Abstract
The molecular circadian clock mechanism is highly conserved between mammalian and avian species. Avian circadian timing is regulated at multiple oscillatory sites, including retina, pineal and the hypothalamic suprachiasmatic nucleus (SCN). Based on our previous studies in the rat ovary, we hypothesized that ovarian clock timing is regulated by the luteinizing hormone (LH) surge. We used the chicken as a model to test this hypothesis because the timing of the endogenous LH surge is accurately predicted from the time of oviposition. Therefore, tissues can be removed before and after the LH surge, allowing one to determine the effect of LH on specific clock genes. We first examined the 24-h expression patterns of the avian circadian clock genes, Bmal1, Cry1 and Per2 in primary oscillatory tissues (hypothalamus and pineal), as well as peripheral tissues (liver and ovary). Second, we determined changes in clock gene expression after the endogenous LH surge. Clock genes were rhythmically expressed in each tissue, but LH influenced expression of these clock genes only in the ovary. The data suggest that expression of ovarian circadian clock genes may be influenced by the LH surge in vivo and directly by LH in cultured granulosa cells. LH induced rhythmic expression of Per1 and Bmal1 in arrhythmic, cultured granulosa cells. Furthermore, LH altered the phase and amplitude of clock gene rhythms in serum-shocked granulosa cells. Thus, the LH surge may be a mechanistic link for communicating circadian timing information from the central pacemaker to the ovary.
Keywords: Circadian clock, pineal, ovary, luteinizing hormone, Per2, Bmal1, Cry1
INTRODUCTION
The ovulatory cycle of the domestic hen is characteristically 24-25 h in duration. This unique cycle is regulated by a daily rhythm in the sensitivity of the hypothalamus, which enables it to respond to ovarian steroids, resulting in the preovulatory LH surge (Fraps, 1954). Ovulation of the largest follicle, the F1, occurs approximately 3 h later. The time of oviposition can be used to predict the time of the previous LH surge and ovulation, because oviposition occurs 15-30 minutes before ovulation. The ovulatory cycle results from complex interactions between two regulatory systems within the hypothalamic-pituitary-gonadal axis. The hypothalamus and pituitary work in concert to produce an ovulatory signal, encoded as an LH surge. The ovary regulates follicular maturation; the presence of a mature ovum is critical for ovulation (Yoshimura et al., 1994; Yoshimura and Bahr, 1995). Towards the conclusion of follicular maturation, the F1 follicle, which is destined to ovulate, increases its production of progesterone to trigger the LH surge. Reciprocally, release of the ovum from the follicle is dependent upon the LH surge. Under standard lighting conditions, the LH surge, which directs the timing of ovulation and oviposition, is restricted to an 8-10 h window commonly referred to as the “open period”.
The “open period” for the LH surge, the driving force for the ovulatory cycle, is generally considered to be under control of a circadian oscillator. Evidence for circadian control includes the fact that both the LH surge and oviposition respond to changes in the lighting schedule by shifting their occurrence to maintain a specific phase relationship to the time of lights-off. Furthermore, the ovulatory cycle can be entrained to photoperiods as short as 21 h or as long as 30 h, which is consistent with circadian regulation. Finally, the ovulatory cycle is free-running with 27 h periodicity under constant illumination in Japanese quail (Underwood et al., 1997). However, the anatomical site and the mechanism of action of the oscillator that controls reproductive processes have yet to be determined.
Circadian rhythms are generated by the interactions of a core group of clock genes. In mammals, the positive regulators of the clock gene network, Clock and Bmal1 interact with each other and together drive transcription of the Period (Per1 and 2) and Cryptochrome (Cry1 and 2) genes. As PER and CRY proteins accumulate, they heterodimerize and feedback to suppress CLK/BMAL1 activity, thereby inhibiting their own transcription and completing the circadian cycle (Tischkau, 2008). Orthologs of the mammalian clock genes, including Clock, Per2, Per3, Bmal1, Cry1 and Cry2 have been described in birds (Yoshimura et al., 2000; Okano et al., 2001; Yamamoto et al., 2001; Fu et al., 2002).
Although the molecular components of the clockworks are highly conserved between birds and mammals, significant disparity exists in the organization of the systemic circadian timing systems. A single, central pacemaker, located in the suprachiasmatic nucleus of the hypothalamus is primarily responsible for regulation of organismic rhythmicity in mammals. In contrast, the avian circadian timing system is regulated by multiple pacemakers, located in the retina, SCN and pineal gland (Gwinner and Brandstatter, 2001; Bell-Pedersen et al., 2005). The contribution of each of these pacemakers to circadian organization of the whole organism remains unclear. However, consensus dictates that an important function of master pacemakers, including the mammalian SCN and each of the avian pacemakers, is the regulation of oscillators located in peripheral tissues.
Recent studies indicate that the ovary contains a circadian oscillator (Fahrenkrug et al., 2006; Karman and Tischkau, 2006; Nakao et al., 2007; Yoshikawa et al., 2009). Furthermore, the ovulatory cycle in both birds and mammals is clearly under circadian control. We hypothesize that the LH surge acts as a circadian signal to communicate circadian timing information from a primary pacemaker to the ovary. We examined expression of avian clock genes in primary pacemakers (pineal and hypothalamus), as well as peripheral tissues (liver and ovary). Prediction of the endogenous LH surge using oviposition times allowed in vivo testing of the hypothesis that the LH surge regulates clock gene expression in the ovary.
EXPERIMENTAL PROCEDURES
Animals and Zeitgeber Time (ZT)
Single-comb white Leghorn hens were caged individually and provided feed and water ad libitum. Adult birds (26-36 wks of age) with regular clutches of at least ten eggs were used for these experiments. Hens were exposed to 16 h of light and 8 h of darkness with lights-on at 0400 h. Zeitgeber time (ZT) 0 is defined as the time of lights-on. Thus, for these experiments, lights were on from ZT0-ZT16 and off from ZT16-ZT24. Ovipositions were monitored by visual inspection at 30-min intervals from ZT2 to ZT12, and again at ZT14, to time late ovipositions. Oviposition accurately predicts the time of the subsequent ovulation (30 min after oviposition) and the preceding LH surge (4 h before oviposition) (Tischkau et al., 1996). All animal experiments were approved by the Institutional Animal Care and Use Committee of the University of Illinois, were conducted in accordance with the NIH Guide for the Care and Use of Laboratory Animals and conform to the international ethical standards required for publication in this journal (Portaluppi et al., 2008).
Experiment 1: Daily Rhythms in Clock Gene Expression
This experiment was designed to determine whether clock gene expression displays 24-h rhythmicity in the domestic hen. Oviposition was monitored and animals were sacrificed on the day when ovulation was predicted to occur at ZT6 (6 h after lights-on) on the day of the experiment. Animals were sacrificed at ZT0, 6, 12, 18 and 24. Liver, anterior hypothalamus containing the SCN, pineal, F1 granulosa and F1 theca (Tischkau et al., 1996) were quickly dissected from birds, placed into RNAlater (Ambion) and stored until use.
