Abstract
A common form of bacterial quorum sensing involves the production and release of acyl homoserine lactone (AHL) signal metabolites. The nitrogen-fixing symbiont Rhizobium leguminosarum reportedly produces at least six different AHLs, but little is known about the regulation of biosynthesis of these molecules. We used a radiolabeling protocol to quantify the relative amounts of AHLs synthesized over time by R. leguminosarum cells with and without the symbiosis plasmid pRL1JI. Cells containing pRL1JI were found to produce three predominant signals. In decreasing order of abundance, these were N-(3-oxo)octanoyl homoserine lactone [(3-O)C8HSL], N-octanoyl homoserine lactone, and N-hexanoyl homoserine lactone. Cells without pRL1JI produced only two major signals, N-(3-hydroxy-7-cis)tetradecanoyl homoserine lactone [(3-OH)C14:1HSL] and (3-O)C8HSL. Each AHL exhibited a distinct temporal pattern of synthesis, suggesting that each AHL is subject to unique regulatory mechanisms. While (3-O)C8HSL was produced in both cultures, the patterns of synthesis were different in cells with and without pRL1JI, possibly as a result of redundant gene functions that are present on both the chromosome and the symbiosis plasmid. None of the AHLs appeared to regulate its own biosynthesis, although exogenous (3-OH)C14:1HSL did activate synthesis of the three AHLs made by cells containing pRL1JI. These results indicate that the synthesis of multiple AHLs in R. leguminosarum is regulated by complex mechanisms that operate independently of quorum sensing itself but that (3-OH)C14:1HSL can supersede these controls in pRL1JI-containing cells. This work provides an important global perspective for AHL regulation that both complements and contrasts with the results of previous studies performed with isolated gene systems.
Quorum sensing is a type of intercellular communication found in many gram-negative bacteria. Communication between cells by quorum sensing is mediated by the production and release of specific N-acyl homoserine lactone (AHL) signal metabolites. The individual cells in a population are able to sense the population density through extracellular accumulation of these signals (7, 8, 10, 21). Two families of transcriptional regulators, called the LuxI and LuxR protein families, control this communication system. LuxI proteins catalyze the synthesis of the AHL signals, which then interact with LuxR proteins to activate or repress specific target genes. This process was first identified as a process that controls bioluminescence in the marine bacterium Vibrio fischeri but has since been found to regulate a variety of functions in many different bacteria (21).
Many bacteria produce more than one type of AHL signal, thus creating the opportunity for interactions among different quorum sensing systems in the same organism (1, 16). One such organism is the symbiotic nitrogen-fixing bacterium Rhizobium leguminosarum, which produces at least six different AHLs (13, 17). These molecules have been identified as N-hexanoyl homoserine lactone (C6HSL), N-(3-oxo)hexanoyl homoserine lactone [(3-O)C6HSL], N-heptanoyl homoserine lactone (C7HSL), N-octanoyl homoserine lactone (C8HSL), N-(3-oxo)octanoyl homoserine lactone [(3-O)C8HSL], and N-(3-hydroxy-7-cis)tetradecanoyl homoserine lactone [(3-OH)C14:1HSL] (9, 13, 17, 19).
The abilities to produce these different signals are encoded by at least four luxI homologs in this species. The chromosomal gene cinI encodes a protein that synthesizes (3-OH)C14:1HSL, while the rhiI gene of the symbiosis (Sym) plasmid pRL1JI encodes an enzyme that produces both C6HSL and C8HSL (13, 17). Two additional loci for AHL synthesis, one each on pRL1JI and the chromosome, have been identified but have not been described yet (13).
The only R. leguminosarum genes known to be regulated directly at the transcriptional level by quorum sensing are those composing the rhiABC operon. This operon is found on the Sym plasmid pRL1JI and is controlled by the LuxR homolog RhiR. The RhiR protein activates rhiABC transcription in response to the presence of C6HSL and other AHLs (4, 17). It was previously found that addition of the chromosomally encoded signal (3-OH)C14:1HSL also resulted in increased expression of the rhiABC operon (9), but subsequent work showed that the activation is indirect, possibly a result of increased synthesis of other AHLs (13, 17). Because (3-OH)C14:1HSL also activates genes that inhibit the growth of cells containing pRL1JI (9), uncharacterized regulatory functions encoded by the Sym plasmid typically suppress (3-OH)C14:1HSL synthesis to levels that are about 5% of normal levels (13). These low levels of (3-OH)C14:1HSL still appear to be required for normal production of the other AHLs made by cells containing pRL1JI (13).
The complex effects of (3-OH)C14:1HSL suggest that there may be communication among different quorum sensing loci and thus a hierarchy of AHL signals within R. leguminosarum. The purpose of this research was to examine possible signal interactions in R. leguminosarum by measuring the temporal pattern of AHL synthesis at a global level and the response of this pattern to exogenous addition of individual AHLs.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
The bacterial strains used in this research are shown in Table 1. All cultures were maintained on TY medium (2) at 30°C. Cultures to be radiolabeled were grown at 30°C in RK minimal medium (22) supplemented with a 10% (vol/vol) addition of TY broth. In vivo labeling experiments were performed in the same supplemented RK minimal medium at 25°C. When appropriate, antibiotics were added at the following final concentrations: chloramphenicol, 25 μg/ml; kanamycin, 50 μg/ml; streptomycin, 50 μg/ml; and tetracycline, 5 μg/ml.
