Abstract
Pneumocystis carinii remains an important opportunistic fungal pathogen causing life-threatening pneumonia in patients with AIDS and malignancy. Currently, little is known about how the organism adapts to environmental stresses and maintains its cellular integrity. We recently discovered an open reading frame approximately 600 bp downstream of the region coding GSC-1, a gene mediating β-glucan cell wall synthesis in P. carinii. The predicted amino acid sequence of this new gene, termed P. carinii PHR1, exhibited 38% homology to Saccharomyces cerevisiae GAS1, a glycosylphosphatidylinositol-anchored protein essential to maintaining cell wall integrity, and 37% homology to Candida albicans PHR1/PHR2, pH-responsive genes encoding proteins recently implicated in cross-linking β-1,3- and β-1,6-glucans. In view of its homology to these related fungal genes, the pH-dependent expression of P. carinii PHR1 was examined. As in C. albicans, P. carinii PHR1 expression was repressed under acidic conditions but induced at neutral and more alkaline pH. PHR1-related proteins have been implicated in glucan cell wall stability under various environmental conditions. Although difficulties with P. carinii culture and transformation have traditionally limited assessment of gene function in the organism itself, we have successfully used heterologous expression of P. carinii genes in related fungi to address functional correlates of P. carinii-encoded proteins. Therefore, the potential role of P. carinii PHR1 in cell wall integrity was examined by assessing its ability to rescue an S. cerevisiae gas1 mutant with absent endogenous Phr1p-like activity. Interestingly, P. carinii PHR1 DNA successfully restored proliferation of S. cerevisiae gas1 mutants under lethal conditions of cell wall stress. These results indicate that P. carinii PHR1 encodes a protein responsive to environmental pH and capable of mediating fungal cell wall integrity.
Pneumocystis carinii remains an important fungal agent causing life-threatening pneumonia in patients with impaired immunity (19). Other fungi such as Aspergillus nidulans and Candida albicans alter gene expression as an adaptive response to environmental pH changes in order to maintain cell wall integrity and promote viability under various conditions (6, 11, 21, 23, 32–35). Specifically, it has been demonstrated that C. albicans expresses a unique family of pH-regulated genes required for virulence. These genes nominally include PHR1, a gene expressed maximally at pH 5.5 to 8.0 which encodes a protein promoting systemic infection of mice. Alternatively, C. albicans also expresses PHR2, whose transcription is greatest at acidic pH (4 to 5) values (24). If PHR2 is rendered inactive, C. albicans mutants exhibit decreased pathogenesis in a mouse model of vaginal infection (6, 24, 32).
Recent studies indicate that Phr1p and Phr2p act on β-1,3-glucans of the C. albicans cell walls, elongating the β-1,3 polysaccharide backbone and potentially mediating the attachment of β-1,6 glucosyl side chains and β-1,6-glycosylated mannoproteins (8, 23). The role of glucan cell wall structure in pathogenesis is not completely understood, but has been postulated to participate in maintenance of cell wall integrity during environmental stress.
The mechanisms by which P. carinii assembles its cell wall have only recently been elucidated. Initial studies largely focused on the prominent surface glycoprotein complexes termed glycoprotein A or major surface glycoproteins, which have been demonstrated to participate in P. carinii attachment to type I alveolar epithelial cells and alveolar macrophages (4, 12, 19). More recently, our group has focused on generation of β-glucan cell wall components, which represent major structural constituents of the cystic form. The cystic form has been postulated to represent a transmissible agent capable of surviving the harsh environmental conditions outside of the mammalian host (3).
To this end, we recently characterized P. carinii GSC1, a gene responsible for assembly of the 1,3-β-d-glucan core carbohydrate of the cyst cell wall (16). During screening of the P. carinii genomic DNA library for GSC1, we isolated an open reading frame downstream of P. carinii GSC1 with considerable homology to C. albicans PHR1/PHR2 and Saccharomyces cerevisiae GAS1. Here we report the identification and initial characterization of this gene, termed P. carinii PHR1. We demonstrate pH-dependent expression of P. carinii PHR1 with greatest expression under physiologic (pH 7.0 to 7.5) conditions present in the lung. We further show that P. carinii PHR1 participates in the maintenance of fungal cell wall integrity, as assessed by the ability of P. carinii PHR1 to complement growth of S. cerevisiae gas1 mutants with defective Gas1p (Phr1p-like) activity.
MATERIALS AND METHODS
Materials.
