Abstract
High-throughput sequencing and functional characterization of the cancer transcriptome have uncovered cancer-specific dysregulation of RNA splicing across a variety of cancers. Alterations in the cancer genome and dysregulation of RNA splicing factors lead to missplicing, splicing alteration-dependent gene expression and, in some cases, generation of novel splicing-derived proteins. Here, we review recent advances in our understanding of aberrant splicing in cancer pathogenesis and present strategies to harness cancer-specific aberrant splicing for therapeutic intent.
RNA splicing is an evolutionarily conserved nuclear enzymatic process whereby precursor mRNA (pre-mRNA) is transformed into mature mRNA for translation to protein. This fundamental eukaryotic process is an essential regulator of gene expression and proteome diversity1,2. Alternative RNA splicing regulates a multitude of cellular processes, including development, differentiation, cell cycle and cell death. As such, dysregulation of alternative splicing can alter fundamental cellular processes to promote neoplastic transformation, cancer progression or therapeutic resistance. Here, we review normal splicing mechanisms and splicing dysregulation in cancer. We highlight current and developing strategies to target cancer-specific aberrant splicing and discuss advances in chemical modulation of splicing and targeting of post-translational modifications (PTMs) of splicing proteins.
Molecular regulation of RNA splicing
RNA splicing, whereby noncoding segments (introns) of pre-mRNA are removed to produce mature mRNA, largely occurs in parallel with transcription by RNA polymerase II. Splicing is performed by a large highly dynamic RNA–protein macromolecular complex known as the spliceosome3. Over 99.5% of splicing reactions are performed by the major (or U2-dependent) spliceosome, which is composed of five small nuclear ribonucleoproteins (snRNPs) U1, U2, U4, U5 and U6 as well as >200 non-snRNP protein components4. The remaining (<0.5%) splice sites are recognized by the minor (or U12-dependent) spliceosome, which is made up of the U5 snRNP as well as distinct functional analogs of the major spliceosome’s U11, U12, U4atac and U6atac snRNPs (reviewed recently in ref. 5).
The spliceosome machinery assembles on introns by the stepwise recognition of three core consensus sequences that help distinguish intronic from exonic regions (Fig. 1a,b). These include the 5′ splice site (5′ss), which contains the GU dinucleotide at the 5′ end of the intron, the branch site residue, which is typically an adenosine nucleotide, and the 3′ splice site (3′ss), which is comprised of the polypyrimidine tract (which promotes spliceosomal recognition of the branch site residue) and the AG dinucleotide at the 3′ end of the intron. The recognition of diverse regulatory sequences including splice sites relies on RNA–RNA, RNA–protein and protein–protein interactions.
Fig. 1 |. Mechanisms of RNA splicing and dysregulation in cancer.
a, Sequential transesterification reactions involved in removal of an intron with resultant splicing product. During splicing catalysis, the branch site adenosine (A) nucleotide carries out nucleophilic attack of the 5′ss, forming a lariat, and the 3′ OH of the released 5′ exon performs a second nucleophilic attack on the last nucleotide of the intron at the 3′ss, joining the exons and releasing the intron lariat. b, Key sequence features and early splicing factors that govern splicing are shown. Sequence elements required for spliceosome assembly include the 5′ss and 3′ss, the polypyrimidine (poly(Y)) tract and the branch site residue that often follows the illustrated consensus motifs. The U1 snRNP (green) initiates splicing by recognizing the 5′ss consensus sequence. The U2 snRNP complex (brown) consisting of SF3B1 and other proteins is recruited to the branch site residue by the U2AF heterodimer (orange), which recognizes the 3′ss. Enhancers (ESE and ISE (intronic splicing enhancer)) and silencers (ESS (exonic splicing silencer) and ISS (intronic splicing silencer)) are recognized by specific trans-acting RNA-binding proteins, including SR proteins and hnRNPs. SR proteins commonly serve as enhancers of splicing, whereas hnRNPs commonly repress splicing. c, The following are mechanisms by which RNA splicing is altered in cancer: (1) cis-acting mutations affecting splicing regulatory sequences, (2) trans-acting mutations in the U1 snRNA, (3) mutations in RNA splicing factors and (4) changes in splicing factor expression. d, Alternative splicing of BCL-2 family of cell death factors BCL2L1 and MCL1 and alternative transcripts with resultant protein domain structures. Alternative splicing of BCL2L1 leads to generation of proapoptotic BCL-xS (green), which inhibits antiapoptotic BCL-xL (red) function, allowing BAX/BAK activation of mitochondrial outer membrane permeabilization (MOMP) and induction of apoptosis. Similarly, alternative splicing of MCL1 leads to generation of proapoptotic MCL1-S (green), inducing apoptosis through inhibition of antiapoptotic MCL1-L (green) function. Ex, exon. e, Protein diagrams (colored regions) of four splicing factors (SF3B1, U2AF1, SRSF2 and ZRSR2) and secondary RNA structure of the U1 and U2 snRNAs, with depictions of the most frequently reported hotspot mutations in red. ZnF, zinc-finger; RS, serine/arginine-rich domain; RRM, RNA recognition motif; UHM, U2AF homology motif.
Splicing catalysis is initiated by the U1 snRNP after binding of the 5′ss consensus sequence, followed by splicing factor 1 (SF1) binding of the branch site sequence and the U2 auxiliary factor (U2AF) complex binding of the polypyrimidine tract and the AG dinucleotide at the 3′ss4. Removal of SF1 from the branch site allows for U2AF-mediated guidance of U2 snRNP and its components, including splicing factor 3b subunit 1 (SF3B1), to the branch site sequence. The preassembled U4/U6.U5 tri-snRNP is then added to the growing spliceosome machinery followed by U1/U4 snRNP release, leading to formation of a catalytically active complex of the spliceosome. Pre-mRNA intron removal then proceeds by two sequential transesterification reactions that are initiated by nucleophilic attack of the 5′ss by the branch site nucleotide that results in the formation of an intron lariat (Fig. 1a). The lariat is a temporary structure that is subsequently removed by 5′ss-mediated attack on the 3′ss, producing a mature mRNA product followed by spliceosome disassembly. Development of single-particle cryo-electron microscopy has revolutionized our understanding of the spliceosome, allowing for visualization of highly dynamic fully assembled spliceosomes, including the U4/U6.U5 tri-snRNP in humans6,7 and other species7,8.
Alternative splicing and splicing regulation.
Splice sites are generally classified based on whether they are always (‘constitutive’) or only sometimes (‘alternative’) recognized as splice sites. High-throughput RNA sequencing studies have revealed that >95% of pre-mRNAs in humans undergo alternative splicing in which different sets of exons are spliced together with the potential to generate non-productive transcripts or multiple mRNA isoforms allowing for a diverse array of translated proteins from a single gene1,2,9,10. Long-read RNA sequencing (RNA-seq) and high-throughput single-cell sequencing can now be applied to quantify genome-wide splicing changes at the individual cell level and to identify a more accurate compendium of full-length transcripts (Fig. 2). Splicing can regulate gene expression through generation of unproductive mRNAs that are targeted to the non-sense-mediated decay (NMD) pathway. Moreover, RNA splicing is tightly linked to nuclear export, subcellular localization, translation and stability of mRNA transcripts. Each of these processes provide means by which splicing regulates expression of a gene to protein.
Fig. 2 |. Recent advances in genomic analysis of RNA splicing.
Schematic of available tools used to assess RNA splicing alterations. One gene (exons in gray) can produce multiple transcripts (different colors represent different exons) through alternative splicing, which allows for variation in RNA modifications, including m6A methylation and alterations in poly(A) tail sequence. While detection of splicing events and isoforms from conventional RNA-seq is the most mature method, short-read lengths (100–200 base pairs) rarely span splice junctions, requiring methods to infer full-length RNA transcripts138–140. Importantly, short-read RNA-seq cannot differentiate intermediate splicing products from final splicing products and is unable to accurately quantitate the efficacy of the two-step enzymatic splicing reaction. Two distinct long-read RNA-seq technologies have been commercialized by Pacific Biosciences (PacBio) and Oxford Nanopore Technologies (ONT). PacBio uses fluorescently labeled dNTPs and DNA polymerase to create average read lengths of >20 kilobases. By contrast, ONT uses biological nanopores within a membrane that translocate nucleic acid under an electric current and has no length limit141–143. Furthermore, ONT nanopore sequencing has the ability for direct RNA sequencing and detection of epigenetic modifications, including RNA modifications. Both long-read RNA-seq systems can generate millions of reads, allowing for comprehensive expression profiling. Such third-generation sequencing technologies have been used in characterizing isoforms in organisms with poorly annotated transcriptomes144,145 as well as for novel isoform discovery146,147. There is substantial heterogeneity in RNA splicing and gene expression among individual cells, even within a clonal population, which highlights limitations to the sequencing of bulk cell populations for gaining insight into splicing regulation and function. While single-cell RNA-seq is high throughput, detection and quantification of splicing changes from single cells remain major challenges, as the most widely used platforms for single-cell RNA-seq rely primarily on sequencing of 5′ and 3′ ends of transcripts. However, several studies have recently performed long-read sequencing of RNA from individual cells148,149. Further efforts to probe splicing at the single-cell level using similar approaches may be enlightening as well as efforts to merge spatial transcriptomics with analysis of RNA splicing.
