Abstract
The DNA region upstream of katG in Mycobacterium smegmatis was cloned and sequenced. The furA gene, highly homologous to Mycobacterium tuberculosis furA, mapped in this region. The furA-katG organization appears to be conserved among several mycobacteria. The transcription pattern of furA and katG in M. smegmatis upon oxidative stress was analyzed by Northern blotting and primer extension. Although transcription of both furA and katG was induced upon oxidative stress, transcripts covering both genes could not be identified either by Northern blotting or by reverse transcriptase PCR. Specific transcripts and 5′ ends were identified for furA and katG, respectively. By cloning M. smegmatis and M. tuberculosis DNA regions upstream of a reporter gene, we demonstrated the presence of two promoters, pfurA, located immediately upstream of the furA gene, and pkatG, located within the terminal part of the furA coding sequence. Transcription from pfurA was induced upon oxidative stress. A 23-bp sequence overlapping the pfurA −35 region is highly conserved among mycobacteria and streptomycetes and might be involved in controlling pfurA activity. Transcription from a cloned pkatG, lacking the upstream pfurA region, was not induced upon oxidative stress, suggesting a cis-acting regulatory role of this region.
Despite the seriousness of tuberculosis in the world, efforts in prevention and control of this disease suffer from the lack of detailed knowledge of the mechanisms used by pathogenic mycobacteria for survival within host cell macrophages. A variety of mechanisms contribute to the survival of Mycobacterium tuberculosis in the host, and protection against oxidative stress is one of the primary defenses (1). However, little is known about regulation and expression of the genes involved in oxidative stress resistance. Most bacteria, exposed to toxic oxygen metabolites, exhibit an adaptive response, expressing several genes involved in detoxification of oxygen species (4). The pathogenic mycobacteria have a limited, nonprotective oxidative stress response, which in the case of M. tuberculosis is restricted to induction of a single gene, katG, encoding a catalase-peroxidase (14, 28). Consistently, the KatG protein is an important virulence factor in M. tuberculosis, since its expression is required for growth and persistence in mouse and guinea pig model systems (17). The importance of a better understanding of the elements involved in katG regulation is increased by its implication in the innate susceptibility and acquired resistance to the front-line antituberculosis drug isoniazid (isonicotinic acid hydrazide) (9, 33). Indeed, inactivation of katG is frequently found among isoniazid-resistant M. tuberculosis strains (32).
Response to oxidative stress in mycobacteria differs from the OxyR-dependent induction observed in gram-negative bacteria (25). Indeed, both M. tuberculosis and Mycobacterium smegmatis lack a functional oxyR gene (6, 8, 31). In all the mycobacterial species studied, the DNA region immediately upstream of the katG genes is highly conserved and encodes the furA gene, suggesting a putative involvement of the FurA protein in katG regulation. Recently, Zahrt et al. (31) demonstrated that in M. smegmatis a knockout furA mutant overexpressed the catalase-peroxidase KatG and that this altered phenotype is complemented by expression of FurA; thus, FurA appears to be a negative regulator of katG.
Fur-like proteins are transcriptional repressors that exhibit a Fe2+-dependent DNA binding activity and regulate several genes involved in iron metabolism (10). There is an intimate relationship between iron metabolism and oxidative stress: indeed, the cytotoxic effects of reactive oxygen species are largely mediated by iron, and it has been shown elsewhere that regulators of Escherichia coli involved in oxidative stress response, OxyR and SoxRS, activate the expression of Fur, the global repressor of ferric iron uptake (34). E. coli Δfur mutants are more sensitive to hydrogen peroxide than are Fur-proficient strains (30). In M. smegmatis, hydrogen peroxide sensitivity is increased by iron starvation (18); in Staphylococcus aureus, Fur is necessary for oxidative stress resistance (15); and in Streptomyces spp., Fur-like proteins regulate catalase-peroxidase genes in an iron-dependent manner (13, 35).
In this work, using M. smegmatis as a model system, we investigated transcription of furA and katG after oxidative stress. Specific transcripts and promoters were identified for furA and katG, respectively. Although transcription of both genes was induced upon oxidative stress, an inducible promoter could be identified only upstream of furA. A similar transcriptional regulation was found in the furA-katG region of M. tuberculosis.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
E. coli XL1-Blue (3) was grown in LD medium (26), supplemented when necessary with kanamycin (50 μg/ml). M. smegmatis mc2155 was grown in LD medium containing 0.2% (vol/vol) glycerol and 0.05% (vol/vol) Tween 80 and supplemented when necessary with kanamycin (20 μg/ml) or spectinomycin (100 μg/ml).
Cloning and sequencing of M. smegmatis furA gene.
A cosmid library of M. smegmatis mc2155 was produced by standard procedures in the vector Tropist4 (7). The library was screened using a 380-bp DNA internal to the katG coding region, obtained by PCR amplification with a couple of primers (218 and 219 [see Fig. 1]) based on the published sequence (GenBank accession no. U46844).
FIG. 1.
