Abstract
Many crowded biomolecular structures in cells and tissues are inaccessible to labelling antibodies. To understand how proteins within these structures are arranged with nanoscale precision therefore requires that these structures be decrowded before labelling. Here, we show that an iterative variant of expansion microscopy (the permeation of cells and tissues by a swellable hydrogel followed by isotropic hydrogel expansion, so as to allow for enhanced imaging resolution with ordinary microscopes) enables the imaging of nanostructures in expanded yet otherwise intact tissues at a resolution of about 20 nm. The method, which we named ‘expansion revealing’ and validated with DNA-probe-based super-resolution microscopy, involves gel-anchoring reagents, and the embedding, expansion and re-embedding of the sample in homogeneous swellable hydrogels. Expansion revealing enabled us to use confocal microscopy to image the alignment of presynaptic calcium channels with postsynaptic scaffolding proteins in intact brain circuits, and to uncover periodic amyloid nanoclusters containing ion-channel proteins in brain tissue from a mouse model of Alzheimer’s disease. Expansion revealing will enable the further discovery of previously unseen nanostructures within cells and tissues.
One-sentence editorial summary:
Iterative expansion microscopy enables the use of confocal microscopes to uncover nanostructures in expanded yet otherwise intact tissues at a resolution of about 20 nm, as shown with the imaging of periodic amyloid nanoclusters in brain tissue.
There is great desire to understand how proteins are arranged, with nanoscale precision, within cells and tissues. Super-resolution microscopy has offered the capability of combining biomolecular recognition, through staining with target protein-specific antibodies, with nanoscale optical resolution1. However, such techniques are limited by the accessibility of labels to the biomolecules to be imaged, because it is the labels that are visualized, and not the biomolecules themselves. Many crowded, biomolecule-rich structures are at the core of biological functions and disease states, with inter-protein distances smaller than the size of antibodies, and which thus may prohibit access by labels to biomolecules of interest. Overcoming this issue requires a technology which can not only achieve super-resolution down to the low tens of nanometres, but also enables decrowding of biomolecules, in cells and tissues.
We reasoned that in order to decrowd proteins from each other, we could leverage the spatial expansion property of expansion microscopy2,3 (in which cells and tissues are densely permeated by an even mesh of swellable hydrogel, and then expanded, to obtain enhanced resolution on ordinary microscopes). To establish decrowding and at the same time achieve resolution on par with the best classical super-resolution techniques (down to ~20 nm resolution), we combined the expansion principle with a scheme for biomolecular preservation, to build a technology that we here call expansion revealing (ExR). By direct comparison of biomolecular structures where labelling was done in the normal, crowded environment vs. after the decrowding effect of ExR, with both being imaged at the same level of resolution, we demonstrate that ExR indeed is revealing previously unknown biological information, and indeed, can lead to the discovery of nanostructures in brain tissue, which would otherwise have remained invisible. We anticipate ExR will enable the visualization of a variety of previously undescribed biological nanostructures. We illustrate the capabilities of ExR by unmasking key components of synaptic nanocolumns as well as periodic nanostructures potentially of relevance to Alzheimer’s disease, both in mouse brain tissue.
Results
Expansion revealing enables simultaneous super-resolution and decrowding
To achieve super-resolution down to the low tens of nanometres, two rounds of expansion (or 15-20x expansion factor) are required. One would ideally minimally alter the proteins during the expansion process, while decrowding them from one another as much as possible. To satisfy these conflicting requirements, ExR (Fig. 1a-c) achieves this with a new scheme. We reasoned that one could expand a brain specimen through one round of gelation and expansion, and that this first swellable hydrogel could then be further expanded if we formed a second swellable hydrogel in the space opened up by the first expansion, and then iteratively swelled the specimen a second time (see Methods for details). The proteins, being anchored to the first hydrogel throughout the entire process, would be retained because the original hydrogel would be further expanded by the second swellable hydrogel, and not cleaved or discarded. Thus, ExR imposes no extra processing steps on the proteins than is required for the first expansion. In the final step, the proteins can be antibody labelled (Fig. 1c), after decrowding and before imaging, enabling visualization of proteins that would have been missed if stained in the crowded state (Fig. 1b).
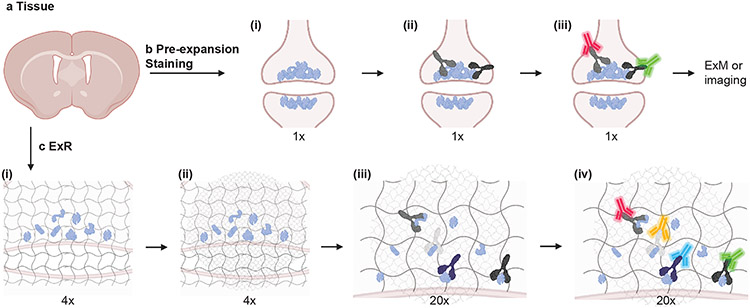
Fig. 1 ∣. Expansion Revealing (ExR), a technology for decrowding of proteins through isotropic protein separation.
(a) Coronal section of mouse brain before staining or expansion. (b) Conventional antibody staining may not detect crowded biomolecules, shown here in pre- and post-synaptic terminals of cortical neurons. (b i) Crowded biomolecules before antibody staining. (b ii) Primary antibody (Y-shaped proteins) staining in non-expanded tissue. Antibodies cannot access interior biomolecules, or masked epitopes of exterior biomolecules. (b iii) Secondary antibody (fluorescent green and red Y-shaped proteins) staining in non-expanded tissue. After staining, tissue can be imaged or expanded using earlier ExM protocols, but inaccessible biomolecules will not be detected. (c) Post-expansion antibody staining with ExR. (c i) Anchoring and first gelation step. Specimens are labelled with gel-anchoring reagents to retain endogenous proteins, with acrylamide included during fixation to serve as a polymer-incorporatable anchor, as in refs56,58. Subsequently, the specimen is embedded in a swellable hydrogel that permeates densely throughout the sample (gray wavy lines), mechanically homogenized via detergent treatment and heat treatment, and expanded in water. (c ii) Re-embedding and second swellable gel formation gelation. The fully expanded first gel (expanded 4x in linear extent) is re-embedded in a charge-neutral gel (not shown), followed by the formation of a second swellable hydrogel (light grey wavy lines). (c iii) 20x expansion and primary antibody staining. The specimen is expanded by another factor of 4x, for a total expansion factor of ~20x, via the addition of water, then incubated with conventional primary antibodies. Because expansion has decrowded the biomolecules, conventional antibodies can now access interior biomolecules and additional epitopes of exterior molecules. (c iv) Post-expansion staining with conventional fluorescent secondary antibodies (fluorescent blue and yellow Y-shaped proteins, in addition to the aforementioned red and green ones) to visualize decrowded biomolecules. Schematic created with BioRender.com.
We first quantified the global isotropy of the expansion process in ExR, and found a similar low distortion (i.e., of a few percent over length scales of tens of microns, Supplementary Fig. 1) as we did for previous ExM protocols3-7. To measure effective resolution, we focused on synapses, given both their importance for neural communication and utility as a super-resolution testbed, staining post-expansion cortical synapses (Fig. 2a) with antibodies against the pre-synaptic protein Bassoon and the post-synaptic proteins PSD95 and Homer1, amongst other synaptic proteins (Fig. 2b-d). We found that when we measured the mean distance between domains containing the proteins PSD95, Homer1, and Shank3, we obtained values (Fig. 2e) that matched classical results found using STORM microscopy8 (note that our study focused on Shank3 and this earlier study focused on Shank1), in the low tens of nanometres. Thus, ExR exhibits effective resolution on the order of ~20 nm, comparable to our earlier iterative expansion microscopy (iExM) protocol, which did not retain proteins4.
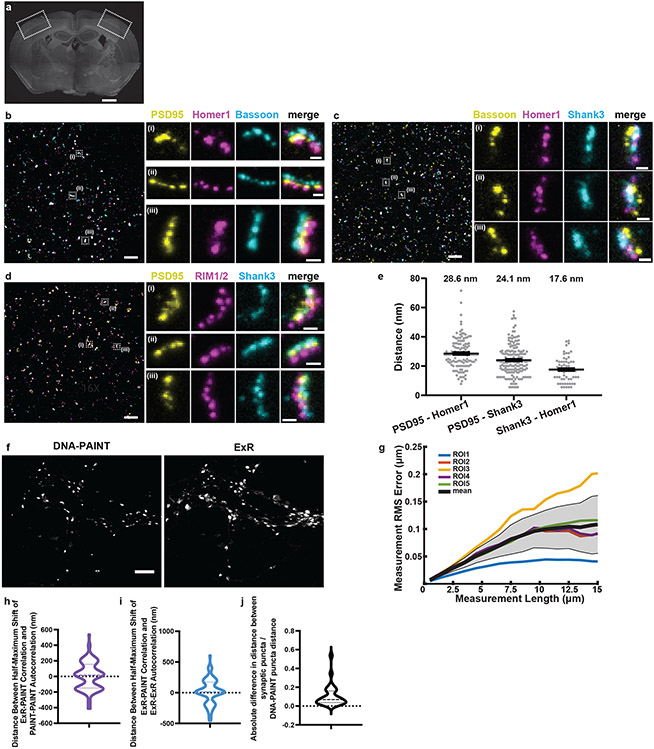
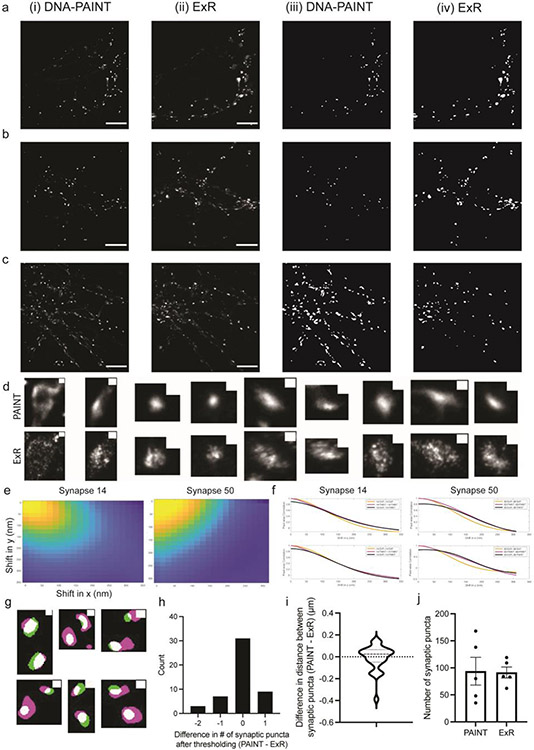
Fig. 2. Validation of ExR using synapses and comparison with DNA-PAINT.
(a) Low-magnification widefield image of a mouse brain slice stained with DAPI, with box highlighting the somatosensory cortex used in subsequent figures for synapse staining (Scale bar, 500 μm), and (b-d) confocal images (max intensity projections) of representative fields of view (cortical L2/3) and specific synapses after ExR expansion and subsequent immunostaining using antibodies against (b) PSD95, Homer1, Bassoon; (c) Bassoon, Homer1, Shank3; and (d) PSD95, RIM1/2, Shank3. Scale bar, 1 μm, left image; 100 nm, right images; in biological units, i.e. the physical size divided by the expansion factor, throughout the paper unless otherwise indicated. Shown are images from one representative experiment from two independent replicates. (e) Measured distance between centroids of protein densities of PSD95 and Homer1, PSD95 and Shank3, and Shank3 and Homer1, in synapses such as those in panels b-d. The mean distance (again, in biological units) between PSD95 and Homer1 is 28.6 nm (n = 126 synapses from 3 slices from 1 mouse), between PSD95 and Shank3 is 24.1 nm (n=172 synapses from 3 slices from 1 mouse), and between Shank3 and Homer1 is 17.6 nm (n=70 synapses from 3 slices from 1 mouse). Plotted is mean +/− standard error; individual grey dots represent the measured distance for individual synapses. (f) A pre-expansion DNA-PAINT image (left) and a registered confocal ExR image (max intensity projection, right) of the same field of cultured neurons after immunostaining with antibodies against Synapsin 1. Scale bar, 4 μm. Shown are images from one representative experiment from two independent replicates. (h) Estimated population distribution (violin plot of density, with a dashed line at the median and dotted lines at the quartiles) of the shift (in nm) at which the correlation is half-maximal for PAINT-PAINT autocorrelation and ExR-PAINT correlation (calculated pixel-wise between intensity values normalized to the minimum and maximum of the image, see Methods; see Supplementary Table 1 for statistics. n = 101 synaptic ROIs from 5 fields of view from 3 wells of cultured neurons from 1 culture batch). (i) Same as (h), for ExR-ExR autocorrelation vs. ExR-PAINT correlation. (j) Estimated population distribution (violin plot of density, with a dashed line at the median and dotted lines at the quartiles) of the normalized absolute difference in radial distance between neighbouring synaptic puncta centroids (Absolute value of PAINT-ExR, normalized to PAINT distance; see Supplementary Table 1 for statistics. n = 27 cropped synaptic pair ROIs from 5 fields of view of 30 μm x 30 μm each, 1 culture batch). (g) Root mean square error vs. measurement length (in biological units), calculated via a non-rigid registration algorithm of DNA-PAINT vs. ExR-processed cultured neurons (n=5 fields of view from 3 wells from 1 culture batch). Black line, mean; gray shading, standard deviation.
To validate the nanoscale precision of ExR further, we compared pre-expansion DNA points accumulation for imaging in nanoscale topography (DNA-PAINT, a classical super-resolution method) and ExR, using the same specimen and the same field of view for comparison across the two technologies. We examined synaptic nanostructures stained with antibodies against synapsin, using cultured neurons, which are amenable to classical high-precision super-resolution methods like DNA-PAINT (see Methods). Both ExR and DNA-PAINT images show qualitatively similar staining patterns and resolution (Fig. 2f, Extended Data Fig. 1a-c). To assess the nanoscale precision of ExR quantitatively, we calculated, as a linearized distortion measure between ExR and PAINT, the shifts (in nm) between the half-maxima of the auto-correlation (ExR-ExR or PAINT-PAINT) and cross-correlation (ExR-PAINT) functions for single synaptic puncta imaged using both modalities (Extended Data Fig. 1d-f). Our analysis of 5 regions of interest from 3 wells of cultured neurons (1 culture batch) showed mean linearized distortion of 4.91 nm for the ExR-PAINT cross-correlation vs. PAINT-PAINT autocorrelation (Fig. 2h, 95% CI of the mean [−29.93, 39.75]) and 18.78 nm for the ExR-PAINT cross-correlation vs. ExR-ExR autocorrelation (Fig. 2i, 95% CI of the mean [−20.10, 57.66]). This analysis is a conservative one: ExR could in principle, through the decrowding effect (Fig. 1a-c), additionally alter the synapsin staining vs. that of PAINT, but note that here, even if we conservatively lump all such decrowding-related changes into the category of distortion (as the above analysis does), we get an upper bound on the distortion in the low double-digit nanometres.