Alternative splicing is controlled by both cis-acting regulatory elements and trans-acting splicing factors. As splicing is a cotranscriptional process, changes in the rate of RNA polymerase II elongation can also influence splicing outcomes by adjusting the chance that splice sites may be recognized by the spliceosome11. Trans-acting splicing regulatory factors, such as serine/arginine-rich (SR) proteins and heterogeneous nuclear ribonucleoproteins (hnRNPs), can recognize and interact with cis-acting regulatory sequences, including exonic and intronic splicing enhancers and silencers, which can strongly affect spliceosome assembly to promote or repress splicing activity, respectively12,13 (Fig. 1b). SR proteins have been shown to primarily promote splice site usage, whereas hnRNPs act primarily as repressors of splicing; however, their actions are context dependent. The ENCODE datasets evaluating the functional impact of RNA-binding proteins on RNA splicing, gene expression and binding to RNA and chromatin have clarified the cell- and site-specific effects of many splicing regulatory proteins14. As described in detail below, certain SR proteins and hnRNPs have been shown to be mutated and or dysregulated in cancer, acting as both oncoproteins and tumor suppressors (Fig. 1c).
Alternative pre-mRNA splicing and regulation of cell death pathways.
Apoptosis is a key cellular process that is tightly regulated by alternative pre-mRNA splicing. The function of many apoptotic proteins is regulated by alternative splicing through generation of mRNA transcripts, producing protein isoforms with distinct apoptosis regulatory activities15–17. Apoptosis can be induced through either the extrinsic (death receptor) or intrinsic (mitochondrial) cell death pathways. Alternative splicing is thereby used to alter the functions of apoptosis proteins at multiple stages of the extrinsic or intrinsic pathways via changes in protein subcellular localization, ratios between pro- and antiapoptotic splice variants, dominant-negative effects and antagonistic functions. Consistent with this, trans-acting splicing regulatory proteins, including SR proteins and hnRNPs, have been shown to substantially modify cell death pathways in cancer (reviewed in ref. 18).
Alternative splicing regulation of the BCL-2 family of cell death factors, which are commonly upregulated in cancer, has been previously described. Traditionally, members of the BCL-2 family function as antiapoptotic factors and possess several shared protein domains, including four BCL-2 homology (BH) domains, one transmembrane (TM) domain and in some cases a polypeptide sequence enriched in proline, glutamate, serine and threonine amino acids (PEST) domain. A striking example is the generation of multiple isoforms of the BCL-2 family member BCL-x due to alternative splicing of its BH domains, which have distinct functions and localizations within the cell. BCL-xL contains four BH domains and functions as an antiapoptotic factor. However, alternative splicing can produce a proapoptotic isoform called BCL-xS, which only contains the BH3 and BH4 domains (Fig. 1d). BCL-xS antagonizes the inhibitory functions of BCL-xL as well as BCL-2 and is widely expressed in tissues with high turnover, such as developing lymphocytes, whereas BCL-xL is found primarily in postmitotic tissues19. Importantly, aberrant BCL-x splicing has been implicated in several cancers, and novel therapeutic strategies, including splice-switch oligonucleotides, small molecular modulators and BH3 mimetics, are being investigated to promote BCL-x splicing correction20. Similarly, the antiapoptotic factor MCL1 has a PEST domain, four BH domains and one TM domain, but alternative splicing events can generate proapoptotic isoforms, including MCL-1S, which only possesses the PEST, BH4, BH3 and TM domains (Fig. 1d), and MCL-1ES, which possesses BH1–BH3 and the TM domain21.
Many annotated alternative splicing variants in apoptotic genes have only been confirmed at the RNA level, and further investigation of these predicted isoforms at the protein level and their functions will be crucial for mechanistic understanding of cell death regulation and exploitation for therapeutic targeting.
Dysregulation of RNA splicing in cancer.
Over the last decade, genomic DNA and RNA sequencing have revealed numerous means by which splicing is pathologically altered in cancer, including mutations in regulatory sequences of tumor suppressor genes22,23, recurrent somatic mutations in genes encoding core spliceosomal proteins and regulatory components24–30 and tumor-specific changes in the expression of specific RNA splicing factors31,32. In parallel, systematic transcriptomic analyses of cancer cells have revealed widespread changes in alternatively spliced transcripts compared to normal tissues, leading to skewing of isoform usage of annotated isoforms and the generation of aberrant, unannotated RNA isoforms in cancer22,23,33,34. A comprehensive pan-cancer analysis combining whole-exome sequencing and RNA-seq showed that tumors harbor up to 30% more alternative splicing events than normal tissues35. These studies highlight the commonality of splicing dysregulation in cancer and provide a rationale for potential therapeutic intervention.
Mutations in RNA splicing factors
In 2011, groundbreaking work by Yoshida and colleagues identified recurrent somatic mutations in genes encoding core spliceosomal proteins in individuals with myelodysplastic syndromes (MDS)24. These findings were supported by additional studies in MDS25,27 and chronic lymphocytic leukemia (CLL)26,29, providing a striking direct connection between splicing misregulation and cancer pathogenesis. Since that time, splicing factor mutations have been identified in a variety of cancer subtypes36, including acute myeloid leukemia (AML), and solid tumors, such as uveal melanoma (15–20%)28,30, breast cancer (5.6%)37,38, pancreatic ductal adenocarcinoma (4%)39, lung adenocarcinoma (3%)40 and bladder cancer36.
Alterations affecting splicing regulation in cancer generally fall into two categories: (1) cis-acting mutations that occur within or outside (intronic mutation altering splicing) the mRNA sequence that is being spliced and (2) trans-acting alterations, whereby mutations, changes in expression and/or PTM of splicing factors promote aberrant splicing of pre-mRNA (Fig. 1c).
The most frequent and well-characterized trans-acting recurrent splicing mutations in cancer primarily occur in four genes (Fig. 1e): SF3B1, serine/arginine-rich splicing factor 2 (SRSF2), U2 snRNA auxiliary factor 1 (U2AF1) and zinc-finger, RNA-binding motif and serine/arginine-rich 2 (ZRSR2)24,25,27. Several features of mutations in these genes, including their occurrence as heterozygous hotspot mutations at specific amino acid residues in SF3B1, SRSF2 and U2AF1, suggest gain or alteration of function. By contrast, mutations in the X chromosome-encoded ZRSR2 occur throughout the gene as stop codons or disruptions in the reading frame, which are more consistent with loss of function24. Importantly, mutations in these four splicing factors occur in a statistically mutually exclusive manner. Below, we discuss the current understanding of the functional implications in the most commonly mutated RNA splicing factors in cancer.
SF3B1.
SF3B1 is the most commonly mutated splicing factor across cancers, with the highest frequency in hematological malignancies41. In MDS, SF3B1 mutations define a distinct clinical entity known as MDS with ring sideroblasts (MDS-RS) and confer an overall favorable prognosis24,25. Alternatively, SF3B1 mutations in CLL are more common in unmutated IGHV CLL, which is a well-established adverse prognostic subset of CLL26,29.
SF3B1 is a subunit of SF3b, which in combination with SF3a and the core U2 snRNP comprise the 17S holo U2 snRNP complex. SF3B1 facilitates binding of the U2 snRNA to the pre-mRNA branch site sequence through N-terminal interactions with the U2AF heterodimer (Fig. 1b). The SF3B1 C-terminal Huntington, elongation factor 3, PR65/A, TOR (HEAT) domain is responsible for a variety of protein–protein and protein–RNA interactions, including the branch site and surrounding pre-mRNA sequences42,43. Importantly, the majority of cancer-associated SF3B1 mutations occur within the HEAT domain as heterozygous point mutations, wherein mutations at distinct residues are associated with different cancer subtypes28,44 (Fig. 1e). For example, substitutions at R625 are restricted to melanomas, whereas K700E substitutions occur in 97% of individuals with MDS-RS28,30,36. K666 substitutions are rare in MDS-RS, at ~1.5%; however, they are distinctly associated with high-risk MDS and AML45. The association of specific SF3B1 point mutations with different cancer types and clinical outcomes warrants further investigation into the mechanism underlying allele-specific effects of distinct SF3B1 mutations.
SF3B1 hotspot mutations are associated with recognition of aberrant branch site residues, which most commonly leads to use of an aberrant intron-proximal 3′ss located 10–30 base pairs upstream of the canonical 3′ss46,47. The transcripts generated from such missplicing most commonly contain premature termination codons predicted to trigger NMD48,49. However, there are many mutant SF3B1-dependent splicing changes that are not aberrant 3′ss events. As a key example, inclusion of a novel unannotated exon within BRD9 occurs across SF3B1-mutant hotspots and cancers and appears to be critical for SF3B1-mutant pathogenesis in the case of uveal melanoma50. Currently, it appears that there are a small number of SF3B1 mutation-induced aberrant splicing events, which appear to be central to development of distinct SF3B1-mutant cancers. For example, recent work dissecting the role of individual misspliced transcripts in MDS-RS identified that aberrant splicing of three to five distinct gene products can recapitulate MDS-RS in human cells51. Furthermore, Lieu and colleagues demonstrated a functional role for MAP3K7 missplicing and subsequent downregulation in MDS anemia52.
