Abstract
Biofilms are considered to be highly resistant to antimicrobial agents. Strictly speaking, this is not the case—biofilms do not grow in the presence of antimicrobials any better than do planktonic cells. Biofilms are indeed highly resistant to killing by bactericidal antimicrobials, compared to logarithmic-phase planktonic cells, and therefore exhibit tolerance. It is assumed that biofilms are also significantly more tolerant than stationary-phase planktonic cells. A detailed comparative examination of tolerance of biofilms versus stationary- and logarithmic-phase planktonic cells with four different antimicrobial agents was performed in this study. Carbenicillin appeared to be completely ineffective against both stationary-phase cells and biofilms. Killing by this β-lactam antibiotic depends on rapid growth, and this result confirms the notion of slow-growing biofilms resembling the stationary state. Ofloxacin is a fluoroquinolone antibiotic that kills nongrowing cells, and biofilms and stationary-phase cells were comparably tolerant to this antibiotic. The majority of cells in both populations were eradicated at low levels of ofloxacin, leaving a fraction of essentially invulnerable persisters. The bulk of the population in both biofilm and stationary-phase cultures was tolerant to tobramycin. At very high tobramycin concentrations, a fraction of persister cells became apparent in stationary-phase culture. Stationary-phase cells were more tolerant to the biocide peracetic acid than were biofilms. In general, stationary-phase cells were somewhat more tolerant than biofilms in all of the cases examined. We concluded that, at least for Pseudomonas aeruginosa, one of the model organisms for biofilm studies, the notion that biofilms have greater resistance than do planktonic cells is unwarranted. We further suggest that tolerance to antibiotics in stationary-phase or biofilm cultures is largely dependent on the presence of persister cells.
Biofilm infections are difficult to eradicate with antimicrobial treatment, and in vitro susceptibility tests show considerable resistance of biofilm cells to killing (for reviews, see references 8, 16, and 22). Up to 60% of all human infections are caused by biofilms. It is commonly accepted that biofilms are more resistant to antibiotics than are planktonic cells. This, however, is a misconception—the biofilm literature does not point to the ability of biofilms to grow at higher concentrations of antimicrobials compared to planktonic cells (20, 21). Rather, biofilms are highly resistant to killing by bactericidal antibiotics. This should properly be referred to as phenotypic tolerance or tolerance, for short. Several factors have been suggested to account for biofilm tolerance—slow growth (15), the presence of an exopolysaccharide matrix that can slow the diffusion of antibiotics, and the presence of unknown resistance mechanisms (8). Slow growth undoubtedly contributes to resistance to killing by antimicrobials, but this is not a unique feature of biofilm cells. With the exception of that of aminoglycosides (17, 19, 26, 33), the exopolysaccharide matrix has not been found to notably retard the diffusion of antibiotics. For example, fluoroquinolones readily penetrate the biofilm (1, 19, 33, 37). Multidrug resistance pumps represent a generalized resistance mechanism and have been considered as candidates for a biofilm resistance mechanism. However, we and two other groups reported that, at least in Pseudomonas aeruginosa, multidrug resistance pumps do not noticeably contribute to biofilm survival (6, 10, 22). In retrospect, this is not surprising—biofilms are not usually more resistant to growth inhibition than are planktonic cells, so there does not seem to be a need to invoke special drug resistance mechanisms.
Much progress has been made in the understanding of biofilm development (9, 13, 28–30, 38; see reference 27 for a recent comprehensive review), but the molecular basis of biofilm tolerance remains elusive. Our previous work suggested a general mechanism of biofilm resistance to antimicrobials. In a study of P. aeruginosa biofilms, we found that the vast majority of cells were eliminated by fairly low, clinically achievable concentrations of fluoroquinolones, but a small fraction of persisters remained essentially invulnerable to killing (6). Biphasic killing kinetics revealing similar persister populations can be found in papers describing treatment of biofilms of various bacterial species with a range of bactericidal antibiotics (for reviews, see references 20 and 21). We suggested that persisters are largely responsible for the resistance of biofilms to killing and explain, in principle, the nature of biofilm resistance (6, 20, 21). Persisters are not mutants. Reculturing of persisters produces a wild-type population with a new population of persisters (3, 6). Are persisters produced primarily by biofilms? In this paper, we report the results of a detailed comparison of tolerance between biofilms and stationary-state populations. We found that, contrary to popular belief, planktonic stationary-phase cells are somewhat more tolerant than biofilms. This increased resistance to killing is due to slow growth and high levels of persisters produced in stationary-phase planktonic populations.