Experiment 2: Effects of the Endogenous LH Surge on Clock Gene Levels
This experiment was designed to determine if the endogenous LH surge alters circadian clock gene transcripts in the chicken. Oviposition was used to predict the times of the LH surge. Oviposition was carefully monitored and animals were divided into treatment groups (Fig. 1). To assure that time-of-day was not a factor in determining expression levels of transcripts for this experiment, all animals were sacrificed at ZT 6. In group 1 (before LH), oviposition occurred at ZT11, and the LH surge occurred at ZT 7. In group 2 (after LH), oviposition occurred at ZT8, and the LH surge occurred at ZT4. After sacrifice, liver, medio-basal hypothalamus containing the SCN, pineal, F1 granulosa and F1 theca were quickly dissected from birds, placed into RNAlater (Ambion) and stored until use.
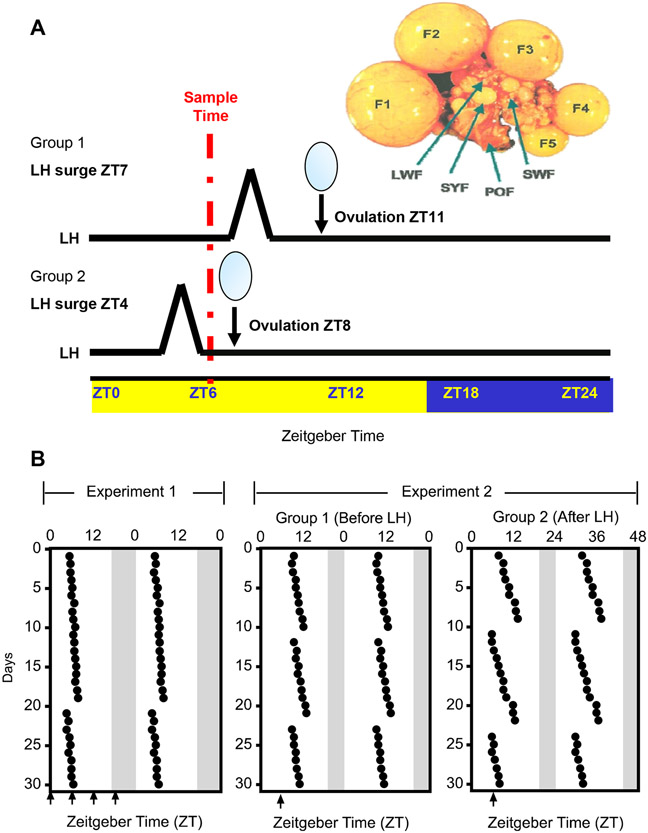
Fig. 1. Ovulation records and experimental design.
Animals were housed in a 16:8 LD cycle with lights on at ZT0 and off at ZT16. A) Representative oviposition record for hens used in experiments 1 and 2. Ovipositions were monitored for 30 days prior to sacrifice. Hens were placed on a 16:8 light:dark schedule. The time of oviposition is indicated by the black dot. Arrows indicates the time of sacrifice on the day of the experiment. Data are double-plotted. Shaded area indicates the time of lights-off. B) The chicken ovary showing the hierarchy of follicles and a schematic of the experimental design for experiment 2. To account for time-of-day dependent change in clock gene transcript expression, all animals were sacrificed at ZT6. Oviposition was monitored at 30-min intervals from ZT3 to ZT12, and again at ZT14, to time late ovipositions. The oviposition cycle in this group of hens was approximately 24.25 h. The time of the previous oviposition was used to predict the time of the LH surge, which reliably occurs ~4 h prior to oviposition (Tischkau et al., 1996). Animals in Group 1 were sacrificed at ZT6, but prior to the LH surge based on predicted oviposition at ZT11 on the day of the experiment. Animals in Group 2 were sacrificed at ZT6, but after the LH surge, based on a predicted oviposition at ZT8. Also depicted is the chicken ovary, with hierarchical follicles (F1-F5), small yellow follicles (SYF), large white follicles (LWF), small white follicles (SWF) and a postovulatory follicle (POF). Only F1 follicles were used for this experiment.
Experiment 3: Effects of LH on Rhythms of Circadian Clock Gene Transcripts in Immortalized Granulosa Cell Cultures
This experiment was designed to determine if spontaneously immortalized granulosa cells (SIGC) express LH receptors and to explore the effects of LH on the phase and amplitude of Per1 and Bmal1 rhythms. Four groups of cells were included in this experiment as follows: Control cells were cultured in serum-free starting and samples were collected every for h up to 52 h; LH only cells were treated with 50 ng/ml for 2 h and samples were harvested every 4 h starting at the end of treatment and continuing until 52 h; Serum Shock cells were treated with 50% horse serum for 2 h and harvested every 4 h for 52 h starting at the end of the serum shock; Serum Shock + LH cells were serum shocked with 50% horse serum, followed by addition of LH to achieve a final concentration of 50 ng/ml and samples were harvested every 4 h until 52 h. Per1 and Bmal1 transcripts were assessed using quantitative real time PCR (qPCR).
Experiment 4: Acute effects of LH is cAMP/PKA-dependent
This experiment was designed to determine whether the acute increase of Per1 transcript levels after gonadotropin treatment occurs through the cAMP/PKA pathway. SIGC were pretreated with SQ22536 (adenylyl cyclase inhibitor) for 30 minutes. The SIGC cells were then treated with Forskolin (1uM), ovine LH (50 ng/ml), SQ22536 (100 uM) or SQ22536 (100uM)/LH (50 ng/ml). Cells were harvested 1 h after treatment. Per1, LHr, and Egr-1 mRNA were assessed using qPCR.
