Abstract
The present study aimed to explore latent markers for identifying primary Sjogren's syndrome (pSS), as well as their expression and molecular mechanism. Hub inducible T cell co-stimulator genes were retrieved from the Gene Expression Omnibus. A total of 95 patients with pSS and 68 healthy individuals from Affiliated Hospital of Nantong University (Nantong, China) were included in the study. The expression of inducible T cell co-stimulator (ICOS) in whole blood and saliva was evaluated using ELISA. Western blotting was performed to investigate aquaporin 5 (AQP5) protein expression, as well as the inflammatory effects of ICOS in patients with pSS compared with healthy individuals. Differentially expressed mRNAs were analyzed in whole blood (GSE84844) and salivary gland (GSE40611) of patients with pSS. In total, 15 hub genes were identified, among which ICOS was indicated to serve a role in the most pivotal immunity pathways. pSS was markedly associated with inflammatory pathways. Results from the present study found that ICOS was upregulated in the salivary gland, whole blood and saliva of patients with pSS. Salivary weight was negatively correlated with the levels of ICOS in the saliva of patients with pSS. The expression of AQP5 was markedly lower in patients with pSS. The expression of AQP5 was negatively associated with ICOS. Compared with that of healthy individuals, the expression of ICOS and inflammatory factors was higher and the expression of AQP5 was lower in pSS patients as assessed by western blotting. These data demonstrated that ICOS may affect AQP5 expression by promoting inflammation in the salivary glands of patients with pSS.
Keywords: inducible T cell co-stimulator, saliva, primary Sjögren's syndrome
Introduction
Primary Sjögren's syndrome (pSS) is a systemic autoimmune disease characterized by the infiltration of periductal lymph cells in the lacrimal and salivary gland tissues (1). Abnormal activation of T and B cells is vital for the progression of pSS. T cell stimulative and differentiative activities require two signals. The first signal being T cell receptor recognition of the major histocompatibility complex on antigen-presenting cells (APCs) (2); the second signal is a co-stimulatory signal mediated by auxiliary biomolecules on APCs (3–6). Costimulatory molecule has importance in treatment of autoimmune disease, since manipulating co-stimulatory signals may offer a way to reinforce or halt immune responses. B7/CD28/CTLA4 pathway has the ability to both positively and negatively regulate immune responses. The CD28/CTLA4/B7 pathway signaling pathway is the most explored T cell co-stimulatory pathway and is pivotal for T cell stimulation and tolerance (7).
Inducible T cell co-stimulator (ICOS) is a component of the CD28 superfamily, which is predominantly expressed on the surface of stimulated T cells (8,9). ICOS supports CD4+ T follicular helper (Tfh) cell differentiation and function during the germinal center (GC) response (10–13). As such, the absence of ICOS or ICOS ligand in humans and mice induces serious abnormalities (14–18). It is known that ICOS is predominantly expressed on both follicular and regulatory follicular T helper cells (Tfh and Tfr cells) which regulate antagonistically the quantity and quality of humoral immunity (19,20). The expression of ICOS lead to dysfunction of T-cell in SLE patients (21) Researcher first focused on T cells and evaluated the key activation molecules, HLA-DR, and costimulatory molecules such as ICOS on CD3+ T cells (22). The ICOS expression in SLE was correlated to the anti-DNA antibodies. Enhanced expression of ICOS-expressing CD3+ T cells may suggest that the systemic lupus erythematosus (SLE) patients are at a more severe inflammatory status (22). Previous studies have reported increased ICOS protein and mRNA expression levels in the peripheral blood of patients with pSS (23,24). The present study found that the expression of ICOS was shown to affect saliva weight and aquaporin 5 (AQP5) expression; however, the specific downstream signal transduction constituents involved remain unknown.
Previous studies indicate that ICOS is pivotal for stimulating the PI3K/Akt/mTOR signaling pathway (25,26), which is also found to be associated with inflammatory activation (27). Sicca symptoms, including dry eyes and mouth, are characteristic features of pSS. AQP5 affects saliva secretion; it is distributed in the salivary gland tissues and functions as a water channel in the secretory glands (28). The expression mechanism of co-stimulatory molecules and their receptors in pSS is still unclear, and further research is needed. It was hypothesized that the expression level and equilibrium of ICOS, as well as relevant expression of AQP5, may markedly affect the sicca symptoms identified in pSS. Thus, the present study aimed to explore whether ICOS affected AQP5 expression by promoting salivary gland inflammation and to offer a prospective candidate for pSS diagnosis.
Materials and methods
Dataset access and information
The Gene Expression Omnibus (GEO; http://www.ncbi.nlm.nih.gov/gds) datasets GSE40611 and GSE84844 were used. These datasets came from the GPL570 (HG-U133_Plus_2) Affymetrix Human Genome U133 Plus 2.0 Array and were analyzed by R packages (http://www.R-project.org/). GSE40611 comprises 17 healthy salivary gland samples and 18 pSS salivary gland samples; GSE84844 contains 30 normal whole blood samples and 30 pSS whole blood. The Perl command ‘strawberry-perl-5.32.1.1-64bit’ (strawberryperl.com/releases.html) was used to transform the genetic probe IDs in the matrix files into the genetic symbols in the dataset files to acquire a matrix file of formal symbols. All datasets were normalized using the limma R package (version 4.1.0 for Windows, R Core Team, Vienna, Austria http://www.R-project.org/) (29). The genetic expression information was log2 transformed.
Dataset analyses
The Perl (strawberry-perl-5.32.1.1-64bit) command is used to convert the genetic probe IDs in the matrix document to the genetic symbols of the platform to obtain the matrix document containing the official symbols (https://strawberryperl.com/releases.html strawberry perl.com project).
The pSS expression datasets were standardized by impute R package and limma R package (R for Windows 4.1.0 Setup and http://www.rstudio.com). Differentially expressed genes (DEGs) between patients and healthy individuals were identified by volcano plot (−https://www.rstudio.com) using the limma R package. All P-value (false discovery rate) <0.05) and |log2 (FC)| >0.75 were considered to indicate a statistically significant difference by comparing pSS with healthy controls using the limma R package.
Functional enrichment analyses
Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) analyses were completed based on the GEO database (30–32). The outcomes of the R analyses were submitted to determine the putative underlying roles of the selected genes. False discovery rate <0.05 and P<0.05 were considered to indicate a statistically significant difference.
Protein-protein interaction (PPI) network establishment
The identified DEGs were uploaded to the Search Tool for the Retrieval of Interacting Genes/Proteins (STRING) database (https://string-db.org) (33). The interaction score was set to 0.8. The hub genes were identified and Cytoscape (Cytoscape_3_8_2_windows_64bit; http://cytoscape.org/download.html) was used to plot the network diagram; nodal points denoted a gene or protein (34); the number of edges between nodal points denoted the molecular interplay.
Patients
A total of 95 patients with pSS and 68 healthy individuals admitted to Affiliated Hospital of Nantong University between September 2017 and September 2018 who met the inclusion criteria (as described below) were selected by the random number table method. The patients with pSS were diagnosed according to the 2016 American-European Consensus Group SS categorization standards (35). Salivary gland biopsy samples were harvested, and the pathological status of 95 pSS patients was evaluated to determine the pathological findings. Donors bearing inflammatory diseases, cancer or contagious diseases were excluded from the study. The study was approved by the ethical board of Affiliated Hospital of Nantong University (Jiangsu, China; approval no. 2017-K003). All patients were >18 years old and provided written informed consent. Features of healthy individuals and pSS patients are given in Table I.