Recent studies have highlighted a critical role for the spliceosomal protein SURP and G-patch domain-containing 1 (SUGP1) in wild-type SF3B1 splicing. MDS-associated mutations in SF3B1 were found to weaken the interaction between mutant SF3B1 and SUGP1, resulting in defects in branch site recognition and increased cryptic 3′ss usage53. Reduction of SUGP1 expression could partially recapitulate mutant SF3B1 splicing defects, and overexpression of SUGP1 in SF3B1-mutant cells partially rescued SF3B1-associated aberrant splicing53. Furthermore, pan-cancer genomic analyses identified SUGP1 mutations in SF3B1 wild-type cancers and showed that SUGP1 mutants induced use of cryptic 3′ss similar to mutant SF3B1 aberrant splicing54.
U2AF mutations.
U2AF is a heterodimeric complex composed of U2AF1 and U2AF2 that function primarily to delineate the 3′ss during a splicing reaction (Fig. 1b). U2AF2 is the larger subunit that binds to the polypyrimidine tract, whereas the small subunit U2AF1 recognizes the AG dinucleotide consensus sequence at the 3′ss and intron–exon boundary55–58. U2AF1 and U2AF2 are recurrently mutated in myeloid malignancies and are associated with high-risk MDS and AML, with U2AF1 mutations being significantly more common27,59,60. U2AF1 is also mutated in solid tumors, with the highest frequency in lung cancer40. U2AF1 mutations occur within the first and second zinc-finger domains at residues S34 and Q157, respectively24,27 (Fig. 1e). The global effects of U2AF1 mutation on RNA splicing are allele specific and lead to changes in cassette exon usage that is dependent on nucleotide sequences surrounding the 3′ss AG dinucleotide61. While U2AF1-mutant-dependent splicing alterations affect a number of genes implicated in myeloid malignancies, including ASXL1, BCOR and DNMT3B61, the role of any of these specific alterations in MDS pathogenesis is not clear. Recent data reveal that U2AF1 mutations promote use of specific isoforms of GNAS and IRAK4, which may promote malignant transformation62,63; however, these are annotated alternative splicing events that are not specific to U2AF1-mutant cells.
U2AF2 mutations are much less common, but recurrent hotspot U2AF2 mutations (G176 and L187) have been identified and cluster within the first of two RNA recognition motif (RRM) domains. U2AF2 mutations are predicted to affect binding to the polypyrimidine tract; however, further functional studies are needed to determine the downstream implications of this change in protein–RNA interaction.
SRSF2.
SRSF2 is an auxiliary splicing factor mutated in ~50% of individuals with chronic myelomonocytic leukemia, 20–30% of individuals with MDS and 10–14% of individuals with AML24,60. Mutations in SRSF2 are enriched in high-risk MDS and are associated with increased risk of transformation to AML in the setting of MDS as well as clonal hematopoiesis64,65. SRSF2 is a member of the SR protein family and is involved in splicing regulation by promoting exon recognition through binding of exonic splicing enhancer (ESE) sequences within pre-mRNAs via its RRM domain66–68. Wild-type SRSF2 efficiently recognizes both C- and G-rich mRNA sequences69; however, the hotspot mutation at the P95 residue alters SRSF2’s ability to bind RNA in a sequence-dependent manner (Fig. 1e), leading to a skewing in which G-rich ESE sequence recognition is reduced while C-rich sequence recognition is increased, ultimately enhancing inclusion of exons with C-rich ESEs70,71.
As with SF3B1, several differentially spliced genes in mutant SRSF2 cells have been implicated in the pathogenesis of myeloid malignancies. Interestingly, SRSF2 mutation-dependent splicing promotes an EZH2 isoform with inclusion of a highly conserved poison exon that undergoes NMD, leading to impaired hematopoietic differentiation70. Consistent with this change in EZH2 expression in SRSF2-mutant cells, EZH2 loss-of-function mutations are mutually exclusive with SRSF2 mutations in MDS60. Of note, mutant SRSF2 splicing changes go beyond cassette exon splicing and include effects on intron retention70,72.
ZRSR2.
ZRSR2 is encoded on the X chromosome and functions as an RNA-binding protein where it primarily interacts with the 3′ss of U12-type introns73. Mutations in ZRSR2 result in the retention of minor introns73,74. The splicing consequences of ZRSR2 genetic alterations, therefore, appear quite different from those in SF3B1, U2AF1 or SRSF2. Moreover, while there is no functional evidence that SF3B1, U2AF1 or SRSF2 mutations confer any advantage in vivo in mouse models without additional genetic aberrations, loss of Zrsr2 in mice results in a strong competitive self-renewal advantage to hematopoietic stem cells74. This unique impact of Zrsr2 mutations on hematopoiesis in mice may be related to the remarkable conservation in minor intron sequences between species (which is not the case with the vast majority of U2-type introns)75,76.
snRNA mutations.
Beyond mutations in protein-coding genes, recurrent hotspot mutations in the U1, U2 and U11 RNA components of the spliceosome (known as snRNAs) have been identified in several cancers77–79. U1 and U2 snRNAs are primarily responsible for recognition of the 5′ss and branch site, respectively, in major introns, and recurrent U1 mutations occur at the third base of U1 snRNA within the 5′ss-binding region. The U1 3A > C mutation was discovered across multiple cancer subtypes, including CLL, other B cell non-Hodgkin lymphomas, hepatocellular carcinoma and pancreatic adenocarcinoma. By contrast, the U1 3A > G mutation is restricted to individuals with medulloblastoma (MB), with a significant preference for sonic hedgehog-type MB77. Hotspot mutations in the fifth nucleotide of U11 snRNA, which is responsible for 5′ss recognition of minor introns, were also reported in MB77. Tumors with mutant U1 and U11 snRNAs have significant aberrant splicing and increased cryptic 5′ss events. Hotspot mutations within the 28th nucleotide of the U2 snRNA (c.28 > T/G substitutions) have been discovered in B cell malignancies as well as prostate and pancreatic cancers79. This unexpected finding was complicated by the fact that there are numerous copies of nearly identical snRNA genes and pseudogenes across the genome, which makes identification of true somatic mutations in genes encoding snRNAs exceedingly difficult. Moreover, it is important to note that the functional impact of U2 snRNA mutations are currently unclear, as there have not been detectable alterations in gene expression or splicing in cell lines or primary tumors with this U2 snRNA mutation to date.
Other mutations and splicing modifications.
In addition to the mutations described above, whole-exome sequencing data from the 33 tumor types within The Cancer Genome Atlas have identified somatic mutations in PRPF8, PHF5A (a component of the SF3b complex that interacts with SF3B1), HNRNPCL1, RBM10, SFPQ, PCBP1, PCBP2, FUBP1, FUBP3 and QKI among others36,80,81. Loss-of-function mutations in the tumor suppressor gene RBM10 are present in lung and bladder adenocarcinomas and are primarily associated with exon inclusion events36,40. For example, mutations and changes in the expression of RBM10 promote alternative splicing of the Notch signaling inhibitor NUMB via an exon inclusion event at exon 9, leading to sustained Notch activation in lung cancer82,83. Outside of RBM10, there has been little functional characterization of the aforementioned mutations to date.
Additional studies have found that changes in the expression or activity of splicing factors, especially SR and hnRNP proteins, can lead to cancer-associated splicing and transformation (reviewed in ref. 81). As splicing factors tend to operate in a concentration-dependent manner, changes in expression levels and PTMs can promote oncogenic transformation. For example, upregulation of SRSF1 is prevalent in multiple cancers and transforms cells by promoting alternative splicing of target genes, including MST1R84 and RPS6KB1 (ref. 31). Additionally, phosphorylation of SRSF1 by SRPK1 can promote tumor-specific isoform expression of Rac1b in colorectal cells85. PTMs of SR proteins have been shown to regulate spliceosome formation and catalysis by regulating the shuttling of SR proteins in and out of the nucleus86,87.
Beyond trans-acting mutations in splicing factors, mutations within introns and exons are well established to have established effects on splicing regulation, including cis-acting DNA mutations that affect the 5′ss, 3′ss, branch site or splicing enhancer or silencer elements. Importantly, even synonymous mutations may alter splicing regulatory elements to cause missplicing22. Both somatic exonic and driver noncoding splice site mutations have been implicated in the inactivation of tumor suppressor genes, including recurrent synonymous mutations and noncoding intronic mutations, disrupting splice sites within TP53 and other genes important in tumorigenesis23,88.