Cell Culture
SIGC (provided by Dr. RC Burghardt, Texas A&M University (Stein et al., 1991)) were seeded at 200,000 cells/well on 6-well plates and were grown to 80-90% in Dulbecco’s Modified Eagle’s Medium (DMEM-reduced serum) supplemented with 7.5% fetal bovine serum or serum-free DMEM-reduced serum (experiment 3). Forskolin, LH and SQ 22536 were dissolved in culture media. Cells were treated as described above before harvesting cells and isolating RNA.
qPCR
For in vivo experiments, tissues were homogenized in cell lysis buffer (RNeasy Kit, Qiagen) using 1.0 mm zirconia beads in a mini-Bead Beater (Cole-Palmer). RNA was isolated using the RNeasy kit (Qiagen), according to the manufacturer’s protocol. RNA was extracted from cultured cells using the Trizol method according to the manufacturer’s protocol. Concentration, purity, and quality of the RNA were determined spectrophotometrically at 260 nm and 280 nm (NanoDrop ND-1000) and by gel electrophoresis. RNA was reverse transcribed with 200 ng random hexamers, 200U SSII reverse transcriptase, and 10mM DTT (Invitrogen) at 43°C for one hour in a volume of 20 μL (BioRad MyCycler). Negative controls included omission of reverse transcriptase and omission of template. Rat primers for Per1 and Bmal1 have been published previously (Karman and Tischkau, 2006). Chicken clock gene primers were chosen using Primer Express 2.0 software (Applied Biosystems, Foster City, CA) for optimum use in qPCR. A BLASTN search was performed against GenBank to ensure their uniqueness to the gene of interest. All primer pairs spanned a large exon-intron-exon junction to prevent amplification from genomic DNA (see Table 1). Rat LHr and Egr-1 primers were purchased from Qiagen. qPCR reactions were performed as previously described (Karman and Tischkau, 2006). The cycle at which amplification of the product exceeded threshold was determined and designated as the ct (cycle at which threshold is crossed) value. Dissociation curves, gel analysis and sequencing confirmed gene specific product amplification.
Table 1.
Primer sequences and amplicon size for chicken clock genes.
Standard Curves and Calculations
Relative standard curves were created with serially diluted total RNA samples from chicken liver RNA or from SIGC (modified from (Bustin, 2000)). Three sets of pooled liver RNA samples (obtained at different times of day) were serially diluted by a factor of two starting at 2 μg and ending at 62.5 ng. This range was determined to be well within the detection level and sensitivity of the qPCR assay. RNA samples used for standard curves are representative of 12 animals. Following qPCR, average Ct values were plotted against the log2 of the total RNA concentration. Amplicons demonstrated a linear relationship with total RNA concentration with good doubling efficiency. The resulting standard curves were used to calculate relative amount of transcripts within each reaction. Standard curves for avian clock gene transcripts were as follows: Bmal1 y = −1.0324x + 34.04, R2 = 0.9843, E=1.95; Per2 y = −1.0261x + 32.053, R2 = 0.9838, E=1.9650; Cry1 y = −1.0387x + 29.16, R2=0.9699, E=1.9490 (E=efficiency of amplicon doubling with each cycle, 2 (−1/slope); and R2=correlation coefficient). Primer sets and standard curve data for rat Per1 has been published previously (Karman and Tischkau, 2006). Resulting standard curves were used to calculate relative amounts of transcripts according to the equation: relative amount = 2((Ct-Intercept)/(slope)). The average relative amount for each experiment was used to normalize individual values. Data are represented as the mean ± SEM.
Statistical Analysis
The least squares ANOVA as implemented by SYSTAT (version 11) was used to analyze all data. Bonferroni’s post-hoc analysis was used and P≤0.05 was considered significant.
RESULTS
Expression of Circadian Clock Genes in the Chicken
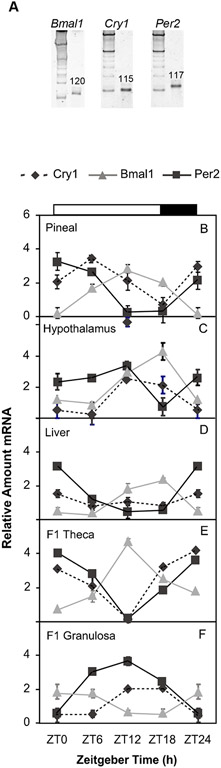
Clock gene products known to be components of the core oscillator in mammalian systems (Bmal1, Per2, and Cry1) were expressed in the chicken ovary (Fig. 2A, PCR products), as well as the pineal gland, medio-basal hypothalamus containing the SCN and the liver (data not shown). PCR products of appropriate sizes predicted from the primer sequences were amplified for Bmal1 (120 bps), Cry1 (115 bps) and Per2 (117 bps). Oviposition records were monitored and birds carefully selected such that ovulation occurred near ZT6 on the day of sacrifice (Fig. 1B). Under a 16:8 light/dark cycle, significant variations in transcript levels were observed for Bmal1, Cry1 and Per2 (Fig. 2, n=3-4 per time point). First, we examined expression patterns of Bmal1, Cry1 and Per2 in tissues considered to house primary oscillators in the bird, namely the pineal gland and the medio-basal hypothalamus (Fig. 2B and C). In the pineal gland, Bmal1 displayed 24-h variation, with peak levels at ZT12 (Fig. 2A, p<0.001 vs. minimal values at ZT24/0). Cry1 and Per2 levels also changed over the 24 h period in the pineal, with peak expression observed at ZT0/24 (p<0.01) for Per2 and at ZT 6 (p<0.001) for Cry1. The medio-basal hypothalamus displayed a robust oscillation in the Bmal1 transcript, with peak expression observed at ZT18 (Fig. 2C, p<0.001 compared to minimal expression at ZT6 (ANOVA with Bonferroni’s post-hoc analysis). Cry1 and Per2 peak expression in the medio-basal hypothalamus occurred at ZT12 (p<0.001 for Cry1 and p<0.05 for Per2, ANOVA with Bonferroni’s post-hoc analysis).
Fig. 2. Daily mRNA levels of circadian clock-related genes in the chicken.
Samples were collected every 6 hours under a light/darkness (L16:D8) schedule (n=3-4 per timepoint). A) Polyacrylamide gel electrophoresis of real-time quantitative PCR products demonstrates amplification of single bands corresponding to the predicted size for chicken Bmal1, Cry1 and Per2. Each sample was run on a separate gel with a 100 bp marker. Quantitative RT-PCR was used to measure Bmal1 (▲), Cry1 (♦) and Per2 (■) transcripts in chicken pineal gland (B), medio-basal hypothalamus (C), liver (D), F1 theca (E) and F1 granulosa (F) tissues. Results are shown as mean fold-change to lowest (trough) level ± SEM (top to bottom). Results of statistical analysis (ANOVA) are described in the results section. White and black bars at the top of the figure indicates the time of lights on and off, respectively.
Next, we examined expression of Bmal1, Cry1 and Per2 transcripts in a typical peripheral clock tissue, the liver. Both Bmal1 and Per2 varied significantly over the 24-h period, with peak expression levels occurring at ZT12-18 (Fig. 2D, p<0.01) and ZT0/24 (Fig. 2D, p<0.05), respectively. Cry1 transcript levels did not change over time in the liver.