Table I.
Clinicopathological characteristics of healthy control individuals and patients with pSS.
| Clinicopathological characteristic | Patients with pSS | Healthy control patients | P-value |
|---|---|---|---|
| Cases, n | 95 | 68 | |
| Sex, Female | 95 (100%) | 68 (100%) | |
| Age, yearsa | 48.82±1.33 | 47.31±1.425 | 0.4472 |
| Disease duration (years)a | 1.54±0.221 | 0 | NA |
| ESSDAIc | 4 (0–7) | 0 (0–2) | <0.001 |
| Number of cariesc | 4 (1–8) | 0 | NA |
| Autoantibody positivity: | |||
| Anti-SSA and/or anti-SSBb | 95 (100) | 0 (0) | NA |
| Rheumatoid factorb | <0.001 | ||
| ≤20 | 31 (33%) | 68 (100%) | |
| ≥20 | 64 (67%) | 0 (0%) |
Mean ± standard error of the mean, analyzed by independent samples t-test;
number (percentage), analyzed by χ2 test;
Median (interquartile range), analyzed by the Mann-Whitney U test. C3, complement 3; C4, complement 4; ESR, erythrocyte sedimentation rate; ESSDAI, EULAR primary Sjögren's syndrome disease activity; ESSPRI, EULAR primary Sjögren's syndrome patient-reported indexes; NA, not applicable; pSS, primary Sjögren's syndrome.
Inclusion and exclusion criteria of patients with pSS
The inclusion criteria were as follows: Diagnosis of pSS, >18 years old, no known cognitive defects and of Chinese nationality. The exclusion criteria included: Secondary Sjögren's syndrome, physiopsychological problems (psychiatric disorders) that may affect the results, pulmonary injury not associated with pSS (such as lung infections, asthmatic disease, persistent obstructive lung illness, lung tuberculosis, bronchodilation and pulmonary carcinoma), interstitial lung disease induced by organic matter or pneumoconiosis, long-term utilization of medicines causing lung fibrosis, and persistent cardiac, hepatic or renal function disorders.
Disease activity
Disease activity was assessed by the European League Against Rheumatism Sjögren's Syndrome Disease Activity Index (ESSDAI) (36). The ESSDAI includes measures systematic activity and 12 domains. Each domain is divided into 3–4 levels (0, no activity; 1, low activity; 2, moderate activity; 3, high activity). According to the ESSDAI, low activity, <5; moderate activity, 5–13; and high activity, >13 (37).
Healthy individuals' inclusion criteria
The inclusion criteria were as follows: >18 years old, no known cognitive defects, not known any disease and of Chinese nationality.
Saliva collection
Patients were not allowed to stimulate the salivary flow. Patients were asked to hold the tongue against the roof of the mouth (the palate) until there was adequate saliva. Salivary samples were pooled and harvested in specimen cups for 300 sec. Between 9:00 and 12:00 a.m., salivary sample harvesting was completed to decrease the impact of circadian fluctuation. The salivary specimens were centrifuged at 4,500 × g for 5 min at 4°C and the debris and cells were discarded (38). After centrifugation, the weight of the saliva was measured. Specimens were stored at −80°C until subsequent analyses.
Blood collection
Blood from 95 pSS patient and 68 controls was collected at 9–12 a.m. The patients did not have strenuous exercise and fasting state. Peripheral blood (5 ml) was collected from each patient and controls in the Rheumatic Immunology Department. Blood samples were centrifuged at 978 × g for 1 min at 4°C. The uppermost transparent liquid is extracted and stored at −80°C. All patients and controls signed informed consent.
ELlSA
A human ICOS ELISA kit (XG-K3005; Suzhou Bright Scistar Biotechnology Company Ltd.) was utilized to quantify the ICOS content in saliva and in blood. A human TNF-α ELISA kit (cat. no. DTA00D; R&D Systems, Inc.) was used to measure the levels of TNF-α, according to the manufacturer's instructions. 100 µl blood and 100 µl saliva from 95 pSS patients and 68 controls were incubated in 96-well plates at 37°C for 60 min. Subsequently, the absorbance was measured in a microplate reading device at 450 nm. All experiments were performed in triplicate.
Immunofluorescence analyses
Salivary gland samples were fixed in 4% paraformaldehyde at 4°C for 180 min, followed by incubation at 4°C for 12 h in 30% sucrose. Next, the specimens were placed in optimal cutting temperature compound and sectioned into 5-µm slices. The salivary gland samples were blocked with 0.1% BSA (cat. no. A8010; Beijing Solarbio Science & Technology Co., Ltd.) at 37°C for 60 min and then incubated with antibodies against ICOS (cat. no. ab175401; Abcam; 1:100) at 4°C for 12 h. Subsequently, the sections were incubated with goat anti-rabbit IgG antibody (H+L; cat. no. BA-5000-1.5; Vector Laboratories, Inc.; 1:1,000) for 30 min at room temperature. Staining was developed with red fluorescent dyes (cat. no. FP1494001KT; PerkinElmer, Inc.; 1:3,000) for 10 min at room temperature and DAPI (cat. no. C0065; Beijing Solarbio Science & Technology Co., Ltd.) for 5 min at room temperature. After all incubation steps, the samples were washed with PBS for 5 min. An Olympus BX53 fluorescence microscope (Olympus Corporation) was used to detect the fluorescence expression of samples. Images were analyzed using Cellsens Standard version 1.9 software (Olympus Corporation).
Western blot analysis
Salivary gland tissues were lysed in RIPA lysis buffer (Beyotime Institute of Biotechnology) containing PMSF (Beyotime Institute of Biotechnology). Protein concentrations were determined by BCA assay (Beyotime Institute of Biotechnology). A total of 5 µg protein samples were separated by 10% SDS-PAGE and then transferred to PVDF membranes (Beyotime Institute of Biotechnology). The membranes were blocked with 5% BSA (cat. no. SW3015; Beijing Solarbio Science & Technology Co., Ltd) for 1 h at room temperature and then incubated with the following primary antibodies at 4°C for 12 h: anti-ICOS (cat. no. ab224644; Abcam; 1:1,000), anti-AQP5 (cat. no. ab191061; Abcam; 1:1,000), anti-phosphorylated (p)-PI3K (cat. no. ab278545; Abcam; 1:1,000), anti-p-AKT (cat. no. ab8933; Abcam; 1;1000), anti-p-mTOR (cat. no. ab109268; Abcam; 1:1,000), PI3K (cat. no. ab32089; Abcam, 1:1,000), AKT (cat. no. ab8805; Abcam; 1:1,000), mTOR (cat. no. ab134903; Abcam; 1:1,000), anti-Bcl-2 (cat. no. ab59348; Abcam; 1:1,000) and anti-TNF-α (cat. no. ab183218; Abcam; 1:1,000). GAPDH (cat. no. ab8245, Abcam, 1:1000) was used as an internal control for normalization. Rabbit anti-mouse or goat anti-rabbit IgG H&L at 37°C for 60 min (cat. nos. ab6728 and ab6721, respectively; Abcam, 1:5,000). Proteins were visualized using an Enhanced ECL Chemiluminescence Detection kit (cat. no. E411-03; Vazyme Biotech Co., Ltd.). Densitometric analysis was conducted using Image Lab System version 6.1 (Bio-Rad Laboratories, Inc.). The assays were repeated at least times.