To compare the ability to distinguish between neighbouring nanostructures using DNA-PAINT and ExR, we examined pairs of neighbouring synaptic puncta (Extended Data Fig. 1g) and found the absolute value of the difference in synaptic puncta distances as measured using ExR vs. DNA-PAINT, normalized to the distance measured using DNA-PAINT, was ~11% (Fig. 2j, 95% CI of normalized absolute difference, [0.06228, 0.1548]; Extended Data Fig. 1i, 95% CI of difference between synaptic puncta centroid distance, PAINT - ExR: [−0.05419,0.03649]; see Supplementary Table 1 for detailed statistics; n = 27 pairs from 5 ROIs from 3 wells of cultured neurons from one culture batch). The coefficient of variation for distance between synaptic puncta imaged using ExR, calculated as the standard deviation of the difference in distance between pairs of synaptic puncta (PAINT - ExR) divided by the mean distance of the same pairs from DNA-PAINT, was 0.1367 (n = 27 cropped synaptic puncta pairs, 5 ROIs from 3 wells of neurons from one culture batch). Furthermore, we found no significant difference in the number of unique puncta detected after thresholding (Extended Data Fig. 1h, 95% CI of difference between number of synaptic puncta, PAINT-ExR: [−0.2936, 0.1336], n = 50 pairs, 5 ROIs from 3 wells of neurons from one culture batch). Similarly, we found no significant difference in the total number of synaptic puncta detected after thresholding across the entire imaging field of view (Extended Data Fig. 1j, paired t-test between ExR and DNA-PAINT images, p = 0.9271, t = 0.09735, df = 4, n = 5 fields of view from 3 wells of neurons from one culture batch). Finally, analysis of the root mean square error (see Methods) between ExR and DNA-PAINT revealed few-percent error over a typical region of interest size, comparable to prior expansion microscopy methods (i.e., < 50-nm error for measurement scales of ~4 μm and < 150 nm error for measurement scales of ~15 μm; Fig. 2g). Thus, ExR exhibits a high degree of precision, and low distortion, compared to a classical high-resolution super-resolution method, DNA-PAINT.
High-fidelity enhancement of synaptic protein visualization via ExR
To gauge whether ExR could reveal nanostructures in the brain that were not visible without decrowding, and to further probe whether it incurred any costs in terms of decreased resolution or added distortion relative to classical staining (i.e. pre-expansion staining and thus, no decrowding), we devised a strategy where we would stain brain slices pre-expansion with an antibody against a synaptic protein, in a fashion so that antibodies would be anchored to the expansion hydrogel for later visualization, then perform ExR, and then re-stain the same proteins with the same antibody a second time, thus enabling a within-sample comparison of a given protein across both conditions (with and without decrowding) at the same level of resolution, which could reveal any nanostructural differences. For pre-expansion staining (see Methods for details), we immunostained mouse brain slices containing somatosensory cortex (Fig. 3a) with primary antibodies followed by 6-((acryloyl)amino)hexanoic acid, succinimidyl ester (abbreviated AcX)-conjugated secondary antibodies, so that these antibodies could be attached to the swellable hydrogel for post-expansion tertiary antibody staining and visualization. This allowed us to compare pre-ExR staining to post-ExR staining, at the same resolution level, and for the same field of view, important for noticing any changes in nanostructural detail. We noted that Homer1 and Shank3 exhibited very similar visual appearances when we compared pre- vs. post-ExR staining (quantified below), so we designated these two stains as “reference channels,” that is, co-stains that could help define synapses for the purposes of technological comparison, and that we could use to help us gauge whether other proteins were becoming more visible at synapses.
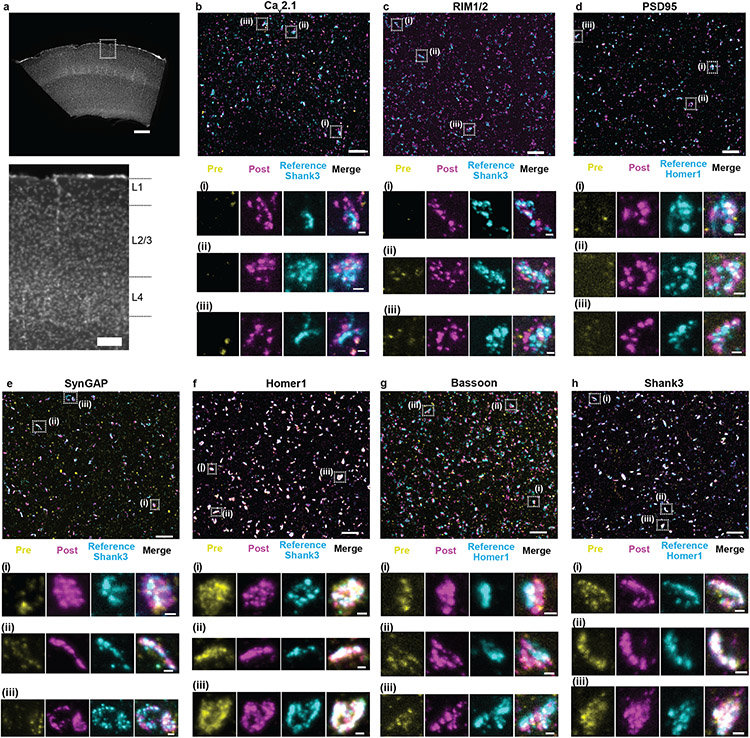
Fig. 3 ∣. Validation of ExR enhancement and effective resolution in synapses of mouse cortex.
(a) Low-magnification widefield image of a mouse brain slice with DAPI staining showing somatosensory cortex (top) and zoomed-in image (bottom) of boxed region containing L1, L2/3, and L4, which are imaged and analyzed after expansion further in panels b-h. (Scale bar, 300 μm (top) and 100 μm (bottom)). (b-h) Confocal images of (max intensity projections) of specimen after immunostaining with antibodies against Cav2.1 (Ca2+ channel P/Q-type) (b), RIM1/2 (c), PSD95 (d), SynGAP (e), Homer1 (f), Bassoon (g), and Shank3 (h), in somatosensory cortex L2/3. For pre-expansion staining, primary and secondary antibodies were stained before expansion, the stained secondary antibodies anchored to the gel, and finally fluorescent tertiary antibodies applied after expansion to enable visualization of pre-expansion staining. For post-expansion staining, the same primary and secondary antibodies were applied after ExR. Antibodies against Shank3 (b, c, e, f) or Homer1 (d, g, h) were applied post-expansion as a reference channel. Confocal images of cortex L2/3 (top) show merged images of pre- and post-expansion staining, and the reference channel. Zoomed-in images of three regions boxed in the top image (i-iii, bottom) show separate channels of pre-expansion staining (yellow), post-expansion staining (magenta), reference staining (cyan), and merged channel. (Scale bar, 1.5 μm (upper panel); 150 nm (bottom panel of i-iii).) Shown are images from one representative experiment from two independent replicates.
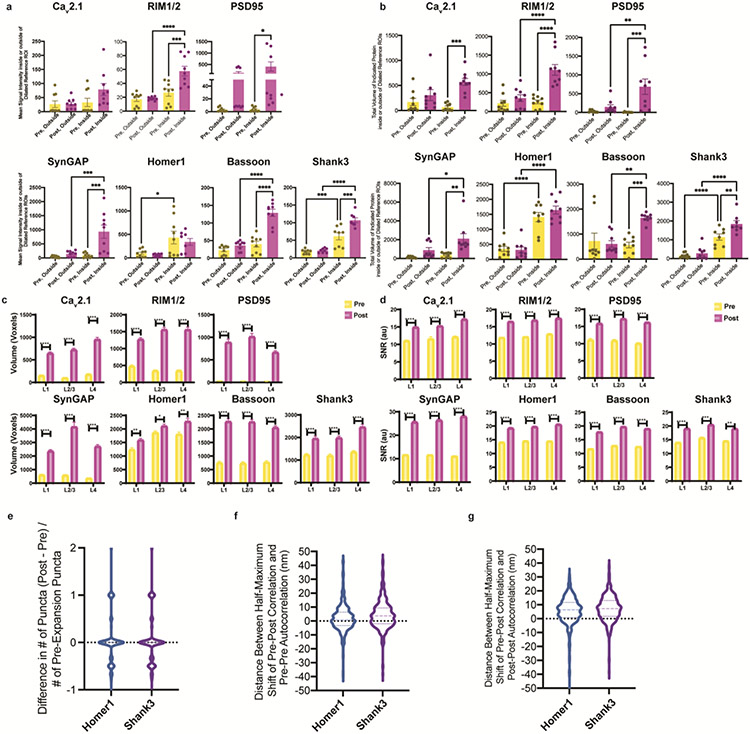
We chose 7 synaptic proteins important for neural architecture and transmission for this experiment – the presynaptic proteins Bassoon, RIM1/2, and the P/Q-type Calcium channel Cav2.1 alpha 1A subunit, and the postsynaptic proteins Homer1, Shank3, SynGAP, and PSD95 (Fig. 3b-h), staining for each protein along with a reference channel stain (note, when we imaged Homer1, we used Shank3 as the reference channel, and vice versa). These proteins were chosen based on their frequent use as synaptic markers, their known interaction with one another, and/or their functional importance at the synapse9-15. Specifically, Shank3 is a key post-synaptic scaffolding protein that interacts with PSD95 and Homer1, amongst other proteins, in the post-synaptic density9, and Shank3 mutant mice display autistic-like behaviours and striatal changes16; RIM1/2 promotes calcium channel localization at the presynaptic active zone17, controls synaptic vesicle release18, and enhances the size of the readily-releasable pool17; Bassoon and calcium channels contribute to vesicle release and promote vesicle reloading19; PSD95, another key post-synaptic scaffolding protein, drives maturation of excitatory synapses20, and PSD95 mutant mice display enhanced long-term potentiation (LTP) and impaired learning21; SynGAP regulates spine formation14; Homer and Shank form a higher-order complex that forms a binding platform for postsynaptic proteins22.
All seven proteins exhibited well-defined images when post-expansion stained, with the geometry reflecting characteristic synaptic shapes. For example, a presynaptic and a postsynaptic stain (Fig. 3b,c,g) revealed parallel regions with a putative synaptic cleft in between, although note that this was only visible if the synapse was being imaged from the side; if a synapse was being imaged with the axial direction of the microscope perpendicular to the synaptic cleft, then the image may look more disc-shaped4,8,23. In many cases, however, post-ExR staining revealed more detailed structures of synapses compared to what was visualized with pre-expansion staining – for example, calcium channels (Fig. 3b), RIM1/2 (Fig. 3c), PSD95 (Fig. 3d), SynGAP (Fig. 3e), and Bassoon (Fig. 3g) appeared more prominent post-expansion than pre-expansion. Applying a conventional antigen retrieval protocol did not result in such improvements (Supplementary Fig. 2), suggesting that the decrowding effect observed via ExR was indeed due to expansion, and not simply due to denaturation or antigen retrieval-like effects associated with other aspects of the ExR process.
To quantitate the improvement in staining enabled by ExR, we measured the amplitude and volume of each synaptic protein stain, both within, and just outside of, identified synapses. First, we manually identified between 49-70 synapses (see Supplementary Table 2 for exact numbers) per ~350x350x20 μm3 (in physical units, e.g. what is actually seen through the microscope lens) field of view, choosing the largest and brightest synapses based on reference channel staining (i.e., Homer1 or Shank3). We developed an automated method to segment synaptic puncta from nearby background. Briefly, we created binary image stacks for each channel using a threshold equal to a multiple of the average measured standard deviation of five manually-identified background regions (not blinded to condition), filtered 3D connected components based on size, and used dilated reference channel ROIs to segment putative synaptic puncta (Supplementary Fig. 3 and Methods). We dilated reference channel-defined ROIs to relax the requirement of exact colocalization of pre-synaptic proteins with a post-synaptic reference. We found that post-expansion staining increased signal intensity and mean total volume of signal within dilated reference-channel defined ROIs, without meaningfully affecting background (i.e., signal just outside the dilated reference channel-defined ROIs) staining (Extended Data Fig. 2a,b). In summary, all proteins except Homer1 and Cav2.1 showed significantly increased signal intensity of post-expansion staining within dilated reference ROIs, with minimal increase in the background (Extended Data Fig. 2a; Supplementary Table 3 for full statistical analysis). (We note that there was no change in signal intensity in the background for PSD95, but a bimodal distribution of signal intensity increases in the foreground and background, due to abnormally high signal in one animal.) Similarly, all proteins except Homer1 exhibited increased total volume of post-expansion staining signals within dilated references ROIs, and minimal or no increase in the volume of background ROIs (Extended Data Fig. 2b; Supplementary Table 4 for full statistical analysis).
Because Homer1 and Shank3 had the smallest changes in pre- vs. post-expansion staining, they were chosen as reference channels to indicate the locations of putative synapses for pre- vs. post-expansion test stain comparisons, as mentioned above. Of course, different antibodies may bind to different sites on a target protein, and we found that a different antibody against PSD95 (from Cell Signaling Technology, product number CST3450S) than the one used in Figure 3d (Thermofisher, product number MA1-046) showed similar signal intensity and volume when compared pre- vs. post-staining (Supplementary Fig. 4); perhaps, in future studies, the use of multiple antibodies against different parts of a single protein, in pre- vs. post-expansion comparisons, could be used to gauge the density of the environment around different parts of that protein.
We further analysed synaptic protein signals pre- vs. post-expansion in the context of different cortical layers. We quantified volume and signal-to-noise ratio (SNR; signal intensity divided by standard deviation of the background) of each protein in 3D synaptic structures by binarizing signals over a threshold (a multiple of the standard deviation of the intensity within a manually-selected background region; see Methods) and selecting putative synapses in each ~350x350x20 μm3 field of view imaged above, comparing the values of pre- vs. post-expansion staining in each layer of somatosensory cortex (L1, L2/3 and L4, respectively) (Extended Data Fig. 2c,d). Post-expansion images exhibited larger volumes and improved SNR in each layer for all synaptic proteins (see Supplementary Tables 6 and 7 for full statistics).
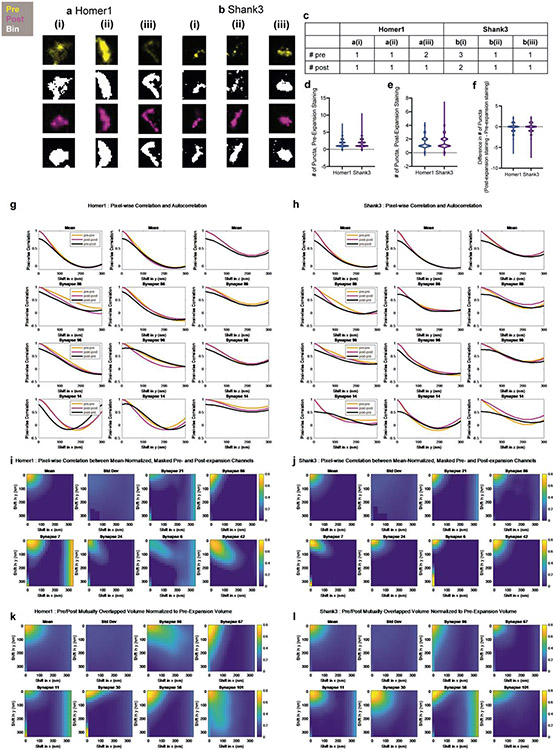
In previous work, we showed using the original iterative expansion microscopy (iExM) protocol, which uses pre-expansion staining, that we could achieve effective resolutions of ~20 nm, with low distortion due to the expansion process4. As another independent way to gauge the potential distortion obtained by staining with antibodies post-expansion, we compared the shapes of synaptic puncta as seen with the pre-expansion stain, with the shapes as seen with the new post-expansion stain, using the within-sample dual staining method used in Figure 3b-h. We compared various properties of synaptic puncta between pre- and post-expansion staining conditions, using the reference proteins Homer1 and Shank3 since they had similar intensities and volumes when comparing pre- and post-expansion datasets, and therefore might be appropriate for comparing shape features across these conditions (Extended Data Fig. 3). In summary, we did not see any substantial distortion being introduced by post-expansion staining; for example, we found no significant difference in the number of synaptic puncta when we compared pre- vs. post-expansion staining in the same sample (Extended Data Fig. 2e), and when we measured the shift in puncta positions between pre- and post-stained conditions, we observed average shifts of <10nm (in biological units, i.e. physical size divided by the expansion factor) between post- and pre-expansion staining for both Homer1 and Shank3 (Extended Data Fig. 2f,g; see Supplementary Table 5 for full statistics). Thus, ExR preserves, relative to classical pre-expansion staining, the locations of proteins with high fidelity for the purposes of post-expansion staining.