Finally, we determined expression patterns of Bmal1, Cry1 and Per2 transcripts in the ovary of the largest preovulatory follicle (F1). Granulosa and theca layers were analyzed separately (Fig. 2E and F). Significant changes in transcript levels of all three genes were observed in the theca layer. The Bmal1 transcript was significantly elevated at ZT12 compared to ZT6 and ZT18 (Fig. 2E, p<0.001). In contrast, expression of both Cry1 and Per2 transcripts were significantly lower at ZT12 compared to ZT6 and ZT18 (Fig. 2E, p<0.01). Interestingly, the expression of these same clock genes in the F1 granulosa was opposite of the pattern observed in the F1 theca. Bmal1 (Fig. 2F, p<0.05, ANOVA with Bonferroni’s post-hoc analysis), transcripts in F1 granulosa displayed peak expression at ZT24/0, whereas Cry1 and Per2 were elevated at ZT12 (Fig. 2F, p<0.01 for Per2 and p>0.01 for Cry1, ANOVA with Bonferroni’s post-hoc analysis).
Acute Effects of the LH Surge on Circadian Clock Genes in the Chicken
To determine whether clock gene levels are affected by the endogenous LH surge, we examined expression levels of Bmal1, Cry1 and Per2 in two groups of birds (Fig. 3). To remove variability associated with endogenous 24-h changes in clock gene expression (as shown in Fig. 2), we obtained all samples at ZT6. Oviposition was carefully monitored. In the first group of birds (before LH), the LH surge occurred at ZT7 and ovulation at ZT11, thus sacrifice at ZT6 was before the LH surge (See Fig. 1). In the second group (after LH), the LH surge was expected to occur at ZT4 and ovulation at ZT8, thus sacrifice at ZT6 was after the LH surge. Although the LH surge is not rapid, the time frame used in these studies is similar to what we have previously used to perform studies in the chicken before and after the LH surge (Krishnan et al., 1993; Tischkau et al., 1996). Our previous experience dictates that this timing gives results similar to those obtained after culturing granulosa cells and applying LH (Jackson et al., 1994; Tischkau et al., 1997). Thus, we are confident that the LH surge had no effect on Bmal1, Cry1 or Per2 transcript levels in medio-basal hypothalamus or liver (Fig. 3, n=4). Similarly, Bmal1 was not increased in F1 theca layers after the LH surge (data not shown). Both Bmal1 (Fig. 3, p<0.05) and Per2 transcripts were significantly increased after the LH surge in F1 granulosa (Fig. 6, p<0.001, ANOVA with Bonferroni’s post-hoc analysis). Furthermore, Cry1 transcripts were significantly decreased after the LH surge (Fig. 3, p<0.05, ANOVA with Bonferroni’s post-hoc analysis).
Fig. 3. Effects of the endogenous LH surge on circadian clock gene transcripts in the chicken.
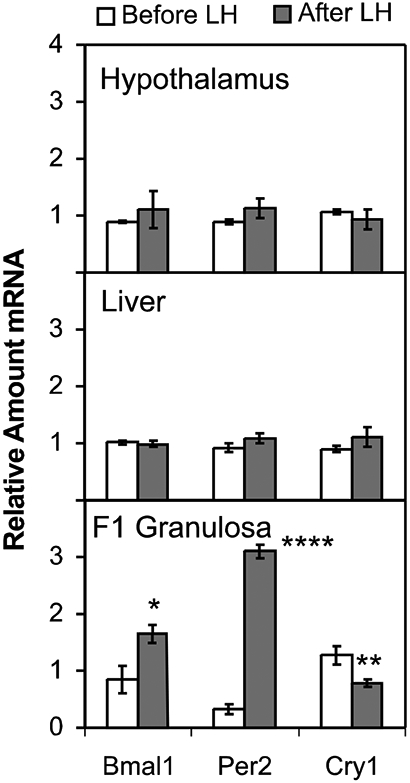
Animals were sacrificed either before or after an endogenous LH surge, predicted from the time of the previous oviposition (see Fig. 1). All tissues were obtained at ZT6 to control for time-of day dependent changes in gene expression. Transcript levels for Bmal1, Per2 and Cry1 were determined by quantitative PCR. Data represent mean ± SEM with n=4 for each group. Statistical significance was determined using ANOVA with Bonferroni’s post-hoc analysis (*, ** and **** represent p < 0.05, p < 0.01 and p < 0.0001, respectively).
Effects of LH on Rhythms of Per1 and Bmal1 in Cultured Granulosa Cells
To determine whether LH can affect the amplitude and phase of clock gene rhythms in granulosa cells, cultured granulosa cells were treated with LH. When cultured without any treatment, no rhythms in either Per1 or Bmal1 transcripts were observed (Fig. 4). LH treatment induced a rhythm of both Per1 and Bmal1 in these cells. In arrhythmic cells, LH induced a transient increase in Per1 transcripts followed by a rhythm that persisted for the duration of the culture (Fig. 4A). Peaks of Per1 expression occurred at 1 h (p<0.001), 16 h (p<0.01) and 40 h (p<0.01) after LH treatment. There was no acute change (at 1 h) in Bmal1 transcript levels after LH treatment. LH did, however, induce a rhythm in Bmal1 transcripts in the arrhythmic cells, with peak levels observed at 8 h (p < 0.01) and 32 h (p <0.01) after LH treatment.
Fig. 4. Effects of LH on Bmal1 and Per1 rhythms in cultured granulosa cells.
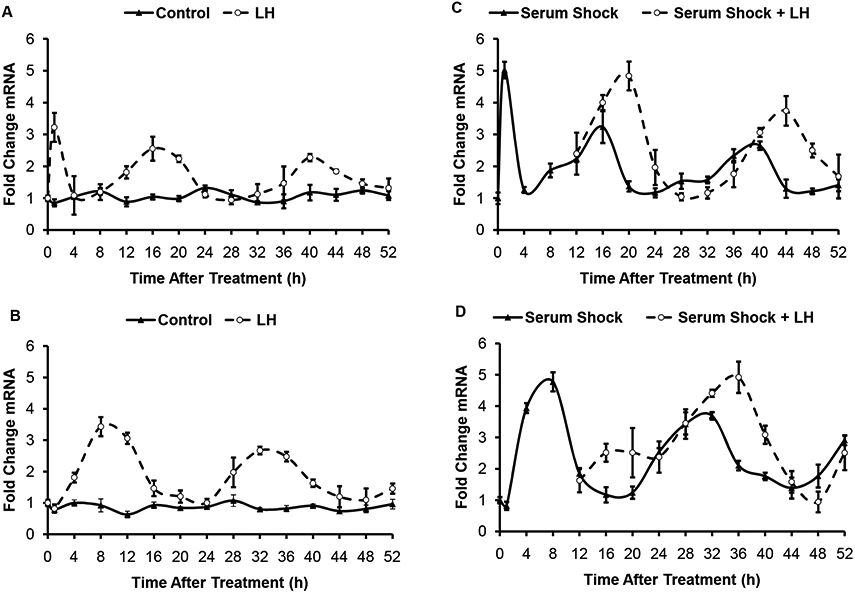
Per1 (A and C) and Bmal1 (B and D) transcripts were assessed after LH treatment of unsynchronized (A and B) or serum shocked (C and D) SIGC. Data means ± SEM of three independent experiments. Statistical significance was determined using ANOVA with Bonferroni’s post-hoc analysis but is not indicated on the graphs. Peaks for Per1 transcript levels occurred at 1h, 16h and 40h after LH treatment or serum shock. Peaks in Bmal1 transcripts occurred at 8h and 32 h after LH treatment or serum shock. LH treatment caused a significant 4-8 h phase delay in transcript expression peaks for both Per1 and Bmal1.