Targeting RNA splicing in cancer
Splicing factor mutations represent attractive therapeutic targets, as they frequently occur as early initiating events, are present in dominant clones and are found in cancers with few effective treatment options81,89. Spliceosomal factors SF3B1, SRSF2 and U2AF1 typically harbor mutually exclusive heterozygous mutations, and coexpression of these mutations is intolerable to cells90,91. Furthermore, several studies have shown that splicing-mutant cells are preferentially dependent on wild-type spliceosome function, where deletion of the wild-type allele leads to cell death across different cancer subtypes with mutations in SF3B1 (ref. 92), SRSF2 (ref. 93) or U2AF1 (ref. 94). These data highlight the vulnerability of splicing factor-mutant cells to global perturbations in splicing catalysis and provide a therapeutic rationale for targeting splicing to trigger cell death. Over the last decade, several different therapeutic strategies targeting splicing have been developed and are discussed below (Fig. 3).
Fig. 3 |. Chemical inhibitors of RNA splicing.
Diagram of key sequence features (5′ss and 3′ss) and splicing factors (U1 snRNP, green; U2 snRNP, brown; U2AF heterodimer, orange) involved in splicing catalysis with accompanying splicing targeting mechanisms with natural products and small-molecule inhibitors. Top left, SF3b complex inhibitors bind to the branch site residue-binding pocket of SF3B1, blocking U2 snRNP recognition of the branch site, leading to intron retention and cassette exon skipping. Top right, protein domains of select UHM protein family members and drugs that have been shown to bind and inhibit these proteins. NSC 194308 targets U2AF2 through binding between its RRM domains to enhance U2AF2’s binding to RNA. Alternatively, phenothiazines target U2AF2 through binding of the UHM domains of several different UHM protein-containing proteins, including U2AF1, U2AF2, PUF60 and SPF45. Bottom right, RBM39 degraders induce an interaction between the E3 ubiquitin ligase adaptor protein DCAF15 and RBM39, leading to polyubiquitination and proteasomal degradation of RBM39. Bottom left, several post-translational modifications are known to regulate splicing function, including lysine phosphorylation and arginine methylation. Splicing factors are the most abundant arginine-methylated substrates in cells, and PRMT inhibitors are currently under clinical investigation. CLK, Cdc-like kinase; SRPK, serine/arginine protein kinase; DYRK, dual-specificity tyrosine-regulated kinases.
Small-molecule modulators of splicing in cancer.
The first class of small molecules to target splicing catalysis were derived from natural products and their derivatives that bind directly to the SF3b complex and abrogate its interactions with the branch site residue. These drugs include spliceostatin A95, sudemycin96, pladienolide97, E7107 (analog of pladienolide), H3B-8800 (orally bioavailable analog of E7107) and herboxidiene98. Functional genomic studies support the specificity of these molecules for the SF3b complex through the identification of resistance mutations in SF3B1 (SF3B1R1074H)99 and PHF5A (PHF5AY36C)43. Additionally, structural analysis of the SF3b complex with pladienolide B100 and E7107 (ref. 43) provided mechanistic insight that these molecules interact at the branch site binding pocket of SF3B1, blocking U2 snRNP recognition of RNA and leading to increased intron retention and cassette exon skipping throughout the genome93.
Phase I trials of E7107 in solid tumors were complicated by unexpected ocular toxicity and precluded further clinical evaluation of this agent101,102. While preclinical studies of the oral SF3b inhibitor H3B-8800 showed great preferential killing of spliceosome-mutant cancers103,104, the phase I dose escalation study of H3B-8800 showed no partial or complete responses and only a mild treatment effect, with 5 of 15 individuals with MDS with missense SF3B1 mutations experiencing red blood cell transfusion independence105. Importantly, several individuals who experienced red blood cell transfusion independence showed downregulation of aberrantly spliced TMEM14C, suggesting an on-target effect of H3B-8800 (ref. 105). Additional trials of H3B-8800 in individuals with SF3B1-mutant myeloid malignancies and exploration of other dosing schedules are currently ongoing.
RBM39 degraders.
Concerns surrounding the safety and therapeutic window of directly targeting core spliceosome components has led to investigation of alternative strategies of mutant splicing factor modulation through targeting accessory splicing factors. Recently, aryl sulfonamide molecules, such as indisulam, tasisulam, E7820 and chloroquinolxaline sulfonamide, have been shown to link accessory RNA splicing factors RBM39 and RBM23 to CRL4/DCAF15 E3 ubiquitin ligases, leading to proteasomal degradation of RBM39 and dose-dependent splicing alterations106,107 (Fig. 3). Functional genomic studies106,107 and structural studies108–110 confirm the targeting of RBM39 with these small molecules. RBM23 is also degraded by these drugs, but it appears dispensable for cell survival. Preclinical data demonstrate that splicing factor-mutant leukemias show increased sensitivity to genetic depletion and pharmacological inhibition of RBM39 compared to wild type111. Interestingly, several phase I and II (ref. 112) clinical trials with RBM39/RBM23 degraders have already been completed with good safety profiles in individuals with cancer. Unfortunately, these trials were completed before knowledge of the mechanism of action of these agents, and data to confirm if RBM39 degradation and/or splicing changes occurred in individuals treated with these drugs is lacking. In future studies, it will be crucial to explore these compounds in splicing factor-mutant malignancies with proper assessment of target engagement in individuals. A phase II trial of the orally bioavailable RBM39 degrader E7820 is ongoing in splicing factor-mutant individuals with myeloid neoplasms refractory to standard therapy (NCT05024994).
Small molecules targeting U2AF–RNA interactions.
New small-molecule splicing inhibitors, such as phenothiazine derivatives, have been shown in vitro to disrupt U2AF homology motifs (UHMs) and U2AF ligand motifs, which are common protein interaction domains among splicing factors and are critical for early spliceosome assembly113. Under normal circumstances, the C-terminal UHM domain of U2AF2 interacts with the N-terminal U2AF ligand motif of SF1 to recognize 3′ss before recruitment of the SF3b complex. These small molecules disrupt the function of an entire family of UHM domain-containing proteins, including U2AF2, RBM39, SPF45 and PUF60. Additionally, Kobayashi and colleagues discovered a small molecule (UHMCP1) targeting the U2AF2 UHM domain, preventing SF3B1/U2AF2 interaction and leading to changes in RNA splicing and cell viability114.
In contrast to the above agents that block U2AF interactions to perturb splicing, Chatrikhi and colleagues have discovered a hit compound (NSC 194308) that specifically enhances RNA binding by a U2AF2 subunit115. This compound stalls pre-mRNA splicing by binding an inter-RRM interface and enhances U2AF2 association with the splice site RNA. This proof-of-principle study identified a therapeutic strategy whereby stabilization of precatalytic splicing intermediates could be applied for therapeutic intervention.
Protein arginine methyltransferase (PRMT) inhibitors.
Given the importance of PTMs in the regulation of spliceosome assembly, subcellular localization and protein–protein interactions, an alternative approach for targeting mutant splicing changes incorporates the use of small molecules that inhibit placement or removal of these modifications. Several PTMs are known to regulate splicing function, including lysine phosphorylation116,117 performed by SR protein kinases and arginine methylation118,119 mediated by type I and II PRMTs. Importantly, splicing factors are the most abundant arginine-methylated substrates in cells, and preclinical studies have highlighted the importance of arginine demethylation mediated by PRMT5 and type 1 PRMT enzymes for splicing factor-mutant leukemia cell survival119,120. PRMT5 inhibitors are under clinical investigation, including two phase I/II trials in advanced MDS/AML (NCT03614728) and advanced solid tumors/hematological malignancies (NCT03886831).
Oligonucleotide-based therapeutic approaches.
Antisense oligonucleotide (ASO) therapy uses modified nucleic acids to base pair with pre-mRNA and modifies splicing by inhibiting RNA–RNA or splicing factor–RNA interactions and has met clinical success in the non-cancer setting with the Food and Drug Administration approval of nusinersen121 and eteplirsen122 for the treatment of spinal muscular atrophy and Duchenne muscular dystrophy, respectively. Two formidable challenges of ASO treatment in cancers include the molecular complexity of cancer (which typically contains hundreds of altered splicing events in addition to many genetic mutations) and the difficulty of delivering therapeutic oligonucleotides systemically. Nonetheless, one proof-of-concept study showed that correcting aberrant BRD9 splicing in SF3B1-mutant uveal melanoma cells using ASOs restored BRD9 protein levels and demonstrated therapeutic effects in vitro and in vivo50. Unfortunately, translation of ASO therapy to human care has proven difficult, and a recent clinical trial of a BCL-2-modifying ASO in combination with chemotherapy showed safety but lacked efficacy in improving outcomes123.
Pharmacologic induction of neoantigens by modulating RNA splicing.
Checkpoint inhibitor-based immunotherapies have greatly improved clinical outcomes across an array of cancer subtypes. Response to checkpoint blockade is associated with high tumor DNA mutational burden124,125 and mismatch repair deficiency126, which contribute to increased tumor neoantigen presentation127,128. While coding mutation-derived neoantigens have been thoroughly investigated, RNA-seq analysis of The Cancer Genome Atlas data provides evidence that tumor-specific alternative splicing events are abundant and produce neoantigens that are predicted to be more immunogenic than missense mutations35,129. Furthermore, two additional studies have shown that noncoding regions are the main sources of targetable tumor-specific antigens in cell lines and human samples130 and that retained intron neoepitopes are presented on major histocompatibility complex class I (MHC class I) on the surface of cancer cell lines131. Additionally, tumor-associated epitopes presented on MHC class I are typically predicted based on cancer-specific mutations in previously annotated protein-coding regions; however, several studies have highlighted that the source of cancer antigens presented on MHC class I may be more diverse and possibly derived from translation of unannotated open reading frames (nuORFs)132,133. The extent to which nuORFs contribute to the diversity of immunopeptidomes of cancer cells was unknown. Recent work coupling ribosome profiling, hierarchical ORF prediction and mass spectrometry of primary healthy and cancer samples and cell lines showed that peptides that originate from nuORFs are displayed on MHC class I of cancer cells and represent a new unexplored pool of MHC class I tumor-specific peptides with potential for therapeutic targeting134.