Synaptic nanocolumns coordinated with calcium channel distributions
Coordinating pre- and post-synaptic protein arrangement in a nanocolumn structure which aligns molecules within the two neurons contributes to precision signalling from presynaptic release sites to postsynaptic receptor locations23,24, as well as to the long-term plasticity of synaptic function25. Given that ExR is capable of unmasking synaptic proteins that are otherwise not detectable, with nanoscale precision, we next sought to explore the nanocolumnar architecture of pre- and postsynaptic proteins, with a focus on important molecules that have not yet been explored in this transsynaptic alignment context. As noted above, ExR greatly helps with visualization of calcium channels, which of course are amongst the most important molecules governing the activation of synaptic release machinery, with nanometre-scale signalling contributing to the precision of synaptic vesicle fusion. However, the nanoscale mapping of calcium channels in the context of nanocolumnar alignment in brain tissue remains difficult26-28 (Fig. 3b). We thus applied ExR to investigate whether calcium channels occupy nanocolumns with other pre- and post-synaptic proteins, such as the critical pre- and post-synaptic proteins RIM1/2 and PSD95, respectively, across the layers of the cortex (Fig. 4a-d).
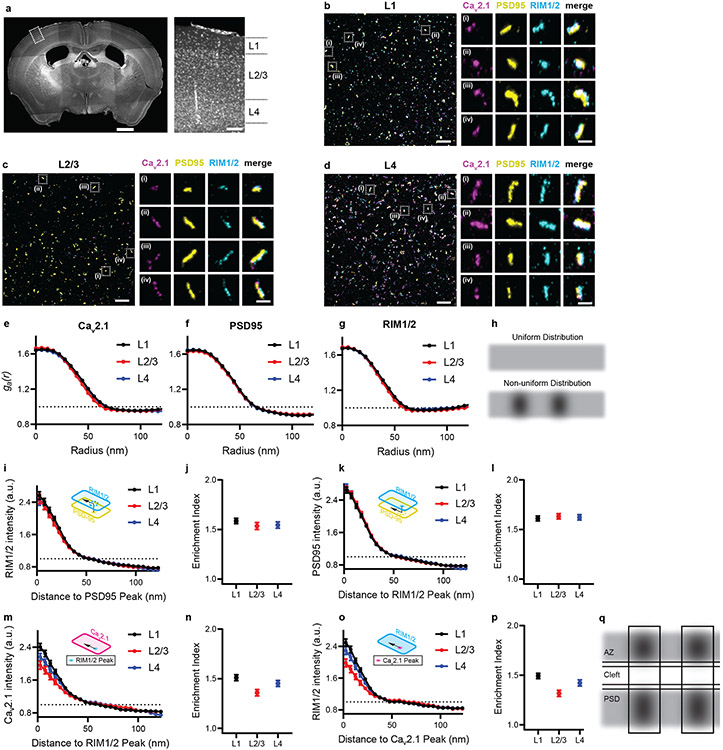
Fig. 4 ∣. ExR reveals how calcium channel distributions participate in transsynaptic nanoarchitecture.
(a) Low-magnification widefield image of DAPI stained mouse brain slice (left) and zoomed in view (right) of the boxed region showing layers 1-4 of the cortex. (Scale bar = 1000 μm (left panel of whole brain) and 100 μm (right panel of the layers 1-4). (b-d) show confocal images (max intensity projections) of layers 1, 2/3 and 4 respectively, after performing ExR and immunostaining with antibodies against Cav2.1 (calcium channel) (magenta), PSD95 (yellow) and RIM1/2 (cyan). In each of (b), (c) and (d), low magnification images are shown on the left while zoomed-in images of four regions, i-iv are shown on the right with separated channels for each antibody along with the merged image. (Scale bar = 1 μm (left panel) and 100 nm (right panels labelled i-iv)). Shown are images from one representative experiment from two independent replicates. (e-g) show the autocorrelation analysis for Cav2.1, PSD95 and RIM1/2 respectively for different layers. (h) shows schematic illustration of protein distribution based on interpretation of autocorrelation results in (e-g). A uniform distribution (h, top) would be predicted if ga(r) = 1 at all radii (dotted lines in e-g), whereas a non-uniform distribution with one or more regions of high local intensity (h, bottom) would be predicted if ga(r) was greater than 1 at short radii and decayed as the radius is increased. (i), (k), (m) and (o) show the enrichment analysis that calculates the average molecular density for RIM1/2 to the PSD95 peak, PSD95 to the RIM1/2 peak, Cav2.1 to the RIM1/2 peak and RIM1/2 to the Cav2.1 peak respectively while (j), (l), (n) and (p) show the corresponding mean enrichment indexes respectively (see Methods for exact n - values). Error bars indicate SEM. (q) shows schematic illustration of protein distribution based on interpretation of enrichment analysis results in (i-p). Enrichment values above 1 represent regions of high local intensity in the measured channel, so the enrichment profiles in (i), (k), (m) and (o) suggest the peak of the reference channel closely aligns with regions of high intensity in the measured channel for each of the four comparisons. Therefore, this suggests that enriched regions of any two proteins (RIM1/2, PSD95 and Cav2.1) are aligned in nanoscale precision with each other. Calcium channels located close to vesicle fusion sites (dictated by RIM1/2) may enhance the calcium-sensitivity of fusion. Additionally, postsynaptic receptors may be exposed to higher, faster peaks of neurotransmitter concentration when vesicle release sites are located directly opposite receptor nanoclusters (PSD95 being a receptor-anchoring protein). (AZ: active zone, PSD: postsynaptic density).
We first performed a 3D autocorrelation function (ga(r))-based test which provides information about the intensity distribution within a defined structure (see Methods). Any heterogeneity in the intensity distribution within the cluster will result in a ga(r)>1, and the distance at which the ga(r) crosses 1 can be used to estimate the size of the internal heterogeneity, here termed a nanodomain23. In all cortical layers, our auto-correlation analysis shows that all 3 proteins explored exhibited a non-uniform arrangement forming nanodomains with average diameters of about 60-70nm (in biological units; Fig. 4e-h). To analyse the spatial relationship between the two distributions and the average molecular density of RIM1/2, PSD95 and Cav2.1 relative to each other, we performed a protein enrichment analysis, which is a measure of volume-averaged intensity of one channel as a function of distance from the peak intensity of another channel (see Methods). To more easily compare the extent of the enrichment between these proteins in each layer, we also calculated the enrichment index, which is an average of all enrichment values within 60 nm (biological units) of the peak of a designated channel. Our analysis shows that the centres of nanodomains of RIM1/2, PSD95 and Cav2.1 are enriched with respect to each other (Fig. 4i-p).
Of particular interest, the nanoscale colocalization of Cav2.1 with RIM1/2 (and thus the vesicle site) likely minimizes the distance between the channels and the molecular Ca sensors that trigger vesicle fusion (Fig. 4q), consistent with the physiological concept of nanodomain coupling that tunes the efficacy and frequency-dependence of neurotransmission29. Furthermore, the precise alignment between RIM1/2 and PSD95 may reduce the distance that released neurotransmitter needs to diffuse before reaching postsynaptic receptors (Fig. 4q). Thus, these nanoscale arrangements may help to optimize the speed, strength, and plasticity of synaptic transmission. To the best of our knowledge, the 3D nanoarchitecture of the distribution of voltage-gated calcium channels within the transsynaptic framework in brain tissue had not been previously probed.
Periodic amyloid nanostructures in Alzheimer’s model mouse brain
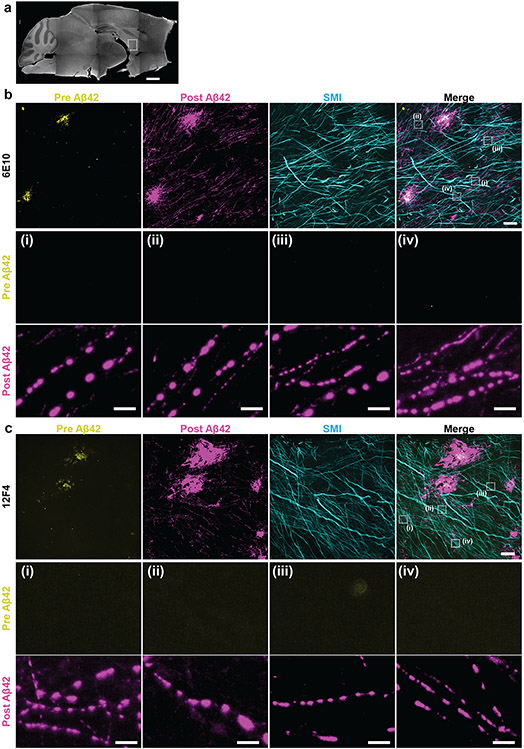
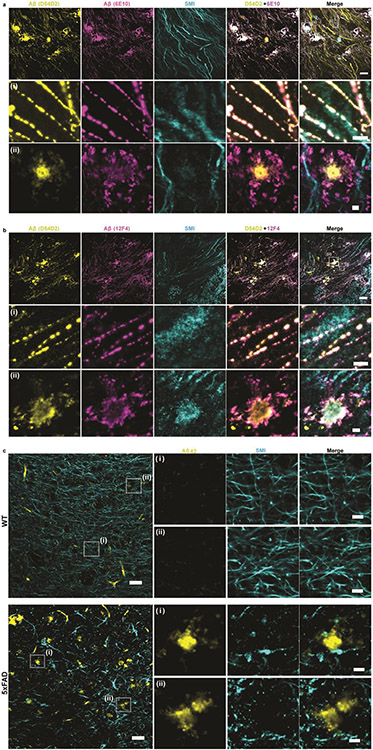
In addition to densely crowded proteins in healthy functioning compartments like synapses, densely crowded proteins appear in pathological states like Alzheimer’s disease. Protein aggregates known as β-amyloid are thought to play roles in synaptic dysfunction, neurodegeneration and neuroinflammation30. However, the densely packed nature of these aggregates may make the nanoscale analysis of their ultrastructure within brain tissue difficult to understand. To understand the nanoarchitecture of β-amyloid in the cellular context of brain tissue, we applied ExR to the brains of 5xFAD Alzheimer’s model mice (Fig. 5a), as this widely used animal model of Alzheimer’s exhibits an aggressive amyloid pathology31. We employed two different commercially available antibodies for β-amyloid, 6E10 (which binds to amino acid residues 1-16 of human Aβ peptides) and 12F4 (which is reactive to the C-terminus of human Aβ and has specificity towards Aβ42). We were particularly interested in investigating the relationship of amyloid deposits and white matter tracts, as these regions have been implicated in human imaging data32-35, but are less investigated in mouse models. We previously reported the accumulation of Aβ aggregates along the fornix, the major white matter tract connecting the subiculum and mammillary body, early in disease progression35.
Fig. 5 ∣. ExR reveals periodic nanoclusters of Aβ42 peptide in the fornix of Alzheimer’s model 5xFAD mice.
(a) Epifluorescence image showing a sagittal section of a 5xFAD mouse brain with the fornix highlighted. (Scale bar, 1000 μm) (b-c) ExR confocal Images (max intensity projections) showing immunolabelling against Aβ42 peptide with two different monoclonal antibodies 6E10 (b) and 12F4 (c). From left to right: pre-expansion immunolabelling of Aβ42 (yellow), post-expansion labelling (magenta) of Aβ42, post-expansion SMI (neurofilament protein), and merged pre- and post-expansion staining of Aβ42 with post-expansion staining of SMI. Insets (i-iv) show regions of interest highlighted in the merged images in the fourth column; top panels, pre-expansion Aβ42 labelling; middle panels, post-expansion Aβ42 labelling; bottom panels, post-expansion SMI labelling. Post-ExR staining reveals periodic nanostructures of β-amyloid, whereas pre-expansion staining can detect only large plaque centers. (Scale bar = 10 μm (upper panel images); 1 μm (bottom panels, i-iv))
We co-stained the amyloid antibodies along with the axonal marker SMI-312, and compared pre-expansion staining with that obtained after ExR. Plaques appeared to be larger, and to have finer-scaled features in post-expansion staining, than in pre-expansion staining (Fig. 5b,c), when visualized by either 6E10 or 12F4 antibodies, and thus the post-expansion staining may unveil aspects of plaque geometry that are not easily visualized through traditional means. Additionally, post-ExR staining revealed detailed nanoclusters of β-amyloid that were not seen when staining was done pre-expansion (Fig. 5b,c). These nanodomains of β-amyloid appeared to occur in periodic structures (Fig. 5b,c, subpanels (i)-(iv)). Co-staining with 2 different β-amyloid antibodies, D54D2 (which binds to isoforms Aβ37, Aβ38, Aβ40, and Aβ42) along with 6E10 or 12F4 in ExR-processed 5xFAD fornix showed similar patterns of periodic nanostructures, which means the observed periodic nanostructures are likely not composed of specific isoforms (Extended Data Fig. 4a,b). These nanodomains, and periodic structures thereof, were not visualized through pre-expansion staining, which was confirmed using unexpanded tissue as well (Extended Data Fig. 4c).
To see whether the periodic nanostructures of β-amyloid revealed through ExR were just nonspecific staining artifacts of ExR, we performed ExR on wild type (WT) mice as a control, which should not have any labelling for human β-amyloid. No β-amyloid structures were observed in ExR-processed WT brain (Supplementary Fig. 5a). Our quantitative analysis examining the volume of amyloid in ExR-processed WT and 5xFAD mice confirms that there is indeed a large amount of amyloid volume occupied in 5xFAD samples, but essentially no such volume occupied in ExR-processed WT mice (Supplementary Fig. 5b). The lack of amyloid staining in ExR-processed WT mice makes it unlikely that the staining seen in ExR-processed 5xFAD mouse brain is nonspecific. Applying a conventional antigen retrieval protocol recovered some amyloid staining, but less than by ExR (Supplementary Fig. 6), suggesting that the decrowding effect observed via ExR was indeed due to expansion, and not simply due to denaturation or antigen retrieval-like effects associated with other aspects of the ExR process. The intact staining of axonal and myelin markers (Fig. 6, Extended Data Fig. 5) adjacent to the amyloid puncta highlight how, within the same volume of tissue, some structures can exhibit periodicity while others remain smooth and continuous, highlighting the small length scales over which the architecture of nanostructures is regulated in cells and tissues.
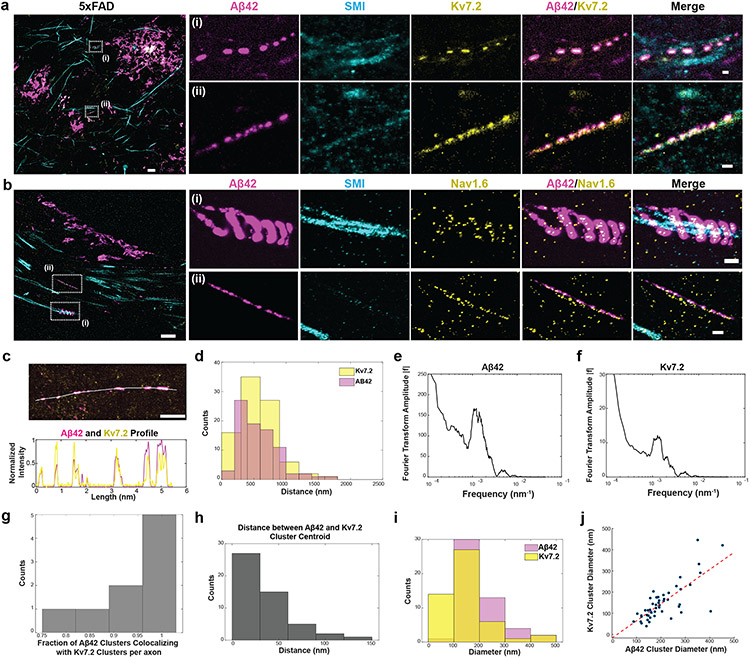
Fig. 6 ∣. ExR reveals co-localized clusters of Aβ42 peptide with potassium and sodium ion channels in the fornix of Alzheimer’s model 5xFAD mice.