All reagents were from Sigma Chemical Company (St. Louis, Mo.) unless otherwise specified. Restriction endonucleases were obtained from Gibco-BRL (Life Technologies, Rockville, Md.), and Pfu polymerase was from Stratagene (La Jolla, Calif.). [α-32P]ATP was obtained from ICN Pharmaceuticals (Costa Mesa, Calif.). The WB2d Saccharomyces cerevisiae strain expressing a mutated and ineffective Gas1p (YEp-gas1 mutant) was the generous gift of Marina Vai, Università degli Studi di Milano, Milan, Italy (38). The plasmid YEp-GAS1, containing the wild-type GAS1 gene used as a complementation control, was also obtained from M. Vai. The yeast expression plasmid p425GAL used in the generation of the p425GAL-PHR1 construct was obtained from the American Type Culture Collection (Rockville, Md.).
Preparation of P. carinii organisms.
P. carinii pneumonia was induced in Harlan Sprague-Dawley rats by immunosuppression with dexamethasone, as reported previously (5, 15, 36). Rats received drinking water containing dexamethasone ad libitum (2 mg/liter). After 1 week of immune suppression, rats were anesthetized with ether and inoculated with ∼106 P. carinii organisms intratracheally. After 6 weeks of additional dexamethasone treatment, the animals were sacrificed, and P. carinii was harvested. Lungs from the rats were minced in Hanks' balanced salt solution and homogenized in a stomacher microbiological blender (Tekmar Inc., Cincinnati, Ohio) for 5 min. P. carinii organisms were passed through a 10-μm filter (Millipore), which retains lung cells but allows passage of P. carinii. These preparations were stained with Wright-Giemsa to confirm that P. carinii organisms were present (19). Preparations containing detectable bacterial or fungal contamination were discarded.
Identification of putative P. carinii PHR1 gene.
In the course of screening a P. carinii λgt11 genomic library for the P. carinii 1,3-β-glucan synthetase (GSC1), an open reading frame was identified ≈600 bp downstream of the GSC1 clone that contained 465 bp of coding sequence. Translation of the open reading frame (BlastX; EMBL) revealed a partial sequence clone with substantial sequence homology to the C. albicans pH-responsive gene PHR1 and containing a start site for the putative P. carinii PHR1 gene. In light of our interest in P. carinii cell wall assembly and because of the reported roles of PHR genes in generation of fungal glucans, we sought to fully identify this new P. carinii gene and characterize its potential functions.
To obtain the remaining 3′ end of P. carinii PHR1, a modified 3′ rapid amplification of genomic ends (RAGE) procedure was used as follows. The λgt11 phage clones that contained the partial P. carinii PHR1 sequences were plaque purified to homogeneity. This λgt11 insert was used as the PCR template. The primers used were 5′-TCGCTTATCAGCCTCCGTTAAG-3′ (gene-specific primer) and 5′-CCAACTGGTAATGGTAGCGACC-3′ (3′ λgt11-specific primer). An initial 5-min hot start at 94°C was followed by 30 cycles of 94°C for 30 s, 60°C for 60 s, 72°C for 90 s, and a final 72°C 15-min extension. A single amplicon of approximately 2.3 kb was generated, subcloned into pGEM-T Easy vector (Promega, Madison, Wis.), and sequenced on both forward and reverse strands. Sequence comparisons to GenBank were performed using the Blast genetic analysis program (NCBI). Sequence analysis was undertaken with MacVector software (Kodak, IBI, New Haven, Conn.). Homology comparisons were conducted with the BlastX (EMBL) algorithms. To perform multiple sequence alignments, the PileUp program of the Genetics Computer Group was used (7).
Confirmation of PHR1 sequences in P. carinii genomic DNA.
To confirm that the PHR1 gene was truly represented within the P. carinii genome and not related to host or other microbial contamination, Southern hybridization was performed using the PHR1 gene as a probe against freshly isolated P. carinii genomic DNA. Genomic DNA from P. carinii was prepared with the IsoQuick nucleic acid extraction kit (Orca Research Inc., Bothell, Wash.). A 32P-labeled P. carinii PHR1 probe was generated using the random primer method (RadPrime; Amersham Pharmacia, Piscataway, N.J.). Twenty micrograms of genomic DNA was digested with either EcoRI or HindIII and separated on a 1% agarose gel. Transfer and hybridization were performed as described (31).
Expression of P. carinii PHR1 mRNA in response to environmental pH.