TABLE 1.
Bacterial strains used in this study
| Strain | Relevant characteristics | Reference or source |
|---|---|---|
| Rhizobium leguminosarum 8401 | Streptomycin-resistant mutant of Sym− strain 8400, Srr | 12 |
| R. leguminosarum 8401 (pRL1JI) | 8401 containing self-transmissible Sym plasmid pRL1JI of R. leguminosarum bv. viciae, pRL1JI represses normal synthesis of (3-OH)C14:1HSL and inhibits cell growth in the presence of this AHL, Srr | 11 |
| R. leguminosarum 8401(pIJ1089) | 8401 containing a 30-kb cosmid fragment of pRL1JI that includes all of the known rhi genes, used as a source of C6HSL, Srr Tcr | 5 |
| R. leguminosarum 14482Sr | Streptomycin-resistant mutant of R. leguminosarum bv. phaseoli ATCC 14482, used as a source of (3-O)C8HSL, C8HSL, and (3-OH)C14:1HSL, Srr | This study |
| R. leguminosarum 8401(pIJ1642) | 8401 with plasmid pIJ1089 containing a Tn3::lacZ reporter fusion in rhiA that is activated in response to C6HSL, Srr Tcr | 6 |
| R. leguminosarum 8401(pRL1JI/pIJ1769) | 8401 containing both pRL1JI and a rhiA′::lacZ reporter plasmid (pIJ1769) that is activated in response to (3-OH)C14:1HSL, Srr Tcr | 9 |
| Agrobacterium tumefaciens NT1(pDCI41E33) | Ti− strain that does not produce AHL, plasmid pDCI41E33 contains the luxR homolog traR and a traG::lacZ reporter that is activated in response to diverse AHLs, Kmr | S. K. Farrand |
| Chromobacterium violaceum CV0blu(pSB403) | Tn5 mutant strain that does not produce AHL, contains the luxR homolog cviR that activates violacein synthesis in response to diverse AHLs, plasmid pSB403 also encodes LuxR from V. fischeri, Cpr Tcr | 3 |
In vivo labeling of AHLs.
The radiolabeling procedure used to measure de novo synthesis of AHLs was adapted from the method of Schaeffer et al. (18). Cultures to be labeled were grown overnight in TY broth to an optical density at 660 nm (OD660) of more than 1.0. In order to downregulate any genes expressed in an overnight culture as a result of quorum sensing, the cells were diluted to an OD660 of 0.1 in fresh TY medium and grown for 1 h at 30°C with shaking. After this, the culture was diluted to an OD660 of 0.01 in supplemented RK minimal medium and incubated at 30°C with shaking. At each labeling time, the OD660 of the culture was recorded, and 1.5 ml was removed and incubated with 1.5 μCi of [α-14C]methionine (specific activity, 55 mCi/mmol; American Radiolabeled Chemicals, Inc., St. Louis, Mo.) for 3 h at 25°C with shaking. After this, the labeled sample was centrifuged at a high speed in a microcentrifuge, and the cell-free supernatant was collected. This supernatant was sequentially extracted with two equal volumes of ethyl acetate containing 0.01% (final concentration) glacial acetic acid. The ethyl acetate phase, which contained the radiolabeled AHLs, was collected and stored at −20°C.
Analysis of labeled AHLs.
Different AHLs labeled by the method described above were separated and quantified by reverse-phase high-performance liquid chromatography (HPLC). Samples of labeled extracts were dried under a stream of sterile air, and each residue was redissolved in 50% methanol. The labeled AHLs were separated on a C18 column (0.46 by 25 cm; Phase Separations, Norwalk, Conn.) by using a 20 to 100% methanol gradient at a flow rate of 0.5 ml/min for 133 min. One-milliliter fractions were collected, scintillation fluid was added to each fraction, and the fractions were counted with a liquid scintillation counter.
The count per minute value for each peak of AHL activity was multiplied by the appropriate dilution factor to obtain a value for the total counts per minute incorporated into AHLs by an entire labeled culture. The total number of cells present in the culture was estimated by using a standard curve of OD660 versus CFU per milliliter. The total number of counts per minute incorporated into AHLs was then divided by the number of CFU present to obtain a final value for counts per minute per CFU. This number represented the amount of label incorporated into a given AHL per bacterial cell and allowed direct comparisons of AHL synthesis between cultures of different ages and cell densities.
AHL purification and bioassay.