(a-b) ExR confocal image (max intensity projections) showing post-expansion (a) Aβ42 (magenta), SMI (cyan) and Kv7.2 (yellow) staining, and (b) Aβ42 (magenta), SMI (cyan) and Nav1.6 (yellow) staining in the fornix of a 5xFAD mouse. Leftmost panel, merged low magnification image. (Scale bar = 4 μm); insets (i-ii) show close-up views of the boxed regions in the leftmost image for (A) Aβ42, SMI, Kv7.2, Aβ42-Kv7.2 merged and Aβ42-Kv7.2-SMI merged respectively, and (b) Aβ42, SMI, Nav1.6, Aβ42-Nav1.6 merged and Aβ42-Nav1.6-SMI merged respectively (scale bar = 400 nm; n = 5 fields of view from 2 slices from 2 mice). (c) ExR confocal image (max intensity projections) showing Aβ42 (magenta) and Kv7.2 (yellow) clusters in a 5xFAD mouse (top) with the indicated cross-section profile shown (bottom). (Scale bar = 1 μm). Shown are images from one representative experiment from four independent replicates. (d) Histograms showing distances between adjacent Aβ42 (magenta) and Kv7.2 (yellow) clusters in 5xFAD mice along imaged segments of axons (n=97 Aβ42 clusters, 92 Kv7.2 clusters from 9 axonal segments from 2 mice). (e-f) Fourier transformed plots of Aβ42 (e) and Kv7.2 (f) showing the same peak position (from the same data set as (d)). (g) Histogram showing the fraction of Aβ42 clusters colocalizing with Kv7.2 clusters along individual segments of axons (n=50 cluster pairs from 9 axonal segments from 2 mice). (h) Histogram showing the distance between the centroids of colocalized Aβ42 and Kv7.2 clusters (same data set as (d)). (i) Histograms showing the diameters of Aβ42 clusters (magenta) and Kv7.2 clusters (yellow) (n=50 cluster pairs from 9 axonal segments from 2 mice). (j) Scatter plot showing the diameters of colocalized Aβ42 and Kv7.2 clusters.
Co-clustering of amyloid nanodomains and ion channels
To understand the biological context of these periodic Aβ nanostructures, we stained brain slices with antibodies against the ion channels Nav1.6 and Kv7.2. Alzheimer’s disease is associated with altered neuronal excitability and alterations in ion channels36. We stained ExR-processed 5xFAD fornix-containing brain slices with antibodies against potassium channels (Kv7.2) and against β-amyloid (12F4) (Fig. 6a, Supplementary Fig. 7a). The periodic β-amyloid nanostructures colocalized with periodic nanoclusters of potassium channels. In ExR-processed WT fornix, such β-amyloid clusters, and frequent clusters of potassium or sodium channel staining were not found (Supplementary Fig. 7b,c). Co-staining of β-amyloid (12F4) and sodium channels (Nav1.6) showed colocalization as well (Fig. 6b, Supplementary Fig. 7d). Specifically, over half of β-amyloid nanoclusters co-localized with Nav1.6 (Supplementary Fig. 7d(i)), and within these colocalized regions, Nav1.6 puncta were much smaller than β-amyloid nanoclusters (Supplementary Fig. 7d(ii)). We also found other interesting amyloid patterns, such as helical structures along axons (Fig. 6b).
To probe the periodic amyloid/Kv7.2 patterns further, we obtained a cross-sectional profile of a stretch of axon, showing that Aβ42 (magenta) and Kv7.2 (yellow) were highly overlapping and with similar periodicities (Fig. 6c). A further histogram analysis showed the periodicity of both repeated protein structures to be 500-1000 nm (Fig. 6d), which we confirmed by Fourier analysis (Fig. 6e,f). This periodicity is notably higher in spatial frequency, by orders of magnitude, than the inter-node-of-Ranvier distances of brain white matter, which range from 20-100μm37-39.
Our analysis along individual segments of axons showed that a high fraction of Aβ42 clusters contained Kv7.2 clusters (Fig. 6g), a result that we confirmed by measuring the distance between the centroids of overlapping Aβ42 and Kv7.2 nanoclusters, finding extremely tight colocalization, within a few tens of nanometres (Fig. 6h). Median sizes of nanoclusters were around 150nm for both Aβ42 and Kv7.2 (Fig. 6i); indeed, overlapping nanodomains of Aβ42 and Kv7.2 were highly correlated in size (Fig. 6j), suggesting a potential linkage between how they are formed and organized as multiprotein complexes.
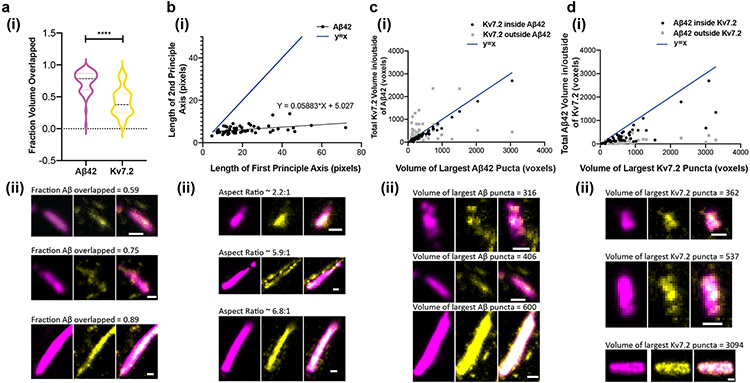
To facilitate visualization of the 3D shapes of these Aβ42 and Kv7.2 puncta, we show three orthogonal slices in the x-y, x-z, and y-z planes (x- and y-directions being transverse directions, and the z-direction being axial) that intersect the centre of each puncta. Qualitatively, we observed that the majority of these puncta are oblong, with smooth, continuous Aβ42 ellipsoids and more punctate Kv7.2 puncta, with some, but not all, Kv7.2 puncta found within Aβ42 puncta. A larger volume of Aβ42 (as a fraction of total Aβ42 puncta volume) was found inside Kv7.2 puncta compared to the fraction of Kv7.2 volume inside Aβ42 puncta (Fig. 7, Supplementary Fig. 8a). On average, the mean volume of an Aβ42 puncta was slightly larger than that of a Kv7.2 puncta (Supplementary Fig. 8b, paired t-test, p = 0.0001, t=4.116, df=54). Quantification of shape characteristics confirmed these observations and revealed more subtle patterns. Representative images illustrating the observed trends are shown below each plot (Fig. 7a(ii)-d(ii)). Despite the larger volume of Aβ42 puncta, the fraction of Aβ42 volume mutually overlapped with (inside of) Kv7.2 puncta was larger than the fraction of Kv7.2 mutually overlapped with (inside of) Aβ42 puncta (Fig. 7a; p < 0.0001, t=10.94, df=54). When considering the ellipsoidal shapes of these puncta, the relationship between the second and first principal axis lengths was highly sublinear (Fig. 7b), and the average aspect ratio (ratio of first to second principal axis length) was ~3.5:1 (mean 3.464, standard deviation 1.911, n = 55 puncta), indicating a highly oblong shape (slope of best-fit line from simple linear regression = 0.05883, p = 0.0020, F = 10.59, df = 53). While the number of Kv7.2 present in each manually cropped ROI was not correlated with the volume of the largest Aβ42 puncta within the cropped ROI (Supplementary Fig. 8c; simple linear regression, 95% CI for slope [−0.003940, 0.005947]), the mean Kv7.2 puncta volume was significantly correlated with the mean Aβ42 puncta volume (Supplementary Fig. 8d; simple linear regression, 95% CI of slope [0.09878-0.2437], R2 = 0.2977, p < 0.0001, F = 22.47, df = 53). Thus, as the size of Aβ42 aggregates increases, Kv7.2 aggregates increase in size, but not number. We found that the volume of Kv7.2 puncta inside Aβ42 puncta is highly correlated with the volume of the Aβ42 puncta (Fig. 7c; simple linear regression, 95% CI of slope [0.8353-9037], R2 = 0.98, p < 0.0001, F = 2,593, df = 53), but the volume of Kv7.2 puncta outside of Aβ42 puncta is not correlated to Aβ42 puncta volume (Fig. 7c; simple linear regression, 95% CI of slope [−0.03919, 0.4503], R2 = 0.05082, p = 0.0929, F = 2.838, df = 53). Conversely, the total volume of Aβ42 puncta inside of Kv7.2 puncta was not as strongly correlated (Fig. 7d; simple linear regression, 95% CI of slope [0.4198, 0.6307], R2 = 0.6531, p < 0.0001, F = 99.29, df = 53) with the volume of the largest Kv7.2 puncta in the cropped ROI. The volume of Aβ42 puncta outside of Kv7.2 was weakly but significantly correlated to the size of the largest Kv7.2 puncta (Fig. 7d; 95% CI of slope [0.0250, 0.09245] R2 = 0.1876, p = 0.0010, F = 12.24, df = 53). Finally, we found that the non-overlapped volume as a function of overlapped volume was much larger, on average, for Kv7.2 than for Aβ42 (Supplementary Fig. 8e; paired t-test, p < 0.0001, t=5.985, df=54). Taken together, these results show that Kv7.2 and Aβ42 puncta are correlated in size only when physically colocalized in a tightly registered fashion, perhaps pointing to new potential hypothesized mechanisms of aggregation.
Fig. 7 ∣. Analysis of Aβ42 peptide and potassium ion channels nanoclusters in shapes and their relationships.
(a)(i) Fraction of total Aβ42 and Kv7.2 puncta volume overlapped with one another in cropped Aβ42 clusters (n = 55 clusters from 2 5xFAD mice; p < 0.0001, two-sided pairwise t-test). ****P < 0.0001. (a)(ii) Representative images illustrating the difference in the proportion of mutually overlapped volume between Aβ42 and Kv7.2 as a fraction of total Aβ42 or Kv7.2 volume (scale bar = 100nm). (b)(i) Length of the second principal axis of the ellipsoid that has the same normalized second central moments as the largest Aβ42 punctum in an ROI, vs. the length of the first principal axis of this ellipsoid (in pixels, 1 pixel = 11.44 nm in x- and y-directions (transverse) and 26.67nm in the z-direction (axial); black points represent individual manually cropped ROIs; slope of best-fit line from simple linear regression = 0.05883, p = 0.0020, F = 10.59, df = 53). Compare to the line y = x (blue). (b)(ii) Representative images illustrating the oblong shape of Aβ42 puncta, for three first principal axis lengths: shorter (top row), medium (middle row) and longer (bottom row). While the length of the first principal axis varies significantly between these examples, the length of the second principal axis remains similar between the three clusters. (c)(i) Total volume (in voxels, 1 voxel = 11.44x11.44x26.67 or 3,490 nm3) of Kv7.2 puncta overlapped/inside (black, R2 = 0.980, p < 0.0001) and outside (gray, R2 = 0.0582, p = 0.0979) Aβ42 puncta, as a function of the volume of the largest Aβ42 puncta within an ROI, compared to the line y = x (blue). (c)(ii) Representative images illustrating that the volume of Kv7.2 outside of Aβ42 is relatively constant as Aβ42 puncta size increases. As in (b)(ii), the three clusters shown are ordered by increasing size. (d) The converse of (c): total volume of Aβ42 puncta overlapped/inside (black, R2 = 0.6531, p < 0.0001) and outside (gray, R2 = 0.1876, p = 0.0010) of Kv7.2 puncta, as a function of the volume of the largest Kv7.2 puncta within an ROI, compared to the line y = x (blue). (d)(ii). Representative images illustrating that the volume of Aβ42 outside of Kv7.2 is smaller than the volume of Aβ42 co-localized with Kv7.2, and both values are positively correlated with the volume of the largest Kv7.2 puncta. Scale bar of (a-d ii) = 100 nm. Shown are images from one representative experiment from four independent replicates.
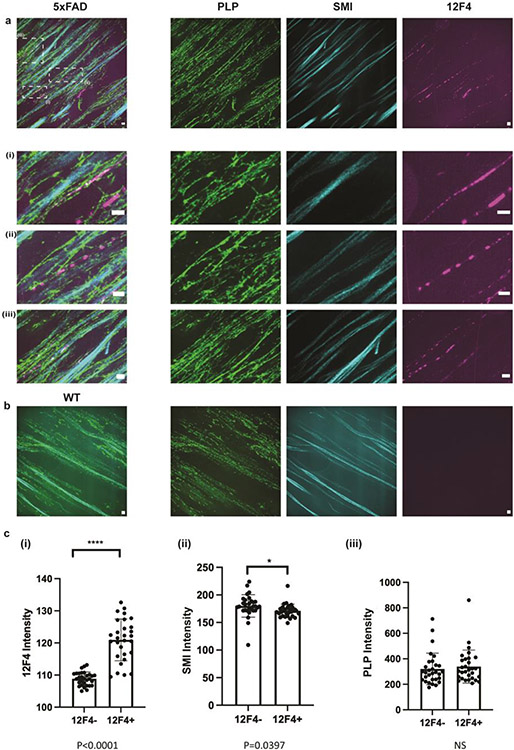
In myelinated axons, Kv7 potassium channels and voltage gated sodium (Nav) channels colocalize tightly with nodes of Ranvier periodically along axons40. We investigated whether the distribution of beta-amyloid is related to the myelination state of the axons. In 5xFAD mouse fornix, PLP staining was intact and partly overlapped with both SMI and Aβ42 clusters (Extended Data Fig. 5a), with the PLP-SMI relationship similar that between PLP and SMI in WT animals (Extended Data Fig. 5b). Quantification of both PLP and SMI in Aβ42+ axons did not show major changes in either SMI or PLP staining intensity as a function of Aβ42 presence (Extended Data Fig. 5c).
While the biological relevance of this periodicity and colocalization of β-amyloid with Kv7.2 and Nav1.6 needs further investigation, it is interesting to note that Nav1.6 and Kv7.2 ion channels can regulate neural excitability41-43, and Aβ peptides have also been known to influence excitability44. As these Aβ structures were often, but not always, colocalized with SMI-positive axons, and are highly reactive for Aβ42, we interpret these structures as periodic amyloid depositions. In the future, it will be interesting to see whether these structures play direct roles in neural hyperexcitability in Alzheimer’s. It is curious to reflect here that periodicity and order45,46,55,47-54 are often thought of as associated with healthy biological systems, whose functionality may be supported by such crystallinity. On the other hand, disorder, misalignments and misfoldings are often tied to pathological states. Here we find a curious mixture of the two – a periodicity that seems to be associated with a pathological state, and that may have implications for new hypotheses related to Alzheimer’s pathology. We are excited to see how ExR might reveal many kinds of previously invisible nanopatterns in healthy and disease states, due to its ease of use and applicability to multiple contexts, as here seen.
Discussion
The ability to optically study the crowded and complex three-dimensional molecular nanoarchitecture of cells and tissues, such as in brain tissue, is challenging, because optical super-resolution methods visualize fluorescent tags (such as antibodies) that are bound to target biomolecules, and these tags may not be able to access all the biomolecules within a nanostructure. Here we present expansion revealing (ExR), a new expansion microscopy method that reveals previously invisible biological nanostructures by enabling 20-nm resolution imaging and protein decrowding. ExR leverages the spatial expansion property of expansion microscopy2,3 and the 20-nm resolution of iterative expansion4 to physically decrowd densely packed biomolecules, providing conventional antibodies with better access to epitopes, revealing more detailed, and even previously unseen, structures when compared to pre-expansion staining, including in intact tissues.