In light of the strong homology of P. carinii PHR1 to pH-regulated genes in other fungi, we next evaluated the expression of P. carinii PHR1 mRNA in response to environmental pH. To address this, P. carinii organisms were placed in 1.0 ml of Ham's F-12 tissue culture medium supplemented with 10% fetal bovine serum at the indicated pH ranges for 1.0 h at 37°C. Total RNA was isolated with hot acidic phenol and separated on 1.0% formaldehyde–agarose gels (1). RNA was transferred to Nytran Plus membranes (Schleicher & Schuell, Keene, N.H.), and hybridization to the radiolabeled PHR1 probe performed at 68°C using Clontech Express hybridization solution (Clontech, Inc., Palo Alto, Calif.). Total RNA loading was verified by ethidium bromide staining and by further stripping the final blots and repeat probing with a Pneumocystis actin probe (16, 17).
Role of P. carinii PHR1 in maintaining fungal cell wall integrity.
Studies of P. carinii gene function have long been hindered by the inability to culture and transform the organism. To circumvent these obstacles, we have recently undertaken analysis of P. carinii gene function by heterologous expression of P. carinii proteins in phylogenetically related fungi which lack endogenous homologues under selective conditions (17). Using such a strategy, we analyzed the ability of P. carinii Phr1p to restore cell wall integrity by transforming Gas1p-deficient S. cerevisiae mutants with P. carinii PHR1 DNA. S. cerevisiae gas1 mutants have increased cellular fragility in the presence of osmotic destabilizing agents such as sodium dodecyl sulfate (SDS), a stringent assessment of cell wall stability. Previous studies have shown that S. cerevisiae gas1 mutants are unable to grow in the presence of 0.01% SDS (38).
P. carinii PHR1 DNA was excised from pGEM-T Easy by digestion with HindIII and NdeI and directionally cloned into the yeast expression vector p425GAL. The p425GAL plasmid has a LEU2 gene that permits growth on medium lacking leucine (25). S. cerevisiae gas1 mutants were grown to mid-log phase in YPD broth at 30°C and transformed by electroporation using 10.0 ng of p425GAL/P. carinii PHR1 DNA or 10.0 ng of p425GAL vector alone without insert (31). Transformed S. cerevisiae cells were plated onto minimal medium plates deficient in leucine at 30°C. After 48 h, yeast transformants were streaked onto minimal medium leucine-deficient plates containing 0.005% SDS, placed at 30°C, and assessed for growth. Transformed yeast colonies proliferating at these conditions were cultured to mid-log phase in leucine-deficient broth, and plasmid DNA was extracted and sequenced to confirm the presence of P. carinii PHR1 DNA.
RESULTS AND DISCUSSION
P. carinii contains a putative pH-responsive gene, PHR1.
The mechanisms through which P. carinii responds to alterations in the environment remain largely unknown. In an effort to better understand the mechanisms by which P. carinii assembles its cyst wall and maintains integrity under various environmental conditions, we fully cloned and sequenced the putative P. carinii PHR1 gene. The initial 465-bp partial P. carinii PHR1 open reading sequence revealed a substantial degree of homology to its C. albicans pH-responsive counterpart (BlastX, 50%). After redundant sequencing of this partial clone to its putative start site, a modified 3′ RAGE procedure was used to obtain the remaining portion of the gene by using a 5′ PHR1 gene-specific primer and a 3′ primer matching the λgt11 cloning site of the P. carinii genomic library. The DNA template used for PCR was obtained by purifying λgt11 plaques that initially hybridized with GSC1 sequences.
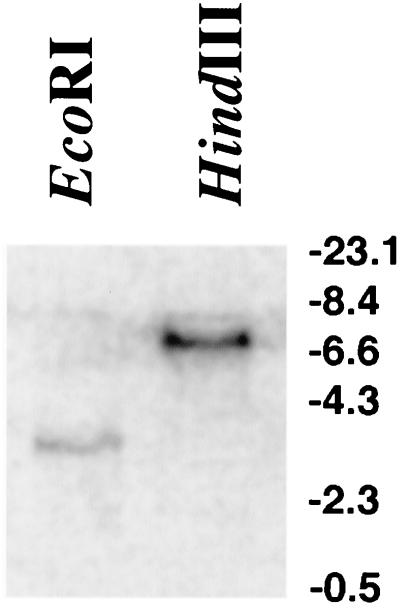
After screening a number of colonies obtained by this strategy, a clone containing the remaining genomic P. carinii PHR1 sequence was isolated. To verify that the DNA sequence obtained by PCR was not a result of amplification of host cell DNA or other foreign DNA and was specifically represented within the P. carinii genome, the full-length PHR1 sequence was used as a probe for Southern hybridization studies against digested P. carinii genomic DNA. The full-length P. carinii PHR1 probe strongly hybridized to a single band on both the EcoRI and HindIII digests, indicating its presence in the P. carinii genome (Fig. 1). As anticipated, an identical pattern of hybridization had previously been observed with the adjacent P. carinii GSC1 DNA sequences (16).