AHLs were isolated from conditioned medium extracts of stationary-phase R. leguminosarum 14482Sr and 8401(pIJ1089) cultures by using HPLC as described above. Samples of individual HPLC fractions were assayed for AHL activity by one of four methods having the following AHL specificities: R. leguminosarum 8401(pRL1JI/pIJ1769) responded to (3-OH)C14:1HSL, R. leguminosarum 8401(pIJ1642) responded to C6HSL, Chromobacterium violaceum CV0blu(pSB403) responded to C6HSL, C7HSL, and C8HSL and more weakly to (3-O)C6HSL and (3-O)C8HSL, and Agrobacterium tumefaciens NT1(pDCI41E33) responded to C8HSL, (3-O)C6HSL, (3-O)C8HSL, and (3-OH)C14:1HSL. Bioassays with R. leguminosarum 8401(pRL1JI/pIJ1769) and 8401(pIJ1642) and C. violaceum CV0blu(pSB403) were performed as previously described (3, 9). Bioassays with A. tumefaciens NT1(pDCI41E33) were performed by diluting overnight cultures of the strain to an OD660 of 0.01 in TY medium and incubating the cultures with samples from individual HPLC fractions for 3 h at 30°C with shaking. Activation of the traG::lacZ reporter in response to AHLs was then measured by performing a β-galactosidase assay (14). HPLC fractions that tested positive for AHL activity were dried by rotary evaporation, resuspended in acidified ethyl acetate, and stored at −20°C. The molecular structures of the individual compounds isolated by this method and by 14C labeling were inferred based on comigration with known AHL structures normally produced by R. leguminosarum (9, 13, 17, 19) and by the previously described specificities of the different bioassays for individual AHLs.
Addition of purified AHLs to R. leguminosarum.
Samples of C6HSL, (3-O)C8HSL, C8HSL, and (3-OH)C14:1HSL, prepared as described above, were added to sterile flasks and dried under a stream of sterile air. R. leguminosarum cells were inoculated into supplemented RK minimal medium as described above and added to the flasks so that the final volume of each culture equaled the volume of supernatant medium from which the dried AHL sample was originally purified. The cultures were then incubated, and individual samples were pulse-labeled with [14C]methionine as described above. HPLC analysis of the resulting labeled AHLs was performed as described above.
RESULTS
Temporal patterns of AHL synthesis in R. leguminosarum.
Bacterial cells incorporate [α-14C]methionine into protein and into the homoserine lactone portion of any AHL molecules that are synthesized (18, 20). As a result, diverse AHL structures are all equally labeled with a single 14C atom. Organic extraction of cell-free supernatants of labeled cultures removes the labeled proteins and unincorporated [14C]methionine from the labeled AHLs. The AHLs present in the extracts can then be separated by HPLC, and the relative amounts of these AHLs can be quantified by liquid scintillation counting.
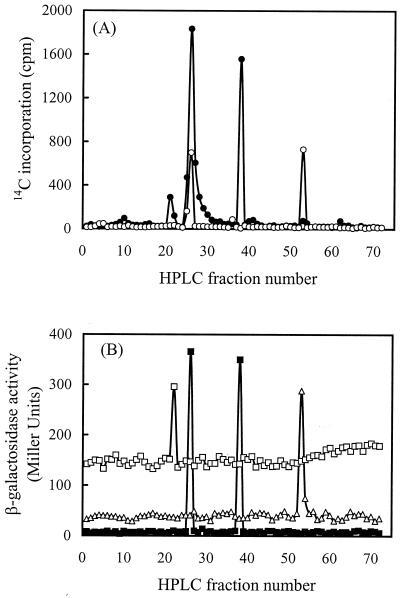
When this method was applied to growing cultures of R. leguminosarum cells with and without Sym plasmid pRL1JI, cells containing pRL1JI yielded individual peaks corresponding to C6HSL, (3-O)C8HSL, and C8HSL, while cells without pRL1JI yielded peaks corresponding to (3-O)C8HSL and (3-OH)C14:1HSL (Fig. 1A). The profiles invariably duplicated the patterns of AHL activities identified by bioassays of unlabeled culture extracts (Fig. 1B). The compounds shown in Fig. 1 were the only signals reliably identified in any of our R. leguminosarum cultures.
FIG. 1.
HPLC analysis of AHLs produced by R. leguminosarum 8401 and 8401(pRL1JI). (A) AHL-containing extracts from 14C-labeled cultures with pRL1JI (●) and without pRL1JI (○) were eluted by using a 20 to 100% methanol gradient. Individual AHLs collected in 1-ml fractions were quantified by liquid scintillation counting. (B) Unlabeled culture extracts were similarly eluted, and the fractions were bioassayed for AHL activity. Bioassays with R. leguminosarum 8401(pIJ1642) (□) and A. tumefaciens NT1 (pDCI41E33) (■) are shown for the 8401 (pRL1JI) extract, and bioassays with R. leguminosarum 8401(pRL1JI/pIJ1769) (▵) are shown for the 8401 extract. Individual peaks correspond to C6HSL (centered at fraction 22), (3-O)C8HSL (fraction 26), C8HSL (fraction 38), and (3-OH)C14:1HSL (fraction 54). AHL structures were inferred from comigration with known standards and from the specificities of individual bioassays.