MATERIALS AND METHODS
Bacterial strain and culture conditions.
The bacterial strain used in this study was P. aeruginosa wild-type strain PAO1 (6). Mueller-Hinton broth (MHB; Difco, Detroit, Mich.) was used to culture PAO1 in all experiments.
Antibiotics.
Tobramycin and carbenicillin were obtained from Sigma. Ofloxacin and peracetic acid were obtained from the R. W. Johnson Research Institute and Aldrich, respectively.
Susceptibility testing.
The MIC of each antibiotic was determined by the standard NCCLS broth microdilution method (25).
Testing of planktonic and biofilm cell resistance to killing.
Planktonic stationary-phase cultures were prepared by inoculating 105 cells/ml and incubating them for 18 h at 37°C with aeration. Cells were then sedimented, washed twice, and resuspended in fresh MHB at the original concentration (∼109 cells/ml). Planktonic logarithmic-phase cultures were prepared by inoculating 105 cells/ml and incubating them for 3 h at 37°C with aeration. For dose-dependent determination of killing, 200 μl of either stationary- or logarithmic-phase cells was dispensed into microtiter plates and incubated with an antibiotic for 6 h at 37°C. Following the challenge, the number of live cells was determined by colony counting.
Biofilms were grown essentially by the method of Ceri et al. (7) as previously described (6). The device used for biofilm formation in this study is a platform carrying 96 polystyrene pegs (Nunc no. 445497) that fits as a microtiter plate lid with a peg hanging into each microtiter plate well (Nunc no. 269787). For biofilm formation, the device was placed in its original sterile tray filled with MHB and cells (104/ml) and incubated for 18 h at 37°C on a tilting shaker that provides a shearing force. After biofilms were formed on the pegs, they were washed in MHB and the device with intact biofilms was placed in a microtiter plate with fresh MHB for drug susceptibility testing. Following a 6-h incubation in the presence of an antimicrobial agent, the pegs were washed twice in MHB and the device was placed in a microtiter plate with MHB and incubated for 10 min in a water bath sonicator (Branson Ultrasonic Cleaner; Branson Cleaning Equipment Company). For each antimicrobial concentration tested, cells were collected from three parallel pegs and plated for colony counting.
RESULTS
Several different bactericidal antimicrobials were chosen to test the relative resistance to killing of planktonic and biofilm cells—ofloxacin, a fluoroquinolone; tobramycin, an aminoglycoside; carbenicillin, a β-lactam; and peracetic acid, an oxidant.
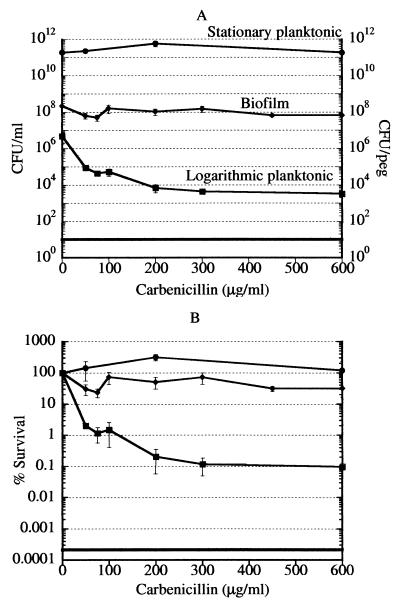
Carbenicillin is a bactericidal antibiotic that, similarly to other β-lactams, kills only rapidly growing cells (35). If biofilm cells were primarily slow growing, they would be expected to be resistant to killing by carbenicillin. Logarithmic-phase, stationary-phase, and biofilm cultures were challenged with carbenicillin over a wide range of concentrations, from 1.67 times the MIC (50 μg/ml) to 20 times the MIC (600 μg/ml). After a 6-h incubation with the antibiotic, viability was determined by colony counting. As expected, carbenicillin produced little killing in stationary-phase cells while the majority of logarithmic-phase cells were killed at 1.67 times the MIC (Fig. 1). The amount of killing of logarithmic-phase cells dropped off significantly at concentrations above 1.67 times the MIC, indicating the presence of a persister subpopulation. This 0.1% of the cells in the rapidly growing logarithmic-phase culture were invulnerable to killing by carbenicillin at 600 μg/ml. Biofilm cells were resistant to killing by carbenicillin. This indicates that the biofilms used in this study are made of slow-growing, essentially stationary-phase cells, in agreement with previous reports (15).
FIG. 1.