FIG. 1.
P. carinii PHR1 homologue is represented in the organism's genome. The full-length P. carinii PHR1 cDNA homologue was radiolabeled and hybridized to P. carinii genomic DNA digested with EcoRI or HindIII, as indicated. PHR1 was present in one location on each Southern analysis.
Thus, a 2.3-kb genomic DNA fragment containing the complete PHR1 gene was available for further study. Similar to C. albicans PHR1, a single uninterrupted open reading frame was identified in the P. carinii sequence (32). This open reading frame of 1,632 bp was predicted to encode a 544-amino-acid peptide. The deduced amino acid sequence of P. carinii PHR1 is shown in Fig. 2 (GenBank accession no. AF191097). Computer analysis of the GenBank database and P. carinii genome project website (http://www.uky.edu/Projects/Pneumocystis/) revealed the entire PHR1 sequence to be unique and yet most homologous to a protein encoded by S. cerevisiae GGP1/GAS1 (38%; BlastX) and to the C. albicans pH-responsive gene products PHR1 and PHR2 (37% each) (24, 32, 37). All three proteins are thought to encode major glycoproteins localized to the plasma membrane through a glycosylphosphatidylinositol (GPI) anchor and important for the maintenance of cell wall integrity (28, 30, 32, 38).
FIG. 2.
Alignment of the predicted amino acid sequences of P. carinii Phr1p and related proteins derived from heterologous fungi. S. cere., S. cerevisiae.
The predicted P. carinii Phr1p protein contains a potential GPI attachment site (G518) followed by a polar region at amino acid residues 519 to 526 (KAIYPDWR) and finally a hydrophobic carboxy terminus encompassing residues 527 to 543 (LIFGIMTYFFGIIIVIA) (2, 13, 18, 22, 27). The putative P. carinii PHR1 exhibited substantial similarity to S. cerevisiae GGP1/GAS1 and C. albicans PHR1/PHR2 over the majority of its length, including conservation in two of the three N-glycosylation sites and 11 of 13 conserved cysteine residues. The greatest divergence of the predicted P. carinii Phr1p to the three previously described proteins was noted in both the amino- and carboxyl-terminal regions. Alignments of fungal GPI-anchored proteins with homology to P. carinii PHR1, including EPD1, which encodes a protein thought to be involved in pseudohyphal growth of Candida maltosa (26), are also demonstrated in Fig. 2.
P. carinii PHR1 expression is regulated by environmental pH.
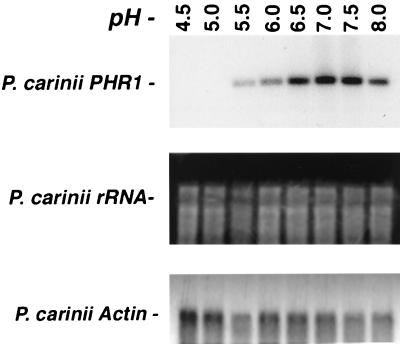
In light of the substantial homology of P. carinii PHR1 to genes responsive to environmental pH in C. albicans, we further evaluated whether expression of this P. carinii gene was similarly responsive to changes in the ambient pH. To test this, P. carinii organisms were removed from rat lung and placed in medium at a range of ambient pHs for 1 h. We have previously used such a short-term medium exposure system to monitor gene expression of P. carinii GSC1 (16). Parallel to C. albicans, differential expression of P. carinii PHR1 steady-state mRNA was observed on Northern analysis (Fig. 3). P. carinii PHR1 mRNA was virtually absent at pHs ranging between 4.5 and 5.0. Over the pH range of 5.5 to 7.5, incremental increases in P. carinii PHR1 mRNA were seen, with optimal expression at physiological pH (7.0 to 7.5). This pH range has also been reported previously to induce expression of C. albicans PHR1 (32). Such a pH is compatible with the environmental pH of the alveolar space under basal and stress conditions (9). Environmental pH may provide one signal to P. carinii organisms that the milieu is conducive for life cycle progression and proliferation (20). To our knowledge, this represents the first description of an environmentally regulated gene in this important opportunistic fungus.
FIG. 3.