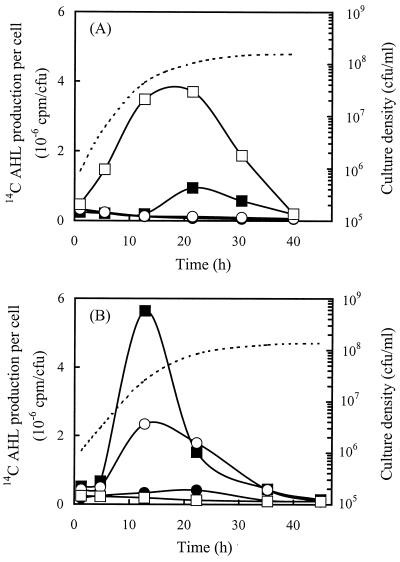
By performing this pulse-labeling procedure at various times during culture growth, we could elucidate the temporal pattern of AHL synthesis in R. leguminosarum. Strain 8401, which lacks a Sym plasmid, produced the AHLs (3-OH)C14:1HSL and (3-O)C8HSL at different times during growth (Fig. 2A). The maximal rate of (3-OH)C14:1HSL synthesis was almost four times that of (3-O)C8HSL, and synthesis of (3-OH)C14:1HSL occurred over a much longer period of culture growth. The rate of (3-OH)C14:1HSL synthesis in the cells increased steadily during growth, reaching a maximum just before the onset of the stationary phase. The rates of (3-O)C8HSL synthesis remained near the lower limits of detection throughout culture growth and then increased briefly at the end of exponential growth. Any other AHLs produced by the cultures were presumed to have been synthesized in amounts less than the amount that could be detected by the assay.
FIG. 2.
AHL biosynthesis in R. leguminosarum with and without pRL1JI. Cells of R. leguminosarum 8401 (A) and 8401(pRL1JI) (B) were pulse-labeled with [14C]methionine at various times, and the labeled AHLs were separated by HPLC and quantified by liquid scintillation counting. The amounts of C6HSL (●), C8HSL (○), (3-O)C8HSL (■), and (3-OH)C14:1HSL (□) produced per cell were determined. Culture growth is indicated by the dashed lines. All values are averages based on three replicate experiments. The standard deviations were ≤11% for all values.
Cells of the same R. leguminosarum strain containing Sym plasmid pRL1JI produced the AHLs (3-O)C8HSL, C8HSL, and C6HSL, in decreasing order of abundance (Fig. 2B). As expected, synthesis of the chromosomally encoded AHL (3-OH)C14:1HSL was inhibited below detectable levels by regulatory functions encoded by the plasmid (9, 13). Production of (3-O)C8HSL was strongly induced in these cells in the mid-exponential phase and was quickly shut off at the onset of the stationary phase. Synthesis of C8HSL was induced at about the same time as synthesis of (3-O)C8HSL, but the rate was less than one-half the maximal rate. Despite the lower level of production, the subsequent decline in C8HSL synthesis was also slower than the decline in (3-O)C8HSL synthesis, so that cells continued to synthesize C8HSL at near-maximal rates during the transition to the stationary phase. Only very small amounts of C6HSL were produced during culture growth, and the rate of synthesis increased gradually until the onset of the stationary phase. Overall, the results of these assays demonstrated that the individual AHLs produced by R. leguminosarum were each synthesized in different amounts in batch culture and that each exhibited a unique temporal pattern of induction and subsequent shutoff.
Surprisingly, maximal production of (3-O)C8HSL in cells containing pRL1JI occurred approximately 10 h earlier than maximal production of (3-O)C8HSL in cells that lacked the plasmid, and the amounts produced by the former cells were sixfold greater than the amounts produced by the latter cells (Fig. 2). These differences in the timing and magnitude of AHL synthesis may be due to the presence of a redundant AHL synthase gene on the Sym plasmid. A gene that encodes (3-O)C8HSL synthesis has been identified on pRL1JI, but it has not been described yet (13). We hypothesize that this gene may be activated earlier and at higher levels than the corresponding gene responsible for (3-O)C8HSL synthesis in Sym− cells.
It should also be noted that extremely hydrophobic AHLs, such as (3-OH)C14:1HSL, can potentially remain associated with cell membranes (15). As a result, a disproportionately high level of these AHLs may be retained with the cell material compared to the level in the extracellular medium. Although no information about the import or efflux of hydrophobic AHLs in R. leguminosarum is available, it is possible that the methods used in this study may underestimate the total amounts of (3-OH)C14:1HSL synthesized.
Addition of purified AHLs to R. leguminosarum 8401(pRL1JI).
The patterns of AHL synthesis observed in cells containing pRL1JI might have resulted from some form of hierarchical control in which the production of each signal was dependent on prior synthesis of a different AHL. Separate additions of purified AHLs to newly inoculated cultures thus allowed us to determine the effect of each of the signals on the overall pattern of AHL synthesis. In every case, the quantities of purified compounds added were equivalent to those naturally produced by other R. leguminosarum cultures. This was accomplished by adding AHLs isolated from a culture sample whose volume was equal to that of the experimental cultures.