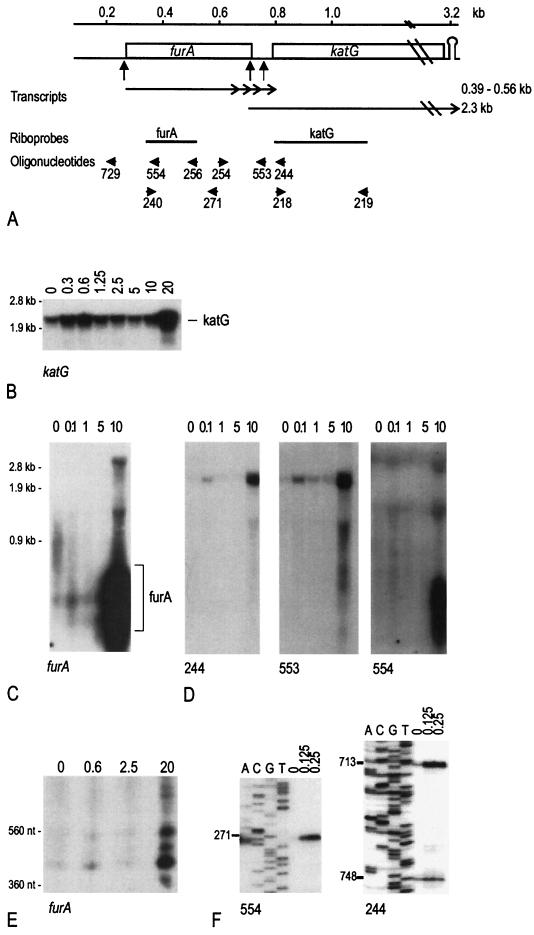
Transcription of the furA-katG region in M. smegmatis. (A) Map of the region. The furA-katG map of M. smegmatis is drawn to scale according to the sequence in the GenBank database with accession no. AF196484 (coordinate 1 corresponds to nt 245 of the deposited sequence). The genes are indicated by boxes. The potential Rho-independent termination site, downstream of katG, is shown. The transcripts are indicated by arrows, and the 5′ ends are indicated by vertical arrows below the map. The positions of the riboprobes and oligonucleotides used for Northern blotting, primer extension analysis, and RT-PCR are reported. (B to E) Northern blotting analysis of furA-katG transcription upon oxidative stress. RNAs were extracted from a culture of M. smegmatis mc2155 treated for 1 h with the millimolar concentration of hydrogen peroxide indicated on the top of the lanes, and equal amounts of each sample (20 μg) were separated by agarose (B, C, and D) or polyacrylamide (E) gel electrophoresis, blotted, and hybridized to the different probes. (B) Riboprobe katG (coordinates 724 to 1113); (C and E) riboprobe furA (coordinates 332 to 511); (D) the same filter used in panel C was hybridized successively to oligonucleotides 554 (coordinates 366 to 385), 553 (coordinates 715 to 742), and 244 (coordinates 803 to 822). Molecular size markers, run in the same gel, are indicated on the left. For panels C and D, probe 554 was exposed about three times longer. In agarose gels, two nonspecific bands (about 1.5 and 3 kb) were observed after prolonged exposure of the filters (particularly visible in panels C and D, oligonucleotide 554). These bands are likely to be furA-katG RNA entrapped by rRNA (5). (F) Primer extension of 5′ ends of furA and katG transcripts. The primer extension experiments were performed with the 554 (furA) and 244 (katG) oligonucleotides. Sequence reactions, performed with the same oligonucleotides on MS3 DNA (Table 1), are reported in the first four lanes of the two panels. The coordinates of the 5′ ends are reported on the left of each panel.
Construction of plasmids.
The pSG10ter vector was derived from pSG10 (11) by inserting the E. coli Rho-independent Ω terminator (24) upstream of the reporter gene luxAB, in order to reduce basal luciferase expression. Plasmid pSM128 is an integrative vector carrying the lacZ reporter gene (29). Plasmid pSG10ter and pSM128 derivatives, containing DNA fragments from either M. smegmatis (MS3, MYS42, MYS44, MYS48, MYS65, and MYS66) or M. tuberculosis (MT3, MYT43, and MYT45), were obtained by PCR amplification of the regions (the oligonucleotide sequences will be provided upon request). All the inserted fragments were sequenced.
RNA extraction, Northern blotting, and primer extension analysis.
Fifteen milliliters of M. smegmatis mc2155 cultures (optical density at 600 nm = 0.8), untreated or treated with H2O2 for 1 h at 37°C at the concentrations indicated in Fig. 1, was pelleted and resuspended in 100 μl of Tris-EDTA buffer; 75 μl of RNA lysis buffer (4 M guanidinium thiocyanate, 0.01 M Tris [pH 7.5], 0.97% β-mercaptoethanol) was added; and the suspension was sonicated (20 s at 40 W). The RNA was then purified using the SV Total RNA Isolation System (Promega). Twenty micrograms of each RNA sample was fractionated, blotted, and hybridized as described previously (5). The 32P-labeled riboprobes furA and katG were prepared by SP6 polymerase transcription of pGEMT-easy (Promega) derivatives, carrying the M. smegmatis 349–509 (oligonucleotides 240 and 256) and 724-1113 (oligonucleotides 218 and 219) DNA regions, respectively (see Fig. 1). The oligonucleotide probes (indicated in Fig. 1) were 5′ end labeled with T4 polynucleotide kinase as described by Sambrook et al. (27). Potential transcription start sites in the M. smegmatis furA-katG region were identified by primer extension analysis (27), using the oligonucleotides indicated.