P. carinii PHR1 exhibits pH-regulated mRNA expression. To examine whether P. carinii PHR1 expression responded to environmental pH, organisms were freshly isolated and maintained in medium at the indicated pH prior to isolation of RNA and Northern analysis. Optimal steady-state mRNA expression of P. carinii PHR1 was observed at physiological pH of 7.0 to 7.5. RNA loading was verified by ethidium bromide staining and reprobing of the blot with P. carinii actin.
Recent investigations in C. albicans document the importance of PHR activity in disease pathogenesis (6, 8, 11). C. albicans deleted of PHR1 exhibits aberrant morphology and is less virulent than PHR1+ strains in mice with disseminated systemic candidiasis. Specifically, the growth of C. albicans PHR1+ strains is favored in the neutral to slightly alkaline pH of the bloodstream (11). Whether P. carinii similarly expresses other pH-related genes under alternative conditions is an attractive hypothesis, which merits further investigation. Review of the evolving P. carinii project genome database (http://www.uky.edu/Projects/Pneumocystis/) has not yet revealed any additional candidate pH-responsive genes. Identification of genes active under alternative environmental conditions might provide important insights into alternative niches used by this intractable organism.
P. carinii PHR1 DNA confers cell wall integrity on Gas1p-deficient S. cerevisiae
PHR class genes have been implicated in cell wall generation, cross-linking, and stability under adverse conditions. Heterologous expression of P. carinii genes in culturable fungal species has recently proven a useful experimental approach to infer P. carinii gene function (10, 17, 36). Accordingly, we evaluated the ability of P. carinii PHR1 to growth complement an S. cerevisiae mutant strain deficient in the PHR1 analogue GAS1 in the presence of a potent osmotic destabilizing agent, SDS (38).
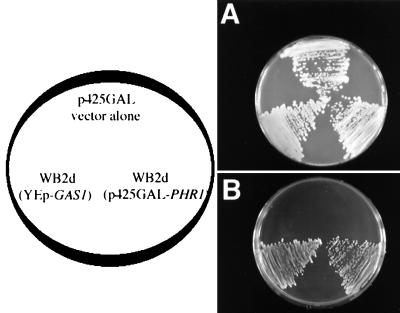
P. carinii PHR1 sequences were subcloned into the S. cerevisiae expression vector p425GAL (25). S. cerevisiae gas1 mutants susceptible to growth inhibition by SDS were transformed with P. carinii PHR1 DNA in pGAL425 and selected by proliferation at 30°C on medium containing 0.005% SDS. A number of colonies were isolated in which the P. carinii PHR1 DNA strongly restored proliferation of the S. cerevisiae mutant parent (Fig. 4). Complemented colonies were propagated at 30°C under the selective conditions, and plasmid DNA from P. carinii PHR1-complemented colonies was isolated to verify the presence of the P. carinii PHR1 gene in the selected transformants. S. cerevisiae gas1 mutant strains transformed with the pGAL425 vector alone failed to grow on selective medium containing 0.005% SDS. In contrast, both the S. cerevisiae strains transformed with either the GAS1 gene (37) or the P. carinii PHR1 gene displayed normal growth in the presence of SDS. Thus, P. carinii PHR1 is fully capable of functioning in fungal cell wall integrity when expressed heterologously in this tractable fungal species.
FIG. 4.
P. carinii PHR1 complements growth of S. cerevisiae gas1 mutants in the presence of SDS. S. cerevisiae gas1 mutants lack the ability to grow in the presence of the osmotic destabilizing agent SDS due to cell wall instability. The Gas1p-deficient mutants were cultured following transformation with vector alone, with the wild-type gene present in YEp-GAS1 (positive control), or with the vector containing P. carinii PHR1 (p245GAL-PHR1). Identical plates were cultured in (A) selective minimal medium or (B) minimal plates supplemented with 0.005% SDS. P. carinii PHR1 successfully rescued the cell wall instability of the yeast mutant and restored growth in the presence of SDS.
Biochemical analyses in C. albicans reveal that Phr1p strongly influences the stability and solubility of glucans under alkaline conditions. Specifically, Phr1+ strains exhibit 50% reduction in alkaline-insoluble glucan at pH 8.0 compared to null mutant strains, likely related to inefficient β-1,3–β-1,6 cross-linking (29). This defective cell wall structure may play a role in the decreased virulence of the PHR-deleted mutants. Furthermore, mutations of the homologous GAS1 gene in S. cerevisiae have been demonstrated to cause rounded, abnormal budding, resulting in decreased fungal proliferation and decreased cell wall integrity, with loss of β-1,3-glucans from the cell wall into the surrounding environment (28, 30, 38).