The above studies provide proof of concept that novel spliced-derived peptides could serve as neoepitopes; however, whether splicing-derived neoepitopes could elicit an endogenous immune response remained unanswered. Recent work demonstrated that pharmacologic modulation of splicing using clinical-grade compounds with distinct mechanisms can boost immune checkpoint blockade by inducing MHC class I-presented neopeptides135 (Fig. 4a). Such splicing-derived neoepitopes were able to trigger an antitumor T cell response in vivo. This provides a means to quickly and reversibly induce neoantigen generation without genomic changes. More broadly, these data suggest that modulation of splicing acts as a novel source of immunogenic peptides, which may have increased immunogenicity compared to mutation-derived neoantigens.
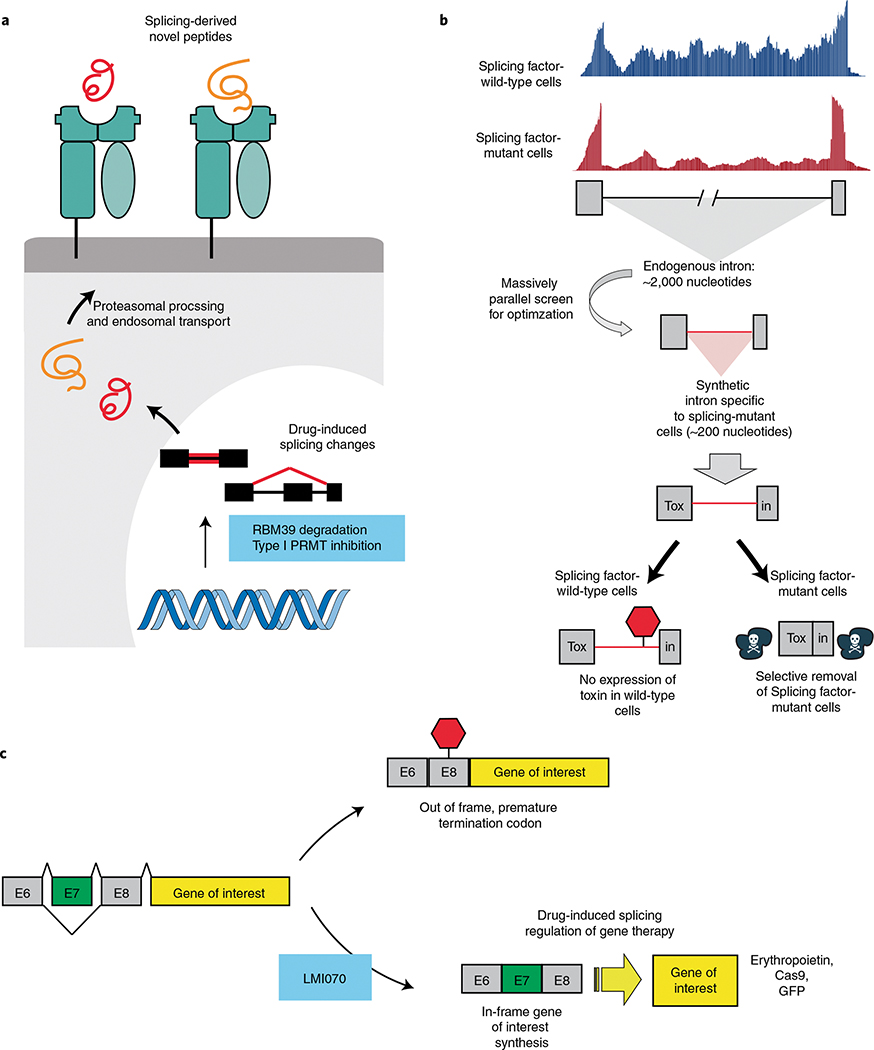
Fig. 4 |. Novel uses of splicing modulator drugs and synthetic introns responsive to splicing factor mutations.
a, Tumor-specific alternative splicing events are abundant in cancer and produce immunogenic neoantigens. Pharmacologic modulation of splicing using RBM39 degraders or PRMT inhibition induces novel splicing-derived neoepitopes that are presented on MHC class I. Splicing inhibition can improve response to immune checkpoint blockade through increased neoantigen generation. b, Diagram depicting the methodology and concept of synthetic intronic sequences to drive selective gene expression in cells with cancer-associated mutations in the RNA-splicing machinery, as described recently in ref. 136. In brief, endogenous splicing events, which are differentially used in splicing factor-mutant cells, are used to generate optimized shortened synthetic sequences. These optimized introns are then used to interrupt the protein-coding sequence of a gene of interest such that the gene of interest is only expressed in cells with an altered RNA splicing machinery. Such an approach may eventually be used to discover drugs that specifically regulate mutant splicing activity. c, Diagram depicting a splicing switch element. The switch element allows for precise control of gene replacement or gene editing after exposure to the small molecule LMI070. In the absence of exon 7 (E7) a premature stop codon blocks translation of the gene of interest. LMI070 regulates usage of exon 7, thereby regulating translation of the gene of interest to a protein product (such as erythropoietin, Cas9 or a fluorescent protein, such as green fluorescence protein (GFP)). Diagram reproduced from ref. 137, Springer Nature.
While the above approach appears promising, whether combination treatment with splicing modulatory compounds and immune checkpoint blockade will be tolerated in a clinical setting, given the possibility of increased inflammatory side effects, is unknown. Furthermore, it is unclear whether splicing modulation will result in long-lasting immune responses and whether maintenance splicing inhibitor drug therapy will be required for a continued antitumor effect.
Synthetic introns for mutation-dependent gene expression.
A limitation of currently available splicing inhibitors is that they target the core spliceosome machinery, which is an essential component of both normal and malignant cells. Indiscriminate inhibition of both wild-type and mutant splicing, even if preferentially affecting malignant cells, increases the potential risk of adverse side effects, and the therapeutic window of these therapies is not well defined. As discussed above, however, recurrent change-of-function mutations in RNA splicing factor genes induce sequence-specific changes in RNA splicing. Thus, the neomorphic functions of mutant RNA splicing factors highlights these mutations as exciting therapeutic targets. To date, however, there have been no therapeutic approaches identified that specifically target splicing factor-mutant cells.
A recent novel method harnesses the change in RNA splicing activity between mutant and wild-type cells to drive spliceosomal mutation-dependent gene expression in cancers136 (Fig. 4b). After identifying endogenous genes that were alternatively spliced in individuals with SF3B1-mutant cancer compared to normal controls, SF3B1-mutant-specific synthetic introns were generated for intron-dependent delivery of a suicide gene that preferentially led to elimination of leukemia, melanoma and breast and pancreatic cancer cells bearing SF3B1 mutations in vitro and in vivo while leaving wild-type cells unaffected. Importantly, any gene of interest can be expressed in a mutant or wild-type splicing factor-dependent manner, allowing for genotype-specific expression of florescent reporters, immunotherapies or cell surface receptors among many other applications.
Conclusion
Decades of research have identified means by which cancer initiation, progression or maintenance is fueled by alterations to the process of RNA splicing. While functional genomic studies continue to dissect the causal links between altered RNA splicing and cancer, a burgeoning toolbox of chemicals is being developed that may provide new means to probe and harness altered RNA splicing in cancer. Even if drugs that perturb splicing do not have direct therapeutic impact on their own, it will be exciting to identify if the enzymatic changes to splicing induced by these agents can be leveraged for novel uses. For example, exciting recent work used splicing regulation induced by the oral drug branaplam (LMI070) to regulate expression of a gene therapy vector in vivo137 (Fig. 4c). In addition, one current major area of interest is understanding whether alterations in RNA splicing, either derived from cancer-specific changes to the splicing process or via drugs that perturb splicing, can generate therapeutically meaningful novel antigens, with relevance for cellular immunotherapies (such as T cells bearing transgenic T cell antigen receptors or chimeric antigen receptor T cells against the misspliced products). Furthermore, it will be important to extend these efforts from RNA splicing in cancer to other abundant and functionally important RNA-processing enzymatic processes, such as RNA polyadenylation and NMD.
Acknowledgements
R.F.S. was supported, in part, by MSK-ICTTP T32-CA009207 and an American Society of Hematology Research Training Award for Fellows. O.A.-W. is supported by the Leukemia & Lymphoma Society, NIH R01s CA251138, HL128239 and CA242020, NIH/NCI 1P50 254838-01 and the Edward P. Evans MDS Foundation.