Reverse transcriptase PCR (RT-PCR).
Two micrograms of RNA extracted from M. smegmatis mc2155 mock treated and treated for 1 h with either 0.6 or 20 mM H2O2 was retrotranscribed with Moloney murine leukemia virus reverse transcriptase (Promega). Samples (50 ng) were amplified with oligonucleotides 254 and 219 and, as a control, with oligonucleotides 218 and 219 and oligonucleotides 240 and 271. The presence of an amplified DNA was evaluated both by ethidium bromide staining and by Southern blot hybridization.
Luciferase assay.
Independent cultures of M. smegmatis mc2155 carrying the plasmids indicated in Table 1 were grown in LD medium at 37°C. The turbidity of the cultures at 600 nm was adjusted to 0.03, and the cultures were sampled and either mock treated or treated with H2O2 (final concentration, 0.125 mM) for 1 h at 37°C. One hundred microliters of 1% (vol/vol) decyl aldehyde solution (Sigma-Aldrich) in ethanol was then added to 1 ml of a 1:10 dilution of the culture, and luminescence was measured over 10 s, using a Stratec luminometer. At least three independent clones of each construct were tested.
TABLE 1.
Expression of luciferase and β-galactosidase from M. smegmatis and M. tuberculosis DNA fragments
| Plasmida | Coordinates of mycobacterial fragmentb | Luciferase (relative units)c
|
β-Galactosidase (relative units)c
|
||||
|---|---|---|---|---|---|---|---|
| −H2O2 | +H2O2 | Relative increment | −H2O2 | +H2O2 | Relative increment | ||
| pSG10ter | 1.5 ± 0.6 | 1.9 ± 0.8 | 0.95 ± 0.04 | NTd | NT | NT | |
| pSM128* | NT | NT | NT | 1.2 ± 0.3 | 1.3 ± 0.3 | 1.11 ± 0.02 | |
| MS3 | 137–821 | 430 ± 127 | 1,782 ± 297 | 4.26 ± 0.76 | NT | NT | NT |
| MYS42/MYS65* | 222–821 | 68 ± 8 | 77 ± 20 | 1.58 ± 0.06 | 172 ± 15 | 241 ± 5.2 | 1.32 ± 0.005 |
| MYS44/MYS66* | 258–821 | 51 ± 7 | 35 ± 0.1 | 0.70 ± 0.09 | 15.4 ± 0.7 | 15.3 ± 1.5 | 0.99 ± 0.045 |
| MYS48 | 669–821 | 17 ± 8 | 13 ± 5 | 0.78 ± 0.10 | NT | NT | NT |
| MT3 | 157–766° | 106 ± 12 | 308 ± 36 | 2.91 ± 0.01 | NT | NT | NT |
| MYT43 | 264–766° | 81 ± 62 | 97 ± 65 | 1.40 ± 0.30 | NT | NT | NT |
| MYT45 | 296–766° | 137 ± 33 | 92 ± 38 | 0.63 ± 0.12 | NT | NT | NT |
pSG10ter derivatives; pSM128 derivatives are indicated by an asterisk.
Coordinates of the M. smegmatis sequence are arbitrary, with coordinate 1 corresponding to position 245 in the sequence deposited in the GenBank database (accession no. AF196484). Coordinates of the M. tuberculosis sequence (°) are arbitrary, with coordinate 1 corresponding to position 2,156,900 in the reverse strand of the M. tuberculosis complete genome sequence (GenBank accession no. AL123456).
Luciferase and β-galactosidase activities expressed by M. smegmatis mc2155 transformed with the plasmids were measured, as described in Materials and Methods, both in the absence of (−H2O2) and after (+H2O2) oxidative stress. Treatment with 0.125 mM and 0.5 mM H2O2 for 1 h was used in luciferase and β-galactosidase assays, respectively, depending on the different cell concentrations used in the two assays. The x̄ values ± standard deviations of three to five independent clones tested are reported. The relative increment of activity upon oxidative stress is also indicated.
NT, not tested.
β-Galactosidase assay.
The cultures were grown as described for the luciferase assay to an optical density at 600 nm of 1.7 and mock treated or treated for 1 h with 0.5 mM H2O2 at 37°C, and the cells were disrupted by sonication. β-Galactosidase activity of the extracts was measured as described in reference 20. The enzyme activity was expressed as nanomoles of o-nitrophenol-β-d-galactopyranoside converted to o-nitrophenol minute−1 milligram of protein−1.
Nucleotide sequence accession number.