Killing of P. aeruginosa cells by carbenicillin. (A) Cells of logarithmic-phase, stationary-phase, and biofilm cultures were treated with carbenicillin for 6 h and then plated for colony counting. The data for biofilm cultures are plotted as CFU per peg of the biofilm device. The limit of detection is indicated by the solid horizontal line. The standard deviation for each point was calculated with n = 3. Biofilm, diamonds; stationary-phase cells, circles; logarithmic-phase cells, squares. (B) Data of panel A replotted as percent survival.
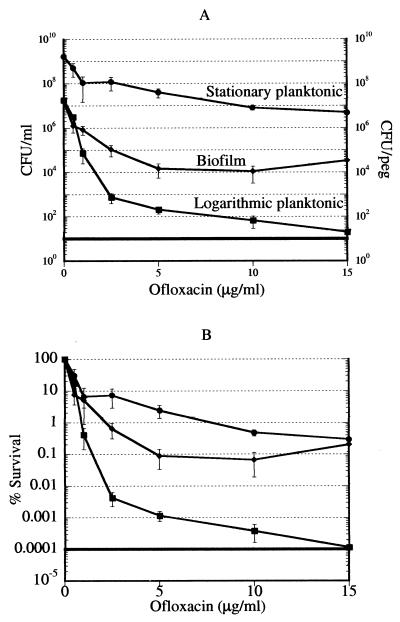
Unlike carbenicillin, ofloxacin can kill nongrowing cells (6), providing a useful tool with which to examine the relative tolerance of stationary-phase and biofilm cultures. Logarithmic-phase, stationary-phase, and biofilm cultures were challenged with ofloxacin over a wide range of concentrations, from the MIC (0.5 μg/ml) to 30 times the MIC (15 μg/ml). After a 6-h incubation with the antibiotic, viability was determined by colony counting. The majority of cells in the three populations examined were killed by low concentrations of ofloxacin (Fig. 2A). The killing in all three cultures was distinctly biphasic, indicating the presence of persister cells. The levels of persisters were dramatically higher in the dense stationary-phase planktonic and biofilm cultures than in the logarithmic-phase cells. Note that the cell concentration of the biofilm is presented as the number of cells per peg and that the density of the biofilm is considerably higher than the density of the stationary-phase culture. Dilution of stationary-phase cells causes a drastic drop in persisters, to the level found in logarithmic-phase cells (N. Kaldalu et al., submitted for publication). (The small persister fraction was not detected in a diluted stationary-phase culture in our previous study [6], apparently due to minor differences in the experimental protocol.) The plateau at increasing antibiotic concentrations shows that persisters are essentially invulnerable to killing by a fluoroquinolone. Note that this experiment was performed with planktonic cells and biofilms that were transferred into fresh medium containing the antibiotic (see Materials and Methods for details). With ofloxacin at 5 μg/ml, which is a clinically achievable concentration (32), the percentage of live cells was 0.001% in the logarithmic-phase population, 0.1% in the biofilm, and 2.5% in the stationary-phase culture (Fig. 2B). Unexpectedly, stationary-phase cells appeared to produce more persisters and were relatively more tolerant than the biofilm (Fig. 2B). Microscopic examination of the suspension of stationary-phase cells prior to antibiotic addition showed that it consisted primarily of individual planktonic cells and a small number of clumps. The total percentage of cells in clumps was ≤0.01%.
FIG. 2.
Killing of P. aeruginosa cells by ofloxacin. (A) Cells of logarithmic-phase, stationary-phase, and biofilm cultures were treated with ofloxacin for 6 h and then plated for colony counting. The conditions were as described in the legend to Fig. 1. Biofilm, diamonds; stationary-phase cells, circles; logarithmic-phase cells, squares. (B) Data of panel A replotted as percent survival.