To determine whether LH can alter the rhythms in a population of cells that are rhythmic, granulosa cell cultures were serum shocked to induce a rhythm, and then treated with LH at 12 h after the serum shock. This treatment time was chosen to most closely mimic the Per2 and Bmal1 levels at the time of the LH surge in experiment 2. Serum shocked cells displayed rhythmic expression of Per1 and Bmal1 with peaks occurring out-of-phase at 16 h and 40 h for Per1 and 8 h and 32 h for Bmal1 (Fig. 4C and D). Similar to the results in the chicken in vivo, LH treatment in vitro increased both Per1 and Bmal1 by 4 h after treatment. Peak expression of both transcripts was altered. After LH, the peaks occurred at 20 h and 44 h, whereas the peaks for Bmal1 occurred at 36 h. Thus, LH induced a phase delay in the expression of both genes. Interestingly, the amplitude of the rhythm also seems to increase in cycling cells treated with LH.
Acute Effects of LH on Per1 and Egr-1 Transcripts in Cultured Granulosa Cells
To determine whether the LH effects on clock gene expression in granulosa cells is mediated by cAMP/PKA, Per1 transcript levels were determined 1 h after treatment of SIGC with forskolin (1uM), ovine luteinizing hormone (LH, 50 ng/ml), SQ22536 (100uM), or SQ22536 (100uM)/LH (50 ng/ml). Egr-1, a gene known to be activated in rat granulosa cells in response to FSH and LH treatment, served as a positive control (Espey et al., 2000; Russell et al., 2003). Treatment with LH and forskolin significantly elevated both Egr-1 and Per1 in SIGC (Fig. 5). In contrast, SQ alone had no significant effect on Per1 transcript levels. Although the levels of LHr are not dramatic we are confident in the presence of LHr at low levels as determined by qPCR (data not shown).
Fig. 5. Acute effects of LH on Egr-1 and Per1 transcripts in cultured granulosa cells.
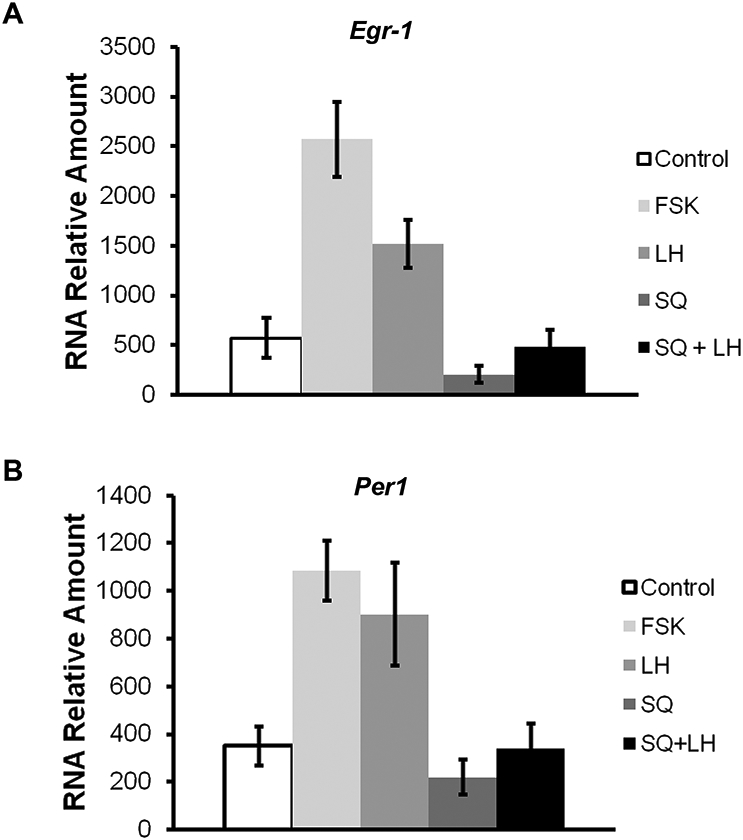
Transcript levels for the LH target gene, Egr-1 (A) Per1 (B) were determined by quantitative PCR in SIGC treated with either forskolin, LH, SQ22536 or LH/SQ22536. Data represent mean ± SEM with n=4-6 for each group. Statistical significance was determined using ANOVA with Bonferroni’s post-hoc analysis (* and ** represent p < 0.05 and p < 0.01, respectively).
DISCUSSION
Molecular clock mechanisms are highly conserved between birds and mammals. Core molecular gene networks are largely the same between these organisms. Major differences reside in the organization of the systemic circadian timing system in avian species. The primary circadian pacemaker in mammals resides in the SCN. The SCN assumes the responsibility of coordinating circadian function throughout the organism and aligning internal rhythmicity with the external environment. The avian circadian timing system is coordinately controlled by three pacemakers, residing in the pineal, the SCN and the retina. Independent oscillations of these “weak” pacemakers diminish over time. Sustained rhythmicity requires either photic stimulation from the environment or internal neuroendocrine input from other components of the system (Cassone and Menaker, 1984). Thus, robust internal rhythmicity requires interactions among the pacemakers. Periodic signals emitted by one pacemaker sustain and amplify oscillations of the other pacemakers, such that rhythmicity of each tissue is dependent on the other tissues (Gwinner et al., 1997; Gwinner and Brandstatter, 2001). Mechanisms by which this inter-related systems of pacemakers act to influence rhythmicity in other tissues remains to be determined. It is possible that each pacemaker acts independently to regulate specific downstream events (Bell-Pedersen et al., 2005). However, it is likely that the SCN and pineal pacemakers are most important in neuroendocrine regulation of rhythms in peripheral clocks. For example, any tissue containing melatonin receptors is a potential target for regulation by the pineal pacemaker. In the ovary, melatonin receptors are present in both granulosa and theca layers. The type 1a subtype of the melatonin receptor is present, however, only in the theca layer and its expression is significantly reduced in the largest preovulatory follicle (Sundaresan et al., 2009).