Footnotes
Competing interests
O.A.-W. has served as a consultant for H3B Biomedicine, Foundation Medicine, Inc., Merck, Prelude Therapeutics and Janssen and is on the Scientific Advisory Board of Envisagenics, Inc., AIChemy, Harmonic Discovery, Inc., and Pfizer Boulder. O.A.-W. has received prior research funding from H3B Biomedicine, Nurix Therapeutics and LOXO Oncology unrelated to the current manuscript. The remaining authors declare no competing interests.
Additional information
Peer review information Nature Cancer thanks James Manley and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Reprints and permissions information is available at www.nature.com/reprints.
References
- 1.Nilsen TW & Graveley BR Expansion of the eukaryotic proteome by alternative splicing. Nature 463, 457–463 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang ET et al. Alternative isoform regulation in human tissue transcriptomes. Nature 456, 470–476 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brody E & Abelson J The ‘spliceosome’: yeast pre-messenger RNA associates with a 40S complex in a splicing-dependent reaction. Science 228, 963–967 (1985). [DOI] [PubMed] [Google Scholar]
- 4.Will CL & Luhrmann R Spliceosome structure and function. Cold Spring Harb. Perspect. Biol. 3, a003707 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.El Marabti E, Malek J & Younis I Minor intron splicing from basic science to disease. Int. J. Mol. Sci. 22, 6062 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Agafonov DE et al. Molecular architecture of the human U4/U6.U5 tri-snRNP. Science 351, 1416–1420 (2016). [DOI] [PubMed] [Google Scholar]
- 7.Fica SM Cryo-EM snapshots of the human spliceosome reveal structural adaptions for splicing regulation. Curr. Opin. Struct. Biol. 65, 139–148 (2020). [DOI] [PubMed] [Google Scholar]
- 8.Nguyen TH et al. The architecture of the spliceosomal U4/U6.U5 tri-snRNP. Nature 523, 47–52 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pan Q, Shai O, Lee LJ, Frey BJ & Blencowe BJ Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat. Genet. 40, 1413–1415 (2008). [DOI] [PubMed] [Google Scholar]
- 10.Gerstein MB et al. Comparative analysis of the transcriptome across distant species. Nature 512, 445–448 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Braberg H et al. From structure to systems: high-resolution, quantitative genetic analysis of RNA polymerase II. Cell 154, 775–788 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Howard JM & Sanford JR The RNAissance family: SR proteins as multifaceted regulators of gene expression. Wiley Interdiscip. Rev. RNA 6, 93–110 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Thibault PA et al. hnRNP A/B proteins: an encyclopedic assessment of their roles in homeostasis and disease. Biology 10, 712 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Van Nostrand EL et al. A large-scale binding and functional map of human RNA-binding proteins. Nature 583, 711–719 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schwerk C & Schulze-Osthoff K Regulation of apoptosis by alternative pre-mRNA splicing. Mol. Cell 19, 1–13 (2005). [DOI] [PubMed] [Google Scholar]
- 16.Jiang ZH, Zhang WJ, Rao Y & Wu JY Regulation of Ich-1 pre-mRNA alternative splicing and apoptosis by mammalian splicing factors. Proc. Natl Acad. Sci. USA 95, 9155–9160 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wu JY, Tang H & Havlioglu N Alternative pre-mRNA splicing and regulation of programmed cell death. Prog. Mol. Subcell. Biol. 31, 153–185 (2003). [DOI] [PubMed] [Google Scholar]
- 18.Kedzierska H & Piekielko-Witkowska A Splicing factors of SR and hnRNP families as regulators of apoptosis in cancer. Cancer Lett. 396, 53–65 (2017). [DOI] [PubMed] [Google Scholar]
- 19.Boise LH et al. bcl-x, a bcl-2-related gene that functions as a dominant regulator of apoptotic cell death. Cell 74, 597–608 (1993). [DOI] [PubMed] [Google Scholar]
- 20.Dou Z et al. Aberrant Bcl-x splicing in cancer: from molecular mechanism to therapeutic modulation. J. Exp. Clin. Cancer Res. 40, 194 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim JH et al. MCL-1ES, a novel variant of MCL-1, associates with MCL-1L and induces mitochondrial cell death. FEBS Lett. 583, 2758–2764 (2009). [DOI] [PubMed] [Google Scholar]
- 22.Supek F, Minana B, Valcarcel J, Gabaldon T & Lehner B Synonymous mutations frequently act as driver mutations in human cancers. Cell 156, 1324–1335 (2014). [DOI] [PubMed] [Google Scholar]
- 23.Jung H et al. Intron retention is a widespread mechanism of tumor-suppressor inactivation. Nat. Genet. 47, 1242–1248 (2015). [DOI] [PubMed] [Google Scholar]
- 24.Yoshida K et al. Frequent pathway mutations of splicing machinery in myelodysplasia. Nature 478, 64–69 (2011). [DOI] [PubMed] [Google Scholar]
- 25.Papaemmanuil E et al. Somatic SF3B1 mutation in myelodysplasia with ring sideroblasts. N. Engl. J. Med. 365, 1384–1395 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang L et al. SF3B1 and other novel cancer genes in chronic lymphocytic leukemia. N. Engl. J. Med. 365, 2497–2506 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Graubert TA et al. Recurrent mutations in the U2AF1 splicing factor in myelodysplastic syndromes. Nat. Genet. 44, 53–57 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Harbour JW et al. Recurrent mutations at codon 625 of the splicing factor SF3B1 in uveal melanoma. Nat. Genet. 45, 133–135 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Quesada V et al. Exome sequencing identifies recurrent mutations of the splicing factor SF3B1 gene in chronic lymphocytic leukemia. Nat. Genet. 44, 47–52 (2011). [DOI] [PubMed] [Google Scholar]
- 30.Martin M et al. Exome sequencing identifies recurrent somatic mutations in EIF1AX and SF3B1 in uveal melanoma with disomy 3. Nat. Genet. 45, 933–936 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Karni R et al. The gene encoding the splicing factor SF2/ASF is a proto-oncogene. Nat. Struct. Mol. Biol. 14, 185–193 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Anczukow O et al. The splicing factor SRSF1 regulates apoptosis and proliferation to promote mammary epithelial cell transformation. Nat. Struct. Mol. Biol. 19, 220–228 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Danan-Gotthold M et al. Identification of recurrent regulated alternative splicing events across human solid tumors. Nucleic Acids Res. 43, 5130–5144 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen L, Tovar-Corona JM & Urrutia AO Increased levels of noisy splicing in cancers, but not for oncogene-derived transcripts. Hum. Mol. Genet. 20, 4422–4429 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kahles A et al. Comprehensive analysis of alternative splicing across tumors from 8,705 patients. Cancer Cell 34, 211–224 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Seiler M et al. Somatic mutational landscape of splicing factor genes and their functional consequences across 33 cancer types. Cell Rep. 23, 282–296 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ellis MJ et al. Whole-genome analysis informs breast cancer response to aromatase inhibition. Nature 486, 353–360 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.The Cancer Genome Atlas Network. Comprehensive molecular portraits of human breast tumours. Nature 490, 61–70 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Biankin AV et al. Pancreatic cancer genomes reveal aberrations in axon guidance pathway genes. Nature 491, 399–405 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Imielinski M et al. Mapping the hallmarks of lung adenocarcinoma with massively parallel sequencing. Cell 150, 1107–1120 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chen S, Benbarche S & Abdel-Wahab O Splicing factor mutations in hematologic malignancies. Blood 138, 599–612 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cretu C et al. Molecular architecture of SF3b and structural consequences of its cancer-related mutations. Mol. Cell 64, 307–319 (2016). [DOI] [PubMed] [Google Scholar]
- 43.Teng T et al. Splicing modulators act at the branch point adenosine binding pocket defined by the PHF5A–SF3b complex. Nat. Commun. 8, 15522 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Malcovati L et al. SF3B1-mutant MDS as a distinct disease subtype: a proposal from the International Working Group for the Prognosis of MDS. Blood 136, 157–170 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dalton WB et al. The K666N mutation in SF3B1 is associated with increased progression of MDS and distinct RNA splicing. Blood Adv. 4, 1192–1196 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Darman RB et al. Cancer-associated SF3B1 hotspot mutations induce cryptic 3′ splice site selection through use of a different branch point. Cell Rep. 13, 1033–1045 (2015). [DOI] [PubMed] [Google Scholar]