Additional biochemical activities of this family of proteins have recently been defined by Mouyna and colleagues, who characterized a novel 1,3-beta-glucanosyltransferase from Aspergillus fumigatus and cloned its corresponding gene, GEL1 (23). This enzyme mediates internal cleavage of the β-1,3-glucan chain and transfers the resulting reducing end to the nonreducing end of another β-1,3-glucan molecule, thereby elongating the polysaccharide. The predicted amino acid sequence of Gel1p was homologous to Gas1p from S. cerevisiae, Phr1p of C. albicans, and Epd from C. maltosa (23). Recombinant Gas1p, Phr1p, and Phr2p were also shown to have similar 1,3-beta-glucanosyltransferase activity in vitro. Additionally, Gel1p appears to be similarly attached to the membrane via a GPI anchor.
It is an attractive hypothesis that P. carinii PHR1 may have similar activities in P. carinii, maintaining proper cell wall glucan structure in the host's lung where a pH of 7 to 7.5 predominates. It is again notable that P. carinii PHR1 is located immediately downstream of the GSC1, gene which directs synthesis of the β-1,3-glucan cyst wall (16). Substantial investigation indicates that generation and maintenance of the β-glucan cell wall of P. carinii are essential for establishment of infection and represent a major target of host recognition and inflammatory response to the organism (14, 34, 39). Further studies aimed at determining the mechanisms by which β-glucan is generated and remodeled under various environmental conditions should further illuminate the life cycle of this intriguing fungus, which afflicts immunocompromised individuals.
ACKNOWLEDGMENTS
This work was supported by National Institutes of Health grants R01-HL55934, R01-HL57125, and R01-HL62150 to A.H.L.
We thank Marina Vai, Università degli Studi di Milano, Milan, Italy, for her generous gift of the WB2d S. cerevisiae gas1 mutant strain and the YEp-GAS1 plasmid used in these studies. We also appreciate the assistance of Kathy Streich in final preparation of the manuscript.
REFERENCES
- 1.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. Vol. 2. New York, N.Y: John Wiley & Sons, Inc.; 1993. pp. 13.12.1–13.12.5. [Google Scholar]
- 2.Berger J, Howard A D, Brink L, Gerber L, Hauber J, Cullen B R, Udenfriend S. COOH-terminal requirements for the correct processing of a phosphatidylinositol-glycan anchored membrane protein. J Biol Chem. 1988;263:10016–10021. [PubMed] [Google Scholar]
- 3.Chin K, Luttrell T D, Roe J D, Shadzi S, Wyder M A, Kaneshiro E S. Putative Pneumocystis dormant forms outside the mammalian host, and long-term culture derived from them: initial characterizations. J Eukaryot Microbiol. 1999;46:95S–99S. [PubMed] [Google Scholar]
- 4.Cushion M T, Harmsen A, Matsumoto Y, Stringer J R, Wakefield A E, Yamada M. Recent advances in the biology of Pneumocystis carinii. J Med Vet Mycol. 1994;32:217–228. [PubMed] [Google Scholar]
- 5.Cushion M T, Kaselis M, Stringer S L, Stringer J R. Genetic stability and diversity of Pneumocystis carinii infecting rat colonies. Infect Immun. 1993;61:4801–4813. doi: 10.1128/iai.61.11.4801-4813.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.De Bernardis F, Muhlschlegel F A, Cassone A, Fonzi W A. The pH of the host niche controls gene expression in and virulence of Candida albicans. Infect Immun. 1998;66:3317–3325. doi: 10.1128/iai.66.7.3317-3325.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Feng D F, Doolittle R F. Progressive sequence alignment as a prerequisite to correct phylogenetic trees. J Mol Evol. 1987;25:351–360. doi: 10.1007/BF02603120. [DOI] [PubMed] [Google Scholar]
- 8.Fonzi W A. PHR1 and PHR2 of Candida albicans encode putative glycosidases required for proper cross-linking of β-1,3- and β-1,6-glucans. J Bacteriol. 1999;181:7070–7079. doi: 10.1128/jb.181.22.7070-7079.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Forster, H. V., J. A. Dempsey, and L. W. Chosy. Incomplete compensation of CSF (H+) in humans during acclimitization to high altitude (4300 m). J. Appl. Physiol. 38:1067–1072. [DOI] [PubMed]