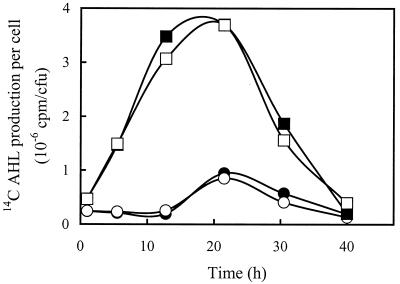
Neither R. leguminosarum 8401 cells grown in the presence of (3-OH)C14:1HSL or (3-O)C8HSL nor cells of R. leguminosarum 8401(pRL1JI) grown in the presence of C6HSL, (3-O)C8HSL, or C8HSL showed any changes in the timing of AHL synthesis or the relative amounts of the compounds synthesized (Fig. 3 and data not shown). These results suggested that there was not a direct regulatory hierarchy for synthesis of these compounds and that a signaling cascade for sequential activation of different AHLs did not occur in these cells. More surprisingly, these results also indicated that none of these signals was subject to positive autoregulation in vivo.
FIG. 3.
Effect of exogenous (3-OH)C14:1HSL on AHL biosynthesis in R. leguminosarum 8401. AHLs synthesized by cells grown in the presence (open symbols) and in the absence (solid symbols) of (3-OH)C14:1HSL were quantified as described in the legend to Fig. 2. The amounts of (3-O)C8HSL (● and ○) and (3-OH)C14:1HSL (■ and □) synthesized per cell were determined. The values are averages based on three experiments. The standard deviations were ≤11% for all values.
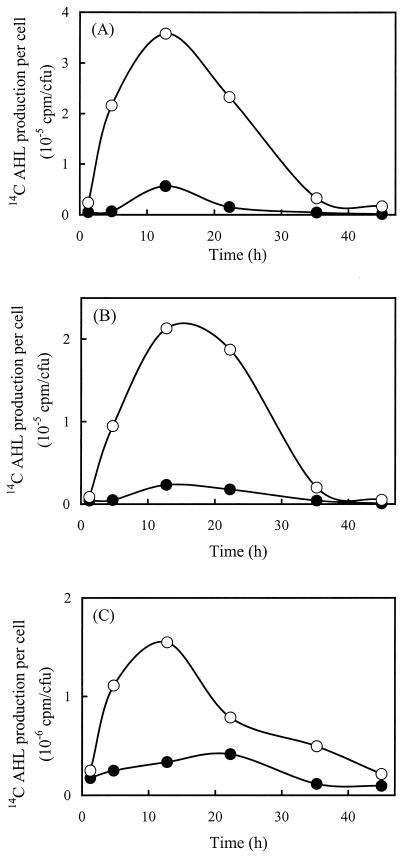
Addition of (3-OH)C14:1HSL to cells containing pRL1JI, however, resulted in marked increases in the amounts of all three of the AHLs normally produced by the cultures (Fig. 4). Maximal production of (3-O)C8HSL increased approximately sixfold, maximal production of C8HSL increased ninefold, and maximal production of C6HSL increased almost fourfold in response to this signal. Despite these changes, addition of (3-OH)C14:1HSL did not derepress or otherwise increase synthesis of (3-OH)C14:1HSL in the cultures.
FIG. 4.
Effect of exogenous (3-OH)C14:1HSL on AHL biosynthesis in R. leguminosarum 8401(pRL1JI). AHLs synthesized by cells grown in the presence (open symbols) and in the absence (solid symbols) of (3-OH)C14:1HSL were quantified as described in the legend to Fig. 2. (A) Amount of (3-O)C8HSL synthesized per cell; (B) amount of C8HSL synthesized per cell; (C) amount of C6HSL synthesized per cell. All values are averages based on three experiments. The standard deviations were ≤11% for all values.
Addition of (3-OH)C14:1HSL not only increased the rates at which the other AHLs were synthesized but also altered the temporal patterns of synthesis. Whereas each AHL previously exhibited a unique pattern of induction and subsequent shutoff (Fig. 2), in the presence of exogenous (3-OH)C14:1HSL similar patterns were observed for all three signals. Cellular rates of AHL synthesis were induced in these cultures within the first 3 h, and the rates continued to increase steadily during 12 h of incubation. The increases were followed by steady decreases in AHL synthesis over the next 24 h (Fig. 4). It should be noted that as expected after addition of (3-OH)C14:1HSL to cells containing pRL1JI, growth in the cultures was inhibited and the cells entered the stationary phase after about 13 h of incubation.
Addition of (3-OH)C14:1HSL thus had no measurable effect on AHL synthesis in Sym− R. leguminosarum cells (Fig. 3) but resulted in four- to ninefold increases in the amounts of all other AHLs synthesized in cells containing pRL1JI (Fig. 4). The corresponding changes in the timing of AHL synthesis by cells containing pRL1JI further suggested that exogenous (3-OH)C14:1HSL was able to override the control mechanism(s) responsible for the unique temporal patterns observed for the individual signals (Fig. 2B).