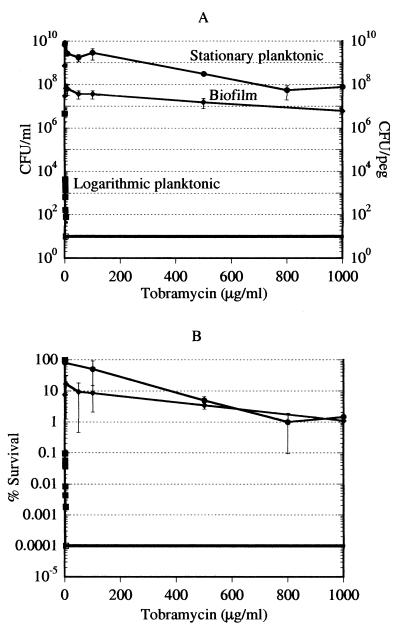
Tobramycin is another bactericidal antibiotic that can kill nongrowing cells. Unlike ofloxacin, tobramycin reportedly binds to the biofilm exopolysaccharide, and we expected biofilms to be considerably more resistant to killing by this antibiotic. Logarithmic-phase, stationary-phase, and biofilm cultures were challenged with tobramycin over a wide range of concentrations, from the MIC (1 μg/ml) to 1,500 times the MIC (1,500 μg/ml). After a 6-h incubation with the antibiotic, viability was determined by colony counting. Tobramycin was exceptionally effective in killing logarithmic-phase cells, and no logarithmic-phase persisters were detected (Fig. 3A). Tobramycin at 50 μg/ml (the maximal clinically achievable concentration is 10 μg/ml [32]) eliminated 90% of the biofilm cells, but the remaining population declined very gradually with increasing amounts of the antibiotic. This relative resistance of the biofilm to killing by tobramycin is in accordance with previous observations in the literature (18). We found tobramycin to be ineffective in killing stationary-phase planktonic cells. Unlike the biofilm, the majority of the cells were not eliminated at low tobramycin concentrations. At a tobramycin concentration of 800 μg/ml, a persistent population of 1% surviving cells became apparent (Fig. 3B).
FIG. 3.
Killing of P. aeruginosa cells by tobramycin. (A) Cells of logarithmic-phase, stationary-phase, and biofilm cultures were treated with tobramycin for 6 h and then plated for colony counting. The conditions were as described in the legend to Fig. 1. Note that the experimental points for logarithmic-phase cells essentially align at the y axis due to the scale of this graph. Complete killing of logarithmic-phase cells was achieved with tobramycin at 4 μg/ml. The open square shows that the data point was at or below the limit of detection. Biofilm, diamonds; stationary-phase cells, circles; logarithmic-phase cells, squares. (B) Data of panel A replotted as percent survival.
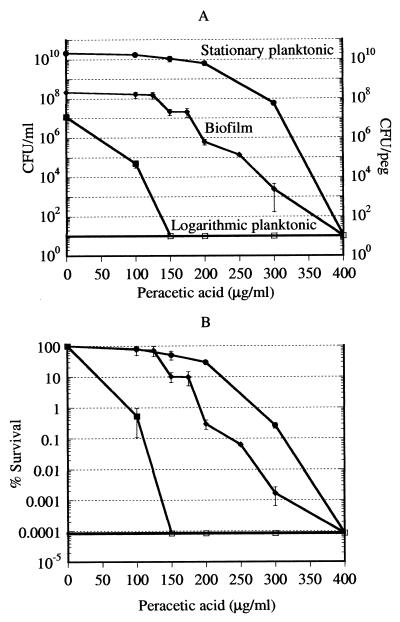
Biofilms have been reported to be tolerant to both specific antibiotics and biocides. It was of interest to learn whether biofilms are indeed more resistant to biocide killing than are planktonic cells. Logarithmic-phase, stationary-phase, and biofilm cultures were challenged with peracetic acid over a range of concentrations, from the MIC (100 μg/ml) to four times the MIC (400 μg/ml). After a 6-h incubation with the antibiotic, viability was determined by colony counting. Biphasic killing by this antimicrobial oxidant was not observed (Fig. 4). Biofilms showed considerable tolerance to this biocide compared to logarithmic-phase planktonic cells. However, stationary-phase planktonic cells were even more tolerant to peracetic acid than were biofilms. Complete killing of all three populations by 400 μg/ml was observed.
FIG. 4.
Killing of P. aeruginosa cells by peracetic acid. (A) Cells of logarithmic-phase, stationary-phase, and biofilm cultures were treated with peracetic acid for 6 h and then plated for colony counting. The conditions were as described in the legend to Fig. 1. The open squares show that the data points were at or below the limit of detection. Biofilm, diamonds; stationary-phase cells, circles; logarithmic-phase cells, squares. (B) Data of panel A replotted as percent survival.
DISCUSSION
Biofilms are commonly viewed as being resistant to killing by a broad range of antimicrobial agents. Indeed, biofilms show more tolerance to antimicrobials than do planktonic logarithmic-phase cells. But this is not surprising—it is well established that rapidly growing cells are more susceptible to both growth inhibition and killing. Some antibiotics, like older β-lactams, absolutely require rapid growth in order to kill cells. However, it is also generally believed that biofilms are more resistant to killing than are stationary-phase planktonic cells. This has led to suggestions that specific resistance mechanisms might be expressed in biofilms (8). But growth of biofilm cells has not been found to be more resistant to antibiotics than that of logarithmic-phase planktonic cells, suggesting that there is no basis for proposing the existence of broadly specific biofilm resistance mechanisms (20, 21). Are biofilms really more resistant to killing than are stationary-phase planktonic cells? A systematic comparison of susceptibility to killing between biofilm and logarithmic- and stationary-phase planktonic cells was undertaken in this study.