- 10.Fox D, Smulian A G. Mitogen-activated protein kinase Mkp1 of Pneumocystis carinii complements the slt2Δ defect in the cell integrity pathway of Saccharomyces cerevisiae. Mol Microbiol. 1999;34:451–462. doi: 10.1046/j.1365-2958.1999.01606.x. [DOI] [PubMed] [Google Scholar]
- 11.Ghannoum M A, Spellberg B, Saporito-Irwin S M, Fonzi W A. Reduced virulence of Candida albicans PHR1 mutants. Infect Immun. 1995;63:4528–4530. doi: 10.1128/iai.63.11.4528-4530.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guadiz G, Haidaris C G, Maine G N, Simpson-Haidaris P J. The carboxyl terminus of Pneumocystis carinii glycoprotein A encodes a functional glycosylphosphatidylinositol signal sequence. J Biol Chem. 1998;273:26202–26209. doi: 10.1074/jbc.273.40.26202. [DOI] [PubMed] [Google Scholar]
- 13.Hemperly J J, Edelman G M, Cunningham B A. cDNA clones of the neural cell adhesion molecule (N-CAM) lacking a membrane-spanning region consistent with evidence for membrane attachment via a phosphatidylinositol intermediate. Proc Natl Acad Sci USA. 1986;83:9822–9826. doi: 10.1073/pnas.83.24.9822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hoffman O A, Standing J E, Limper A H. Pneumocystis carinii stimulates tumor necrosis factor-alpha release from alveolar macrophages through a beta-glucan-mediated mechanism. J Immunol. 1993;150:3932–3940. [PubMed] [Google Scholar]
- 15.Hong S T, Steele P E, Cushion M T, Walzer P D, Stringer S L, Stringer J R. Pneumocystis carinii karyotypes. J Clin Microbiol. 1990;28:1785–1795. doi: 10.1128/jcm.28.8.1785-1795.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kottom T J, Limper A H. Cell wall assembly by Pneumocystis carinii: evidence for a unique GSC-1 subunit mediating beta 1,3-glucan deposition. J Biol Chem. 2000;275:40628–40634. doi: 10.1074/jbc.M002103200. [DOI] [PubMed] [Google Scholar]
- 17.Kottom T J, Thomas C F J, Mubarak K K, Leof E B, Limper A H. Pneumocystis carinii utilizes a functional cdc13 B-type cyclin complex during its life cycle. Am J Respir Cell Mol Biol. 2000;22:722–731. doi: 10.1165/ajrcmb.22.6.3838. [DOI] [PubMed] [Google Scholar]
- 18.Kyte J, Doolittle R F. A simple method for displaying the hydropathic character of a protein. J Mol Biol. 1982;157:105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- 19.Limper A H, Hoyte J S, Standing J E. The role of alveolar macrophages in Pneumocystis carinii degradation and clearance from the lung. J Clin Investig. 1997;99:2110–2117. doi: 10.1172/JCI119384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Limper A H, Thomas C F, Jr, Anders R A, Leof E B. Interactions of parasite and host epithelial cell cycle regulation during Pneumocystis carinii pneumonia. J Lab Clin Med. 1997;130:132–138. doi: 10.1016/s0022-2143(97)90089-5. [DOI] [PubMed] [Google Scholar]
- 21.Maccheroni W, Jr, May G S, Martinez-Rossi N M, Rossi A. The sequence of palF, an environmental pH response gene in Aspergillus nidulans. Gene. 1997;194:163–167. doi: 10.1016/s0378-1119(97)00095-4. [DOI] [PubMed] [Google Scholar]
- 22.Micanovic R, Gerber L D, Berger J, Kodukula K, Udenfriend S. Selectivity of the cleavage/attachment site of phosphatidylinositol-glycan-anchored membrane proteins determined by site-specific mutagenesis at Asp-484 of placental alkaline phosphatase. Proc Natl Acad Sci USA. 1990;87:157–161. doi: 10.1073/pnas.87.1.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mouyna I, Pontaine T, Vai M, Monod M, Fonzi W A, Diaquin M, Popolo L, Hartland R P, Latge J P. Glycophosphatidylinositol-anchored glucanosyltransferases play an active role in the biosynthesis of the fungal cell wall. J Biol Chem. 2000;275:14882–14889. doi: 10.1074/jbc.275.20.14882. [DOI] [PubMed] [Google Scholar]