DISCUSSION
This work represents the first direct, quantitative comparison of the relative amounts of different AHLs synthesized by a bacterial culture. Previous investigations relied on bioassays that can detect only the net accumulation of AHLs during culture growth. In addition, different bioassays have unique affinities for specific AHL structures, thus precluding direct comparisons of the different AHLs that may be present. By contrast, the radiolabeling protocol used here allows the accurate determination of short-term rates of AHL synthesis. Because a single molecule of labeled substrate is incorporated equally into the homoserine lactone portions of diverse AHL structures, it is also possible to determine the relative amounts of different AHLs produced. With this method we attempted to address questions of temporal control and hierarchical regulation of AHL synthesis in R. leguminosarum on a global level.
We found that the Sym− strain R. leguminosarum 8401 produced two major AHLs, (3-O)C8HSL and (3-OH)C14:1HSL, while cells containing Sym plasmid pRL1JI produced (3-O)C8HSL, C8HSL, and C6HSL (Fig. 2). These data are at odds with the results of Lithgow et al. (13), who identified C6HSL, C7HSL, and C8HSL in Sym− strain 8401 but did not detect either of the two signals found in this study. Similarly, when they used pRL1JI-containing cells, Lithgow et al. identified the same three AHLs described in this paper, but they also reported the presence of two more compounds, (3-O)C6HSL and C7HSL.
These discrepancies could reflect differences in the methods used for AHL preparation, separation, and detection. First, the overall amounts of AHLs produced might differ as a result of the different growth conditions employed. Previous studies were performed with cultures grown in TY medium (2), while in our experiments we used cultures grown in TY medium diluted 10-fold with minimal medium. Thus, certain AHLs might have been produced in these experiments at levels below our level of detection as a result of the dilute nutrient conditions. When supernatant extracts from cultures grown in full-strength TY medium were subjected to HPLC, however, no new signals were found by any of the bioassays employed.
Alternatively, Lithgow et al. separated their AHL-containing extracts by thin-layer chromatography (TLC) rather than by HPLC and then used a Chromobacterium-based bioassay to detect active signal compounds on the TLC plates. This bioassay, which is optimal for unsubstituted short-chain acyl homoserine lactone structures (21), responds only weakly to (3-O)C8HSL and does not detect (3-OH)C14:1HSL at all (Blosser Middleton, unpublished data). Thus, the absence of these two signals in the results obtained by Lithgow et al. with the Sym− strain may be attributable to the bioassay itself. Thinking that our inability to detect the additional compounds which Lithgow et al. found in pRL1JI-containing cells might be explained in a similar way, we retested HPLC fractions from cultures grown in both dilute and full-strength media by using a quantitative Chromobacterium-based bioassay (3). The new assays, however, still failed to identify any new peaks of signal activity in any of the cultures tested.
We therefore propose that the remaining discrepancies between our results and those of Lithgow et al. are due either to a greater sensitivity of the TLC method compared to the sensitivity of separations made by HPLC or to the much higher concentrations of AHL-containing extracts being tested in their work. In light of the differences, our data should be interpreted as reflecting only the major AHLs produced by R. leguminosarum cultures and not the signals that may be synthesized at minor levels.
Analysis of the labeled AHLs demonstrated that there was a unique temporal pattern for synthesis of each signal in the bacteria. Interestingly, synthesis of (3-OH)C14:1HSL by Sym− cells began when the numbers of cells were quite low but increased steadily on a per cell basis during most of the culture growth period (Fig. 2A). While we have no explanation for this unusual pattern of regulation, it stands in sharp contrast to the model system of V. fischeri, in which AHL synthesis is rapidly activated at high cell densities through a mechanism of positive autoregulation (8). None of the R. leguminosarum signals appeared to be autoregulated, however, as prior addition of these signals to the culture medium had no effect (Fig. 3 and data not shown).
This last finding is at odds with previous reports that described positive autoregulation of the R. leguminosarum AHL synthase genes cinI and rhiI (13, 17). There are two possible reasons for the differences. First, the amounts of AHLs added may have been very different. The previous investigators added synthetic AHLs at arbitrary concentrations that may have had little relationship to natural levels, whereas we endeavored to add undefined but biologically relevant concentrations of naturally produced AHLs. More importantly, positive autoregulation was demonstrated previously by using plasmid constructs transferred into heterologous hosts in order to avoid the confusing effects of additional regulators or competing signals. Our work, in contrast, was performed in the native host with all other regulatory systems intact specifically so that we could observe global patterns of control. Our results thus suggest that while cinI and rhiI have a demonstrated capacity to autoregulate when they are isolated in heterologous hosts (13, 17), they do not appear to do so in their native hosts when grown in laboratory culture. Instead, the primary control of AHL synthesis in R. leguminosarum seems to be due to other, as-yet-undescribed regulatory mechanisms.