Carbenicillin kills rapidly growing cells. In our experiments, carbenicillin had no effect on biofilm or stationary-phase planktonic cultures. This indicates that biofilm cells are essentially in the stationary state. Slow growth may satisfactorily explain resistance to killing by antibiotics like carbenicillin. We then tested whether biofilms are more resistant to killing by antibiotics that have activity against nongrowing cells.
Ofloxacin at low concentrations produced significant killing of logarithmic-phase planktonic cells. A small percentage (0.001%) of persister cells resistant to killing was evident. The killing of biofilm cells similarly followed biphasic kinetics, but the proportion of persisters was significantly higher than in logarithmic-phase planktonic cells, comprising 0.1% of the population. These results are in agreement with our previous findings (6) and clearly show that the majority of biofilm cells are very sensitive to killing by ofloxacin and that the overall biofilm resistance to killing is due to the presence of persisters. Note that in these experiments, the biofilm was transferred into a fresh, antibiotic-containing growth medium. In order to make a meaningful comparison with the biofilm, stationary-phase cells were also transferred, essentially without dilution, into fresh, antibiotic-containing growth medium. Unexpectedly and contrary to the general assumption, a stationary-state culture produced relatively more persisters than did a biofilm and was more resistant to killing by ofloxacin. In our previous study, we found that a stationary-phase planktonic population was very sensitive to killing by ofloxacin. Cells in that experiment were intentionally diluted (about 100-fold) in order to arrive at a population of a size comparable to that of the biofilm growing on a single peg. That seemingly reasonable procedure produced a misleading result. We now find that the formation and maintenance of persisters depend strongly on the density of the population—dilution of stationary-phase cells leads to a collapse in the number of persisters. This indicates that the dense population of either stationary-phase cells or biofilms favors persister formation. It is possible that a quorum-sensing factor(s) controls persister formation (Kaldalu et al., submitted). To our knowledge, undiluted stationary-phase cell populations have not been used in comparative studies with biofilms in tests with antibiotics prior to this study. The common assumption that biofilms are more resistant to killing than are planktonic cells derives from experiments with either logarithmic-phase or diluted stationary-phase cultures.
Tobramycin was probably the first antibiotic capable of killing nongrowing cells that was reported to be very ineffective in killing biofilms (2). It was subsequently found that the biofilm exopolysaccharide binds and restricts the penetration of cationic aminoglycosides like tobramycin (17, 19, 26, 33). This seemed to be a group of antibiotics for which a biofilm-specific mechanism of resistance was justified. However, our results show that a stationary-phase cell population is even more resistant to killing by tobramycin than is a biofilm. These stationary-phase cells were washed twice and then resuspended in fresh medium, suggesting that exopolysaccharide was probably removed from the culture. Slow growth rather than sequestration of the antibiotic might be the critical contributing factor. Indeed, tobramycin activity was found to be growth rate dependent (15).
The nature of persistence and the mechanism of cell death are interrelated but virtually unexplored. Mutations dramatically increasing the production of persisters in a logarithmic-phase population of Escherichia coli (hip) have been described (4, 5, 12, 23, 24, 31, 39). This observation suggests that in bacteria, death might be a regulated event. We have argued that, similarly to metazoan tissues, populations of kin bacterial cells would benefit from elimination of defective members through a programmed cell death (PCD) mechanism (20). According to this hypothesis, bactericidal antibiotics do not kill cells but inflict damage that activates PCD. This logic is identical to what we know of metazoan cells damaged by toxins that induce apoptosis. The problem with this scenario is that an antibiotic diffusing uniformly through a bacterial population will lead to total suicide, which is counterproductive. We proposed that persisters are cells with a disabled PCD mechanism whose function is survival (20).