- 47.DeBoever C et al. Transcriptome sequencing reveals potential mechanism of cryptic 3′ splice site selection in SF3B1-mutated cancers. PLoS Comput. Biol. 11, e1004105 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Boultwood J et al. The role of the iron transporter ABCB7 in refractory anemia with ring sideroblasts. PLoS ONE 3, e1970 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nikpour M et al. The transporter ABCB7 is a mediator of the phenotype of acquired refractory anemia with ring sideroblasts. Leukemia 27, 889–896 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Inoue D et al. Spliceosomal disruption of the non-canonical BAF complex in cancer. Nature 574, 432–436 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Clough CA et al. Coordinated mis-splicing of TMEM14C and ABCB7 causes ring sideroblast formation in SF3B1-mutant myelodysplastic syndrome. Blood 139, 2038–2048 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lieu YK et al. SF3B1 mutant-induced missplicing of MAP3K7 causes anemia in myelodysplastic syndromes. Proc. Natl Acad. Sci. USA 119, e2111703119 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhang J et al. Disease-causing mutations in SF3B1 alter splicing by disrupting interaction with SUGP1. Mol. Cell 76, 82–95 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Liu Z et al. Pan-cancer analysis identifies mutations in SUGP1 that recapitulate mutant SF3B1 splicing dysregulation. Proc. Natl Acad. Sci. USA 117, 10305–10312 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gozani O, Potashkin J & Reed R A potential role for U2AF–SAP 155 interactions in recruiting U2 snRNP to the branch site. Mol. Cell. Biol. 18, 4752–4760 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Merendino L, Guth S, Bilbao D, Martinez C & Valcarcel J Inhibition of msl-2 splicing by sex-lethal reveals interaction between U2AF35 and the 3′ splice site AG. Nature 402, 838–841 (1999). [DOI] [PubMed] [Google Scholar]
- 57.Wu S, Romfo CM, Nilsen TW & Green MR Functional recognition of the 3′ splice site AG by the splicing factor U2AF35. Nature 402, 832–835 (1999). [DOI] [PubMed] [Google Scholar]
- 58.Reed R The organization of 3′ splice-site sequences in mammalian introns. Genes Dev. 3, 2113–2123 (1989). [DOI] [PubMed] [Google Scholar]
- 59.Haferlach T et al. Landscape of genetic lesions in 944 patients with myelodysplastic syndromes. Leukemia 28, 241–247 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Papaemmanuil E et al. Clinical and biological implications of driver mutations in myelodysplastic syndromes. Blood 122, 3616–3627 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ilagan JO et al. U2AF1 mutations alter splice site recognition in hematological malignancies. Genome Res. 25, 14–26 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wheeler EC et al. Integrative RNA-omics discovers GNAS alternative splicing as a phenotypic driver of splicing factor-mutant neoplasms. Cancer Discov. 12, 836–855 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Smith MA et al. U2AF1 mutations induce oncogenic IRAK4 isoforms and activate innate immune pathways in myeloid malignancies. Nat. Cell Biol. 21, 640–650 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Thol F et al. Frequency and prognostic impact of mutations in SRSF2, U2AF1, and ZRSR2 in patients with myelodysplastic syndromes. Blood 119, 3578–3584 (2012). [DOI] [PubMed] [Google Scholar]
- 65.Desai P et al. Somatic mutations precede acute myeloid leukemia years before diagnosis. Nat. Med. 24, 1015–1023 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Graveley BR & Maniatis T Arginine/serine-rich domains of SR proteins can function as activators of pre-mRNA splicing. Mol. Cell 1, 765–771 (1998). [DOI] [PubMed] [Google Scholar]
- 67.Liu HX, Chew SL, Cartegni L, Zhang MQ & Krainer AR Exonic splicing enhancer motif recognized by human SC35 under splicing conditions. Mol. Cell. Biol. 20, 1063–1071 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Schaal TD & Maniatis T Multiple distinct splicing enhancers in the protein-coding sequences of a constitutively spliced pre-mRNA. Mol. Cell. Biol. 19, 261–273 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Daubner GM, Clery A, Jayne S, Stevenin J & Allain FH A syn-anti conformational difference allows SRSF2 to recognize guanines and cytosines equally well. EMBO J. 31, 162–174 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kim E et al. SRSF2 mutations contribute to myelodysplasia by mutant-specific effects on exon recognition. Cancer Cell 27, 617–630 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zhang J et al. Disease-associated mutation in SRSF2 misregulates splicing by altering RNA-binding affinities. Proc. Natl Acad. Sci. USA 112, E4726–4734 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yoshimi A et al. Coordinated alterations in RNA splicing and epigenetic regulation drive leukaemogenesis. Nature 574, 273–277 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Madan V et al. Aberrant splicing of U12-type introns is the hallmark of ZRSR2 mutant myelodysplastic syndrome. Nat. Commun. 6, 6042 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Inoue D et al. Minor intron retention drives clonal hematopoietic disorders and diverse cancer predisposition. Nat. Genet. 53, 707–718 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Burge CB, Padgett RA & Sharp PA Evolutionary fates and origins of U12-type introns. Mol. Cell 2, 773–785 (1998). [DOI] [PubMed] [Google Scholar]
- 76.Tarn WY, Yario TA & Steitz JA U12 snRNA in vertebrates: evolutionary conservation of 5′ sequences implicated in splicing of pre-mRNAs containing a minor class of introns. RNA 1, 644–656 (1995). [PMC free article] [PubMed] [Google Scholar]
- 77.Suzuki H et al. Recurrent noncoding U1 snRNA mutations drive cryptic splicing in SHH medulloblastoma. Nature 574, 707–711 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Shuai S et al. The U1 spliceosomal RNA is recurrently mutated in multiple cancers. Nature 574, 712–716 (2019). [DOI] [PubMed] [Google Scholar]
- 79.Bousquets-Munoz P et al. PanCancer analysis of somatic mutations in repetitive regions reveals recurrent mutations in snRNA U2. NPJ Genom. Med. 7, 19 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kurtovic-Kozaric A et al. PRPF8 defects cause missplicing in myeloid malignancies. Leukemia 29, 126–136 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Dvinge H, Kim E, Abdel-Wahab O & Bradley RK RNA splicing factors as oncoproteins and tumour suppressors. Nat. Rev. Cancer 16, 413–430 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Hernandez J et al. Tumor suppressor properties of the splicing regulatory factor RBM10. RNA Biol. 13, 466–472 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Bechara EG, Sebestyen E, Bernardis I, Eyras E & Valcarcel J RBM5, 6, and 10 differentially regulate NUMB alternative splicing to control cancer cell proliferation. Mol. Cell 52, 720–733 (2013). [DOI] [PubMed] [Google Scholar]
- 84.Ghigna C et al. Cell motility is controlled by SF2/ASF through alternative splicing of the Ron protooncogene. Mol. Cell 20, 881–890 (2005). [DOI] [PubMed] [Google Scholar]
- 85.Goncalves V et al. Phosphorylation of SRSF1 by SRPK1 regulates alternative splicing of tumor-related Rac1b in colorectal cells. RNA 20, 474–482 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Tacke R, Chen Y & Manley JL Sequence-specific RNA binding by an SR protein requires RS domain phosphorylation: creation of an SRp40-specific splicing enhancer. Proc. Natl Acad. Sci. USA 94, 1148–1153 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Xiao SH & Manley JL Phosphorylation–dephosphorylation differentially affects activities of splicing factor ASF/SF2. EMBO J. 17, 6359–6367 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Cao S et al. Discovery of driver non-coding splice-site-creating mutations in cancer. Nat. Commun. 11, 5573 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Mian SA et al. SF3B1 mutant MDS-initiating cells may arise from the haematopoietic stem cell compartment. Nat. Commun. 6, 10004 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Lee SC et al. Synthetic lethal and convergent biological effects of cancer-associated spliceosomal gene mutations. Cancer Cell 34, 225–241 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Taylor J et al. Single-cell genomics reveals the genetic and molecular bases for escape from mutational epistasis in myeloid neoplasms. Blood 136, 1477–1486 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Zhou Q et al. A chemical genetics approach for the functional assessment of novel cancer genes. Cancer Res. 75, 1949–1958 (2015). [DOI] [PubMed] [Google Scholar]
- 93.Lee SC et al. Modulation of splicing catalysis for therapeutic targeting of leukemia with mutations in genes encoding spliceosomal proteins. Nat. Med. 22, 672–678 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Fei DL et al. Wild-type U2AF1 antagonizes the splicing program characteristic of U2AF1-mutant tumors and is required for cell survival. PLoS Genet. 12, e1006384 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Kaida D et al. Spliceostatin A targets SF3b and inhibits both splicing and nuclear retention of pre-mRNA. Nat. Chem. Biol. 3, 576–583 (2007). [DOI] [PubMed] [Google Scholar]
- 96.Fan L, Lagisetti C, Edwards CC, Webb TR & Potter PM Sudemycins, novel small molecule analogues of FR901464, induce alternative gene splicing. ACS Chem. Biol. 6, 582–589 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Kotake Y et al. Splicing factor SF3b as a target of the antitumor natural product pladienolide. Nat. Chem. Biol. 3, 570–575 (2007). [DOI] [PubMed] [Google Scholar]
- 98.Hasegawa M et al. Identification of SAP155 as the target of GEX1A (herboxidiene), an antitumor natural product. ACS Chem. Biol. 6, 229–233 (2011). [DOI] [PubMed] [Google Scholar]
- 99.Yokoi A et al. Biological validation that SF3b is a target of the antitumor macrolide pladienolide. FEBS J. 278, 4870–4880 (2011). [DOI] [PubMed] [Google Scholar]
- 100.Cretu C et al. Structural basis of splicing modulation by antitumor macrolide compounds. Mol. Cell 70, 265–273 (2018). [DOI] [PubMed] [Google Scholar]