The data presented in this manuscript demonstrate that clock genes are rhythmically expressed both in primary pacemakers, such as pineal and hypothalamus containing the SCN, as well as in peripheral tissues. In mammals, expression of the positive element Bmal1 in an anti-phase relationship to the negative elements, Per1, Per2, Cry1 and Cry2 in any given tissue is considered evidence of a functional clock (Bell-Pedersen et al., 2005). Our data indicate that this anti-phase relationship is present in the avian pineal gland (Fig. 2B). Peak levels of Per2, occurred at ZT0, whereas Bmal1 peaked 12 h later, at ZT12. The expression patterns for Per2 and Bmal1 in the pineal are similar to previous reports (Chong et al., 2003; Yasuo et al., 2003; Herichova et al., 2008). Cry1 peak expression was slightly delayed compared to Per2, with peak expression occurring 6 h after lights on. The pattern for Cry1 was similar to previous reports in the 6-week old chick pineal (Csernus et al., 2007; Nagy and Csernus, 2007), but very different from the adult male Japanese quail, which displayed peak levels of Cry1 at night under the same lighting conditions (Yasuo et al., 2003). Interestingly, the pattern of expression of Cry1 in our studies most closely resembled that of Cry2 in the Japanese quail study, whereas of Cry2 in the 6-week old chick pineal mirrors Cry1 in the Japanese quail (Bailey et al., 2002). Collectively, the data suggest that roles for Cry1 and Cry2 may be reversed in pineal glands between the two galliform species.
We have also demonstrated significant cycling of clock genes in the hypothalamus containing the SCN. Although expression patterns differed between the three clock genes tested, there was not an obvious anti-phase relationship between the negative elements, Cry1 and Per2 and the positive element, Bmal1. In fact, the Cry1 expression pattern was very similar to the Bmal1 pattern. Expression of all three clock genes was similar to in situ hybridization studies that explored patterns of clock gene expression in adult Japanese quail (Yasuo et al., 2003). However, they were significantly different from patterns in medio-basal hypothalamus in that same study, which likely indicates that our samples were highly enriched with SCN tissue, although our dissection certainly included surrounding hypothalamic tissue. Furthermore, peak levels of each gene were delayed by 6 h in SCN/hypothalamus compared to the pineal.
The liver is an important site of peripheral clock function in mammals. Our study is the first to examine expression of both positive and negative clock elements in the liver of the adult hen. Both Bmal1 and Per2 displayed an oscillation. Consistent with a functional clock in this tissue, expression of Per2 was significantly out-of-phase with Bmal1. Per2 peaked at ZT0, whereas the peak for Bmal1 occurred later in the day, at ZT12-18. These patterns most closely resembled those that we observed in the pineal, although the Bmal1 peak occurred later in the liver. Peak expression of Bmal1 was similarly delayed by 4-8 h in livers of 6 week old chicks (Chong et al., 2003). No significant change in the Cry1 transcript level was observed in the liver over the 24-h period. In contrast, Cry1 displays robust oscillatory behavior in the liver of Japanese quail (Fu et al., 2002) and in mice (Miyamoto and Sancar, 1999), but Cry2 does not oscillate (Miyamoto and Sancar, 1999; Fu et al., 2002). These results raise the question of whether functions of Cry1 and Cry2 are potentially reversed under certain circumstances in the chicken.
Significant cycling of Bmal1, Cry1 and Per2 was also observed in the largest follicle (F1) of the chicken ovary. Analysis of granulosa layers separately from the overlying theca layer revealed interesting differences in expression patterns for each of the clock genes examined. In both F1 granulosa and F1 theca, Bmal1 cycled out-of phase with Per2 and Cry1, which were in phase with each other, suggesting that the molecular clock network is functional in the largest follicle. In F1 granulosa, Per2 and Cry1 levels were increasing during the early day, with peak levels observed at ZT12 for both transcripts. A weak, yet significant oscillation was observed for Bmal1 in F1 granulosa, with a peak at ZT0. In F1 theca, time-of-day dependent changes in the clock gene transcripts were also observed. However, peak expression of Bmal1 occurred at ZT12, whereas Cry1 and Per2 levels were very low at this time. A previous study in the Japanese quail has also shown cycling of clock genes in the F1 follicle. In that study, F1 theca and granulosa layers were examined separately at only two times of day. Per2 levels in the granulosa layer were increased during the day (ZT7) compared to night (ZT19); Bmal1 levels were not higher at ZT7 compared to ZT19. In the theca layers, neither Per2 nor Bmal1 appeared different between day and night. After pooling granulosa and theca layers and examining transcript levels at several times across the 24 h period, Per2 peaked at ZT7, with a trough at ZT19 and Bmal1 showed no significant oscillation (Nakao et al., 2007). Comparison of our data with the quail studies reveals some intriguing differences. Our data clearly indicate that Bmal1 is rhythmic. Levels are lower at ZT6 and ZT18 compared to ZT12. If our sampling was restricted to ZT6 and ZT18, as in the quail study, we would not have seen any differences in the theca. Bmal1 was, however, in complete anti-phase in the theca layer compared to the granulosa layer in our study, which may explain why no oscillation was observed in the F1 follicle of the quail ovary when granulosa and theca layers were pooled.
Neither the cause nor the significance of the anti-phase relationship of clock gene expression between granulosa and theca layers can be determined from this study. One might speculate that clock phase is regulated by different signals in the two cell types. Clearly, our data indicate that LH can regulate clock gene phase and amplitude in granulosa cells. In the bird, LH receptors are found on both granulosa and theca cells, but expression is substantially higher in granulosa cells, particularly in the largest follicle. LH increases progesterone production by the granulosa layer through increased expression of the steroidogenic acute regulatory protein (StAR) (Bahr and Johnson, 1984; Johnson and Bridgham, 2001; Johnson et al., 2002; Nakao et al., 2007). StAR is also regulated by BMAL1 in the mouse ovary (Boden et al.). Thus, the increase in Bmal1 observed after the LH surge may provide a mechanism to increase progesterone production through regulating the StAR protein. Alternatively, melatonin receptors are present in both cell types. Melatonin receptor expression is increased, however, in theca cells compared to granulosa cells. Melatonin reduces steroidogenic responsiveness of granulosa cells to LH (Murayama et al., 1997), which might explain why granulosa cells respond to LH and theca cells do not. Together with differential expression of LH receptors between the two cells types, this might provide a mechanism for LH to alter the rhythm in granulosa cells, thereby creating phase separation in clock gene expression between theca and granulosa layers. Further studies are necessary to establish the functional significance of differing clock phases between these cell types.