Unlike conventional antibiotics, biocides are likely to inflict sufficient damage to kill cells directly. Biofilms have been reported to be resistant to killing by biocides. We find that peracetic acid, a strong oxidant and a widely used biocide, is indeed far less effective in killing biofilm cells than in killing logarithmic-phase planktonic cells. However, stationary-phase cells were even more resistant to killing than were biofilms. In this respect, the action of peracetic acid appeared to be similar to that of the bactericidal antibiotics ofloxacin and tobramycin. The apparent lack of persisters in this dose-dependent experiment was a distinct feature of peracetic acid killing. Note that persisters are virtually invulnerable to killing by antibiotics and survive in the presence of antibiotic levels 100- to 1,000-fold higher than the MIC. By contrast, the killing concentration range was very narrow in the case of the biocide and cells in all of the cultures were eradicated at four times the MIC. This comparison further supports the idea that damage from antibiotics is relatively limited and death requires active participation on the part of cells (PCD) while biocides kill cells directly.
The main unexpected conclusion of this study is that biofilms of P. aeruginosa are not different from stationary-phase planktonic cells in their resistance to killing by antibiotics and a biocide. Our experiments were limited to a single bacterial species, but P. aeruginosa has served as a main model organism for biofilm studies. The presence of persisters in other species (20) and the increase in resistance to killing with an increase in cell density in Burkholderia cepacia (11), E. coli, and Staphylococcus aureus (Kaldalu et al., submitted) suggest that this is a general phenomenon. The critical role of persisters in the survival of both biofilm and planktonic populations suggests a new paradigm in our understanding of biofilm infections. A biofilm sheds planktonic cells that are primarily responsible for the manifestation of a disease. Antibiotics like ofloxacin eliminate most planktonic and biofilm cells but leave the persisters intact. The immune system is likely to eliminate the remaining planktonic persisters. However, biofilm cells are physically protected from the components of the immune system by the exopolysaccharide matrix (18, 36) and biofilm persisters will persevere. Once the antibiotic level drops, persisters will recreate the biofilm (20, 21). This model explains the relapsing nature of biofilm infections.
The persister hypothesis provides a satisfactory explanation for the puzzling observation that bacterial biofilms are resistant to killing by all known antibiotics. This hypothesis (6, 20, 21) is gaining acceptance as a general explanation for the phenomenon of biofilm tolerance (14, 34). The challenge is to understand the nature of persistence. Development of drugs that disable the persister phenotype might lead to compounds that can enable conventional antibiotics to eradicate a biofilm. An important practical conclusion from our results is that there might not be a need to study biofilm resistance per se—genes and proteins responsible for persistence can be identified in planktonic populations that are much easier to manipulate. Similarly, planktonic populations can be used for the discovery of antipersistence drugs.
ACKNOWLEDGMENTS
We thank Niilo Kaldalu for helpful discussions and Slava Epstein for help with microscopy.
This research was supported by NIH grant R01 GM61162.
REFERENCES
- 1.Anderl J N, Franklin M J, Stewart P S. Role of antibiotic penetration limitation in Klebsiella pneumoniae biofilm resistance to ampicillin and ciprofloxacin. Antimicrob Agents Chemother. 2000;44:1818–1824. doi: 10.1128/aac.44.7.1818-1824.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anwar H, Dasgupta M, Lam K, Costerton J W. Tobramycin resistance of mucoid Pseudomonas aeruginosa biofilm grown under iron limitation. J Antimicrob Chemother. 1989;24:647–655. doi: 10.1093/jac/24.5.647. [DOI] [PubMed] [Google Scholar]
- 3.Bigger J W. Treatment of staphylococcal infections with penicillin. Lancet. 1944;ii:497–500. [Google Scholar]