The sequence upstream of katG was determined and deposited in the GenBank database under accession number AF196484.
RESULTS
Cloning and sequencing of M. smegmatis furA.
The katG region of M. smegmatis was identified by screening a cosmid library with an internal katG PCR fragment as a probe (see Materials and Methods). Sequence inspection of this region revealed the presence of an open reading frame predicted to encode a polypeptide of 153 amino acids, with a calculated molecular mass of 17 kDa, homologous to Fur-like proteins. The gene was named furA, by analogy with other fur genes located upstream of katG in mycobacteria (23) (Fig. 1A). A Shine-Dalgarno sequence is present upstream of katG, while no typical Shine-Dalgarno sequence was identified upstream of furA.
Comparison of the M. smegmatis and M. tuberculosis furA-katG nucleotide regions by FASTA indicated a 70 to 80% identity in the region starting about 40 bp upstream of the start codon of furA and ending near the end of the katG coding sequence; the adjacent regions diverge completely. Analysis of the sequence upstream of furA indicated the presence of the terminal 149 codons of an open reading frame, encoding a polypeptide highly homologous (129 of 149 amino acids identical) to an M. tuberculosis hypothetical oxidoreductase (GenBank accession no. B70697). However, the corresponding gene in M. tuberculosis maps in a different part of the genome.
Transcription of furA-katG in M. smegmatis upon oxidative stress.
Transcription of katG and furA in M. smegmatis in response to oxidative stress was analyzed by Northern blotting, and the transcripts were mapped by hybridization with different probes. The RNAs were extracted from mc2155 cultures after 1 h of treatment with different concentrations of hydrogen peroxide (Fig. 1B, C, D, and E). The katG coding sequence is 2,223 nucleotides (nt) long. A single transcript, about 2.3 kb long, hybridized with the katG riboprobe (Fig. 1B). The same transcript could be observed with the 244 probe, internal to katG, and with the 553 probe, encompassing the 3′ end of the furA coding sequence (Fig. 1D). With the furA riboprobe and with the 554 oligonucleotide, both mapping in the 5′ half of the furA coding sequence, the 2.3-kb transcript was not detected, even after prolonged exposure of the filter (Fig. 1C and D). Thus, the katG transcript extended to the intergenic region between furA and katG but did not cover the whole furA gene. Immediately downstream of katG, a potential Rho-independent termination site was identified by the TERMINATOR program of the GCG package (data not shown).
The basal level of the katG mRNA was relatively abundant, and the intensity of the signal varied upon oxidative stress: as determined by PhosphorImager analysis, a two- to threefold increase compared to untreated control was detected upon exposure at low doses (0.1 to 0.6 mM H2O2 for 1 h); at higher doses (1 to 10 mM H2O2), a decrease was observed that reduced the intensity of the signal to 1.5-fold the basal level. A strong increase (about 10-fold) was present after severe treatment with hydrogen peroxide (10 to 20 mM; the concentration of H2O2 causing this induction varied by a factor of 2 in different experiments). The induction at low doses is likely to be the adaptive response, whereas the latter increase might be a response to a global alteration of cell metabolism.
The furA riboprobe and the 554 oligonucleotide hybridized to transcripts less than 0.6 kb long, which were better resolved in a polyacrylamide gel (Fig. 1C, D, and E). Although the intensity of these signals was lower than that of katG RNA (a three-times-longer exposure was required for their detection), the induction pattern upon oxidative stress appeared similar: a 2.2-fold increase at low doses, followed by a decrease to the basal level at 1 to 5 mM and an approximately 10-fold increase at ≥10 mM H2O2 (Fig. 1E). In this latter condition, four RNAs, 390, 430, 480, and 560 nt long, were identified.
Transcripts covering both furA and katG could not be identified by Northern blotting, not even after prolonged exposures of the filters. The absence of furA-katG transcripts was confirmed by RT-PCR using RNA extracted from both noninduced and H2O2-induced cells (data not shown; see Materials and Methods).
Identification of promoter regions upstream of katG and furA.
The 5′ ends of the transcripts were mapped by primer extension. Using an oligonucleotide internal to the furA coding sequence (oligonucleotide 554 in Fig. 1A), a unique 5′ end was identified at nt 271 (Fig. 1F), 1 base upstream of the translation start codon of furA. The intensity of the signal increased 9- to 12-fold upon oxidative stress. No other signals could be identified further upstream, neither with this oligonucleotide nor with an oligonucleotide located upstream of furA (oligonucleotide 729 in Fig. 1A; data not shown).
With an oligonucleotide internal to the katG coding sequence (oligonucleotide 244 in Fig. 1A), two bands were identified at 713 and 748 nt (Fig. 1F). The signal at nt 713 is located within the terminal part of the furA coding sequence; the nt 748 end maps in the intergenic region (Fig. 1A). No signals could be identified further upstream; in particular, no signal at nt 271 was observed (data not shown). The position of the two 5′ ends is consistent with the Northern blotting data, which indicated that the 2.3-kb transcript started upstream of katG. The nt 713 band increased about fourfold upon oxidative stress, whereas the nt 748 band showed only a less-than-twofold intensification.