While many expansion papers have mentioned or implied decrowding6,56-59, previous claims have not been accompanied with systematic data demonstrating specificity of signal-to-noise increase in specific biological structures vs. background, minimal distortion between pre- and post-expansion staining, and/or whether decrowding, vs. heat-mediated antigen retrieval, is the dominant contributor to improved staining. Nevertheless, these pioneering efforts are exciting. For example, ultrastructure expansion microscopy (U-ExM)58 combined with confocal microscopy showed higher labeling efficiency than in unexpanded dSTORM images. Indeed, the authors noted inhomogeneous tubulin signal “probably because of epitope masking of antibodies” and in the discussion, stated that the relatively smaller antibody size in post-expansion labeling approaches “unveiled” the chirality of the centriole. As another example, a previously described method that combines post-expansion staining with single-molecule localization microscopy (Ex-SMLM)59 also claimed that expansion of the sample increases epitope accessibility and labelling efficiency. As with U-ExM, the authors claim that post-expansion staining “increases epitope accessibility and thus labelling efficiency.” Here, we showed in comparisons of pre- and post-expansion staining in the same samples of brain tissue, that epitopes were indeed unmasked using ExR. We showed that the decrowding effect is not due to increases in nonspecific signal or antigen retrieval effects, and that ExR does not introduce any substantial distortion relative to the non-expansion super-resolution technique DNA-PAINT.
By comparing pre- and post-expansion staining, we found that enhanced post-expansion staining is observed within synaptic structures without a corresponding increase in nonspecific background staining, and preserves protein labelling abundance and localization with low distortion (<10 nm). ExR revealed biological structures such as previously undescribed nanoscale arrangements of calcium channels within transsynaptic nanocolumns23, and periodic amyloid nanostructures co-clustering with ion channels in a model of Alzheimer’s Disease. Presynaptic functional diversity is maintained in part by how calcium channels are organized with respect to vesicle-resident sensor proteins that mediate calcium-fusion coupling, yet dense active zone complexes have been difficult to map accurately in mammalian brain. The clustering of calcium channels in nanocolumns as we have observed here likely represents just one of many unique organizations of these ion channels to be discovered. The discovery of a unique pattern of periodic depositions of ion channels in amyloid aggregates in AD mouse model brains is another example of the unexpected observations that can be made using this tool. Observing the distribution of ion channels in axons ex vivo has been hindered by the crowded nature of axon tracts, and these results have implications for both saltatory conduction in vivo and the mechanisms of hyperexcitability in AD. We thus ExR to open up opportunities to observe new, previously unidentifiable biological structures via decrowding, which cannot be obtained by other super-resolution techniques.
Going forward, because ExR is an iterative expansion technique, it may be possible to achieve 100x expansion with one more round of re-embedding and gelation to obtain even better resolution and potentially single molecule localization. Furthermore, ExR could in principle be combined with RNA fluorescence in situ hybridization (FISH), as in expansion FISH (ExFISH)60, to label both proteins and RNA molecules, enabling visualization of the precise geometry and spatial relationship between RNA and proteins. Finally, because ExR anchors endogenous proteins to the gel, repeated rounds of antibody staining and stripping could in principle be performed, facilitating highly multiplexed imaging of potentially hundreds of proteins in the same field of view. Multiplexed ExR could in principle be used to create high-dimensional protein-protein colocalization maps, potentially revealing complex putative interactions and biological states that cannot be captured with imaging of only a few proteins at once (e.g., through conventional three-color staining and imaging). Thus, we anticipate that ExR will facilitate discoveries in biology and medicine.
Methods
Brain-tissue preparation
All procedures involving animals were in accordance with the US National Institutes of Health Guide for the Care and Use of Laboratory Animals and approved by the Massachusetts Institute of Technology Committee on Animal Care. Both male and female wild type mice (C57BL/6 or Thy1-YFP, 6-8 weeks of age, from JAX) and 5xFAD mice (12-13 months of age, from the Mutant Mouse Resource and Research Center) were used, because of the current study’s focus on developing and validating a technology. Mice were deeply anesthetized using isoflurane in room air. Mice were transcardially perfused at room temperature with ice cold 10 mL of 2 % (w/v) acrylamide in phosphate buffered saline (PBS) followed by ice cold 10 mL of 30 % acrylamide (w/v) and 4% paraformaldehyde in PBS. Brains were harvested and incubated in 20 mL of the same fixative solution (30 % acrylamide and 4% paraformaldehyde in PBS) at 4°C overnight. Fixed brains were transferred to 100 mM glycine at 4 °C for 6 h, then stored in PBS at 4°C for long term storage or sectioned to 50-100 μm-thick slices with a vibrating microtome (Leica VT1000S).
Cultured-neuron preparation
Cultured mouse hippocampal neurons were prepared from postnatal day ~0 Swiss Webster mice (Taconic) (both male and female mice were used) as previously described61. In summary, imaging chambers (112358 CS16-CultureWell™ Removable Chambered Coverglass, Grace Bio-Lab) were pre-treated with diluted Matrigel, and 5,000–10,000 cells were plated in each well. Neurons were grown at 37 °C and 5% CO2 in a humidified atmosphere for 14 days before fixation. Prior to fixation, the cells were briefly washed with 1x PBS warmed to 37 °C. The cultured cells were then fixed with 4% PFA in 1x PBS for 10 min at room temperature. Immediately thereafter, the fixation solution was replaced with a solution of 0.7% PFA, 1% Acrylamide, in 1x PBS, and the samples were incubated at 37 °C for 6 h. Once incubation was done, samples were washed with 1x PBS. Samples were stored at 4 °C overnight. Prior to expansion, the coverglass from the cell culture well was separated using the coverglass removal tool (103259, Grace Bio-Lab).
Expansion of brain tissue slices and cultured neurons
For the 1st gelling step, brain slices or cultured neurons were incubated in the 1st gelling solution (8.625% (w/v) sodium acrylate (SA), 2.5% (w/v) acrylamide (AA), 0.075% (w/v) N,N’-methylenebisacrylamide (Bis), 0.2% (w/v) ammonium persulfate (APS) initiator, 0.2% (w/v) tetramethylethylenediamine (TEMED) accelerator, 0.2% (w/v), 0.01% 4-Hydroxy-TEMPO (HT)) for 30 min at 4°C. Slices were embedded in 4 well dish gelling chambers on cover glasses surrounded by excess 1st gelling solution, and incubated at 37 °C for 2 h. Cultured neurons were incubated in the 1st gelling solution with coverglass on top at 37 °C for 2 h. After the incubation, gels containing the tissue or cultured neurons were cut out from the chamber and incubated with denaturation buffer (200 mM SDS, 200 mM NaCl, and 50 mM Tris pH 9)5 for 1 h at 95°C. Denatured gels were fully expanded via 4 washes for 15 min each with 5 mL distilled (DI) water in a 6 well-plate.
For the re-embedding step, expanded 1st gels were incubated in re-embedding solution (13.75% (w/v) AA, 0.038% (w/v) Bis, 0.025% (w/v) APS, 0.025% (w/v) TEMED) twice, replacing the first solution with freshly made re-embedding solution for 1 h each time on a shaker at room temperature. The re-embedded gels were transferred to a 4 well dish gelling chamber on cover glasses with a coverglass on top. The gelling chambers were placed in a ziplock bag with nitrogen flushing for 5 min and incubated for 2 h at 45°C. The re-embedded gels were washed 3 times for 15 min each with 5 mL PBS in a 6 well-plate.
For the 3rd gelling step, the re-embedded gels were incubated in 3rd gelling solution (8.625% (w/v) SA, 2.5% (w/v) AA, 0.038% (w/v) Bis, 0.025% (APS), 0.025% (w/v) TEMED) twice, replacing the first solution with fresh made 3rd gelling solution for 1 h each time on a shaker at room temperature. The 3rd gels were placed in a 4 well dish gelling chamber on cover glasses with a coverglass on top. The gelling chambers were placed in a ziplock bag with nitrogen flushing for 5 min and incubated at 60 °C for 1 h.
Gels were fully expanded in DI water by changing excess water 5 times for 2 h each and trimming axially to reduce thickness to 1 mm to facilitate subsequent immunostaining and imaging.
Immunostaining of expanded tissues and cultured neurons
Expanded gels were incubated in blocking solution (0.5% Triton X-100, 5% normal donkey serum (NDS) in PBS) for 2 h at room temperature. Gels were then incubated with primary antibodies (see Methods Antibody list) in ‘0.25T’ blocking buffer (0.25% Triton X-100, 5% NDS in PBS) overnight at 4°C. Gels were washed in washing buffer (0.1% Triton X-100 in PBS) 6 times for 1 h each time on a shaker at room temperature. Gels were then incubated with secondary antibodies in blocking solution overnight at 4 °C, and washed in washing buffer 6 times for 1 h each on a shaker at room temperature. ExR-processed samples images were acquired using a Nikon CSU-W1 confocal microscope with 100% laser power and 1s exposure time. Z-steps are varied between 0.250 and 0.500 μm.
Super-resolution imaging (DNA-PAINT)
Antibody conjugation:
The antibody (Jackson ImmunoResearch Laboratories, 711-005-152) was conjugated with thiolated DNA following the published protocol62. Briefly, 250 μM 5’ thiol-modified DNA oligonucleotides were reduced by 100 mM DTT (dithiothreitol) in 1× PBS + 1 mM EDTA for 1 hour at room temperature. Then, excessive DTT was removed using NAP5 columns (GE Healthcare Life Sciences, 17-0853-02). Carrier-free antibody (without BSA or sodium azide) formulated in 1× PBS was concentrated to 2 mg/ml using 50 KDa Amicon Ultra Filters (EMDMillipore, UFC505096). Then, crosslinkers (PEGylated SMCC crosslinker, ThermoFisher Scientific, A35397) were added to antibody tube at a 5:1 molar ratio (crosslinker: Antibody) and reacted for 2 h at 4°C. Next, antibody was purified using 0.5ml 7 kDA Zeba desalting columns (LifeTechnologies, 89883) to remove excess crosslinker. Reduced thiol-DNA oligonucleotide was then mixed with antibody at a 5:1 molar ratio (DNA: Antibody) and reacted overnight at 4 °C. Final conjugated antibodies were washed five times with 1× PBS (100ug/ml) in Amicon Ultra Filters (50k) to remove unreacted DNA oligonucleotides. Conjugated antibody was kept at 4 °C until use.
DNA sequences (5’ to 3’): /5ThioMC6-D/ATACATCT (ID, Docking; vendor, Integrated DNA Technologies); AGATGTAT /Atto-655/ (ID, Imager; vendor, Biosynthesis).
Immunostaining
Neuron cultures were blocked and permeabilized in 0.1% Triton X-100, 0.1% Tween20, 3% IgG-free BSA and 5% NDS for 2 h at room temperature. Cell cultures were incubated with primary antibodies (diluted in 0.1% Triton X-100, 0.1% Tween20, 3% IgG-free BSA) overnight at 4 °C and washed with 0.1% Tween20, 1% IgG-free BSA five times (10 min each). Secondary antibodies were diluted in 0.1% Triton X-100, 0.1% Tween20, 3% IgG-free BSA and incubated with samples for 2 h at room temperature and then washed as for primary antibodies. 1:100 dilution of 100 nm gold nanoparticles (753688-25ML, Sigma-Aldrich) was then added to wells and gently spun down in a centrifuge (500g, 5 mins). Finally, the cells were briefly washed three times with 1× PBS+500 mM NaCl.
Super-resolution imaging (DNA-PAINT)
Super-resolution imaging was accomplished on a Nikon Eclipse Ti microscope which was (1) operated by Nikon Elements software, (2) used a 1.49 NA CFI Apo 100x objective, (3) had a perfect focus system, and (4) had a total internal reflection fluorescence (TIRF) laser (488nm, 561nm, 647 nm). Lasers were operated in TIRF mode for all acquisitions. For image acquisition, an electron multiplying charge coupled device (iXon X3 DU-897, Andor Technologies) was used. Cameras were operated at 5-MHz refresh rate with 100 EM gain and a 50 ms exposure time. A 1.5x lens was introduced into the optical path allowed for imaging with a pixel size of 110 nm. Images were acquired using RAM capture via Nikon Fast Timelapse acquisition. 15000-20000 frames of single molecule image stacks were then acquired and analyzed in Picasso. To correct for image drift, 100 nm gold nanoparticles were used as fiducial markers in the imaging process. DNA imager stock solution were diluted and used in 1× PBS + 500 mM NaCl at a working concentration of 2 nM.
Decrowding experiments
Nanoscale-resolution imaging of synapses in somatosensory cortex (Fig 3b-h).
For pre-expansion antibody staining, brain slices were incubated with primary antibodies in blocking solution overnight at 4°C. Stained tissues were washed in washing buffer 6 times for 1 hr each time on a shaker at room temperature. Secondary antibodies (100 μL, 1 mg/mL) were incubated with 6-((acryloyl)amino)hexanoic acid, succinimidyl ester (AcX) (2 μL, 1 mg/mL) overnight at room temperature to prepare AcX conjugated secondary antibodies. Primary antibody stained tissues were then incubated with AcX-secondary antibodies in blocking solution overnight at 4°C, and washed in washing buffer 6 times for 1 h each time on a shaker at room temperature. Tissue expansion was carried out the same as previously described. The tertiary antibodies were stained after expansion to bind against AcX-secondary antibodies to visualize the pre-expansion staining.
For post-expansion antibody staining, expanded gels were incubated with the same primary and secondary antibodies without AcX conjugation. Antibodies against Shank3 or Homer1 were provided as a reference channel after expansion.
Quantification and validation of the decrowding effect
Comparison of ExR and DNA-PAINT synapsin staining in cultured neurons (Fig. 2h,i).
2-dimensional DNA-PAINT images were rendered using Picasso in Python63 using a blur width of 0.0, oversampling of 4.74 – 5.42 (corresponding to a pixel resolution of 20.31 – 21.10 nm/pixel), and max density of 350. The oversampling parameter was calculated on a per-well basis to match the pixel resolution given by the measured expansion factor of 7.7-8.0X for ExR images. The expansion factor for each well was calculated by measuring the distances between identical pairs of Synapsin puncta in pre- and post-expansion images, and dividing the physical units in the post-expansion image by the physical units in the pre-expansion image. The mean value over five such distances was calculated: 7.9X for well 1, 7.7X for well 2, 8.0X for well 3. Post-expansion ExR stacks were background subtracted using Fiji’s Rolling Ball algorithm with radius 50 pixels and collapsed to two dimensions using a maximum intensity projection. DNA-PAINT and ExR images were registered using either custom MATLAB scripts employing automatic intensity-based rigid body registration. Images were first min-max normalized, passed through a 2-D Gaussian filter (sigma = 5), and binarized using Otsu’s thresholding method (MATLAB’s graythresh). The geometric transform was estimated using MATLAB’s configurations for multimodal intensity-based registration, with the following optimizer parameters: initial radius, 0.004, epsilon = 1.5x10−4, growth factor = 1.01, maximum iterations = 300. An initial rigid body transformation was estimated prior to calculation of the final transform using the same parameters.