Statistical analysis
An independent samples t-test was conducted to determine differences between two groups. χ2 test was used to assess differences in proportion. SPSS 19.0 software (IBM Corp.) was used for statistical analysis. All experiments were performed in triplicate. Shapiro-Wilk test was utilized to assess the normal distribution of the data. Spearman correlation analysis to determine relationships between variables. Comparisons were made using one-way ANOVA followed by Tukey's honestly significant difference post hoc test. Data are expressed as mean ± SEM. χ2 test was used to assess the differences in proportion. Median (interquartile range), analyzed by the Mann-Whitney U test. P<0.05 was considered to indicate a statistically significant difference.
Results
Identification of DEGs in patients with pSS
GSE40611 genetic expression matrix and corresponding annotated files were acquired from GEO database. The dataset, which included 17 healthy salivary gland and 18 pSS salivary gland was analyzed using the GPL570 Affymetrix Human Genome U133 Plus 2.0 Array. A total of 118 DEGs genes were identified as consistently differentially expressed in pSS, and these are represented using volcano plots (Fig. 1A). Among these, 13 were downregulated and 105 were upregulated. GSE84844 genetic expression matrix and corresponding annotated files were acquired from GEO database. GSE84844 microarray included 30 healthy blood and 30 pSS blood was analyzed using the GPL570 Affymetrix Human Genome U133 Plus 2.0 Array. Similarly, DEGs were identified using volcano plots of the Differentially expressed mRNAs were analyzed in whole blood (GSE84844) (Fig. 1B). Here, a total of 244 genes were identified that were consistently differentially expressed in pSS compared with the control group data. Among these, 5 were downregulated and 239 were upregulated. The GSE40611 and GSE84844 data were merged to establish an intersection of DEGs and 15 overlapping DEGs were found (Fig. 1C), including CXCL10, OAS3, MX1, MS4A1, GZMA, ICOS, CENPK, TCL1A, EVI2A, IFI44L, IFIT1, CD69, FAM26F, CKS2 and TRAT1. Among these, 0 were downregulated and 15 were upregulated.
Figure 1.
Identification of DEGs involved in ICOS expression in patients with pSS. (A) Volcano plots of the Gene Expression Omnibus datasets (A) GSE40611 (B) GSE84844 were used to identify DEGs. Red dots represent upregulated genes; green dots represent downregulated genes; and black dots represent genes with no significant difference according to the. (C) Venn diagram showing the intersection of the DEGs identified in the GSE40611 and GSE84844 datasets. (D) Protein-protein interaction network. (E) GSE40611 data was analyzed by GO enriched functions. The circle shows of GO enrichment data for the biological process (BP) involved in ICOS. (F) GSE40611 databases was analyzed by KEGG enriched functions. The circle shows KEGG pathways involved in ICOS. (G) BP functional enrichment GO terms, KEGG pathways and DEGs involving ICOS participation were obtained using Cytoscape in GSE40611 databases. Red represents KEGG pathway; yellow represents DEGs; and green represent GO enriched functions. DEGs, differentially expressed genes; FC, fold change; GO, Gene Ontology; ICOS, inducible T cell co-stimulator; KEGG, Kyoto Encyclopedia of Genes and Genomes; p.adj., adjusted P-value; pSS, primary Sjogren's syndrome.
The STRING database was used for PPI network analysis of these DEGs, and Cytoscape software was utilized to visualize the outcomes of the GSE40611 database (Fig. 1D). In the PPI analyses, the association between nodal points was analyzed to determine the interactions between the proteins encoded by DEGs in patients with pSS. According to the PPI results, 14 immune-related genes were found, including CD22, CD38, CXCL10, CD163, CXCL13, CD48, CD19, IL2RB, CCL21, CXCL9, ICOS, CD69, CCL19, CXCL11. Based on the PPI results and 15 overlapping DEGs, co-expressed genes CXCL10, CD69 and ICOS were identified. A previous study reported that the expression of ICOS is associated with EULAR primary Sjögren's syndrome disease activity (ESSDAI) score and gradation of focus scoring, indicating that ICOS may be associated with the severity of pSS (23). A modification based on focus score (FS) calculation was proposed in 1974 (39) and further work established FS ≥1 (i.e., ≥1 focus per 4 mm2) for use in classification criteria (40–44). Therefore, ICOS was identified as a core gene for further research.
The 105 upregulated genes were uploaded for GO term and KEGG pathway enrichment analyses. In the GO analysis, the biological process (BP) term in which ICOS participated in were selected (Fig. 1E). The results revealed that ‘leukocyte cell-cell adhesion’ (GO:0007159; P=2.20×10−10) was the most significant for BP, followed by ‘T cell activation’ (GO:0042110; P=1.19×10−9), ‘regulation of leukocyte cell-cell adhesion’ (GO:1903037; P=4.75×10−9), ‘positive regulation of cell-cell adhesion’ (GO:0022409; P=5.03×10−9), ‘positive regulation of leukocyte cell-cell adhesion’ (GO:1903039; P=7.89×10−9), ‘regulation of cell-cell adhesion’ (GO:0022407; P=2.21×10−8), ‘positive regulation of T cell activation’ (GO:0050870; P=3.05×10−8), ‘regulation of T cell activation’ (GO:0050863; P=3.77×10−8), ‘positive regulation of cell activation’ (GO:0050867; P=7.69×10−8) and ‘positive regulation of lymphocyte activation’ (GO:0051251; P=1.02×10−7).
In addition, KEGG analysis revealed that ‘cell adhesion molecules’ (hsa04514; P=3.25×10−9), ‘intestinal immune network for IgA production’ (hsa04672; P=6.00×10−9), ‘primary immunodeficiency’ (hsa05340; P=0.000280497) and ‘T cell receptor signaling pathway’ (hsa04660; P=0.001814358) were shown. (Fig. 1F).
Biological annotation of the DEGs in patients with pSS identified from the integrated analysis of the microarray data was performed using Cytoscape. BP functional enrichment GO terms (GO:0007159 leukocyte cell-cell adhesion; GO:0042110 T cell activation; GO:1903037 regulation of leukocyte cell-cell adhesion; GO:0022409 positive regulation of cell-cell adhesion; GO:1903039 positive regulation of leukocyte cell-cell adhesion; GO:0022407 regulation of cell-cell adhesion; GO:0050870 positive regulation of T cell activation; GO:0050863 regulation of T cell activation; GO:0050867 positive regulation of cell activation; GO:0051251 positive regulation of lymphocyte activation), KEGG pathways (hsa04514 cell adhesion molecules; hsa04672 intestinal immune network for IgA production; hsa05340 primary immunodeficiency; hsa04660 T cell receptor signaling pathway) and DEGs (HLA-DPB1, IGHG3, CD22, CXCL13, VCAM1, CD3D, TRAT1, HLA-B, HLA-DRB5, CD74, ITK, FCGR2B, CD38, HLA-DRA, HLA-DMB, PLEK, PECAM1, THEMIS, FYN, CD19, CCL19, SELL, HLA-DPA1, HLA-DQA1, CCL21, ICOS, ALQX5, RHOH, LAPTM5, RASGRP1, AICDA.) with P<0.05 involving ICOS participation were obtained (Fig. 1G).