In order to identify promoter regions, we cloned fragments of the M. smegmatis furA-katG DNA in pSG10ter upstream of the bacterial luxAB reporter gene and measured luciferase expressed by the constructs. The cloned fragments, obtained by PCR amplification, are indicated in Table 1. The constructs were tested for luciferase activity in M. smegmatis mc2155 both in the absence of and upon oxidative stress. Consistent with the results obtained by Northern blotting, MS3 and MYS42, whose cloned fragments extend, respectively, from nt −135 and nt −50 upstream of furA to the first codons of katG, expressed a high level of luciferase that was increased upon peroxide treatment. (In MYS42 the increase was about three times lower than that in MS3.)
In contrast, luciferase expression by MYS44, whose DNA region extends from nt −14 upstream of furA to the first codons of katG, did not increase upon oxidative stress (the slight reduction of luciferase activity observed upon 1 h of H2O2 treatment was probably caused by cell lethality). Moreover, the DNA fragment cloned in MYS48, which covers the terminal part of furA, also expressed a low noninducible luciferase activity. These results suggest the following.
(i) MS3 and MYS42 contain an inducible promoter, pfurA, which requires the region upstream of −50 for oxidative stress activation. Sequence inspection of this region revealed the presence of potential −35 (TTGACT) and −10 (TAGCCT) consensus sequences. pfurA might be responsible for the 5′ end at nt 271 identified by primer extension.
(ii) In the DNA fragment cloned in MYS44, the −35 region of pfurA is deleted. The noninducible activity expressed by MYS44 and MYS48 might be due to a weak promoter, pkatG, located in the terminal region of furA. This promoter might be responsible for the 5′ ends at nt 713 and 748 identified by primer extension. However, transcription from pkatG was not induced upon oxidative stress in either MYS44 or MYS48.
To confirm these results, we cloned the same DNA fragments in a different reporter system, using the integrative vector pSM128 and lacZ as a reporter gene (Table 1). β-Galactosidase activity expressed by the constructs clearly indicated the presence of two promoters, pfurA and pkatG, and confirmed that pkatG was not induced upon oxidative stress in the absence of the region upstream of −50.
Promoters of furA and katG in M. tuberculosis.
Fragments of M. tuberculosis DNA of the furA-katG region were cloned in pSG10ter. M. smegmatis mc2155 was transformed with the plasmids, and luciferase activity was determined (Table 1). The DNA regions cloned were homologous to the corresponding M. smegmatis fragments, except for the first 113 nt of MT3. The expression of luciferase activity from the different plasmids indicated that in M. tuberculosis furA-katG transcription is regulated in a way similar to that found in M. smegmatis: an inducible pfurA promoter is located immediately upstream of furA, where −35 (TTGACT) and −10 (TATTGT) sequences were identified. Deletion of the −35 region eliminated inducibility upon oxidative stress (compare MYT43 and MYT45), although a basal level of luciferase activity was still observed.
DISCUSSION
Expression of the furA-katG region in M. smegmatis.
We have cloned and sequenced the M. smegmatis region upstream of katG. It contains the furA gene, which codes for FurA, one of the two Fur-like proteins identified in the M. tuberculosis genome. Zahrt et al. (31) have recently sequenced the same region of M. smegmatis (GenBank accession no. AF012631). The FurA proteins deduced from the two sequences differ for some residues, with FurA deduced from our sequence presenting a higher similarity to M. tuberculosis FurA. The M. smegmatis FurA protein conserves both the helix-turn-helix DNA binding domain (35) and the metal binding domain (2), suggesting that this protein might carry out regulatory functions in response to stress conditions such as iron depletion or oxidative stress.
The furA-katG organization is conserved in many mycobacteria (23) as well as in the closely related Streptomyces coelicolor (13) and Streptomyces reticuli (35), and a regulatory role for the FurS protein on expression of catalase-peroxidase has been demonstrated previously in streptomycetes (13, 22). Moreover, it has been demonstrated elsewhere that katG expression is enhanced in an M. smegmatis Δfur mutant strain, suggesting that FurA negatively regulates katG expression (31).
Our transcription analysis in M. smegmatis indicates that both furA and katG are induced upon oxidative stress, although, contrary to what was previously reported for mycobacteria and streptomycetes (13, 19, 22, 23), distinct messengers for each gene were found.
Transcription of furA.
Four transcripts covering furA were detected with a riboprobe encompassing the 5′ half of furA. The two longest ones cover the entire furA coding sequence, whereas the two shorter terminate before the end of the gene. Thus, they may either represent stable intermediates of the processed furA transcript or be produced by premature transcription termination.
The transcription from pfurA starts at nt 271, just 1 base upstream of the GTG initiation codon of furA. The leaderless mRNA is consistent with the lack of a Shine-Dalgarno sequence upstream of the furA gene (12, 16). Upstream of the transcription start point, −35 (TTGACT) and −10 (TAGCCT) consensus sequences are found. Corresponding sequences are present in M. tuberculosis upstream of furA (TTGACT and TATTGT, respectively). The expression of luciferase and β-galactosidase in the reporter systems used confirmed the presence of a promoter, induced upon oxidative stress, both in M. smegmatis and in M. tuberculosis. The presence of pfurA in M. tuberculosis has been recently demonstrated (19).