A third possible explanation for the apparent lack of autoregulation in the experiments performed is that the strains to which the AHLs were added were already producing the same signals. It is thus possible that the receptor proteins in these cells were already responding to endogenously produced AHLs, so that the exogenous signals had no additional effect. The weakness of this hypothesis is that because the purified AHLs were added immediately upon transfer of the cultures, endogenous AHLs would have to be abundant enough to saturate the cellular response even in very dilute suspensions. However, bioassays of supernatant media from newly inoculated cultures revealed undetectable levels of AHL activity (data not shown). In addition, labeling experiments revealed extremely low levels of de novo AHL synthesis in the first few hours after culture transfer (Fig. 2).
Alternatively, one might hypothesize that AHLs already bound to the receptors could be carried over during culture transfer as stable complexes that are not removed by simple dilution of the cell suspensions. This was why all cultures were routinely preincubated for 1 h at a low cell density before the cells were diluted further at the beginning of each experiment. Although the possibility of retained activity cannot be ruled out completely, the time allowed should have been more than adequate for the turnover of any active signal-receptor complexes.
While none of the AHLs exhibited positive autoregulation in our experiments, addition of (3-OH)C14:1HSL to cultures of R. leguminosarum 8401(pRL1JI) cells did promote synthesis of all three other AHLs normally produced by these cells (Fig. 4). This finding confirms previous suggestions that (3-OH)C14:1HSL activates the synthesis of other AHLs (13, 17), but with the unexpected twist that this activation appears to occur only in cells containing pRL1JI. Insertional inactivation of the cinI gene, which is solely responsible for (3-OH)C14:1HSL synthesis, has been shown to reduce the synthesis of all other AHLs in R. leguminosarum 8401(pRL1JI) (13). This has led to the hypothesis that while cells containing pRL1JI repress (3-OH)C14:1HSL production to very low levels, these levels are nonetheless required for the synthesis of other AHLs (13).
Activation of AHL synthesis to levels above the normal level by exogenous (3-OH)C14:1HSL may be a simple extension of the same process. Our hypothesis is that increased amounts of (3-OH)C14:1HSL promote AHL synthesis to an even greater extent than normal while simultaneously activating growth inhibition functions. Presumably, both growth inhibition and enhanced AHL synthesis are mediated by the interaction of (3-OH)C14:1HSL with LuxR-like receptors encoded by pRL1JI. Such a model accounts for the absence of any changes in growth or AHL synthesis when (3-OH)C14:1HSL is added to cells that lack pRL1JI. It is interesting that while (3-OH)C14:1HSL appears to play a dominant role in regulating the synthesis of pRL1JI-encoded AHLs, at low concentrations its effects are seemingly modulated by other regulatory functions so that each signal is produced in a unique pattern. Whether this modulation can be accounted for by differential receptor and/or promoter affinities or whether it involves additional regulatory networks remains to be seen.
In summary, this study provided the first quantitative comparisons of the global rates of synthesis of multiple AHL signals in a single bacterial species. The results indicate that of the six AHLs reported to be synthesized by R. leguminosarum, only two or three are produced in abundance. The predominant AHLs that we identified for R. leguminosarum were synthesized with distinct and occasionally unexpected patterns that require further investigation to explain. Finally, our data suggest that primary control of quorum sensing signal generation in R. leguminosarum may not be due to autoregulation or AHL signal hierarchies. Discrepancies between some of our results and the results of previous studies provide a valuable perspective on the differences that may exist between the global patterns of gene expression and the measured responses of isolated genes.
ACKNOWLEDGMENTS
We thank M. R. Parsek and E. P. Greenberg for information and advice about the [14C]AHL labeling method in advance of publication.
This research was supported by CAREER grant MCB-9600766 from the National Science Foundation.
