholds during Myxococcus development. J Bacteriol. 1991;173:1722–1728. doi: 10.1128/jb.173.5.1722-1728.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim S K, Kaiser D, Kuspa A. Control of cell density and pattern by intercellular signaling in Myxococcus development. Annu Rev Microbiol. 1992;46:117–139. doi: 10.1146/annurev.mi.46.100192.001001. [DOI] [PubMed] [Google Scholar]
- 17.Kroos L, Kaiser D. Expression of many developmentally regulated genes in Myxococcus depends on a sequence of cell interactions. Genes Dev. 1987;1:840–854. doi: 10.1101/gad.1.8.840. [DOI] [PubMed] [Google Scholar]
- 18.Kroos L, Kuspa A, Kaiser D. Defects in fruiting body development caused by Tn5 lac insertions in Myxococcus xanthus. J Bacteriol. 1990;172:484–487. doi: 10.1128/jb.172.1.484-487.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kroos L, Kuspa A, Kaiser D. A global analysis of developmentally regulated genes in Myxococcus xanthus. Dev Biol. 1986;117:252–266. doi: 10.1016/0012-1606(86)90368-4. [DOI] [PubMed] [Google Scholar]
- 20.Kruse T, Lobedanz S, Berthelsen N M S, Sogaard-Anderson L. C-signal: a cell surface-associated morphogen that induces and coordinates fruiting body morphogenesis and sporulation in Myxococcus xanthus. Mol Microbiol. 2001;40:156–168. doi: 10.1046/j.1365-2958.2001.02365.x. [DOI] [PubMed] [Google Scholar]
- 21.Kuner J M, Kaiser D. Fruiting body morphogenesis in submerged cultures of Myxococcus xanthus. J Bacteriol. 1982;151:458–461. doi: 10.1128/jb.151.1.458-461.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kuspa A, Kaiser D. Genes required for developmental signaling in Myxococcus xanthus: three asg loci. J Bacteriol. 1989;171:2762–2772. doi: 10.1128/jb.171.5.2762-2772.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kuspa A, Kroos L, Kaiser D. Intercellular signaling is required for developmental gene expression in Myxococcus xanthus. Dev Biol. 1986;117:267–276. doi: 10.1016/0012-1606(86)90369-6. [DOI] [PubMed] [Google Scholar]
- 24.Manoil C, Kaiser D. Accumulation of guanosine tetraphosphate and guanosine pentaphosphate in Myxococcus xanthus during starvation and myxospore formation. J Bacteriol. 1980;141:297–304. doi: 10.1128/jb.141.1.297-304.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Manoil C, Kaiser D. Guanosine pentaphosphate and guanosine tetraphosphate accumulation and induction of Myxococcus xanthus fruiting body development. J Bacteriol. 1980;141:305–315. doi: 10.1128/jb.141.1.305-315.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McBride M J, Kohler T, Zusman D R. Methylation of FrzCD, a methyl-accepting taxis protein of Myxococcus xanthus, is correlated with factors affecting cell behavior. J Bacteriol. 1992;174:4246–4257. doi: 10.1128/jb.174.13.4246-4257.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McBride M J, Weinberg R A, Zusman D R. “Frizzy” aggregation genes of the gliding bacterium Myxococcus xanthus show sequence similarities to the chemotaxis genes of enteric bacteria. Proc Natl Acad Sci USA. 1989;86:424–428. doi: 10.1073/pnas.86.2.424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McBride M J, Zusman D R. FrzCD, a methyl-accepting taxis protein from Myxococcus xanthus, shows modulated methylation during fruiting body formation. J Bacteriol. 1993;175:4936–4940. doi: 10.1128/jb.175.15.4936-4940.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McCleary W R, McBride M J, Zusman D R. Developmental sensory transduction in Myxococcus xanthus involves methylation and demethylation of FrzCD. J Bacteriol. 1990;172:4877–4887. doi: 10.1128/jb.172.9.4877-4887.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nelson D R, Zusman D R. Transport and localization of protein S, a spore coat protein, during fruiting body formation by Myxococcus xanthus. J Bacteriol. 1983;154:547–553. doi: 10.1128/jb.154.2.547-553.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shi W, Kohler T, Zusman D. Motility and chemotaxis in Myxococcus xanthus. In: Adolph K W, editor. Molecular microbiology techniques. Vol. 3. San Diego, Calif: Academic Press; 1994. pp. 258–269. [Google Scholar]
- 32.Shi W, Kohler T, Zusman D R. Chemotaxis plays a role in the social behaviour of Myxococcus xanthus. Mol Microbiol. 1993;9:601–611. doi: 10.1111/j.1365-2958.1993.tb01720.x. [DOI] [PubMed] [Google Scholar]
- 33.Shi W, Kohler T, Zusman D R. Isolation and phenotypic characterization of Myxococcus xanthus mutants which are defective in sensing negative stimuli. J Bacteriol. 1994;176:696–701. doi: 10.1128/jb.176.3.696-701.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shimkets L J, Gill R E, Kaiser D. Developmental cell interactions in Myxococcus xanthus and the spoC locus. Proc Natl Acad Sci USA. 1983;80:1401–1410. doi: 10.1073/pnas.80.5.1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shimkets L J, Rafiee H. CsgA, an extracellular protein essential for Myxococcus xanthus development. J Bacteriol. 1990;172:5299–5306. doi: 10.1128/jb.172.9.5299-5306.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Singer M, Kaiser D. Ectopic production of guanosine penta- and tetraphosphate can initiate early developmental gene expression in Myxococcus xanthus. Genes Dev. 1995;9:1633–1644. doi: 10.1101/gad.9.13.1633. [DOI] [PubMed] [Google Scholar]
- 37.Sogaard-Andersen L, Kaiser D. C factor, a cell-surface-associated intercellular signaling protein, stimulates the cytoplasmic Frz signal transduction system in Myxococcus xanthus. Proc Natl Acad Sci USA. 1996;93:2675–2679. doi: 10.1073/pnas.93.7.2675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sun H, Shi W. Genetic studies of mrp, a locus essential for cellular aggregation and sporulation of Myxococcus xanthus. J Bacteriol. 2001;183:4786–4795. doi: 10.1128/JB.183.16.4786-4795.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Teintze M, Thomas R, Furuichi T, Inouye M, Inouye S. Two homologous genes coding for spore-specific proteins are expressed at different times during development of Myxococcus xanthus. J Bacteriol. 1985;163:121–125. doi: 10.1128/jb.163.1.121-125.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Thony-Meyer L, Kaiser D. devRS, an autoregulated and essential genetic locus for fruiting body development in Myxococcus xanthus. J Bacteriol. 1993;175:7450–7462. doi: 10.1128/jb.175.22.7450-7462.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ward M J, Zusman D R. Regulation of directed motility in Myxococcus xanthus. Mol Microbiol. 1997;24:885–893. doi: 10.1046/j.1365-2958.1997.4261783.x. [DOI] [PubMed] [Google Scholar]