Deletion of the 222–258 region in M. smegmatis and of the equivalent region of M. tuberculosis (264–296 [Table 1]) abolished H2O2 inducibility. These deletions cover the −35 pfurA sequences. An AT-rich region of 23 bp overlaps the −35 sequence of pfurA in M. smegmatis (ATTCTTGACTAATTCCAGAAAAG) and in M. tuberculosis (AGTCTTGACTGATTCCAGAAAAG). A highly homologous region was found in several mycobacteria upstream of furA (Mycobacterium bovis, Mycobacterium fortuitum, and Mycobacterium leprae) and in S. reticuli upstream of furS, and it has been demonstrated elsewhere that FurS, the protein corresponding to FurA in Streptomyces, binds to this region in a redox-dependent manner (22). Similarly, FurA might be a redox-sensing regulator both in M. smegmatis and in M. tuberculosis. In its reduced form, the mycobacterial FurA protein might bind to the AT-rich region, acting as a repressor of transcription initiation. Upon oxidative stress, the oxidized FurA protein might lose its DNA binding activity, allowing transcription from pfurA. Thus, FurA might autoregulate its own expression. Consistently, purified M. tuberculosis FurA changed its electrophoretic mobility in sodium dodecyl sulfate-polyacrylamide gel electrophoresis upon addition of increasing amounts of dithiothreitol (data not shown).
BLAST analysis indicated that the 23-bp AT-rich region is unique in the M. tuberculosis genome. Thus, either FurA, unlike other Fur proteins, is not a global regulator or different DNA sequences might be bound by FurA. It appears noteworthy that the high homology between M. smegmatis and M. tuberculosis furA-katG DNA regions starts exactly at the 23-bp sequence.
Transcription of katG.
Two possible 5′ ends were identified for the 2.3-kb katG transcript. This suggests the presence of two mRNAs, differing by 35 bp in length, that were not resolved by agarose gel electrophoresis. The transcript starting at nt 713 might be partially processed, generating the nt 748 5′ end. Alternatively, transcription may start at both sites. Similarly, for M. tuberculosis Mulder et al. (21) identified three potential 5′ ends for katG transcripts, two of them internal to the furA coding sequence and one in the intergenic region, whereas Master et al. (19) identified a single 5′ end internal to furA, 54 nt upstream of the katG coding sequence.
The recent work of Master et al. (19), based on S1 nuclease protection experiments, reports that in M. tuberculosis furA and katG are cotranscribed. Our data, for M. smegmatis, support a different hypothesis, with independent transcription of the two genes. It cannot be excluded that transcription in the two mycobacterial species differs. However, S1 protection experiments did not detect a transcript covering both furA and katG; rather, they indicated the presence of RNAs that cover furA and extend within the first few nucleotides of the katG coding sequence (the probe used extends to the first 22 nt of katG). An M. tuberculosis RNA equivalent to the 560-nt-long furA transcript that we observed by Northern analysis might be responsible for the 5′ end at pfurA.
Upon mild oxidative stress, the 2.3-kb transcript and the intensity of the nt 713 signal increased about three- to fourfold, whereas the nt 748 signal appeared to be less inducible (less than twofold). No consensus promoter sequences were identified upstream of either 5′ signal. However, by cloning this M. smegmatis DNA region (MYS44 and MYS66) and the corresponding M. tuberculosis DNA region (MYT45) upstream of the luciferase and the β-galactosidase genes, a low promoter activity was detected, which did not increase upon oxidative stress. Thus, in the reporter system used pkatG was not inducible.
The response to oxidative stress of pkatG might require the presence of the upstream region, encompassing pfurA. Indeed, Master et al. (19) demonstrated that M. tuberculosis pkatG was induced upon oxidative stress by using a construct that includes the upstream furA region (similar to our MT3 [Table 1]). We suggest that basal katG expression depends on the activity of pkatG and that cis-acting regulatory sites, necessary for activation by oxidative stress, are located in the upstream region. In agreement with the idea that this region contains a promoter activator, Mulder et al. (21) identified in M. tuberculosis an upstream activation region required for pkatG expression. The upstream activation region, which includes pfurA, enhanced transcription when cloned upstream of the exogenous Mycobacterium paratuberculosis PAN promoter.
Recent results (31) demonstrated that katG expression is greatly activated in a furA knockout mutant, whereas overexpression is reduced by the FurA protein provided in trans. Thus, FurA may act as a katG repressor either directly (by interacting with a specific regulatory region upstream of katG) or indirectly (by repressing expression of a katG activator). Furthermore, the possibility that FurA controls catalase-peroxidase expression at the posttranscriptional level cannot be excluded. The identification of possible targets recognized by FurA is under investigation.