To assess the accuracy/resolution and distortion of ExR compared to DNA-PAINT, we calculated linear distortion of nanoscale synaptic puncta, the difference in pairwise distances between neighbouring synaptic puncta, and the differences in mean synaptic puncta number in identical cultured neuron samples imaged using the two technologies. To analyse nanoscale error introduced by ExR relative to DNA-PAINT, ROIs containing single synaptic puncta were manually segmented from DNA-PAINT and corresponding ExR images, taken from the same fields of view in the same specimens. Pixel intensity was min-max normalized separately for ExR and DNA-PAINT channels. Manually cropped individual synaptic ROIs from ExR images were automatically registered to their DNA-PAINT counterparts (note, after the global registration) using a rigid body transformation calculated using custom MATLAB scripts as previously described, with MATLAB’s default configurations for monomodal intensity-based registration and no initial transformation estimation. This automatic registration failed for 6 out of 107 ROIs, which were excluded from subsequent analyses. We calculated pixel-wise autocorrelation and cross-correlation as a function of distance as follows. Images were shifted by one voxel in each direction and padded using the intensity values of the pixels that were shifted out at that step. The pairwise linear correlation coefficient between pixel intensity values in DNA-PAINT and ExR staining channels, or PAINT-PAINT or ExR-ExR staining channels for autocorrelation, was calculated using MATLAB’s “corr” function. To calculate the half-maximal shift distance, we fit a third-degree polynomial (MATLAB’s “fit” with “poly3”) to the correlation or autocorrelation function, and used the best-fit curve to estimate the shift distance at which the correlation or autocorrelation reached 50% of its maximum value. Each of these calculations was repeated for each shift in the x- and y-dimensions. Plotted in Fig. 2h,i is the mean difference in half-maximal shifts over x- and y-dimensions.
The total number of synaptic puncta for each image was calculated using custom MATLAB scripts (Extended Data Fig. 1j). Images were first passed through a 2-D median filter of size [10, 10], binarized using Otsu’s method, and filtered to exclude puncta smaller than 150nm in size. Synaptic puncta number was calculated from 2D connected components in the binary image with connectivity 26. Corresponding pairs of neighbouring synaptic puncta (within ~1 μm from one another) and single synaptic puncta were manually cropped from DNA-PAINT and ExR images and binarized as described above (n = 50 ROIs from 5 fields of view, 6 ROIs each, 1 culture batch). Puncta centroids were calculated from 2-D connected components in the binary image (connectivity = 26) using MATLAB’s regionprops function. For cropped ROIs in which two synaptic puncta were counted after thresholding for both ExR and DNA-PAINT (Extended Data Fig. 1g-i) 27/50), the radial distance between puncta centroids (x1, y1) and (x2, y2) was calculated as sqrt((x2 – x1)2 + (y2 – y1)2). In Fig. 2j, the absolute value of the difference in distances between neighbouring synaptic puncta was normalized to the DNA-PAINT image. The coefficient of variation for distance between synaptic puncta imaged using ExR was calculated as the standard deviation of the difference in distance between pairs of synaptic puncta (PAINT - ExR) divided by the mean distance of the same pairs from DNA-PAINT.
Root mean squared error (RMS) between ExR and DNA-PAINT images was calculated as previously described (3). Briefly, a custom MATLAB script was used to implement a B-spline based non-rigid registration between pre and post expansion images, yielding vector fields for deformation within the images. These vector fields were then used to calculate root-mean-square length distortions across varying lengths. For plotting, RMS error values were binned into measurement length increments of 1 μm, and the mean RMS error within each bin is shown.
Decrowding analysis of manually segmented synapses (Extended Data Fig. 2a,b).
We compared the amplitude of signal intensity in the foreground (putative synapses) and background (everything else). First, we manually identified, based on brightness and size of reference channel staining, 47-70 of the largest, brightest synapses per ~350x350x20 micron (physical units) field of view (see Supplementary Table 2 for exact numbers of synapses; one field of view per cortical layer, three cortical layers per sample, two mice per synaptic protein). We developed an automated method to segment putative synaptic puncta from background. First, background was subtracted from image stacks using ImageJ/Fiji’s Rolling Ball algorithm with a radius of 50 pixels. Images were then binarized using a threshold calculated as seven times the standard deviation of the average intensity of manually-identified background regions selected every 10th slice of the z-stack. Binary images were passed through a 3-D median filter of radius 5x5x3 pixels to remove small puncta of non-specific staining. We then identified 3-D connected components from the filtered binary stack using MATLAB’s “bwconncomp” function, with a pixel connectivity of 26, meaning that pixels are connected if their faces, edges, or corners touch. Connected components smaller than 100x100x100nm3 (biological units) were removed, as most synapses are larger than this volume. ROIs, or putative synaptic puncta, were defined as 3-D connected components of the filtered, binary reference stack dilated using a disk structuring element with a radius of six pixels. A radius of six pixels (~100nm, in biological units) was chosen because both pre- and post-synaptic proteins of the same synapse, but not other synapses, fall within this range (the synaptic cleft is ~12-20nm64, which we confirmed by manual inspection. Segmented synapses with zero filtered connected components (synaptic puncta) in the reference channel were excluded from further analysis. We calculated the average intensity of background-subtracted images either within or outside of these dilated reference ROIs (Extended Data Fig. 2a) to measure signal increase within putative synapses relative to signal increase in the background. All images were acquired under the same microscope conditions to allow for comparison of mean signal intensity. The total volume of pre- or post-expansion staining test puncta located within dilated reference channel ROIs (Extended Data Fig. 2b) was calculated from binarized stacks of pre- and post-expansion channels (thresholded and filtered as described for the reference channel) after multiplying the dilated binary reference stack (for inside dilated reference ROIs) or its inverse (for just outside dilated reference ROIs, but still within the manually-cropped synaptic area) by the binary pre- or post-expansion stack and calculating the sum of nonzero voxels for each product. Data are shown as the mean of each measure across the ~50 synapses per field of view, and the deviation was calculated as the standard error of these means across the nine fields of view (three fields of view for three animals) for each protein. We identified measured signal from one field of view from one animal as an outlier using the ROUT method65 with Q=1% in GraphPad Prism and excluded it from further analysis and visualization.
Quantification of synaptic properties (Extended Data Fig. 2c,d).
To compare volume and signal-to-noise ratio (SNR) of pre- and post-expansion staining, we used the same dataset for Extended Data Fig. 2a,b analysis. First, the background was subtracted from image stacks using ImageJ/Fiji’s Rolling Ball algorithm with a radius of 50 pixels. Images were then binarized using a threshold calculated as seven times the standard deviation of the average intensity of manually-identified background regions selected every 10th slice of the z-stack. We then identified and selected the biggest 3-D connected components in pre- and post-staining test channels separately in each layer of somatosensory cortex (L1, L2/3 and L4, respectively), as these are the most likely to be synapses. We calculated the voxel and signal intensity in the largest 3-D connected components from 49-70 manually selected synapses (see Supplementary Table 2 for exact numbers for each layer, protein, and mouse). The signal intensity was divided by the standard deviation of the background intensity to calculate SNR.
Analysis of distortion introduced by ExR relative to pre-expansion staining (Extended Data Fig. 2e-g and Extended Data Fig. 3).
To calculate the number of synaptic puncta, background-subtracted images were first thresholded as described previously (based on a multiple of the standard deviation of manually-identified background regions) and passed through a 3x3x5 voxel (1 voxel = 17.16x17.16x40nm3) median filter. MATLAB’s “bwconncomp” function was used to find connected components (putative synaptic puncta, connectivity of 26), and connected components with fewer than 30 voxels of volume were excluded from further analysis. Distortion between pre- and post-expansion images was calculated as was for distortion between DNA-PAINT and ExR images. Specifically, for the plots shown in Extended Data Fig. 3g-l, images were shifted by one voxel in each direction and padded using the intensity values of the pixels that were shifted out at that step. To calculate pixel-wise correlations and autocorrelations (Extended Data Fig. 3g-h), images were first normalized to their minimum and maximum intensity values. From these, we calculated the pairwise linear correlation coefficient (MATLAB’s “corr”) between pixel intensity values in the pre- and post-expansion staining channels, or pre-(/post-) and pre(/post)-expansion staining channels for autocorrelation. To calculate the half-maximal shift distance (Extended Data Fig. 2f,g), we fit a third-degree polynomial (MATLAB’s “fit” with “poly3”) to the correlation or autocorrelation as a function of shift distance, and used the best-fit curve to estimate the shift distance at which the correlation or autocorrelation reached 50% of its maximum value. For the plots shown in Extended Data Fig. 3i,j, the correlation was calculated with a slight modification to account for differences in puncta volume. First, background-subtracted images were masked based on the corresponding binary image. Second, nonzero pixels were divided by the mean intensity value in the nonzero regions. Finally, the correlation was calculated as the pairwise linear correlation coefficient (MATLAB’s “corr”) between masked, mean-normalized intensity values in the pre- and post-expansion staining channels. Mutually overlapped volume was calculated as the sum of nonzero pixels in intersection of the binary pre- and post-expansion staining z-stacks, and normalized to the total puncta volume (sum of nonzero pixels in the binary z-stack) in the pre-expansion staining channel (Extended Data Fig. 3k,l). Each of these calculations was repeated for each shift in the x-, y-, and z-directions. Synapses with zero puncta in the pre- or post-expansion staining channels were excluded from analysis.
Comparison between antigen retrieval and decrowding effect
Confocal images after immunostaining with antibodies against (Supplementary Fig. 2) Cav2.1, PSD95 and Homer1 and (Supplementary Fig. 6) 6E10, 12F4 and SMI with or without antigen retrieval treatment to compare signal quality for antigen retrieval vs. ExR treatment. To determine whether antigen retrieval by heat denaturation alone is the dominant factor underlying increased signal quality afforded by ExR, we treated one group of tissues with a standard antigen-retrieval step (placing tissues in 20 mM sodium citrate at pH 8 and incubating at 100°C for 30 sec and 60°C for 30 min)11. Tissues with or without this antigen-retrieval step were processed by ExR. Then, we compared the amplitude of signal intensity in foreground (putative synapses) and background (everything else). First, we manually identified, based on brightness and size of reference channel staining (Homer1 for Cav2.1, Shank3 for PSD95 and Homer1), 30 of the largest and brightest synapses per ~350x350x20 micron (physical units) field of view (n = 30 synapses from 1 field of view from 1 mouse). We used the automated segmentation procedure and calculated mean signal intensity and volume as described above (see “Decrowding analysis of manually segmented synapses”).
Protein distance measurement and Synaptic nanocolumn results analysis
For analysis, potential synapses were manually identified and selected based on 1) the juxtaposition of presynaptic clusters and postsynaptic clusters, and 2) the colocalization of clusters on the same side of the synapse (Fig 2e, Fig 4e-g, Fig 4i-p). As camera pixel size was 167 nm (physical units) and the step size of the z-stack was 250 nm (physical units), the voxel size was not equivalent in all dimensions. Because isometric voxels were necessary for subsequent analysis, each voxel was then subdivided into 12 smaller isometric voxels, each 83.3 nm (physical units) in all three dimensions. For comparisons of RIM1/2 and PSD95, one cluster was shifted in space to optimally overlap with the other cluster, as previously described13,64. The vector of this shift was determined by cross-correlation of the two clusters, and defined both the transsynaptic axis and the distance between the two clusters. For comparisons of RIM1/2 and Cav2.1, the shift distance was set as 0, and for comparisons of RIM1/2 and PSD95, putative synapses with a RIM1/2 to PSD95 peak-to-peak distance of less than 20 or greater than 180 nm (biological units) were rejected from further analysis, consistent with the dimensions of the active zone and PSD. Any synapses that extended beyond the z-range of the imaged stack were also excluded.
The autocorrelation (ga(r)) and the protein enrichment analyses were adapted from previously described localization data-based analyses13,66. The 3D autocorrelation function (ga(r)) reports the increased probability of finding a similar signal at a distance (r) away from a given signal, and thus can be used to quantify the heterogeneity of the measured signal within a defined volume. The autocorrelation of each synaptic cluster was normalized by the autocorrelation of an object with the same shape and volume and that has a homogenous voxel intensity set to the average intensity of the synaptic cluster. Therefore, a synaptic cluster with a homogenous intensity will give a ga(r) = 1 at all radii, and local intensity peaks within a synaptic cluster will result in a ga(r) > 1 over a radius of the size of the region of high intensity. The cluster boundary was defined based on fluorescent intensity after convolution with a spherical kernel (r ~ 300 nm).
The molecular distribution of two different protein clusters relative to one another was characterized using a cross-enrichment analysis. The cross-enrichment analysis was performed by measuring the angularly averaged voxel intensity of one protein cluster (measured cluster) as a function of the distance from the point of peak intensity in the other protein cluster (reference cluster, shifted in space as described above). This value was then normalized by the angularly averaged intensity (as a function of the distance from the point of peak intensity in the reference cluster) of an object of the same shape and volume as the measured cluster with voxels set to the average intensity of the measured cluster. Regions of high local intensity in the measured cluster will result in values (normalized intensity) > 1. The enrichment index was calculated by taking the average of the enrichment values within a radius of 60 nm from the peak of the reference cluster.
Synapse numbers (n) for the analysis from 2 mice:
Autocorrelations
Cav2.1 (Fig. 4e): n = 144 synapses (Layer 1), 101 synapses (Layer 23), 103 synapses (Layer 4)
PSD95 (Fig. 4f): n = 144 synapses (Layer 1), 101 synapses (Layer 23), 103 synapses (Layer 4)
RIM1/2 (Fig. 4g): n = 144 synapses (Layer 1), 101 synapses (Layer 23), 103 synapses (Layer 4)
Enrichment analysis
RIM1/2 enrichment to PSD95 peak (Fig. 4i,j): n = 153 synapses (Layer 1), 103 synapses (Layer 23), 108 synapses (Layer 4)
PSD95 enrichment to RIM1/2 peak (Fig. 4k,l): n = 152 synapses (Layer 1), 102 synapses (Layer 23), 108 synapses (Layer 4)
Cav2.1 enrichment to RIM1/2 peak (Fig. 4m,n): n = 150 synapses (Layer 1), 103 synapses (Layer 23), 107 synapses (Layer 4)
RIM1/2 enrichment to Cav2.1 peak (Fig. 4o,p): n = 153 synapses (Layer 1), 99 synapses (Layer 23), 108 synapses (Layer 4)
Enrichment Index values (mean +/− S.D.):
RIM1/2 to PSD-95 peak (Fig. 4j): 1.585 +/− 0.330 (Layer 1), 1.535 +/− 0.358 (Layer 23), 1.545 +/− 0.332 (Layer 4)
PSD-95 to RIM1/2 peak (Fig. 4l): 1.611 +/− 0.308 (Layer 1), 1.632 +/− 0.269 (Layer 23), 1.622 +/− 0.285 (Layer 4)
Cav2.1 to RIM1/2 peak (Fig. 4n): 1.510 +/− 0.364 (Layer 1), 1.359 +/− 0.330 (Layer 23), 1.452 +/− 0.314 (Layer 4)
RIM1/2 to Cav2.1 peak (Fig. 4p): 1.493 +/− 0.330 (Layer 1), 1.317 +/− 0.311 (Layer 23), 1.422 +/− 0.322 (Layer 4)
Analysis of the Alzheimer’s results
Comparison of Aβ42 volume in WT vs 5xFAD (Supplementary Fig. 5b).
3D image stacks of Aβ42 and SMI312 staining were background subtracted via rolling-ball background subtraction with a 200 pixel radius using ImageJ/Fiji. For each color channel, the standard deviation for the background was calculated using a 75x75 pixel window. Subsequently, each color channel was binarized by applying a threshold of 28 times the standard deviation of the background. This value was determined by evaluating the amount of thresholding required to remove putative non-specific staining spots. Finally, after binarization, the volume of Aβ42 and SMI312 for each field of view (FOV) was determined by adding up the segmented pixels of each color channel.
Distance Measurement between Clusters (Fig. 6d).