- 24.Muhlschlegel F A, Fonzi W A. PHR2 of Candida albicans encodes a functional homolog of the pH-regulated gene PHR1 with an inverted pattern of pH-dependent expression. Mol Cell Biol. 1997;17:5960–5967. doi: 10.1128/mcb.17.10.5960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mumberg D, Muller R, Funk M. Regulatable promoters of Saccharomyces cerevisiae: comparison of transcriptional activity and their use for heterologous expression. Nucleic Acids Res. 1994;22:5767–5768. doi: 10.1093/nar/22.25.5767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nakazawa T, Horiuchi H, Ohta A, Takagi M. Isolation and characterization of EPD1, an essential gene for pseudohyphal growth of a dimorphic yeast, Candida maltosa. J Bacteriol. 1998;180:2079–2086. doi: 10.1128/jb.180.8.2079-2086.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nuoffer C, Jeno P, Conzelmann A, Riezman H. Determinants for glycophospholipid anchoring of the Saccharomyces cerevisiae Gas1 protein to the plasma membrane. Mol Cell Biol. 1991;11:27–37. doi: 10.1128/mcb.11.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Popolo L, Gilardelli D, Bonfante P, Vai M. Increase in chitin as an essential response to defects in assembly of cell wall polymers in the ggp1Δ mutant of Saccharomyces cerevisiae. J Bacteriol. 1997;179:463–469. doi: 10.1128/jb.179.2.463-469.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Popolo L, Vai M. Defects in assembly of the extracellular matrix are responsible for altered morphogenesis of a Candida albicans phr1 mutant. J Bacteriol. 1998;180:163–166. doi: 10.1128/jb.180.1.163-166.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ram A F, Kapteyn J C, Montijn R C, Caro L H, Douwes J E, Baginsky W, Mazur P, van den Ende H, Klis F M. Loss of the plasma membrane-bound protein Gas1p in Saccharomyces cerevisiae results in the release of beta-1,3-glucan into the medium and induces a compensation mechanism to ensure cell wall integrity. J Bacteriol. 1998;180:1418–1424. doi: 10.1128/jb.180.6.1418-1424.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 32.Saporito-Irwin S M, Birse C E, Sypherd P S, Fonzi W A. PHR1, a pH-regulated gene of Candida albicans, is required for morphogenesis. Mol Cell Biol. 1995;15:601–613. doi: 10.1128/mcb.15.2.601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sarkar S, Caddick M X, Bignell E, Tilburn J, Arst H N., Jr Regulation of gene expression by ambient pH in Aspergillus: genes expressed at acid pH. Biochem Soc Trans. 1996;24:360–363. doi: 10.1042/bst0240360. [DOI] [PubMed] [Google Scholar]
- 34.Schmatz D M, Romancheck M A, Pittarelli L A, Schwartz R E, Fromtling R A, Nollstadt K H, Vanmiddlesworth F L, Wilson K E, Turner M J. Treatment of Pneumocystis carinii pneumonia with 1,3-beta-glucan synthesis inhibitors. Proc Natl Acad Sci USA. 1990;87:5950–5954. doi: 10.1073/pnas.87.15.5950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sentandreu M, Elorza M V, Sentandreu R, Fonzi W A. Cloning and characterization of PRA1, a gene encoding a novel pH-regulated antigen of Candida albicans. J Bacteriol. 1998;180:282–289. doi: 10.1128/jb.180.2.282-289.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Thomas C F, Anders R A, Gustafson M P, Leof E B, Limper A H. Pneumocystis carinii contains a functional cell-division-cycle Cdc2 homologue. Am J Respir Cell Mol Biol. 1998;18:297–306. doi: 10.1165/ajrcmb.18.3.3122. [DOI] [PubMed] [Google Scholar]
- 37.Vai M, Gatti E, Lacana E, Popolo L, Alberghina L. Isolation and deduced amino acid sequence of the gene encoding gp115, a yeast glycophospholipid-anchored protein containing a serine-rich region. J Biol Chem. 1991;266:12242–12248. [PubMed] [Google Scholar]
- 38.Vai M, Orlandi I, Cavadini P, Alberghina L, Popolo L. Candida albicans homologue of GGP1/GAS1 gene is functional in Saccharomyces cerevisiae and contains the determinants for glycosylphosphatidylinositol attachment. Yeast. 1996;12:361–368. doi: 10.1002/(SICI)1097-0061(19960330)12:4%3C361::AID-YEA920%3E3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- 39.Vassallo R, Standing J E, Limper A H. Isolated Pneumocystis carinii cell wall glucan provokes lower respiratory tract inflammatory responses. J Immunol. 2000;164:3755–3763. doi: 10.4049/jimmunol.164.7.3755. [DOI] [PubMed] [Google Scholar]