REFERENCES
- 1.Atkinson S, Throup J P, Stewart G S A B, Williams P. A hierarchical quorum-sensing system in Yersinia pseudotuberculosis is involved in the regulation of motility and clumping. Mol Microbiol. 1999;33:1267–1277. doi: 10.1046/j.1365-2958.1999.01578.x. [DOI] [PubMed] [Google Scholar]
- 2.Beringer J E. R factor transfer in Rhizobium leguminosarum. J Gen Microbiol. 1974;84:188–198. doi: 10.1099/00221287-84-1-188. [DOI] [PubMed] [Google Scholar]
- 3.Blosser R S, Gray K M. Extraction of violacein from Chromobacterium violaceum provides a new bioassay for N-acyl homoserine lactone autoinducers. J Microbiol Methods. 2000;40:47–55. doi: 10.1016/s0167-7012(99)00136-0. [DOI] [PubMed] [Google Scholar]
- 4.Cubo M T, Economou A, Murphy G, Johnston A W B, Downie J A. Molecular characterization and regulation of the rhizosphere-expressed genes rhiABCR that influence nodulation by Rhizobium leguminosarum biovar viciae. J Bacteriol. 1992;174:4026–4035. doi: 10.1128/jb.174.12.4026-4035.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Downie J A, Ma Q S, Knight C D, Hombrecher G, Johnston A W B. Cloning of the symbiotic region of Rhizobium leguminosarum: the nodulation genes are between the nitrogenase genes and the mifA-like gene. EMBO J. 1983;2:947–952. doi: 10.1002/j.1460-2075.1983.tb01526.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Economou A, Hawkins F K L, Downie J A, Johnston A W B. Transcription of rhiA, a gene on a Rhizobium leguminosarum bv. viciae Sym plasmid, requires rhiR and is repressed by flavonoids that induce nod genes. Mol Microbiol. 1989;3:87–93. doi: 10.1111/j.1365-2958.1989.tb00107.x. [DOI] [PubMed] [Google Scholar]
- 7.Fuqua C, Winans S C, Greenberg E P. Census and consensus in bacterial ecosystems: the LuxR-LuxI family of quorum sensing transcriptional regulators. Annu Rev Microbiol. 1996;50:727–751. doi: 10.1146/annurev.micro.50.1.727. [DOI] [PubMed] [Google Scholar]
- 8.Gray K M. Intercellular communication and group behavior in bacteria. Trends Microbiol. 1997;5:184–188. doi: 10.1016/S0966-842X(97)01002-0. [DOI] [PubMed] [Google Scholar]
- 9.Gray K M, Pearson J P, Downie J A, Boboye B E A, Greenberg E P. Cell-to-cell signaling in the symbiotic nitrogen-fixing bacterium Rhizobium leguminosarum: autoinduction of a stationary phase and rhizosphere-expressed genes. J Bacteriol. 1996;178:372–376. doi: 10.1128/jb.178.2.372-376.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Greenberg E P. Acyl-homoserine lactone quorum sensing in bacteria. J Microbiol. 2000;38:117–121. [Google Scholar]
- 11.Hirsch P R. Plasmid-determined bacteriocin production by Rhizobium leguminosarum. J Gen Microbiol. 1979;113:219–228. [Google Scholar]
- 12.Lamb J W, Hombrecher G, Johnston A W B. Plasmid-determined nodulation and nitrogen-fixing abilities in Rhizobium phaseoli. Mol Gen Genet. 1982;186:449–452. [Google Scholar]
- 13.Lithgow J K, Wilkinson A, Hardman A, Rodelas B, Wisniewski-Dye F, Williams P, Downie J A. The regulatory locus cinRI in Rhizobium leguminosarum controls a network of quorum-sensing loci. Mol Microbiol. 2000;37:81–97. doi: 10.1046/j.1365-2958.2000.01960.x. [DOI] [PubMed] [Google Scholar]
- 14.Miller J A. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1972. pp. 352–355. [Google Scholar]
- 15.Pearson J P, Van Delden C, Iglewski B H. Active efflux and diffusion are involved in transport of Pseudomonas aeruginosa cell-to-cell signals. J Bacteriol. 1999;181:1203–1210. doi: 10.1128/jb.181.4.1203-1210.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pesci E C, Pearson J P, Seed P C, Iglewski B H. Regulation of las and rhl quorum sensing in Pseudomonas aeruginosa. J Bacteriol. 1997;179:3127–3132. doi: 10.1128/jb.179.10.3127-3132.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rodelas B, Lithgow J K, Wisniewski-Dye F, Hardman A, Wilkinson A, Economou A, Williams P, Downie J A. Analysis of quorum-sensing-dependent control of rhizosphere-expressed (rhi) genes in Rhizobium leguminosarum bv. viciae. J Bacteriol. 1999;181:3816–3823. doi: 10.1128/jb.181.12.3816-3823.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schaeffer A L, Greenberg E P, Parsek M R. Acylated homoserine lactone detection in Pseudomonas aeruginosa biofilms by radiolabel assay. Methods Enzymol. 2001;336:41–47. doi: 10.1016/s0076-6879(01)36576-x. [DOI] [PubMed] [Google Scholar]
- 19.Schripsema J, de Rudder K E E, van Vliet T B, Lankhorst P P, de Vroom E, Kijne J W, van Brussel A A N. Bacteriocin small of Rhizobium leguminosarum belongs to the class of N-acyl-l-homoserine lactone molecules known as autoinducers and as quorum sensing co-transcription factors. J Bacteriol. 1996;178:366–371. doi: 10.1128/jb.178.2.366-371.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Singh P K, Schaeffer A L, Parsek M R, Moninger T O, Welsh M J, Greenberg E P. Quorum-sensing signals indicate that cystic fibrosis lungs are infected with bacterial biofilms. Nature. 2000;407:762–764. doi: 10.1038/35037627. [DOI] [PubMed] [Google Scholar]
- 21.Swift S, Williams P, Stewart G S A B. N-Acyl homoserine lactones and quorum sensing in proteobacteria. In: Dunny G M, Winans S C, editors. Cell-cell signaling in bacteria. Washington, D.C.: American Society for Microbiology; 1999. pp. 291–313. [Google Scholar]
- 22.Zhang L, Kerr A. A diffusible compound can enhance conjugal transfer of the Ti plasmid in Agrobacterium tumefaciens. J Bacteriol. 1991;173:1867–1872. doi: 10.1128/jb.173.6.1867-1872.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]