The Per2 profile in F1 granulosa cells in our studies indicate that Per2 transcripts peak after the LH surge. Furthermore, both Bmal1 and Per2 transcripts are increased in F1 granulosa cells harvested after the LH surge. Selecting groups of hens that laid either early (oviposition at ZT8) or late (oviposition at ZT11), allowed us to harvest samples either before or after the endogenous LH surge, yet at a single time-of-day, to control for 24 h variation in clock gene expression. Under these conditions, Bmal1 and Per2 were elevated subsequent to the LH surge. Whether the effects of LH on Per2 are mediated solely via actions on Bmal1 or through activation of cAMP signaling, remains to be tested. The adenylyl cyclase agonist forskolin acts independent of Clock/Bmal1 to drive transcription of mammalian Per1 and Per2 promoters (Travnickova-Bendova et al., 2002). LH may act directly on Bmal1, which in turn will drive an increase in Per2. In addition, it is likely that LH acts both directly and in concert with Bmal1 to promote increased Per2 levels because the effects of Clock/Bmal1 and LH on Per2 may be additive (Nakao et al., 2007). Our results in cultured granluosa cells indicate that gonadotropin-induced Per1 elevation is mediated by cAMP. The cells responded to gonadotropin despite expressing only modest levels of LHr, as indicated by elevation of the known LH target gene, Egr-1. Furthermore, the adenylyl cyclase antagonist blocked Per1 induction by gonadotropins in these cells.
LH altered both the phase and amplitude of clock gene rhythms in cultured granulosa cells. LH promoted rhythmic expression of Per1 and Bmal1 in cultures similar to the effects of serum shock. Bmal1 and Per1 were rhythmic, but out of phase in serum shocked and LH treated cultures. LH treatment of synchronized cultures caused the peak expression of both Per1 and Bmal1 to be delayed by 4 to 8 h in cultures, which is similar to the effects of LH on Per-luciferase rhythms in ovarian explants cultures (Yoshikawa et al., 2009). Thus it appears that LH has a classic resetting effect on the circadian clock that can be observed by the first 24 h after treatment.
Our aggregate data reveal an interesting relationship between clock gene expression and the LH surge in the chicken ovary, which are consistent with the hypothesis that the LH surge acts as a zeitgeber for the ovarian clock. Furthermore, the data raise the intriguing hypothesis that this clock plays a critical role in regulating the ovulatory cycle. It is not unreasonable to speculate that an internal coincidence timer governs the interaction of the ovarian clock with the LH surge in order to produce the open window of the ovulatory cycle. The ovarian clock may restrict the effects of LH such that they occur only when both LH and Bmal1 can act in concert to drive the ovulatory process. By acting on Bmal1 and StAR (Nakao et al., 2007), LH may stimulate, or at least amplify the production of progesterone required for generation of the surge. By acting on Per2, LH may regulate ovarian clock function. If the ovary is not prepared to receive LH, perhaps because the clock in the newly formed F1 follicle is not yet functional, or because LH arrives at a time when the clock cannot respond, ovulation will not occur. These data provide evidence that the ovary is more than a passive player in the ovulatory cycle. The important role of ovarian hormones in directing the period of the ovulatory cycle under constant lighting conditions (Kadono et al., 1981) and the fact that the body temperature rhythm in the hen is abolished in ovariectomized hens (Underwood et al., 1997; Zivkovic et al., 2000) suggest a pivotal role for the ovary in the complex network that is the avian circadian timing system.
REFERENCES
- Bahr JM, Johnson AL (1984) Regulation of the follicular hierarchy and ovulation. J Exp Zool 232:495–500. [DOI] [PubMed] [Google Scholar]
- Bailey MJ, Chong NW, Xiong J, Cassone VM (2002) Chickens' Cry2: molecular analysis of an avian cryptochrome in retinal and pineal photoreceptors. FEBS Lett 513:169–174. [DOI] [PubMed] [Google Scholar]
- Bell-Pedersen D, Cassone VM, Earnest DJ, Golden SS, Hardin PE, Thomas TL, Zoran MJ (2005) Circadian rhythms from multiple oscillators: lessons from diverse organisms. Nat Rev Genet 6:544–556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boden MJ, Varcoe TJ, Voultsios A, Kennaway DJ Reproductive biology of female Bmal1 null mice. Reproduction 139:1077–1090. [DOI] [PubMed] [Google Scholar]
- Bustin SA (2000) Absolute quantification of mRNA using real-time reverse transcription polymerase chain reaction assays. J Mol Endocrinol 25:169–193. [DOI] [PubMed] [Google Scholar]
- Cassone VM, Menaker M (1984) Is the avian circadian system a neuroendocrine loop? J Exp Zool 232:539–549. [DOI] [PubMed] [Google Scholar]
- Chong NW, Chaurasia SS, Haque R, Klein DC, Iuvone PM (2003) Temporal-spatial characterization of chicken clock genes: circadian expression in retina, pineal gland, and peripheral tissues. J Neurochem 85:851–860. [DOI] [PubMed] [Google Scholar]
- Csernus VJ, Nagy AD, Faluhelyi N (2007) Development of the rhythmic melatonin secretion in the embryonic chicken pineal gland. Gen Comp Endocrinol 152:148–153. [DOI] [PubMed] [Google Scholar]
- Espey LL, Ujioka T, Russell DL, Skelsey M, Vladu B, Robker RL, Okamura H, Richards JS (2000) Induction of early growth response protein-1 gene expression in the rat ovary in response to an ovulatory dose of human chorionic gonadotropin. Endocrinology 141:2385–2391. [DOI] [PubMed] [Google Scholar]
- Fahrenkrug J, Georg B, Hannibal J, Hindersson P, Gras S (2006) Diurnal rhythmicity of the clock genes Per1 and Per2 in the rat ovary. Endocrinology 147:3769–3776. [DOI] [PubMed] [Google Scholar]
- Fraps R (1954) Neural basis of diurnal periodicity in release of ovulation-inducing hormone in fowl. Proc Natl Acad Sci U S A 40:348–356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Z, Inaba M, Noguchi T, Kato H (2002) Molecular cloning and circadian regulation of cryptochrome genes in Japanese quail (Coturnix coturnix japonica). J Biol Rhythms 17:14–27. [DOI] [PubMed] [Google Scholar]
- Gwinner E, Brandstatter R (2001) Complex bird clocks. Philos Trans R Soc Lond B Biol Sci 356:1801–1810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gwinner E, Hau M, Heigl S (1997) Melatonin: generation and modulation of avian circadian rhythms. Brain Res Bull 44:439–444. [DOI] [PubMed] [Google Scholar]