- 101.Eskens FA et al. Phase I pharmacokinetic and pharmacodynamic study of the first-in-class spliceosome inhibitor E7107 in patients with advanced solid tumors. Clin. Cancer Res. 19, 6296–6304 (2013). [DOI] [PubMed] [Google Scholar]
- 102.Hong DS et al. A phase I, open-label, single-arm, dose-escalation study of E7107, a precursor messenger ribonucleic acid (pre-mRNA) splicesome inhibitor administered intravenously on days 1 and 8 every 21 days to patients with solid tumors. Invest. New Drugs 32, 436–444 (2014). [DOI] [PubMed] [Google Scholar]
- 103.Crews LA et al. RNA splicing modulation selectively impairs leukemia stem cell maintenance in secondary human AML. Cell Stem Cell 19, 599–612 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Seiler M et al. H3B-8800, an orally available small-molecule splicing modulator, induces lethality in spliceosome-mutant cancers. Nat. Med. 24, 497–504 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Steensma DP et al. Phase I first-in-human dose escalation study of the oral SF3B1 modulator H3B-8800 in myeloid neoplasms. Leukemia 35, 3542–3550 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Ting TC et al. Aryl sulfonamides degrade RBM39 and RBM23 by recruitment to CRL4–DCAF15. Cell Rep. 29, 1499–1510 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Han T et al. Anticancer sulfonamides target splicing by inducing RBM39 degradation via recruitment to DCAF15. Science 356, eaal3755 (2017). [DOI] [PubMed] [Google Scholar]
- 108.Du X et al. Structural basis and kinetic pathway of RBM39 recruitment to DCAF15 by a sulfonamide molecular glue E7820. Structure 27, 1625–1633 (2019). [DOI] [PubMed] [Google Scholar]
- 109.Faust TB et al. Structural complementarity facilitates E7820-mediated degradation of RBM39 by DCAF15. Nat. Chem. Biol. 16, 7–14 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Bussiere DE et al. Structural basis of indisulam-mediated RBM39 recruitment to DCAF15 E3 ligase complex. Nat. Chem. Biol. 16, 15–23 (2020). [DOI] [PubMed] [Google Scholar]
- 111.Wang E et al. Targeting an RNA-binding protein network in acute myeloid leukemia. Cancer Cell 35, 369–384 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Assi R et al. Final results of a phase 2, open-label study of indisulam, idarubicin, and cytarabine in patients with relapsed or refractory acute myeloid leukemia and high-risk myelodysplastic syndrome. Cancer 124, 2758–2765 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Jagtap PKA et al. Identification of phenothiazine derivatives as UHM-binding inhibitors of early spliceosome assembly. Nat. Commun. 11, 5621 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Kobayashi A et al. Identification of a small molecule splicing inhibitor targeting UHM domains. FEBS J. 289, 682–698 (2021). [DOI] [PubMed] [Google Scholar]
- 115.Chatrikhi R et al. A synthetic small molecule stalls pre-mRNA splicing by promoting an early-stage U2AF2–RNA complex. Cell Chem. Biol. 28, 1145–1157 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Mathew R et al. Phosphorylation of human PRP28 by SRPK2 is required for integration of the U4/U6-U5 tri-snRNP into the spliceosome. Nat. Struct. Mol. Biol. 15, 435–443 (2008). [DOI] [PubMed] [Google Scholar]
- 117.Edmond V et al. Acetylation and phosphorylation of SRSF2 control cell fate decision in response to cisplatin. EMBO J. 30, 510–523 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Musiani D et al. Proteomics profiling of arginine methylation defines PRMT5 substrate specificity. Sci. Signal. 12, eaat8388 (2019). [DOI] [PubMed] [Google Scholar]
- 119.Fong JY et al. Therapeutic targeting of RNA splicing catalysis through inhibition of protein arginine methylation. Cancer Cell 36, 194–209 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Radzisheuskaya A et al. PRMT5 methylome profiling uncovers a direct link to splicing regulation in acute myeloid leukemia. Nat. Struct. Mol. Biol. 26, 999–1012 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Finkel RS et al. Nusinersen versus sham control in infantile-onset spinal muscular atrophy. N. Engl. J. Med. 377, 1723–1732 (2017). [DOI] [PubMed] [Google Scholar]
- 122.McDonald CM et al. Open-label evaluation of eteplirsen in patients with Duchenne muscular dystrophy amenable to exon 51 skipping: PROMOVI trial. J. Neuromuscul. Dis. 8, 989–1001 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Walker AR et al. Phase 3 randomized trial of chemotherapy with or without oblimersen in older AML patients: CALGB 10201 (Alliance). Blood Adv. 5, 2775–2787 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Sha D et al. Tumor mutational burden as a predictive biomarker in solid tumors. Cancer Discov. 10, 1808–1825 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Marabelle A et al. Association of tumour mutational burden with outcomes in patients with advanced solid tumours treated with pembrolizumab: prospective biomarker analysis of the multicohort, open-label, phase 2 KEYNOTE-158 study. Lancet Oncol. 21, 1353–1365 (2020). [DOI] [PubMed] [Google Scholar]
- 126.Le DT et al. Mismatch repair deficiency predicts response of solid tumors to PD-1 blockade. Science 357, 409–413 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Turajlic S et al. Insertion-and-deletion-derived tumour-specific neoantigens and the immunogenic phenotype: a pan-cancer analysis. Lancet Oncol. 18, 1009–1021 (2017). [DOI] [PubMed] [Google Scholar]
- 128.Rizvi NA et al. Cancer immunology. Mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer. Science 348, 124–128 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Jayasinghe RG et al. Systematic analysis of splice-site-creating mutations in cancer. Cell Rep. 23, 270–281 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Laumont CM et al. Noncoding regions are the main source of targetable tumor-specific antigens. Sci. Transl. Med. 10, eaau5516 (2018). [DOI] [PubMed] [Google Scholar]
- 131.Smart AC et al. Intron retention is a source of neoepitopes in cancer. Nat. Biotechnol. 36, 1056–1058 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Wang RF et al. A breast and melanoma-shared tumor antigen: T cell responses to antigenic peptides translated from different open reading frames. J. Immunol. 161, 3598–3606 (1998). [PubMed] [Google Scholar]
- 133.Robbins PF et al. The intronic region of an incompletely spliced gp100 gene transcript encodes an epitope recognized by melanoma-reactive tumor-infiltrating lymphocytes. J. Immunol. 159, 303–308 (1997). [PubMed] [Google Scholar]
- 134.Ouspenskaia T et al. Unannotated proteins expand the MHC-I-restricted immunopeptidome in cancer. Nat. Biotechnol. 40, 209–217 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Lu SX et al. Pharmacologic modulation of RNA splicing enhances anti-tumor immunity. Cell 184, 4032–4047 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.North K et al. Synthetic introns enable splicing factor mutation-dependent targeting of cancer cells. Nat. Biotechnol. 10.1038/s41587-022-01224-2 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Monteys AM et al. Regulated control of gene therapies by drug-induced splicing. Nature 596, 291–295 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Steijger T et al. Assessment of transcript reconstruction methods for RNA-seq. Nat. Methods 10, 1177–1184 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Soneson C, Love MI & Robinson MD Differential analyses for RNA-seq: transcript-level estimates improve gene-level inferences. F1000Res 4, 1521 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Zhang C, Zhang B, Lin LL & Zhao S Evaluation and comparison of computational tools for RNA-seq isoform quantification. BMC Genomics 18, 583 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.De Paoli-Iseppi R, Gleeson J & Clark MB Isoform age—splice isoform profiling using long-read technologies. Front. Mol. Biosci. 8, 711733 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Simpson JT et al. Detecting DNA cytosine methylation using nanopore sequencing. Nat. Methods 14, 407–410 (2017). [DOI] [PubMed] [Google Scholar]
- 143.Lorenz DA, Sathe S, Einstein JM & Yeo GW Direct RNA sequencing enables m6A detection in endogenous transcript isoforms at base-specific resolution. RNA 26, 19–28 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Chen SY, Deng F, Jia X, Li C & Lai SJ A transcriptome atlas of rabbit revealed by PacBio single-molecule long-read sequencing. Sci Rep. 7, 7648 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Kim JA et al. Genome-wide transcriptome profiling of the medicinal plant Zanthoxylum planispinum using a single-molecule direct RNA sequencing approach. Genomics 111, 973–979 (2019). [DOI] [PubMed] [Google Scholar]
- 146.Roach NP et al. The full-length transcriptome of C. elegans using direct RNA sequencing. Genome Res. 30, 299–312 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Lagarde J et al. High-throughput annotation of full-length long noncoding RNAs with capture long-read sequencing. Nat. Genet. 49, 1731–1740 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Gupta I et al. Single-cell isoform RNA sequencing characterizes isoforms in thousands of cerebellar cells. Nat. Biotechnol. 36, 1197–1202 (2018). [DOI] [PubMed] [Google Scholar]
- 149.Singh M et al. High-throughput targeted long-read single cell sequencing reveals the clonal and transcriptional landscape of lymphocytes. Nat. Commun. 10, 3120 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]