Features of healthy individuals and patients with pSS
To verify the role of ICOS, saliva, salivary gland and whole blood were collected from 95 patients with pSS and 68 healthy individuals. There was no statistically significant difference in the age (P=0.4472). In order to avoid gender differences, all female patients were selected as both the pSS patients and the healthy individuals. Compared with that of healthy individuals, the positive rate of anti-Sjogren's syndrome A antibody (anti-SSA) and/or anti-Sjogren's syndrome B antibody (anti-SSB) was higher. The number of caries diagnosed by two dentists and the amount of dental caries in patients with pSS was greater. In addition, compared with that of healthy individuals, the ESSDAI and rheumatoid factor was higher in pSS patients (Table I)
ICOS expression in salivary glands, whole blood and saliva of healthy individuals and patients with pSS
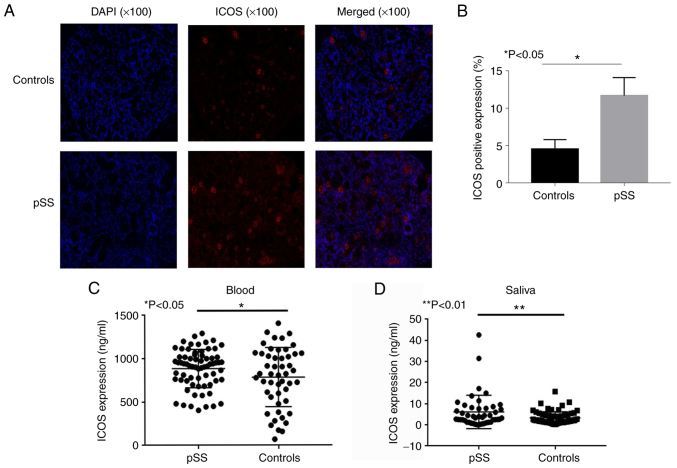
Immunofluorescence analysis of salivary gland samples from patients with pSS revealed that these samples displayed a higher expression level of ICOS compared with that of controls (Fig. 2A and B). The positive ratio of ICOS in the salivary gland tissues of controls and pSS patients was 3.76 and 14.4%, respectively (P<0.05). Compared with that of healthy individuals, the levels of ICOS in peripheral blood (P<0.05; Fig. 2C) and salivary samples (P<0.01; Fig. 2D) were increased in patients with pSS, as determined by ELISA.
Figure 2.
ICOS expression in salivary gland, whole blood and saliva of healthy individuals and patients with pSS. (A) ICOS immunofluorescence expression in the salivary gland tissues of patients with pSS and healthy controls, and (B) quantitative analysis of the results showing significantly higher ICOS stating in pSS tissues compared with that in controls. (C) Patients with pSS (n=95) had higher levels of ICOS in whole blood compared with those of healthy controls (n=68). (D) Patients with pSS (n=95) had higher levels of ICOS in saliva compared with those of controls (n=68). *P<0.05, **P<0.01 vs. controls. ICOS, inducible T cell co-stimulator; pSS, primary Sjogren's syndrome.
ICOS expression affects the expression of AQP5
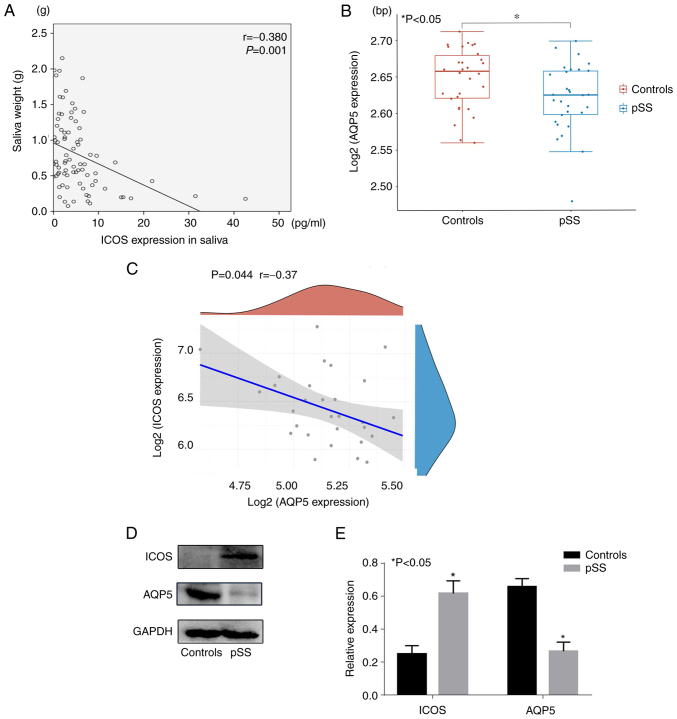
Saliva was collected from patients with pSS and controls. Salivary weight was negatively associated with the expression of ICOS in patients with pSS (r=−0.380; P=0.001; Fig. 3A). The GSE84844 dataset comprises 30 normal and 30 pSS whole blood samples. Compared with those in healthy individuals, the levels of AQP5 in salivary gland tissues were lower in patients with pSS (P<0.05; Fig. 3B) in GSE84844. According to the data, AQP5 was negatively correlated with the expression of ICOS in patients with pSS (r=−0.37; P=0.044 Fig. 3C). The salivary gland tissues of 95 patients and 68 healthy individuals were harvested and used for western blot analysis. The results demonstrated that the expression of AQP5 in patients with pSS was significantly lower compared with that in healthy individuals (P<0.05; Fig. 3D and E), whereas the expression of ICOS in patients with pSS was significantly higher compared with that in healthy individuals (P<0.05). Based on these results, it was hypothesized that the expression of ICOS may affect the expression of AQP5 and thus the secretion of saliva.
Figure 3.
ICOS expression may affect the expression of AQP5. (A) Correlation between saliva weight and the expression of ICOS in saliva. (B) Compared with those of the controls, the levels of AQP5 in salivary gland tissues were lower in pSS patients in the GSE84844 database. (C) The expression of ICOS had a negative correlation with the expression of AQP5 in patients with pSS in the GSE84844 dataset. (D and E) Compared with healthy controls, the expression of AQP5 was significantly lower and the expression of ICOS was higher in patients with pSS. GAPDH was used as an internal control. *P<0.05 vs. controls. AQP5, aquaporin 5; ICOS, inducible T cell co-stimulator; pSS, primary Sjogren's syndrome.