- Herichova I, Monosikova J, Zeman M (2008) Ontogeny of melatonin, Per2 and E4bp4 light responsiveness in the chicken embryonic pineal gland. Comp Biochem Physiol A Mol Integr Physiol 149:44–50. [DOI] [PubMed] [Google Scholar]
- Jackson JA, Tischkau SA, Zhang P, Bahr JM (1994) Plasminogen activator production by the granulosa layer is stimulated by factor(s) produced by the theca layer and inhibited by the luteinizing hormone surge in the chicken. Biol Reprod 50:812–819. [DOI] [PubMed] [Google Scholar]
- Johnson AL, Bridgham JT (2001) Regulation of steroidogenic acute regulatory protein and luteinizing hormone receptor messenger ribonucleic acid in hen granulosa cells. Endocrinology 142:3116–3124. [DOI] [PubMed] [Google Scholar]
- Johnson AL, Solovieva EV, Bridgham JT (2002) Relationship between steroidogenic acute regulatory protein expression and progesterone production in hen granulosa cells during follicle development. Biol Reprod 67:1313–1320. [DOI] [PubMed] [Google Scholar]
- Kadono H, Besch EL, Usami E (1981) Body temperature, oviposition, and food intake in the hen during continuous light. J Appl Physiol 51:1145–1149. [DOI] [PubMed] [Google Scholar]
- Karman BN, Tischkau SA (2006) Circadian clock gene expression in the ovary: Effects of luteinizing hormone. Biol Reprod 75:624–632. [DOI] [PubMed] [Google Scholar]
- Krishnan KA, Proudman JA, Bolt DJ, Bahr JM (1993) Development of an homologous radioimmunoassay for chicken follicle-stimulating hormone and measurement of plasma FSH during the ovulatory cycle. Comp Biochem Physiol Comp Physiol 105:729–734. [DOI] [PubMed] [Google Scholar]
- Miyamoto Y, Sancar A (1999) Circadian regulation of cryptochrome genes in the mouse. Brain Res Mol Brain Res 71:238–243. [DOI] [PubMed] [Google Scholar]
- Murayama T, Kawashima M, Takahashi T, Yasuoka T, Kuwayama T, Tanaka K (1997) Direct action of melatonin on hen ovarian granulosa cells to lower responsiveness to luteinizing hormone. Proc Soc Exp Biol Med 215:386–392. [DOI] [PubMed] [Google Scholar]
- Nagy AD, Csernus VJ (2007) Cry1 expression in the chicken pineal gland: effects of changes in the light/dark conditions. Gen Comp Endocrinol 152:144–147. [DOI] [PubMed] [Google Scholar]
- Nakao N, Yasuo S, Nishimura A, Yamamura T, Watanabe T, Anraku T, Okano T, Fukada Y, Sharp PJ, Ebihara S, Yoshimura T (2007) Circadian clock gene regulation of steroidogenic acute regulatory protein gene expression in preovulatory ovarian follicles. Endocrinology 148:3031–3038. [DOI] [PubMed] [Google Scholar]
- Okano T, Yamamoto K, Okano K, Hirota T, Kasahara T, Sasaki M, Takanaka Y, Fukada Y (2001) Chicken pineal clock genes: implication of BMAL2 as a bidirectional regulator in circadian clock oscillation. Genes Cells 6:825–836. [DOI] [PubMed] [Google Scholar]
- Portaluppi F, Touitou Y, Smolensky MH (2008) Ethical and methodological standards for laboratory and medical biological rhythm research. Chronobiol Int 25:999–1016. [DOI] [PubMed] [Google Scholar]
- Russell DL, Doyle KM, Gonzales-Robayna I, Pipaon C, Richards JS (2003) Egr-1 induction in rat granulosa cells by follicle-stimulating hormone and luteinizing hormone: combinatorial regulation by transcription factors cyclic adenosine 3',5'-monophosphate regulatory element binding protein, serum response factor, sp1, and early growth response factor-1. Mol Endocrinol 17:520–533. [DOI] [PubMed] [Google Scholar]
- Stein LS, Stoica G, Tilley R, Burghardt RC (1991) Rat ovarian granulosa cell culture: a model system for the study of cell-cell communication during multistep transformation. Cancer Res 51:696–706. [PubMed] [Google Scholar]
- Sundaresan NR, Marcus Leo MD, Subramani J, Anish D, Sudhagar M, Ahmed KA, Saxena M, Tyagi JS, Sastry KV, Saxena VK (2009) Expression analysis of melatonin receptor subtypes in the ovary of domestic chicken. Vet Res Commun 33:49–56. [DOI] [PubMed] [Google Scholar]
- Tischkau SA (2008) Circadian Cycle. In: Encyclopedia of Neuroscience (Binder MD, Hirokawa N, Windhorst U, eds). Berlin Heidelberg New York: Springer. [Google Scholar]
- Tischkau SA, Jackson JA, Finnigan-Bunick C, Bahr JM (1996) Granulosa layer: primary site of regulation of plasminogen activator messenger ribonucleic acid by luteinizing hormone in the avian ovary. Biol Reprod 55:75–79. [DOI] [PubMed] [Google Scholar]
- Tischkau SA, Neitzel LR, Walsh JA, Bahr JM (1997) Characterization of the growth center of the avian preovulatory follicle. Biol Reprod 56:469–474. [DOI] [PubMed] [Google Scholar]
- Travnickova-Bendova Z, Cermakian N, Reppert SM, Sassone-Corsi P (2002) Bimodal regulation of mPeriod promoters by CREB-dependent signaling and CLOCK/BMAL1 activity. Proc Natl Acad Sci U S A 99:7728–7733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Underwood H, Siopes T, Edmonds K (1997) Eye and gonad: role in the dual-oscillator circadian system of female Japanese quail. Am J Physiol 272:R172–182. [DOI] [PubMed] [Google Scholar]
- Yamamoto K, Okano T, Fukada Y (2001) Chicken pineal Cry genes: light-dependent up-regulation of cCry1 and cCry2 transcripts. Neurosci Lett 313:13–16. [DOI] [PubMed] [Google Scholar]
- Yasuo S, Watanabe M, Okabayashi N, Ebihara S, Yoshimura T (2003) Circadian clock genes and photoperiodism: Comprehensive analysis of clock gene expression in the mediobasal hypothalamus, the suprachiasmatic nucleus, and the pineal gland of Japanese Quail under various light schedules. Endocrinology 144:3742–3748. [DOI] [PubMed] [Google Scholar]
- Yoshikawa T, Sellix M, Pezuk P, Menaker M (2009) Timing of the ovarian circadian clock is regulated by gonadotropins. Endocrinology 150:4338–4347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimura T, Suzuki Y, Makino E, Suzuki T, Kuroiwa A, Matsuda Y, Namikawa T, Ebihara S (2000) Molecular analysis of avian circadian clock genes. Brain Res Mol Brain Res 78:207–215. [DOI] [PubMed] [Google Scholar]
- Yoshimura Y, Bahr JM (1995) Atretic changes of follicular wall caused by destruction of the germinal disc region of an immature preovulatory follicle in the chicken: an electron microscope study. J Reprod Fertil 105:147–151. [DOI] [PubMed] [Google Scholar]
- Yoshimura Y, Tischkau SA, Bahr JM (1994) Destruction of the germinal disc region of an immature preovulatory follicle suppresses follicular maturation and ovulation. Biol Reprod 51:229–233. [DOI] [PubMed] [Google Scholar]
- Zivkovic BD, Underwood H, Siopes T (2000) Circadian ovulatory rhythms in Japanese quail: role of ocular and extraocular pacemakers. J Biol Rhythms 15:172–183. [DOI] [PubMed] [Google Scholar]