To calculate the distance between adjacent clusters for either Aβ42 or Kv7.2, clusters that line along SMI312 neurofilaments were manually cropped out in 3D. Then, after applying rolling-ball background subtraction with a 100 pixel radius, the centroid of each cluster was annotated manually using ImageJ/Fiji in 3D. Given that the spacing between clusters is much larger than the size of each cluster, we reasoned that manual labeling of the centroids incurs minimal error. Finally, the distance between adjacent clusters was calculated in 3D.
Calculation of Aβ42 and Kv7.2 Cluster Diameter (Fig. 6g-i).
After applying rolling-ball background subtraction with a 100 pixel radius to 3D FOVs of Aβ42 and KV7.2 staining, overlapping KV7.2 and Aβ42 clusters were manually cropped out. After calculating the standard deviation of the background of each channel, the cropped images were binarized by applying a threshold ten times the standard deviation of the background. The volume of each cluster was then identified via connected component analysis using MATLAB’s “bwconncomp” function. Finally, the centroid and principal axis length of each cluster were determined using the associated “regionprops” function, which models each connected component region as an ellipsoid. The centroid values were then used to calculate the distance between overlapped Aβ42 and Kv7.2 clusters.
Aβ42 and Kv7.2 Cluster Shape Analysis (Fig. 7a-d).
ROIs containing single Aβ42 puncta that were part of a periodic chain-like structure were manually identified (n = 55 ROIs, 5 ROIs per field of view, from 11 fields of view from two mice) from background-subtracted images (ImageJ/Fiji’s Rolling Ball algorithm, radius of 50 pixels). To visualize the 3D shape of Aβ42 and Kv7.2 puncta within these ROIs, we resliced the image stack along both transverse dimensions, at equal spacing to the axial dimension. We display the middle slice in each stack in the x-y plane in Fig. 7a(ii)-d(ii) and the middle slice in each stack in the x-y, y-z, and x-z planes in Supplementary Fig. 8a (where x- and y-directions are transverse, and z-direction is axial). To quantify shape features, we used CellProfiler’s67 Watershed68 segmentation module to segment puncta within manually extracted ROIs, using a footprint of 30 pixels for each channel. A custom MATLAB script was deployed to calculate the number of puncta in each channel, mean and maximum volume and surface area of these puncta, length of the three principal axes of the ellipsoid that have the same normalized second central moments as the region for the largest puncta, and the total volume of puncta overlap between Aβ42 and Kv7.2 as the number of non-zero pixels in the intersection of the binary image stacks. To quantify the statistical significance of the relationships between these measures, either two-tailed paired t-tests or simple linear regression were used as described in the text.
Expansion factor and measurement of the root mean square error
A Thy1-YFP mouse was perfused as described above and 50 μm coronal sections were prepared using a vibratome. Before expansion, YFP fluorescence was imaged in six fields of view from the cortex of three cortical slices. Subsequently, these slices were processed with the ExR protocol as described above. Expanded slices were then labelled with a primary antibody against GFP (thermoFisher A-11122) and a secondary antibody (see Supplementary Table 10. List of antibodies). The same fields of view imaged pre-expansion were identified and confocal images were acquired of the antibody staining. Pre and post ExR images were acquired on an Andor spinning disk (CSU-X1 Yokogawa) confocal microscope with a 40 × 1.15 numerical aperture water objective.
To determine distortion arising from the process of ExR, pre and post expansion images were aligned and deformations in images were determined as described previously (3). Briefly, pre and post ExR images were background subtracted with a Rolling Ball background subtraction algorithm (ImageJ/Fiji) with a 200 pixel radius. Then, corresponding confocal planes from pre and post images were identified and registered using Fiji’s Turboreg method allowing for scaling and rigid rotation. Then, a custom MATLAB script was used to implement a B-spline based non-rigid registration between pre and post expansion images, yielding vector fields for deformation within the images. These vector fields were then used to calculate root-mean-square length distortions across varying lengths.
To calculate the expansion factor, the physical distances between feature-containing YFP structures (e.g. dendrites, axons) in fluorescent protein containing specimens were measured in pre- and post- ExR images. In cases of 5xFAD or C57BL/6 mice, DAPI staining of neighbouring cells was used to measure corresponding distances in pre- and post- ExR images, and the expansion factor was calculated by dividing the distances.
Tables listing the chemicals and antibodies used are available as Supplementary Information.
ExR protocol
1. Mouse tissue slices
i. Anesthetize mice using isoflurane in oxygen and perfuse with 10 mL of 2% acrylamide in PBS followed by 10 mL of 30% acrylamide and 4% paraformaldehyde in PBS.
ii. Harvest brains and incubate in 20 mL of the same fixative solution (30% acrylamide and 4% formaldehyde in PBS) at 4°C overnight.
iii. Transfer fixed brains to 100 mM Glycine at 4°C for 6 h.
iv. Store tissues in PBS at 4°C for long term storage.
v. Slice tissues on a vibrating microtome to a thickness of 50-100 μm.
2. Gelation
A. Gelling for 1st expansion
i. Incubate brain slices in the 1st gelling solution for 30 min at 4°C.
ii. Place brain slices with excess 1st gelling solution between two #1.5 coverglass separated by two pieces of #1.5 coverglass, and then incubate at 37°C for 2 h.
iii. Cut out gels from the chamber and incubate with denaturation buffer (200 mM SDS, 200 mM NaCl, and 50 mM Tris pH 9) for 1 h at 95°C.
iv. Wash gels 4 times with DI water in shaker and expand gels in DI water at 4°C overnight.
B. Re-embedding
i. Incubate expanded 1st gels in re-embedding solution twice for 1 h each time in shaker at room temperature.
ii. Transfer gels between #1.5 coverglass separated by slide glass and incubate with excess re-embedding solution at 45°C for 2 h.
iii. Wash gels 3 times with PBS in shaker.
C. 3rd gelling
i. Incubate the re-embedded gels in the 3rd gelling solution twice for 1 h each time in shaker at room temperature.
ii. Transfer gels between #1.5 coverglass separated by slide glass and incubate at 60°C for 1 h.
iii. Wash gels 4 times with DI water in shaker and expand gels in DI water at 4°C overnight.
iv. Trim gels axially to 1 mm thickness.
3. Staining
i. Incubate gels in blocking solution (0.5% Triton X-100, 5% normal donkey serum (NDS) in PBS) for 2 h at room temperature.
ii. Incubate gels with primary antibodies in ‘0.25T’ blocking buffer (0.25% Triton X-100, 5% NDS in PBS) overnight at 4°C.
iii. Wash gels with washing buffer (0.1% Triton X-100 in PBS) 6 times for 1 h each time.
iv. Incubate gels with secondary antibodies in blocking solution at 4°C overnight.
v. Wash gels with washing buffer (0.1% Triton X-100 in PBS) 6 times for 1 h each time and expand gels in DI water for 20x expansion or 0.05x PBS for 15x expansion.
Extended Data
Extended Data Fig. 1. Comparison of synaptic nanostructures imaged using DNA-PAINT and ExR in cultured neurons.
(a-c) Three representative fields of view imaged using DNA-PAINT (i) and ExR (ii) after rigid body registration to DNA-PAINT images. Scale bar = 5 μm. (iii-iv) Processed, binary versions of (i) and (ii) used to automatically count synaptic puncta number and pairwise distances. Scale bar, 5μm. (d) Representative manually-cropped matched synaptic ROIs for DNA-PAINT (top row) and ExR (bottom row), used for the distortion analysis shown in Fig. 2h,i (scale bar = 250 nm). (e) Pixel-wise correlation between min-max normalized ExR and DNA-PAINT channels as a function of shift distance in x- and y-directions for two randomly selected synaptic ROIs. (f) Pixel-wise autocorrelation between min-max normalized DNA-PAINT (PAINT-PAINT, magenta), ExR (ExR-ExR, yellow), and pixel-wise correlation between DNA-PAINT and ExR (PAINT-ExR, black) as a function of shift distance in x- and y-directions for the synaptic ROIs shown in (e). (g) Representative manually-cropped pairs of synaptic puncta used to generate the data shown in Fig. 2j and panel i. Shown is an overlay of DNA-PAINT (green) and ExR (magenta) binary masks (scale bar = 250 nm). (h) Histogram of difference in number of synaptic puncta counted after thresholding pairs of synaptic puncta. (i) Difference in radial distance between pairs of synaptic puncta, DNA-PAINT – ExR (mean = −0.008854, 95% CI [−0.05419, 0.03649]). (j) Total number of synaptic puncta for the five fields of view imaged using DNA-PAINT and ExR (two-sided paired t-test, p = 0.9271, t = 0.09735, df = 4). All data are from 5 ROIs from 3 wells of one cultured neuron batch. Shown are images from one representative experiment from two independent replicates.
Extended Data Fig. 2. Analysis of the ExR decrowding effect.
(a, b) Quantification of decrowding in a set of manually identified synapses. Statistical significance was determined using Sidak’s multiple comparisons test on two-sided t-tests following ANOVA (*P ≤ 0.05, **P ≤ 0.01, ***P < 0.001, ****P < 0.0001, here and throughout the paper, and plotted is the mean, with error bars representing standard error of the mean (SEM), here and throughout the paper). (a) Mean signal intensity inside and outside of (that is, nearby to) dilated reference ROIs for pre- and post-expansion stained manually-identified synapses (Supplementary Table 2 for numbers of technical and biological replicates)). Data points represent the mean across all synapses from a single field of view. (b) Total volume (in voxels; 1 voxel = 17.16 × 17.15 × 40, or 11,779, nm3) of signals inside and outside of dilated reference ROIs, in both cases within cropped images containing one visually identified synapse (Supplementary Fig. 3h), for pre- and post-expansion stained manually-identified synapses (Supplementary Table 2 for numbers of biological and technical replicates). Data points represent the mean across all synapses for 3 fields of view from 3 biological replicates (n = 9 fields of view total per protein). (c) Mean voxel size and (d) mean signal-to-noise (SNR) ratio of pre- and post-expansion immunostaining showing 7 proteins in somatosensory cortex regions L1, L2/3, and L4 of 3 mice. Plotted is mean and SEM. To compare the 3D voxel size and SNR of pre- and post-expansion stained synapses for each of the seven proteins, three two-sided t-tests (one for each layer) were run (n = 49-70 puncta per layer from 3 mice; Supplementary Table 2 for exact n values). Statistical significance was determined using multiple t-tests corrected using the Holm-Sidak method, with alpha = 0.05. (e) Population distribution (violin plot of density, with a dashed line at the median and dotted lines at the quartiles) of the fractional difference in the number of synaptic puncta between post- and pre-expansion staining channels for Homer1 and Shank3 (n = 480 synapses from 3 mice). (f) Population distribution of the difference in distance (in nm) between the shift at which the correlation is half maximal half-maximal shift for pre-pre autocorrelation and post-pre correlation (calculated pixel-wise between intensity values normalized to the minimum and maximum of the image, see Methods) for x-, y-, and z-directions (x- and y-directions being transverse, z-direction being axial) for Homer1 and Shank3 (n = 458 synapses, 3 directions each, from 3 mice). (g) Same as (f), for post-post autocorrelation and pre-post correlation.
Extended Data Fig. 3. Analysis of the distortion caused by post-expansion staining, as compared to classical pre-expansion staining.
(a-b) Representative background-subtracted and binary images of Homer1 (a) and Shank3 (b) in pre- and post-expansion staining channels (top row (yellow): pre-expansion channel, second row (black/white): binary pre-expansion channel, third row (magenta): post-expansion channel, bottom row (black/white): binary post-expansion channel). (c) Number of synaptic puncta for pre- and post-expansion staining channels, after filtration, for the images in (a-b). (d) Population distribution (violin plot of density, with a dashed line at the median and dotted lines at the quartiles) of the number of synaptic puncta in the pre-expansion staining channel for Homer1 and Shank3 (see Supplementary Table 5 for statistics for this figure). (e) Population distribution of the number of synaptic puncta in the post-expansion staining channel for Homer1 and Shank3. (f) Difference in the number of synaptic puncta between post- and pre-expansion staining channels normalized to the number of synaptic puncta in the pre-expansion staining channel. (g-h) Pixel-wise autocorrelation between pre-expansion (pre-pre, yellow), post-expansion (post-post, magenta), and pixel-wise correlation between pre- and post-expansion (pre-post, black) as a function of shift distance in x- (left column), y- (middle column), and z- (right column) directions for Homer1 (g) and Shank3 (h). The mean across all synapses is shown in the top row, and representative synapses are shown in the second through fourth rows. These values were used to calculate the linearized error measure shown in Extended Data Fig. 2f, g. (i-j) Pixel-wise correlation between mean-normalized, masked pre- and post-expansion channels as a function of shift distance in x- and y-directions (z = 1) for Homer1 (i) and Shank3 (j). The mean across all synapses is shown in the top left, standard deviation across all synapses shown in second from the top left, and representative synapses are shown in the remaining plots. (k-l) Mutually overlapped volume between pre- and post-expansion stained synaptic puncta, normalized to total puncta volume in the pre-expansion staining channel, as a function of shift distance in x- and y-directions (z = 1) for Homer1 (k) and Shank3 (l). The mean across all synapses is shown in the top left, standard deviation across all synapses shown in second from the top left, and representative synapses are shown in the remaining plots. Analysis was conducted on 480 (before exclusion based on size) synapses for Shank3 and Homer1 from 3 mice (see Supplementary Table 5 for exact numbers).
Extended Data Fig. 4. ExR and unexpanded tissue confocal images showing immunolabeling of Aβ42.
ExR confocal images (single z-slices) showing immunolabeling of Aβ42 with two different monoclonal antibodies (a) D54D2 + 6E10 and (b) D54D2 + 12F4 with SMI co-staining in the fornix of 5xFAD mouse (n = 3 fields of view of 2 slices from 2 mice). Scale bar, 10 μm (top row), 1 μm (i, ii panels). (c) Unexpanded tissue confocal image, a single z-slice, showing pre-expansion Aβ42 (yellow) and SMI (cyan) staining in the fornix of WT (upper panel) and 5xFAD mice (lower panel) (n = 3 fields of view of 1 slice from 1 mouse per WT and 5xFAD, respectively). Scale bar, 30 μm (left panel) and 6 μm (panels i, ii)).
Extended Data Fig. 5. ExR confocal images showing immunolabeling of PLP, SMI, and 12F4 in 5xFAD and WT fornix.
ExR reveals relative localization of Aβ42 peptide and myelin in the fornix of Alzheimer’s model 5xFAD and WT mice (n = 2 fields of view of 1 slice from 2 mice per WT and 5xFAD, respectively). (a) ExR confocal image (max intensity projections, 900–1000 nm thickness) showing post-expansion Aβ42 (magenta), SMI (cyan) and PLP (green) staining in the fornix of 5xFAD mice. Leftmost panel, merged low-magnification image; right images show individual channels. Insets (i-iii) show close-up views of the boxed regions highlighted in the upper left image. (b) ExR confocal image (max intensity projections, 1.72 μm thickness) showing post-expansion Aβ42 (magenta), SMI (cyan) and PLP (green) staining in the fornix of wild-type mice. Leftmost panel, merged image. All images were subtracted background using Fiji’s Rolling Ball algorithm with radius 50 pixels, and adjusted with auto-contrast. Scale bar = 500 nm. (c) Comparison of 12F4, SMI, and PLP intensity levels along axons in 5xFAD fornix with and without 12F4. To analyze axonal amyloid beta deposition with myelination and SMI intensity, we measure the (i) 12F4, (ii) SMI and (iii) PLP intensity levels along 10 axons with (12F4+) and without 12F4 (12F4−) from the same field of view (n = 3 fields of view of 2 slices from 2 5xFAD). All images were subtracted background with 50 pixels, and adjusted with auto-contrast for analysis by ImageJ. On each axon, three lines were drawn cross-sectionally across each axon in Image J and averaged intensity levels of PLP, 12F4 and SMI from different channels were measured respectively along these lines. For 12F4 + axons, each line was drawn across the centroid of amyloid beta deposition. For 12F4− axons, lines were positioned randomly along the axon. We then compared PLP, 12F4 and SMI312 intensity levels between 12F4 + and 12F4− axons. Plotted is the mean, with error bars representing standard error of the mean (SEM). Two-sided paired t-test, (i) ****p < 0.0001, t = 6.112, df = 18, (ii) p = 0.0595, t = 2.012, df = 18, (iii) p = 0.6580, t = 0.4502, df = 18.