Relative protein expression in pSS patients
A previous study reported that ICOS signaling can affect the expression of inflammatory cytokine through the PI3K-Akt-mTOR pathway. ICOS signaling appears to be critical for Ab production, but its role in the maintenance phase may involve inflammatory cytokines such as TNF-α (44). To investigate a putative association between the PI3K/AKT/mTOR signaling pathway and the expression of relative protein in pSS patients the expressions of ICOS, AQP5, p-PI3K, p-AKT, p-mTOR, TNF-α and Bcl-2 were determined. Compared with those of healthy individuals, the levels of TNF-α in serum were elevated in patients with pSS, as determined by ELISA (P<0.05; Fig. 4A). Western blotting analysis showed that p-PI3K/PI3k, p-AKT/AKT, p-mTOR/mTOR ratios, as well as TNF-α protein expression level were significantly increased, whereas the expression of Bcl-2 and AQP5 were significantly decreased in patients with pSS (all P<0.05; Fig. 4B and C).
Figure 4.
Relative protein expression in pSS patients. (A) Expression of TNF-α in the serum of patients with pSS and controls. (B and C) The protein expression levels of ICOS, AQP5, PI3K, p-PI3K, AKT, p-AKT, mTOR, p-mTOR, TNF-α and Bcl-2 in patients with pSS and the control group were evaluated by western blotting. GAPDH served as a positive control. *P<0.05. AQP5, aquaporin 5; ICOS, inducible T cell co-stimulator; p-, phosphorylated; pSS, primary Sjogren's syndrome.
Discussion
pSS is a systemic autoimmune disease characterized by periductal lymphocyte infiltration in the lacrimal and salivary glands, resulting in low tear and saliva secretion, dry mouth and eyes, early tooth loss (45,46), atypical and/or serious dental caries (47–51), onset of oral moniliasis, atrophic oral mucous damage and tongue lobulation (52). Complications, such as variations in sense of taste, dysphagia (53,54), inability to eat or speak, sensation of burning and persistent fatigue (54), may decrease the quality of life of patients. However, the cellular and body fluid causal links involved in the etiopathogenesis of pSS remain to be elucidated.
It has been demonstrated that ICOS is important in T-cell-driven multiorgan inflammatory events in autoimmunity illnesses (55). A previous study reported that ICOS signaling can affect the expression of inflammatory cytokine through the PI3K-Akt-mTOR pathway. ICOS signaling appears to be critical for Ab production, but its role in the maintenance phase may involve inflammatory cytokines such as TNF-a. A previous study also reported that the expression of ICOS is markedly associated with ESSDAI score and gradation of focus scoring, indicating that ICOS may be associated with the severity of pSS (23). However, the mechanism by which the expression of ICOS affects the secretion of saliva has not been studied to date. The present study compared pSS salivary tissue expression dataset (GSE40611) and whole blood expression dataset (GSE84844) from GEO. In addition, saliva, salivary gland tissues and whole blood were collected from 95 patients with pSS and 68 healthy individuals. The positive ratio of ICOS in the salivary gland tissues of controls and pSS patients was 3.76 and 14.4%, respectively. Compared with that of healthy individuals, the levels of ICOS in peripheral blood and salivary samples were increased in patients with pSS, as determined by ELISA. As with blood, saliva is a complex fluid containing a variety of enzymes, hormones, antibodies, antimicrobial constituents, and growth factors (56). Saliva and tears drain the main targets of autoimmune response, which in the case of pSS are the salivary and lacrimal glands (57). Saliva has been used as a prospective biofluid for identifying specific markers for pSS with underlying functions and clinical impact (58,59). In the present study, in the saliva of patients with pSS, a negative correlation was detected between the expression of ICOS and the salivary weight.
AQPs are membrane proteins that mediate water movement across the lipid bilayer in response to an osmotic gradient (60). AQPs have been identified in nearly every living organism (61). A previous study revealed that disordered salivary secretion was partially due to the aberrant distributional status of AQP5 in human salivary gland epithelial cells (62). AQP5 is affected leading to the reduction of salivary secretion by apoptotic biomolecules and inflammatory cell factors (62). However, the association of cellular factors with pSS remains elusive in terms of immunopathogenesis. To verify the expression of AQP5, salivary gland tissues were collected from normal and pSS patients. Compared with those in healthy individuals, the levels of AQP5 in salivary gland tissues were lower in patients with pSS. The GSE84844 dataset showed AQP5 was negatively correlated with the expression of ICOS in patients with pSS. The salivary gland tissues of 95 patients and 68 healthy individuals were harvested and used for western blot analysis. The results demonstrated that the expression of AQP5 in patients with pSS was significantly lower compared with that in healthy individuals whereas the expression of ICOS in patients with pSS was significantly higher compared with that in healthy individuals. The effect of ICOS on AQP5 expression is unclear.
In the present study, the salivary weight was negatively correlated with the expression of ICOS in patients with pSS. Compared with that of healthy individuals, the expression of ICOS was elevated in patients with pSS in whole blood and salivary gland tissue. A previous study revealed that ICOS/PI3K signal transduction was pivotal for the production of Tfh cells (25). Similar defects in antibody generation are detected in immunized mice lacking ICOS or selectively dysregulated ICOS/PI3K signal transduction (44).
A previous study reported that ICOS signaling can affect the expression of inflammatory cytokine through the PI3K-Akt-mTOR pathway (26). Another study indicated that ICOS increased the expression of TNF-α (44). The high expression of TNF-α is vital for the development of collagen-induced arthritis (CIA) in human rheumatoid arthritis (63,64). Increased circulation of IL-6 and TNF-α in patients with pSS has also been reported (65–70). The expression of TNF-α can caused epithelial cell impairment such as salivary gland epithelial cells in pSS (71). A previous study revealed that the overexpression of TNF-α in murine models decreases the expression of Bcl-2 and induces the downstream caspase cascade, causing inherent inflammatory pathway stimulation (72) In the present study, the expression of TNF-α in the serum of patients with pSS was greater compared with that in healthy individuals. Compared with healthy individuals, the expression of ICOS, TNF-α, p-PI3K, p-AKT and p-mTOR was increased, whereas the expression of Bcl-2 and AQP5 was reduced in patients with pSS. It was hypothesized that ICOS may lead to salivary gland inflammatory responses and may affect saliva secretion. In summary, ICOS may serve a role in salivary gland inflammatory events, which may be an underlying causal link for the reduction in saliva secretion exhibited by patients with pSS.
However, owing to the rarity of pSS disease and the limited research time, the sample size of the present study was small. In the future, the sample size should be increased to validate the conclusions of the present study. It is a matter for regret that no mature cell model or animal model has been found to detect the expression of AQP5 by overexpression or knockout of ICOS. In the future, once there is a mature cell model or animal model, the verification of relevant experiments will be performed.
Acknowledgements
Not applicable.
Funding Statement
The present study was supported by The National Natural Science Foundation of China (grant no. 81801610).
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
JJ and ZX conceived and designed the experiments; PL, YJ and RZ performed the experiments; PL, YJ and RZ analyzed the data, provided reagents/materials/analysis tools and wrote the manuscript. ZX and JJ confirm the authenticity of all the raw data. All authors read and approved the final version of manuscript.