- 4.Black D S, Irwin B, Moyed H S. Autoregulation of hip, an operon that affects lethality due to inhibition of peptidoglycan or DNA synthesis. J Bacteriol. 1994;176:4081–4091. doi: 10.1128/jb.176.13.4081-4091.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Black D S, Kelly A J, Mardis M J, Moyed H S. Structure and organization of hip, an operon that affects lethality due to inhibition of peptidoglycan or DNA synthesis. J Bacteriol. 1991;173:5732–5739. doi: 10.1128/jb.173.18.5732-5739.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brooun A, Liu S, Lewis K. A dose-response study of antibiotic resistance in Pseudomonas aeruginosa biofilms. Antimicrob Agents Chemother. 2000;44:640–646. doi: 10.1128/aac.44.3.640-646.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ceri H, Olson M E, Stremick C, Read R R, Morck D, Buret A. The Calgary Biofilm Device: new technology for rapid determination of antibiotic susceptibilities of bacterial biofilms. J Clin Microbiol. 1999;37:1771–1776. doi: 10.1128/jcm.37.6.1771-1776.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Costerton J W, Stewart P S, Greenberg E P. Bacterial biofilms: a common cause of persistent infections. Science. 1999;284:1318–1322. doi: 10.1126/science.284.5418.1318. [DOI] [PubMed] [Google Scholar]
- 9.Davies G D, Parsek M R, Pearson J P, Iglewski B H, Costerton J W, Greenberg E P. The involvement of cell-to-cell signals in the development of a bacterial biofilm. Science. 1998;280:295–298. doi: 10.1126/science.280.5361.295. [DOI] [PubMed] [Google Scholar]
- 10.De Kievit T R, Parkins M D, Gillis R J, Srikumar R, Ceri H, Poole K, Iglewski B H, Storey D G. Multidrug efflux pumps: expression patterns and contribution to antibiotic resistance in Pseudomonas aeruginosa biofilms. Antimicrob Agents Chemother. 2001;45:1761–1770. doi: 10.1128/AAC.45.6.1761-1770.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Desai M, Buhler T, Weller P H, Brown M R. Increasing resistance of planktonic and biofilm cultures of Burkholderia cepacia to ciprofloxacin and ceftazidime during exponential growth. J Antimicrob Chemother. 1998;42:153–160. doi: 10.1093/jac/42.2.153. [DOI] [PubMed] [Google Scholar]
- 12.Falla T J, Chopra I. Joint tolerance to beta-lactam and fluoroquinolone antibiotics in Escherichia coli results from overexpression of hipA. Antimicrob Agents Chemother. 1998;42:3282–3284. doi: 10.1128/aac.42.12.3282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Genevaux P, Muller S, Bauda P. A rapid screening procedure to identify mini-Tn10 insertion mutants of Escherichia coli K-12 with altered adhesion properties. FEMS Microbiol Lett. 1996;142:27–30. doi: 10.1111/j.1574-6968.1996.tb08402.x. [DOI] [PubMed] [Google Scholar]
- 14.Gilbert P, Alison D G, Rickhard A, Sufya N, Whyte F, McBain A J. Do biofilms present a nidus for the evolution of antibacterial resistance? In: Gilbert D G P, Allison M B, Verran J, Walker J, editors. Biofilm community development: chance or necessity? Cardiff, Wales: Bioline Press; 2001. pp. 341–351. [Google Scholar]
- 15.Gilbert P, Collier P J, Brown M R. Influence of growth rate on susceptibility to antimicrobial agents: biofilms, cell cycle, dormancy, and stringent response. Antimicrob Agents Chemother. 1990;34:1865–1868. doi: 10.1128/aac.34.10.1865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gilbert P, Das J, Foley I. Biofilm susceptibility to antimicrobials. Adv Dent Res. 1997;11:160–167. doi: 10.1177/08959374970110010701. [DOI] [PubMed] [Google Scholar]
- 17.Gordon C A, Hodges N A, Marriott C. Antibiotic interaction and diffusion through alginate and exopolysaccharide of cystic fibrosis-derived Pseudomonas aeruginosa. J Antimicrob Chemother. 1988;22:667–674. doi: 10.1093/jac/22.5.667. [DOI] [PubMed] [Google Scholar]
- 18.Hoyle B D, Jass J, Costerton J W. The biofilm glycocalyx as a resistance factor. J Antimicrob Chemother. 1990;26:1–5. doi: 10.1093/jac/26.1.1. [DOI] [PubMed] [Google Scholar]
- 19.Ishida H, Ishida Y, Kurosaka Y, Otani T, Sato K, Kobayashi H. In vitro and in vivo activities of levofloxacin against biofilm-producing Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1998;42:1641–1645. doi: 10.1128/aac.42.7.1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lewis K. Programmed death in bacteria. Microbiol Mol Biol Rev. 2000;64:503–514. doi: 10.1128/mmbr.64.3.503-514.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lewis K. The riddle of biofilm resistance. Antimicrob Agents Chemother. 2001;45:999–1007. doi: 10.1128/AAC.45.4.999-1007.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Maira-Litran T, Allison D G, Gilbert P. An evaluation of the potential of the multiple antibiotic resistance operon (mar) and the multidrug efflux pump acrAB to moderate resistance towards ciprofloxacin in Escherichia coli biofilms. J Antimicrob Chemother. 2000;45:789–795. doi: 10.1093/jac/45.6.789. [DOI] [PubMed] [Google Scholar]