ACKNOWLEDGMENTS
A. Milano and F. Forti equally contributed to the work.
We thank S. Gordon and F. Bigi for providing plasmids pSG10 and pSM128. We thank G. Dehò for stimulating discussions and for reading the manuscript.
This work was supported by grant COFIN99 from the Ministero dell'Università e della Ricerca Scientifica e Tecnologica, Rome, and by Università di Pavia FAR 2001.
REFERENCES
- 1.Aderem A, Underhill D M. Mechanisms of phagocytosis in macrophages. Annu Rev Immunol. 1999;17:593–623. doi: 10.1146/annurev.immunol.17.1.593. [DOI] [PubMed] [Google Scholar]
- 2.Bagg A, Neilands J B. Molecular mechanism of regulation of siderophore-mediated iron assimilation. Microbiol Rev. 1987;51:509–518. doi: 10.1128/mr.51.4.509-518.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bullock W, Fernandez J, Short J. XL1-Blue: a high efficiency plasmid transforming recA E. coli strain with β-galactosidase selection. BioTechniques. 1987;5:376–379. [Google Scholar]
- 4.Christman M F, Morgan R W, Jacobson F S, Ames B N. Positive control of a regulon for defences against oxidative stress and some heat-shock protein in Salmonella typhimurium. Cell. 1985;41:753–762. doi: 10.1016/s0092-8674(85)80056-8. [DOI] [PubMed] [Google Scholar]
- 5.Dehò G, Zangrossi S, Sabbattini P, Sironi G, Ghisotti D. Bacteriophage P4 immunity controlled by small RNAs via transcription termination. Mol Microbiol. 1992;6:3415–3425. doi: 10.1111/j.1365-2958.1992.tb02209.x. [DOI] [PubMed] [Google Scholar]
- 6.Deretic V, Song J, Pagàn-Ramos E. Loss of oxyR in Mycobacterium tuberculosis. Trends Microbiol. 1997;5:367–372. doi: 10.1016/S0966-842X(97)01112-8. [DOI] [PubMed] [Google Scholar]
- 7.De Smet K A, Jamil S, Stoker N G. Tropist3: a cosmid vector for simplified mapping of both G+C-rich and A+T-rich genomic DNA. Gene. 1993;136:215–219. doi: 10.1016/0378-1119(93)90467-h. [DOI] [PubMed] [Google Scholar]
- 8.Dhandayuthapani S, Zhang Y, Mudd M H, Deretic V. Oxidative stress response and its role in sensitivity to isoniazid in mycobacteria: characterization and inducibility of ahpC by peroxides in Mycobacterium smegmatis and lack of expression in M. aurum and M. tuberculosis. J Bacteriol. 1996;178:3641–3649. doi: 10.1128/jb.178.12.3641-3649.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dussurget O, Smith I. Interdependence of mycobacterial iron regulation, oxidative-stress response and isoniazid resistance. Trends Microbiol. 1998;6:354–358. doi: 10.1016/s0966-842x(98)01307-9. [DOI] [PubMed] [Google Scholar]
- 10.Escolar L, Pèrez-Martin J, de Lorenzo V. Opening the iron box: transcriptional metalloregulation by the Fur protein. J Bacteriol. 1999;181:6223–6229. doi: 10.1128/jb.181.20.6223-6229.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gordon S, Parish T, Roberts I S, Andrew P W. The application of luciferase as a reporter of environmental regulation of gene expression in mycobacteria. Lett Appl Microbiol. 1994;19:336–340. doi: 10.1111/j.1472-765x.1994.tb00469.x. [DOI] [PubMed] [Google Scholar]
- 12.Grill S, Gualerzi C O, Londei P, Blasi U. Selective stimulation of translation of leaderless mRNA by initiation factor 2: evolutionary implications for translation. EMBO J. 2000;19:4101–4110. doi: 10.1093/emboj/19.15.4101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hahn J S, Oh S Y, Roe J H. Regulation of the furA and catC operon, encoding a ferric uptake regulator homologue and catalase-peroxidase, respectively, in Streptomyces coelicolor A3(2) J Bacteriol. 2000;182:3767–3774. doi: 10.1128/jb.182.13.3767-3774.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Heym B, Zhang Y, Poulet S, Young D, Cole S T. Characterization of the katG gene encoding a catalase-peroxidase required for the isoniazid susceptibility of Mycobacterium tuberculosis. J Bacteriol. 1993;175:4255–4259. doi: 10.1128/jb.175.13.4255-4259.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Horsburgh M J, Ingham E, Foster S J. In Staphylococcus aureus, Fur is an interactive regulator with PerR, contributes to virulence, and is necessary for oxidative stress resistance through positive regulation of catalase and iron homeostasis. J Bacteriol. 2001;183:468–475. doi: 10.1128/JB.183.2.468-475.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Janssen G R. Eubacterial, archaebacterial, and eukaryotic genes that encode leaderless mRNA. In: Baltz R H, Hegeman G D, Skatrud P L, editors. Industrial microorganisms: basic and applied molecular genetics. Washington, D.C.: American Society for Microbiology; 1993. pp. 59–67. [Google Scholar]