Ethics approval and consent to participate
Written informed consent for the use of saliva, whole blood samples and salivary gland tissues was obtained from all patients. The present study was approved by the ethics committee of the Affiliated Hospital of Nantong University (Nantong, China; approval no. 2017-K003).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Manoussakis MN, Moutsopoulos HM. Sjögren's syndrome: Autoimmune epithelitis. Baillieres Best Pract Res Clin Rheumatol. 2000;14:73–95. doi: 10.1053/berh.1999.0078. [DOI] [PubMed] [Google Scholar]
- 2.Norcross MA. A synaptic basis for T-lymphocyte activation. Ann Immunol (Paris) 1984;135D:113–134. doi: 10.1016/s0769-2625(84)81105-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Janeway CA, Jr, Bottomly K. Signals and signs for lymphocyte responses. Cell. 1994;76:275–285. doi: 10.1016/0092-8674(94)90335-2. [DOI] [PubMed] [Google Scholar]
- 4.Jenkins MK. The ups and downs of T cell costimulation. Immunity. 1994;1:443–446. doi: 10.1016/1074-7613(94)90086-8. [DOI] [PubMed] [Google Scholar]
- 5.Li X, Yu D, Yu N, Wang X, Li X, Harris DCH, Wang Y. B7-H4 deficiency in salivary gland of patients with primary Sjögren's syndrome impairs the regulatory effect on T cells. Int J Rheum Dis. 2017;20:474–480. doi: 10.1111/1756-185X.13041. [DOI] [PubMed] [Google Scholar]
- 6.Mozaffarian N, Wiedeman AE, Stevens AM. Active systemic lupus erythematosus is associated with failure of antigen-presenting cells to express programmed death ligand-1. Rheumatology (Oxford) 2008;47:1335–1341. doi: 10.1093/rheumatology/ken256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Greenfield EA, Nguyen KA, Kuchroo VK. CD28/B7 costimulation: A review. Crit Rev Immunol. 1998;18:389–418. doi: 10.1615/CritRevImmunol.v18.i5.10. [DOI] [PubMed] [Google Scholar]
- 8.Hutloff A, Dittrich AM, Beier KC, Eljaschewitsch B, Kraft R, Anagnostopoulos I, Kroczek RA. ICOS is an inducible T-cell co-stimulator structurally and functionally related to CD28. Nature. 1999;397:263–266. doi: 10.1038/16717. [DOI] [PubMed] [Google Scholar]
- 9.Sharpe AH, Freeman GJ. The B7-CD28 superfamily. Nat Rev Immunol. 2002;2:116–126. doi: 10.1038/nri727. [DOI] [PubMed] [Google Scholar]
- 10.Crotty S. T follicular helper cell differentiation, function, and roles in disease. Immunity. 2014;41:529–542. doi: 10.1016/j.immuni.2014.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Suh WK. Life of T follicular helper cells. Mol Cells. 2015;38:195–201. doi: 10.14348/molcells.2015.2331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu D, Xu H, Shih C, Wan Z, Ma X, Ma W, Luo D, Qi H. T-B-cell entanglement and ICOSL-driven feed-forward regulation of germinal centre reaction. Nature. 2015;517:214–218. doi: 10.1038/nature13803. [DOI] [PubMed] [Google Scholar]
- 13.Xu H, Li X, Liu D, Li J, Zhang X, Chen X, Hou S, Peng L, Xu C, Liu W, et al. Follicular T-helper cell recruitment governed by bystander B cells and ICOS-driven motility. Nature. 2013;496:523–527. doi: 10.1038/nature12058. [DOI] [PubMed] [Google Scholar]
- 14.Grimbacher B, Hutloff A, Schlesier M, Glocker E, Warnatz K, Dräger R, Eibel H, Fischer B, Schäffer AA, Mages HW, et al. Homozygous loss of ICOS is associated with adult-onset common variable immunodeficiency. Nat Immunol. 2003;4:261–268. doi: 10.1038/ni902. [DOI] [PubMed] [Google Scholar]
- 15.Tafuri A, Shahinian A, Bladt F, Yoshinaga SK, Jordana M, Wakeham A, Boucher LM, Bouchard D, Chan VS, Duncan G, et al. ICOS is essential for effective T-helper-cell responses. Nature. 2001;409:105–109. doi: 10.1038/35051113. [DOI] [PubMed] [Google Scholar]
- 16.Dong C, Juedes AE, Temann UA, Shresta S, Allison JP, Ruddle NH, Flavell RA. ICOS co-stimulatory receptor is essential for T-cell activation and function. Nature. 2001;409:97–101. doi: 10.1038/35051100. [DOI] [PubMed] [Google Scholar]
- 17.Mak TW, Shahinian A, Yoshinaga SK, Wakeham A, Boucher LM, Pintilie M, Duncan G, Gajewska BU, Gronski M, Eriksson U, et al. Costimulation through the inducible costimulator ligand is essential for both T helper and B cell functions in T cell-dependent B cell responses. Nat Immunol. 2003;4:765–772. doi: 10.1038/ni947. [DOI] [PubMed] [Google Scholar]
- 18.McAdam AJ, Greenwald RJ, Levin MA, Chernova T, Malenkovich N, Ling V, Freeman GJ, Sharpe AH. ICOS is critical for CD40-mediated antibody class switching. Nature. 2001;409:102–105. doi: 10.1038/35051107. [DOI] [PubMed] [Google Scholar]
- 19.Li DY, Xiong XZ. ICOS+ Tregs: A functional subset of tregs in immune diseases. Front Immunol. 2020;11:2104. doi: 10.3389/fimmu.2020.02104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhu Y, Zou L, Liu YC. T follicular helper cells, T follicular regulatory cells and autoimmunity. Int Immunol. 2016;28:173–179. doi: 10.1093/intimm/dxv079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gang C, Jiahui Y, Huaizhou W, Qing C, Dongbao Z, Qian S. Defects of mitogen-activated protein kinase in ICOS signaling pathway lead to CD4(+) and CD8(+) T-cell dysfunction in patients with active SLE. Cell Immunol. 2009;258:83–89. doi: 10.1016/j.cellimm.2009.03.016. [DOI] [PubMed] [Google Scholar]
- 22.Zhou H, Li B, Li J, Wu T, Jin X, Yuan R, Shi P, Zhou Y, Li L, Yu F. Dysregulated T cell activation and aberrant cytokine expression profile in systemic lupus erythematosus. Mediators Inflamm. 2019;2019:8450947. doi: 10.1155/2019/8450947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Luo J, Liao X, Zhang L, Xu X, Ying S, Yu M, Zhu L, Lin S, Wang X. Transcriptome sequencing reveals potential roles of ICOS in primary Sjögren's syndrome. Front Cell Dev Biol. 2020;8:592490. doi: 10.3389/fcell.2020.592490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.García-Espinoza JA, Muñoz-Valle JF, García-Chagollán M, Hernández-Bello J, Palafox-Sánchez CA, López-Villalobos EF, Sánchez-Zuno GA, Martínez-Bonilla GE, Cerpa-Cruz S, Carrillo-Ballesteros FJ, Oregon-Romero E. ICOS gene polymorphisms (IVS1 + 173 T/C and c. 1624 C/T) in Primary Sjögren's syndrome patients: Analysis of ICOS expression. Curr Issues Mol Biol. 2022;44:764–776. doi: 10.3390/cimb44020053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gigoux M, Shang J, Pak Y, Xu M, Choe J, Mak TW, Suh WK. Inducible costimulator promotes helper T-cell differentiation through phosphoinositide 3-kinase. Proc Natl Acad Sci USA. 2009;106:20371–20376. doi: 10.1073/pnas.0911573106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gigoux M, Lovato A, Leconte J, Leung J, Sonenberg N, Suh WK. Inducible costimulator facilitates T-dependent B cell activation by augmenting IL-4 translation. Mol Immunol. 2014;59:46–54. doi: 10.1016/j.molimm.2014.01.008. [DOI] [PubMed] [Google Scholar]