- 23.Moyed H S, Bertrand K P. hipA, a newly recognized gene of Escherichia coli K-12 that affects frequency of persistence after inhibition of murein synthesis. J Bacteriol. 1983;155:768–775. doi: 10.1128/jb.155.2.768-775.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Moyed H S, Broderick S H. Molecular cloning and expression of hipA, a gene of Escherichia coli K-12 that affects frequency of persistence after inhibition of murein synthesis. J Bacteriol. 1986;166:399–403. doi: 10.1128/jb.166.2.399-403.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.NCCLS. Minimum inhibitory concentration (MIC) interpretive standards (μg/ml) for Pseudomonas aeruginosa and other non-Enterobacteriaceae. NCCLS document M7–A5. Villanova, Pa: NCCLS; 2000. [Google Scholar]
- 26.Nichols W W, Dorrington S M, Slack M P, Walmsley H L. Inhibition of tobramycin diffusion by binding to alginate. Antimicrob Agents Chemother. 1988;32:518–523. doi: 10.1128/aac.32.4.518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.O'Toole G, Kaplan H B, Kolter R. Biofilm formation as microbial development. Annu Rev Microbiol. 2000;54:49–79. doi: 10.1146/annurev.micro.54.1.49. [DOI] [PubMed] [Google Scholar]
- 28.O'Toole G A, Kolter R. Flagellar and twitching motility are necessary for Pseudomonas aeruginosa biofilm development. Mol Microbiol. 1998;30:295–304. doi: 10.1046/j.1365-2958.1998.01062.x. [DOI] [PubMed] [Google Scholar]
- 29.O'Toole G A, Kolter R. Initiation of biofilm formation in Pseudomonas fluorescens WCS365 proceeds via multiple, convergent signaling pathways: a genetic analysis. Mol Microbiol. 1998;28:449–461. doi: 10.1046/j.1365-2958.1998.00797.x. [DOI] [PubMed] [Google Scholar]
- 30.Pratt L A, Kolter R. Genetic analysis of Escherichia coli biofilm formation: roles of flagella, motility, chemotaxis and type I pili. Mol Microbiol. 1998;30:285–293. doi: 10.1046/j.1365-2958.1998.01061.x. [DOI] [PubMed] [Google Scholar]
- 31.Scherrer R, Moyed H S. Conditional impairment of cell division and altered lethality in hipA mutants of Escherichia coli K-12. J Bacteriol. 1988;170:3321–3326. doi: 10.1128/jb.170.8.3321-3326.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schulz M, Schmoldt A. Therapeutic and toxic blood concentrations of more than 500 drugs. Pharmazie. 1997;52:895–911. [PubMed] [Google Scholar]
- 33.Shigeta M, Tanaka G, Komatsuzawa H, Sugai M, Suginaka H, Usui T. Permeation of antimicrobial agents through Pseudomonas aeruginosa biofilms: a simple method. Chemotherapy. 1997;43:340–345. doi: 10.1159/000239587. [DOI] [PubMed] [Google Scholar]
- 34.Stewart P S, Costerton J W. Antibiotic resistance of bacteria in biofilms. Lancet. 2001;358:135–138. doi: 10.1016/s0140-6736(01)05321-1. [DOI] [PubMed] [Google Scholar]
- 35.Tuomanen E, Cozens R, Tosch W, Zak O, Tomasz A. The rate of killing of Escherichia coli by beta-lactam antibiotics is strictly proportional to the rate of bacterial growth. J Gen Microbiol. 1986;132:1297–1304. doi: 10.1099/00221287-132-5-1297. [DOI] [PubMed] [Google Scholar]
- 36.von Eiff C, Heilmann C, Peters G. New aspects in the molecular basis of polymer-associated infections due to staphylococci. Eur J Clin Microbiol Infect Dis. 1999;18:843–846. doi: 10.1007/s100960050417. [DOI] [PubMed] [Google Scholar]
- 37.Vrany J D, Stewart P S, Suci P A. Comparison of recalcitrance to ciprofloxacin and levofloxacin exhibited by Pseudomonas aeruginosa biofilms displaying rapid-transport characteristics. Antimicrob Agents Chemother. 1997;41:1352–1358. doi: 10.1128/aac.41.6.1352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Watnick P I, Kolter R. Steps in the development of a Vibrio cholerae El Tor biofilm. Mol Microbiol. 1999;34:586–595. doi: 10.1046/j.1365-2958.1999.01624.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wolfson J S, Hooper D C, McHugh G L, Bozza M A, Swartz M N. Mutants of Escherichia coli K-12 exhibiting reduced killing by both quinolone and beta-lactam antimicrobial agents. Antimicrob Agents Chemother. 1990;34:1938–1943. doi: 10.1128/aac.34.10.1938. [DOI] [PMC free article] [PubMed] [Google Scholar]