- 17.Li Z, Kelley C, Collins F, Rouse D, Morris S. Expression of katG in Mycobacterium tuberculosis is associated with its growth and persistence in mice and guinea pigs. Infect Immun. 1998;67:74–79. doi: 10.1086/515254. [DOI] [PubMed] [Google Scholar]
- 18.Lundrigan M D, Arceneaux J E, Zhu W, Byers B R. Enhanced hydrogen peroxide sensitivity and altered stress protein expression in iron-starved Mycobacterium smegmatis. Biometals. 1997;10:215–225. doi: 10.1023/a:1018355928990. [DOI] [PubMed] [Google Scholar]
- 19.Master S, Zahrt T C, Song J, Deretic V. Mapping of Mycobacterium tuberculosis katG promoters and their differential expression in infected macrophages. J Bacteriol. 2001;183:4033–4039. doi: 10.1128/JB.183.13.4033-4039.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Miller J M. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1972. [Google Scholar]
- 21.Mulder M A, Zappe H, Steyn L M. The Mycobacterium tuberculosis katG promoter region contains a novel upstream activator. Microbiology. 1999;145:2507–2518. doi: 10.1099/00221287-145-9-2507. [DOI] [PubMed] [Google Scholar]
- 22.Ortiz de Orué Lucana D, Schrempf H. The DNA-binding characteristics of the Streptomyces reticuli regulator FurS depend on the redox state of its cysteine residues. Mol Gen Genet. 2000;264:341–353. doi: 10.1007/s004380000328. [DOI] [PubMed] [Google Scholar]
- 23.Pagàn-Ramos E, Song J, McFalone M, Mudd M H, Deretic V. Oxidative stress response and characterization of the oxyR-ahpC and furA-katG loci in Mycobacterium marinum. J Bacteriol. 1998;180:4856–4864. doi: 10.1128/jb.180.18.4856-4864.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Prentki P, Krisch H M. In vitro insertional mutagenesis with a selectable DNA fragment. Gene. 1985;29:303–313. doi: 10.1016/0378-1119(84)90059-3. [DOI] [PubMed] [Google Scholar]
- 25.Rosner J L, Storz G. Regulation of bacterial responses to oxidative stress. Curr Top Cell Regul. 1997;35:163–177. doi: 10.1016/s0070-2137(97)80007-6. [DOI] [PubMed] [Google Scholar]
- 26.Sabbattini P, Forti F, Ghisotti D, Dehò G. Control of transcription termination by an RNA factor in bacteriophage P4 immunity: identification of the target sites. J Bacteriol. 1995;177:1425–1434. doi: 10.1128/jb.177.6.1425-1434.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 28.Sherman D R, Sabo P J, Hickey M J, Arain T M, Mahairas G G, Yuan Y, Barry III C E, Stover C K. Disparate responses to oxidative stress in saprophytic and pathogenic mycobacteria. Proc Natl Acad Sci USA. 1995;92:6625–6629. doi: 10.1073/pnas.92.14.6625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Timm J, Lim E M, Gicquel B. Escherichia coli-mycobacteria shuttle vectors for operon and gene fusions to lacZ: the pJEM series. J Bacteriol. 1994;176:6749–6753. doi: 10.1128/jb.176.21.6749-6753.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Touati D, Jacques M, Tardat B, Bouchard L, Despied S. Lethal oxidative damage and mutagenesis are generated by iron in Δfur mutants of Escherichia coli: protective role of superoxide dismutase. J Bacteriol. 1995;177:2305–2314. doi: 10.1128/jb.177.9.2305-2314.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zahrt T C, Song J, Siple J, Deretic V. Mycobacterial FurA is a negative regulator of catalase-peroxidase gene katG. Mol Microbiol. 2001;39:1174–1185. doi: 10.1111/j.1365-2958.2001.02321.x. [DOI] [PubMed] [Google Scholar]
- 32.Zhang Y, Heim B, Allen B, Young D, Cole S. The catalase-peroxidase gene and isoniazid resistance of Mycobacterium tuberculosis. Nature. 1992;358:591–593. doi: 10.1038/358591a0. [DOI] [PubMed] [Google Scholar]
- 33.Zhang Y, Dhandayuthapani S, Deretic V. Molecular basis for the exquisite sensitivity of Mycobacterium tuberculosis to isoniazid. Proc Natl Acad Sci USA. 1996;93:13212–13216. doi: 10.1073/pnas.93.23.13212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zheng M, Storz G. Redox sensing by prokaryotic transcription factors. Biochem Pharmacol. 2000;59:1–6. doi: 10.1016/s0006-2952(99)00289-0. [DOI] [PubMed] [Google Scholar]
- 35.Zou P, Borovok I, Ortiz de Orué Lucana D, Muller D, Schrempf H. The mycelium-associated Streptomyces reticuli catalase-peroxidase, its gene and regulation by FurS. Microbiology. 1999;145:549–559. doi: 10.1099/13500872-145-3-549. [DOI] [PubMed] [Google Scholar]