- 27.Bonifazi P, D'Angelo C, Zagarella S, Zelante T, Bozza S, De Luca A, Giovannini G, Moretti S, Iannitti RG, Fallarino F, et al. Intranasally delivered siRNA targeting PI3K/Akt/mTOR inflammatory pathways protects from aspergillosis. Mucosal Immunol. 2010;3:193–205. doi: 10.1038/mi.2009.130. [DOI] [PubMed] [Google Scholar]
- 28.King LS, Agre P. Pathophysiology of the aquaporin water channels. Annu Rev Physiol. 1996;58:619–648. doi: 10.1146/annurev.ph.58.030196.003155. [DOI] [PubMed] [Google Scholar]
- 29.Farzan N, Vijverberg SJ, Arets HG, Raaijmakers JA, Maitland-van der Zee AH. Pharmacogenomics of inhaled corticosteroids and leukotriene modifiers: A systematic review. Clin Exp Allergy. 2017;47:271–293. doi: 10.1111/cea.12844. [DOI] [PubMed] [Google Scholar]
- 30.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. Gene ontology: Tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kanehisa M, Goto S, Sato Y, Furumichi M, Tanabe M. KEGG for integration and interpretation of large-scale molecular data sets. Nucleic Acids Res. 2012;40:D109–D114. doi: 10.1093/nar/gkr988. (Database issue) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Huang DW, Sherman BT, Tan Q, Kir J, Liu D, Bryant D, Guo Y, Stephens R, Baseler MW, Lane HC, Lempicki RA. DAVID Bioinformatics Resources: Expanded annotation database and novel algorithms to better extract biology from large gene lists. Nucleic Acids Res. 2007;35:W169–W175. doi: 10.1093/nar/gkm415. (Web Server issue) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Szklarczyk D, Gable AL, Lyon D, Junge A, Wyder S, Huerta-Cepas J, Simonovic M, Doncheva NT, Morris JH, Bork P, et al. STRING v11: Protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019;47((D1)):D607–D613. doi: 10.1093/nar/gky1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Doncheva NT, Morris JH, Gorodkin J, Jensen LJ. Cytoscape StringApp: Network analysis and visualization of proteomics data. J Proteome Res. 2019;18:623–632. doi: 10.1021/acs.jproteome.8b00702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vitali C, Bombardieri S, Jonsson R, Moutsopoulos HM, Alexander EL, Carsons SE, Daniels TE, Fox PC, Fox RI, Kassan SS, et al. Classification criteria for Sjögren's syndrome: A revised version of the European criteria proposed by the American-European Consensus Group. Ann Rheum Dis. 2002;61:554–558. doi: 10.1136/ard.61.6.554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nocturne G. Sjögren's syndrome update: Clinical and therapeutic aspects. Rev Med Interne. 2019;40:433–439. doi: 10.1016/j.revmed.2019.03.329. (In French) [DOI] [PubMed] [Google Scholar]
- 37.Rosas J, Sánchez-Piedra C, Fernández-Castro M, Andreu JL, Martínez-Taboada V, Olivé A, SJÖGRENSER Group and part of the Spanish Society of Rheumatology Systemic Autoimmune Diseases Study Group (EASSER) ESSDAI activity index of the SJÖGRENSER cohort: Analysis and comparison with other European cohorts. Rheumatol Int. 2019;39:991–999. doi: 10.1007/s00296-019-04285-w. [DOI] [PubMed] [Google Scholar]
- 38.Chen X, Aqrawi LA, Utheim TP, Tashbayev B, Utheim ØA, Reppe S, Hove LH, Herlofson BB, Singh PB, Palm Ø, et al. Elevated cytokine levels in tears and saliva of patients with primary Sjögren's syndrome correlate with clinical ocular and oral manifestations. Sci Rep. 2019;9:7319. doi: 10.1038/s41598-019-43714-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Greenspan JS, Daniels TE, Talal N, Sylvester RA. The histopathology of Sjögren's syndrome in labial salivary gland biopsies. Oral Surg Oral Med Oral Pathol. 1974;37:217–229. doi: 10.1016/0030-4220(74)90417-4. [DOI] [PubMed] [Google Scholar]
- 40.Vitali C, Moutsopoulos HM, Bombardieri S. The European Community Study Group on diagnostic criteria for Sjögren's syndrome. Sensitivity and specificity of tests for ocular and oral involvement in Sjögren's syndrome. Ann Rheum Dis. 1994;53:637–647. doi: 10.1136/ard.53.10.637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vitali C, Bombardieri S, Moutsopoulos HM, Balestrieri G, Bencivelli W, Bernstein RM, Bjerrum KB, Braga S, Coll J, de Vita S, et al. Preliminary criteria for the classification of Sjögren's syndrome. Results of a prospective concerted action supported by the European Community. Arthritis Rheum. 1993;36:340–347. doi: 10.1002/art.1780360309. [DOI] [PubMed] [Google Scholar]
- 42.Vitali C, Bombardieri S, Moutsopoulos HM, Coll J, Gerli R, Hatron PY, Kater L, Konttinen YT, Manthorpe R, Meyer O, et al. Assessment of the European classification criteria for Sjögren's syndrome in a series of clinically defined cases: Results of a prospective multicentre study. The European Study Group on Diagnostic Criteria for Sjögren's Syndrome. Ann Rheum Dis. 1996;55:116–121. doi: 10.1136/ard.55.2.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Daniels TE, Whitcher JP. Association of patterns of labial salivary gland inflammation with keratoconjunctivitis sicca. Analysis of 618 patients with suspected Sjögren's syndrome. Arthritis Rheum. 1994;37:869–877. doi: 10.1002/art.1780370615. [DOI] [PubMed] [Google Scholar]
- 44.Panneton V, Bagherzadeh Yazdchi S, Witalis M, Chang J, Suh WK. ICOS signaling controls induction and maintenance of collagen-induced arthritis. J Immunol. 2018;200:3067–3076. doi: 10.4049/jimmunol.1701305. [DOI] [PubMed] [Google Scholar]
- 45.Daniels TE, Silverman S, Jr, Michalski JP, Greenspan JS, Sylvester RA, Talal N. The oral component of Sjögren's syndrome. Oral Surg Oral Med Oral Pathol. 1975;39:875–885. doi: 10.1016/0030-4220(75)90108-5. [DOI] [PubMed] [Google Scholar]