Data availability
The main data supporting the results in this study are available within the paper and its Supplementary Information. The raw and analysed datasets generated during the study are too large to be publicly shared, yet they are available for research purposes from the corresponding authors on reasonable request.
Code availability
Accompanying code is available on Zenodo at https://doi.org/10.5281/zenodo.6383293 and on Github at https://github.com/schroeme/ExR.
Supplementary Material
Acknowledgments
We thank T. Biederer, C. Zhang and Y. Liu for antibody advice, S. Alon and K. Piatkevich for trainings and discussions, B. Kang for decrowding analysis advice, and D. Goodwin for instructions and scripts for manual image registration. D. S. acknowledges funding from NIH K99/R00 Pathway to independence Award (K99GM126277 and R00GM126277), M.E.S was supported by the National Science Foundation Graduate Research Fellowship under Grant No. 1745302 and the MathWorks Science Fellowship, A.H.T. acknowledges funding from NARSAD, E.D.N. acknowledges funding from Alana Down Syndrome Center and Robert A. and Renee E. Belfer Foundation, P.Y. acknowledges funding from NIH RF1 MH124606 and NIH Director’s Pioneer Award DP1 GM133052, L.H.T. acknowledges funding from Ludwig Family Foundation, the JPB Foundation, NIH Grants RO1 NS102730 and RF1 AG054321, T.A.B acknowledges funding from NIMH R37MH080046 and R01MH119826, E.S.B. acknowledges funding from the Ludwig Family Foundation, the Open Philanthropy Project, John Doerr, Lisa Yang and the Tan-Yang Center for Autism Research at MIT, U. S. Army Research Laboratory and the U. S. Army Research Office under contract/grant number W911NF1510548, Charles Hieken, Tom Stocky, Kathleen Octavio, Lore McGovern, Good Ventures, NIH Director's Pioneer Award 1DP1NS087724, NIH R01MH110932, NIH R01EB024261, NIH U24NS109113, NIH R37MH08004613, NIH 1RM1HG008525, NIH 1R01AG070831, NIH 1R56AG069192, NIH R01MH124606 and the HHMI-Simons Faculty Scholars Program. We also thank Brian Chow and Eric Betzig, as well as many current and past members of the Synthetic Neurobiology group, for discussions. The funders had no role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnotes
Reporting Summary. Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Competing interests
D.S., A.T.W., J.K. and E.S.B. are co-inventors on a patent application for ExR. E.S.B. is co-founder of a company seeking to deploy medical applications of ExM-related technologies. The other authors declare no competing interests.
Supplementary information The online version contains supplementary material available at https://doi.org/10.1038/s41551-01X-XXXX-X.
References
- 1.Sydor AM, Czymmek KJ, Puchner EM & Mennella V Super-Resolution Microscopy: From Single Molecules to Supramolecular Assemblies. Trends in Cell Biology vol. 25 730–748 (2015). [DOI] [PubMed] [Google Scholar]
- 2.Wassie AT, Zhao Y & Boyden ES Expansion microscopy: principles and uses in biological research. Nature Methods vol. 16 33–41 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen F, Tillberg PW & Boyden ES Expansion microscopy. Science 347, 543–548 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chang JB et al. Iterative expansion microscopy. Nat. Methods 14, 593–599 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen F et al. Nanoscale imaging of RNA with expansion microscopy. Nat. Methods 13, 679–684 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tillberg PW et al. Protein-retention expansion microscopy of cells and tissues labeled using standard fluorescent proteins and antibodies. Nat. Biotechnol 34, 987–992 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhao Y et al. Nanoscale imaging of clinical specimens using pathology-optimized expansion microscopy. Nat. Publ. Gr 35, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dani A, Huang B, Bergan J, Dulac C & Zhuang X Superresolution Imaging of Chemical Synapses in the Brain. Neuron 68, 843–856 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Monteiro P & Feng G SHANK proteins: Roles at the synapse and in autism spectrum disorder. Nature Reviews Neuroscience vol. 18 147–157 (2017). [DOI] [PubMed] [Google Scholar]
- 10.Dolphin AC & Lee A Presynaptic calcium channels: specialized control of synaptic neurotransmitter release. Nature Reviews Neuroscience vol. 21 213–229 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xiao B, Cheng Tu J & Worley PF Homer: A link between neural activity and glutamate receptor function. Current Opinion in Neurobiology vol. 10 370–374 (2000). [DOI] [PubMed] [Google Scholar]
- 12.Szumlinski KK, Kalivas PW & Worley PF Homer proteins: implications for neuropsychiatric disorders. Current Opinion in Neurobiology vol. 16 251–257 (2006). [DOI] [PubMed] [Google Scholar]
- 13.Zhu F et al. Architecture of the Mouse Brain Synaptome. Neuron 99, 781–799.e10 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vazquez LE, Chen HJ, Sokolova I, Knuesel I & Kennedy MB SynGAP regulates spine formation. J. Neurosci 24, 8862–8872 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Davydova D et al. Bassoon specifically controls presynaptic P/Q-type Ca2+ channels via RIM-binding protein. Neuron 82, 181–194 (2014). [DOI] [PubMed] [Google Scholar]
- 16.Peça J et al. Shank3 mutant mice display autistic-like behaviours and striatal dysfunction. Nature 472, 437–442 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Graf ER et al. RIM promotes calcium channel accumulation at active zones of the Drosophila neuromuscular junction. J. Neurosci 32, 16586–16596 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kiyonaka S et al. RIM1 confers sustained activity and neurotransmitter vesicle anchoring to presynaptic Ca2+ channels. Nat. Neurosci 10, 691–701 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Frank T et al. Bassoon and the synaptic ribbon organize Ca2+ channels and vesicles to add release sites and promote refilling. Neuron 68, 724–738 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.El-Husseini AE, Schnell E, Chetkovich DM, Nicoll RA & Bredt DS PSD-95 involvement in maturation of excitatory synapses. Science 290, 1364–8 (2000). [PubMed] [Google Scholar]
- 21.Migaud M et al. Enhanced long-term potentiation and impaired learning in mice with mutant postsynaptic density-95 protein. Nature 396, 433–439 (1998). [DOI] [PubMed] [Google Scholar]
- 22.Hayashi MK et al. The Postsynaptic Density Proteins Homer and Shank Form a Polymeric Network Structure. Cell 137, 159–171 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tang AH et al. A trans-synaptic nanocolumn aligns neurotransmitter release to receptors. Nature 536, 210–214 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Heine M & Holcman D Asymmetry Between Pre- and Postsynaptic Transient Nanodomains Shapes Neuronal Communication. Trends in Neurosciences vol. 43 182–196 (2020). [DOI] [PubMed] [Google Scholar]
- 25.Hruska M, Henderson N, Marchand S. J. Le, Jafri H & Dalva MB Synaptic nanomodules underlie the organization and plasticity of spine synapses. Nat. Neurosci 21, 671–682 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rebola N et al. Distinct Nanoscale Calcium Channel and Synaptic Vesicle Topographies Contribute to the Diversity of Synaptic Function. Neuron 104, 693–710.e9 (2019). [DOI] [PubMed] [Google Scholar]
- 27.Brockmann MM et al. RIM-BP2 primes synaptic vesicles via recruitment of Munc13-1 at hippocampal mossy fiber synapses. Elife 8, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Holderith N et al. Release probability of hippocampal glutamatergic terminals scales with the size of the active zone. Nat. Neurosci 15, 988–997 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Eggermann E, Bucurenciu I, Goswami SP & Jonas P Nanodomain coupling between Ca 2+ channels and sensors of exocytosis at fast mammalian synapses. Nature Reviews Neuroscience vol. 13 7–21 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Canter RG, Penney J & Tsai LH The road to restoring neural circuits for the treatment of Alzheimer’s disease. Nature vol. 539 187–196 (2016). [DOI] [PubMed] [Google Scholar]
- 31.Oakley H et al. Intraneuronal β-amyloid aggregates, neurodegeneration, and neuron loss in transgenic mice with five familial Alzheimer’s disease mutations: Potential factors in amyloid plaque formation. J. Neurosci 26, 10129–10140 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chao LL et al. Associations between White Matter Hyperintensities and β Amyloid on Integrity of Projection, Association, and Limbic Fiber Tracts Measured with Diffusion Tensor MRI. PLoS One 8, e65175 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Song SK, Kim JH, Lin SJ, Brendza RP & Holtzman DM Diffusion tensor imaging detects age-dependent white matter changes in a transgenic mouse model with amyloid deposition. Neurobiol. Dis 15, 640–647 (2004). [DOI] [PubMed] [Google Scholar]
- 34.Dong JW et al. Diffusion MRI biomarkers of white matter microstructure vary nonmonotonically with increasing cerebral amyloid deposition. Neurobiol. Aging 89, 118–128 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gail Canter R et al. 3D mapping reveals network-specific amyloid progression and subcortical susceptibility in mice. Commun. Biol 2, 1–12 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dunn AR & Kaczorowski CC Regulation of intrinsic excitability: Roles for learning and memory, aging and Alzheimer’s disease, and genetic diversity. Neurobiol. Learn. Mem 164, 107069 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chong SYC et al. Neurite outgrowth inhibitor Nogo-A establishes spatial segregation and extent of oligodendrocyte myelination. Proc. Natl. Acad. Sci. U. S. A 109, 1299–1304 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Brohawn SG et al. The mechanosensitive ion channel traak is localized to the mammalian node of ranvier. Elife 8, 1–22 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dupree JL et al. Oligodendrocytes assist in the maintenance of sodium channel clusters independent of the myelin sheath. Neuron Glia Biol. 1, 179–192 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lubetzki C, Sol-Foulon N & Desmazières A Nodes of Ranvier during development and repair in the CNS. Nat. Rev. Neurol 16, (2020). [DOI] [PubMed] [Google Scholar]
- 41.Shah NH & Aizenman E Voltage-Gated Potassium Channels at the Crossroads of Neuronal Function, Ischemic Tolerance, and Neurodegeneration. Transl. Stroke Res 5, 38–58 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hessler S et al. β-secretase BACE1 regulates hippocampal and reconstituted M-currents in a β-subunit-like fashion. J. Neurosci 35, 3298–3311 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ciccone R et al. Amyloid β-Induced Upregulation of Nav1.6 Underlies Neuronal Hyperactivity in Tg2576 Alzheimer’s Disease Mouse Model. Sci. Rep 9, 1–18 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ghatak S et al. Mechanisms of hyperexcitability in alzheimer’s disease hiPSC-derived neurons and cerebral organoids vs. Isogenic control. Elife 8, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lim CJ, Lee SY, Teramoto J, Ishihama A & Yan J The nucleoid-associated protein Dan organizes chromosomal DNA through rigid nucleoprotein filament formation in E. coli during anoxia. Nucleic Acids Res. 41, 746–753 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Xu K, Zhong G & Zhuang X Actin, spectrin, and associated proteins form a periodic cytoskeletal structure in axons. Science 339, 452–456 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Winardhi RS, Castang S, Dove SL & Yan J Single-molecule study on histone-like nucleoid-structuring protein (H-NS) paralogue in Pseudomonas aeruginosa: MvaU Bears DNA organization mode similarities to MvaT. PLoS One 9, (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Leterrier C et al. Nanoscale Architecture of the Axon Initial Segment Reveals an Organized and Robust Scaffold. Cell Rep. 13, 2781–2793 (2015). [DOI] [PubMed] [Google Scholar]
- 49.Chiang YL et al. Atomic force microscopy characterization of protein fibrils formed by the amyloidogenic region of the bacterial protein MinE on mica and a supported lipid bilayer. PLoS One 10, (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Prakash K et al. Superresolution imaging reveals structurally distinct periodic patterns of chromatin along pachytene chromosomes. Proc. Natl. Acad. Sci. U. S. A 112, 14635–14640 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Makky A, Bousset L, Polesel-Maris J & Melki R Nanomechanical properties of distinct fibrillar polymorphs of the protein α-synuclein. Sci. Rep 6, (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.D’Este E, Kamin D, Göttfert F, El-Hady A & Hell SW STED Nanoscopy Reveals the Ubiquity of Subcortical Cytoskeleton Periodicity in Living Neurons. Cell Rep. 10, 1246–1251 (2015). [DOI] [PubMed] [Google Scholar]
- 53.D’Este E et al. Subcortical cytoskeleton periodicity throughout the nervous system. Sci. Rep 6, (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Qu Y, Hahn I, Webb SED, Pearce SP & Prokop A Periodic actin structures in neuronal axons are required to maintain microtubules. Mol. Biol. Cell 28, 296–308 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bose K, Lech CJ, Heddi B & Phan AT High-resolution AFM structure of DNA G-wires in aqueous solution. Nat. Commun 9, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ku T et al. Multiplexed and scalable super-resolution imaging of three-dimensional protein localization in size-adjustable tissues. Nat. Biotechnol 34, 973–981 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.M’Saad O & Bewersdorf J Light microscopy of proteins in their ultrastructural context. Nat. Commun 1–15 (2020) doi: 10.1101/2020.03.13.989756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gambarotto D et al. Imaging cellular ultrastructures using expansion microscopy (U-ExM). Nat. Methods 16, 71–74 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zwettler FU et al. Molecular resolution imaging by post-labeling expansion single-molecule localization microscopy (Ex-SMLM). Nat. Commun 11, 1–11 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chen F et al. Nanoscale imaging of RNA with expansion microscopy. Nat. Methods 13, 679–684 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Klapoetke NC, K. et al. Independent optical excitation of distinct neural populations. Nat. Methods 11, 338–346 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Saka SK et al. Immuno-SABER enables highly multiplexed and amplified protein imaging in tissues. Nat. Biotechnol 2019 379 37, 1080–1090 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Schnitzbauer J et al. Super-resolution microscopy with DNA-PAINT. Nat. Protoc 12, 1198–1228 (2017). [DOI] [PubMed] [Google Scholar]
- 64.Savtchenko LP & Rusakov DA The optimal height of the synaptic cleft. Proc. Natl. Acad. Sci. U. S. A 104, 1823–1828 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Motulsky HJ & Brown RE Detecting outliers when fitting data with nonlinear regression - A new method based on robust nonlinear regression and the false discovery rate. BMC Bioinformatics 7, 123 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Chen JH, Blanpied TA & Tang AH Quantification of trans-synaptic protein alignment: A data analysis case for single-molecule localization microscopy. Methods 174, 72–80 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.McQuin C et al. CellProfiler 3.0: Next-generation image processing for biology. PLOS Biol. 16, e2005970 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Mangan AP & Whitaker RT Partitioning 3D surface meshes using watershed segmentation. IEEE Trans. Vis. Comput. Graph 5, 308–321 (1999). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The main data supporting the results in this study are available within the paper and its Supplementary Information. The raw and analysed datasets generated during the study are too large to be publicly shared, yet they are available for research purposes from the corresponding authors on reasonable request.
Accompanying code is available on Zenodo at https://doi.org/10.5281/zenodo.6383293 and on Github at https://github.com/schroeme/ExR.