- 46.Baudet-Pommel M, Albuisson E, Kemeny JL, Falvard F, Ristori JM, Fraysse MP, Sauvezie B. Early dental loss in Sjögren's syndrome. Histologic correlates. European Community Study Group on Diagnostic Criteria for Sjögren's Syndrome (EEC COMAC) Oral Surg Oral Med Oral Pathol. 1994;78:181–186. doi: 10.1016/0030-4220(94)90143-0. [DOI] [PubMed] [Google Scholar]
- 47.Daniels TE, Fox PC. Salivary and oral components of Sjögren's syndrome. Rheum Dis Clin North Am. 1992;18:571–589. doi: 10.1016/S0889-857X(21)00317-3. [DOI] [PubMed] [Google Scholar]
- 48.Greenspan D. Xerostomia: Diagnosis and management. Oncology (Williston Park) 1996;10((3 Suppl)):S7–S11. [PubMed] [Google Scholar]
- 49.Lilly JP, Fotos PG. Sjögren's syndrome: Diagnosis and management of oral complications. Gen Dent. 1996;44:404–408. 419–420. quiz. [PubMed] [Google Scholar]
- 50.Ravald N, List T. Caries and periodontal conditions in patients with primary Sjögren's syndrome. Swed Dent J. 1998;22:97–103. [PubMed] [Google Scholar]
- 51.Soto-Rojas AE, Villa AR, Sifuentes-Osornio J, Alarcón-Segovia D, Kraus A. Oral manifestations in patients with Sjögren's syndrome. J Rheumatol. 1998;25:906–910. [PubMed] [Google Scholar]
- 52.Hernandez YL, Daniels TE. Oral candidiasis in Sjögren's syndrome: Prevalence, clinical correlations, and treatment. Oral Surg Oral Med Oral Pathol. 1989;68:324–329. doi: 10.1016/0030-4220(89)90218-1. [DOI] [PubMed] [Google Scholar]
- 53.Rhodus NL, Colby S, Moller K, Bereuter J. Quantitative assessment of dysphagia in patients with primary and secondary Sjögren's syndrome. Oral Surg Oral Med Oral Pathol Oral Radiol Endod. 1995;79:305–310. doi: 10.1016/S1079-2104(05)80224-0. [DOI] [PubMed] [Google Scholar]
- 54.Pedersen AM, Reibel J, Nauntofte B. Primary Sjögren's syndrome (pSS): Subjective symptoms and salivary findings. J Oral Pathol Med. 1999;28:303–311. doi: 10.1111/j.1600-0714.1999.tb02045.x. [DOI] [PubMed] [Google Scholar]
- 55.Odegard JM, DiPlacido LD, Greenwald L, Kashgarian M, Kono DH, Dong C, Flavell RA, Craft J. ICOS controls effector function but not trafficking receptor expression of kidney-infiltrating effector T cells in murine lupus. J Immunol. 2009;182:4076–4084. doi: 10.4049/jimmunol.0800758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Deepa T, Thirrunavukkarasu N. Saliva as a potential diagnostic tool. Indian J Med Sci. 2010;64:293–306. doi: 10.4103/0019-5359.99854. [DOI] [PubMed] [Google Scholar]
- 57.Katsiougiannis S, Wong DT. The Proteomics of Saliva in Sjögren's Syndrome. Rheum Dis Clin North Am. 2016;42:449–456. doi: 10.1016/j.rdc.2016.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Giusti L, Baldini C, Bazzichi L, Bombardieri S, Lucacchini A. Proteomic diagnosis of Sjögren's syndrome. Expert Rev Proteomics. 2007;4:757–767. doi: 10.1586/14789450.4.6.757. [DOI] [PubMed] [Google Scholar]
- 59.Hu S, Wang J, Meijer J, Ieong S, Xie Y, Yu T, Zhou H, Henry S, Vissink A, Pijpe J, et al. Salivary proteomic and genomic biomarkers for primary Sjögren's syndrome. Arthritis Rheum. 2007;56:3588–3600. doi: 10.1002/art.22954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Verkman AS. Aquaporins at a glance. J Cell Sci. 2011;124((Pt 13)):2107–2112. doi: 10.1242/jcs.079467. [DOI] [PubMed] [Google Scholar]
- 61.Hoffert JD, Chou CL, Knepper MA. Aquaporin-2 in the ‘-omics’ era. J Biol Chem. 2009;284:14683–14687. doi: 10.1074/jbc.R900006200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yoshimura S, Nakamura H, Horai Y, Nakajima H, Shiraishi H, Hayashi T, Takahashi T, Kawakami A. Abnormal distribution of AQP5 in labial salivary glands is associated with poor saliva secretion in patients with Sjögren's syndrome including neuromyelitis optica complicated patients. Mod Rheumatol. 2016;26:384–390. doi: 10.3109/14397595.2015.1083146. [DOI] [PubMed] [Google Scholar]
- 63.Brennan FM, McInnes IB. Evidence that cytokines play a role in rheumatoid arthritis. J Clin Invest. 2008;118:3537–3545. doi: 10.1172/JCI36389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Williams RO, Feldmann M, Maini RN. Anti-tumor necrosis factor ameliorates joint disease in murine collagen-induced arthritis. Proc Natl Acad Sci USA. 1992;89:9784–9788. doi: 10.1073/pnas.89.20.9784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Liu C, Guan Z, Zhao L, Song Y, Wang H. Elevated level of circulating CD4+Helios+FoxP3+ cells in primary Sjogren's syndrome patients. Mod Rheumatol. 2017;27:630–637. doi: 10.1080/14397595.2016.1226470. [DOI] [PubMed] [Google Scholar]
- 66.Szodoray P, Papp G, Horvath IF, Barath S, Sipka S, Nakken B, Zeher M. Cells with regulatory function of the innate and adaptive immune system in primary Sjögren's syndrome. Clin Exp Immunol. 2009;157:343–349. doi: 10.1111/j.1365-2249.2009.03966.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Roescher N, Tak PP, Illei GG. Cytokines in Sjogren's syndrome: Potential therapeutic targets. Ann Rheum Dis. 2010;69:945–948. doi: 10.1136/ard.2009.115378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Baturone R, Soto MJ, Márquez M, Macías I, de Oca MM, Medina F, Chozas N, García-Pérez S, Girón-González JA. Health-related quality of life in patients with primary Sjögren's syndrome: Relationship with serum levels of proinflammatory cytokines. Scand J Rheumatol. 2009;38:386–389. doi: 10.1080/03009740902973821. [DOI] [PubMed] [Google Scholar]
- 69.Szodoray P, Alex P, Brun JG, Centola M, Jonsson R. Circulating cytokines in primary Sjögren's syndrome determined by a multiplex cytokine array system. Scand J Immunol. 2004;59:592–599. doi: 10.1111/j.0300-9475.2004.01432.x. [DOI] [PubMed] [Google Scholar]
- 70.Garcíc-Carrasco M, Font J, Filella X, Cervera R, Ramos-Casals M, Sisó A, Aymamí A, Ballesta AM, Ingelmo M. Circulating levels of Th1/Th2 cytokines in patients with primary Sjögren's syndrome: Correlation with clinical and immunological features. Clin Exp Rheumatol. 2001;19:411–415. [PubMed] [Google Scholar]
- 71.Fox RI, Kang HI, Ando D, Abrams J, Pisa E. Cytokine mRNA expression in salivary gland biopsies of Sjögren's syndrome. J Immunol. 1994;152:5532–5539. [PubMed] [Google Scholar]
- 72.Xu H, Li J, Zhao Y, Liu D. TNFα-induced downregulation of microRNA-186 contributes to apoptosis in rat primary cardiomyocytes. Immunobiology. 2017;222:778–784. doi: 10.1016/j.imbio.2017.02.005. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.